Open Access Article
Open Access ArticleA near-infrared fluorescence probe for imaging of pantetheinase in cells and mice in vivo†
Yuantao
Yang‡
ab,
Yiming
Hu‡
a,
Wen
Shi
 *ab and
Huimin
Ma
*ab and
Huimin
Ma
 ab
ab
aBeijing National Laboratory for Molecular Sciences, Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China. E-mail: shiwen@iccas.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 25th September 2020
Abstract
Pantetheinase is an amidohydrolase that cleaves pantetheine into pantothenic acid and cysteamine. Functional studies have found that ubiquitous expression of this enzyme is associated with many inflammatory diseases. However, the lack of near-infrared fluorescence probes limits the better understanding of the functions of the enzyme. In this work, we have developed a new near-infrared fluorescence probe, CYLP, for bioimaging of pantetheinase by using pantothenic acid with a self-immolative linker as a recognition group. The probe produces a sensitive fluorescence off–on response at 710 nm to pantetheinase with a detection limit of 0.02 ng mL−1 and can be used to image the intraperitoneal pantetheinase activity in mice in vivo. Moreover, with the probe we have observed that pantetheinase is significantly increased in the tissues of mouse inflammatory models as well as in the intestines of mice with inflammatory bowel disease. Therefore, CYLP may provide a convenient and intuitive tool for studying the role of pantetheinase in diseases.
Introduction
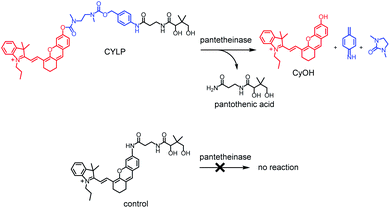
Pantetheinase, mainly known as vascular non-inflammatory molecule-1 (vanin-1), is highly expressed in many organs, such as the liver, intestines, kidneys and lungs.1 The initial inspections on pantetheinase focused on its function in coenzyme A (CoA) metabolism and lipid metabolism, since it can break down pantetheine into cysteamine and pantothenic acid (vitamin B5), a precursor of CoA.2 In recent years, many studies have intensely elucidated the latent role of pantetheinase in relation to inflammation and diseases. For example, vanin 1 knockout mice showed higher susceptibility to drug-induced hepatic injury and a decreased capacity to detoxify xenobiotics, suggesting enzymic protective effects.3 Moreover, pantetheinase has displayed a promising capacity as a disease marker since it is upregulated in the early phase of kidney injury and inflammatory bowel diseases.4,5 On the other hand, pantetheinase's inhibitor can attenuate the oxidative stress and inflammation in both acute and chronic intestinal diseases, implying that pantetheinase is a pro-inflammatory factor.6,7 The diverse and seemingly paradoxical results reflect the poor understanding of pantetheinase biofunction. Obviously, this is closely related to the lack of more methods to analyze the activity of pantetheinase in complex biological samples under physiological and pathological conditions.8–10 In particular, an imaging method that can be used to study pantetheinase activity in vivo is still lacking. Our group reported a fluorescent probe for imaging pantetheinase activity in living cells.11 It was comprised of pantothenic acid as a recognition moiety and cresyl violet as a fluorochrome, both of which were connected through an acyl amide bond directly. However, the analytical wavelength of the previous probe is relatively short and unsuitable for in vivo imaging study due to the poor penetration depth in a biological matrix. After this, Li et al. developed a bioluminogenic probe for pantetheinase by using aminoluciferin as the bioluminophore;12 however, costly transgenic mice are needed. As is known, a near-infrared (NIR) fluorescence probe would possess significant advantages in imaging pantetheinase activity in vivo on account of the simple operation and deep tissue penetration,13–15 but such a NIR probe has not been reported yet.Herein we have developed the first fluorescent probe, CYLP, towards pantetheinase with NIR analytical wavelengths (Scheme 1). Initially, we constructed a probe (control; Scheme 1) by directly coupling pantothenic acid to the fluorophore of amino hemicyanine, also expecting that the enzymatic cleavage of the acyl amide bond would release the fluorophore and generate NIR fluorescence. Nevertheless, the control probe showed almost no response toward pantetheinase (Fig. S11†). A possible reason for this phenomenon might be that the narrow substrate channel in the enzyme was incompatible with the bulky fluorophore, preventing the substrate from accessing the active center.16 To facilitate the recognition moiety of the probe approaching the catalytic site, a self-immolative linker, 4-aminobenzyl methyl(2-(methylamino)-ethyl)carbamate, was inserted between the bulky fluorophore and pantothenic acid to obtain the probe CYLP.17,18 To our delight, this indirect construction strategy significantly improved the analytical performance of the resulting probe, i.e., CYLP exhibited outstanding selectivity and sensitivity toward pantetheinase with NIR emission wavelength (>700 nm). Therefore, the fluorescence imaging of pantetheinase activity could be achieved in vivo.
Results and discussion
Fluorescence response of CYLP to pantetheinase
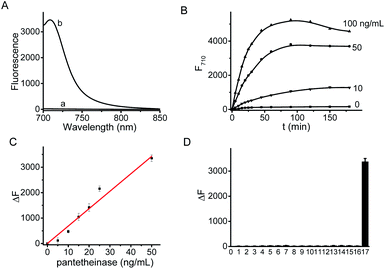
The fluorescence spectra of CYLP in the absence and presence of pantetheinase are shown in Fig. 1. As is seen, CYLP (10 μM) itself has almost no fluorescence in the NIR region (quantum yield <0.1%); upon reaction with pantetheinase (50 ng mL−1), the system showed 40-fold fluorescence enhancement at 710 nm. Meanwhile, the maximum absorption peak of the reaction system is red-shifted from 590 to 680 nm (Fig. S12†). These observations suggest that the reaction of CYLP with pantetheinase led to the release of the fluorophore CyOH (quantum yield 21%), which was clearly verified by ESI-MS analysis with the generation of a mass peak of CyOH at m/z = 412 [M]+ (Fig. S13†).The fluorescence kinetic study results of CYLP reacting with different concentrations of pantetheinase are shown in Fig. 1B. The probe showed a dose-dependent response toward the addition of pantetheinase and the fluorescence reached an approximate plateau in about 1 h, which is much faster than that with our previous probe.11 Meanwhile, no significant fluorescence variation could be observed in the absence of pantetheinase, suggesting the excellent stability of CYLP. Furthermore, the Michaelis constant (Km) of CYLP was determined to be 84 μM and the Kcat was 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 s−1 according to the Lineweaver–Burk plot, which exhibited high affinity and rapid catalytic rate toward pantetheinase (Fig. S14†).
000 s−1 according to the Lineweaver–Burk plot, which exhibited high affinity and rapid catalytic rate toward pantetheinase (Fig. S14†).
Then the effects of pH ranging from 4 to 10 and temperature between 25 °C and 42 °C were assessed (Fig. S15†). The probe itself maintained consistently low fluorescence in the tested pH range, and the reaction system displayed the maximum fluorescence enhancement in neutral media of about pH 7.4. On the other hand, a modestly elevated temperature (e.g., 37 °C) facilitated the enzymatic reaction slightly compared to room temperature (about 25 °C). The above observations indicated that the detection of pantetheinase using CYLP can be performed under physiological conditions.
The fluorescence titration of CYLP with pantetheinase exhibited good linearity (Fig. 1C) in the concentration range of 0–50 ng mL−1 pantetheinase, with an equation of ΔF = 68.5 × [C (ng mL−1)] (R = 0.99), where ΔF is the fluorescence enhancement of CYLP at 710 nm with and without pantetheinase. The detection limit was determined to be 0.02 ng mL−1 pantetheinase, which is much lower than that obtained with a bioluminescent probe. Thus, probe CYLP constructed by coupling the recognition moiety through the self-immolative linker showed higher efficacy in the enzymatic reaction compared to those probes from the direct strategy.
The specificity of CYLP was investigated by reaction with various substances, including inorganic salts (KCl, MgCl2, CaCl2, ZnCl2, CuCl2 and FeCl3), small biomolecules [glucose, cysteine, lysine, glutamic acid, aspartic acid and lipopolysaccharides (LPS)], and some enzymes [superoxide dismutase (SOD), leucine amino peptidase (LAP) and carbonic anhydrase]. The results (Fig. 1D) showed that CYLP exhibited excellent selectivity for pantetheinase over the other biosubstances tested, indicating the reliability of the detection.
Fluorescence imaging of pantetheinase in living cells
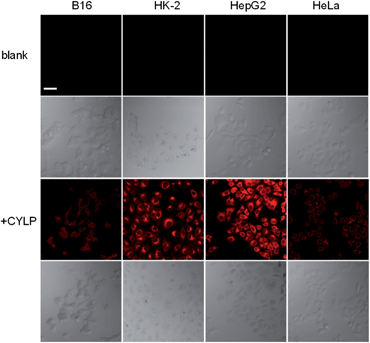
To test whether probe CYLP can be used to image pantetheinase in living cells, four cell lines were chosen as models. Firstly, the cytotoxicity of CYLP was evaluated with HK-2 and HepG2 cells by the standard MTT assay, and no significant toxicity was observed for 10 μM CYLP with 24 h incubation, indicating the good biocompatibility of the probe (Fig. S16†).In cell imaging, the cells (including B16, HK2, HepG2 and HeLa cells) themselves displayed no fluorescence under an excitation wavelength of 635 nm. After incubation with CYLP for 1 h, the cells exhibited obvious fluorescence (Fig. 2). However, if we pretreated the cells with the specific inhibitor of pantetheinase, RR6, the fluorescence in the cells was largely suppressed (Fig. S17†). Moreover, dose positive correlation inhibition can be observed for these cell lines, suggesting that the intracellular fluorescence may result from the activity of pantetheinase. In addition, RR6 itself was proved to have no obvious influence on both the cell viability (Fig. S18†) and the fluorescence of the released fluorophore in cells (Fig. S19†). On the other hand, these cell lines were subjected to western blot analyses to assess the pantetheinase level (Fig. S20†), revealing that HepG2 cells displayed the most abundant pantetheinase, which is consistent with the fluorescence imaging result. Besides, HK-2 and HeLa cells had similar pantetheinase levels from the western blot assay, but they showed obviously different fluorescence intensities, possibly due to the different enzymatic reactivity or uptake efficiency of the probe in different cells. Even so, the above data suggest that the probe is capable of imaging intracellular pantetheinase.
Fluorescence imaging of pantetheinase in mice in vivo
Due to the excellent analytical performance of CYLP in vitro, we subsequently applied the probe for fluorescence imaging of pantetheinase activity in vivo. Pantetheinase's primary function is the recycling of pantothenic acid, an important precursor of CoA, which is the key cofactor in fat metabolism. Besides, pantetheinase is often expressed in organs with high CoA turnover, such as the liver and intestines. Hence, we attempted to image the endogenous pantetheinase activity in these organs. Also, negative controls with the pantetheinase inhibitor RR6 were studied.6 Specifically, Kunming mice were intraperitoneally injected with saline or RR6 (30 mg kg−1, negative control) first. After 1 h, all mice were injected intraperitoneally with CYLP (400 μM × 200 μL), and then their abdominal fluorescence was imaged every 10 min. The fluorescence increase in the mice of the experimental group continued and became relatively slow in 50 min. Meanwhile, the fluorescence in the mice of the control group did not show noticeable enhancement. In a representative example shown in Fig. 3, at 50 min after intraperitoneal injection, the experimental mouse showed a 2.5-fold brighter fluorescence than that in the control group with the inhibitor, which indicated that CYLP can image the endogenous pantetheinase in living animals.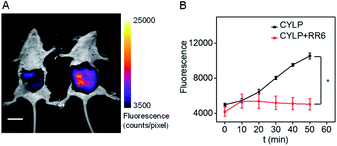 | ||
| Fig. 3 (A) The representative image at 50 min of mice after intraperitoneal injection of CYLP. Left: mouse pretreated with RR6 as the negative control; right: mouse pretreated with only saline (NaCl 0.9%). The full images of the mice at different times are given in Fig. S21.† Scale bar, 2 cm. (B) Time dependent change of the fluorescence in the mice captured at different times (n = 3 pairs of mice). *p < 0.05, two-sided Student's t-test. | ||
Fluorescence imaging of pantetheinase in mice with inflammation
Inflammation, a common defense mechanism, can protect our body from infection and injury using immune cells and the substances they produce, including cytokines and reactive oxygen species (ROS).19 However, abnormal ROS levels would cause certain diseases. Since pantetheinase can lead to the release of cysteamine, which can further regulate oxidative stress of the cells, this enzyme has been implicated in many inflammatory diseases by serving as a pro-inflammatory factor or upregulated serum biomarker.1 However, the in situ pantetheinase level in the inflammatory diseases has not been studied due to the lack of suitable tools. Therefore, encouraged by the analytical performance of CYLP, we further explored the probe's application in imaging the pantetheinase activity change in an inflammation model. Namely, hind limb inflammation of Kunming mice was induced by injecting 200 μL of 1 mg mL−1 LPS into the right hind leg of the mice (Fig. 4A and S22†). In the meantime, an equal amount of saline was injected into the left leg as a control. After 14 h, CYLP (400 μM × 20 μL) was injected into both the legs of the mice subcutaneously. Fluorescence images of the mice were captured at different time points (Fig. S22†). As can be seen from Fig. 4A, the mice have stronger fluorescence in the right hind leg than in the left hind leg. The analyses from 3 mice showed that the fluorescence intensity of the inflammation in the leg was significantly higher than that in the control leg and the increased pantetheinase was confirmed by western blot analysis (Fig. S23†). Further, we conducted a negative control experiment in which the inhibitor RR6 was injected for 30 min prior to the administration of CYLP in the inflammation leg. As shown in Fig. S24,† the RR6 pretreatment can suppress the fluorescence in the inflammation leg substantially (Fig. S24†). This also supports the fact that the fluorescence enhancement is mainly due to pantetheinase activity.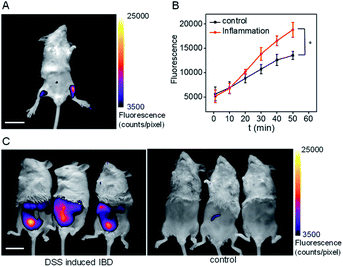 | ||
| Fig. 4 (A) The representative image at 50 min of the mouse after the administration of CYLP. Left leg: injected with saline as a control; right leg: injected with 200 μL of 1 mg mL−1 LPS as an inflammation model. The full images of the mouse at different time points are shown in Fig. S22.† Scale bar, 2 cm. (B) Time dependent change of the fluorescence of the control leg and inflammation leg captured at different times (n = 3 mice). *p < 0.05, two-sided Student's t-test. (C) The images of the IBD mice and control mice after the administration of CYLP through the anus into the colon. | ||
Finally, we demonstrated the feasibility and practicability of the probe CYLP for imaging pantetheinase in the inflammatory bowel disease (IBD) model of mice. IBD is a chronic inflammatory syndrome of the digestive tract, and pantetheinase has been regarded as a therapeutic target or fecal biomarker in the disease.20 IBD mice were induced by dextran sulfate sodium (DSS) via drinking water for 8 days.21 Then, CYLP (400 μM × 100 μL) was administrated through the anus. As a result, the IBD mice displayed stronger fluorescence in the abdomen compared to the control mice (Fig. 4C). It should be mentioned that the fluorescence appeared mostly in the small intestines rather than at the injection point of the colon region (Fig. S25†), probably implying the upregulation of pantetheinase in this area. Thus, CYLP may be used as a convenient and intuitive tool to illustrate the change of pantetheinase activity in living animal models.
Conclusions
In summary, by inserting a carbamate-based self-immolative linker between the recognition group (pantothenic acid) and the fluorophore (CyOH), we have developed CYLP as a new near-infrared fluorescent probe for pantetheinase assay. The probe displays high selectivity and sensitivity, with a detection limit of 0.02 ng mL−1 pantetheinase, and has been used to image pantetheinase in cells and mice. Moreover, with the probe, we demonstrate that the local inflammation can indeed increase the pantetheinase level in the tissue. We believe the probe CYLP may serve as an effective tool for studying the biological functions of pantetheinase and related diseases.Conflicts of interest
There are no conflicts to declare.Acknowledgements
All animal care and experimental protocols complied with the Animal Management Rules of the Ministry of Health of the People's Republic of China and were approved by the Institute of Process S12 Engineering, Chinese Academy of Sciences. This work was supported by grants from the NSF of China (No. 21922412, 21675159, 21820102007, and 21775152) and Youth Innovation Promotion Association of CAS (2016027).Notes and references
- R. Bartucci, A. Salvati, P. Olinga and Y. L. Boersma, Int. J. Mol. Sci., 2019, 20, 3891 CrossRef CAS.
- G. Pitari, F. Malergue, F. Martin, J. M. Philippe, M. T. Massucci, C. Chabret, B. Maras, S. Duprcè, P. Naquet and F. Galland, FEBS Lett., 2000, 483, 149–154 CrossRef.
- D. W. Ferreira, M. J. Goedken, S. Rommelaere, L. Chasson, F. Galland, P. Naquet and J. E. Manautou, Biochim. Biophys. Acta, Mol. Basis Dis., 2016, 1862, 662–669 CrossRef CAS.
- K. Hosohata, H. Ando and A. Fujimura, J. Pharmacol. Exp. Ther., 2012, 341, 656–662 CrossRef CAS.
- T. Gensollen, C. Bourges, P. Rihet, A. Rostan, V. Millet, T. Noguchi, V. Bourdon, H. Sobol, L. Dubuquoy, B. Bertin, M. Fumery, P. Desreumaux, J. F. Colombel, X. Hebuternee, P. Hofman, P. Naquet and F. Gallandt, Inflammatory Bowel Dis., 2013, 19, 2315–2325 CrossRef.
- P. A. M. Jansen, J. A. Diepen, B. Ritzen, P. L. J. M. Zeeuwen, I. Cacciatore, C. Cornacchia, I. M. J. J. Vlijmen-Willems, E. Heuvel, P. N. M. Botman, R. H. Blaauw, P. H. H. Hermkens, F. P. J. T. Rutjes and J. Schalkwijk, ACS Chem. Biol., 2013, 8, 530–534 CrossRef CAS.
- F. Martin, M. F. Penet, F. Malergue, H. Lepidi, A. Dessein, F. Galland, M. Reggi, P. Naquet and B. Gharib, J. Clin. Invest., 2004, 113, 591–597 CrossRef CAS.
- C. Wittwer, B. Wyse and R. G. Hansen, Anal. Biochem., 1982, 122, 213–222 CrossRef CAS.
- S. Duprè, R. Chiaraluce, M. Nardini, C. Cannella, G. Ricci and D. Cavallini, Anal. Biochem., 1984, 142, 175–181 CrossRef.
- B. H. Ruan, D. C. Cole, P. Wu, A. Quazi, K. Page, J. F. Wright, N. Huang, J. R. Stock, K. Nocka, A. Aulabaugh, R. Krykbaev, L. J. Fitz, N. M. Wolfman and M. L. Fleming, Anal. Biochem., 2010, 399, 284–292 CrossRef CAS.
- Y. M. Hu, H. Y. Li, W. Shi and H. M. Ma, Anal. Chem., 2017, 89, 11107–11112 CrossRef CAS.
- Y. X. Lin, Y. Q. Gao, Z. Ma, Z. Z. Li, C. C. Tang, X. J. Qin, Z. Zhang, G. K. Wang, L. P. Du and M. Y. Li, Anal. Chem., 2018, 90, 9545–9550 CrossRef CAS.
- (a) X. H. Li, X. H. Gao, W. Shi and H. M. Ma, Chem. Rev., 2014, 114, 590–659 CrossRef CAS; (b) Y. C. Liu, L. L. Teng, L. L. Chen, H. C. Ma, H. W. Liu and X. B. Zhang, Chem. Sci., 2018, 9, 5347–5353 RSC; (c) P. H. Cheng, J. J. Zhang, J. G. Huang, Q. Q. Miao, C. J. Xu and K. Y. Pu, Chem. Sci., 2018, 9, 6340–6347 RSC.
- (a) Z. Q. Guo, S. Park, J. Y. Yoon and I. Shin, Chem. Soc. Rev., 2014, 43, 16–29 RSC; (b) P. Li, J. J. Wang, X. Wang, Q. Ding, X. Y. Bai, Y. D. Zhang, D. Su, W. Zhang, W. Zhang and B. Tang, Chem. Sci., 2019, 10, 2805–2810 RSC; (c) H. D. Li, Y. Q. Li, Q. C. Yao, J. L. Fan, W. Sun, S. Long, K. Shao, J. J. Du, J. Y. Wang and X. J. Peng, Chem. Sci., 2019, 10, 1619–1625 RSC.
- (a) Z. Q. Mao, W. Q. Feng, Z. Li, L. Y. Zeng, W. J. Lv and Z. H. Liu, Chem. Sci., 2016, 7, 5230–5235 RSC; (b) S. Xu, H. W. Liu, X. Yin, L. Yuan, S. Y. Huan and X. B. Zhang, Chem. Sci., 2019, 10, 320–325 RSC; (c) Z. Q. Mao, M. T. Ye, W. Hu, X. X. Ye, Y. Y. Wang, H. J. Zhang, C. Y. Li and Z. H. Liu, Chem. Sci., 2018, 9, 6035–6040 RSC.
- Y. L. Boersma, J. Newman, T. E. Adams, N. Cowieson, G. Krippner, K. Bozaoglu and T. S. Peat, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2014, 70, 3320–3329 CrossRef CAS.
- Q. Q. Miao, D. C. Yeo, C. Wiraja, J. J. Zhang, X. Y. Ning, C. J. Xu and K. Y. Pu, Angew. Chem., Int. Ed., 2018, 130, 1270–1274 CrossRef.
- J. Ning, T. Liu, P. P. Dong, W. Wang, G. B. Ge, B. Wang, Z. L. Yu, L. Shi, X. G. Tian, X. K. Huo, L. Feng, C. Wang, C. P. Sun, J. N. Cui, T. D. James and X. C. Ma, J. Am. Chem. Soc., 2019, 141, 1126–1134 CrossRef CAS.
- M. Mittal, M. R. Siddiqui, K. Tran, S. P. Reddy and A. B. Malik, Antioxid. Redox Signaling, 2014, 20, 1126–1167 CrossRef CAS.
- C. Ballet, M. S. P. Correia, L. P. Conway, T. L. Locher, L. C. Lehmann, N. Garg, M. Vujasinovic, S. Deindl, J. M. Löhr and D. Globisch, Chem. Sci., 2018, 9, 6233–6239 RSC.
- S. Wirtz, V. Popp, M. Kindermann, K. Gerlach, B. Weigmann, S. Fichtner-Feigl and M. F. Neurath, Nat. Protoc., 2017, 12, 1295–1309 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc04537c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |