Open Access Article
Open Access ArticleEngineering photoautotrophic carbon fixation for enhanced growth and productivity
Feiyan
Liang
,
Pia
Lindberg
and
Peter
Lindblad
 *
*
Microbial Chemistry, Department of Chemistry-Ångström, Uppsala, Sweden. E-mail: peter.lindblad@kemi.uu.se
First published on 16th October 2018
Abstract
Oxygenic photosynthesis is the origin of most organic carbon compounds on Earth and an essential part of the natural carbon cycle. Cyanobacteria, the only oxygenic photoautotrophic prokaryotes, are important in several natural processes: as primary sustainable producers, in providing oxygen to the atmosphere, and in nitrogen fixation. From a biotechnological perspective, cyanobacteria are ideal cell factories since (i) the required energy and carbon source, sunlight and CO2, are abundant and freely available, (ii) cyanobacteria are capable of producing a variety of natural products, which can be used as fuels, medicines, cosmetics etc., and (iii) metabolic engineering and synthetic biology tools of cyanobacteria are being developed rapidly, making them feasible as host organisms for heterologous production of interesting compounds. However, compared to commercially employed heterotrophic microorganisms, the growth and productivity of cyanobacteria are currently not competitive. Therefore, improving cyanobacterial growth and productivity is an important task to enable commercialization of cyanobacterial bioproducts. Such studies also offer important clues for increasing the photosynthesis and yield of crop plants, which is important in view of providing food for a rapidly increasing world population. There are many strategies targeting this task, such as optimizing cultivation conditions, engineering native pathways, and introducing synthetic pathways based on an understanding of overall metabolic networks. One major limitation of cyanobacterial productivity, however, is the low efficiency of carbon fixation through the Calvin–Benson–Bassham (CBB) cycle. In this review, we introduce and discuss the possibilities to enhance growth and productivity by engineering the CBB cycle. We also give a brief discussion of options to further extend the capabilities of cells to fix inorganic carbon by the introduction of other native and artificial carbon fixation cycles.
1. Introduction
It is predicted that by 2050, the world population will increase by at least 30% compared to 2015. Together with increased daily food consumption per individual, a 50% increase in food production will be required. It is impossible to meet this demand only by expanding the arable lands. Therefore, high yield crops and/or non-crop food will be needed. Similar to the situation with food, a much larger energy supply will also be required in the future. The energy used today is mainly fossil fuels, usage of which must be diminished due to air pollution and global warming. Therefore, increasing the crop yield and finding alternatives to fossil fuels are two of the major tasks that modern society is facing.Food and fossil fuels both originate from oxygenic photosynthesis, which consists of a photochemical phase and a second biosynthetic phase. In the photochemical phase, solar energy is absorbed and converted into chemical energy. During this process, water is split, releasing oxygen and protons into the thylakoid lumen. At the same time, the released electrons enter the photosynthetic electron transfer chain. In the electron transfer chain, plastoquinone is reduced to plastoquinol, transferring electrons to the cytochrome b6f complex. When plastoquinol is oxidized back to plastoquinone, protons are transferred from the cytoplasm to the thylakoid lumen.1,2 Finally, the electrons are used to reduce NADP+ into NADPH, and the generated proton gradient between the thylakoid lumen and cytoplasm drives ATP synthesis with an ATP synthase. NADPH is subsequently used in the so-called biosynthetic phase, where CO2 is assimilated to generate the building blocks of amino acids and other carbon-containing molecules, at the same time consuming ATP and NADPH.
One method to obtain food and renewable fuels from the oxygenic photosynthetic process is to supply plant biomass to Escherichia coli (E. coli), yeasts or other well understood microorganisms for fermentation to get the desired compound(s). However, due to the multiple steps in this process, from energy absorption by plants to fermentation by heterotrophic organisms, the solar energy-to-product efficiency is very low, for example 0.2% in ethanol production from sugarcane fermentation.3 In addition, E. coli and yeasts are not able to ferment all types of plant biomass components. For example, the most abundant carbohydrate, lignocellulose from plant materials, needs to be broken down into smaller molecules before it can be used. This breaking down process requires enzymes, high temperature, or/and high pressure.4 Besides, using crop plants as the source of sugar for fermentation is not a viable alternative on a large scale, as it would lead to less arable land available for food production.
Another method to generate renewable fuels would be to directly obtain them from autotrophic microorganisms like cyanobacteria or microalgae. Using such organisms as green cell factories or cell catalysts has many advantages over traditional fermentation technologies. Cyanobacteria and microalgae use cheap and abundant energy (sunlight) and carbon (CO2) sources, and also show a higher energy conversion efficiency compared to land plants (e.g. 1.5% in oil production from microalgae).5 Cyanobacteria, being prokaryotes, are relatively easy to engineer compared to eukaryotic organisms. Ideal cyanobacterial cell factories are those that can maintain the cell population, distribute the remaining energy and carbon to the designed product(s), and possess the capability to excrete the product(s) out of the cells. Excreting products will minimize any potential toxicities or other negative effects on the cell metabolism, simplify recovery, and may also increase the yield.6,7
While the main advantage of cyanobacteria as cell factories compared to heterotrophic microorganisms is their capability for oxygenic photosynthesis, the main drawback is the relatively low yields achieved per volume and time in bioreactors, requiring large scale facilities and a large surface area.8–10 The main reason for this is the limitations inherent to photosynthesis, and thus, increasing the photosynthetic efficiency is necessary to enhance the yield and to realize commercialization of cyanobacterial products. The photosynthetic efficiency is determined as the sum efficiency of the photochemical and biosynthetic phases. In this review, we will focus on the biosynthetic phase, discussing the control enzymes of the Calvin–Benson–Bassham (CBB) carbon fixation cycle and other natural and artificial carbon fixation cycles. Some other possible strategies to enhance growth and productivity will also be briefly discussed.
2. The Calvin–Benson–Bassham (CBB) cycle
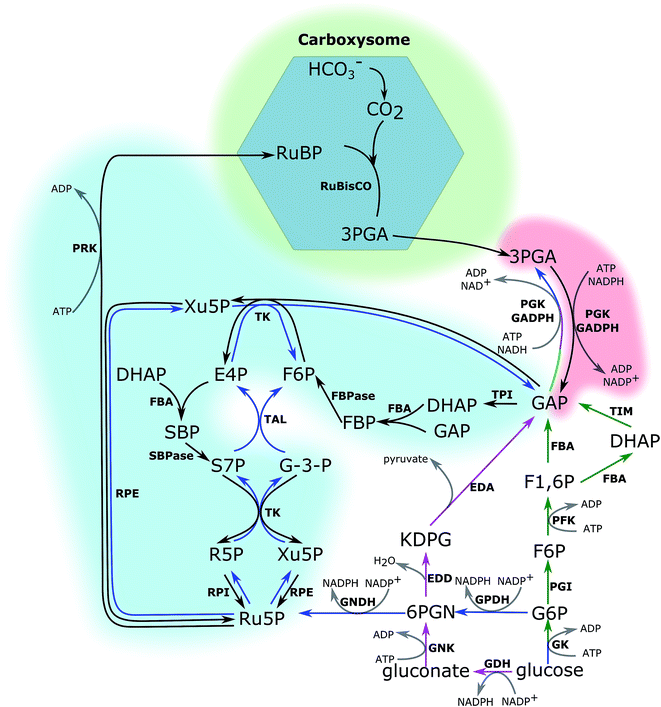
The CBB cycle, employing one of the most abundant but also one of the least efficient enzymes in nature, ribulose-1,5-bisphosphate carboxylase (RuBisCO), to fix CO2, is the major carbon fixation cycle in nature. It consists of 13 reactions catalysed by 11 enzymes (Fig. 1). The entire cycle is divided into 3 stages: (i) ribulose-1,5-bisphosphate (RuBP) carboxylation, (ii) 3-phosphoglycerate (PGA) reduction, and (iii) RuBP regeneration. In the carboxylation stage, RuBisCO catalyses one molecule of CO2 and one molecule of RuBP to form two molecules of PGA. The PGA is converted into glyceraldehyde-3-phosphate (GAP), consuming ATP and NADPH generated in the photochemical phase in the reduction stage. Finally, RuBP is regenerated from GAP through several steps, also consuming ATP in the RuBP regeneration stage. Without considering the side reaction of RuBisCO where the substrate is oxygen (photorespiration), nine molecules of ATP and six molecules of NADPH are required to assimilate three molecules of CO2 and to generate one molecule of GAP in the CBB cycle.The cyanobacterial CBB cycle partially overlaps the oxidative pentose phosphate (OPP) pathway, the main NADPH supplier, under heterotrophic growth conditions. However, three steps are specific in the CBB cycle (Fig. 1). They are (i) fixing CO2 into PGA by RuBisCO (consumption of RuBP), (ii) phosphorylating ribulose-5-phosphate (Ru5P) to RuBP by phosphoribulokinase (PRK, regeneration of RuBP), and (iii) dephosphorylating sedoheptulose-1,7-bisphosphate (SBP) into sedoheptulose-7-phosphate (S7P) by the bi-functional enzyme FBP/SBPase, a step in which photoautotrophic eukaryotes are catalysed by SBPase. Generally, chloroplast FBPase in plants is able to catalyse conversions between SBP and S7P; however, the catalytic rate is too low to support a normal autotrophic growth.11 Introducing RuBisCO and PRK into a heterotrophic microorganism would theoretically complete the CBB cycle. Together with an appropriate energy supply, introducing RuBisCO and PRK would allow heterotrophic microorganisms to fix CO2 into carbohydrates.12 Re-fixing the CO2 released during the carbon metabolism is a promising strategy to increase the carbon usage efficiency in heterotrophic microorganisms. Therefore, studying the CBB cycle and increasing its efficiency will potentially have beneficial effects on the efficiencies of both autotrophic and heterotrophic cell factories.
The CBB cycle is one of the limitations to higher yield and some of its enzymes are more critical than others for the overall efficiency. Therefore, identifying these steps and the corresponding enzymes is the first step towards enhancing the CBB cycle efficiency and subsequently productivity, and technologies to achieve this have been developed. Generally, increasing or decreasing the catalytic efficiency of enzyme(s) will accordingly increase or decrease the flux through the pathway. Analysis of the metabolism following enzyme engineering is one of the methods to determine the critical step(s). In this method, the term “flux control co-efficient” is used, which is determined by calculating the ratio between the change of flux and the change of specific enzyme activity under certain growth conditions.13 The value varies between 0 (no control) and 1 (full control). In many cases, the full control comes from several enzymes, of which one of them is critical. Two things should be kept in mind: (i) specific growth conditions may be required for an enzyme to show control on the flux through the pathway, and the flux control co-efficient varies under different cultivation conditions; (ii) significant changes of the enzyme are required to detect the flux change when the capacity of the enzyme is much higher than the natural capacity of the pathway. In this case, a small flux control coefficient is expected; in other words, the enzyme is not a flux-controlling enzyme under many cultivation conditions. In the early 1990s, numerous studies on antisense plants with deduced CBB cycle enzymes explored the flux control coefficient and physiological features under different growth conditions. Table 1 summarises these studies.
| Enzyme | Organisms | Conditions | Effects or flux control coefficient | Reference |
|---|---|---|---|---|
| RuBisCO | Tobacco (Nicotiana tabacum L.) | 15 h/9 h light/dark cycle (25 °C/20 °C), 300 μmol photons m−2 s−1 | Antisense lines (20% RuBisCO left) have longer senescence phase | 14 |
| Tobacco (Nicotiana tabacum L.) | 500 μmol photons m−2 s−1 irradiance (12 h light/12 h dark) | Decreased photosynthesis and nitrogen metabolism especially under higher nitrate (12 mM) or ammonium nitrate (6 mM) conditions with less than 60% RuBisCO left | 15 | |
| Tobacco (Nicotiana tabacum L.) | Lower than 1000 μmol m−2 s−1 irradiance, ambient CO2 | >0.2 flux control coefficient; 0.01–0.03 flux control coefficient with 100 μmol m−2 s−1 irradiance | 16 | |
| Higher than 1500 μmol m−2 s−1 irradiance, ambient CO2 | 0.8–0.9 flux control coefficient | |||
| Limited inorganic nitrogen | 0.5 flux control coefficient; photosynthesis was largely reduced while growth was hardly affected | |||
| Tobacco (Nicotiana tabacum L.) | 12 h day/l2 h night cycle (irradiance 340 μmol m−2 s−1, 20 °C, 65% relative humidity) | Reduced growth and photosynthesis, increased leaf area ratio and increased shoot-to-root ratio when less than 50% RuBisCO is left | 17 | |
| Rice (Oryza sativa L.) | Saturated light conditions | Strains with 65% RuBisCO activity showed a 20% lower photosynthesis rate in ambient CO2 and a 5% to 15% higher photosynthesis rate in 100–115 Pa CO2 | 18 | |
| Rice (Oryza sativa L.) | 16–22 °C and 28 Pa intercellular CO2 | Flux control coefficient (>0.88) detected | 19 | |
| Flaveria bidentis | Day/night growth temperatures were 28/15 °C | With less than 40% RuBisCO left, CO2 assimilation and photosynthesis was reduced at high light irradiance but unaffected under low light condition (less than 100 μmol m−2 s−1) on a wide range of CO2 concentration | 20 | |
| Tobacco (Nicotiana tabacum) | 1000 μmol m−2 s−1, 350 microbars CO2, and 25 °C | With 18% RuBisCO left, soluble protein content reduced the same level as RuBisCO, little change on other photosynthesis proteins. 63% reduction of CO2 assimilation | 21 | |
| Arabidopsis thaliana | Low (200 μmol m−2 s−1) and high (600 μmol m−2 s−1) | With 30 to 40% RuBisCo activity, photosynthesis, growth and above ground biomass are all reduced | 22 | |
| Flaveria bidentis | 28/20 °C day/night temperature, 700 μmol m−2 s−1, and a 16 h photoperiod or a naturally lit greenhouse | With 40% RuBisCO activity left, photosynthesis decreased but the H2O exchange rate was similar to that of the wild type | 23 | |
| Flaveria bidentis | 28/20 °C day/night temperature, 700 μmol m−2 s−1, and a 16 h photoperiod or a naturally lit greenhouse | Quantum yield of PSI to PSII, PSI to CO2 fixation and PSII to CO2 fixation increased with enhanced irradiance in the RuBisCO decreased plant | 24 | |
| SBPase | Tobacco (Nicotiana tabacum) | 14 h light/10 h dark, 26 °C/18 °C, 600–1200 μmol m−2 s−1 | With less than 20% SBPase left, there were no changes in other CBB cycle enzymes but starch accumulation was reduced. With 57% SBPase left, the CO2 assimilation and quantum yield of PSII reduced | 25 |
| Tobacco (Nicotiana tabacum) | Saturated light conditions | Flux control coefficient: 0.31 under ambient CO2 conditions and 0.54 under saturated CO2 conditions | 26 | |
| Tobacco (Nicotiana tabacum) | Greenhouse in light levels between 400 and 1500 μmol m−2 s−1, a 16 h photoperiod at a minimum of 25 °C light/18 °C dark and a maximum of 35 °C light/25 °C dark | Reduced growth and shoot biomass with 75% SBPase activity. SBPase decreased resulting in a shorter plant | 27 | |
| Tobacco (Nicotiana tabacum L.) | Greenhouse with a 14 h photoperiod at 25 °C light/18 °C dark, 700 to 1200 μmol m−2 s−1 | −0.2 flux control coefficient in the youngest developing leaves, 0.3–0.5 flux control coefficient in mature leaves, and decreased photosynthesis and starch accumulation in mature leaves | 28 | |
| Tobacco (Nicotiana tabacum L.) | Greenhouse 14 h/10 h (26 °C/18 °C) light/dark period, 1000–1400 μmol m−2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s−1 s−1 |
Reductions in SBPase activity between 9% and 60%, maximum RuBP regeneration capacity declined linearly (r2 = 0.79) and no significant change in Rubisco activity | 29 | |
| Rice (Oryza sativa L. ssp. japonica) | Greenhouse at 25 ± 2 °C with a photosynthetic photon flux density of 300 μmol m−2 s−1, a relative humidity of 70% to 80%, and a photoperiod of 14 h/10 h light/dark | Stronger effects on reducing growth, photosynthesis and starch accumulation under low N than high N conditions when SBPase was deduced | 30 | |
| Chloroplast FBPase | Potato (Solanum tuberosum) | 16 h light (250 μmol m−2 s−1, 22 °C)/8 h dark (15 °C) under greenhouse conditions | Unchanged tuber yield and reduced photosynthetic rate when FBPase activity decreased to 36% of that of the wild type. Reduced growth and photosynthetic rate when FBPase activity is below 15% of that of the wild type. <0.2 flux control coefficient | 31 |
| PRK | Tobacco (Nicotiana tabacum L.) | 330 μmol photons m−2 s−1, 350 ppm CO2, 25 °C | With less than 85% PRK activities left, CO2 assimilation was reduced. Almost 0 flux control coefficient under low light conditions | 32 |
| Tobacco (Nicotiana tabacum L.) | 330 μmol photons m−2 s−1, 350 μmol CO2 mol−1, 25 °C | 94% decrease in PRK has stronger inhibition on photosynthesis (by 35%) with 0.4 mM NH4NO3 than with 0.5 mM NH4NO3 (by 20%) | 33 | |
| Tobacco (Nicotiana tabacum L.) | 330 μmol photons m−2 s−1 | 0.25 flux control coefficient to CO2 assimilation | 34 | |
| 800 μmol photons m−2 s−1 | 0 flux control coefficient to CO2 assimilation | |||
| GAPDH | Tobacco (Nicotiana tabacum L.) | Ambient CO2 | With less than 35% GAPDH left, photosynthesis was inhibited (900 and 1500 μmol m−2 ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s−1). RuBP was reduced linearly with reduction of GAPDH. <0.2 flux control coefficient s−1). RuBP was reduced linearly with reduction of GAPDH. <0.2 flux control coefficient |
35 |
| TK | Tobacco (Nicotiana tabacum L.) | 170 μmol m−2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s−1, ambient CO2 s−1, ambient CO2 |
0.07 flux control coefficient | 36 |
700 μmol m−2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s−1, ambient CO2 s−1, ambient CO2 |
0.32 flux control coefficient | |||
| Saturating light and CO2 | Almost 1 flux control coefficient | |||
| FBA | Potato (Solanum tuberosum) | 70 μmol m−2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s−1 or 390 μmol m−2 s−1 or 390 μmol m−2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s−1, ambient CO2 or 800 ppm CO2 s−1, ambient CO2 or 800 ppm CO2 |
Decrease of FBA has stronger inhibition on photosynthesis and growth under higher light and CO2 conditions than lower light and CO2 conditions | 37 |
| Potato (Solanum tuberosum) | 70 μmol m−2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s−1, ambient CO2 s−1, ambient CO2 |
0.15 flux control coefficient | 38 | |
| 390 μmol m−2 s−1 light and 400 ppm CO2 | 0.21 flux control coefficient | |||
| 390 μmol m−2 s−1 light and 800 ppm CO2 | 0.55 flux control coefficient |
2.1 RuBP carboxylation
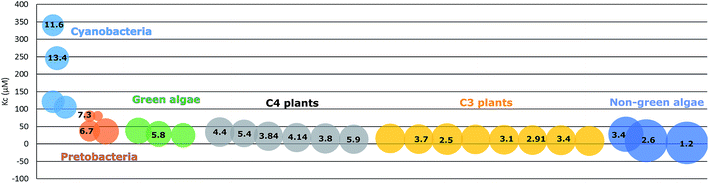
Photoautotrophic organisms have developed other strategies to deal with the issue of the changed atmospheric ratio of CO2 and O2, adapted to different habitation conditions. Green algae and cyanobacteria have developed special carbon concentrating mechanisms, pyrenoids in green algae and carboxysomes in cyanobacteria, to increase the CO2 concentration near RuBisCO. Pyrenoids are also found in hornworts.47,48 In C4 and crassulacean acid metabolism (CAM) plants, the carbon fixation process uses the relatively more efficient carboxylase phosphoenolpyruvate carboxylase (PEPc) to fix atmospheric CO2 into a C4 acid. This C4 acid then in a later step undergoes decarboxylation to release CO2 near RuBisCO, resulting in a high concentration of CO2 in the vicinity of the enzyme. In another word, the initial CO2-fixation step is separated from the RuBisCO step either in space, as is the case of C4 plants with different cell types, or in time, as in CAM plants where the reactions take place at different phases of the day/night cycles.49 In contrast, C3 plants do not have similar compartments or mechanisms to concentrate CO2. They instead contain a very high concentration of RuBisCO, about a quarter of the soluble proteins in leaves, consuming almost half of the available nitrogen in the cells, to compensate for the low catalytic performance.
Due to the adaption to different growth environments and available CO2 concentrations in cells, RuBisCO enzymes purified from different sources have varied carboxylation velocity and CO2 affinity (Fig. 2). For instance, RuBisCO in C3 plants (without carbon-concentrating mechanisms) has relatively higher CO2 affinity and a lower carboxylation velocity compared to RuBisCO in organisms with carbon-concentrating mechanisms (C4 plants, CAM plants, hornworts, green algae and cyanobacteria).50 Since the kinetic parameters are so different, heterologous RuBisCO expression will be a promising strategy to improve RuBisCO performance and to increase the carbon fixation efficiency, for example, introducing a higher CO2 affinity RuBisCO into cyanobacteria. However, RuBisCO folding and assembly are complex processes involving chaperons that vary between species.51,52 Therefore, this strategy is highly restricted.53 In one example, a functional RuBisCO from Synechococcus elongatus PCC 7942 was heterologously expressed in tobacco. Even though the transformed lines could only grow autotrophically at an elevated CO2 concentration (3% CO2), the CO2 fixation rate per RuBisCO protein was significantly increased.54 Later, it was further demonstrated that co-expression of the chaperone RbcX or the carboxysome protein CcmM35 from Synechococcus elongatus PCC 7942 was not essential for a functional cyanobacterial RuBisCO in tobacco.55 This indicates that Synechococcus elongatus PCC 7942 RuBisCO is able to fold and assemble properly with tobacco chaperons. In contrast, RuBisCO from the rhodophyte Galdieria sulphuraria and the diatom Phaeodactylum tricornutum failed to fold and assemble into functional enzymes in tobacco plants.53 This may be mainly due to the incompatibility of RuBisCO with the assembly chaperon RbcX, since RbcX requires specific interactions with its corresponding RuBisCO and acts as a gatekeeper for formation of the holoenzyme. In contrast, folding chaperons are nonspecific and did not show negative effects on mutated RuBisCO.46Synechococcus elongatus PCC 7942 is evolutionarily closer to tobacco than the other two species. This indicates that choosing a more closely related RuBisCO to transfer to host cells may be beneficial. Following this line, heterologous expression of RuBisCO in the same groups e.g. from cyanobacteria to cyanobacteria is a strategy worth trying. Many studies have shown positive effects on target bioproduct formation using this strategy in host cells (Table 2). For example, overexpressing Synechococcus elongatus PCC 6301 RuBisCO in the very closely related strain Synechococcus elongatus PCC 7942 resulted in increased RuBisCO activity and isobutyraldehyde production.56 Heterologous expression may avoid some of the negative regulations of protein expression and/or activity, but at the same time still use the chaperones of the host cells to fold and assemble into a functional enzyme.
| Enzyme | Host | Source | Main effects | Reference |
|---|---|---|---|---|
| RuBisCO | Synechococcus elongatus PCC 7942 | Allochromatium vinosum | 1.5 to 4 fold increase of RuBisCO activity; 1.6 times higher total photosynthesis activity | 77 |
| Synechococcus elongatus PCC 7942 | Synechococcus elongatus PCC 6301 | 1.4 fold increase in total RuBisCO activity; unchanged photosynthetic O2 production; 2 fold higher isobutyraldehyde production | 56 | |
| Synechococcus elongatus PCC 7942 | Synechococcus elongatus 7942 | Unchanged oxygen evolution and free fatty acid production | 78 | |
| Synechococcus PCC 7002 | Synechococcus elongatus PCC 7942 | 3 fold increase of free fatty acid production | 79 | |
| Tobacco, Nicotiana tabacum | Synechococcus elongatus PCC 7942 | Higher rates of CO2 fixation per unit of enzyme, autotrophic growth only rely on the cyanobacterial RuBisCO under 3% CO2 conditions | 54 | |
| Synechococcus elongatus PCC 7942 | Synechococcus PCC 7002 | Positive effects on growth and 2,3-butanediol production using 10 g L−1 glucose in the BG11 medium when cooverexpressed with PRK and OPP enzymes, and the galactose transporter | 80 | |
| Synechococcus elongatus PCC 7942 | Synechococcus elongatus PCC 7942 | No significant effects on growth and 2,3-butanediol production using 10 g L−1 glucose in the BG11 medium | 80 | |
| Synechocystis PCC 6803 | Synechocystis PCC 6803 | Increased RuBisCO content when the FLAG tag or ccmM gene was used; increased growth, biomass accumulation and oxygen evolution under 100 μmol photons m−2 s−1 light intensity; increased ethanol production with air | 81,82 | |
| Synechococcus PCC 7002 | Synechococcus PCC 7002 | No impact on growth and O2 evolution, increased protein content of pyruvate metabolism and fatty acid biosynthesis | 83 | |
| Chloroplastic FBPase | Anabaena PCC 7120 | Wheat | 1.4 fold higher FBPase activity, increased net photosynthesis (117.2%) and true photosynthesis (122.5%), faster growth and more chlorophyll a under atmospheric conditions (360 μmol mol−1 CO2) | 84 |
| Chlamydomonas reinhardtii | Chlamydomonas reinhardtii | 1.4 fold higher FBPase activity and slower photoautotrophic growth with or without elevated CO2 levels | 85 | |
| Form II FBPase | Tobacco, Nicotiana tabacum cv. Xanthi | Synechococcus elongatus PCC 7942 | Transformant line with 2.3 fold higher FBPase activity had higher dry matter under atmospheric conditions, photosynthetic activity at saturated light intensity, RuBP level, in vivo RuBisCO activation state, and hexose, sucrose and starch concentrations in upper and lower leaves | 86 |
| FBP/SBPase | Tobacco | Synechococcus elongatus PCC 7942 | Increased photosynthetic CO2 fixation, photosynthesis, growth, RuBP and GAP concentrations, initial RuBisCO activity (probably due to the higher RuBP concentration) under atmospheric conditions | 87 |
| Euglena gracilis |
Synechococcus elongatus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PCC 7942 PCC 7942 |
Larger cell volume at 100 μmol photons m−2 s−1 and 0.04% CO2, enhanced biomass and photosynthetic activity under high light and high CO2 conditions, increased wax ester production on anaerobiosis | 88 | |
| Soybean, Glycine max | Cyanobacteria | Increased carbon assimilation (4–14%), RuBP regeneration capacity (4–8%), and maximum carboxylation rate of the RuBisCO (5–8%); maintained seed yield at an elevated CO2 level (600 ppm) and higher temperature | 89 | |
| Synechocystis PCC 6803 | Synechocystis PCC 6803, Synechococcus elongatus 7942, and Synechococcus PCC 7002 | Increased growth, biomass accumulation and oxygen evolution at 100 μmol photons m−2 s−1 light intensity; increased ethanol production with air | 72,82 | |
| Synechococcus PCC 7002 | Synechococcus PCC 7002 | Enhanced FBP/SBPase, RuBisCO, and FBA activity, increased cell volume, growth, O2 evolution rate, CO2 assimilation and non-storage carbohydrate accumulation | 83 | |
| SBPase | Tobacco, Nicotiana tabacum | Arabidopsis thaliana | Higher photosynthetic capacity, RuBP regeneration capacity, sucrose and starch accumulation, leaf area at the 4–5 leaf stage, and carbon fixation (6–12%) | 90 |
| Tobacco, Nicotiana tabacum cv. Xanthi | Chlamydomonas | Increased SBPase activity (1.6 or 4.3 fold), dry matter, photosynthetic CO2 fixation, growth rate, RuBP contents and RuBisCO activation state | 86 | |
| Rice, Oryza sativa L. | Rice, Oryza sativa L. | Increased CO2 assimilation at 30 °C and maintained CO2 assimilation above 30 °C | 91 | |
| Tobacco, Nicotiana tabacum | Arabidopsis thaliana | Enhanced carbon assimilation and RuBP regeneration capacity under elevated CO2 conditions (585 ppm) | 92 | |
| Arabidopsis thaliana | Arabidopsis thaliana | Increased SBPase activity, leaf area, and photosynthetic carbon fixation rate | 93 | |
| FBA | Tobacco, Nicotiana tabacum | Arabidopsis thaliana | Enhanced growth and increased biomass especially at a high CO2 concentration | 94 |
| Arabidopsis thaliana | Arabidopsis thaliana | Increased FBA and FBA activity, photosynthetic CO2 assimilation rate and more seeds and aerial biomass | 95 | |
| Tobacco, Nicotiana tabacum | Arabidopsis thaliana | Enhanced CO2 assimilation rate, electron transporting rate, photosynthesis and leaf area (even combined with SBPase overexpression) | 96 | |
| Arabidopsis thaliana | Arabidopsis thaliana | Increased FBA activity, PSII efficiency, maximum CO2 fixation efficiency, and leaf area | 93 | |
| Chlorella vulgaris | Synechocystis PCC 6803 | FBA activity increased about 1.3 fold; increased CO2 fixation rate, growth rate and biomass accumulation | 97 | |
| Synechocystis PCC 6803 | Synechocystis PCC 6803 | Increased growth, biomass accumulation and oxygen evolution at 100 μmol photons m−2 s−1 light intensity; increased ethanol production with air | 72,82 | |
| TK | Tobacco, Nicotiana tabacum | Arabidopsis thaliana | Increased TK activity, decreased thiamine levels and leaf area | 98 |
| Synechocystis PCC 6803 | Synechocystis PCC 6803 | Increased biomass accumulation at 15 and 100 μmol photons m−2 s−1 light intensity; increased ethanol production with air | 72,82 | |
| RuBisCO (the small subunit) and TK | Rice, Oryza sativa | Rice, Oryza sativa | Increased RuBisCO and TK amounts,; similar CO2 assimilation rate to the wild type | 99 |
In cyanobacteria, carbon-concentrating mechanisms include the inorganic carbon transporters (of CO2 and HCO3−) and the special compartment named carboxysome. There are five inorganic carbon transporters in cyanobacteria; BCT1, SbtA, BicA, NDH-I4 and NDH-I3. The five transporters are different in expression, substrate affinity, and location57 (Table 3). Among the five inorganic transporters, native BicA has been overexpressed in Synechocystis PCC 6803. It was found that in the engineered strains, the growth rate was two times faster than that of the wild-type strain, and a higher amount of biomass was accumulated.58 When supplied with a higher CO2 concentration (0.5%), growth of the BicA-overexpressing strains was further enhanced. This means that under atmospheric CO2 conditions, the growth was still restricted by the carbon concentration even though BicA was overexpressed. This may be due to the low CO2 affinity of BicA.58 Therefore, overexpressing higher CO2 affinity transporters may be a better way to increase growth under ambient CO2 conditions. However, each of the other four transporters is encoded by a long operon consisting of several genes, increasing the cloning difficulties. Another promising strategy is to change low Ci induced transporters, like SbtA, BCT1 and NDH-I3, into constitutively expressed transporters. This does not only involve controlling the transcription of the corresponding genes, but may also be related to translation and/or post-translation modifications, depending on the induction mechanisms.
| Transporter | Category | Expression | Features | Location |
|---|---|---|---|---|
| BCT1 | Traffic ATPase, HCO3− | Induced under Ci limited conditions, enhanced transcription under high light | High affinity (5 μM Synechococcus elongatus PCC 7942 BCT1); medium to low flux rate | Plasma membrane |
| SbtA | Na+-dependent HCO3− transporter | Induced under Ci limited conditions | High affinity (2 μM Synechococcus PCC 7002 SbtA); low flux rate | Plasma membrane |
| BicA | Na+-dependent HCO3-transporter | Induced under Ci limited conditions in Synechococcus PCC 7002; constitutively expressed in Synechocystis PCC 6803 | Low affinity (≈38 μM of Synechococcus PCC 7002 BicA); fast flux rate | Plasma membrane |
| NDH-I4 | CO2 uptake | Constitutively expressed | Relatively high affinity (≈10 μM) | Plasma membrane |
| NDH-I3 | CO2 uptake | Induced under Ci limited conditions | High affinity (1–2 μM) | Thylakoid membrane |
In cyanobacteria, another crucial parameter in the carbon-concentrating mechanism is the special micro-compartment called carboxysome.59 Carboxysomes encapsulate RuBisCO and carbonic anhydrase. Carbonic anhydrase converts bicarbonate into CO2, leading to an order of magnitude enhanced CO2 concentration near RuBisCO. Since C3 plants do not have similar compartments, transplanting carboxysomes or compartments with similar functions into C3 plants is regarded as one potential way to increase carbon fixation and plant productivity. Cyanobacteria have two types of carboxysomes, α and β carboxysomes, encapsulating form 1A and 1B RuBisCOs, respectively. Since plants have form 1B RuBisCO, it would make sense to attempt transplanting β carboxysomes into plants. Even though there are reports of successfully assembling carboxysomes in E. coli cells,60 doing the same thing in plant chloroplasts is still challenging given the higher complexity of an eukaryotic cell. The assembly process of β carboxysomes in Synechococcus elongatus PCC 7942 has been elucidated, increasing the chance to engineer a functional carboxysome in foreign cells.61 Functional carboxysomes with truncated proteins have also been reported.62 This work benefits carboxysome transplanting, since shorter and/or fewer genes are required for a functional carboxysome in this case. A smaller carboxysome will also conserve carbon, nitrogen and other resources for other usages in the cell. Carbonic anhydrases also localize differently in plants and cyanobacteria, in the cytosol in plants and in the carboxysome in cyanobacteria. Therefore, how to direct a cytosolic carbonic anhydrase into a carboxysome is another issue. One possible strategy is to inactivate or delete the native cytosolic carbonic anhydrase and at the same time express a carboxysome carbonic anhydrase together with the carboxysome.
2.2 3-Phosphoglycerate (PGA) reduction
Two enzymes are involved in the process of PGA reduction, a phosphoglycerate kinase (PGK, EC 2.7.2.3) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, EC 1.2.1.13). In the CBB cycle, NADPH and two thirds of the ATP used in the CBB cycle are consumed in these two steps. The product of this process, glyceraldehyde-3-phosphate (GAP), is one of the main outputs of the CBB cycle. In the cyanobacterial model strain Synechocystis PCC 6803, there are two types of GAPDH, GAPDH1 and GAPDH2, encoded by gap1 and gap2, respectively.66 GAPDH2 catalyses the reaction in the CBB cycle and is redox regulated through associating and dissociating with the small redox-sensitive protein CP12 and phosphoribulokinase (PRK, EC 2.7.1.19).67 The three proteins form a complex when the NAD(P)H level is low (for example in darkness).67 Formation of the complex is essential, specifically in mixotrophic growth or light/dark cycles due to the competition between the CBB cycle and OPP pathway. Ribulose-5-phosphate is an intermediate in both the CBB cycle (substrate of PRK) and the OPP pathway (substrate of ribulose-5-phosphate epimerase/isomerase). This means that PRK competes for ribulose-5-phosphate with ribulose-5-phosphate epimerase/isomerase. In darkness, cyanobacteria rely on the OPP pathway to generate reducing equivalents (mainly NADPH). If the OPP pathway is inhibited in darkness (by PRK assimilating ribulose-5-phosphate in this case), there will not be enough reducing equivalents and cell growth will be hampered. However, it is not the same situation when cells are grown in continuous light, since the photochemical phases will supply cells with enough NADPH. Therefore, the role of GAPDH should attract attention, especially when cells are exposed to light/dark cycles. However, in anti-sense tobacco plants with reduced levels of chloroplast GAPDH, carbon assimilation under ambient conditions was not inhibited unless less than 30–40% of the wild-type GAPDH activity was left.35 This means that GAPDH does not have a significant flux control co-efficient for the CBB cycle.2.3 Ribulose-1,5-biphosphate (RuBP) regeneration
As discussed above, there are 11 enzymes in the CBB cycle and most of them are in the stage called “RuBP regeneration”. This stage is crucial to maintain the carbon fixation rate especially under relatively low light conditions. This concept was first suggested in the 1980s, from a CBB cycle model of C3 plants.68 In this stage, the last reaction, catalysed by PRK, consumes the remaining one-third of the ATP used in the whole CBB cycle. Interestingly, reducing the expression levels of PRK in anti-sense tobacco plants did not affect carbon assimilation and growth until the activity of PRK was decreased by more than 85%32 even though it catalyses an irreversible reaction. This indicates only minor control on the carbon flux by PRK.Three enzymes which have been reported to play important roles in controlling the CBB cycle flux are aldolase, fructose-1,6/sedoheptulose-1,7-biphosphatase and transketolase.69,70 Aldolase (FBA, EC 4.1.2.13) catalyses the aldol condensation of dihydroxyacetone phosphate (DHAP) and GAP to fructose-1,6-bisphosphate (FBP) or DHAP and erythrose-4-phosphate (E4P) into sedoheptulose-1,7-bisphosphate (SBP) in the CBB cycle. Since SBP is not an OPP pathway intermediate, one can say that FBA catalyses a reversible reaction between FBP and DHAP and GAP and an irreversible reaction between SBP and DHAP and E4P. There are two classes (I and II) of FBA, defined based on the catalytic mechanisms and distributions in the biosphere. Their respective genes do not show DNA sequence homology, which implies separate evolution paths. Class II FBA catalyses reactions in the CBB cycle in both Synechocystis PCC 6803 and plants. The function of class II FBA cannot be replaced by class I FBA in Synechocystis PCC 6803 under autotrophic growth conditions, indicating that class I FBA is not able to catalyse the reactions in the CBB cycle.71
Early studies of FBA in controlling photosynthesis and growth of potato plants showed that reduction of plastid FBA activity by more than 30% inhibits photosynthesis under 400 μmol photons m−2 s−1 irradiance and ambient carbon conditions.37,38 The slow regeneration of RuBP is responsible for the phenomenon. Activities of plastid FBPase and PRK are also reduced in FBA antisense potato lines. These results indicate that even though FBA catalyses reversible reactions, it has significant control over the CBB cycle carbon flux. This is further confirmed in other plants, microalgae and cyanobacteria (Table 2). When class II FBA is overexpressed, growth, biomass accumulation, and/or CO2 assimilation are increased under certain growth conditions. Recently, a report showed that overexpressing FBA in engineered Synechocystis PCC 6803 increases the production of ethanol.72
The next metabolic steps in the CBB cycle after FBA are catalyzed by plastid FBPase (EC 3.1.3.12), dephosphorylating FBP into fructose-6-phosphate (F6P) and SBPase (EC 3.1.3.37), and dephosphorylating SBP into sedoheptulose-7-phosphate (S7P) in plants. Plastid FBPase and SBPase have very different evolutionary origins. Plastid FBPase originated from bacteria and was transferred into plant chloroplasts by endosymbiosis, while SBPase originated from archaea.11 Regardless of the different evolution paths, the two enzymes share high structural homology, with differences only on the solvent-exposed surface area. Plant SBPase and most plastid FBPases, except some later emerging plastid FBPases in land plants,73 contain the redox regulation target, cysteine.74 The major redox regulator is thioredoxin f, which is reduced by ferredoxin. Therefore, the activities of FBPase and SBPase are regulated by light. They are also inhibited by glycerate.75 These regulatory mechanisms allow dedicated regulation of plastid FBPase and SBPase. In cyanobacteria, the bi-functional enzyme FBP/SBPase plays the same role as plastid FBPase and SBPase. However, cyanobacterial FBP/SBPase does not share amino acid homology with plant plastid FBPase and SBPase. Therefore, existence and mechanisms of the redox-regulations, if there are any, of cyanobacterial FBP/SBPase are unclear.
Products of FBPase and SBPase are points where carbon leaves the CBB cycle in the form of carbohydrate. This makes these enzymes important in keeping the input and output of the CBB cycle balanced. A C3 plant model shows that the plastid FBPase and SBPase contents are very low compared to levels of other CBB cycle enzymes.68 Concentrations of most of the CBB cycle enzymes are higher than those of their corresponding substrates, while FBPase and SBPase are present in similar concentrations to their substrates in Chlamydomonas reinhardtii.76 It has been shown that dramatic reduction of the levels of plastid FBPase in potato (less than 15% activity remained) and SBPase in tobacco (less than 20% activity remained) have negative effects on plant growth.25,31
To summarize, plastid FBPase and SBPase are redox regulated, catalyse irreversible reactions, and occur at low levels compared to the other CBB cycle enzymes, and these reduced levels result in growth deficiencies. Therefore, plastid FBPase and SBPase may be crucial enzymes in controlling the carbon flux through the CBB cycle, maybe by affecting the RuBP regeneration. Furthermore, there are plenty of experimental reports where increasing FBPase and/or SBPase through genetic engineering in plants, algae or cyanobacteria led to enhanced growth and/or photosynthesis (Table 2). In Synechocystis PCC 6803, increasing the content of FBP/SBPase, either a native or a non-native version, resulted in increased ethanol production in the engineered strains (Table 2).
The next step in regeneration of RuBP is catalysed by transketolase (TK, EC 2.2.1.1). In general, reactions catalysed by transketolase are reversible. It uses GAP and S7P to produce xylulose-5-phosphate (Xu5P) and ribose-5-phosphate (R5P), or GAP and F6P to produce Xu5P and erythrose 4-phosphate (E4P). Activation of TK requires the cofactor thiamine diphosphate (TPP), the active form of thiamine (vitamin B1). The synthesis of TPP in turn requires GAP and R5P as substrates. Therefore, there may be some circuit regulation on TK activity and TPP levels. E4P is also one of the precursors of the shikimic acid pathway, which is one of the connections between central and secondary carbon metabolisms. Thus, TK is another branch point where fixed carbon leaves the CBB cycle. The indirect regulation by its own product (R5P) and substrate (GAP) and the role in connecting the secondary metabolism to the central carbon metabolism make TK one of the key enzymes in controlling the carbon flux through the CBB cycle. In tobacco anti-sense plants, a 20% reduction of TK activity resulted in decreased photosynthesis and growth. Formation of aromatic amino acids and the phenylpropanoid metabolism pathway are also inhibited.36 Under saturated light (700 μmol photons m−2 s−1) and CO2 (5000 ppm) conditions, it was found that TK had full control over the maximum photosynthesis rate, meaning that photosynthesis was linear with TK activity under these conditions. In contrast, RuBisCO does not exert any control over the carbon flux under these conditions. Even though it is unexpected that TK would have a strong control over the CBB cycle carbon flux since it catalyses reversible reactions and is regarded as a non-finely regulated enzyme, positive effects on carbohydrate accumulation, leaf size or end product yield are observed by overexpressing this enzyme (Table 2).
3. Alternative carbon fixation cycles
Organisms that utilize CO2 (or HCO3−) as the sole carbon source are defined as autotrophic organisms. The energy source of autotrophic organisms can be solar energy (like cyanobacteria and plants, photoautotrophs) or chemical energy (chemolithoautotrophs). The CBB cycle is found in most photoautotrophs. Since efforts at optimizing RuBisCO performance and improving the CBB cycle efficiency have shown only limited success, interest has been redirected towards other carbon fixation cycles and carboxylases. Alternative carbon fixation cycles found in archaea include the reductive acetyl-CoA cycle, the reductive TCA (rTCA) cycle, the 3-hydroxypropionate (3-HP) cycle, and the 4-hydroxybutyrate cycles (3-hydroxypropionate/4-hydroxybutyrate, 3-HP/4-HB cycle, and dicarboxylate–4-hydroxybutyrate, DC/4-HB cycle100) (Table 4). In some archaea, there is form III RuBisCO. However, form III RuBisCO does not support autotrophic growth and no complete CBB cycle has been found in these archaea.| Pathway | Electron donor | Carbon fixation enzyme(s) | Oxygen sensitivity |
|---|---|---|---|
| CBB cycle | H2O | RuBisCO | − |
| Reductive acetyl-CoA pathway | H2 | Formate dehydrogenase | + |
| CO dehydrogenase/acetyl-CoA synthase | |||
| Reductive TCA cycle | H2S | Isocitrate dehydrogenase | − |
| α-Ketoglutarate:ferredoxin oxidoreductase | + | ||
| Pyruvate:ferredoxin oxidoreductase | + | ||
| Phosphoenolpyruvate carboxylase | − | ||
| 3-HP bicycle | S2−, H2 | Acetyl-CoA/propionyl-CoA carboxylase | − |
| 3-HP/4-HB cycle | FeS2 | Acetyl-CoA/propionyl-CoA carboxylase | − |
| DC/4-HB cycle | H2 | Pyruvate:ferredoxin oxidoreductase | + |
| Phosphoenolpyruvate carboxylase | − |
One standard to evaluate carbon fixation cycles is the carbon fixation efficiency, which is mainly estimated from reaction kinetics and ATP requirements. ATP requirements in a carbon fixation cycle are largely affected by the carbon fixation enzyme(s).101 Some carboxylases directly or indirectly catalyse the assimilation of CO2 associated with ATP hydrolysis. For example, in the CBB cycle, the CO2 assimilation using RuBisCO is associated with ATP usage in the step catalysed by phosphoglycerate kinase. In contrast, α-ketoglutarate:ferredoxin oxidoreductase in the reductive TCA cycle assimilates CO2 associated with reduction of ferredoxin. This will reduce the overall ATP requirement in the cycle. Therefore, to synthesize one molecule of acetyl-CoA, 7 versus 2 ATPs are required in the CBB cycle and the reductive TCA cycle.102 Among all the native cycles, the reductive acetyl-CoA pathway is special in respect to the carbon fixation reactions. In the reductive acetyl-CoA pathway, five sixths of the carbon is assimilated by formate dehydrogenase and CO dehydrogenase/acetyl-CoA synthase. Therefore, the carbon assimilation in this cycle is more reduction dependent, which makes this cycle more energy efficient, and it is predicted to have a higher biomass yield.103 In general, the ATP efficient cycles are naturally found under anaerobic conditions while aerobic archaea usually have the ATP inefficient cycles. This makes it challenging to transplant ATP efficient cycles into aerobic organisms, such as cyanobacteria or E. coli. Of course, intermediate compatibility is another challenge, since foreign intermediates due to non-native gene expression may interfere with the native cell metabolism.
Recently, another potential CO2 assimilation cycle was identified in the dissimilatory phosphite oxidation (DPO) bacterium Candidatus phosphitivorax.104 In this process, phosphite is oxidized, thereby providing reducing equivalents, and NADP-dependent formate dehydrogenase assimilates CO2 into the formate. Afterwards, the formate is converted into pyruvate through the reductive glycine pathway.105 Since phosphite oxidation to phosphate is an efficient electron providing process, this process may drive cell growth under energy limited conditions. The carbon fixation during this process is mainly reduction dependent, which means it is more ATP efficient than the CBB cycle. If an oxygen insensitive formate dehydrogenase was applied, the reductive glycine pathway will be oxygen tolerant. This makes it promising to be transplanted into aerobic organisms.
Instead of understanding, transplanting and engineering native carbon fixation cycles, assembling an artificial carbon fixation cycle with potentially more efficient carbon fixation enzymes is another attractive strategy to improve the carbon fixation efficiency. Bar-Even et al. examined 5000 native existing enzymes for alternative synthetic carbon fixation cycles.101 Focusing on the cycle kinetics, thermodynamic feasibility, oxygen tolerance, and high energy efficiency (the amount of NADPH equivalents and ATP equivalents to produce one molecule of the product), they modelled a pathway using PEPc to fix CO2, generating glyoxylate, which was named the malonyl-CoA-oxaloacetate-glyoxylate (MOG) pathway. The starting reactions of this pathway overlap with the C4 carbon fixation pathway in the plant mesophyll cell, but the MOG pathway does include the reactions in the bundle-sheath cells, therefore avoiding the usage of RuBisCO. Computational modelling indicated that this pathway is potentially 2- to 3-fold faster at fixing carbon than the CBB cycle.101 Although the whole cycle has not been tested experimentally yet, it has been reported that overexpressing PEPc in Synechocystis PCC 6803 enhanced cell growth under low light conditions.106
There is another synthetic carbon fixation cycle, crotonyl-coenzyme A (CoA)/ethylmalonyl-CoA/hydroxybutyryl-CoA (CETCH) cycle, which has been examined in vitro.107 The CETCH cycle uses enoyl-CoA carboxylases/reductases, which are not present in autotrophic CO2 fixing organisms, to carboxylate CO2. After enzyme modification or replacement, a well functional version of this cycle, CETCH 5.4, was generated and tested in vitro. This cycle includes 13 enzymes in the carbon fixation cycle and 5 enzymes for auxiliary proofreading (converting the CO2 fixation product glyoxylate into malate) and cofactor regeneration. CETCH 5.4, in which two CO2 molecules were fixed into glyoxylate at the expense of two ATPs and three NADPH, is more energy-efficient than the CBB cycle.107
Both the MOG cycle and the CETCH 5.4 are promising CBB cycle alternatives. However, introducing synthetic pathways into living organisms is challenging because of the unpredictable interference between the artificial metabolism and the background native metabolism of the host. Testing the function of the synthetic pathways in vitro while considering and mimicking in vivo conditions can provide useful guides on how to introduce complicated synthetic pathways into living organisms. Even though there are no reports about introducing new complete CO2 fixation pathways into photoautotrophic organisms, there is one example of an artificial pathway to compensate the CO2 loss when pyruvate is converted into acetyl-CoA in Synechococcus elongates PCC 7942.108
4. Further strategies to improve the photosynthetic efficiency
One of the main reasons that oxygenic photoautotrophic organisms (plants, cyanobacteria and algae) are promising cell factories is that they can directly convert solar energy into chemical energy. Solar energy reaching the earth in one hour is enough for global human activities in one year. If sustainable methods are developed to capture, convert and store solar energy, it will be the infinite fuel for the future. Solar panels are a well-developed technology in capturing and converting solar energy into electricity. However, they do not capture CO2. Photoautotrophic organisms like cyanobacteria and algae are more sustainable in this aspect. However, the solar energy conversion efficiency of even the most efficient photoautotrophic organisms is much lower than that of current commercial solar cells (4.6–6% versus 18%).102In photoautotrophic organisms, the antenna complexes absorb and transfer solar energy to the photochemical reaction centres, photosystems II and I. During this process, excess absorbed solar energy is dissipated as heat. This often happens in the leaves on the top of the canopies and cells in the surface facing the light source of the algal and cyanobacterial cultures, especially under high light conditions. At the same time, leaves at the bottom of the canopies and cells further from the surface of algal and cyanobacterial cultures will not get enough light. As a result, the energy conversion efficiency of the entire population is low. Therefore, a strategy, using “truncated light-harvesting antenna” (TLA) strains, was developed and used in plants, algae and cyanobacteria to increase the total solar energy conversion efficiency.109–111 The strategy is to decrease the antenna size to prevent the top leaves or cells from absorbing excess solar energy. There will be two positive outcomes, one is to reduce light inhibition of the top leaves or surface cells, and the other is to increase light penetration thereby allowing the lower leaves and cells to receive more solar energy. Therefore, the overall solar energy conversion efficiency and production are enhanced. However, it has been reported that the TLA did not enhance Synechocystis PCC 6803 productivity under low light (<200 μmol photons m−2 s−1) and did not enhance cell growth in the early stage (OD735 < 1).112 In contrast, a strain of Chlamydomonas reinhardtii with a truncated antenna showed enhanced oxygen evolution and a higher light saturation point, and strongly enhanced biomass productivities under greenhouse conditions with about 1500 μmol photons m−2 s−1.109 Enhanced growth was also reported in Synechocystis PCC 6803 when 200 μmol photons m−2 s−1 light intensity was applied.113 Besides, the truncated antenna benefits cell growth and biomass accumulation in Synechocystis PCC 6803 only under ambient CO2 and high light (1000 μmol photons m−2 s−1) conditions.114 Furthermore, another interesting strategy to improve light utilization is to introduce pigments that allow cells to absorb light of longer, red-shifted wavelengths.115
In order to convert the output compound(s) of carbon fixation cycles, like PGA in the CBB cycle, to compounds interesting for production, further enzymatic steps are required. Traditional engineering methods, such as introducing genes with known functions from another organism, and new synthetic biology methods like assembling new pathways116 and computational modelling41,117 are available to optimize this process. In addition to genetic engineering strategies, growth conditions, like nitrogen starvation, medium pH adjustment, and salt stress, may also have major effects on product yields. Continuously extracting the desired product out of the cells during cultivation may also benefit photosynthesis or/and carbon fixation.6,7
5. Conclusions
As the only prokaryotic oxygenic photoautotrophic organisms, cyanobacteria are promising green cell factories to produce sustainable bioproducts. During the whole process, from carbon fixation to extraction of products, optimization at every step may contribute to enhancing growth and the product yield (Fig. 3).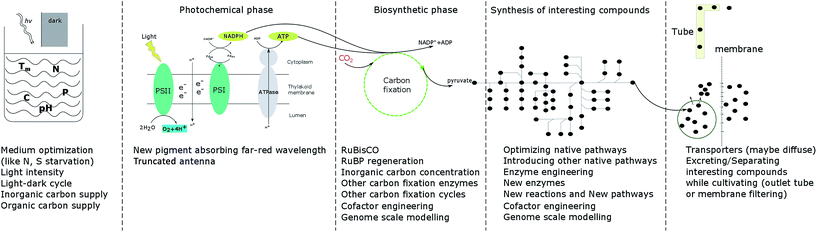 | ||
| Fig. 3 Summary of strategies to enhance cell growth and/or product yields in photoautotrophic organisms. | ||
In the photochemical phase, RuBisCO, FBA, FBP/SBPase and TK are four enzymes investigated in cyanobacteria, algae and plants as targets of engineering to enhance photosynthesis, growth and/or product formation. Since RuBisCO has a low efficiency and since protein engineering has not resulted in significant improvement in RuBisCO performance, increasing the CO2 concentration near RuBisCO is an alternative strategy. Research is also directed towards finding more efficient carbon fixation cycles, natural or artificial, which could be implemented in engineered cells. With the development of new technologies, like cyanobacterial computational modelling and gene editing technologies, scientists have a better and better understanding of the biosynthetic phase, which will give clues to future studies.
Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by the NordForsk NCoE program “NordAqua” (project # 82845).References
- C. W. Mullineaux, Co-existence of photosynthetic and respiratory activities in cyanobacterial thylakoid membranes, Biochim. Biophys. Acta, Bioenerg., 2014, 1837(4), 503–511 CrossRef CAS PubMed.
- D. J. Lea-Smith, P. Bombelli, R. Vasudevan and C. J. Howe, Photosynthetic, respiratory and extracellular electron transport pathways in cyanobacteria, Biochim. Biophys. Acta, Bioenerg., 2016, 1857(3), 247–255 CrossRef CAS PubMed.
- M. R. L. V. Leal, A. S. Walter and J. E. A. Seabra, Sugarcane as an energy source, Biomass Convers. Biorefin., 2013, 3(1), 17–26 CrossRef CAS.
- D. Tilman, R. Socolow, J. A. Foley, J. Hill, E. Larson and L. Lynd, et al., Beneficial Biofuels—The Food, Energy, and Environment Trilemma, Science, 2009, 325(5938), 270–271 CrossRef CAS PubMed.
- R. H. Wijffels and M. J. Barbosa, An Outlook on Microalgal Biofuels, Science, 2010, 329(5993), 796–799 CrossRef CAS PubMed.
- W. Du, F. Liang, Y. Duan, X. Tan and X. Lu, Exploring the photosynthetic production capacity of sucrose by cyanobacteria, Metab. Eng., 2013, 19, 17–25 CrossRef CAS PubMed.
- B. W. Abramson, B. Kachel, D. M. Kramer and D. C. Ducat, Increased photochemical efficiency in cyanobacteria via an engineered sucrose sink, Plant Cell Physiol., 2016, 57(12), 2451–2460 CrossRef CAS PubMed.
- D. C. Ducat, J. C. Way and P. A. Silver, Engineering cyanobacteria to generate high-value products, Trends Biotechnol., 2011, 29(2), 95–103 CrossRef CAS PubMed.
- N. E. Nozzi, J. W. Oliver and S. Atsumi, Cyanobacteria as a Platform for Biofuel Production, Front. Bioeng. Biotechnol., 2013, 1, 7 Search PubMed.
- V. Singh, D. K. Chaudhary, I. Mani and P. K. Dhar, Recent advances and challenges of the use of cyanobacteria towards the production of biofuels, Renewable Sustainable Energy Rev., 2016, 60, 1–10 CrossRef.
- D. D. Gütle, T. Roret, S. J. Müller, J. Couturier, S. D. Lemaire and A. Hecker, et al., Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(24), 6779–6784 CrossRef PubMed.
- N. Antonovsky, S. Gleizer and R. Milo, Engineering carbon fixation in E. coli: from heterologous RuBisCO expression to the Calvin–Benson–Bassham cycle, Curr. Opin. Biotechnol., 2017, 47, 83–91 CrossRef CAS PubMed.
- D. Fell, Understanding the control of metabolism, Front. Metab., 1997, 2, 300 Search PubMed.
- A. Miller, C. Schlagnhaufer, M. Spalding and S. Rodermel, Carbohydrate regulation of leaf development: prolongation of leaf senescence in Rubisco antisense mutants of tobacco, Photosynth. Res., 2000 Jan, 63(1), 1–8 Search PubMed.
- P. Matt, A. Krapp, V. Haake, H. Mock and M. Stitt, Decreased Rubisco activity leads to dramatic changes of nitrate metabolism, amino acid metabolism and the levels of phenylpropanoids and nicotine in tobacco antisense RBCS transformants, Plant J., 2002, 30(6), 663–677 CrossRef CAS PubMed.
- M. Stitt and D. Schulze, Does Rubisco control the rate of photosynthesis and plant growth? An exercise in molecular ecophysiology, Plant, Cell Environ., 1994, 17(5), 465–487 CrossRef CAS.
- W. P. Quick, U. Schurr, K. Fichtner, E. -D. Schulze, S. R. Rodermel and L. Bogorad, et al., The impact of decreased Rubisco on photosynthesis, growth, allocation and storage in tobacco plants which have been transformed with antisense rbcS, Plant J., 1991, 1(1), 51–58 CrossRef CAS.
- A. Makino, T. Shimada, S. Takumi, K. Kaneko, M. Matsuoka and K. Shimamoto, et al., Does Decrease in Ribulose-1,5-Bisphosphate Carboxylase by Antisense RbcS Lead to a Higher N-Use Efficiency of Photosynthesis under Conditions of Saturating CO2 and Light in Rice Plants?, Plant Physiol., 1997, 114(2), 483–491 CrossRef CAS PubMed.
- A. Makino and R. F. Sage, Temperature response of photosynthesis in transgenic rice transformed with “sense” or “antisense” rbcS, Plant Cell Physiol., 2007, 48(10), 1472–1483 CrossRef CAS PubMed.
- R. T. Furbank, J. A. Chitty, S. Von Caemmerer and C. Jenkins, Antisense RNA inhibition of RbcS gene expression reduces Rubisco level and photosynthesis in the C4 plant Flaveria bidentis, Plant Physiol., 1996, 111(3), 725–734 CrossRef CAS PubMed.
- G. S. Hudson, J. R. Evans, S. von Caemmerer, Y. B. Arvidsson and T. J. Andrews, Reduction of ribulose-1,5-bisphosphate carboxylase/oxygenase content by antisense RNA reduces photosynthesis in transgenic tobacco plants, Plant Physiol., 1992, 98(1), 294–302 CrossRef CAS PubMed.
- N. A. Eckardt, G. W. Snyder, A. R. Portis Jr and W. L. Ogren, Growth and Photosynthesis under High and Low Irradiance of Arabidopsis thaliana Antisense Mutants with Reduced Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase Activase Content, Plant Physiol., 1997, 113(2), 575–586 CrossRef CAS PubMed.
- S. Von Caemmerer, A. Millgate, G. D. Farquhar and R. T. Furbank, Reduction of Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase by Antisense RNA in the C4 Plant Flaveria bidentis Leads to Reduced Assimilation Rates and Increased Carbon Isotope Discrimination, Plant Physiol., 1997, 113(2), 469–477 CrossRef CAS PubMed.
- K. Siebke, S. von Caemmerer, M. Badger and R. T. Furbank, Expressing an RbcS Antisense Gene in Transgenic Flaveria bidentis Leads to an Increased Quantum Requirement for CO2 Fixed in Photosystems I and II, Plant Physiol., 1997, 115(3), 1163–1174 CrossRef CAS PubMed.
- E. P. Harrison, N. M. Willingham, J. C. Lloyd and C. A. Raines, Reduced sedoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate accumulation, Planta, 1998, 204(1), 27–36 CrossRef CAS.
- C. A. Raines, E. P. Harrison and J. C. Lloyd, Minireview: investigating the role of the thiol-regulated enzyme sedoheptulose-1,7-bisphosphatase in the control of photosynthesis, Physiol. Plant., 2000, 110(3), 303–308 CrossRef CAS.
- T. Lawson, B. Bryant, S. Lefebvre, J. C. Lloyd and C. A. Raines, Decreased SBPase activity alters growth and development in transgenic tobacco plants, Plant, Cell Environ., 2006, 29(1), 48–58 CrossRef CAS.
- H. Olçer, J. C. Lloyd and C. A. Raines, Photosynthetic capacity is differentially affected by reductions in sedoheptulose-1,7-bisphosphatase activity during leaf development in transgenic tobacco plants, Plant Physiol., 2001, 125, 982–989 CrossRef.
- E. P. Harrison, Small decreases in SBPase cause a linear decline in the apparent RuBP regeneration rate, but do not affect Rubisco carboxylation capacity, J. Exp. Bot., 2001, 52(362), 1779–1784 CrossRef CAS PubMed.
- L. Feng, H. Li, J. Jiao, D. Li, L. Zhou and J. Wan, et al., Reduction in SBPase activity by antisense RNA in transgenic rice plants: effect on photosynthesis, growth, and biomass allocation at different nitrogen levels, J. Plant Biol., 2009, 52(5), 382–394 CrossRef.
- J. Kobmann, U. Sonnewald and L. Willmitzer, Reduction of the chloroplastic fructose-l,6-bisphosphatase in transgenic potato plants impairs photosynthesis and plant growth, Plant J., 1994, 6(5), 637–650 CrossRef.
- M. J. Paul, J. S. Knight, D. Habash, M. A. J. Parry, D. W. Lawlor and S. A. Barnes, et al., Reduction in phosphoribulokinase activity by antisense RNA in transgenic tobacco: effect on CO2 assimilation and growth in low irradiance, Plant J., 1995, 7(4), 535–542 CrossRef CAS.
- F. Banks, S. Driscoll, M. Parry, D. Lawlor, J. Knight and J. Gray, et al., Decrease in phosphoribulokinase activity by antisense RNA in transgenic tobacco. Relationship between photosynthesis, growth, and allocation at different nitrogen levels, Plant Physiol., 1999, 119(3), 1125–1136 CrossRef CAS PubMed.
- M. J. Paul, S. P. Driscoll, P. J. Andralojc, J. S. Knight, J. C. Gray and D. W. Lawlor, Decrease of phosphoribulokinase activity by antisense RNA in transgenic tobacco: definition of the light environment under which phosphoribulokinase is not in large excess, Planta, 2000, 211(1), 112–119 CrossRef CAS PubMed.
- G. D. Price, J. R. Evans, S. von Caemmerer, J. W. Yu and M. R. Badger, Specific reduction of chloroplast glyceraldehyde-3-phosphate dehydrogenase activity by antisense RNA reduces CO2 assimilation via a reduction in ribulose bisphosphate regeneration in transgenic tobacco plants, Planta, 1995, 195(3), 369–378 CrossRef CAS PubMed.
- S. Henkes, U. Sonnewald, R. Badur, R. Flachmann and M. Stitt, A Small Decrease of Plastid Transketolase Activity in Antisense Tobacco Transformants Has Dramatic Effects on Photosynthesis and Phenylpropanoid Metabolism, Plant Cell, 2001, 13, 535–551 CrossRef CAS.
- V. Haake, M. Geiger, P. Walch-Liu, C. Engels, R. Zrenner and M. Stitt, Changes in aldolase activity in wild-type potato plants are important for acclimation to growth irradiance and carbon dioxide concentration, because plastid aldolase exerts control over the ambient rate of photosynthesis across a range of growth condition, Plant J., 1999, 17(5), 479–489 CrossRef CAS.
- V. Haake, R. Zrenner, U. Sonnewald and M. Stitt, A moderate decrease of plastid aldolase activity inhibits photosynthesis, alters the levels of sugars and starch, and inhibits growth of potato plants, Plant J., 1998, 14(2), 147–157 CrossRef CAS PubMed.
- M. Eisenhut, W. Ruth, M. Haimovich, H. Bauwe, A. Kaplan and M. Hagemann, The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(44), 17199–17204 CrossRef CAS PubMed.
- D. M. Kramer and J. R. Evans, The Importance of Energy Balance in Improving Photosynthetic Productivity, Plant Physiol., 2011, 110, 166652 Search PubMed.
- H. Knoop, M. Gründel, Y. Zilliges, R. Lehmann, S. Hoffmann and W. Lockau, et al., Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp, PCC 6803, PLoS Comput. Biol., 2013, 9(6), e1003081 CrossRef CAS PubMed.
- Y. Savir, E. Noor, R. Milo and T. Tlusty, Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(8), 3475–3480 CrossRef CAS PubMed.
- G. G. B. Tcherkez, G. D. Farquhar and T. J. Andrews, Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(19), 7246–7251 CrossRef CAS PubMed.
- R. A. Studer, P.-A. Christin, M. A. Williams and C. A. Orengo, Stability-activity tradeoffs constrain the adaptive evolution of RubisCO, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(6), 2223–2228 CrossRef CAS PubMed.
- P. L. Cummins, B. Kannappan and J. E. Gready, Directions for Optimization of Photosynthetic Carbon Fixation: RuBisCO's Efficiency May Not Be So Constrained After All, Front. Plant. Sci., 2018, 9 Search PubMed.
- P. Durão, H. Aigner, P. Nagy, O. Mueller-Cajar, F. U. Hartl and M. Hayer-Hartl, Opposing effects of folding and assembly chaperones on evolvability of Rubisco, Nat. Chem. Biol., 2015, 11(2), 148–155 CrossRef PubMed.
- M. Faulkner, J. Rodriguez-Ramos, G. F. Dykes, S. V. Owen, S. Casella and D. M. Simpson, et al., Direct characterization of the native structure and mechanics of cyanobacterial carboxysomes, Nanoscale, 2017, 9(30), 10662–10673 RSC.
- F. W. Li, J. C. Villarreal and P. Szövényi, Hornworts: An Overlooked Window into Carbon-Concentrating Mechanisms, Trends Plant Sci., 2017, 22(4), 275–277 CrossRef CAS PubMed.
- R. F. Sage, Photosynthetic efficiency and carbon concentration in terrestrial plants: the C4 and CAM solutions, J. Exp. Bot., 2014, 65(13), 3323–3325 CrossRef CAS PubMed.
- S. M. Whitney, R. L. Houtz and H. Alonso, Advancing Our Understanding and Capacity to Engineer Nature's CO2-Sequestering Enzyme, Rubisco, Plant Physiol., 2011, 155(1), 27–35 CrossRef CAS PubMed.
- T. Hauser, L. Popilka, F. U. Hartl and M. Hayer-Hartl, Role of auxiliary proteins in Rubisco biogenesis and function, Nat. Plants, 2015, 1(6), 15065 CrossRef CAS PubMed.
- H. Aigner, R. H. Wilson, A. Bracher, L. Calisse, J. Y. Bhat and F. U. Hartl, et al., Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2, Science, 2017, 358(6368), 1272–1278 CrossRef CAS PubMed.
- S. M. Whitney, P. Baldet, G. S. Hudson and T. J. Andrews, Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts, Plant J., 2001, 26(5), 535–547 CrossRef CAS PubMed.
- M. T. Lin, A. Occhialini, P. J. Andralojc, M. A. J. Parry and M. R. Hanson, A faster Rubisco with potential to increase photosynthesis in crops, Nature, 2014, 513(7519), 547–550 CrossRef CAS PubMed.
- A. Occhialini, M. T. Lin, P. J. Andralojc, M. R. Hanson and M. A. J. Parry, Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO2, Plant J., 2016, 85(1), 148–160 CrossRef CAS PubMed.
- S. Atsumi, W. Higashide and J. C. Liao, Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde, Nat. Biotechnol., 2009, 27(12), 1177–1180 CrossRef CAS PubMed.
- G. D. Price, Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism, Photosynth Res, 2011, 109(1–3), 47–57 CrossRef CAS PubMed.
- N. A. Kamennaya, S. E. Ahn, H. Park, R. Bartal, K. A. Sasaki and H. Y. Holman, et al. Installing extra bicarbonate transporters in the cyanobacterium Synechocystis sp. PCC6803 enhances biomass production, Metab. Eng., 2015, 29, 76–85 CrossRef CAS PubMed.
- C. A. Kerfeld and M. R. Melnicki, Assembly, function and evolution of cyanobacterial carboxysomes, Curr. Opin. Plant Biol., 2016, 31, 66–75 CrossRef CAS PubMed.
- W. Bonacci, P. K. Teng, B. Afonso, H. Niederholtmeyer, P. Grob and P. A. Silver, Modularity of a carbon-fixing protein organelle, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(2), 478–483 CrossRef CAS PubMed.
- J. C. Cameron, S. C. Wilson, S. L. Bernstein and C. A. Kerfeld, Biogenesis of a bacterial organelle: the carboxysome assembly pathway, Cell, 2013, 155(5), 1131–1140 CrossRef CAS PubMed.
- F. Cai, S. L. Bernstein, S. C. Wilson and C. A. Kerfeld, Production and Characterization of Synthetic Carboxysome Shells with Incorporated Luminal Proteins, Plant Physiol., 2016, 170(3), 1868–1877 CAS.
- W. W. Cleland, T. J. Andrews, S. Gutteridge, F. C. Hartman and G. H. Lorimer, Mechanism of Rubisco: The Carbamate as General Base, Chem. Rev., 1998, 98(2), 549–562 CrossRef CAS PubMed.
- J. Y. Bhat, G. Miličić, G. Thieulin-Pardo, A. Bracher, A. Maxwell and S. Ciniawsky, et al., Mechanism of Enzyme Repair by the AAA+ Chaperone Rubisco Activase, Mol. Cell, 2017, 67(5), 744–756 CrossRef CAS PubMed.
- L. A. Li, L. Janet and R. T. F. Gibson, The Rubisco activase (rca) gene is located downstream from rbcS in Anabaena sp. strain CA and is detected in other Anabaena/Nostoc strains, Plant Mol. Biol., 1993, 21(5), 753–764 CrossRef CAS PubMed.
- O. Koksharova, M. Schubert, S. Shestakov and R. Cerff, Genetic and biochemical evidence for distinct key functions of two highly divergent GAPDH genes in catabolic and anabolic carbon flow of the cyanobacterium Synechocystis sp. PCC 6803, Plant Mol. Biol., 1998, 36(1), 183–194 CrossRef CAS PubMed.
- M. Tamoi, T. Miyazaki, T. Fukamizo and S. Shigeoka, The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions, Plant J., 2005, 42(4), 504–513 CrossRef CAS PubMed.
- S. V. Caemmerer and J. A. Berry, A Biochemical Model of Photosynthetic CO2 Assimilation in Leaves of C3 Species, Planta, 1980, 149(1), 78–90 CrossRef PubMed.
- C. A. Raines, The Calvin cycle revisited, Photosynth. Res., 2003, 75(1), 1–10 CrossRef CAS PubMed.
- X.-G. Zhu, E. de Sturler and S. P. Long, Optimizing the Distribution of Resources between Enzymes of Carbon Metabolism Can Dramatically Increase Photosynthetic Rate: A Numerical Simulation Using an Evolutionary Algorithm, Plant Physiol., 2007, 145(2), 513–526 CrossRef CAS PubMed.
- A. Flechner, W. Gross, W. Martin and C. Schnarrenberger, Chloroplast class I and class II aldolases are bifunctional for fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate cleavage in the Calvin cycle, FEBS Lett., 1999, 447, 200–202 CrossRef CAS PubMed.
- F. Liang, E. Englund, P. Lindberg and P. Lindblad, Engineered cyanobacteria with enhanced growth show increased ethanol production and higher biofuel to biomass ratio, Metab. Eng., 2018, 46, 51–59 CrossRef CAS PubMed.
- A. J. Serrato, E. M. Yubero-Serrano, L. M. Sandalio, J. MuÑoz-Blanco, A. Chueca and J. L. Caballero, et al. CpFBPaseII, a novel redox-independent chloroplastic isoform of fructose-1,6-bisphosphatase, Plant, Cell Environ., 2009, 32(7), 811–827 CrossRef CAS PubMed.
- M. Chiadmi, A. Navaza, M. Miginiac-Maslow, J. P. Jacquot and J. Cherfils, Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase, EMBO J., 1999, 18(23), 6809–6815 CrossRef CAS PubMed.
- D. Schimkat, D. Heineke and H. W. Heldt, Regulation of sedoheptulose-l,7-bisphosphatase by sedoheptulose-7-phosphate and glycerate, and of fructose-l,6-bisphosphatase by glycerate in spinach chloroplasts, Planta, 1990, 181, 97–103 CrossRef CAS PubMed.
- T. Mettler, T. Muhlhaus, D. Hemme, M.-A. Schottler, J. Rupprecht and A. Idoine, et al., Systems Analysis of the Response of Photosynthesis, Metabolism, and Growth to an Increase in Irradiance in the Photosynthetic Model Organism Chlamydomonas reinhardtii, Plant Cell, 2014, 26(6), 2310–2350 CrossRef CAS PubMed.
- T. Iwaki, K. Haranoh, N. Inoue, K. Kojima, R. Satoh and T. Nishino, et al., Expression of foreign type I ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39) stimulates photosynthesis in cyanobacterium Synechococcus PCC7942 cells, Photosynth. Res., 2006, 88(3), 287–297 CrossRef CAS PubMed.
- A. M. Ruffing, Borrowing genes from Chlamydomonas reinhardtii for free fatty acid production in engineered cyanobacteria, J. Appl. Phycol., 2013, 25(5), 1495–1507 CrossRef CAS.
- A. M. Ruffing, Improved Free Fatty Acid Production in Cyanobacteria with Synechococcus sp. PCC 7002 as Host, Front. Bioeng. Biotechnol., 2014, 2, 17 Search PubMed.
- M. Kanno, A. L. Carroll and S. Atsumi, Global metabolic rewiring for improved CO2 fixation and chemical production in cyanobacteria, Nat. Commun., 2017, 8, 14724 CrossRef CAS PubMed.
- F. Liang and P. Lindblad, Synechocystis PCC 6803 overexpressing RuBisCO grow faster with increased photosynthesis, Metab. Eng. Commun., 2017, 4, 29–36 CrossRef PubMed.
- F. Liang and P. Lindblad, Effects of overexpressing photosynthetic carbon flux control enzymes in the cyanobacterium Synechocystis PCC 6803, Metab. Eng., 2016, 38, 56–64 CrossRef CAS PubMed.
- A. J. De Porcellinis, H. Nørgaard, L. M. F. Brey, S. M. Erstad, P. R. Jones and J. L. Heazlewood, et al., Overexpression of bifunctional fructose-1,6-bisphosphatase/sedoheptulose-1,7-bisphosphatase leads to enhanced photosynthesis and global reprogramming of carbon metabolism in Synechococcus sp. PCC 7002, Metab. Eng., 2018, 47, 170–183 CrossRef CAS PubMed.
- W. Ma, D. Shi, Q. Wang, L. Wei and H. Chen, Exogenous expression of the wheat chloroplastic fructose-1,6-bisphosphatase gene enhances photosynthesis in the transgenic cyanobacterium, Anabaena PCC7120, J. Appl. Phycol., 2005, 17(3), 273–280 CrossRef CAS.
- W. Dejtisakdi and S. M. Miller, Overexpression of Calvin cycle enzyme fructose 1,6-bisphosphatase in Chlamydomonas reinhardtii has a detrimental effect on growth, Algal Res., 2016, 14, 116–126 CrossRef.
- M. Tamoi, M. Nagaoka, Y. Miyagawa and S. Shigeoka, Contribution of fructose-1,6-bisphosphatase and sedoheptulose-1,7- bisphosphatase to the photosynthetic rate and carbon flow in the Calvin cycle in transgenic plants, Plant Cell Physiol., 2006, 47(3), 380–390 CrossRef CAS PubMed.
- Y. Miyagawa, M. Tamoi and S. Shigeoka, Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth, Nat. Biotechnol., 2001, 19, 965–969 CrossRef CAS PubMed.
- T. Ogawa, M. Tamoi, A. Kimura, A. Mine, H. Sakuyama and E. Yoshida, et al., Enhancement of photosynthetic capacity in Euglena gracilis by expression of cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase leads to increases in biomass and wax ester production, Biotechnol. Biofuels, 2015, 8, 80 CrossRef PubMed.
- I. H. Köhler, U. M. Ruiz-Vera, A. VanLoocke, M. L. Thomey, T. Clemente and S. P. Long, et al., Expression of cyanobacterial FBP/SBPase in soybean prevents yield depression under future climate conditions, J. Exp. Bot., 2017, 68(3), 715–726 Search PubMed.
- S. Lefebvre, T. Lawson, M. Fryer, O. V. Zakhleniuk, J. C. Lloyd and C. a Raines, Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development, Plant Physiol., 2005, 138(1), 451–460 CrossRef CAS PubMed.
- L. Feng, Y. Han, G. Liu, B. An, J. Yang and G. Yang, et al. Overexpression of sedoheptulose-1,7-bisphosphatase enhances photosynthesis and growth under salt stress in transgenic rice plants, Funct. Plant Biol., 2007, 34(9), 822 CrossRef CAS.
- D. M. Rosenthal, A. M. Locke, M. Khozaei, C. A. Raines, S. P. Long and D. R. Ort, Over-expressing the C(3) photosynthesis cycle enzyme sedoheptulose-1-7 bisphosphatase improves photosynthetic carbon gain and yield under fully open air CO(2) fumigation (FACE), BMC Plant Biol., 2011, 11(1), 123 CrossRef CAS PubMed.
- A. J. Simkin, P. E. Lopez-Calcagno, P. A. Davey, L. R. Headland, T. Lawson and S. Timm, et al. Simultaneous stimulation of sedoheptulose 1,7-bisphosphatase, fructose 1,6-bisphosphate aldolase and the photorespiratory glycine decarboxylase-H protein increases CO2 assimilation, vegetative biomass and seed yield in Arabidopsis, Plant Biotechnol. J., 2017, 15(7), 805–816 CrossRef CAS PubMed.
- K. Uematsu, N. Suzuki, T. Iwamae, M. Inui and H. Yukawa, Increased fructose 1,6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants, J. Exp. Bot., 2012, 63(8), 3001–3009 CrossRef CAS PubMed.
- A. Hatano-Iwasaki and K. Ogawa, Biomass production is promoted by increasing an aldolase undergoing glutathionylation in Arabidopsis thaliana, Int. J. Plant Dev. Biol., 2012, 1–8 Search PubMed.
- A. J. Simkin, L. McAusland, L. R. Headland, T. Lawson and C. A. Raines, Multigene manipulation of photosynthetic carbon assimilation increases CO2 fixation and biomass yield in tobacco, J. Exp. Bot., 2015, 66(13), 4075–4090 CrossRef CAS PubMed.
- B. Yang, J. Liu, X. Ma, B. Guo, B. Liu and T. Wu, et al. Genetic engineering of the Calvin cycle toward enhanced photosynthetic CO2 fixation in microalgae, Biotechnol. Biofuels, 2017, 10(1), 229 CrossRef PubMed.
- M. Khozaei, S. Fisk, T. Lawson, Y. Gibon, R. Sulpice and M. Stitt, et al. Overexpression of plastid transketolase in tobacco results in a thiamine auxotrophic phenotype, Plant Cell, 2015, 27(2), 432–447 CrossRef CAS PubMed.
- Y. Suzuki, E. Kondo and A. Makino, Effects of co-overexpression of the genes of Rubisco and transketolase on photosynthesis in rice, Photosynth Res., 2017, 131(3), 281–289 CrossRef CAS PubMed.
- I. A. Berg, D. Kockelkorn, W. H. Ramos-Vera, R. F. Say, J. Zarzycki and M. Hügler, et al. Autotrophic carbon fixation in archaea, Nat. Rev. Microbiol., 2010, 8(6), 447–460 CrossRef CAS PubMed.
- A. Bar-Even, E. Noor, N. E. Lewis and R. Milo, Design and analysis of synthetic carbon fixation pathways, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(19), 8889–8894 CrossRef CAS PubMed.
- N. J. Claassens, D. Z. Sousa, V. A. P. M. Dos Santos, W. M. De Vos and J. Van Der Oost, Harnessing the power of microbial autotrophy, Nat. Rev. Microbiol., 2016, 14(11), 692–706 CrossRef CAS PubMed.
- C. A. Cotton, C. Edlich-Muth and A. Bar-Even, Reinforcing carbon fixation: CO2 reduction replacing and supporting carboxylation, Curr. Opin. Biotechnol., 2018, 49, 49–56 CrossRef CAS PubMed.
- I. A. Figueroa, T. P. Barnum, P. Y. Somasekhar, C. I. Carlström, A. L. Engelbrektson and J. D. Coates, Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway, Proc. Natl. Acad. Sci. U. S. A., 2017,(5), 201715549 Search PubMed.
- A. Bar-Even, Formate Assimilation: The Metabolic Architecture of Natural and Synthetic Pathways, Biochemistry, 2016, 55(28), 3851–3863 CrossRef CAS PubMed.
- C. Durall, N. Rukminasari and P. Lindblad, Enhanced growth at low light intensity in the cyanobacterium Synechocystis PCC 6803 by overexpressing phosphoenolpyruvate carboxylase, Algal Res., 2016, 16, 275–281 CrossRef.
- T. Schwander, L. Schada von Borzyskowski, S. Burgener, N. S. Cortina and T. J. Erb, A synthetic pathway for the fixation of carbon dioxide in vitro, Science, 2016, 354(6314), 900–904 CrossRef CAS PubMed.
- H. Yu, X. Li, F. Duchoud, D. S. Chuang and J. C. Liao, Augmenting the Calvin–Benson–Bassham cycle by a synthetic malyl-CoA-glycerate carbon fixation pathway, Nat. Commun., 2018, 9(1), 1–10 CrossRef PubMed.
- J. E. W. Polle, S.-D. Kanakagiri and A. Melis, tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size, Planta, 2003, 217(1), 49–59 CAS.
- H. Kirst, C. Formighieri and A. Melis, Maximizing photosynthetic efficiency and culture productivity in cyanobacteria upon minimizing the phycobilisome light-harvesting antenna size, Biochim. Biophys. Acta, Bioenerg., 2014, 1837(10), 1653–1664 CrossRef CAS PubMed.
- T. Gabilly, K. K. Niyogi and P. G. Lemaux, Photosynthetic antenna engineering to improve crop yields, Planta, 2017, 245(5), 1009–1020 CrossRef PubMed.
- L. E. Page, M. Liberton and H. B. Pakrasi, Reduction of photoautotrophic productivity in the cyanobacterium Synechocystis sp. strain PCC 6803 by phycobilisome antenna truncation, Appl. Environ. Microbiol., 2012, 78(17), 6349–6351 CrossRef CAS PubMed.
- J. H. Kwon, G. Bernát, H. Wagner, M. Rögner and S. Rexroth, Reduced light-harvesting antenna: consequences on cyanobacterial metabolism and photosynthetic productivity, Algal Res., 2013, 2(3), 188–195 CrossRef.
- D. J. Lea-Smith, P. Bombelli, J. S. Dennis, S. A. Scott, A. G. Smith and C. J. Howe, Phycobilisome-Deficient Strains of Synechocystis sp. PCC 6803 Have Reduced Size and Require Carbon-Limiting Conditions to Exhibit Enhanced Productivity, Plant Physiol., 2014, 165(2), 705–714 CrossRef CAS PubMed.
- M. Chen, M. Schliep, R. D. Willows, Z. Cai, B. A. Beilan and H. Scheer, A red-shifted chlorophyll, Science, 2010, 329, 1318–1320 CrossRef CAS PubMed.
- T. J. Erb, P. R. Jones and A. Bar-Even, Synthetic metabolism: metabolic engineering meets enzyme design, Curr. Opin. Chem. Biol., 2017, 37, 56–62 CrossRef CAS PubMed.
- J. T. Broddrick, B. E. Rubin, D. G. Welkie, N. Du, N. Mih and S. Diamond, et al. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(51), E8344–E8353 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |