An “off–on” phosphorescent aptasensor for the detection of thrombin based on PRET†
Yan
Xiong
,
Meiyu
Liang
,
Yue
Cheng
,
Jiarui
Zou
and
Yan
Li
 *
*
Key Laboratory of Inorganic–Organic Hybrid Functional Materials Chemistry (Tianjin Normal University), Tianjin Key Laboratory of Structure and Performance for Functional Molecule, College of Chemistry, Tianjin Normal University, Tianjin, 300387, P.R. China. E-mail: nkliyan398@gmail.com
First published on 2nd October 2018
Abstract
Thrombin plays an important role in the blood coagulation cascade and it stimulates the process of platelet aggregation. Herein, we developed a highly efficient and sensitive phosphorescent aptasensor system for the quantitative analysis of thrombin. The phosphorescence of 3-mercaptopropionic acid capped Mn-doped ZnS quantum dots (MPA-Mn:ZnS QDs) was gradually quenched with the addition of thrombin binding aptamers-BHQ2 (TBA-BHQ2) based on phosphorescence resonance energy transfer (PRET). With the addition of the target analyte thrombin into the system, TBA-BHQ2 could change its spatial structure from a random coil to an antiparallel G-quadruplex which resulted from the combination of thrombin and TBA-BHQ2, leading to the phosphorescence recovery. Finally, the concentration of thrombin could be accurately determined by means of measuring the phosphorescence intensity change value (ΔP). The limit of detection (LOD) was obtained as low as 15.26 pM with wide linear ranges both from 60 to 2000 pM and from 2 to 900 nM. The proposed strategy was also successfully applied for thrombin detection in human serum samples and plasma samples with satisfactory recoveries from 96% to 99% and 95% to 104%, respectively. The long lifetime of phosphorescent QDs possessed a suitable time delay to eliminate autofluorescence and scattered light interference from biological matrices effectively. Thus, the signal to noise ratio of the phosphorescent aptasensor was improved visibly for the analysis of target analytes.
Introduction
Aptamers are single stranded DNA, RNA, nucleotide analogues or peptides obtained from the systematic evolution of ligands by the exponential enrichment (SELEX) procedure, which was established in the 1990s for the first time.1,2 Aptamers have a predetermined recognition ability for target molecules such as small molecule ligands or proteins leading to them folding into a unique tertiary structure, and they exhibit great possibilities for the development of new biosensors and clinical application.3,4 In addition, aptamers can be easily functionalized with a variety of functional groups and dyeing groups.5,6 In general, the applications of aptamers are similar to those of an antibody, and aptamers exhibited higher sensitivity and specificity than antibodies in analytical detection.7,8 Antibodies, as sensors, are susceptible to denaturation by environmental factors such as pH and temperature. In contrast, aptamers have the characteristics of enduring different temperatures and salt concentrations, repeated degeneration and renaturing by the metal chelating agent, and are extensively applied as excellent biorecognition elements in bioanalysis and sensor design.9,10One of the most commonly studied proteins for molecular targets is thrombin in aptamer-based assays. Thrombin, as a serine protease, is produced by the proteolytic process of the non-active thrombin in the prothrombin complex under the action of Xa factor (FXa).11,12 The molecule plays an important role in the blood coagulation cascade where it converts fibrinogen to insoluble fibrin that forms a fibrin gel, and it stimulates the process of platelet aggregation.13,14 It also plays an increasingly significant part in various physical and pathological activities, such as the blood coagulation cascade, cardiovascular disease, inflammatory reactions and tissue recovery.15,16 After the local application of thrombin, the blood on the lesion surface forms the stable sludged blood and then quickly controls capillary and venous hemorrhage, adhesion and fixation of skin and tissue grafts.17 Generally, the normal concentration of thrombin in blood ranges from nM to μM during the coagulation process. The bodily injury or disease leads to coagulation and anticoagulation imbalance and the thrombin activity changes in vivo.18–20 Consequently, the quantitative detection of thrombin based on aptamers is a viable diagnostic tool for monitoring the thrombin level in plasma or blood in the clinical areas.21
Current methodologies of using thrombin binding aptamers (TBA) to detect thrombin have been extensively reported over the past few decades, including fluorescence spectroscopy,22 inductively coupled plasma mass spectrometry (ICP-MS),23 colorimetric approaches,24 chemiluminescence,25,26 affinity chromatography,27 surface enhanced resonance Raman scattering (SERRS) spectroscopy28 and localized surface plasmon resonance spectroscopy (LSPR).29 However, the practical application of these detection methods is limited by their low sensitivity and selectivity, serious interference, slow response, false positive readings, operational complexity, difficult real-time monitoring, and severe background and scattering light interference originating from real biological samples or environmental samples.30,31 Consequently, it is crucial to develop a rapid and reliable detection method for thrombin.
Recently, the room temperature phosphorescent (RTP) detection method has gained considerable attention and has been widely applied in sensors, such as ratiometric phosphorescent probes,32 time-resolved phosphorescence (TRP) sensors,33 aggregation-induced phosphorescence enhancement (AIPE),34 the combination of molecularly imprinted polymers and phosphorescence,35 and the phosphorescent inner filter effect (PIFE).36 The energy transfer of phosphorescence emission shifts from the triplet state (4T1) to the ground state (6A1) leading to a longer lifetime,37,38 which can avoid autofluorescence and scattered light interference from biological matrices.39,40 Moreover, RTP biosensors do not require complex pretreatments and any deoxidant or inducer.41 To the best of our knowledge, RTP can be employed in “turn-off” or “turn-on” modes.42 In the “turn-off” mode, all sorts of reasons can give rise to the ultimate phosphorescent “off” state. In contrast, the phosphorescent “turn-on” mode seems to be more reliable and attractive because it can reduce the opportunity of false positives, making it more amenable to multiplexing.43,44 Generally speaking, the “off–on” phosphorescence aptasensor mode has been applied to experiments which can overcome the false positive signals caused by the nonspecific adsorption of fluorescent nanoprobes on the surface of non-target cells,45 can improve the signal response mode of the signal/background ratio, and can also achieve the specific detection by using the specific combination of the aptamer and the target.46
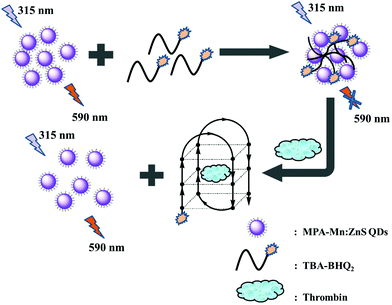
In this work, an “off–on” phosphorescence aptasensor system for detecting thrombin was developed based on phosphorescence QDs and a specific TBA tagged with BHQ2 dye (TBA-BHQ2). Phosphorescence aptasensor detection methods have garnered a lot of attention in recent years owing to their high specificity and affinity toward their targets. The “off” signal was attributed to the phosphorescence quenching of QDs effectively, where phosphorescent QDs served as the energy donors and TBA-BHQ2 served as the energy acceptors. The close proximity of TBA-BHQ2 and the QDs constituted a phosphorescence resonance energy transfer (PRET) system. The high sensitivity of the phosphorescent “turn-on” response signal indicated the great affinity between thrombin and TBA-BHQ2. In this time, TBA-BHQ2 moved away from the TBA-BHQ2/QDs complex and then formed the TBA-BHQ2/thrombin hybrid system where TBA-BHQ2 formed an antiparallel G-quadruplex. As a result, the concentration of thrombin could be quantitatively determined by means of measuring the restoring phosphorescence intensity change value (ΔP).
Experimental section
Materials and chemicals
All chemicals were of analytical grade and were used without further purification. TBA-BHQ2 was synthesized by Sangon Biotechnology Co., Ltd (Shanghai, China). The sequence of TBA-BHQ2 was 5′ BHQ2-GGTTGGTGTGGTTGG-3′. ZnSO4·7H2O, Mn(CH3COO)2, Na2S·9H2O, KCl, NaCl and CH3CH2OH were obtained from Tianjin Guangfu fine chemical research institute (Tianjin, China). 3-Mercaptopropyl acid (MPA) was supplied by J&K Scientific (Beijing, China). (NH4)2SO4 was purchased from Fengchuan Chemical Reagent Technologies Co., Ltd (Tianjin, China). Ultrapure water was purchased from Wahaha Group Co., Ltd (Hangzhou, China). CaCl2 was purchased from Damao Chemical Reagent Factory (Tianjin, China). Thrombin was the product of Beijing solarbio technology Co., Ltd (Beijing, China). Human factor Xa was provided by Sino Biological Inc. (Beijing, China). Tris-(hydroxymethyl) aminomethane (Tris) was provided by Beijing Dingguo Biological Technology Co., Ltd (Beijing, China). Isoleucine (Ile), tryptophan (Trp), methionine (Met), leucine (Leu), threonine (Thr), phenylalanine (Phe), lysine (Lys) and histidine (His) were obtained from Sangon Biotechnology Co., Ltd (Shanghai, China). Alkaline phosphatase (Alp) was purchased from Yuanye Bio-Technology Co., Ltd (Shanghai, China). Alanine transaminase (Alt) was purchased from Sun-Shine Chemical Technology Co., Ltd (Shanghai, China).Apparatus
Transmission electron microscopy (TEM) was performed using a Tecnai G2 F20 (FEI). The X-ray diffraction (XRD) spectrum was recorded with a Bruker D8 diffractometer in the 2θ range from 5° to 80°. Fourier transform infrared (FT-IR) spectra (4000–500 cm−1) were recorded by using a Nicolet 6700 FT-IR spectroscope with KBr pellets (Nicolet, USA). UV-vis absorption spectra were recorded using a UV-2600 UV-vis spectrophotometer (Shimadzu, Japan) with 1 cm path-length cells. The RTP measurement was performed by using a Cary Eclipse Fluorescence spectrophotometer (Agilent Technologies, USA) equipped with a plotter unit and a micro-quartz cell in the phosphorescence mode where the delay time and gate time were 0.2 ms and 5.0 ms, respectively. Under the excitation at 315 nm, phosphorescence emission spectra were obtained in the wavelength range of 500–700 nm. The phosphorescence lifetimes were surveyed with an excitation wavelength of 315 nm and an emission wavelength of 590 nm. Circular dichroism (CD) spectra were recorded on a Jasco J-715 spectrophotometer in a 10 mm quartz cell. And all spectra were obtained with a response time of 1 s and a scan speed of 200 nm min−1 in the wavelength range of 200–320 nm.Preparation of MPA-Mn:ZnS QDs
MPA-Mn:ZnS QDs were synthesized in an aqueous solution according to a previous publication with minor modifications.47,48 In a typical experiment, 5 mL of 0.1 M ZnSO4, 5 mL of 0.02 M Mn(CH3COO)2 and 45 mL of 0.05 M MPA were mixed together, and the solution was adjusted to 11 (pH) with 1 M NaOH. Then, the purging of air with nitrogen bubbles was carried out for 30 min under stirring, ensuring that the stabilizer MPA, Zn2+ and Mn2+ were fully integrated and the oxygen was removed. Afterwards, 5 mL of 0.1 M Na2S was quickly injected into the mixture. The solution was continuously exposed to nitrogen for 20 min. The crude solution showed nearly no phosphorescence emission. The phosphorescence emission was observed after the aging procedure at 50 °C in air for 2 h. The MPA-Mn:ZnS QDs could be precipitated with ethanol. The precipitate was centrifuged and washed with ethanol, and then dried for 24 h under vacuum. The obtained powder was highly soluble in water.Process of detecting thrombin
To study the quenching effect of TBA-BHQ2 on the phosphorescence intensity of MPA-Mn:ZnS QDs, 50 μL of 0.1 M Tris-HCl (pH = 7.4), 25 μL of 500 mg L−1 MPA-Mn:ZnS QDs and a series of different concentrations of TBA-BHQ2 were successively added to the reaction system (37 °C), which was diluted to 500 μL with ultrapure water and mixed thoroughly. After 60 min, the reaction system formed a MPA-Mn:ZnS QDs/TBA-BHQ2 hybrid system. The phosphorescence spectra were obtained from 500 nm to 700 nm upon excitation at 315 nm. And then the phosphorescence intensity of the maximum emission peak at 590 nm was used for quantitative analysis. All values were collected from at least three independent measurements.The detection of the thrombin assay was carried out as follows: 50 μL of 0.1 M Tris-HCl (pH = 7.4), 25 μL of 500 mg L−1 MPA-Mn:ZnS QDs and 50 μL of 1 M TBA-BHQ2 were diluted to 450 μL with ultrapure water and reacted for 60 min (37 °C). Whereafter, various concentrations of thrombin (50 μL) were sequentially added to the reaction system, shaken thoroughly and equilibrated for 60 min. The phosphorescence spectra were recorded from 500 nm to 700 nm upon excitation at 315 nm. And then the phosphorescence intensity of the maximum emission peak at 590 nm was applied to quantitative analysis of thrombin. All data were measured from at least three individual measurements.
Real sample collection and pretreatment
Fresh human serum samples and plasma samples were received from Nanjing SenBeiJia Biological Technology Co., Ltd. In order to satisfy the calibration curve and decrease the matrix effects, human serum samples were diluted 50 times with Tris-HCl buffer solution (pH 7.4). No other pretreatment process was performed. Human plasma samples (2.5 mL) were quickly mixed with 1 mL of 0.1 M NaCl and 1.25 mL of 2 M (NH4)2SO4. The mixture was centrifuged for 4 min and the supernatant was used for further experiment. Whereafter, 8 nM human factor Xa (Ca2+ of 0.03 M) was added to the plasma to promote the transformation of prothrombin into thrombin. The as-obtained plasma containing thrombin was immediately introduced to the phosphorescent aptasensor for the detection of thrombin.11Results and discussion
Characterization of MPA-Mn:ZnS QDs
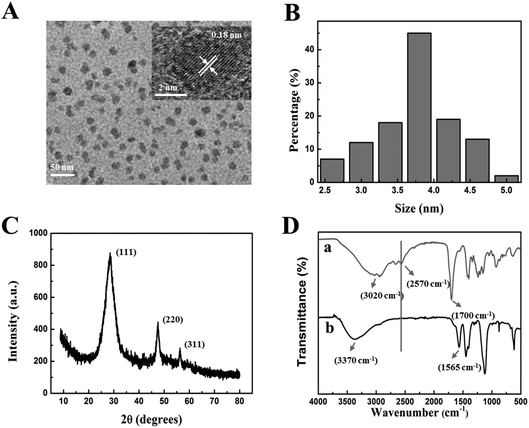
The characteristics, morphologies and optical properties of MPA-Mn:ZnS QDs were obtained by TEM, FT-IR, XRD, UV-vis absorption and phosphorescence spectroscopy, respectively. By TEM measurement, the crystallization and microstructure of nanoparticles for observing the morphology, dispersity and particle size of nanoparticles were analysed. The as-prepared MPA-Mn:ZnS QDs particles showed a roughly spherical morphology with a relatively good monodispersity and crystallinity as shown in Fig. 1A. The crystal of MPA-Mn:ZnS QDs was illustrated by the clear lattices. The distance between the adjacent lattices was 0.18 nm (inset of Fig. 1A) corresponding to the (111) planes of MPA-Mn:ZnS QDs. The average diameter of these nanocrystals was estimated to be 3.8 ± 1.3 nm obtained from statistics of approximately 100 particles (Fig. 1B). X-ray diffraction (XRD) accurately measured the crystal structure and analyzed the material phase by using the principle of diffraction. Fig. 1C shows the wide angle X-ray diffraction patterns of MPA-Mn:ZnS nanocrystals. The XRD spectrum was scanned over the 2 theta (θ) range from 5° to 80°, and the diffractogram of the QDs showed the (111), (220) and (311) planes, indicating that the nanocrystals structure were cubic (zinc blende phase). From this diffractogram, the QDs size was estimated to be 3.1 nm through the Scherrer equation.49To confirm the existence of MPA on the surface of the as-prepared MPA-Mn:ZnS QDs, we compared the FT-IR spectra of the pure MPA and MPA-Mn:ZnS QDs (Fig. 1D, curves a and b). Compared to the pure MPA, the carboxyl stretching vibration peak red shifted from 1700 cm−1 to 1560 cm−1 and free hydroxyl blue shifted from 3020 cm−1 to 3370 cm−1. Furthermore, the thiol group vibration band (2570 cm−1) disappeared because of the covalent affinity between the surface of the QDs and the -SH groups of MPA and the introduction of MPA can greatly improve the efficiency and reproducibility of QDs. The UV-vis absorption and phosphorescence emission spectra of MPA-Mn:ZnS QDs are shown in Fig. S1.† The maximum excitation peak was identified at 295 nm on the basis of the UV-vis absorption spectrum (Fig. S1,† curve a). The phosphorescent MPA-Mn:ZnS QDs showed barely phosphorescence emission before the aging procedure (Fig. S1,† curve b), but exhibited strong phosphorescence emission at 590 nm after the aging procedure for 2 h at 50 °C under open air conditions (Fig. S1,† curve c) when excited at 315 nm, which was attributed to the energy transfer from the band gap of ZnS to the dopant Mn2+ and subsequently from the triplet state (4T1) to the ground state (6A1) of the Mn2+ involved in the ZnS host lattices. (Fig. S2†).
Strategy of the phosphorescent aptasensor for the detection of thrombin
The aptamer was characterized to have high specificity and affinity to the target, which could distinguish the target analyte from analogues. TBA emerged to have a stable quadruplex conformation with the addition of thrombin. The detection of thrombin has not yet been reported based on phosphorescent aptamer probes or sensors in the literature. To test the ability of the phosphorescent aptasensor based on the MPA-Mn:ZnS QDs/TBA-BHQ2 hybrid system for detecting thrombin, we first investigated whether thrombin could interact with MPA-Mn:ZnS QDs and cause a phosphorescence change. The results showed that the phosphorescence emission of QDs was little changed with thrombin at a concentration as high as 5 μM, (Fig. S3,† curves of a and b) demonstrating that thrombin had negligible influences on the phosphorescence of QDs.In another way, the phosphorescence intensity of MPA-Mn:ZnS QDs was quenched effectively by TBA-BHQ2via donor–acceptor recognition (Fig. S3,† curve of c). The QDs grew closer to TBA obviously by forming hydrogen bonds and then constituting a PRET mechanism due to energy transfer from QDs (as the energy donors) to BHQ2 (as the energy acceptors). With the addition of thrombin to the QDs/TBA-BHQ2 system, the phosphorescence was “turned on” due to the formation of TBA-BHQ2/thrombin complex. The target analyte (thrombin) could be specifically recognized by TBA-BHQ2, which led to the release of the quencher (TBA-BHQ2) from the MPA-Mn:ZnS QDs surface and then the phosphorescence intensity was recovered (Fig. S3,† curve of d).
Possible mechanism of the phosphorescent aptasensor for the detection of thrombin
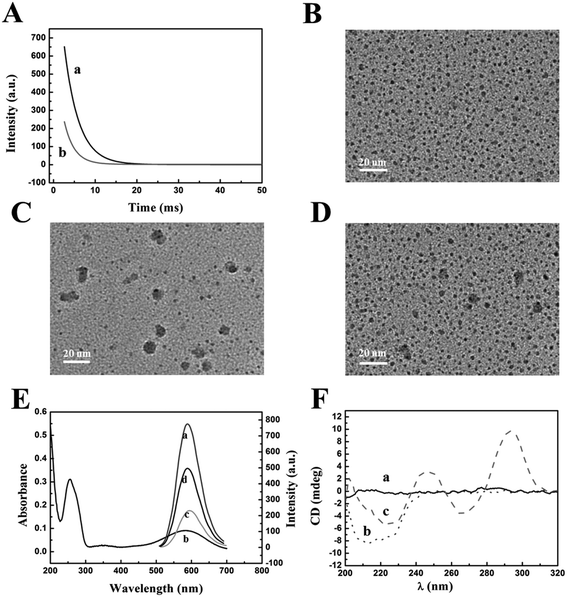
The reaction mechanism of the “off–on” phosphorescent aptasensor for the detection of thrombin based on PRET is shown in Scheme 1. It had been known that the phosphorescence of QDs could be generally quenched via either the dynamic quenching or static quenching process.50,51 To investigate the potential quenching mechanism between MPA-Mn:ZnS QDs and TBA-BHQ2, we measured the phosphorescence lifetime of the QDs before and after the interaction with TBA-BHQ2 in aqueous solution because the measurement of the phosphorescence lifetime is the most appropriate method to distinguish between static quenching and dynamic quenching processes. As shown in Fig. 2A, the phosphorescence lifetime of the QDs was estimated to be 3.45 ms (Fig. 2A, curve a) and that of the TBA-BHQ2/QDs complex was 2.42 ms (Fig. 2A, curve b), which suggested that the “turn-off” procedure was a dynamic quenching possibly.52As is known to all, two important parameters are essential for a PRET system. For one thing, the energy donor and energy receptor must be drawn to the distance close to each other. Afterwards, the emission spectrum of the energy donor and the adsorption spectrum of the energy receptor must be adequately overlapped, which manifested that an energy donor–acceptor pair could be manufactured. Here in this work, the phosphorescent QDs containing H and O atoms on the surface of the nanocrystal could form hydrogen bonds with TBA containing N, H, and O atoms, making QDs and TBA get closer, and then generating the TBA-BHQ2/QDs complex (Fig. 2B and C).53 Whereafter, as shown in Fig. 2B, the emission peak of QDs and the maximum absorption peak of TBA-BHQ2 were 590 and 585 nm, respectively (Fig. 2E, curves a and b). The excellent spectral overlap between the absorption spectrum of TBA-BHQ2 and the emission spectrum of QDs could ensure efficient energy transfer from QDs to BHQ2 based on the PRET mechanism, leading to quenching of phosphorescence (Fig. 2E, curve c).54–56 Finally, with the addition of thrombin into the TBA-BHQ2/QDs complex solution, the phosphorescence was recovered gradually (Fig. 2E, curve d) due to the release of QDs from the QDs/TBA-BHQ2 system (Fig. 2D). It was reasonable that TBA-BHQ2 in the TBA-BHQ2/QDs complex would combine with thrombin to form a quadruplex because of the higher affinity between TBA-BHQ2 and thrombin than TBA-BHQ2 and the QDs.
Information on structural changes in proteins and DNA induced by the binding of ligands is an essential part of the mechanism of action and regulation of biological activity. CD provides an experimentally convenient means of detecting such changes in different spectral regions.57 As shown in Fig. 2F, the CD spectra of TBA-BHQ2/QDs, thrombin, and the products resulting from the interaction between QDs/TBA-BHQ2 and thrombin were obtained at pH 7.4 (37 °C). There is no observable peak in the CD spectra of TBA-BHQ2/QDs (curve a). The feature peak of the secondary structure was observed as an intense negative band with two shoulders located at around 212 nm and 225 nm in the CD spectra of native thrombin (curve b). With the addition of thrombin into the TBA-BHQ2/QDs complex solution, the peak of thrombin appeared at 225 nm and the typical antiparallel quadruplex structure peaks were observed containing the major positive peak at around 295 nm with a minimum at around 265 nm (curve c).58 This CD evidence shows that TBA-BHQ2 could change its spatial structure from a random coil to an antiparallel G-quadruplex with thrombin.
Factors affecting the sensitivity of the phosphorescent aptasensor for the detection of thrombin
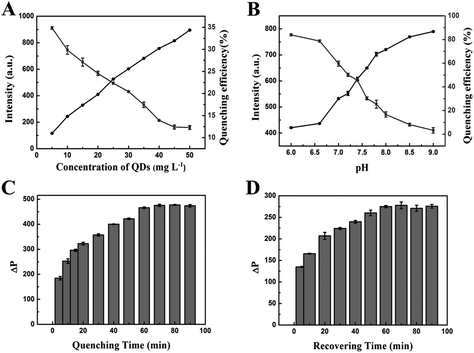
To achieve high sensitivity in the assay, we optimized analytical parameters such as the concentration of MPA-Mn:ZnS QDs, the pH of the solution, the reaction time, the temperature of the system and the concentration of TBA-BHQ2, which could affect the response of the reaction system. At first, we surveyed the effect of the concentration of MPA-Mn:ZnS QDs. As shown in Fig. 3A, the phosphorescence intensity of the QDs increased linearly with the concentrations of the QDs increasing from 5 to 50 mg L−1. Meanwhile, the quenching efficiency of QDs decreased immediately in the presence of 0.1 μM TBA-BHQ2. Considering the phosphorescence intensity and the quenching efficiency, 25 mg L−1 was chosen as the optimum reaction concentration of MPA-Mn:ZnS QDs in subsequent experiments.The pH of the solution was an important parameter to detect thrombin. The pH affected not only the phosphorescence intensity of QDs, but also the subsequent quenching efficiency. In this work, a pH less than 6 was not studied owing to the instability of MPA-Mn:ZnS QDs in acidic media. Along with the increase of pH, the phosphorescence intensity increased proportionally, but the quenching efficiency of QDs decreased obviously. In consideration of the phosphorescence intensity and quenching efficiency, pH 7.4 was regarded as the optimal pH value for the phosphorescent aptasensor response (Fig. 3B).
To examine the kinetic behavior of the phosphorescent aptasensor for detecting thrombin, the influence of the reaction time was investigated. ΔP gradually increased with the reaction time between the TBA-BHQ2 (as the quencher) and MPA-Mn:ZnS QDs. On increasing up to 60 min, ΔP remained unchanged with a further increase of the reaction time (Fig. 3C). As thrombin (as the phosphorescence recover) reacted with TBA-BHQ2/MPA-Mn:ZnS QDs complex, ΔP tended to be relatively flat when the reaction time was up to 60 min (Fig. 3D). Therefore, 60 min was chosen as the optimal reaction time for both the quenching and recovering processes.
In the optimum temperature range, the enzyme activity was the strongest and the reaction speed was the greatest, so temperature was also one of the important parameters. As shown in Fig. S4,† the activity of the enzyme was affected by temperature, and the activity of the enzyme increased gradually with the increase of temperature in a certain range of temperature. At the optimum temperature, the enzyme activity was the strongest. Over a certain temperature range, the activity of the enzyme decreased and even deactivated with the increase of temperature. Therefore, 37 °C was chosen as the optimal temperature in the reaction system.
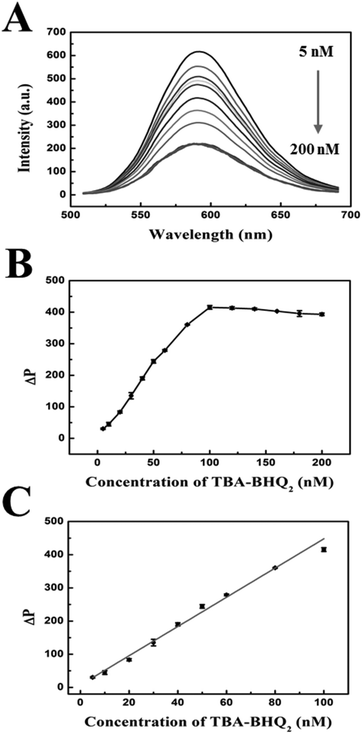
The concentration of TBA-BHQ2 was another important parameter for the sensitive detection of thrombin based on the phosphorescence aptasensor. Fig. 4A shows the phosphorescence emission spectra of MPA-Mn:ZnS QDs with different concentrations of TBA-BHQ2, in which it could be seen that the phosphorescence intensity of MPA-Mn:ZnS QDs gradually decreased with the increase of TBA-BHQ2 concentration. As shown in Fig. 4B, ΔP increased with TBA-BHQ2 increasing from 5 to 100 nM. However, when the TBA-BHQ2 concentration exceeded 100 nM, ΔP reached equilibrium. The phosphorescence intensity of QDs was quenched by about 61.5% in 60 min after the addition of 100 nM TBA-BHQ2. Fig. 4C shows that there was a good linear relationship between ΔP and the concentration of TBA-BHQ2 in the range of 5–100 nM. The regression equation is as follows: ΔP = 4124.43 + 32.13C (C is the concentration of TBA-BHQ2, and the corresponding regression coefficient (R2) = 0.992). Because an excessive amount of quencher would hamper the responsive efficiency and sensitivity for the detection of thrombin subsequently, the concentration of TBA-BHQ2 in solution was set at 100 nM in order to obtain a lower detection limit.
Figures of merit of the phosphorescent aptasensor for the detection of thrombin
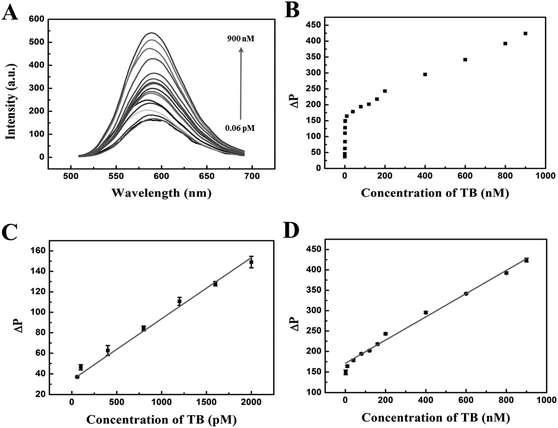
Under the optimum experimental conditions, we explored the quantitative detection of the phosphorescent aptasensor assay. In this work, the phosphorescence intensity of the QDs/TBA-BHQ2 complex was recovered due to the formation of the quadruplex/thrombin complex in the presence of thrombin. It was rational that TBA-BHQ2 in TBA-BHQ2/QDs complex combined with thrombin to form an antiparallel G-quadruplex because of the high affinity between TBA-BHQ2 and thrombin. Therefore, after the addition of thrombin to the QDs/TBA-BHQ2 system, TBA-BHQ2 bound to thrombin preferentially, which resulted in phosphorescence intensity recovery of the mixed system due to the release of QDs from the QDs/TBA-BHQ2 system.As shown in Fig. 5A, the phosphorescence intensity increased with the increase of thrombin concentrations. The plot of phosphorescence intensity with increasing thrombin concentrations from 0.06 to 900 nM is shown in Fig. 5B. There was a good linear relationship between ΔP and thrombin concentrations in the concentration range from 60 to 2000 pM (Fig. 5C). The regression equation could be described as follows: ΔP = 33.97 + 0.060C (C is the concentration of thrombin in pM). Additionally, R2 was 0.994 and the LOD was estimated to be as low as 15.26 pM according to 3SD/k in which SD is the standard deviation of the blank sample (n = 11) and k is the slope of the obtained equation. Then, in the concentration range from 2 to 900 nM, the regression equation could be described as follows: ΔP = 170.83 + 0.28C (C is the concentration of thrombin in nM, and R2 = 0.995) (Fig. 5D).
There were two linear ranges in the calibration line for the detection of thrombin in solution. In detail, at low thrombin levels from 60 pM to 2 nM, the local concentration was rapidly depleted as TBA-BHQ2 could change its spatial structure from a random coil to an antiparallel G-quadruplex which resulted from the special combination between thrombin and TBA-BHQ2, resulting in a high sensitivity of the phosphorescence recovery with a large slope. At high thrombin levels from 2 nM to 900 nM, the QDs/TBA-BHQ2 complex was supplied with thrombin for a longer period of time and the reaction proceeds over a larger time window. Thus, the combination between thrombin and TBA-BHQ2 to form an antiparallel G-quadruplex and simultaneously the release of QDs could be hampered, resulting in a lower slope. Also, it attained a saturation level at higher concentration of thrombin as for the reaction. Thus, the “off–on” phosphorescent aptasensor showed different linear correlations in different concentration ranges.59,60
Furthermore, some analytical methods of detecting thrombin are listed in Table 1. Compared with those analytical methods, our method presents a simple and convenient phosphorescent aptasensor detection method to offer a relatively good detection limit and a wide linear range, and then, the method does not require complex pretreatments and can effectively eliminate the autofluorescence and scattered light interference from biological matrices.
| Method | Detection limit | Linear range | Ref. |
|---|---|---|---|
| Phosphorescence detection based on Mn-doped ZnS quantum dots/carbon nanodots | 13 pM | 0–40 nM | 11 |
| Colorimetric detection based on AuNP oligomers | 5 fM | 13 fM–0.13 nM | 21 |
| Fluorescence detection based on a graphene fluorescence resonance energy transfer aptasensor | 31.3 pM | 62.5–187.5 pM | 22 |
| Colorimetric detection based on aptamer–AuNPs | 100 pM | 0.1–15 nM | 24 |
| Chemiluminescence detection based on heparin-mimicking hyperbranched polyester | 0.031 pM | 2.70 pM–27 nM and 1.35 μM–27.0 μM | 27 |
| Electrochemiluminescence detection based on multiwalled carbon nanotubes | 0.23 pM | 1 pM–1 nM | 28 |
| Colorimetric detection based on GO-AuPtNP-Apt15/G-quadruplex/hemin | 150 pM | 0.3–100 nM | 61 |
| Ultrasensitive and universal fluorescent aptasensor based on a DNA/Ag nanocluster fluorescence light-up system | 8400 pM | 50–800 nM | 16 |
| Localized surface plasmon resonance based on a label-free anti-thrombin aptasensor | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pM 000 pM |
0–1.9 μM | 62 |
| Solid phase microextraction-liquid chromatography/tandem mass spectrometry based on aptamer-functionalized probes | 300 pM | 0.5–50 nM | 63 |
| Phosphorescence detection based on a phosphorescent aptasensor | 15.26 pM | 60–2000 pM and 2–900 nM. | In this work |
Selectivity and anti-interference ability of the phosphorescent aptasensor for the detection of thrombin
Selectivity and anti-interference ability are important parameters to evaluate the performance of a new analytical method, especially those with potential applications in biomedical samples. Thus a highly selective response to the target over other potentially competing species was necessary. We evaluated the selectivity of our method with the addition of various foreign interfering substances. Under the optimal conditions, the selectivity of the proposed method to detect thrombin was investigated and a number of common proteins and amino acids including lysozyme, Alt, Alp, Phe, Arg, His, Try, Lys, Leu, Met, Cys, Gly and Ile were adopted in the assay system. As shown in Fig. 6A, the concentrations of other proteins and amino acids were equal to thrombin (600 nM), and only thrombin could cause an obvious phosphorescence recovery of the QDs in the assay system because the detection system had good selectivity and specificity for thrombin detection.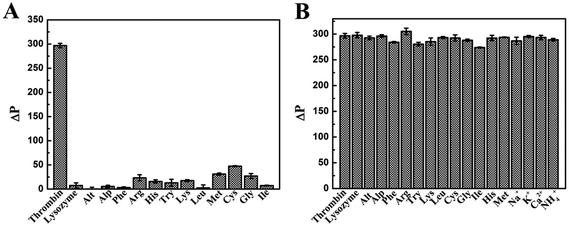 | ||
| Fig. 6 (A) Selectivity and (B) interference evaluation of the phosphorescent aptasensor for thrombin detection by comparison with common potentially interfering species. | ||
The detection of thrombin will be subject to interference by other substances in organisms, such as ions, amino acids and proteases. Thrombin, which acts as the main protease in the blood coagulation cascade, was most affected by other proteins during the monitoring. The tolerable concentrations of these interfering proteases with the thrombin concentration of 600 nM were 20 μM for lysozyme, Alt, and Alp. As amino acids are the building blocks of proteins for animal nutrition, the effect of detecting thrombin was less than that of proteins. The tolerable concentrations were 40 μM for Phe, Arg, His, Try, Lys, Leu, Met, Cys and Gly. Ions, such as K+ and Na+, had a little effect on the process of detecting thrombin, because they maintain the osmotic pressure in the body as inorganic matter. The tolerable concentrations were 400 μM for K+, Na+, Ca2+ and NH4+. As shown in Fig. 6B, the response of this phosphorescent aptasensor system to detect thrombin was almost unaffected by the presence of other tested proteins. The results suggested that our assay had excellent selectivity and specificity for the detection of thrombin.
Application of the phosphorescent aptasensor for the detection of thrombin in biological samples
To investigate the ability of the QDs/TBA-BHQ2 aptasensor system to detect thrombin in the complex biological environment, the detection of thrombin in human serum samples and plasma samples was next performed by the standard addition method. The QDs/TBA-BHQ2 aptasensor system and various amounts of thrombin were incubated in human serum and plasma samples. These serum samples and plasma samples were used for phosphorescence measurement. As shown in Table 2, no thrombin was found in three different human serum samples, and the recovery ranged from 96% to 99% at concentrations of 1 nM to 200 nM, which suggested a good accuracy. Then, the concentration of thrombin was determined from 2.46 to 5.65 nM for three different human plasma samples, and the quantitative spiked recoveries of thrombin at concentrations of 10 nM and 200 nM ranged from 95% to 104%, respectively. These results demonstrated that the QDs/TBA-BHQ2 aptasensor system was appropriate for thrombin detection in biological samples, which might provide great potential for further use in clinics.| Sample | Spiked thrombin/nM | Concentration (mean ± σ, n = 3)/nM | Recovery/% |
|---|---|---|---|
| a Not detected. | |||
| Serum-1 | 0 | nda | |
| 1 | 0.99 ± 0.01 | 99 ± 1 | |
| 200 | 198.49 ± 4.08 | 99 ± 2 | |
| Serum-2 | 0 | nd | |
| 1 | 0.98 ± 0.02 | 98 ± 2 | |
| 200 | 197.80 ± 3.54 | 98 ± 1 | |
| Serum-3 | 0 | nd | |
| 1 | 0.99 ± 0.01 | 99 ± 1 | |
| 200 | 191.95 ± 0.98 | 96 ± 1 | |
| Plamsa-1 | 0 | 3.77 | |
| 10 | 13.56 ± 0.12 | 95 ± 3 | |
| 200 | 203.91 ± 0.05 | 104 ± 1 | |
| Plamsa-2 | 0 | 2.46 | |
| 10 | 12.57 ± 0.02 | 104 ± 1 | |
| 200 | 202.35 ± 0.11 | 96 ± 4 | |
| Plamsa-3 | 0 | 5.65 | |
| 10 | 15.91 ± 0.01 | 104 ± 1 | |
| 200 | 205.39 ± 0.28 | 95 ± 5 | |
Conclusion
In conclusion, an “off–on” phosphorescent aptasensor system was used for sensitive detection of thrombin based on the phosphorescence energy transfer mechanism. The “off” signal was attributed to the effective phosphorescence quenching by QDs, where phosphorescence QDs served as the energy donors and TBA-BHQ2 served as the energy acceptors. Then, phosphorescence recovery of the QDs was achieved by specific interaction between TBA-BHQ2 and thrombin to form a G-quadruplex structure. The proposed method provided a proof-of-concept of an “off–on” phosphorescent aptasensor for the detection of thrombin at trace levels.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful for the financial support from the National Natural Science Foundation of China (21375095 and 20975054).Notes and references
- V. Pavlov, B. Shlyahovsky and I. Willner, J. Am. Chem. Soc., 2005, 127, 6522–6523 CrossRef CAS PubMed.
- Y. Xiao, B. D. Piorek, K. W. Plaxco and A. J. Heeger, J. Am. Chem. Soc., 2005, 127, 17990–17991 CrossRef CAS PubMed.
- L. Romhildt, C. Pahlke, F. Zorgiebel, H. G. Braun, J. Opitz, L. Baraban and G. Cuniberti, ACS Appl. Mater. Interfaces, 2013, 5, 12029–12035 CrossRef PubMed.
- Y. H. Lao, C. W. Chi, S. M. Friedrich, K. Peck, T. H. Wang, K. W. Leong and L. C. Chen, ACS Appl. Mater. Interfaces, 2016, 8, 12048–12055 CrossRef CAS PubMed.
- L. Ge, H. N. Li, X. J. Du, M. Y. Zhu, W. Chen, T. Y. Shi, N. Hao, Q. Liu and K. Wang, Biosens. Bioelectron., 2018, 111, 131–137 CrossRef CAS PubMed.
- J. Qian, C. C. Ren, C. Q. Wang, W. Chen, X. T. Lu, H. A. Li, Q. Liu, N. Hao, H. M. Li and K. Wang, Anal. Chim. Acta, 2018, 1019, 119–127 CrossRef CAS PubMed.
- Z. E. Hughes and T. R. Walsh, ACS Sens., 2017, 2, 1602–1611 CrossRef CAS PubMed.
- B. E. F. de Ávila, M. A. Lopez-Ramirez, D. F. Báez, A. Jodra, V. V. Singh, K. Kaufmann and J. Wang, ACS Sens., 2016, 1, 217–221 CrossRef.
- V. Gedi, C. K. Song, G. B. Kim, J. O. Lee, E. Oh, B. S. Shin, M. Jung, J. Shim, H. Lee and Y. P. Kim, Sens. Actuators, B, 2018, 256, 89–97 CrossRef CAS.
- X. F. Liu, X. X. Hua, Q. L. Fan, J. Chao, S. Su, Y. Q. Huang, L. H. Wang and W. Huang, ACS Appl. Mater. Interfaces, 2015, 7, 16458–16465 CrossRef CAS PubMed.
- L. Zhang, P. Cui, B. C. Zhang and F. Gao, Chem. – Eur. J., 2013, 19, 9242–9250 CrossRef CAS PubMed.
- M. Liu, J. Li and B. X. Li, Talanta, 2017, 175, 224–228 CrossRef CAS PubMed.
- C. Riccardi, I. R. Krauss, D. Musumeci, F. Morvan, A. Meyer, J. J. Vasseur, L. Paduano and D. Montesarchio, ACS Appl. Mater. Interfaces, 2017, 9, 35574–35587 CrossRef CAS PubMed.
- D. Y. Zhao, Y. Peng, L. H. Xu, W. Zhou, Q. Wang and L. Guo, ACS Appl. Mater. Interfaces, 2015, 7, 23418–23422 CrossRef CAS PubMed.
- Y. Y. Liu, Y. H. Zhao, Q. Fan, M. S. Khan, X. J. Li, Y. Zhang, H. M. Ma and Q. Wei, Mikrochim. Acta, 2018, 185, 85 CrossRef PubMed.
- Y. Zhu, X. C. Hu, S. Shi, R. R. Gao, H. L. Huang, Y. Y. Zhu, X. Y. Lv and T. M. Yao, Biosens. Bioelectron., 2016, 79, 205–212 CrossRef CAS PubMed.
- G. Shen, H. Zhang, C. R. Yang, Q. F. Yang and Y. L. Tang, Anal. Chem., 2017, 89, 548–551 CrossRef CAS PubMed.
- Y. Z. Liu, X. K. Jiang, W. F. Cao, J. Y. Sun and F. Gao, Sensors, 2018, 18, 589 CrossRef PubMed.
- R. Yang, S. W. Liu, Z. J. Wu, Y. Tan and S. Q. Sun, Talanta, 2018, 182, 348–353 CrossRef CAS PubMed.
- G. L. Wang, X. L. Hu, X. M. Wu, Y. M. Dong and Z. J. Li, Microchim. Acta, 2016, 183, 765–771 CrossRef CAS.
- J. J. Li, Y. F. Jiao, Q. Y. Liu and Z. B. Chen, ACS Sustainable Chem. Eng., 2018, 6, 6738–6745 CrossRef CAS.
- H. X. Chang, L. H. Tang, Y. Wang, J. H. Jiang and J. H. Li, Anal. Chem., 2010, 82, 2341–2346 CrossRef CAS PubMed.
- Q. Zhao, X. F. Lu, C. G. Yuan, X. F. Li and X. C. Le, Anal. Chem., 2009, 81, 7484–7489 CrossRef CAS PubMed.
- Z. B. Chen, L. L. Tan, L. Y. Hu, Y. M. Zhang, S. X. Wang and F. Y. Lv, ACS Appl. Mater. Interfaces, 2016, 8, 102–108 CrossRef CAS PubMed.
- Y. L. Niu, M. L. Chu, P. Xu, S. S. Meng, Q. Zhou, W. B. Zhao, B. Zhao and J. Shen, Biosens. Bioelectron., 2018, 101, 174–180 CrossRef CAS PubMed.
- D. Wu, X. Xin, X. H. Pang, M. Pietraszkiewicz, R. Hozyst, X. G. Sun and Q. Wei, ACS Appl. Mater. Interfaces, 2015, 7, 12663–12670 CrossRef CAS PubMed.
- Q. Zhao, X. F. Li, Y. H. Shao and X. C. Le, Anal. Chem., 2008, 80, 7586–7593 CrossRef CAS PubMed.
- H. S. Cho, B. R. Baker, S. Wachsmann-Hogiu, C. V. Pagba, T. A. Laurence, S. M. Lane, L. P. Lee and J. B. H. Tok, Nano Lett., 2008, 8, 4386–4390 CrossRef CAS PubMed.
- W. Kotkowiak, J. Lisowiec-Wachnicka, J. Grynda, R. Kierzek, J. Wengel and A. Pasternak, Mol. Ther.–Nucleic Acids, 2018, 10, 304–316 CrossRef CAS PubMed.
- C. F. Jiang, T. T. Zhao, S. Li, N. Y. Gao and Q. H. Xu, ACS Appl. Mater. Interfaces, 2013, 5, 10853–10857 CrossRef CAS PubMed.
- S. Y. Liu, W. D. Na, S. Pang, F. P. Shi and X. G. Su, Analyst, 2014, 139, 3048–3054 RSC.
- X. M. Lu, J. Y. Zhang, Y. N. Xie, X. F. Zhang, X. M. Jiang, X. D. Hou and P. Wu, Anal. Chem., 2018, 90, 2939–2945 CrossRef CAS PubMed.
- W. J. Jing, Y. X. Lu, F. Y. Wang, L. Y. He, J. W. Sun and Y. Y. Liu, Mikrochim. Acta, 2018, 185, 298 CrossRef PubMed.
- J. Y. Zhang, D. D. Tang, Y. D. Yao, X. D. Hou and P. Wu, Nanoscale, 2018, 10, 9236–9244 RSC.
- X. Wei, M. M. Yu, C. Li, X. H. Gong, F. Qin and Z. H. Wang, Mikrochim. Acta, 2018, 185, 208 CrossRef PubMed.
- D. D. Tang, J. Y. Zhang, R. X. Zhou, Y. N. Xie, X. D. Hou, K. L. Xu and P. Wu, Nanoscale, 2018, 10, 8477–8482 RSC.
- N. Chang, J. P. Mao, Y. X. Lu, J. E. Yang, Y. X. Pu, S. C. Zhang and Y. Y. Liu, Sens. Actuators, B, 2016, 233, 17–24 CrossRef CAS.
- E. Sotelo-Gonzalez, M. T. Fernandez-Arguelles, J. M. Costa-Fernandez and A. Sanz-Medel, Anal. Chim. Acta, 2012, 712, 120–126 CrossRef CAS PubMed.
- J. Z. Lv, Y. M. Miao, J. J. Yang, J. Qin, D. X. Li and G. Q. Yan, Biosens. Bioelectron., 2017, 91, 560–565 CrossRef CAS PubMed.
- Q. Jin, Y. Li, J. Z. Huo and X. J. Zhao, Sens. Actuators, B, 2016, 227, 108–116 CrossRef CAS.
- Y. M. Miao, J. Z. Lv and G. Q. Yan, Biosens. Bioelectron., 2017, 94, 263–270 CrossRef CAS PubMed.
- W. Bian, F. Wang, Y. L. Wei, L. Wang, Q. L. Liu, W. J. Dong, S. M. Shuang and M. M. F. Choi, Anal. Chim. Acta, 2015, 856, 82–89 CrossRef CAS PubMed.
- Y. T. Liu, J. P. Lei, Y. Huang and H. X. Ju, Anal. Chem., 2014, 86, 8735–8741 CrossRef CAS PubMed.
- E. Farjami, L. Clima, K. Gothelf and E. E. Ferapontova, Anal. Chem., 2011, 83, 1594–1602 CrossRef CAS PubMed.
- D. Feng, Y. C. Song, W. Shi, X. H. Li and H. M. Ma, Anal. Chem., 2013, 85, 6530–6535 CrossRef CAS PubMed.
- S. Umrao, V. Jain, Anusha, B. Chakraborty and R. Roy, Sens. Actuators, B, 2018, 267, 294–301 CrossRef CAS.
- J. Q. Zhuang, X. D. Zhang, G. Wang, D. M. Li, W. S. Yang and T. J. Li, J. Mater. Chem., 2003, 13, 1853–1587 RSC.
- P. Wu, Y. He, H. F. Wang and X. P. Yan, Anal. Chem., 2010, 82, 1427–1433 CrossRef CAS PubMed.
- Y. Chen, G. H. Chen, X. Y. Liu and T. Yang, J. Lumin., 2018, 195, 314–320 CrossRef CAS.
- H. Yang, G. H. Ran, J. J. Yan, H. Zhang and X. L. Hu, Luminescence, 2018, 33, 349–355 CrossRef CAS PubMed.
- Y. Wang, J. Liu, M. Q. Zhu, L. J. Wang, X. Y. Zen, S. S. Fan, Z. Wang, H. L. Li, R. S. Na, X. Zhao and Q. X. Li, J. Lumin., 2018, 200, 111–119 CrossRef CAS.
- T. T. Gan, N. J. Zhao, G. F. Yin, M. D. Tu, J. G. Liu and W. Q. Liu, New J. Chem., 2017, 41, 13425–13434 RSC.
- Y. Xiong, Y. Chen, L. Wang and Y. Li, Talanta, 2018, 190, 226–234 CrossRef CAS PubMed.
- Q. Yan, Z. H. Chen, S. F. Xue, X. Y. Han, Z. Y. Lin, S. Q. Zhang, G. Y. Shi and M. Zhang, Sens. Actuators, B, 2018, 268, 108–114 CrossRef CAS.
- Y. P. Zhao, Y. Y. Ma and W. Y. Lin, Sens. Actuators, B, 2018, 268, 157–163 CrossRef CAS.
- J. J. Yang, Z. F. Zhang and G. Q. Yan, Anal. Biochem., 2017, 539, 127–133 CrossRef CAS PubMed.
- S. M. Kelly, T. J. Jess and N. C. Price, Biochim. Biophys. Acta, 2005, 1751, 119–139 CrossRef CAS PubMed.
- D. M. Kong, Y. E. Ma, J. H. Guo, W. Yang and H. X. Shen, Anal. Chem., 2009, 81, 2678–2684 CrossRef CAS PubMed.
- A. Gholizadeh, S. Shahrokhian, A. I. Zad, S. Mohajerzadeh, M. Vosoughi, S. Darbari, J. Koohsorkhi and M. Mehran, Anal. Chem., 2012, 84, 5932–5938 CrossRef CAS PubMed.
- X. X. Dong, M. Y. Li, N. N. Feng, Y. M. Sun, C. Yang and Z. L. Xu, RSC Adv., 2015, 5, 86485–86489 RSC.
- L. Wang, W. Yang, T. F. Li, D. Li, Z. M. Cui, Y. Wang, S. L. Ji, Q. X. Song, C. Shu and L. Ding, Microchim. Acta, 2017, 184, 3145–3151 CrossRef CAS.
- Y. Jalit, F. A. Gutierrez, G. Dubacheva, C. Goyer, L. Coche-Guerente, E. Defrancq, P. Labbe, G. A. Rivas and M. C. Rodriguez, Biosens. Bioelectron., 2013, 41, 424–429 CrossRef CAS PubMed.
- F. Y. Du, M. N. Alam and J. Pawliszyn, Anal. Chim. Acta, 2014, 845, 45–52 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an01571f |
| This journal is © The Royal Society of Chemistry 2019 |