Computational approaches leveraging integrated connections of multi-omic data toward clinical applications
Habibe Cansu
Demirel†
 a,
Muslum Kaan
Arici†
a,
Muslum Kaan
Arici†
 ab and
Nurcan
Tuncbag
ab and
Nurcan
Tuncbag
 *cde
*cde
aGraduate School of Informatics, Middle East Technical University, Ankara, 06800, Turkey
bFoot and Mouth Diseases Institute, Ministry of Agriculture and Forestry, Ankara, 06044, Turkey
cChemical and Biological Engineering, College of Engineering, Koc University, Istanbul, 34450, Turkey
dSchool of Medicine, Koc University, Istanbul, 34450, Turkey
eKoc University Research Center for Translational Medicine (KUTTAM), Istanbul, Turkey. E-mail: ntuncbag@ku.edu.tr
First published on 19th October 2021
Abstract
In line with the advances in high-throughput technologies, multiple omic datasets have accumulated to study biological systems and diseases coherently. No single omics data type is capable of fully representing cellular activity. The complexity of the biological processes arises from the interactions between omic entities such as genes, proteins, and metabolites. Therefore, multi-omic data integration is crucial but challenging. The impact of the molecular alterations in multi-omic data is not local in the neighborhood of the altered gene or protein; rather, the impact diffuses in the network and changes the functionality of multiple signaling pathways and regulation of the gene expression. Additionally, multi-omic data is high-dimensional and has background noise. Several integrative approaches have been developed to accurately interpret the multi-omic datasets, including machine learning, network-based methods, and their combination. In this review, we overview the most recent integrative approaches and tools with a focus on network-based methods. We then discuss these approaches according to their specific applications, from disease-network and biomarker identification to patient stratification, drug discovery, and repurposing.
Introduction
With the recent developments in high-throughput omic technologies, “big data” in biological and health sciences, which includes genomic, transcriptomic, proteomic, and metabolomic data at the molecular level, have been accumulating at a fast pace.1–3 Each omic data type provides different aspects of the cellular state. Yet, these are not isolated layers. Therefore, integrative approaches can uncover the causal relationships between each omic entity, such as proteins, genes, and metabolites.4 Within and across different data types, biomolecules may closely interact and tightly regulate each other.5 These biomolecular interactions are tissue-, context- and disease-specific and form multiple dynamic networks.6 Abnormal interactions may alter the cellular networks and eventually lead to pathological signaling output. Therefore, multi-omic data integration plays a central role to fully understand the disease etiology.1,7In recent studies, integration of multi-omic data elucidated transcriptional dysregulation of pathways in Alzheimer's disease,8 comprehensive molecular profiles of SARS-CoV-2 infection to propose drug candidates,9 pathway modulation by drugs in breast cancer cell lines,10 and novel alcoholism-related genes that are associated with neurodegenerative diseases.11 Additionally, integration approaches utilize the prior knowledge about the connectivity of the omic entities, such as a reference interactome collated from several databases, which may potentially reveal perturbed networks.12 Several initiatives have been established to study genetic variants, protein/gene expression profiles within and across different tissues, including Human Proteome Atlas,13 GTEx,14 and ENCODE.15 Additionally, many efforts have been put to explore the etiology of complex diseases through multi-omic data for the same group of tumors, patients, or perturbations. Among them, The Cancer Genome Atlas (TCGA),16 the International Cancer Genome Consortium (ICGC),17 Clinical Proteomic Tumor Analysis Consortium (CPTAC),18,19 and TARGET20 for pediatric cancers span multiple layers of omic data from thousands of tumor tissues in human cancers. The Cancer Cell Line Encyclopedia (CCLE)21 and Cancer Dependency Map (DepMap)22 harbor genomic, transcriptomic data, genetic dependency, and small molecule sensitivities of cancer cell lines that can be used for drug response studies.23,24 In today's world, patient- or condition-specific multi-omic data storage and determination of treatment strategies based on the findings obtained from the integrative analysis are proceeding at a considerable pace.
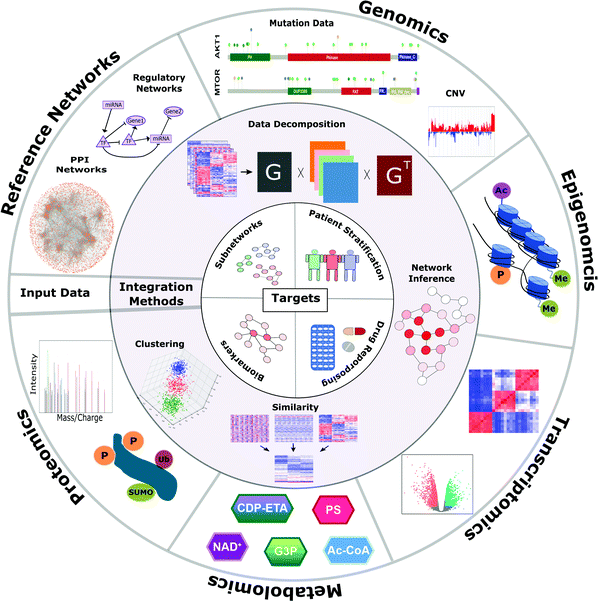
As the multi-omic data accumulates, novel computational integrative approaches are also developed with an overarching aim of transforming the data into clinically interpretable knowledge. Some examples that help clinical interpretations are survival analysis,25 identifying biomarkers,26 patient stratification,27 and precision medicine.28 Computational integrative approaches have been previously reviewed based on either the technical details or the targeted disease.7,29–32 Integrative approaches include machine learning strategies, network-based applications, or their combination depending on the condition or disease to be studied. In this review, we give a technical overview of the multi-omic data integration approaches, mainly network-based and learning-based data integration, with a focus on network-based approaches. Then, we dive into more details of their applications, namely identification of disease-associated subnetworks, patient stratification, biomarker identification, and leveraging them for drug discovery and repurposing. We mostly focus on applications in cancer research, but some examples from infectious and neurodegenerative diseases are also included. We conceptually summarize the multi-omic data integration methods in Fig. 1 in three layers: input data types, integration methods, and aims.
Multi-omic data integration approaches
The main challenge in multi-omic data integration is how to develop efficient methods to reverse engineer from this big data to explain the molecular basis of a disease or a perturbation.33–35 There are many network-based approaches and multidimensional techniques to integrate multi-omic data.36,37 We tabulate a comprehensive list of these approaches, including their aims, algorithms, and the omic data types they integrate in Table 1. These techniques are classified as horizontal or vertical based on their application.38–40 In horizontal integration, the same data type from multiple samples is used, such as transcriptomic data from multiple patients. On the other hand, multiple layers of omic data are leveraged in vertical integration, such as linking gene expression and mutation profiles. One example of horizontal integration is an application of hierarchical HotNet to pan-cancer somatic mutation profiles and eventually finding cancer-driver subnetworks.41 On the other hand, iCell has an application of a vertical integration that uses tissue-specific protein–protein interaction, gene co-expression, and gene interaction networks to obtain rewired genes in the network, which are potential cancer biomarkers.42| Tool | Year | Data | Accessibility | Algorithm | Aim |
|---|---|---|---|---|---|
| iCluster52 | 2009 | Genomics, Transcriptomics | Tool: https://www.mskcc.org/departments/epidemiology-biostatistics/biostatistics/icluster | Joint latent variable model-based clustering | Subgroup identification, biomarker discovery |
| Source code: https://github.com/cran/iCluster | |||||
| CONEXIC134 | 2010 | Genomics, Transcriptomics | Source code: https://github.com/dpeerlab/CONEXIC | A Bayesian network based algorithm | Biomarker discovery, subnetwork construction |
| CNAmet135 | 2011 | Genomics, Epigenomics, Transcriptomics | Tool: https://csbi.ltdk.helsinki.fi/CNAmet/ | Correlation based method | Biomarker discovery |
| DriverNet136 | 2012 | Genomics, Transcriptomics | Tool: http://compbio.bccrc.ca/software/drivernet/ | Stochastic resampling | Biomarker discovery |
| Source code: https://github.com/shahcompbio/drivernet | |||||
| iClusterPlus99 | 2013 | Genomics, Epigenomics, Transcriptomics | Tool: https://bioconductor.org/packages/release/bioc/html/iClusterPlus.html | Joint multivariate regression | Subgroup identification, biomarker discovery |
| TieDIE49 | 2013 | Genomics, Transcriptomics | Tool: https://sysbiowiki.soe.ucsc.edu/tiedie | Modified heat diffusion algorithm | Subnetwork construction |
| Source code: https://github.com/epaull/TieDIE | |||||
| AMARETTO137 | 2020 | Genomics, Epigenomics, Transcriptomics | Tool: https://bitbucket.org/gevaertlab/pancanceramaretto | Univariate beta mixture models, a linear regression model, k-means clustering | Subnetwork construction, biomarker discovery |
| Source code: https://github.com/gevaertlab/AMARETTO | |||||
| iBAG55 | 2013 | Genomics, Epigenomics, Transcriptomics | Integrative bayesian analysis | Biomarker discovery | |
| MCIA138 | 2014 | Transcriptomics, Proteomics | Tool: https://rdrr.io/github/mengchen18/omicade4/man/mcia.html | Multiple co-inertia analysis | Subgroup identification, biomarker discovery |
| Source code: https://github.com/mengchen18/omicade4/ | |||||
| SNF94 | 2014 | Epigenomics, Transcriptomics | Source code: https://github.com/maxconway/SNFtool | Similarity network fusion | Subgroup identification |
| FEM104 | 2014 | Epigenomics, Transcriptomics | Tool: https://sourceforge.net/projects/funepimod/ | Empirical Bayesian framework | Subnetwork construction, biomarker discovery |
| Source code: https://sourceforge.net/projects/funepimod/ | |||||
| Joint Bayesian Factor139 | 2014 | Genomics, Epigenomics, Transcriptomics | Source code: https://sites.google.com/site/jointgenomics/ | Non-parametric Bayesian factor | Biomarker discovery |
| rMKL-LPP97 | 2015 | Epigenomics, Transcriptomics | Tool: executable is available upon request. | Regularized multiple kernel learning | Subgroup identification |
| LRACluster101 | 2015 | Genomics, Transcriptomics | Tool: https://rdrr.io/github/xlucpu/MOVICS/man/LRAcluster.html | Low-rank approximation based integrative probabilistic model | Subgroup identification |
| Lemon-Tree124 | 2015 | Genomics, Transcriptomics | Source code: https://github.com/erbon7/lemon-tree | Tight-clustering and decision tree | Biomarker discovery |
| rJIVE140 | 2016 | Epigenomics, Transcriptomics | Tool: https://cran.r-project.org/web/packages/r.jive/ | An extension of PCA | Subgroup identification |
| Source code: https://github.com/cran/r.jive | |||||
| Omics Integrator73 | 2016 | Genomics, Transcriptomics, Proteomics, Phosphoproteomics | Source code: https://github.com/fraenkel-lab/OmicsIntegrator2 | Prize collecting Steiner forest Tree | Subnetwork construction |
| Web server: http://fraenkel-nsf.csbi.mit.edu/omicsintegrator/ | |||||
| PIUMet80 | 2016 | Proteomics, Lipidomics | Web server: http://fraenkel-nsf.csbi.mit.edu/piumet2/ | Prize collecting Steiner forest Tree | Subnetwork construction |
| mixOmics54 | 2017 | Genomics, Transcriptomics, Epigenomics | Source code: https://github.com/cran/mixOmics | Multivariate projection-based | Subgroup identification, viomarker discovery |
| mixKernel98 | 2017 | Genomics, Transcriptomics | Tool: https://cran.r-project.org/web/packages/mixKernel/index.html | Multiple kernel learning | Subgroup identification |
| Source code: https://github.com/cran/mixKernel | |||||
| PINS95 | 2017 | Genomics, Transcriptomics, Epigenomics | Perturbation clustering | Subgroup identification, biomarker discovery | |
| iClusterBayes56 | 2018 | Genomics, Transcriptomics, Epigenomics | Tool: https://rdrr.io/bioc/iClusterPlus/man/iClusterBayes.html | Bayesian integrative clustering | Subgroup identification, biomarker identification |
| MOFA100 | 2018 | Genomics, Transcriptomics | Source code: https://github.com/bioFAM/MOFA | Multi-omics factor analysis | Subgroup identification, subnetwork construction |
| Web server: http://www.ebi.ac.uk/shiny/mofa/ | |||||
| Ding et al.129 | 2018 | Transcriptomics, Genomics | Deep learning | Drug discovery and repurposing | |
| PINSPlus96 | 2019 | Epigenomics, Transcriptomics | Tool: https://rdrr.io/cran/PINSPlus/ | Perturbation clustering | Subgroup identification |
| Source code: https://github.com/cran/PINSPlus | |||||
| NEMO102 | 2019 | Epigenomics, Transcriptomics | Tool: https://rdrr.io/github/xlucpu/MOVICS/man/nemo.clustering.html | Similarity based clustering | Subgroup identification |
| Source code: https://github.com/Shamir-Lab/NEMO | |||||
| Web server: https://nemoanalytics.org/ | |||||
| ModulOmics48 | 2019 | Genomics, Transcriptomics | Source code: https://github.com/danasilv/ModulOmics | Integer linear programming (ILP) and stochastic searches | Subnetwork construction |
| Web server: http://anat.cs.tau.ac.il/ModulOmicsServer/ | |||||
| iOmicsPass81 | 2019 | Transcriptomics, Proteomics, Genomics | Source code: https://github.com/cssblab/iOmicsPASS | A modified nearest shrunken centroid classification | Subnetwork construction, subgroup identification |
| MOSClip57 | 2019 | Genomics, Transcriptomics | Tool: https://cavei.github.io/ | Principal component analysis | Subnetwork construction, biomarker discovery |
| Source code: https://github.com/cavei/MOSClip | |||||
| iProFun141 | 2019 | Transctiptomics, Proteomics, Phophoproteomics, Epigenomics | Source code: https://github.com/songxiaoyu/iProFun | Multiple Linear Regression | Biomarker discovery |
| MOLI130 | 2019 | Genomics, Transcriptomics | Source code: https://github.com/hosseinshn/MOLI | Deep neural networks | Drug discovery and repurposing |
| DrugComboExplorer131 | 2019 | Genomics, Transcriptomics | Source code: https://github.com/Roosevelt-PKU/drugcombinationprediction | Non-parametric bootstrapping-based simulated annealing (NPBSA) | Drug discovery and repurposing, subnetwork construction |
| SALMON25 | 2019 | Genomics, Transcriptomics | Source code: https://github.com/huangzhii/SALMON/ | Neural network, Cox proportional hazards regression networks | Biomarker discovery |
| fMKL-DR142 | 2020 | Genomics, Transcriptomics, Epigenomics | Fast multiple kernel learning | Subgroup identification | |
| HC-fused143 | 2021 | Epigenomics, Transcriptomics | Source code: https://github.com/pievos101/HC-fused | Hierarchical data fusion and integrative clustering | Subgroup identification |
| DeepDRK93 | 2021 | Transcriptomics, Genomics, Epigenomics | Source code: https://github.com/wangyc82/DeepDRK | Kernel-adapted deep neural network | Drug discovery and repurposing |
| COSMOS84 | 2021 | Transcriptomics, Phosphoproteomics, Metabolomics | Source code: https://github.com/saezlab/COSMOS_MSB. | Causal network inference | Drug discovery and repurposing, subnetwork construction |
| CausalPath82 | 2021 | Phosphoproteomics, Proteomics | Source code: https://github.com/PathwayAndDataAnalysis/causalpath | Causal network inference | Subnetwork construction |
| Web server: http://causalpath.org/ | |||||
| MOGONET89 | 2021 | Genomics, Transcriptomics | Source code: (https://github.com/txWang/MOGONET) | Convolutional neural networks | Biomarker discovery, subgroup identification |
Integration methods can also be classified based on the order of the data usage as sequential and simultaneous integration approaches.43,44 Omic datasets are evaluated and optimized separately in sequential approaches.45–47 Each sequential step improves the output of the previous data by pruning the search space and extends data size. Yet, this process causes a loss of sensitivity as omitted weak signals may contain useful information.48 For example, TieDIE integrates mutations and differential gene expression profiles with the PPI-proximity test to find the final subnetworks using two consecutive heat diffusion steps.49 Firstly, the heat is diffused from the significantly mutated genes to other genes in the directed reference interactome. Then, the same is applied in the reverse direction in the reference interactome. These results are combined to obtain the final subnetwork. As the dimension increases in the multi-omic studies, the data sparsity also increases that causes a problem known as “the curse of dimensionality”.50 Besides, the high dimensional data structure makes integration approaches prone to overfitting in learning-based methods, especially in fitting supervised models.36,50 Overfitting is a problem for both sequential and simultaneous approaches. To overcome it, sequential approaches separately conduct dimensionality reduction on each omic data set.51 On the other hand, simultaneous integration methods handle all features at the same time. They commonly utilize learning-based approaches including non-negative matrix factorization (iCluster,52 JIVE53), multivariate analysis (mixOmics54), Bayesian framework (iBAG,55 iClusterBayes56), and component analyses (MOSClip57) for dimensionality reduction. These methods potentially overcome biases in the multi-omic data and lead to information loss.58–60
In the following subsections, we review the network-based and learning-based approaches in detail. We need to note that there may not be a strict borderline between these categories for several tools. Usually, they complement each other in many approaches and may belong to more than one category. For example, we reviewed iCell as a network-based approach, but it uses machine-learning to integrate multiple networks and omic data to obtain a final subnetwork.42
Network-based data integration
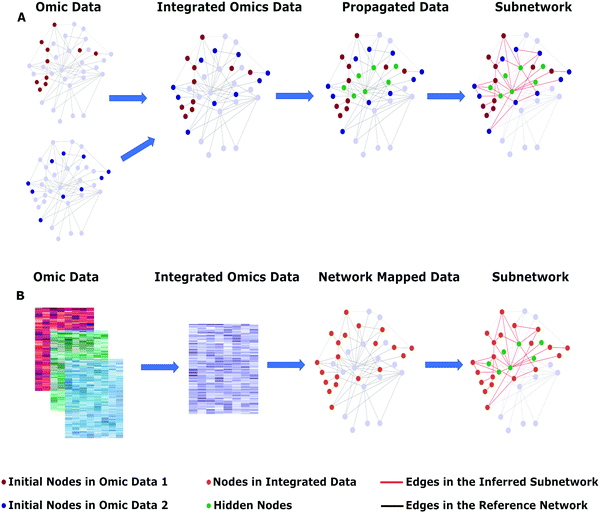
Network-based approaches aim to reveal the dependencies between the omic entities by leveraging the graph theory where proteins, genes, and transcription factors are nodes, and their interactions are edges.35,61,62 Usually, a reference interactome is used during data integration which may consist of protein–protein interactions, gene co-expression, metabolite interactions, and regulatory interactions.63–65 A reference interactome may bring false positive and false negative interactions. Therefore, a tremendous effort has been spent to score the interactions based on their confidence. These scoring schemes consider the experimental detection method, the number of publications, interologs, and many other gold-standard properties of PPIs.66–69 Some hub proteins (proteins that have a high number of interactions) such as TP53 and EGFR have hundreds of high-confidence connections because of being well-studied, which leads to a bias in the interactomes.70–72 Several network-based algorithms such as Omics Integrator,73 TieDIE,49 and Hierarchical HotNet41 penalize these hub nodes and additionally use context-specific interactions to overcome this bias.The direct mapping of multi-omic data to a reference interactome, such as considering only the interactions between the omic hits or their first neighbor proximity, may result in either an incomplete subnetwork or a hairball-like structure.74–76 Therefore, network-based approaches aim at one side to reveal hidden nodes, on the other side, to find the optimal connections (Fig. 2A). The initial node sets (the set of differentially expressed genes/proteins, highly mutated genes, transcription factors, etc.) are propagated over a reference interactome with different approaches such as random walk (MEXCOwalk,77 uKIN,78 and Hierarchical HotNet41), heat diffusion (TieDIE,49 NetICS,61 and HotNet79), and prize-collecting Steiner forest (Omics Integrator73 and PIUMet80). Network-based integration methods primarily focus on the heterogeneous union among diverse omic data at different molecular levels to overcome their incompleteness. For example, the Forest module of Omics Integrator integrates multi-omic datasets with a reference interactome to construct an optimal network by solving the prize-collecting Steiner forest problem.73 In these approaches, the user is able to configure the reference interactome to be multi-layered and the initial set to include different omic data types so that multi-omic data can be simultaneously analyzed with a single integrated reference network.
Another approach, that is conceptually shown in Fig. 2B, first integrates multi-omic data and then maps it to the reference interactome. In this method, the data integration part is separately implemented from the subnetwork construction part so that a consensus matrix from the multi-omic data is obtained with combined scores. iOmicsPASS belongs to this class where it first integrates the multi-omic data to obtain edge scores and then predicts the subnetworks using the nearest shrunken centroid (NSC) classification algorithm.81 ModulOmics uses a protein–protein interaction, transcription factor-gene regulatory and gene co-expression networks, and mutual exclusivity of the molecular alterations simultaneously to find cancer driver modules with the help of a two-stage optimization, namely Integer Linear Programming followed by a stochastic search.48 iCell simultaneously leverages protein–protein interaction, gene co-expression, and genetic interaction networks to represent tissue-specific cells uniquely.42 The core technique in iCell is non-negative matrix factorization, and it finally ranks the most rewired cancer genes. The advantage of ModulOmics and iCell is their simultaneous integration capabilities so that multi-layered information about each gene can be incorporated.
Besides the direct integration of multi-omic data, adding the literature-curated mechanistic details of cellular signaling can elucidate the cause-and-effect relationships. CausalPath uses prior knowledge of biochemical reactions and proteomic data to construct causal pathways.82,83 Similarly, COSMOS constructs a causal network, but it uses transcriptomic, proteomic and metabolomic data together with curated prior knowledge about pathways to identify disease mechanisms.84 CausalPath is comparison and correlation-based, while COSMOS applies network optimization to identify causal relationships.
Overall, the performance of the network-based approaches is highly dependent on the reference interactome, the parameter set selection, and integrated biological intuition. Therefore, context-specific usage of interactomes and extensive parameter tuning may increase the quality of the final integrated network.85–88
Learning-based data integration
Learning-based approaches are frequently used to gain biological knowledge from large multi-omic datasets. Novel multi-omic integration models can be utilized for classification,89 clustering,90 and ranking.91 These algorithms are grouped as supervised and unsupervised methods at the top level.88 Supervised learning algorithms require data labels, and the aim is to predict the labels, such as predicting cancer driver genes, disease-associated pathways, or drug response. For example, CapsNetMMD is a supervised deep-learning-based method that uses the multi-omic data as the input in a two-layer convolutional neural network to rank breast cancer-associated genes.92 Another example is DeepDRK that trains a classification model using the deep neural networks (DNNs) by using multi-omic datasets from multiple drug-treated cell lines and drug properties to predict cell line drug sensitivity.93 Another recent deep learning tool, MOGONET, learns from each omic data as well as across different omic data, using convolutional neural networks to classify patients and discover biomarkers.89A wide range of multi-omic integration tools leverages similarity metrics, kernels, and statistical methods to develop unsupervised learning approaches. Similarity-based integration is rather commonly applied to the patient stratification problem as it provides a grouping factor based on the distances in the multi-omic data between the patients. The example tools of this group are SNF94 (Similarity network fusion), PINS95 (perturbation clustering for data integration and disease subtyping), and PINSPlus.96 In similarity-based integration, the contribution of the original data points is hard to identify in the prediction performance.90
rMKL-LPP97 and MixKernel98 adopt multiple kernel learning methods to integrate multi-omic datasets in a flexible manner. rMKL-LPP uses Locality Preserving Projections (LPP) algorithm to project the data to a lower dimension by preserving the similarities and nearest neighbors. Some methods can only work with continuous data, and mixKernel overcomes this limitation.
Statistical approaches model the relationships between features associated with the highest biological variation based on correlation formulas, regression formulas, and probability distribution assumptions. Most of the recent tools are able to integrate different data types such as binary (somatic mutation), categorical (copy number gain, normal, loss), and continuous (gene expression) that follow different probabilistic distributions, while examples including iCluster46 and JIVE53 cannot work with discrete and continuous data at the same time. iClusterPlus99 uses a generalized regression model where the latent variables represent the underlying disease-driving factors. The model requires a large sample space and grid search for optimum regression and meaningful variables. Thus, statistical inference with direct regression is a computationally intensive approach because of the data dimensionality. Statistical solutions such as generalized principal component analysis (MOFA100 and mixOmics54) and low-rank approximation methods (iClusterBayes,56 JIVE,53 and LRAcluster101) decompose the data sets to explain shared variation, individual variation, and noise. Neighborhood-based Multi-Omics Clustering (NEMO102) is a hybrid approach that can integrate partial sets with missing data values without applying data imputation. NEMO first creates similarity matrices for each data set and then merges them into a single matrix to cluster into subgroups using a spectral clustering variant.
Overall, high-throughput technologies generate various data modalities, and integrative approaches are the key to transforming these data sets into biologically meaningful knowledge. The following sections exemplify the applications of integrative approaches in disease-associated subnetworks, patient stratification, biomarker discovery, and drug repurposing.
Applications of integrative methods based on their aims
Disease-associated subnetwork identification
Finding disease-associated subnetworks provides a causal relationship between altered omic entities and gives insights into the perturbed pathways in complex diseases. Some of the integrative techniques that we reviewed in the previous sections were successfully applied to discover disease-associated networks, including iOmicsPASS,81 ModulOmics,48 and TieDIE.49 These tools were previously applied to identify breast cancer associated-subnetworks using multi-omic data in TCGA or CPTAC.103 iOmicsPASS inferred subtype-specific networks that were enriched in several up- and down-regulated pathways.81 Similarly, ModulOmics integrated single nucleotide variants and transcriptomic data with a PPI network to find driver modules.48 The top modules distinguished luminal A subtype of breast cancer from normal tissues and identified functional relations between multiple tumor suppressors such as TP53, BRCA1, RB1, and PTEN for the triple-negative subtype. TieDIE used the mutation profiles of several patient tumors as the initial set and identified a core signaling pathway representing the known differences between the luminal A and basal subtypes of breast cancer.49Different cancer types or pan-cancer analyses have been previously demonstrated in the discovery of disease-associated networks. For example, Omics Integrator was used with Glioblastoma Multiforme (GBM) mutations from TCGA to create patient-specific subnetworks.73 A grouping of patients based on the inferred subnetworks was significantly associated with survival and used for possible drug sensitivity assessments. FEM identified functional epigenetic modules as subnetworks by integrating methylation and expression data of endometrial cancer from TCGA with a PPI network.104 One of the top modules focused around HAND2, a driver gene for endometrial cancer, successfully indicated its deregulation in the cancer samples, which has been previously identified as a biomarker.105 MOSClip utilized an ovarian cancer dataset from TCGA to discover survival-associated pathways and modules where expression, methylation, copy number variation, and mutation data were integrated.57 Most of the identified pathways and modules contained known ovarian cancer drivers and processes, and the results indicated the presence of a circuit that can be used for survival prediction. Overall, all these approaches aim to integrate multi-layered omic datasets to reveal the pathway level alterations in tumors that may be a signature in diagnosis, prognosis, and treatment of the disease.
Apart from cancer, some integration tools were used to infer subnetworks associated with different diseases. For example, focusing on host–pathogen interactions, Omics Integrator integrated transcriptomic and metabolomic data from Kaposi's Sarcoma associated Herpesvirus infection.106 Among the identified pathways, peroxisome biogenesis was highlighted as lipid metabolism in the peroxisome is crucial for infected cells. PIUMet revealed Huntington's disease-associated pathways by integrating untargeted lipidomic and phosphoproteomic data with protein–protein and protein-metabolite interactions.80
Patient stratification and subtype discovery
Due to the heterogeneous nature of cancer, tumors from the same cancer type may exhibit different biological features, which eventually leads to differences in treatment responses. Hence, stratifying patients and finding cancer subtypes have the potential to reveal hidden similarities across patients, which can be utilized to gain insights for personalized medicine and optimization of treatment strategies.107,108 In general, three common approaches are used to illustrate the performances of patient stratification tools based on their ability to detect groups with (i) significant survival differences, (ii) known subtypes, and (iii) different cancer types. Among the tools adopting the first approach, PINS,95 PINSPlus,96 NEMO,102 SNF,94 and rMKL-LPP97 identified patient clusters with significant survival differences for most of the tested cancer types, integrating mRNA expression, DNA methylation, and miRNA expression data. Depending on the availability, prior subtype information can also be incorporated for an evaluation. rMKL-LPP97 and iClusterBayes56 grouped GBM patients by integrating mRNA expression with DNA methylation and miRNA data; and with mutation and copy number data from TCGA, respectively. Illustrating the advantage of multi-omic integration, the clusters identified by rMKL-LPP represented both of the established subtypes previously found based on expression109 or methylation.110 In addition, survival analysis showed that some clusters had a better response to Temozolomide, a drug used for GBM.111 iClusterBayes identified biologically meaningful subtypes with oncogenes and tumor suppressors that show significantly different genomic profiles.56 Chaudhary et al. obtained two groups of hepatocellular carcinoma with significant survival differences where the more aggressive subtype was bearing increased Tp53 inactivation mutations and tumor marker expression.112 An integration expanded with clinical data failed to improve the performance, hinting that the model already captured this information from multi-omic data. iClusterplus integrated copy number, gene expression, and mutation data belonging to hundreds of CCLE cell lines of various cancer types.99 Interestingly, not all clusters obtained using this pan-cancer dataset were lineage-dependent. Instead, cross-cancer similarities were also revealed. In another pan-cancer application where methylation data is incorporated, LRACluster reported similar results as samples from the same cancer types are not always present in the same cluster.101Biomarker identification
Biomarkers refer to a single mutation,113 any altered gene/protein module,114 or specific subnetworks115 that can predict the disease progression such as being an indicator of survival rate,116 biological processes on disease development,117 and drug response.118 Predicting novel breast cancer biomarkers, CapsNetMMD integrated mRNA expression, DNA methylation, and copy number alterations data. Candidate cancer driver genes are identified after training with known breast cancer-related genes.92 According to cancer survival prognosis assessments, most of the top-ranked genes were selected as candidate biomarkers. However, CapsNetMMD can only be applied to diseases having prior information about the associated genes.Complex diseases are caused by the combination of alterations in several molecules rather than being dependent on a single molecule. Thus, elucidating a module of proteins/genes that can distinguish the disease from healthy samples can more realistically represent the disease-association and be utilized to mechanistically explore molecular complexes and signaling pathways mechanisms.115,119,120 For instance, SALMON integrated gene expression, miRNA, copy number burden, and tumor mutation burden, and ranked modules to better predict survival in breast cancer.25 As a result, a significant relationship between CD8+ and CD4+ in T cells, regulation of T cell function by MST1 kinase, and the different roles of multiple breast cancer-related genes were found. MOFA combined drug response measurements with mutation profiles, transcriptome, and DNA methylation data and identified important clinical markers to predict drug response.100 MOFA was also applied to single-cell multi-omic data, including DNA methylation and gene expression datasets, to identify modules affecting pluripotency states in cellular differentiation.
Beyond the modules, subnetworks are also used as biomarkers for survival analysis and biological interpretations.121–123 Lemon-Tree constructed subnetworks by integrating somatic copy-number alterations and gene expression datasets from GBM samples and identified oncogenes and tumor suppressor genes as potential biomarkers for survival analyses.124 iCell vertically integrated tissue or single-cell omic data with the interactome to identify biomarkers in several cancer types.42 It distinguished cancer cells from healthy cells based on the comparison of the final network and illustrated the structure, heterogeneity, and dynamics of tumor progression.
Drug discovery and repurposing
Integrative approaches were successfully applied to drug discovery and repurposing studies for several diseases including cancer and infectious diseases by leveraging pharmacogenomic datasets.125–128 Drug repurposing studies aim to discover novel usages for approved drugs and provide some advantages like reduced cost and risk enabling faster development. In a very recent study, Tomazou et al. integrated transcriptomic, proteomic and metabolomic data from patients, cell lines and databases to repurpose drugs for COVID-19.9 Ding et al. applied a learning-based approach to integrate mutation, copy number alteration, and gene expression data to predict drug sensitivities of different cancer cell lines.129 Predicted drug response values were validated with observed response data in CCLE. MOLI predicted drug responses using somatic mutation, CNA, and gene expression data.130 After the model was trained on a pan-drug input for the epidermal growth factor receptor (EGFR) inhibitor, the predicted responses were significantly associated with the expression of the genes in the EGFR pathway. In addition to predicting the drug effects, DeepDRK repurposed drugs via training their model on genomic, epigenomic, and transcriptomic data as well as the chemical features of the drugs and clinical response data.93 DeepDRK performance was highly dependent on the cohort size. Therefore, it performed well in breast cancer and head–neck squamous cell carcinoma but not others. DrugComboExplorer discovered potential synergic drug combinations by exploring cancer-driver networks for each drug treatment.131 The perturbed driver networks were extracted with genomic data, known mutations, and expression profiles. Additionally, DrugComboExplorer identified cross-talks between effector signaling pathways which reveal how cancer cells survive and develop resistance to targeted therapy.Conclusion
In this review, we overview multi-omic data integration approaches. These tools circumvent the constraints of single-level data utilization by employing various integration methods for various intents, including but not limited to patient stratification, biomarker discovery, subtype and subgroup identification, and drug discovery and repurposing. At the same time, this vast availability leads to the major challenge of selecting the most appropriate method to address the chosen biological question. Difficulties in setting criteria for the performance assessments due to the lack of gold-standard datasets make this selection even more challenging. Inevitably, the ever-growing biological big data, thanks to the increasing availability of techniques such as sequencing technologies, bring along a need for tools that can exploit it in a fast, effective, accurate, and user-friendly manner. These tools need to efficiently address common problems like high dimensionality and the noise of the datasets. The ability to integrate different data types without a requirement of matched samples or utilizing the not-so-common data types like metabolomics alongside the frequently used transcriptomic and genomic datasets could be an important advantage for the forthcoming approaches. On top of multi-omic data, trans-omic studies also utilize clinical information to uncover underlying disease mechanisms that cannot be revealed based on the omic data itself.132,133 In this review, we only included the approaches integrating bulk multi-omic datasets. Omic data in single-cell resolution and spatial omic technologies emerge as well. Some of the reviewed approaches have already been adopted to single-cell omics datasets. Therefore, tools integrating spatial and single-cell multi-omic data and elucidating the cell–cell communications from single-cell data started to be developed, and there will be more in the future.Conflicts of interest
There are no conflicts to declare.Acknowledgements
NT has received support from the Career Development Program of TUBITAK under the project number 117E192. MA has been financially supported with the TUBITAK-2211 fellowship. NT acknowledges the support from the UNESCO-L*Oreal National for Women in Science Fellowship and the UNESCO-L*Oréal International Rising Talent Fellowship and TUBA-GEBIP.References
- Y. Hasin, M. Seldin and A. Lusis, Genome Biol., 2017, 18, 1–15 CrossRef.
- S. Shilo, H. Rossman and E. Segal, Nat. Med., 2020, 26, 29–38 CrossRef CAS PubMed.
- Z. Zhang, W. Zhao, J. Xiao, Y. Bao, F. Wang, L. Hao, J. Zhu, T. Chen, S. Zhang and X. Chen, et al. , Nucleic Acids Res., 2019, 47, D8–D14 CrossRef CAS.
- S. Graw, K. Chappell, C. Washam, A. Gies, J. Bird, M. Robeson and S. Byrum, Mol. Omi., 2021, 17, 170–185 RSC.
- H. De Jong, J. Comput. Biol., 2004, 9, 67–103 CrossRef PubMed.
- E. Yeger-Lotem and R. Sharan, Front. Genet., 2015, 257 Search PubMed.
- G. de Anda-Jáuregui and E. Hernández-Lemus, Front. Oncol., 2020, 423 CrossRef PubMed.
- R. Nativio, Y. Lan, G. Donahue, S. Sidoli, A. Berson, A. R. Srinivasan, O. Shcherbakova, A. Amlie-Wolf, J. Nie, X. Cui and S. L. Berger, et al. , Nat. Genet., 2020, 52, 1024–1035 CrossRef CAS PubMed.
- M. Tomazou, M. M. Bourdakou, G. Minadakis, M. Zachariou, A. Oulas, E. Karatzas, E. M. Loizidou, A. C. Kakouri, C. C. Christodoulou and G. M. Spyrou, et al. , Brief. Bioinform., 2020, 1–24, DOI:10.1093/bib/bbab114.
- M. Oh, S. Park, S. Lee, D. Lee, S. Lim, D. Jeong, K. Jo, I. Jung and S. Kim, Front. Genet., 2020, 1053 Search PubMed.
- M. Kapoor, M. J. Chao, E. C. Johnson, G. Novikova, D. Lai, J. L. Meyers, J. Schulman, J. I. Nurnberger, B. Porjesz, Y. Liu, T. Foroud, H. J. Edenberg, E. Marcora, A. Agrawal and A. Goate, Nat. Commun. 2021 121, 2021, 12, 1–12 Search PubMed.
- M. Vidal, M. E. Cusick and A.-L. Barabási, Cell, 2011, 144, 986–998 CrossRef CAS.
- E. Uhlén, M. Fagerberg, L. Hallström, B. M. Lindskog, C. Oksvold, P. Mardinoglu, A. Sivertsson, Å. Kampf, C. Sjöstedt and F. Pontén, et al. , Science, 2015, 347, 6220, DOI:10.1126/SCIENCE.1260419.
- J. Lonsdale, J. Thomas, M. Salvatore, R. Phillips, E. Lo, S. Shad, R. Hasz, G. Walters, F. Garcia and H. F. Moore, et al. , Nat. Genet., 2013, 45, 580–585 CrossRef CAS PubMed.
- E. K. Silverman, H. H. H. W. Schmidt, E. Anastasiadou, L. Altucci, M. Angelini, L. Badimon, J. L. Balligand, G. Benincasa, G. Capasso and J. Baumbach, et al. , Wiley Interdiscip. Rev.: Syst. Biol. Med., 2020, 12, 1489 Search PubMed.
- J. N. Weinstein, E. A. Collisson, G. B. Mills, K. R. M. Shaw, B. A. Ozenberger, K. Ellrott, I. Shmulevich, C. Sander and J. M. Stuart, Nat. Genet., 2013, 45, 1113–1120 CrossRef PubMed.
- The International Cancer Genome Consortium, Nature, 2010, 464, 993–998 CrossRef.
- N. J. Edwards, M. Oberti, R. R. Thangudu, S. Cai, P. B. McGarvey, S. Jacob, S. Madhavan and K. A. Ketchum, J. Proteome Res., 2015, 14, 2707–2713 CrossRef CAS PubMed.
- M. J. Ellis, M. Gillette, S. A. Carr, A. G. Paulovich, R. D. Smith, K. K. Rodland, R. R. Townsend, C. Kinsinger, M. Mesri, H. Rodriguez and D. C. Liebler, Cancer Discovery, 2013, 3, 1108–1112 CrossRef CAS PubMed.
- X. Ma, Y. Liu, Y. Liu, L. B. Alexandrov, M. N. Edmonson, C. Gawad, X. Zhou, Y. Li, M. C. Rusch and J. Zhang, et al. , Nature, 2018, 555, 371–376 CrossRef CAS.
- D. P. Nusinow, J. Szpyt, M. Ghandi, C. M. Rose, E. R. McDonald, M. Kalocsay, J. Jané-Valbuena, E. Gelfand, D. K. Schweppe, M. Jedrychowski, J. Golji, D. A. Porter, T. Rejtar, Y. K. Wang, G. V. Kryukov, F. Stegmeier, B. K. Erickson, L. A. Garraway, W. R. Sellers and S. P. Gygi, Cell, 2020, 180, 387–402.e16 CrossRef CAS PubMed.
- A. Tsherniak, F. Vazquez, P. G. Montgomery, B. A. Weir, G. Kryukov, G. S. Cowley, S. Gill, W. F. Harrington, S. Pantel and W. C. Hahn, et al. , G, Cell, 2017, 170, 564–576.e16 CrossRef CAS PubMed.
- J. Ma, S. H. Fong, Y. Luo, C. J. Bakkenist, J. P. Shen, S. Mourragui, L. F. A. Wessels, M. Hafner, R. Sharan, J. Peng and T. Ideker, Nat. Cancer, 2021, 2, 233–244 CrossRef.
- Y.-C. Chiu, H.-I. H. Chen, T. Zhang, S. Zhang, A. Gorthi, L.-J. Wang, Y. Huang and Y. Chen, BMC Med. Genomics, 2019, 12, 143–155 CrossRef PubMed.
- Z. Huang, X. Zhan, S. Xiang, T. S. Johnson, B. Helm, C. Y. Yu, J. Zhang, P. Salama, M. Rizkalla, Z. Han and K. Huang, Front. Genet., 2019, 10, 166 CrossRef CAS PubMed.
- Z. Fan, Y. Zhou and H. W. Ressom, Metab., 2020, 10, 144 CAS.
- H. Yang, R. Chen, D. Li and Z. Wang, Bioinformatics, 2021, 37, 2231–2237 CrossRef CAS PubMed.
- N. Selevsek, F. Caiment, R. Nudischer, H. Gmuender, I. Agarkova, F. L. Atkinson, I. Bachmann, V. Baier, G. Barel and J. Kleinjans, et al. , Commun. Biol., 2020, 3, 1–15 CrossRef PubMed.
- S. Huang, K. Chaudhary and L. X. Garmire, Front. Genet., 2017, 8, 84 CrossRef CAS.
- I. Subramanian, S. Verma, S. Kumar, A. Jere and K. Anamika, Bioinf. Biol. Insights, 2020, 14, 1–24 CrossRef.
- E. I. Vlachavas, J. Bohn, F. Ückert and S. Nürnberg, Int. J. Mol. Sci., 2021, 22, 2822 CrossRef CAS PubMed.
- O. Menyhárt and B. Győrffy, Comput. Struct. Biotechnol., 2021, 19, 949–960 CrossRef.
- B. Palsson and K. Zengler, Nat. Chem. Biol., 2010, 6, 787–789 CrossRef PubMed.
- F. Finotello, E. Calura, D. Risso, S. Hautaniemi and C. Romualdi, Front. Oncol., 2020, 1768 CrossRef.
- T. M. Santiago-Rodriguez and E. B. Hollister, Semin. Perinatol., 2021, 45, 151456 CrossRef PubMed.
- B. Mirza, W. Wang, J. Wang, H. Choi, N. C. Chung and P. Ping, Genes, 2019, 10, 87 CrossRef CAS.
- A. R. Sonawane, S. T. Weiss, K. Glass and A. Sharma, Front. Genet., 2019, 294 CrossRef CAS.
- I. Mihaylov, M. Kańduła, M. Krachunov and D. Vassilev, Biol. Direct, 2019, 14, 1–17 CrossRef CAS PubMed.
- Z. Huo, L. Zhu, T. Ma, H. Liu, S. Han, D. Liao, J. Zhao and G. Tseng, Stat. Biosci., 2019, 12, 1–22 Search PubMed.
- B. Ulfenborg, BMC Bioinf., 2019, 20, 1–10, DOI:10.1186/s12859-019-3224-4.
- M. A. Reyna, M. D. M. Leiserson and B. J. Raphael, Bioinformatics, 2018, 34, i972–i980 CrossRef CAS PubMed.
- N. Malod-Dognin, J. Petschnigg, S. F. L. Windels, J. Povh, H. Hemmingway, R. Ketteler and N. Pržulj, Nat. Commun., 2019, 10, 1–13, DOI:10.1038/s41467-019-08797-8.
- M. Bersanelli, E. Mosca, D. Remondini, E. Giampieri, C. Sala, G. Castellani and L. Milanesi, BMC Bioinf., 2016, 17, 167–177 CrossRef PubMed.
- F. Ahmad, A. Mahmood and T. Muhmood, Biomater. Sci., 2021, 9, 1598–1608 RSC.
- C. Wu, F. Zhou, J. Ren, X. Li, Y. Jiang and S. Ma, High-Throughput, 2019, 8, 1–25, DOI:10.3390/HT8010004.
- S. Kim, S. Oesterreich, S. Kim, Y. Park and G. C. Tseng, Biostatistics, 2017, 18, 165–179 CrossRef PubMed.
- M. D. Ritchie, E. R. Holzinger, R. Li, S. A. Pendergrass and D. Kim, Nat. Rev. Genet., 2015, 16, 85–97 CrossRef CAS PubMed.
- D. Silverbush, S. Cristea, G. Yanovich-Arad, T. Geiger, N. Beerenwinkel and R. Sharan, Cell Syst., 2019, 8, 456–466.e5 CrossRef CAS PubMed.
- E. O. Paull, D. E. Carlin, M. Niepel, P. K. Sorger, D. Haussler and J. M. Stuart, Bioinformatics, 2013, 29, 2757–2764 CrossRef CAS PubMed.
- M. Kim and I. Tagkopoulos, Mol. Omi., 2018, 14, 8–25 RSC.
- M. Picard, M.-P. Scott-Boyer, A. Bodein, O. Périn and A. Droit, Comput. Struct. Biotechnol. J., 2021, 19, 3735–3746 CrossRef CAS PubMed.
- R. Shen, Q. Mo, N. Schultz, V. E. Seshan, A. B. Olshen, J. Huse, M. Ladanyi and C. Sander, PLoS One, 2012, 7, e35236 CrossRef CAS PubMed.
- E. F. Lock, K. A. Hoadley, J. S. Marron and A. B. Nobel, Ann. Appl. Statistics, 2013, 7, 523–542 Search PubMed.
- F. Rohart, B. Gautier, A. Singh and K.-A. Lê Cao, PLoS Comput. Biol., 2017, 13, e1005752 CrossRef PubMed.
- W. Wang, V. Baladandayuthapani, J. S. Morris, B. M. Broom, G. Manyam and K.-A. Do, Bioinformatics, 2013, 29, 149–159 CrossRef CAS PubMed.
- Q. Mo, R. Shen, C. Guo, M. Vannucci, K. S. Chan and S. G. Hilsenbeck, Biostatistics, 2018, 19, 71–86 CrossRef PubMed.
- P. Martini, M. Chiogna, E. Calura and C. Romualdi, Nucleic Acids Res., 2019, 47, e80 CAS.
- C. Grigo and P.-S. Koutsourelakis, SIAM/ASA Journal on Uncertainty Quantification, 2019, 7, 292–323 CrossRef.
- S. Vinga, Brief. Bioinf., 2021, 22, 77–87 CrossRef PubMed.
- A. Holzinger, B. Haibe-Kains and I. Jurisica, Eur. J. Nucl. Med. Mol. Imaging, 2019, 46, 2722–2730 CrossRef PubMed.
- C. Dimitrakopoulos, S. K. Hindupur, L. Häfliger, J. Behr, H. Montazeri, M. N. Hall and N. Beerenwinkel, Bioinformatics, 2018, 34, 2441–2448 CrossRef CAS PubMed.
- B. Güvenç Paltun, H. Mamitsuka and S. Kaski, Brief. Bioinf, 2021, 22, 346–359 CrossRef PubMed.
- T. Ideker, O. Ozier, B. Schwikowski and A. F. Siegel, Bioinformatics, 2002, 18, S233–S240 CrossRef PubMed.
- K. Ozturk, M. Dow, D. E. Carlin, R. Bejar and H. Carter, J. Mol. Biol., 2018, 430, 2875–2899 CrossRef CAS PubMed.
- P. Paci, G. Fiscon, F. Conte, R. S. Wang, L. Farina and J. Loscalzo, NPJ Syst. Biol. Appl., 2021, 7, 1–11, DOI:10.1038/s41540-020-00168-0.
- G. Alanis-Lobato, P. Mier and M. Andrade-Navarro, Bioinformatics, 2018, 34, 2826–2834 CrossRef CAS PubMed.
- A. Kamburov, U. Stelzl and R. Herwig, Nucleic Acids Res., 2012, 40, W140–W146, DOI:10.1093/nar/gks492.
- D. Szklarczyk, A. L. Gable, D. Lyon, A. Junge, S. Wyder, J. Huerta-Cepas, M. Simonovic, N. T. Doncheva, J. H. Morris, P. Bork, L. J. Jensen and C. Von Mering, Nucleic Acids Res., 2019, 47, D607–D613 CrossRef CAS PubMed.
- A. L. Turinsky, S. Razick, B. Turner, I. M. Donaldson and S. J. Wodak, Nat. Biotechnol., 2011, 29, 391–393 CrossRef CAS PubMed.
- M. A. Reyna, U. Chitra, R. Elyanow and B. J. Raphael, J. Comput. Biol., 2021, 28, 469–484 CrossRef CAS PubMed.
- M. H. Schaefer, L. Serrano and M. A. Andrade-Navarro, Front. Genet., 2015, 6, 260, DOI:10.3389/fgene.2015.00260.
- M. A. Skinnider, R. G. Stacey and L. J. Foster, PLoS Comput. Biol., 2018, 14, 1–22, DOI:10.1371/journal.pcbi.1006474.
- N. Tuncbag, S. J. C. Gosline, A. Kedaigle, A. R. Soltis, A. Gitter and E. Fraenkel, PLoS Comput. Biol., 2016, 2, 1–18, DOI:10.1371/journal.pcbi.1004879.
- J. Ma, A. Shojaie and G. Michailidis, Bioinformatics, 2016, 32, 3165–3174 CrossRef CAS PubMed.
- C. Nogales, A. G. B. Grønning, S. Sadegh, J. Baumbach and H. H. H. W. Schmidt, Handb. Exp. Pharmacol., 2020, 264, 49–68 CrossRef PubMed.
- S. Ohsawa, T. Umemura, T. Terada and Y. Muto, Genes, 2020, 11, 1457 CrossRef CAS PubMed.
- R. Ahmed, I. Baali, C. Erten, E. Hoxha and H. Kazan, Bioinformatics, 2020, 36, 872–879 CAS.
- B. H. Hristov, B. Chazelle and M. Singh, Cell Syst., 2020, 10, 470–479.e3 CrossRef CAS PubMed.
- M. D. M. Leiserson, F. Vandin, H. T. Wu, J. R. Dobson, J. V. Eldridge, J. L. Thomas, A. Papoutsaki, Y. Kim and B. J. Raphael, et al. , Nat. Genet., 2015, 47, 106–114 CrossRef CAS.
- L. Pirhaji, P. Milani, M. Leidl, T. Curran, J. Avila-Pacheco, C. B. Clish, F. M. White, A. Saghatelian and E. Fraenkel, Nat. Methods, 2016, 13, 770–776 CrossRef CAS PubMed.
- H. W. L. Koh, D. Fermin, C. Vogel, K. P. Choi, R. M. Ewing and H. Choi, npj Syst. Biol. Appl., 2019, 5, 1–10 CAS.
- Ö. Babur, A. Luna, A. Korkut, F. Durupinar, M. C. Siper, U. Dogrusoz, A. S. Vaca Jacome, R. Peckner, K. E. Christianson, J. D. Jaffe, P. T. Spellman, J. E. Aslan, C. Sander and E. Demir, Patterns, 2021, 100257, 1–12 Search PubMed.
- Z.-R. Anna-Liisa, Y. Jevgenia, Z. Samuel Tassi, P.-I. Tony, M. Iván, M. Jessica, J. T. D. Owen, R. Emek, P. W. Ashok, A. D. Phillip, L. A. Larry and E. Joseph, Blood, 2020, 12, 2346–2358 Search PubMed.
- A. Dugourd, C. Kuppe, M. Sciacovelli, E. Gjerga, A. Gabor, K. B. Emdal, V. Vieira, D. B. Bekker-Jensen, J. Kranz, E. M. J. Bindels and J. Saez-Rodriguez, et al. , Mol. Syst. Biol., 2021, 17, e9730, DOI:10.15252/msb.20209730.
- T. Rubel and A. Ritz, Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, ACM, New York, NY, USA, 2020, vol. 10, pp. 1–10 Search PubMed.
- A. Ritz, C. L. Poirel, A. N. Tegge, N. Sharp, K. Simmons, A. Powell, S. D. Kale and T. M. Murali, NPJ Syst. Biol. Appl., 2016, 2, 1–9 Search PubMed.
- C. S. Magnano and A. Gitter, NPJ Syst. Biol. Appl., 2021, 7, 1–12 CrossRef PubMed.
- R. S. G. Sealfon, A. K. Wong and O. G. Troyanskaya, Nat. Rev. Mater., 2021, 6, 717–729 CrossRef.
- T. Wang, W. Shao, Z. Huang, H. Tang, J. Zhang, Z. Ding and K. Huang, Nat. Commun., 2021, 12, 1–13 CrossRef.
- N. Rappoport and R. Shamir, Nucleic Acids Res., 2018, 46, 10546–10562 CrossRef CAS PubMed.
- D. Veyel, K. Wenger, A. Broermann, T. Bretschneider, A. H. Luippold, B. Krawczyk, W. Rist and E. Simon, Sci. Rep., 2020, 10, 1–14 CrossRef.
- C. Peng, Y. Zheng and D. S. Huang, IEEE/ACM Trans. Comput. Biol. Bioinf., 2020, 17, 1605–1612 CAS.
- Y. Wang, Y. Yang, S. Chen and J. Wang, Brief. Bioinf., 2021, 00, 1–10 Search PubMed.
- B. Wang, A. M. Mezlini, F. Demir, M. Fiume, Z. Tu, M. Brudno, B. Haibe-Kains and A. Goldenberg, Nat. Methods, 2014, 11, 333–337 CrossRef CAS.
- T. Nguyen, R. Tagett, D. Diaz and S. Draghici, Genome Res., 2017, 27, 2025–2039 CrossRef CAS PubMed.
- H. Nguyen, S. Shrestha, S. Draghici and T. Nguyen, Bioinformatics, 2019, 35, 2843–2846 CrossRef CAS PubMed.
- N. K. Speicher and N. Pfeifer, Bioinformatics, 2015, 31, i268–i275 CrossRef CAS.
- J. Mariette and N. Villa-Vialaneix, Bioinformatics, 2018, 34, 1009–1015 CrossRef CAS PubMed.
- Q. Mo, S. Wang, V. E. Seshan, A. B. Olshen, N. Schultz, C. Sander, R. S. Powers, M. Ladanyi and R. Shen, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4245–4250 CrossRef CAS PubMed.
- R. Argelaguet, B. Velten, D. Arnol, S. Dietrich, T. Zenz, J. C. Marioni, F. Buettner, W. Huber and O. Stegle, Mol. Syst. Biol., 2018, 14, 8124 CrossRef PubMed.
- D. Wu, D. Wang, M. Q. Zhang and J. Gu, BMC Genomics, 2015, 16, 1–10 CrossRef.
- N. Rappoport and R. Shamir, Bioinformatics, 2019, 35, 3348–3356 CrossRef CAS PubMed.
- P. Wu, Z. J. Heins, J. T. Muller, L. Katsnelson, I. de Bruijn, A. A. Abeshouse, N. Schultz, D. Fenyö and J. Gao, Mol. Cell. Proteomics, 2019, 18, 1893–1898 CrossRef CAS PubMed.
- Y. Jiao, M. Widschwendter and A. E. Teschendorff, Bioinformatics, 2014, 30, 2360–2366 CrossRef CAS PubMed.
- A. Jones, A. E. Teschendorff, Q. Li, J. D. Hayward, A. Kannan, T. Mould, J. West, M. Zikan, D. Cibula, H. Fiegl and M. Widschwendter, et al. , PLoS Med., 2013, 10, e1001551 CrossRef PubMed.
- Z. E. Sychev, A. Hu, T. A. DiMaio, A. Gitter, N. D. Camp, W. S. Noble, A. Wolf-Yadlin and M. Lagunoff, PLoS Pathog., 2017, 13, e1006256 CrossRef PubMed.
- E. A. Collisson, P. Bailey, D. K. Chang and A. V. Biankin, Nat. Rev. Gastroenterol. Hepatol., 2019, 16, 207–220 CrossRef PubMed.
- Y. Lin, W. Zhang, H. Cao, G. Li and W. Du, Genes, 2020, 11, 1–18 CrossRef.
- R. G. W. Verhaak, K. A. Hoadley, E. Purdom, V. Wang, Y. Qi, M. D. Wilkerson, C. R. Miller, L. Ding, T. Golub, J. P. Mesirov, G. Alexe and D. N. Hayes, et al. , Cancer Cell, 2010, 17, 98–110 CrossRef CAS PubMed.
- H. Noushmehr, D. J. Weisenberger, K. Diefes, H. S. Phillips, K. Pujara, B. P. Berman, F. Pan, C. E. Pelloski, E. P. Sulman and K. Aldape, et al. , Cancer Cell, 2010, 17, 510–522 CrossRef CAS PubMed.
- J. Zhang, M. F. G. Stevens and T. D. Bradshaw, Curr. Mol. Pharmacol., 2011, 5, 102–114 CrossRef PubMed.
- K. Chaudhary, O. B. Poirion, L. Lu and L. X. Garmire, Clin. Cancer Res., 2018, 24, 1248–1259 CrossRef CAS PubMed.
- S. Ogino, P. Lochhead, E. Giovannucci, J. A. Meyerhardt, C. S. Fuchs and A. T. Chan, Oncogene, 2013, 33, 2949–2955 CrossRef PubMed.
- E. Gov and K. Y. Arga, Sci. Rep., 2017, 7, 1–10 CrossRef CAS.
- R. Liu, X. Wang, K. Aihara and L. Chen, Med. Res. Rev., 2014, 34, 455–478 CrossRef PubMed.
- L. Cui, H. Li, W. Hui, S. Chen, L. Yang, Y. Kang, Q. Bo and J. Feng, BMC Bioinf., 2020, 21, 1–14 CrossRef PubMed.
- A. J. Espay, L. V. Kalia, Z. Gan-Or, C. H. Williams-Gray, P. L. Bedard, S. M. Rowe, F. Morgante, A. Fasano, B. Stecher and A. E. Lang, et al. , Neurology, 2020, 94, 481–494 CrossRef.
- W. Yang, J. Soares, P. Greninger, E. J. Edelman, H. Lightfoot, S. Forbes, N. Bindal, D. Beare, J. A. Smith and M. J. Garnett, et al. , Nucleic Acids Res., 2013, 41, D955–D961 CrossRef CAS PubMed.
- R. Yang, B. J. Daigle, L. R. Petzold and F. J. Doyle, BMC Bioinf., 2012, 13, 1–11 Search PubMed.
- T. Ideker and R. Sharan, Genome Res., 2008, 18, 644–652 CrossRef CAS PubMed.
- R. S. Wang and J. Loscalzo, J. Mol. Biol., 2018, 430, 2939–2950 CrossRef CAS PubMed.
- W. Zhang, T. Ota, V. Shridhar, J. Chien, B. Wu and R. Kuang, PLoS Comput. Biol., 2013, 9, e1002975 CrossRef CAS PubMed.
- F. Altieri, T. V. Hansen and F. Vandin, Front. Genet., 2019, 0, 265 CrossRef CAS PubMed.
- E. Bonnet, L. Calzone and T. Michoel, PLoS Comput. Biol., 2015, 11, e1003983 CrossRef PubMed.
- G. Adam, L. Rampášek, Z. Safikhani, P. Smirnov, B. Haibe-Kains and A. Goldenberg, npj Precis. Oncol., 2020, 4, 1–10 CrossRef.
- B. Chen, L. Ma, H. Paik, M. Sirota, W. Wei, M.-S. Chua, S. So and A. J. Butte, Nat. Commun., 2017, 8, 1–12 CrossRef PubMed.
- T. N. Jarada, J. G. Rokne and R. Alhajj, J. Cheminf., 2020, 12, 1–23 Search PubMed.
- S. Z. Mousavi, M. Rahmanian and A. Sami, Infect., Genet. Evol., 2020, 86, 104610 CrossRef CAS PubMed.
- M. Q. Ding, L. Chen, G. F. Cooper, J. D. Young and X. Lu, Genomics, 2018, 16, 269–278 CAS.
- H. Sharifi-Noghabi, O. Zolotareva, C. C. Collins and M. Ester, Bioinformatics, 2019, 35, i501–i509 CrossRef CAS PubMed.
- L. Huang, D. Brunell, C. Stephan, J. Mancuso, X. Yu, B. He, T. C. Thompson, R. Zinner, J. Kim, P. Davies and S. T. C. Wong, Bioinformatics, 2019, 35, 3709–3717 CrossRef CAS PubMed.
- P. Wu, D. Chen, W. Ding, P. Wu, H. Hou, Y. Bai, Y. Zhou, K. Li, S. Xiang, P. Liu and J. G. Chen, et al. , Nat. Commun., 2021, 12, 1–16 CrossRef PubMed.
- X. Wang, Cell Biol. Toxicol., 2018, 34, 163–166 CrossRef CAS PubMed.
- U. D. Akavia, O. Litvin, J. Kim, F. Sanchez-Garcia, D. Kotliar, H. C. Causton, P. Pochanard, E. Mozes, L. A. Garraway and D. Pe’Er, Cell, 2010, 143, 1005–1017 CrossRef CAS PubMed.
- R. Louhimo and S. Hautaniemi, Bioinformatics, 2011, 27, 887–888 CrossRef CAS PubMed.
- A. Bashashati, G. Haffari, J. Ding, G. Ha, K. Lui, J. Rosner, D. G. Huntsman and S. P. Shah, et al. , Genome Biol., 2012, 13, 1–14 CrossRef PubMed.
- O. Gevaert, M. Nabian, S. Bakr, C. Everaert, J. Shinde, A. Manukyan, T. Liefeld, T. Tabor and N. Pochet, et al. , JCO Clin. Cancer Inf., 2020, 1, 421–435 Search PubMed.
- C. Meng, B. Kuster, A. C. Culhane and A. M. Gholami, BMC Bioinf., 2014, 15, 162 CrossRef PubMed.
- P. Ray, L. Zheng, J. Lucas and L. Carin, Bioinformatics, 2014, 30, 1370–1376 CrossRef CAS PubMed.
- M. J. O’Connell and E. F. Lock, Bioinformatics, 2016, 32, 2877–2879 CrossRef PubMed.
- X. Song, J. Ji, K. J. Gleason, F. Yang, J. A. Martignetti, L. S. Chen and P. Wang, Mol. Cell. Proteomics, 2019, 18, S52–S65 CrossRef PubMed.
- T.-T. Giang, T.-P. Nguyen and D.-H. Tran, BMC Med. Inf. Decis. Making, 2020, 20, 1–15 CrossRef PubMed.
- B. Pfeifer and M. G. Schimek, J. Biomed. Inform., 2021, 113, 103636 CrossRef PubMed.
Footnote |
| † Co-first authors. |
| This journal is © The Royal Society of Chemistry 2022 |