Open Access Article
Open Access ArticleA review on the cytotoxicity of graphene quantum dots: from experiment to simulation
Lijun
Liang
 a,
Xiangming
Peng
b,
Fangfang
Sun
a,
Zhe
Kong
*c and
Jia-Wei
Shen
a,
Xiangming
Peng
b,
Fangfang
Sun
a,
Zhe
Kong
*c and
Jia-Wei
Shen
 *d
*d
aCollege of Automation, Hangzhou Dianzi University, Hangzhou, 310018, People's Republic of China. E-mail: zhekong@hdu.edu.cn; Fax: +86 571 87951895
bDepartment of Clinical Laboratory, GuangZhou Red Cross Hospital, 396 Tongfu Zhong Road, Guangzhou 510220, GuangDong, China
cCenter of Advanced Optoelectronic Materials and Devices, College of Materials and Environmental Engineering, Hangzhou Dianzi University, Hangzhou, 310018, People's Republic of China
dSchool of Medicine, Hangzhou Normal University, Hangzhou, 311121, People's Republic of China. E-mail: shen.jiawei@hotmail.com
First published on 26th December 2020
Abstract
Graphene quantum dots (GQDs) generate intrinsic fluorescence and improve the aqueous stability of graphene oxide (GO) while maintaining wide chemical adaptability and high adsorption capacity. Despite GO's remarkable advantages in bio-imaging, bio-sensing, and other biomedical applications, many experiments and simulations have focused on the biosafety of GQDs. Here, we review the findings on the biosafety of GQDs from experiments; then, we review the results from simulated interactions with biological membranes, DNA molecules, and proteins; finally, we examine the intersection between experiments and simulations. The biosafety results from simulations are explained in detail. Based on the literature and our experiments, we also discuss the trends toward GQDs with better biosafety.
1. Introduction
Graphene quantum dots (GQDs), a novel type of zero-dimensional luminescent nanomaterial, are small graphene fragments varying in size from 1 to 100 nm.1,2 Confined by the quantum-size effect, GQDs exhibit extraordinary opto-electronic properties,3–6 because of which they have the potential to replace the well-known metal chalcogenide-based quantum dots. In recent years, GQD-based nanomaterials have become an important research topic and have attracted much attention.7–9 GQDs have achieved rapid growth as breakthrough tools for multiple purposes in various fields of science including photonics, composites, energy,10,11 and electronics.12 In particular, GQD-based nanomaterials have shown much promise in biomedical fields, particularly for diagnostics,13,14 drug delivery,15,16 and bioimaging17,18in vitro and in vivo. At the same time, questions about the biosafety of GQDs have also attracted much attention in recent years.19–23 However, a paradox concerning the biosafety of GQDs has emerged from experiments. Many studies have shown that GQDs have excellent bio-compatibility and low cytotoxicity,24,25 but others have shown them to have high cytotoxicity.26 In particular, the biological interaction mechanism of GQDs with biomolecules including DNA, proteins, and cell membranes is not well understood.GQDs are unique and variable, and many properties including the degree of surface oxidation, surface charge density, doping status, or links with polymers can be dramatically different in different settings, yielding clearly different interaction behaviors with biomolecules. Therefore, experimental studies alone are not sufficient, and they have certain limitations for understanding the cytotoxicity of GQDs (especially at the atomic and molecular levels) which hinder the design and development of GQDs for biomedicine. As a complementary tool to experimentation, theoretical calculation covering the length scale from the atomic to the molecular level has been extensively used to uncover the cytotoxicity mechanism of carbon nanomaterials such as carbon nanotubes,27,28 graphene sheets,29–33 and fullerene.34,35 With the development of supercomputers, theoretical calculations have played a significant role in both providing atomistic pictures of interaction processes and proposing the cytotoxicity mechanism of GQDs at the molecular level.
Excellent reviews have focused on the experimental findings on GQD toxicity.36,37 This review will also be dedicated to the cytotoxicity of GQDs, presenting experimental and theoretical results together with the aim of finding an intersection where experiment and theory can meet. We provide an in-depth view of the contribution of biophysical and theoretical calculations to characterizing the interaction and dynamics of GQDs and biomolecules. The effect of factors including size, geometry, and oxidation degree of GQDs on cytotoxicity will be thoroughly explained. (There are published reviews available on the biosafety of other graphene-based materials as well.) The structure of this review is as follows:
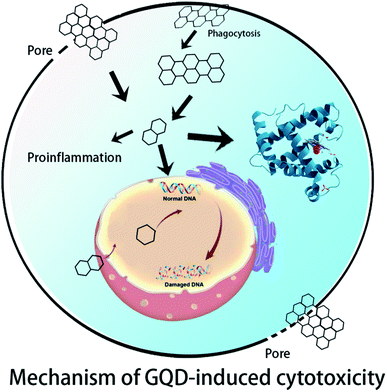
In Section 2, experiments on the cytotoxicity of GQDs in vitro and in vivo are summarized. In Section 3, the simulation results of GQDs with different biomolecules including DNA, proteins, and lipid membranes are summarized. In Section 4, the intersections between experiment and theoretical calculation and the future and outlook of GQD biosafety are discussed (Fig. 1).
2. Experiments on the toxicity of GQDs
Nanomaterials can enter into cells and affect cell division, proliferation, apoptosis, and more. Klopfer et al. found that GQDs of less than 5 nm could enter into E. coli and Bacillus subtilis cells directly and produce toxic effects.38 Chong et al. revealed that the cytotoxicity of GO is less than that of pristine graphene,39 which agreed with Liao's result.40 Akahvan et al. discovered that pristine graphene could greatly affect DNA even at very low concentrations.41 They also pointed out that the genotoxicity of graphene in human stem cells depended on the size of graphene particles.42 This was also confirmed by Ros's experiment, where the small size of GQDs demonstrated good biocompatibility and ability to pass through the ultrafiltration barrier in an in vitro model.43 In addition, due to the different oxidation extent and surface chemistry of GO sheets, GO shows low cytotoxicity in some experiments but high cytotoxicity in others.44,45 As shown in confocal laser scanning microscopy images (Fig. 2B) and consistent with the MTT assay results, cells incubated with a high concentration (200 μg ml−1) of GO and rGO showed bright red fluorescence, indicating that severe cell death was induced; GO seemed to cause more cell death than rGO. In stark contrast, cells treated with GQDs, aminated GQDs (GQD-NH2), carboxyl GQDs (GQD-COOH), and graphene oxide quantum dots (GOQDs) still showed high cell viability. GQD-COOH also showed low systemic toxicity for nanodrug-delivery system in vivo experiments.46 From Wang's experiment, it was also confirmed that GQDs, nitro doped GQDs (GQD-N) and folic acid-modified GQDs (GQD-FA) are all safe by in vitro and in vivo tests.47 There was no significant difference in cell death after treatment with different concentrations of GQD, GQD-NH2, GQD-COOH, and GOQD. Taken together, these results indicate that the concentration and redox state are more important factors for cell viability.48 It was also identified that GQD, GQD-NH2, GQD-COOH, and GOQD could be safely used for biomedical applications as drug carriers.49–52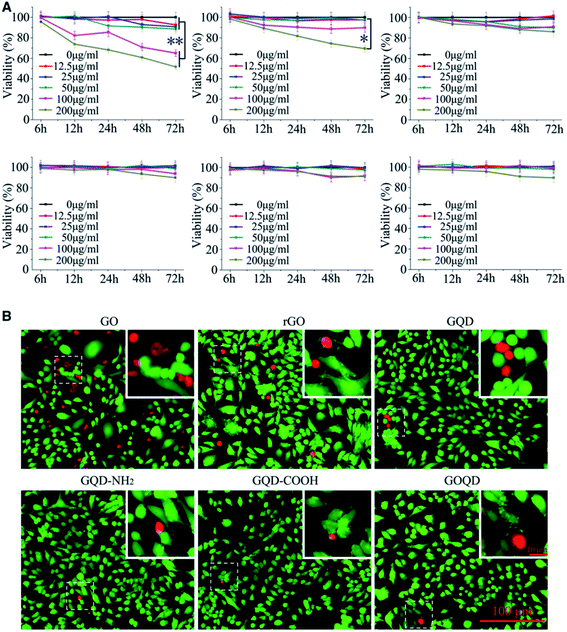 | ||
| Fig. 2 Relative viability of tumor cells after incubation with GQDs. (A) HL-7702 cells incubated with various concentrations of GQDs for 6, 12, 24, 48, and 72 h. (B) Fluorescence images of HL-7702 cells treated with GQDs (200 μg ml−1) for 24 h. Viable cells were stained with calcein AM, while dead cells were stained with PI. Inset: high magnification images of the area within the white dotted square. Reprinted with permission from ref. 48, Copyright© 2018, Oxford University Press. | ||
Several experiments exhibited the interaction of GQDs with proteins. Deng et al. found that nitrogen-doped graphene quantum dots (N-GQDs) could perturb a redox-sensitive system via the selective inhibition of antioxidant enzyme activities in zebrafish.53 However, Jiang et al. discovered that the toxicity of N-GQDs was much lower than that of GO by using surface-enhanced infrared absorption spectroscopy. They found that the adsorption of GO could destroy the integrity of a membrane by extracting the lipid bilayer, and N-GQDs could only disturb the membrane of red blood cells.54 The experiments showed that GQDs exhibit very low cytotoxicity owing to their ultra-small size and high oxygen content in vitro and in vivo.55 Wang et al. found that the permeability and transport mechanism of GQDs across the biological barrier depend on the GQD size.56 Kim et al. pointed out that GQDs could inhibit fibrillization of α-synuclein and trigger their disaggregation.57 This was confirmed with the results of Yang's work.58 Besides membranes and proteins, GQDs also exhibit high affinity to DNA molecules and a certain level of toxicity in DNA molecules in experiments.59,60 Moreover, although the viability of cells shows no significant decrease, the expression level of miR-21, miR-29a, Bax, Bcl2 and PTEN genes was affected greatly after treatment with large sized GQDs (50 nm).23 In Yu's experiment, OH-GQDs, rGQDs, and NH2-GQDs led to global DNA hypermethylation in zebrafish, and surface chemical groups of GQDs were critical for modulating DNA methylation.61
3. Computational studies
Computational studies on the interaction of GQDs and biomolecules including the lipid bilayer, proteins, and DNA molecules have been widely carried out. Li et al. found that the nearly orthogonal orientation of a sharp graphene corner with respect to the bilayer minimized the interactive free energy and is the thermodynamically preferred configuration even before penetration begins when using coarse-grained MD (CG-MD). They also found that pristine GQDs could enter into the lipid membrane spontaneously when they put the closed angle of GQDs in the membrane. To further explore the free energy barrier between GQDs and the membrane, the all-atomic molecular dynamics (AA-MD) simulation was used. As seen in Fig. 3B, the energy barrier of the closed angle of GQDs translocating through the membrane was approximately 5 kBT based on their potential of mean force (PMF) calculation. After the insertion of the sharp corner of GQDs, the interaction between the hydrophobic tail of a lipid and hydrophobic GQD increased. Further studies revealed that the free energy barrier of translocation of GQDs through the membrane depends on four variables: hH and hT (thicknesses of the head and tail groups in the lipid monolayer, as shown in Fig. 3C) and γH and γT (interaction energy densities between one side surface of GQDs and head and tail groups of lipids relative to the concentration of GQDs in the solvent). The energy may be predicted using the following equation:E = 2(1 − γH/γT)hH2 × γH × tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α α | (1) |
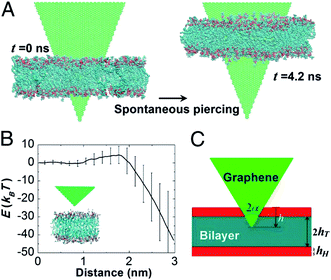 | ||
| Fig. 3 All-atom molecular dynamics simulations of corner piercing of GQDs across a lipid bilayer. (A) Simulations directly showing that the corner piercing proceeds spontaneously. (B) GQD–bilayer interaction energy as a function of the penetration distance, showing the existence of an energy barrier of about 5 kBT associated with corner piercing. The mean value of interaction energy is obtained from 11 independent simulation runs and the error bars show the SD. The relatively large fluctuations of interaction energy at a large penetration distance are mainly due to random translational and rotational movements of GQDs relative to the bilayer membrane and random configurational changes of individual lipids adjacent to the GQD. (C) Analytical model of corner piercing. Reprinted with permission from ref. 62, Copyright (2013) National Academy of Sciences. | ||
Dallavalle et al. also found from CG-MD simulation that small GQDs (<5.2 nm) could enter into a membrane spontaneously. The large GQDs (>11.2 nm) mainly lay flat on the top of the bilayer, where they wreaked havoc on the membrane and created a patch of upturned phospholipids.63 As seen in Fig. 4, the z coordination of phospholipid heads when the membrane is pierced by the GQD is more ordered than when the GQD is adsorbed onto the membrane. As shown in Table 1, the order parameter of the membrane is 0.77 with a 5.2 nm GQD piercing the membrane, and it is only 0.03 with a 5.2 nm GQD adsorbed onto the membrane. This large gap between order parameters showed that GQDs adsorbed onto a lipid membrane can greatly disturb it. In addition, the order parameter of the membrane decreased with increasing GQD size. The parameter was 0.72 with the 0.9 nm GQD piercing the membrane, and it decreased to 0.1 with an 11.2 nm GQD. Thus, the larger GQD induced more local anti-alignment (S was negative for anti-alignment). This also indicated that a GQD adhering on the membrane could disrupt it much more than one piercing the membrane. A hydrophobic GQD adhered to the top of a membrane made the GQDs interact with the hydrophobic tail and the hydrophilic phospholipids of the layer directly under the capsized sheet. The anti-alignment was therefore truly related to the hydrophobic–hydrophobic interaction that allowed the sheet to adhere to the membrane. Importantly, the overturned phospholipids were able to impair cell function and disrupt the functioning of the membrane proteins. This may explain the cytotoxic activity of adhered GQDs, the so-called masking effect in experiments.64–67
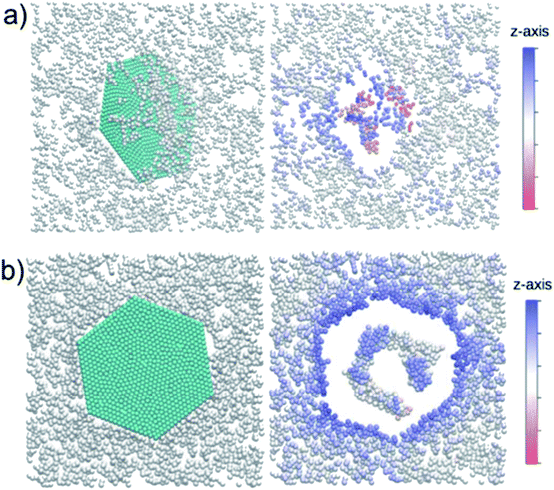 | ||
| Fig. 4 Two views of the interaction of GQDs with the membrane are depicted corresponding to (a) piercing through and (b) adsorbing onto the membrane. The z-axis color code corresponds to the position of the phospholipid heads. The graphene flake locally affects the membrane structure. The empty spaces are occupied by the tails. Phospholipids are displaced with respect to the z-average position. Water molecules are not shown. Reprinted (adapted) with permission from ref. 63. Copyright (2015) American Chemical Society. | ||
| Nanosheet size(nm) | GQD piercing through the membrane | GQD adhering to the membrane | ||
|---|---|---|---|---|
| S local | S global | S local | S global | |
| 0.9 | 0.72 | 0.69 | — | — |
| 2.7 | 0.72 | 0.69 | — | — |
| 5.2 | 0.77 | 0.68 | 0.03 | 0.66 |
| 8.1 | 0.34 | 0.65 | −0.16 | 0.59 |
| 11.2 | 0.1 | 0.57 | −0.16 | 0.52 |
| 13.3 | — | — | −0.13 | 0.45 |
The piercing behavior of small GQDs and the adhering behavior of large GQDs was also found in our research from AA-MD simulation.68 By calculating the PMF of GQDs with different sizes including GQD7, GQD19, and GQD61, the translocation mechanism of GQDs through the lipid membrane was further interpreted. As seen in Fig. 5, the free energy of GQDs decreased as they moved from the water phase into the lipid membrane. This indicates that GQDs preferred to enter into the lipid membrane. The free energy differences between the lowest point in the membrane of GQDs to that of GQDs in the water phase of are −56.3 kJ mol−1, −55.2 kJ mol−1 and −56.1 kJ mol−1 for GQD7, GQD19 and GQD61. The positions corresponding to the lowest free energy in the z direction are −1.10 nm and 0.98 nm for GQD7 and GQD19. However, the position corresponding to lowest free energy in the z direction increased to 1.75 nm for GQD61. This implies that the location of the lowest free energy in the z direction moves farther from the middle of the lipid membrane as the GQD size increases from GQD19 to GQD61. The free energy barrier increased from 35.1 kJ mol−1 to 96.2 kJ mol−1 with the increase of GQD size from GQD19 to GQD61. The free energy barrier for GQDs passing through the lipid membrane was much higher with increased GQD size. The relatively small GQDs (GQD7 and GQD19) could pierce the membrane, and the large GQDs could stably adsorb on top of it.
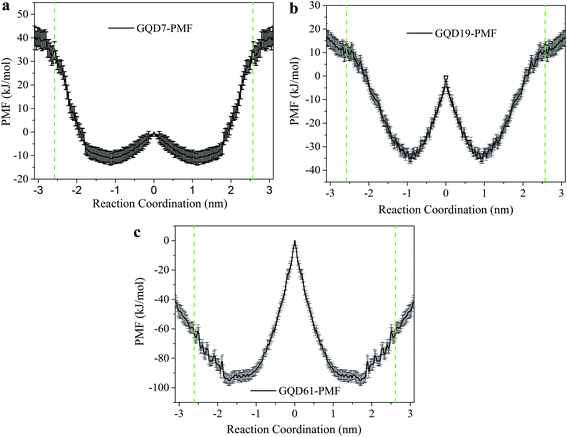 | ||
| Fig. 5 Potential of mean force (PMF) of GQDs with different sizes translocating through the POPC membrane. (a) GQD7; (b) GQD19; (c) GQD61. Green dashed lines represent the location of two ends of the POPC membrane. Reprinted (adapted) with permission from ref. 68. Copyright (2016) American Chemical Society. | ||
Besides the piercing and adsorption models, Tu et al. further revealed that large GQDs could destroy the lipid bilayer by an extraction process.69 As seen in Fig. 6, three distinguishable modes are observed during the process of the GQD spontaneously entering the outer and inner cell membranes. First, the swing mode, the initial stage in which the GQD underwent an unbiased orientation swing, swinging back and forth around the constrained atoms. This process lasted for a short period of time. Second, the insertion mode, in which the strong van der Waals force (vdW) between the GQD and lipid membrane caused the insertion of the GQD into the membrane. In the third mode (the extraction mode), the GQD started to extract phospholipid molecules on the cell membrane surface, destroying the cell membrane structure. This shows that GQDs can not only be inserted into a cell membrane by physical cutting, but can also destructively extract lipid molecules on the surface of the membrane through hydrophobic interaction. GQDs can interact strongly with the phospholipid molecules on the bacterial cell membrane, which causes a large number of phospholipid molecules to break away from the membrane and adsorb to the graphene surface.
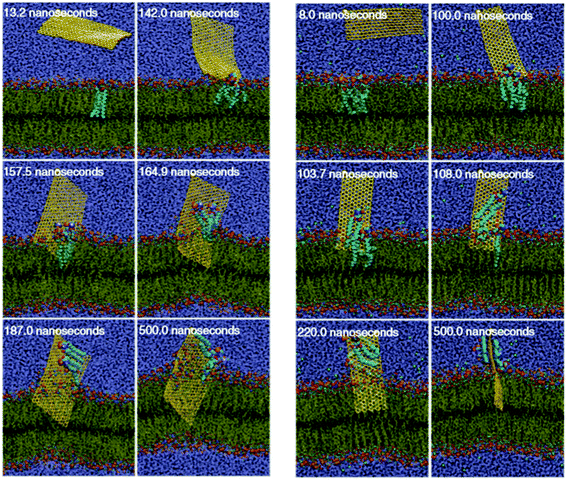 | ||
| Fig. 6 Insertion of a GQD into the cell: (a) outer membrane and (b) inner membrane. Reprinted with permission from ref. 69, Copyright© 2013, Springer Nature. | ||
Besides the effect of GQD size, the effect of GQD concentration on the membrane was also investigated. As seen in Fig. 7a, multiple small GQDs aggregated into clusters in the aqueous solution before translocation into the lipid membrane during the first 10 ns. This is similar to the permeation of fullerene into the POPC membrane.34,70 After this, the aggregates permeated into the POPC membrane at 20 ns, as shown in Fig. 7b. In Fig. 7c, most of the GQDs were still aggregated in one large cluster. However, as opposed to the aggregates at 10 ns, most GQDs in the aggregates tend to be parallel to the POPC lipid at 110 ns. The aggregate of GQD7 tends to dissociate and disperse to the position of energy minimum on both sides of the POPC membrane. The hydrophobic GQDs tend to adsorb on the tail of POPC lipids in the membrane, and GQD7 tends to be parallel to the main chain of POPC molecules. The simulation revealed that GQD7 tends to permeate into the membrane as aggregates but disperses in the membrane after permeation. This may cause disruption of the membrane (pore formation) in the piercing process of GQD7 under high concentration, but not after permeation. It could also explain why small GQDs do not display any cytotoxicity at low concentrations but have high cytotoxicity at high concentrations.
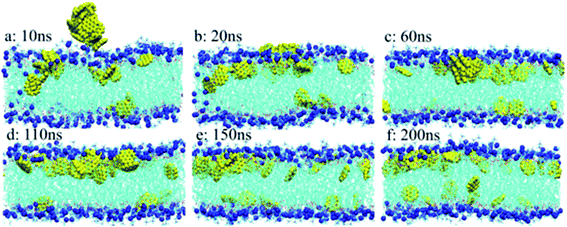 | ||
| Fig. 7 Aggregated structures and distribution of GQD7-H3 during simulation: (a) 10 ns; (b) 20 ns; (c) 60 ns; (d) 110 ns; (e) 150 ns; and (f) 200 ns. GQDs are represented by the yellow vdW model, the POPC membrane is represented by the line model, and the P atoms in the membrane are represented by the blue vdW model. For clarity, water molecules are not shown. Reprinted (adapted) with permission from ref. 68. Copyright (2016) American Chemical Society. | ||
In addition, simulation helps us to predict the average time of entry of nanoparticles in lipid membranes, using a combination of molecular dynamics simulations and statistical approaches.71 From the results of the theoretical model, researchers have found that different sizes and shapes of GQD molecules can affect the translocation time greatly, and this data was confirmed by experiments. Yang et al. found through all-atom molecular dynamics simulation (AAMD) that the dynamics of GQD translocation through a membrane could be modulated by the charge on the GQDs.72 Using coarse-grained MD (CGMD), as seen in Fig. 8, GQDs could be incorporated with the membrane in a sandwiched structure.73Fig. 8a illustrates how the micelle including GQDs and lipids merged. As the micelle slowly merges with the top lipid layer, some lipids become trapped below the attached micelle (Fig. 8b). At t = 120 ns, a neck-like protrusion formed on the bottom part of the bilayer (Fig. 8c). This initiated fusion of GQDs into the membrane at t = 360 ns, as shown in Fig. 8d. The process continued until the GQD was stabilized in the center of the bilayer membrane at t = 516 ns (Fig. 8f). The results were confirmed by both Brownian dynamics simulations and by experiments.74
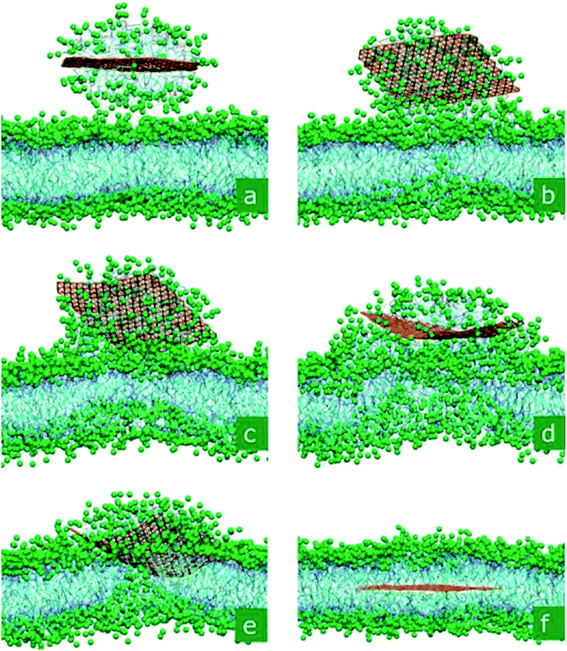 | ||
| Fig. 8 Self-insertion of GQDs inside a phospholipid membrane. In the displayed process, a GQD micelle merges with a membrane and releases the monolayer, which penetrates into the membrane. The snapshots were taken at ta–f = 2.9, 52.4, 120.0, 299.2, 356.4, and 516.4 ns. Reprinted (adapted) with permission from ref. 73. Copyright (2010) American Chemical Society. | ||
It is estimated that there are more than 1 million protein types in the human proteome. The interaction between materials and biomolecules (such as DNA and proteins) can interfere with biological functions and lead to cytotoxicity.75 Graphene can break the structure of polypeptides, protein fragments, and proteins.76,77 Evidently GQDs can also interact with proteins and other biological macromolecules. Fang et al. revealed the interaction mechanism of GQDs and the ubiquitin protein by both experiments and MD simulations.78 Our research group79 used AAMD to reveal the effect of GQD size on the structure of the protein villin headpiece (HP35). With increased GQD size, more protein residues could adsorb on the GQDs and the adsorption capacity of protein on GQDs increased. Meanwhile, the damage to the secondary structure of proteins rose along with GQD size. Moreover, a remarkable capacity of GQDs in regulating aberrant protein expression was revealed through H-bonding and hydrophobic interactions80 from discrete MD. GQDs were able to exhibit potential toxicity by disrupting the protein–protein interaction via the hydrophobic interaction.81Fig. 9b–e illustrate the insertion process of the GQD into the protein–protein interaction from the trajectory defined as sim-1. Initially parallel and close to the dimer's interface, the GQD rotated and came into contact with one monomer after 2 ns (Fig. 9b). Because the graphene surface is hydrophobic, typically a protein molecule with hydrophobic residues on its surface can be adsorbed on the GQD. From 30 ns to 40 ns, the protein monomer began to rotate around its barrel axis, due to the preferred hydrophobic interaction between the GQD and non-polar residues at the dimer interface. After this, the GQD was fully inserted into the dimer and the dimer was separated. Other simulations confirmed that this insertion process was representative. In addition, the amyloid fibrils could be destroyed by penetration and extraction of the peptides by GQDs due to the strong interaction between peptides and GQDs.82,83 In addition to lipid membranes and proteins, interaction with DNA molecules is also an essential topic for the evaluation of GQD toxicity. Pristine GQDs can interact with DNA molecules strongly.84 As GQD oxidation increases, the interaction between DNA and GQDs decreases. For clarity, the simulations of GQD with biomolecules were summarized in Table 2.
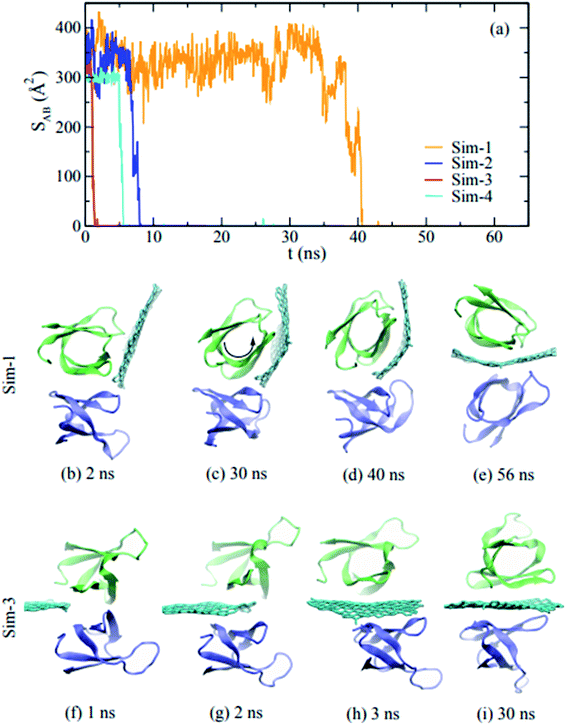 | ||
| Fig. 9 (a) Time-dependent contact areas of the dimer during the insertion of a graphene sheet; (b)–(i) snapshots of the insertion process of a graphene sheet into the dimer. Reprinted (adapted) with permission from ref. 81. Copyright (2015) American Chemical Society. | ||
| Materials | Simulation Methods | Force field | Time scale | Results | References |
|---|---|---|---|---|---|
| GQDs | MD | GROMOS54A7 | Hundreds of ns | GQDs tend to penetrate into the membrane perpendicularly | Vatanparast et al.85 |
| GQDs | MD | CHARMM27 | Hundreds of ns | Graphene quantum dots (GQDs) act as a potent inhibitor against the in vivo aggregation and toxicity of human islet amyloid polypeptide (IAPP) | Wang et al.86 |
| GQDs | MD | AMBER99 | Hundreds of ns | GQDs interact with human peroxidases and are degraded by them | Martín et al.87 |
| GQDs | MD | CHARMM36 | Hundreds of ns | The translocation of GQDs could be modulated by charge on the GQDs. | Tang et al.,72 |
| GQD (sheet) | CGMD | — | Hundreds of ns | Graphene sheets could be hosted in the hydrophobic interior of biological membranes formed by amphiphilic phospholipid molecules | Titov et al.73 |
| GQDs | MD | CHARMM27 | Hundred of ns | The time of entry of nanoparticles in lipid membranes | Liu et al.71 |
| GQDs | MD | CHARMM27 | Hundreds of ns | Small GQDs can translocate through membranes via drugs. | Xue et al.88 |
| Graphene | Brownian MD | — | ∼hundreds of μs | GQDs incorporated with membranes as a sandwiched graphene–cell membrane superstructure | Chen et al.74 |
| GQDs GOQDs | MD | Universal force field | Dozens of ns | GQDs interact more strongly with DNA than GOQDs. | Wang et al.84 |
| GQDs | MD | CHARMM27 | Dozens of ns | The chirality of D-GQDs provides a stronger tendency to accumulate within the cellular membrane than that of L-GQDs. | Suzuki et al.89 |
| GQDs | Discrete MD | CHARMM27 | — | A remarkable capacity of GQDs in regulating aberrant protein expression through H-bonding and hydrophobic interactions | Faridi et al.80 |
| GQDs | MD | CHARMM27 | ∼Hundreds of ns | GQDs can destroy the lipid membrane by extracting the lipid molecules | Tu et al.69 |
| GQDs | MD | CHARMM27 | Hundreds of ns | GQDs can disrupt protein–protein interaction | Luan et al.81 |
| GQDs | MD | CHARMM27 | 1 μs | Destruction of amyloid fibrils by graphene through penetration and extraction of peptides | Yang et al.82 |
| GQDs | MD | CHARMM27 | Hundreds of ns | Small GQDs cannot disturb membranes. | Liang et al.68 |
| GQDs | MD | CHARMM27 | Hundreds of ns | Small GQDs can penetrate membranes | Xue et al.88 |
| GQDs | MD | CHARMM27 | Hundreds of ns | Secondary structure of proteins can be destroyed by GQDs | Zhou et al.79 |
| GQDs | MD | CHARMM27 | Hundreds of ns | DNA fragment | Kong et al.90 |
4. Cross point between experiments and simulations
We have discussed the agreement among the experiments and theoretical work on the interaction of GQDs with biomolecules including lipids, proteins, and DNA molecules. In terms of the effect of GQD size on lipid membranes, the results from the simulations and the experiments correspond well for the lipid bilayer. Small GQDs can only penetrate the membrane, and simulation also gave us the atomic details of the penetration process. Large GQDs were seen to wreak havoc on the lipid bilayer, in both simulations and experiments. Simulation allowed observation of the extraction process of lipids from the membrane by GQDs.In terms of GQD concentration, the toxicity of GQDs increases along with concentration. With higher concentrations of GQDs, the GQDs aggregate into a larger cluster and can form pores in the lipid membrane. However, the time scale for the translocation of GQDs through the membrane increases exponentially with GQD size. Limited computational resources have prevented investigation of the translocation mechanism of large GQDs by AA-MD and CG-MD. However, a simplified model such as DPD could be used to describe the translocation mechanism of large GQDs.
Regarding interactions between GQDs and protein and DNA molecules, strong adsorption between GQDs and biomolecules was observed in both experiments and simulations. Due to the strong hydrophobic interaction between GQDs and proteins, proteins can adsorb on GQDs strongly, and the conformation of proteins on the GQDs changes greatly. In particular, GQDs can insert themselves into the interior of proteins and disrupt protein–protein interactions. Due to the strong π–π interaction between GQDs and DNA molecules, GQDs can also change the structure of DNA molecules.
5. Summary of the intersection and outlook
In summary, the aim of this review was to bring together results from computer simulations and computational models relating to experiments on the cytotoxicity of GQDs. Simulations and experiments also assessed the interaction of lipids and functionalized GQDs. However, the results from these two approaches cannot be compared directly. The first reason is the relative diversity of functionalized GQDs in experiments compared to simulations. Also, the scale of GQDs in the experiments was larger than that in the simulations. Simulation with simplified functionalized GQDs could shed light on the effect of particular functional groups on the translocation of GQDs. To enhance the potential applications of GQDs in biomedicine, surface modification and functionalization are important methods to improve their properties. However, due to the diversity and complexity of surface modifications of GQDs, a systematic evaluation of the effect of surface modification of GQDs on their toxicity and function is still lacking both in experiments and in theoretical simulation. In particular, the simulation of the biosafety effect of polymer modified GQD is still an interesting and challenging task.Thus, the first task is to further elucidate the interaction of functionalized GQDs and biomolecules including lipids, proteins, and DNA molecules based on simulation and experiments. Several challenges also remain to be overcome in the research process itself. One is the scale gap between theoretical work and experiments. In almost all theoretical simulations, the size of GQDs is less than 10 nm, and larger GQDs (10–100 nm) should be investigated with simulation in order to allow comparison with experiments performed with larger GQDs. Another challenge is the time scale; the time scale of GQD cytotoxicity in experiments is based on hours or even days and years, but in simulation the time scale is measured in ns or μs, a tiny fraction in comparison. We recommend further development of accelerated MD simulation to resolve this mismatch. Finally, the force fields generally used in MD simulation are not sufficiently accurate. Most simulations cited in this review used the AMBER and CHARMM force fields to describe the potential energy of biomolecules. The performance of these two force fields is considered superior for describing biomolecules; however, we believe that more accurate force fields need to be developed in order to achieve a more comprehensive understanding of GQD cytotoxicity. Ideally, these force fields would precisely describe the cross-interaction between biomolecules and inorganic materials and consider the polarization effect.
In recent years, our group has attempted to answer open questions remaining in this field and provide a perspective on the future of GQD cytotoxicity research. With advances in nanotechnology and theoretical modeling, we anticipate that a nanoscale biosafety evaluation based on GQDs or other nanomaterials could be developed in the future. The studies we have summarized here represent the first steps toward this pressing goal.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant No. 21978060, 51702073, 21674032, and 11904300) and Key Research and Development Plan of Zhejiang Province (No. 2020C01008). This work was also supported by the start up funding of Hangzhou Dianzi University (No. KYS195618111), the Key Fostering Project of Scientific Research of Hangzhou Normal University (2018PYXML006) and the High Level Returned Overseas Chinese Innovation Projects in Hangzhou.References
- M. O. Valappil, V. K. Pillai and S. Alwarappan, Spotlighting graphene quantum dots and beyond: Synthesis, properties and sensing applications, Appl. Mater. Today, 2017, 9, 350–371 CrossRef.
- H. Sun, L. Wu, W. Wei and X. Qu, Recent advances in graphene quantum dots for sensing, Mater. Today, 2013, 16(11), 433–442 CrossRef CAS.
- P. Das, S. Ganguly, S. Banerjee and N. C. Das, Graphene based emergent nanolights: a short review on the synthesis, properties and application, Res. Chem. Intermed., 2019, 45(7), 3823–3853 CrossRef CAS.
- Y. R. Kumar, K. Deshmukh, K. K. Sadasivuni and S. K. K. Pasha, Graphene quantum dot based materials for sensing, bio-imaging and energy storage applications: a review, RSC Adv., 2020, 10(40), 23861–23898 RSC.
- G. S. Kumar, R. Roy, D. Sen, U. K. Ghorai, R. Thapa, N. Mazumder, S. Saha and K. K. Chattopadhyay, Amino-functionalized graphene quantum dots: origin of tunable heterogeneous photoluminescence, Nanoscale, 2014, 6(6), 3384–3391 RSC.
- K. A. Ritter and J. W. Lyding, The influence of edge structure on the electronic properties of graphene quantum dots and nanoribbons, Nat. Mater., 2009, 8(3), 235–242 CrossRef CAS.
- J. M. Yoo, J. H. Kang and B. H. Hong, Graphene-based nanomaterials for versatile imaging studies, Chem. Soc. Rev., 2015, 44(14), 4835–4852 RSC.
- M. Mehrzad-Samarin, F. Faridbod, A. S. Dezfuli and M. R. Ganjali, A novel metronidazole fluorescent nanosensor based on graphene quantum dots embedded silica molecularly imprinted polymer, Biosens. Bioelectron., 2017, 92, 618–623 CrossRef CAS.
- H. Zhao, R. Ding, X. Zhao, Y. Li, L. Qu, H. Pei, L. Yildirimer, Z. Wu and W. Zhang, Graphene-based nanomaterials for drug and/or gene delivery, bioimaging, and tissue engineering, Drug Discovery Today, 2017, 22(9), 1302–1317 CrossRef CAS.
- X. Li, M. Rui, J. Song, Z. Shen and H. Zeng, Carbon and graphene quantum dots for optoelectronic and energy devices: a review, Adv. Funct. Mater., 2015, 25(31), 4929–4947 CrossRef CAS.
- Q. Chen, Y. Hu, C. Hu, H. Cheng, Z. Zhang, H. Shao and L. Qu, Graphene quantum dots–three-dimensional graphene composites for high-performance supercapacitors, Phys. Chem. Chem. Phys., 2014, 16(36), 19307–19313 RSC.
- J. Feng, H. Dong, L. Yu and L. Dong, The optical and electronic properties of graphene quantum dots with oxygen-containing groups: a density functional theory study, J. Mater. Chem. C, 2017, 5(24), 5984–5993 RSC.
- K. Ghanbari, M. Roushani and A. Azadbakht, Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen, Anal. Biochem., 2017, 534, 64–69 CrossRef CAS.
- N. Chauhan, T. Maekawa and D. N. S. Kumar, Graphene based biosensors—accelerating medical diagnostics to new-dimensions, J. Mater. Res., 2017, 32(15), 2860–2882 CrossRef CAS.
- D. Iannazzo, A. Pistone, M. Salamò, S. Galvagno, R. Romeo, S. V. Giofré, C. Branca, G. Visalli and A. Di Pietro, Graphene quantum dots for cancer targeted drug delivery, Int. J. Pharm., 2017, 518(1–2), 185–192 CrossRef CAS.
- X. Wang, X. Sun, J. Lao, H. He, T. Cheng, M. Wang, S. Wang and F. Huang, Multifunctional graphene quantum dots for simultaneous targeted cellular imaging and drug delivery, Colloids Surf., B, 2014, 122, 638–644 CrossRef CAS.
- X. Tan, Y. Li, X. Li, S. Zhou, L. Fan and S. Yang, Electrochemical synthesis of small-sized red fluorescent graphene quantum dots as a bioimaging platform, Chem. Commun., 2015, 51(13), 2544–2546 RSC.
- W.-S. Kuo, C.-Y. Chang, K.-S. Huang, J.-C. Liu, Y.-T. Shao, C.-H. Yang and P.-C. Wu, Amino-Functionalized Nitrogen-Doped Graphene-Quantum-Dot-Based Nanomaterials with Nitrogen and Amino-Functionalized Group Content Dependence for Highly Efficient Two-Photon Bioimaging, Int. J. Mol. Sci., 2020, 21(8), 2939 CrossRef CAS.
- H. Zheng, R. Ma, M. Gao, X. Tian, Y.-Q. Li, L. Zeng and R. Li, Antibacterial applications of graphene oxides: structure-activity relationships, molecular initiating events and biosafety, Sci. Bull., 2018, 63(2), 133–142 CrossRef CAS.
- M. Chu, Toxicity of Graphene Quantum Dots. World Scientific Encyclopedia of Nanomedicine and Bioengineering Ii, the: Bioimplants, Regenerative Medicine, and Nano-Cancer Diagnosis and Phototherapy (A 3-volume Set), 2017, vol. 9 Search PubMed.
- Z. G. Wang, Z. Rong, D. Jiang, E. S. Jing, X. U. Qian, S. I. Jing, Y. P. Chen, Z. Xin, G. A. N. Lu and J. Z. Li, Toxicity of graphene quantum dots in zebrafish embryo, Biomed. Environ. Sci., 2015, 28(5), 341–351 Search PubMed.
- D. Zhang, Z. Zhang, Y. Wu, K. Fu, Y. Chen, W. Li and M. Chu, Systematic evaluation of graphene quantum dot toxicity to male mouse sexual behaviors, reproductive and offspring health, Biomaterials, 2019, 194, 215–232 CrossRef CAS.
- M. S. Hashemi, S. Gharbi, S. Jafarinejad-Farsangi, Z. Ansari-Asl and A. S. Dezfuli, Secondary toxic effect of graphene oxide and graphene quantum dots alters the expression of miR-21 and miR-29a in human cell lines, Toxicol. in Vitro, 2020, 65, 104796 CrossRef CAS.
- S. Fasbender, L. Zimmermann, R.-P. Cadeddu, M. Luysberg, B. Moll, C. Janiak, T. Heinzel and R. Haas, the Low toxicity of Graphene Quantum Dots is Reflected by Marginal Gene expression changes of primary Human Hematopoietic Stem cells, Sci. Rep., 2019, 9(1), 1–13 CrossRef CAS.
- C. Zhu, Z. Chen, S. Gao, B. L. Goh, I. B. Samsudin, K. W. Lwe, Y. Wu, C. Wu and X. Su, Recent advances in non-toxic quantum dots and their biomedical applications, Prog. Nat. Sci.: Mater. Int., 2019, 29(6), 628–640 CrossRef CAS.
- K. Murugan, D. Nataraj, A. Jaganathan, D. Dinesh, S. Jayashanthini, C. M. Samidoss, M. Paulpandi, C. Panneerselvam, J. Subramaniam and M. Nicoletti, Nanofabrication of graphene quantum dots with high toxicity against malaria mosquitoes, Plasmodium falciparum and MCF-7 cancer cells: impact on predation of non-target tadpoles, odonate nymphs and mosquito fishes, J. Cluster Sci., 2017, 28(1), 393–411 CrossRef CAS.
- C. Ge, J. Du, L. Zhao, L. Wang, Y. Liu, D. Li, Y. Yang, R. Zhou, Y. Zhao and Z. Chai, Binding of blood proteins to carbon nanotubes reduces cytotoxicity, Proc. Natl. Acad. Sci., 2011, 108(41), 16968–16973 CrossRef CAS.
- C. A. Jimenez-Cruz, S. g. Kang and R. Zhou, Large scale molecular simulations of nanotoxicity, Wiley Interdiscip. Rev. Syst. Biol. Med., 2014, 6(4), 329–343 CrossRef CAS.
- J. Chen, G. Zhou, L. Chen, Y. Wang, X. Wang and S. Zeng, Interaction of graphene and its oxide with lipid membrane: a molecular dynamics simulation study, J. Phys. Chem. C, 2016, 120(11), 6225–6231 CrossRef CAS.
- E. Duverger, F. Picaud, L. Stauffer and P. Sonnet, Simulations of a graphene nanoflake as a nanovector to improve ZnPc phototherapy toxicity: from vacuum to cell membrane, ACS Appl. Mater. Interfaces, 2017, 9(43), 37554–37562 CrossRef CAS.
- R. Guo, J. Mao and L.-T. Yan, Computer simulation of cell entry of graphene nanosheet, Biomaterials, 2013, 34(17), 4296–4301 CrossRef CAS.
- L. Zhang, B. Xu and X. Wang, Cholesterol extraction from cell membrane by graphene nanosheets: a computational study, J. Phys. Chem. B, 2016, 120(5), 957–964 CrossRef CAS.
- J. Mao, R. Guo and L.-T. Yan, Simulation and analysis of cellular internalization pathways and membrane perturbation for graphene nanosheets, Biomaterials, 2014, 35(23), 6069–6077 CrossRef CAS.
- J. Wong-Ekkabut, S. Baoukina, W. Triampo, I. M. Tang, D. P. Tieleman and L. Monticelli, Computer simulation study of fullerene translocation through lipid membranes, Nat. Nanotechnol., 2008, 3(6), 363 CrossRef CAS.
- R. Gupta and B. Rai, Molecular dynamics simulation study of translocation of fullerene C 60 through skin bilayer: effect of concentration on barrier properties, Nanoscale, 2017, 9(12), 4114–4127 RSC.
- S. Wang, I. S. Cole and Q. Li, The toxicity of graphene quantum dots, RSC Adv., 2016, 6(92), 89867–89878 RSC.
- X. Zhao; W. Gao; H. Zhang; X. Qiu and Y. Luo, Graphene quantum dots in biomedical applications: recent advances and future challenges, in Handbook of Nanomaterials in Analytical Chemistry, Elsevier: 2020; pp 493–505 Search PubMed.
- J. A. Kloepfer, R. E. Mielke and J. L. Nadeau, Uptake of CdSe and CdSe/ZnS quantum dots into bacteria via purine-dependent mechanisms, Appl. Environ. Microbiol., 2005, 71(5), 2548–2557 CrossRef CAS.
- Y. Chong, C. Ge, Z. Yang, J. A. Garate, Z. Gu, J. K. Weber, J. Liu and R. Zhou, Reduced cytotoxicity of graphene nanosheets mediated by blood-protein coating, ACS Nano, 2015, 9(6), 5713–5724 CrossRef CAS.
- K.-H. Liao, Y.-S. Lin, C. W. Macosko and C. L. Haynes, Cytotoxicity of graphene oxide and graphene in human erythrocytes and skin fibroblasts, ACS Appl. Mater. Interfaces, 2011, 3(7), 2607–2615 CrossRef CAS.
- O. Akhavan, E. Ghaderi, H. Emamy and F. Akhavan, Genotoxicity of graphene nanoribbons in human mesenchymal stem cells, Carbon, 2013, 54, 419–431 CrossRef CAS.
- O. Akhavan, E. Ghaderi and A. Akhavan, Size-dependent genotoxicity of graphene nanoplatelets in human stem cells, Biomaterials, 2012, 33(32), 8017–8025 CrossRef CAS.
- C. Hadad, J. M. González-Domínguez, S. Armelloni, D. Mattinzoli, M. Ikehata, A. Istif, A. Ostric, F. Cellesi, C. M. Alfieri and P. Messa, Graphene quantum dots: From efficient preparation to safe renal excretion, Nano Res., 2020, 1–10 Search PubMed.
- M. Lv, Y. Zhang, L. Liang, M. Wei, W. Hu, X. Li and Q. Huang, Effect of graphene oxide on undifferentiated and retinoic acid-differentiated SH-SY5Y cells line, Nanoscale, 2012, 4(13), 3861–3866 RSC.
- X. Zhang, W. Hu, J. Li, L. Tao and Y. Wei, A comparative study of cellular uptake and cytotoxicity of multi-walled carbon nanotubes, graphene oxide, and nanodiamond, Toxicol. Res., 2012, 1(1), 62–68 CrossRef CAS.
- Z. Li, J. Fan, C. Tong, H. Zhou, W. Wang, B. Li, B. Liu and W. Wang, A smart drug-delivery nanosystem based on carboxylated graphene quantum dots for tumor-targeted chemotherapy, Nanomedicine, 2019, 14(15), 2011–2025 CrossRef CAS.
- S. Zhang, X. Pei, Y. Xue, J. Xiong and J. Wang, Bio-safety assessment of carbon quantum dots, N-doped and folic acid modified carbon quantum dots: A systemic comparison, Chin. Chem. Lett., 2020, 31(6), 1654–1659 CrossRef CAS.
- J. Li, X. Zhang, J. Jiang, Y. Wang, H. Jiang, J. Zhang, X. Nie and B. Liu, Systematic assessment of the toxicity and potential mechanism of graphene derivatives in vitro and in vivo, Toxicol. Sci., 2019, 167(1), 269–281 CrossRef CAS.
- C. Zhao, X. Song, Y. Liu, Y. Fu, L. Ye, N. Wang, F. Wang, L. Li, M. Mohammadniaei and M. Zhang, Synthesis of graphene quantum dots and their applications in drug delivery, J. Nanobiotechnol., 2020, 18(1), 1–32 CrossRef.
- D. Iannazzo, A. Pistone, C. Celesti, C. Triolo, S. Patané, S. V. Giofré, R. Romeo, I. Ziccarelli, R. Mancuso and B. Gabriele, A Smart Nanovector for Cancer Targeted Drug Delivery Based on Graphene Quantum Dots, Nanomaterials, 2019, 9(2), 282 CrossRef CAS.
- M.-H. Chan, R.-S. Liu and M. Hsiao, Graphitic carbon nitride-based nanocomposites and their biological applications: a review, Nanoscale, 2019, 11(32), 14993–15003 RSC.
- C. Cheng, S. Li, A. Thomas, N. A. Kotov and R. Haag, Functional graphene nanomaterials based architectures: biointeractions, fabrications, and emerging biological applications, Chem. Rev., 2017, 117(3), 1826–1914 CrossRef CAS.
- S. Deng, A. Fu, M. Junaid, Y. Wang, Q. Yin, C. Fu, L. Liu, D.-S. Su, W.-P. Bian and D.-S. Pei, Nitrogen-doped graphene quantum dots (N-GQDs) perturb redox-sensitive system via the selective inhibition of antioxidant enzyme activities in zebrafish, Biomaterials, 2019, 206, 61–72 CrossRef CAS.
- T. Wang, S. Zhu and X. Jiang, Toxicity mechanism of graphene oxide and nitrogen-doped graphene quantum dots in RBCs revealed by surface-enhanced infrared absorption spectroscopy, Toxicol. Res., 2015, 4(4), 885–894 CrossRef CAS.
- Y. Chong, Y. Ma, H. Shen, X. Tu, X. Zhou, J. Xu, J. Dai, S. Fan and Z. Zhang, The in vitro and in vivo toxicity of graphene quantum dots, Biomaterials, 2014, 35(19), 5041–5048 CrossRef CAS.
- X.-y. Wang, R. Lei, H.-d. Huang, N. Wang, L. Yuan, R.-y. Xiao, L.-d. Bai, X. Li and L.-m. Li, The permeability and transport mechanism of graphene quantum dots (GQDs) across the biological barrier, Nanoscale, 2015, 7(5), 2034–2041 RSC.
- D. Kim, J. M. Yoo, H. Hwang, J. Lee, S. H. Lee, S. P. Yun, M. J. Park, M. Lee, S. Choi and S. H. Kwon, Graphene quantum dots prevent α-synucleinopathy in Parkinson's disease, Nat. Nanotechnol., 2018, 13(9), 812–818 CrossRef CAS.
- M. Yousaf, H. Huang, P. Li, C. Wang and Y. Yang, Fluorine functionalized graphene quantum dots as inhibitor against hIAPP amyloid aggregation, ACS Chem. Neurosci., 2017, 8(6), 1368–1377 CrossRef CAS.
- L. Xu, Y. Dai, Z. Wang, J. Zhao, F. Li, J. C. White and B. Xing, Graphene quantum dots in alveolar macrophage: uptake-exocytosis, accumulation in nuclei, nuclear responses and DNA cleavage, Part. Fibre Toxicol., 2018, 15(1), 1–17 CrossRef.
- R. S. Li, B. Yuan, J. H. Liu, M. L. Liu, P. F. Gao, Y. F. Li, M. Li and C. Z. Huang, Boron and nitrogen co-doped single-layered graphene quantum dots: a high-affinity platform for visualizing the dynamic invasion of HIV DNA into living cells through fluorescence resonance energy transfer, J. Mater. Chem. B, 2017, 5(44), 8719–8724 RSC.
- J. Hu, W. Lin, B. Lin, K. Wu, H. Fan and Y. Yu, Persistent DNA methylation changes in zebrafish following graphene quantum dots exposure in surface chemistry-dependent manner, Ecotoxicol. Environ. Saf., 2019, 169, 370–375 CrossRef CAS.
- Y. Li, H. Yuan, A. Von Dem Bussche, M. Creighton, R. H. Hurt, A. B. Kane and H. Gao, Graphene microsheets enter cells through spontaneous membrane penetration at edge asperities and corner sites, Proc. Natl. Acad. Sci., 2013, 110(30), 12295–12300 CrossRef CAS.
- M. Dallavalle, M. Calvaresi, A. Bottoni, M. Melle-Franco and F. Zerbetto, Graphene can wreak havoc with cell membranes, ACS Appl. Mater. Interfaces, 2015, 7(7), 4406–4414 CrossRef CAS.
- Y. Chang, S.-T. Yang, J.-H. Liu, E. Dong, Y. Wang, A. Cao, Y. Liu and H. Wang, In vitro toxicity evaluation of graphene oxide on A549 cells, Toxicol. Lett., 2011, 200(3), 201–210 CrossRef CAS.
- J. Russier, E. Treossi, A. Scarsi, F. Perrozzi, H. Dumortier, L. Ottaviano, M. Meneghetti, V. Palermo and A. Bianco, Evidencing the mask effect of graphene oxide: a comparative study on primary human and murine phagocytic cells, Nanoscale, 2013, 5(22), 11234–11247 RSC.
- X. Cheng, Q. Wan and X. Pei, Graphene family materials in bone tissue regeneration: perspectives and challenges, Nanoscale Res. Lett., 2018, 13(1), 289 CrossRef.
- V. Palmieri, G. Perini, M. De Spirito and M. Papi, Graphene oxide touches blood: in vivo interactions of bio-coronated 2D materials, Nanoscale Horiz., 2019, 4(2), 273–290 RSC.
- L. Liang, Z. Kong, Z. Kang, H. Wang, L. Zhang and J.-W. Shen, Theoretical evaluation on potential cytotoxicity of graphene quantum dots, ACS Biomater. Sci. Eng., 2016, 2(11), 1983–1991 CrossRef CAS.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan and H. Fang, Destructive extraction of phospholipids from Escherichia coli membranes by graphene nanosheets, Nat. Nanotechnol., 2013, 8(8), 594 CrossRef CAS.
- G. Rossi, J. Barnoud and L. Monticelli, Partitioning and solubility of C60 fullerene in lipid membranes, Phys. Scr., 2013, 87(5), 058503 CrossRef.
- C. Liu, P. Elvati, S. Majumder, Y. Wang, A. P. Liu and A. Violi, Predicting the Time of Entry of Nanoparticles in Lipid Membranes, ACS Nano, 2019, 13(9), 10221–10232 CrossRef CAS.
- X. Tang, S. Zhang, H. Zhou, B. Zhou, S. Liu and Z. Yang, The role of electrostatic potential polarization in the translocation of graphene quantum dots across membranes, Nanoscale, 2020, 12(4), 2732–2739 RSC.
- A. V. Titov, P. Král and R. Pearson, Sandwiched graphene− membrane superstructures, ACS Nano, 2010, 4(1), 229–234 CrossRef CAS.
- P. Chen, H. Yue, X. Zhai, Z. Huang, G.-H. Ma, W. Wei and L.-T. Yan, Transport of a graphene nanosheet sandwiched inside cell membranes, Sci. Adv., 2019, 5(6), eaaw3192 CrossRef CAS.
- Y. Zhang, S. F. Ali, E. Dervishi, Y. Xu, Z. Li, D. Casciano and A. S. Biris, Cytotoxicity effects of graphene and single-wall carbon nanotubes in neural phaeochromocytoma-derived PC12 cells, ACS Nano, 2010, 4(6), 3181–3186 CrossRef CAS.
- X. Guo and N. Mei, Assessment of the toxic potential of graphene family nanomaterials, J. Food Drug Anal., 2014, 22(1), 105–115 CrossRef CAS.
- A. Bianco, Graphene: safe or toxic? The two faces of the medal, Angew. Chem., Int. Ed., 2013, 52(19), 4986–4997 CrossRef CAS.
- G. Fang, B. Luan, C. Ge, Y. Chong, X. Dong, J. Guo, C. Tang and R. Zhou, Understanding the graphene quantum dots-ubiquitin interaction by identifying the interaction sites, Carbon, 2017, 121, 285–291 CrossRef CAS.
- M. Zhou, Q. Shen, J.-W. Shen, L. Jin, L. Zhang, Q. Sun, Q. Hu and L. Liang, Understanding the size effect of graphene quantum dots on protein adsorption, Colloids Surf., B, 2019, 174, 575–581 CrossRef CAS.
- A. Faridi, Y. Sun, M. Mortimer, R. R. Aranha, A. Nandakumar, Y. Li, I. Javed, A. Kakinen, Q. Fan and A. W. Purcell, Graphene quantum dots rescue protein dysregulation of pancreatic β-cells exposed to human islet amyloid polypeptide, Nano Res., 2019, 12(11), 2827–2834 CrossRef CAS.
- B. Luan, T. Huynh, L. Zhao and R. Zhou, Potential toxicity of graphene to cell functions via disrupting protein–protein interactions, ACS Nano, 2015, 9(1), 663–669 CrossRef CAS.
- Z. Yang, C. Ge, J. Liu, Y. Chong, Z. Gu, C. A. Jimenez-Cruz, Z. Chai and R. Zhou, Destruction of amyloid fibrils by graphene through penetration and extraction of peptides, Nanoscale, 2015, 7(44), 18725–18737 RSC.
- C. Li, J. Adamcik and R. Mezzenga, Biodegradable nanocomposites of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties, Nat. Nanotechnol., 2012, 7(7), 421–427 CrossRef CAS.
- Z. Wang, H. Fang, S. Wang, F. Zhang and D. Wang, Simulating molecular interactions of carbon nanoparticles with a double-stranded DNA fragment, J. Chem., 2015, 2015, 531610 Search PubMed.
- M. Vatanparast and Z. Shariatinia, Revealing the role of different nitrogen functionalities in the drug delivery performance of graphene quantum dots: a combined density functional theory and molecular dynamics approach, J. Mater. Chem. B, 2019, 7(40), 6156–6171 RSC.
- M. Wang, Y. Sun, X. Cao, G. Peng, I. Javed, A. Kakinen, T. P. Davis, S. Lin, J. Liu and F. Ding, Graphene quantum dots against human IAPP aggregation and toxicity in vivo, Nanoscale, 2018, 10(42), 19995–20006 RSC.
- C. Martín, G. Jun, R. Schurhammer, G. Reina, P. Chen, A. Bianco and C. Ménard-Moyon, Enzymatic Degradation of Graphene Quantum Dots by Human Peroxidases, Small, 2019, 15(52), 1905405 CrossRef.
- Z. Xue, Q. Sun, L. Zhang, Z. Kang, L. Liang, Q. Wang and J.-W. Shen, Graphene quantum dot assisted translocation of drugs into a cell membrane, Nanoscale, 2019, 11(10), 4503–4514 RSC.
- N. Suzuki, Y. Wang, P. Elvati, Z.-B. Qu, K. Kim, S. Jiang, E. Baumeister, J. Lee, B. Yeom and J. H. Bahng, Chiral graphene quantum dots, ACS Nano, 2016, 10(2), 1744–1755 CrossRef CAS.
- Z. Kong, W. Hu, F. Jiao, P. Zhang, J.-W. Shen, B. Cui, H. Wang and L. Liang, Theoretical evaluation of graphene quantum dots on DNA genotoxicity: a combination of density functional theory and molecular dynamics simulation, J. Phys. Chem. B, 2020, 124(42), 9335–9342 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |