Biotin-functionalized nanoparticles: an overview of recent trends in cancer detection
Sonia
Fathi-karkan
 ab,
Saman
Sargazi†
ab,
Saman
Sargazi†
 *cd,
Shirin
Shojaei
e,
Bahareh
Farasati Far
*cd,
Shirin
Shojaei
e,
Bahareh
Farasati Far
 f,
Shekoufeh
Mirinejad
c,
Marco
Cordani
gh,
Arezoo
Khosravi
i,
Ali
Zarrabi
f,
Shekoufeh
Mirinejad
c,
Marco
Cordani
gh,
Arezoo
Khosravi
i,
Ali
Zarrabi
 jkl and
Saeid
Ghavami†
jkl and
Saeid
Ghavami†
 *mno
*mno
aNatural Products and Medicinal Plants Research Center, North Khorasan University of Medical Sciences, Bojnurd, 94531-55166 Iran. E-mail: soniafathi92@gmail.com
bDepartment of Advanced Sciences and Technologies in Medicine, School of Medicine, North Khorasan University of Medical Sciences, Bojnurd 9414974877, Iran. E-mail: soniafathi92@gmail.com
cCellular and Molecular Research Center, Research Institute of Cellular and Molecular Sciences in Infectious Diseases, Zahedan University of Medical Sciences, Zahedan, Iran. E-mail: sgz.biomed@gmail.com; shokufemn@gmail.com
dDepartment of Clinical Biochemistry, School of Medicine, Zahedan University of Medical Sciences, Zahedan, Iran. E-mail: sgz.biomed@gmail.com
eNano Drug Delivery Research Center, Health Technology Institute, Kermanshah University of Medical Sciences, Kermanshah, Iran. E-mail: shojaeishirin891@gmail.com
fDepartment of Chemistry, Iran University of Science and Technology, Tehran, Iran. E-mail: Bahar.ferasati@gmail.com
gDepartment of Biochemistry and Molecular Biology, Faculty of Biology, Complutense University, 28040 Madrid, Spain
hInstituto de Investigaciones Sanitarias San Carlos (IdISSC), 28040 Madrid, Spain
iDepartment of Genetics and Bioengineering, Faculty of Engineering and Natural Sciences, Istanbul Okan University, Istanbul 34959, Turkiye. E-mail: arezoo.khosravi@okan.edu.tr
jDepartment of Biomedical Engineering, Faculty of Engineering and Natural Sciences, Istinye University, Istanbul 34396, Turkiye. E-mail: ali.zarrabi@istinye.edu.tr
kDepartment of Research Analytics, Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences, Saveetha University, Chennai - 600 077, India
lGraduate School of Biotechnology and Bioengineering, Yuan Ze University, Taoyuan 320315, Taiwan
mDepartment of Human Anatomy and Cell Science, Max Rady College of Medicine, Rady Faculty of Health Sciences, University of Manitoba, Winnipeg, MB R3T 2N2, Canada. E-mail: saeid.ghavami@umanitoba.ca
nFaculty of Medicine in Zabrze, University of Technology in Katowice, 41-800 Zabrze, Poland
oResearch Institute of Oncology and Hematology, Cancer Care Manitoba, University of Manitoba, Winnipeg, MB R3T 2N2, Canada
First published on 10th June 2024
Abstract
Electrochemical bio-sensing is a potent and efficient method for converting various biological recognition events into voltage, current, and impedance electrical signals. Biochemical sensors are now a common part of medical applications, such as detecting blood glucose levels, detecting food pathogens, and detecting specific cancers. As an exciting feature, bio-affinity couples, such as proteins with aptamers, ligands, paired nucleotides, and antibodies with antigens, are commonly used as bio-sensitive elements in electrochemical biosensors. Biotin–avidin interactions have been utilized for various purposes in recent years, such as targeting drugs, diagnosing clinically, labeling immunologically, biotechnology, biomedical engineering, and separating or purifying biomolecular compounds. The interaction between biotin and avidin is widely regarded as one of the most robust and reliable noncovalent interactions due to its high bi-affinity and ability to remain selective and accurate under various reaction conditions and bio-molecular attachments. More recently, there have been numerous attempts to develop electrochemical sensors to sense circulating cancer cells and the measurement of intracellular levels of protein thiols, formaldehyde, vitamin-targeted polymers, huwentoxin-I, anti-human antibodies, and a variety of tumor markers (including alpha-fetoprotein, epidermal growth factor receptor, prostate-specific Ag, carcinoembryonic Ag, cancer antigen 125, cancer antigen 15-3, etc.). Still, the non-specific binding of biotin to endogenous biotin-binding proteins present in biological samples can result in false-positive signals and hinder the accurate detection of cancer biomarkers. This review summarizes various categories of biotin-functional nanoparticles designed to detect such biomarkers and highlights some challenges in using them as diagnostic tools.
1. Introduction
Cancer remains a leading cause of premature death globally.1–3 Despite progress, new diagnoses (19.3 million) and deaths (nearly 10 million) in 2020 highlight its ongoing burden.4,5 Rising incidence due to population growth and risk factors like tobacco use and obesity demands continued advancements.6–8 Early detection is key to reducing cancer death.9 Modern techniques like magnetic resonance spectroscopy (MRS), X-ray computed tomography (CT), positron emission tomography (PET), and molecular diagnostics offer rapid identification but have limitations like cost, target selection, and artifacts hindering precise diagnosis.10 Numerous cancer markers have been acknowledged as reliable tools for predicting the behavior of different malignancies and assisting clinical scientists in understanding the genetic pathways behind tumor formation.11–14 Enzyme-linked immunosorbent assays (ELISAs), fluorescence-based assays, mass-based assays, and electrochemical assays are some modern methods for finding tumor markers. Although several of these approaches have high selectivity, obstacles to their wider implementation, such as low sample concentrations or measurement difficulties, persist.15–17 Nanotechnology has enabled many studies on designing and utilizing nanomaterials (NMs) in detection techniques to target tumor markers at low concentrations.18,19 The use of nanostructures, such as nanoparticles (NPs), has evolved dramatically during the last several decades. Hyperbranched polymers, up-conversion nanoparticles (UNPs), optical nanosensors, magnetic nanoparticles (MNPs), quantum dots (QDs), metal NPs, carbon nanotubes (CNTs), graphene nanosensors, carbon-based nanosensors, piezoelectric biosensors, etc. are a few examples of these materials that have shown high sensitivity and selectivity in the detection of tumor indicators.15,20,21 This review examines how biotin-functionalized NPs are used in identifying cancer, highlighting the importance of distinguishing between detection and treatment. Detection involves the early recognition of specific biomarkers or cancer cells through these NPs, utilizing their diagnostic capabilities.15,22,23 Conversely, targeted therapy employs these NPs to deliver therapeutic agents directly to cancer cells once detected.24 Although both methods use similar targeting strategies, their objectives are distinctly different, focusing on diagnosis in one instance and treatment in the other. Moreover, NPs can be used to deliver multiple agents simultaneously for combined treatment or to achieve simultaneous diagnostic and therapeutic results.25–30 Identifying disease markers at the molecular level is crucial to develop effective diagnostic strategies using NMs. These markers can be used to design ligands that bind specifically to related proteins on the surface of cancer cells. These ligands may then be conjugated to the NPs’ surface to improve their targeting's effectiveness and specificity.31,322. Comparative overview of functionalized NPs in cancer detection
Nanotechnology is rapidly advancing, introducing diverse functionalized NPs specifically designed for cancer detection. This section delves into the distinct characteristics and applications of prominent NP types, including gold nanoparticles (AuNPs),33 QDs,34 and magnetic nanoparticles (MNPs),35 which are extensively studied for their unique properties. Our focus is to underscore their critical roles in enhancing the precision of diagnostics.AuNPs are renowned for their exceptional optical properties, making them ideal for photothermal therapy and imaging techniques.36 Their ability to efficiently absorb and convert light into heat is utilized in methods targeting the precise destruction of cancer cells while sparing surrounding healthy tissue.37 Furthermore, the surface plasmon resonance feature of AuNPs significantly enhances imaging capabilities, which is crucial for detecting tumor markers across various imaging platforms.38
QDs are highlighted for their outstanding brightness and the ability to emit light at variable wavelengths, facilitating multiplexed imaging. This capability allows for the simultaneous detection of multiple biomarkers, making QDs exceptionally useful for long-term cancer studies, especially in monitoring tumor progression.39
MNPs leverage their magnetic properties to enable not just imaging but also remote manipulation. Typically composed of iron oxides, MNPs are directed to specific bodily locations using external magnetic fields, enhancing targeted therapy applications.40 These NPs are often modified with specific ligands or antibodies to target distinct cancer biomarkers, thereby increasing the selectivity and sensitivity of detection techniques. For example, MNPs attached to antibodies targeting cancer-specific antigens significantly aid in the early detection of several cancers.41
While each NP type presents unique advantages, their potential limitations, such as biocompatibility, toxicity, and nonspecific binding, also pose significant challenges that must be addressed to optimize their clinical utility. These challenges substantially influence the choice of NPs for specific applications in cancer detection and therapy.42 In summary, with a thorough understanding of these NPs, we now shift our focus to biotin-functionalized NPs in the subsequent section. These NPs utilize the robust biotin–avidin interaction, providing a powerful platform for explicitly targeting and detecting cancer cells. This specificity is crucial for developing diagnostic strategies to minimize false positives and enhance detection accuracy.43
3. Biotin in cancer diagnostics and therapeutics: a nanotechnological approach
The B complex vitamins encompass diverse compounds, including biotin, which dissolves in water and is also referred to as vitamin B7, vitamin H, and coenzyme R.44 Humans cannot synthesize biotin; thus, intestinal bacteria and nutrition supply it to the small intestine. Cell growth, proliferation, and differentiation require biotin, an essential nutrient. To develop quickly, tumor cells produce high biotin receptor rates.45 The avidin–biotin non-covalent interaction is one of nature's strongest.46–48 Avidin forms a probe by binding to solid surfaces or biomolecules.49 Biotin may bind to many proteins and nucleotides without changing their main properties due to its small size and tendency to not cross-link the carboxy-containing side chain with avidin.50 A simple construction procedure that preserves both attached portions’ chemical and biological properties makes the avidin–biotin system a versatile nanotechnology platform.51 Beyond their basic use in diagnostics, biotin-functionalized NPs are now being explored for their therapeutic properties, showcasing a two-pronged strategy in battling cancer.52 This investigation underlines their ability to profoundly influence both the detection and treatment methods.Biotin may pre-target several cancer cell lines, allowing the targeted detection of synthetic materials in tumor locations. Due to the preferential biotin uptake by tissues or malignant cells, biotinylated viral vectors, nucleic acids, polypeptides, liposomal chemicals, and artificial materials have been used in biosensing and imaging.44,53–55 The distinctive binding characteristics of avidin–biotin methodologies to diverse biomaterials, such as DNA and RNA, NPs, antibodies, and aptamers.49,54,55 Several biotinylated techniques can identify Hodgkin lymphoma,56 breast,57 lung,58 prostate,59 and cervical60 cancer cells.
Maiti et al. reviewed only polymer-surfaced NPs and medicines with biotin-mediated cancer theranostic methods.61 It did not cover all NPs. Another study compares the efficacy of a single NP vs. two NPs targeted with folic acid and biotin to deliver anti-cancer drugs.62 Biotin conjugation has been extensively reviewed in biological imaging, sensing, and target delivery. We investigated this critical topic and its challenges because biotin-conjugated NPs’ diagnostic applications in cancer care have not been thoroughly studied.
4. Biotin conjugates and their role as targeting molecules in biomedicine
4.1. Biochemical structure and properties of biotin
Biotin, called vitamin B7, is a non-enzymatic with an utterly unique structure involved in routine biochemical activities. It plays a key role in metabolic processes such as carboxylation and is crucial in macronutrient metabolism.63 Structurally, it features a unique ureido ring and a carboxylic group, making it suitable for biotinylation in various biochemical applications.64,65 In an amides ring, tetrahydrothiophene, and the ureido ring are fused to each other. The nitrogen atom, along with a lone pair of electrons in the ureido ring, forms the central core of the biotin molecule's functionality.66 This specific configuration is crucial for binding CO2 to the structure to boost the enzymes’ biotin-dependent reactions, such as the tolerance of urea as a nitrogen source, which is very useful for microorganisms,67 and the breakdown of isoprenoids in bacteria and plants. The ureido ring is bound to two intramolecular hydride transfer reactions at the acyl carbon, where ureido oxygen attack of the acyl group is more preferred when it is attenuated than otherwise. This mechanism points out the significance of the ureido group, which is required for carboxylation reactions as a part of many metabolic pathways.68 Thus, the ureido ring of biotin acts as the carrier of the CO2 during the biotin-dependent enzymes, allowing microorganisms to use urea along with geranyl-coenzyme A carboxylase to occur isoprenoid catabolism in the fungi, plants, and microorganisms.69 However, the chemical structure of biotin, mainly the presence of ureido ring in its structure, makes it unique as it is for the roles in carboxylation rather than any other distinct role. Bearing a distinctive feature of the ureido group, biotin can be a coenzyme for the carboxylase enzymes essential for fatty acid synthesis, that of isoleucine and valine, as well as other metabolic functions.70 Biotin deficiency can affect metabolism and is found in red meat, egg yolks, and nuts, while undiagnosed and non-treated biotinidase deficiency remains a health concern.71–734.2. Avidin–biotin interaction
Avidin–biotin interactions’ affinity for biotinylated cancer cells makes them powerful cancer treatments. Biotin–avidin interactions may be a thousand to a million times stronger than those of avidin and other antibodies.51,74 High-binding affinity links biotin and avidin.75 Avidin is a tetrameric protein found in bird, reptile, and amphibian egg whites.76–78 Esmond Emerson Snell discovered avidin after seeing biotin deficiency in egg white-fed chicks.79 This was found to be caused by avidin, an egg-white glycoprotein that sequesters biotin.78 Unlike streptavidin (SA), avidin has six tyrosine (Tyr) residues per subunit, while SA has one. Tyr-33 at avidin position 43 is similar to Tyr-33 at SA spot 40 (40Thr-Gly-Thr-Tyr-Glu-Ser-Ala-Val).80,81 Biotin changes conformation, stability, and flexibility when strept(avidin) binds. This avidin–biotin interaction allows biotinylated compounds to selectively target cancer cells.82Avidin–biotin interaction can deliver chemotherapeutic agents, radionuclides,83 and NPs84 to cancer cells.51,85,86 Drugs conjugated with biotinylated avidin can enter cancer cells efficiently.87,88 Targeted delivery reduces the off-target effects of chemotherapy and radiation. This increases therapeutic agent concentration at cancerous growth sites. Many researchers have focused on biotin conjugation for bioimaging,89 sensing,90 and targeted delivery (see Fig. 1).91 Cancer imaging with conjugated avidin–biotin agents can help detect and monitor cancer early.92
4.3. Obtaining biotinylated nanosystems for drug and gene delivery
Biotin is covalently added to biomolecules (oligonucleotides, antibodies, proteins, and peptides) or NPs [liposomes, polyethylene glycol (PEG), etc.] to enhance their properties.93–95 Due to biotin's small size, the reaction is fast, specific, and unlikely to disrupt biomolecule or NP functions.96 Biotinylation also targets drugs and genes in particular cells, improving delivery.97 Several methods can be used to obtain biotinylated nanosystems for drug and gene delivery.Biotinylated NPs are produced using various methods, including conjugation with biotin-modified polymers like PEG. This approach allows for injecting drugs or genes within NPs, enabling targeted delivery to cells that overexpress biotin receptors on their surface (Fig. 2A). A recent study98 demonstrated the potential of this strategy by developing a biotin-conjugated PEG-poly(glutamic acid) for in vitro cargo delivery to lung epithelial cells. The success of this approach in both intracellular delivery and linking avidin to fluorescent DNA highlights the broad applicability of biotin-PEG copolymers in various biomedical applications, including drug delivery.
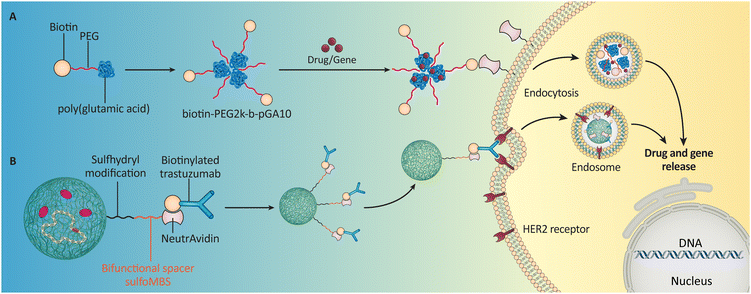 | ||
| Fig. 2 Possible applications of biotinylated NPs in drug and gene delivery; biotins could be attached to the (A) PEG when forming a polymeric NP (Li et al. (2023)55 or B) antibody (Ab) molecules to make the whole particle targetable (Wartlick et al. (2004)101). | ||
An alternative strategy is to use biotinylated liposomes, lipid-based spherical NPs, as shown by Wang et al. (2010).99 Further enhancing their specificity for specific cell types can be achieved by functionalizing biotinylated liposomes with other targeting ligands. This was demonstrated by Gautam et al. (2023), who enhanced the specificity of biotinylated liposomes for targeted cell types using a “nano-on-nano” biotin–SA–biotin system, showcasing advanced precision in drug delivery.
Nanosystems and biotinylated antibodies are another option (Fig. 2B).100 Wartlick et al. (2004) manipulated NP surfaces by covalently attaching biotin-binding protein.101 Herceptin®-conjugated NPs target human epidermal growth factor receptor 2 (HER2)-overexpressing cells using avidin–biotin complexes, demonstrating how biotinylation improves drug delivery systems. Biotin-conjugated polymers, liposomes, or antibodies allow nanosystems to selectively target specific cell types, enhancing efficacy and reducing off-target effects.61
4.4. Internalization of biotinylated nanosystems in tumor cells
The body generally does not metabolize biotin extensively, with over half of the consumed amounts remaining unchanged in the urine.102–104 Vitamin B7 is nontoxic due to its high solubility in water, and the half-life of biotin usually varies from 1.8 hours to 18.8 hours, depending on the ingested dose.105 A sodium-dependent multivitamin transporter (SMVT) and a high-affinity biotin transporter are responsible for biotin's cellular internalization.104,106–108The SLC5A6 gene, located on chromosome 2p23, encodes the SMVT responsible for transporting essential nutrients like biotin into cells. Interestingly, cancer cells often have higher SMVT expression or take up more biotin than healthy cells.109–111 Interestingly, cancer cells often have higher SMVT expression or take up more biotin compared to healthy cells. This characteristic makes SMVT receptors valuable for cancer diagnosis. Researchers can develop tools that target these receptors to identify cancer.112 Additionally, scientists are designing biotin nanocarriers to enhance transporter-targeted nano-drug delivery via regulating SMVT expression.113,114 These nanocarriers can be loaded with drugs or genes and then delivered specifically to cancer cells for targeted therapy.115–117
Biotinylated NPs exploit receptor-mediated endocytosis for cellular uptake by tumor cells (Fig. 3). Like how cells import nutrients, these NPs bind to biotin receptors on the cell surface, triggering an inward folding of the membrane and engulfment into vesicles.118 Inside the cell, various mechanisms like enzymes or acidic environments within endosomes and lysosomes can break down the nanocarriers, releasing their cargo.119,120
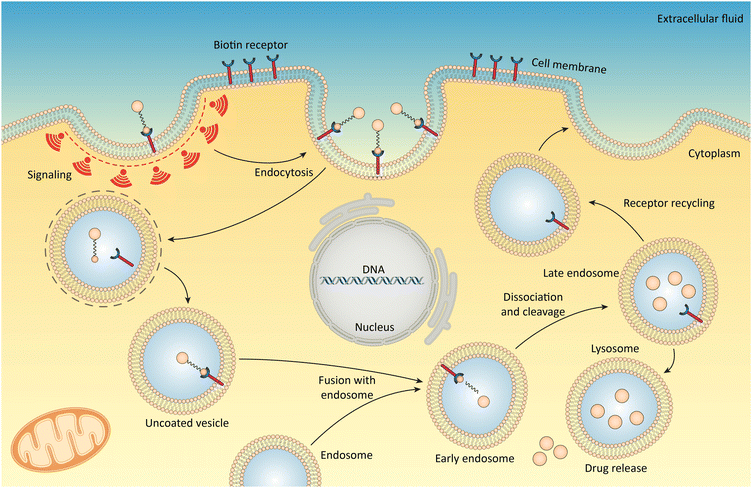 | ||
| Fig. 3 Cellular uptake of biotinylated nanosystems via receptor-mediated endocytosis. Biotin receptors on the cell membrane recognize and bind to biotin-tagged agents, initiating endocytosis. The process is depicted through various stages including vesicle formation, fusion with endosomes, transport to lysosomes, and eventual drug release inside the cell, aiming for precise therapeutic delivery to tumor cells. This mechanism ensures the nanosystems’ targeted entry into cells, enhancing the efficacy of drug delivery. Reproduced from ref. 119 with permission from [American Chemical Society], copyright [2010]. | ||
4.5. Biotinylated antibodies/antigens used in immunoassay
Researchers depend on biotinylated antibodies in immunoassays. Immunoassays detect high-specificity and sensitivity biomolecules using antibodies and antigens (Ag).121,122 However, biotinylated antibodies have revolutionized immunoassay. Khosravi et al. (1991) compared cortisol-labeled SA to unlabeled SA in biotin-binding.123,124 In immunoassays, biotinylated antibodies have many benefits. According to Mishra et al. (2019), biotinylated antibodies first amplify detection assay signals. This study used biotinylated secondary antibodies and SA conjugated with poly-horseradish peroxidase (HRP) to improve western blotting sensitivity.125 Besides being versatile, biotinylated antibodies can be used in ELISA assays,126 western blot analysis,127 and immunohistochemistry.128 Competitive and sandwich immunoassays129 can analyze biotinylated antibodies/Ag.130Competitive assays show how well-biotinylated analytes and the Ag bind to SA molecules at a given concentration. Thus, analytes are displaced from the SA surface and quantified, accurately determining their number in a sample. Additionally, biotinylated antibodies/Ag used in immunoassay cannot bind or conjugate analytes without forming a stable complex with SA. Then, conjugated analyte activity is measured by concentration. Low analyte concentrations increase signal response, while high concentrations decrease it. High biotin levels strongly bind to these free molecules, removing SA from beads. The biotinylated Ab binds the analyte and conjugates during washing. False positives occur when the calibration curve shows a high analyte level, and the signal response is low. This method can target small molecules like triiodothyronine (T3), thyroxine (T4), cortisol, testosterone, hydroxyvitamin D, and steroids. Sandwich or two-site immunoassays use a capture Ab to bind the target molecule and a detection Ab to detect it. These assays detect target analytes with biotinylated antibodies.
Additionally, some Abs are enzyme-conjugated secondary antibodies or detection antibodies. SA and Ag bind differently to biotinylated capture Ab in biotin-free samples. Ag and labeled Ab form a tertiary complex. Ag is quantified and linked to labeled enzyme activity. When biotin is unbound, biotinylated capturing antibodies bind SA to the target. A decrease in ternary compound protein content may cause a false negative or overestimated sample Ag concentration. Clinically, sandwich immunoassays measure large to extremely large molecules, including pituitary, stimulating, parathyroid, glycoprotein, insulin-like growth factor-1, human chorionic gonadotropin (hCG), insulin, thyroglobulin, C-peptide, follicle-stimulating hormone, luteinizing hormone, and thyrotropin.131
4.6. Biotin-functionalized NPs: advancing cancer precision medicine
Precision medicine tailors treatment to each patient's needs. This effort covers genomics, drug discovery, health communication, genetics, and causal inference.132–134 Precision medicine formalizes action recommendations based on current patient data into decision rules per decision point. Precision medicine can be transformed by biotin-functionalized NPs. These particles are a few nanometers wide.135 Biotin-functionalized NPs can deliver drugs directly to disease-causing cells without harming healthy cells.135,136 This section lists recent precision medicine and biotin-functionalized NP results to demonstrate the importance of this field (see Table 1).| NPs | Drug delivered | Advantages | Outcome | Indication | Ref. |
|---|---|---|---|---|---|
| Biotin functionalized fullerenes | Irinotecan | During colon cancer cell invasion, the conjugate (C60-PEI-Biotin/IRI) successfully crosses the cell membrane through overexpressed biotin receptors | This conjugate showed low toxicity to vital organs and high efficacy against tumor cells | Colon tumors | 137 |
| Biotin-enriched dendritic mesoporous Silica | — | A robust Ab enrichment method based on fluorescent signal reporters can simultaneously detect two ovarian cancer biomarkers in human serums | High consistency was obtained by using this method | Ovarian cancer | 138 |
| Biotinylated Ab to poly(chitosan) gold NPs | — | Provide telomerase assays for early cancer prognosis based on a unique method | A novel biosensor platforms for point-of-care diagnostics for telomerase management | Immunosensing of telomerase | 139 |
| Biotinylated fluorescent polymeric NPs | — | Increases the ability to image epidermal growth factor receptors (EGFR) on the surface of cells significantly | Using the developed nanoprobes, disease biomarkers can be detected very efficiently and with high sensitivity | Cancer | 140 |
| SA Fe2O3-gold NPs | Thymol | Highly selective towards bacteria | Effective drug delivery was demonstrated using the model system | Antibiotic-resistant bacteria | 141 |
| Biotin-functionalized silica NPs | — | The use of these NPs also improved protection against photobleaching | Great potential systems for photodynamic therapy applications | Gliobastoma multiforme | 142 |
| Avidin–Biotin functionalized protein NPs | — | Enhancement in the targeting efficiency | Reduced side effects and more apoptosis of cells | Lung cancer | 143 |
| Biotin-functionalized DNA cages | RuII–PtII metal complexes | Effectively suppresses the innate activity of telomerase in cancer cells that are resistant to cisplatin | Potent antitumor activity against tumor cells that are resistant to cisplatin | Cancer | 144 |
5. Biotin-functionalized NPs for cancer detection
Biotin-functionalized NPs have been extensively studied for cancer detection. Biotin-functionalized NPs can target cancer cells, which have more surface receptors than healthy cells. NPs can deliver diagnostic or therapeutic agents directly to cancer cells, improving treatment specificity and efficacy. Cancer imaging with biotin-functionalized NPs is promising. Researchers can attach biotin to NPs to add antibodies, peptides, or aptamers that bind to cancer cell receptors. Targeted binding enhances imaging contrast, improving malignancy detection sensitivity and specificity. Cancer treatment also uses biotin-functionalized NPs.145,146 Researchers can selectively deliver drugs or siRNA molecules to cancer cells while minimizing their toxicity to healthy cells by attaching them to the NPs. Scientists can reduce side effects and maximize efficacy by adjusting NPs’ size, shape, and surface properties to optimize therapeutic agent pharmacokinetics and biodistribution.147 The possible applications of NPs in cancer detection are summarized in Fig. 4.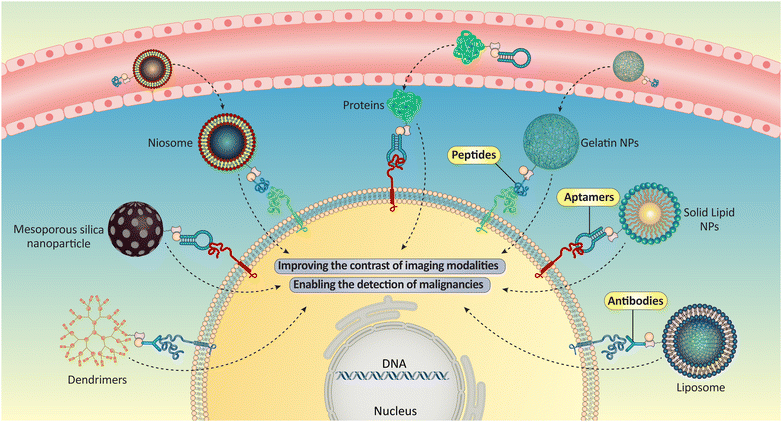 | ||
| Fig. 4 Different biotin-functionalized NPs could be used for cancer detection purposes by connection to the anticancer agents or specific targeting agents such as antibodies, peptides, or aptamers. | ||
5.1. Cancer cell detection in circulation
Circulating cancer cells (CTCs) are essential for cancer detection and treatment. Cancer cells that have spread from primary or metastatic cancers are CTCs. Metastasis—cancer cells detaching, migrating, and invading other body parts—determines disease progression.148 Immunological, molecular, and physical methods detect bloodstream CTCs. CTCs in the bloodstream reveal cancer evolution, prognosis, and treatment response. CTC detection early in the disease's progression improves treatment outcomes.149 CTC prevalence is inversely related to tumor aggressiveness and metastasis. Thus, peripheral blood CTC levels may predict disease progression and lifespan.Monitoring the efficacy of cancer therapies like chemotherapy, targeted therapy, and immunotherapy may be easier with CTC detection. CTC levels in the blood may indicate therapy efficacy and help choose a different treatment.150 Investigating CTCs for molecular characteristics like gene expression patterns, mutations, and protein expression may illuminate the tumor's biology and help develop personalized treatments. CTCs are rare and heterogeneous, making identification difficult. CTCs may change during circulation, making them more elusive. CTC detection technologies must be sensitive and targeted for accurate cancer diagnosis and treatment.151 An intriguing study created a reversible, well-organized bio-interface that captures CTCs without harming them (Fig. 5). Boronic acid moieties at the interface make it reversible and order affinity ligands. Carbon nitride nanosheets lined with boronic acid could support biotinylated aptamers by glycosylating avidin with boronate. This organized arrangement of aptamers reduced inter-strand entanglement and increased cell affinity for CTC capture. The boronate conjugation also delicately discharged highly viable CTCs by treating them with acid fructose. The engineered bio-interface isolated CTCs for mutation analysis and drug susceptibility testing have been done in cancer patients and tumor-bearing mice. These findings show bio-interface potential for early cancer detection and precision medicine.
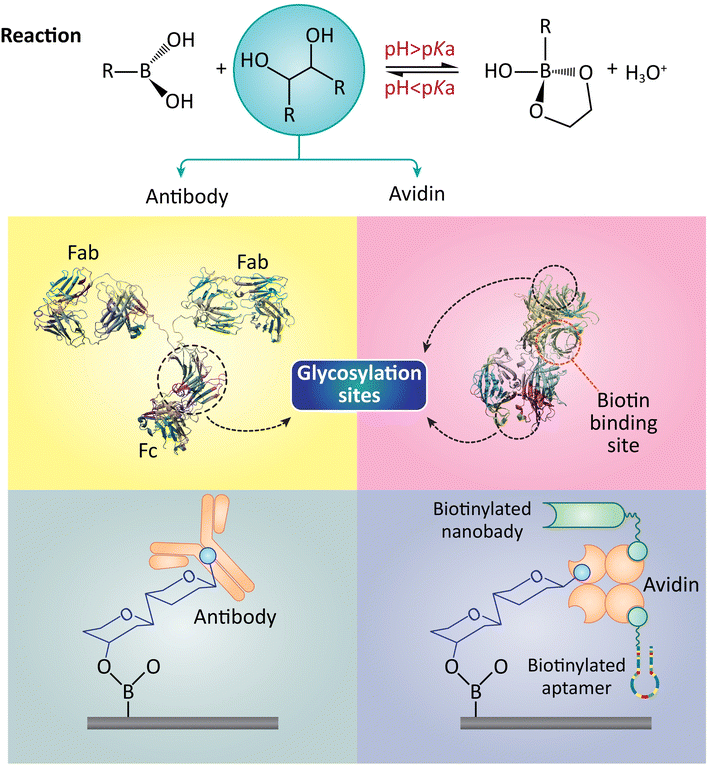 | ||
| Fig. 5 Biochemical interaction where boronic acid forms complexes with diols on antibodies and avidin. The top reaction sequence shows the pH-dependent binding, while the lower images detail the biotinylation of antibodies and nanobodies, highlighting glycosylation sites and biotin-binding sites. This visualization aids in understanding the modification of biomolecules with biotin, which is essential in various biomedical applications, as referenced in this study. Reproduced from ref. 152 with permission from [American Chemical Society], copyright [2022]. | ||
For early cancer diagnosis, biotin-activated immunomagnetic methods can extract rare, pure CTCs. Peptide-tagged antibodies chemically linked to protein-coated magnetic particles catch and release CTCs via biotin-mediated breakage of the modified protein-peptide connection. Peptides help antibodies co-immobilize on protein-coated magnetic beads (MBs), improving CTC capture. Can isolate and grow 79% of whole blood cancer cells. Biotin releases 70% of cells, 85% of which survive. Anti-Epithelial cell adhesion molecule (EpCAM), anti-HER2, and anti-EGFR immune-MNPs identified CTCs in 17 cancer patients’ peripheral blood samples. They found 215–215 pure CTCs in peripheral blood. These findings prove CTCs’ molecular profiling, diagnosis, and treatment reliability.153
In situ overexpression of CTCs allows hybrid nanoparticles (HNPs) collection and analysis. These HNPs allow antibodies, QDs, and biotinylated DNA to capture heterogeneous CTCs, including those lacking EpCAM. This strategy was tested by simultaneously detecting EpCAM, HER2, and EGFR in breast cancer subtype cells. 92.4% of CTCs were detected and 87.5% collected. Restriction enzymes released trapped cells with 86.1% efficiency, and later, research showed that they were alive and could proliferate in vitro (Fig. 6). To improve clinical decision-making, HNPs can be used for molecular and cellular research, including medication screening and tailored treatment.154
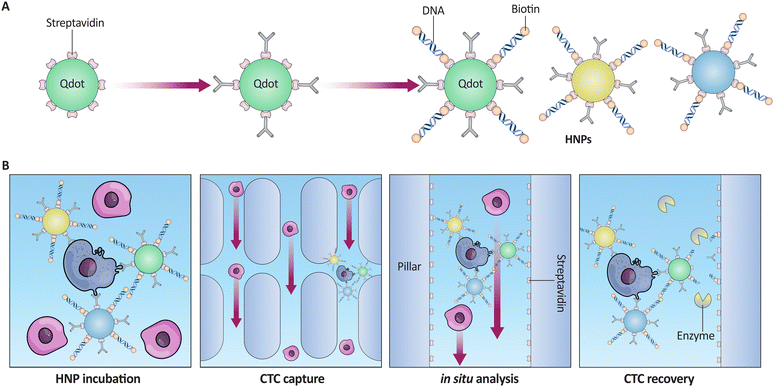 | ||
| Fig. 6 (A) Schematic representation of biotinylated DNA conjugation to SA-coated QDs to create HNPs. (B) Sequential illustration of the CTC capture process using HNPs: HNP incubation with CTCs, capture on an SA-coated cartridge, in situ analysis within the device, and the final release of viable CTCs using restriction enzymes for subsequent proliferation studies. This figure correlates with the described efficiency of CTC detection and recovery in breast cancer subtype analysis. Reproduced from ref. 154 with permission from [American Chemical Society], copyright [2017]. | ||
A novel CTC isolation method uses peptide-functionalized iron oxide MNPs. MNPs were coated with Pep10, a novel EpCAM-targeting peptide. This method isolates CTCs using the Pep10 recognition peptide instead of NMs or nanostructured surfaces functionalized with EpCAM antibodies. Pep10's binding affinity for EpCAM was equal to anti-EpCAM, and biotin–avidin connected the peptides to MNPs. Like anti-EpCAM@MNPs, Pep10@MNPs captured breast, prostate, and liver cancer cells from spiking blood with over 90% efficiency and 93% purity. Molecular biology may be performed on the acquired cells for further study. This peptide-based separation technology can improve stability, reproducibility, cancer prognosis, and metastasis reduction for clinical treatment assessment and research.155
Non-small cell lung cancer (NSCLC) blood samples may have low EpCAM and cytokeratin expression, making CTC detection difficult. Researchers solved this problem by developing MNPs functionalized with biotin peptides with excellent NSCLC therapeutic results. Two NSCLC cell lines were used to compare the novel method to Ab-based detection. The recognition peptide's identical binding affinity to A549 and NCI-H1975 allowed the Tumor Fisher method to capture CTCs in 71.4% of early-stage NSCLC patients, better than Cell Search and with fewer false negatives. The study included seven early-stage cancer patients and 81 NSCLC stages I–IV patients. Tumor Fisher had a 72.8% detection rate across stages in a larger clinical cohort, suggesting it could be used for early-stage NSCLC screening, prognosis, and treatment assessment (Fig. 7). Peptide-MNPs isolated CTCs from NSCLC patients in the first study.156
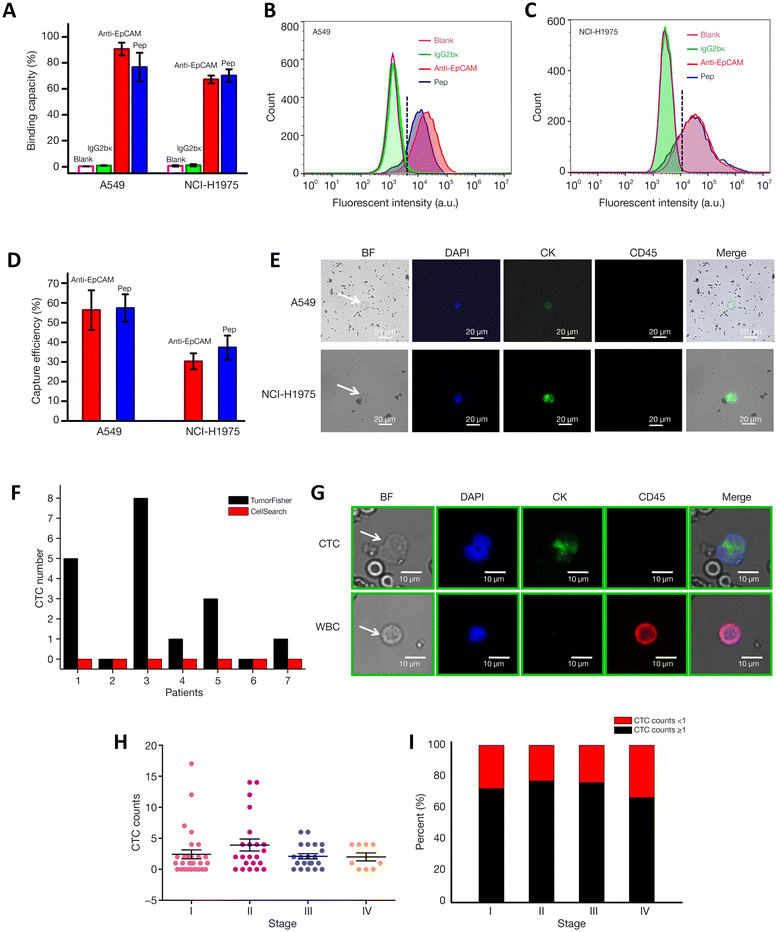 | ||
| Fig. 7 Assessment of biotin-peptide binding and anti-EpCAM efficacy in targeting cancer cells and capturing CTCs with flow cytometry, immunocytochemistry, and fluorescence microscopy. The study delineates the distinction of tumor cells from white blood cells and evaluates CTC detection across cancer stages, demonstrating the diagnostic potential of biotinylated tools. (A) Binding affinity of IgG2bκ, anti-EpCAM, and Pep to A549 and NCI-H1975 cells. (B, C) Fluorescence intensity of A549 and NCI-H1975 by flow cytometry (purple: control, green: IgG2bκ, red: anti-EpCAM, blue: Pep). (D) Capture efficiency of anti-EpCAM and Pep. (E) Immunofluorescence of CTCs (DAPI: blue, CK: green, CD45: red). Clinical data: (F) CTC counts by TumorFisher and CellSearch. (G) Representative CTC and WBC by TumorFisher (same channels as E). (H) CTC counts by NSCLC stage. (I) Percentage of patients with CTCs > 1/2 mL blood by NSCLC stage. Reproduced from ref. 156 under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by-nc-nd/4.0/). | ||
The well-matched hepatocellular carcinoma (HCC) cell lines MHCC97-L with low metastatic potential and HCCLM9 with high metastatic potential were used to find molecular probes for biomedical applications. Using MHCC97-L as subtractive cells and HCCLM9 as target cells, the scientists created aptamers that targeted HCCLM9 surface molecules but not MHCC97-L. The chosen aptamers’ binding affinity and specificity were tested by flow cytometry. The candidate DNA-aptamers were coupled to biotin to create unique bioprobes for cultured cells, animal models, and human HCC tissues. HCC cells were captured by magnetic particles and biotin-conjugated aptamers in peripheral blood-like conditions. In a whole live cells-SELEX method, aptamer LY-1 identified highly metastatic human HCC cells. They proved sensitive and specific as molecular probes. The aptamer LY-1-QDs conjugates could detect highly metastatic HCC cells in liver and lung tissue sections and clinical samples when conjugated with fluorescent QDs. The aptamer LY-1-magnetic particle conjugates isolated and identified metastatic HCC cells from whole blood. This suggests that aptamer LY-1 may be an excellent molecular probe for early HCC metastatic prediction.157
5.2. Protein thiols detection
An increase in oxidative stress often accompanies malignant cells’ aberrant growth and reproduction. Reactive oxygen species (ROS), which are crucial for preserving cellular homeostasis and redox signaling, are created as a consequence of cellular metabolism. However, unlike normal cells, cancer cells can utilize ROS for signaling pathways that promote their survival. Overproduction of ROS may still harm cellular constituents, including DNA, proteins, and lipids, in addition to causing DNA mutations and genomic instability.158 Thiol-containing amino acids, such as glutathione (GSH), cysteine (Cys), and homocysteine (Hcy), control the redox equilibrium in cancer cells.159 These amino acids function as antioxidants and prevent oxidative damage by scavenging ROS. Fig. 8 depicts the complex biochemical reactions involved in amino acid metabolism, highlighting the interplay between different amino acids and their derivatives, as well as enzymes and cofactors involved in these reactions.160 Notably, cancer cells often have greater access to thiol-containing amino acids than normal cells. This difference and the ability to cleave disulfide bonds (–S–S–) create a crucial mechanism for maintaining redox balance and persisting in a highly oxidizing environment.161,162 Disulfide bond cleavage by free thiols regenerates reduced forms like GSH and Cys, further aiding in ROS detoxification. This process is essential for the longevity of cancer cells because it protects them from oxidative stress-induced cell demise. The intricate processes involved in producing, eliminating, and signaling ROS are further illustrated in Fig. 9.163 In conclusion, the preferential availability of thiol-containing amino acids and the ability to cleave disulfide bonds are crucial mechanisms that allow cancer cells to maintain redox balance and survive. Targeting these mechanisms offers a promising avenue for developing novel anticancer drugs with potentially higher specificity and fewer side effects than traditional therapies.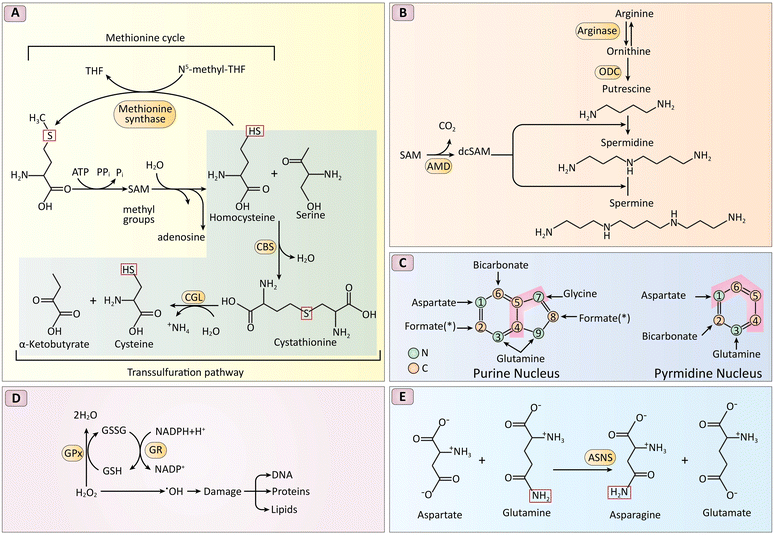 | ||
| Fig. 8 Biochemical pathways crucial for cellular metabolism. It includes the methionine and transsulfuration pathways transforming methionine into Cys, polyamine synthesis from arginine, purine and pyrimidine synthesis utilizing nitrogen and carbon donors, the role of antioxidants like NADPH and GSH in reducing ROS, and the function of ASNS in asparagine synthesis. Enzymes critical to these processes are highlighted, illustrating the complex interactions and conversions that sustain vital cellular functions. (A) Reverse-transsulfuration pathway: Cysteine is synthesized from methionine. Key enzymes (CBS, CGL) are highlighted in red. (B) Polyamine synthesis: Polyamines are synthesized from arginine and require SAM (highlighted enzyme: ODC, AMD). (C) Nitrogen and carbon source for nucleic acids: Aspartate, glycine, and glutamine provide precursors for purine and pyrimidine biosynthesis (C – yellow, N – green). (D) GSH and NADPH as antioxidants: GSH and NADPH neutralize reactive oxygen species (ROS) via enzymes GPX and GR (highlighted in red). (E) Amidation reaction for asparagine synthesis: Asparagine synthetase (ASNS, red circle) catalyzes asparagine formation. The conserved amide nitrogen is boxed in red. Reproduced from ref. 160 under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/). | ||
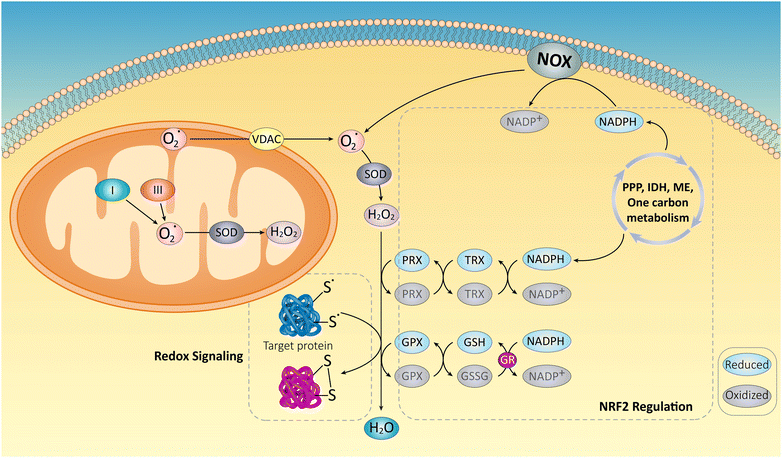 | ||
| Fig. 9 Superoxide (O2˙) is essentially created by mitochondria and NADPH oxidases (NOXs) and is then converted to hydrogen peroxide (H2O2) by superoxide dismutases (SODs). By oxidizing the thiols contained in redox-regulated proteins, the resulting H2O2 may either be transformed to water by antioxidant systems mainly composed of NRF2-regulated enzymes, or it can participate in cellular signaling. Thioredoxins (TRX), NADPH-dependent peroxiredoxins (PRX), GSHs, and glutathione peroxidases (GPX), use a complex network of metabolic pathways and enzymes, including the pentose phosphate route, isocitrate dehydrogenases, malic enzymes, and one-carbon metabolism, as their energy source. Additionally, NOXs that make ROS need NADPH as a substrate, highlighting the crucial functions that both ROS producers and antioxidant systems play in biological processes. Reproduced from ref. 163 under the terms of the Creative Commons CC-BY license with permission from [Elsevier], copyright [2018]. | ||
Recent advancements in fluorescent sensors offer promising tools for monitoring intracellular thiols in living organisms, particularly cancer cells. Jung et al.164 developed a biotin–disulfide–coumarin conjugate that enables real-time monitoring of reduced thiols like GSH, Cys, and Hcy within cells. This approach addresses limitations of earlier probes, such as poor water solubility, high background signals, and restricted applicability in biological systems. The conjugate's design incorporates a disulfide bond that undergoes cleavage upon encountering intracellular thiols. This cleavage triggers the release of a biotinylated coumarin unit, leading to a significant increase in fluorescence intensity. The researchers confirmed that biologically relevant thiols effectively induced this response in cell imaging experiments. Furthermore, the study demonstrated successful cellular uptake of the probe via receptor-mediated endocytosis following thiol-mediated disulfide bond cleavage. This uptake enhanced fluorescence within the cells, particularly in the endoplasmic reticulum (ER) and mitochondria, where disulfide bonds are abundant. These findings demonstrate the potential of this approach for in vivo detection of intracellular thiols like GSH, Cys, and Hcy.
Building on the concept of thiol-mediated activation, another study explored the ranostic prodrug design for targeted delivery and monitoring of the potent anticancer drug SN-38.165,166 This design utilizes a self-immolating linker containing a disulfide bond. The high concentration of intracellular thiols, particularly GSH, within cancer cells, is expected to trigger the cleavage of this linker. This cleavage event would release a fluorophore (N-biotinylated piperazinerhodol) and activate the therapeutic SN-38 molecule. The presence of biotin in both the precursor and the final prodrug allows for potential targeted delivery to cancer cells, which express abundant biotin receptors. This targeted uptake would concentrate the prodrug within cancer cells, maximizing therapeutic effect. The key to this approach lies in the cleavable disulfide bond. This bond breaks apart within the thiol-rich environment of cancer cells, releasing the active SN-38 and the N-biotinylated piperazinerhodol, which acts as a fluorescent reporter for monitoring drug release. The recovered fluorescence signal of the reporter molecule at its peak emission wavelength can be used to track the release of SN-38 within the cells.167 In conclusion, the preferential availability of thiol-containing amino acids and the ability to cleave disulfide bonds are crucial mechanisms for cancer cells to maintain redox balance and survive. Targeting these mechanisms offers a promising avenue for developing novel anticancer drugs with potentially higher specificity and fewer side effects compared to traditional therapies. Recent advancements in fluorescent sensors and theranostic prodrugs that exploit the reducing environment of cancer cells provide valuable tools for researchers to monitor and deliver therapeutic agents more effectively.168
Thiols perform various biological tasks, such as signaling, antioxidant defense, and cell development. GSH, the most abundant thiolated tripeptide in human cells, has been implicated in several diseases, including liver diseases169 and cancers.170 Accurately measuring intracellular thiol levels is crucial for disease progression assessment, early diagnosis, and treatment efficacy evaluation. Researchers have focused on developing bio-probes to visualize and quantify intracellular thiols in this context. A recent study171 described a new cell-specific light-up probe designed to target integrin αvβ3, a protein receptor overexpressed in some cancers. While traditional thiol detection methods often utilize fluorescence sensors and thiol-addition reactions with electrophiles, these probes usually suffer from limitations such as poor water solubility and high background signals due to aggregation.134 The newly developed probe addresses these challenges by incorporating a disulfide bond that selectively reacts with intracellular thiols. The researchers demonstrated that this probe exhibits a significant increase in fluorescence upon encountering GSH, indicating successful thiol-mediated cleavage of the disulfide bond. This finding highlights the potential of such bio-probes for non-invasive, high signal-to-noise ratio imaging of intracellular thiol levels, paving the way for improved diagnosis and treatment strategies for various diseases.
Fluorescence-based methods have gained popularity for biothiol detection due to their non-invasive and sensitive nature. However, traditional probes often suffer from limitations in both sensitivity and selectivity, leading to background noise and inaccurate measurements. Another study135 introduced a novel design concept called DQ Probe 1 (Dual-Quenching Probe 1) to address these limitations. This innovative probe incorporates both dual-quenching and dual-reactive groups on the fluorophore molecule. Traditional mono-quenching probes (MQ-probes), like Probe 2 mentioned in the original text (details likely limited), typically possess a single quenching group and a reactive group for thiol interaction. DQ Probe 1 builds upon this concept by combining two MQ-probes, resulting in a more focused response upon thiol detection due to its dual-quenching mechanism. This dual-quenching mechanism likely involves photoinduced electron transfer and intramolecular charge transfer (ICT). In photoinduced electron transfer, an excited fluorophore transfers an electron to a nearby quencher, reducing fluorescence. ICT involves electron density movement within the molecule, further suppressing fluorescence. Both mechanisms keep the probe in an “off” state until a specific thiol reaction disrupts them, triggering a fluorescence turn-on signal for thiol detection. Experiments demonstrated DQ Probe 1's exceptional sensitivity, detecting Cys concentrations as low as 20 nM. This superior sensitivity is attributed to the dual-quenching mechanism requiring specific interactions for activation. Additionally, DQ Probe 1 displayed superior selectivity compared to earlier probes, differentiating between various amino acids and responding primarily to thiols. Finally, the probe's good cell permeability allows it to effectively target and detect intracellular thiols within living cells. DQ Probe 1 represents a significant advancement in fluorescence-based thiol detection. Its innovative design with dual quenching and dual-reactive groups offers superior sensitivity and selectivity compared to traditional probes. This allows for more accurate and specific detection of crucial thiol molecules within complex biological environments, including living cells. These features make DQ Probe 1 a valuable tool for studying thiol dynamics and their potential roles in various diseases.
Researchers have developed novel biotin–PEG–gold nanoparticle (AuNP) probes for sensitive and specific detection of biomarkers, such as proteins and nucleic acids.172 These probes offer a promising approach for multiplexed analysis, potentially enabling the detection of several biomarkers from a single sample. As illustrated in Fig. 10A, the biotin-PEG linker plays a crucial role in this assay. Biotin facilitates target capture through its strong interaction with SA, while the PEG linker minimizes non-specific interactions between the probe and other biomolecules, enhancing assay specificity. The biotin-PEG linker's design significantly impacts the AuNP probes’ stability. The study compared probes constructed with monothiol, dithiol, and trithiol anchors (Fig. 10B–F). Experiments revealed that dithiol- and trithiol-linked AuNP probes exhibit superior colloidal stability compared to monothiol-linked ones. This enhanced stability is likely due to the formation of multiple covalent linkages between the linker and the AuNP surface, leading to a more robust structure. The assay utilizes a sandwich format, leveraging the high-affinity interaction between biotin and SA to capture and immobilize the target biomolecules. This approach enables sensitive detection of biotinylated targets with a limit of detection (LOD) as low as 50 fM for both nucleic acid targets and the prostate-specific Ag (PSA). Additionally, the high throughput nature of this assay makes it suitable for efficiently analyzing large numbers of samples. Future studies will explore the potential of this platform for the simultaneous detection of multiple biomarkers, allowing for more comprehensive disease diagnosis.
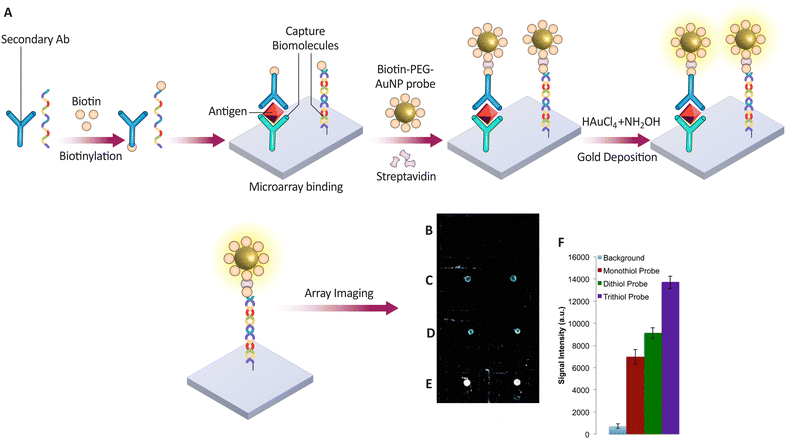 | ||
| Fig. 10 . (A) Method for capturing and detecting nucleic acids and proteins using biotin–PEG AuNP probes on a microarray. The biotinylated targets are amplified for detection after binding to the microarray via nucleic acid sequences or antibodies. (B–F) Comparison of the effectiveness of different PEG-AuNP probes—monothiol, dithiol, and trithiol—in identifying microRNAs, with signal intensities indicating the detection efficiency of each probe type. Reproduced from ref. 172 with permission from [American Chemical Society], copyright [2016]. | ||
In another study, a novel probe named Ac was introduced, which was designed for the specific and sensitive detection of cancer cells. This innovative probe utilizes a fluorescein derivative as the fluorophore, enabling fluorescence-based detection. The efficacy of Ac is attributed to several key features it incorporates. Biotin serves as a targeting moiety in Ac, specifically binding to receptors that are overexpressed on the surface of cancer cells. This targeted approach enhances the selectivity of the probe towards malignant cells. The probe exhibits excellent biocompatibility and water solubility, which allows for its safe and efficient application in biological systems. Ac demonstrates high selectivity for Cys, a biothiol abundantly present in cells, with a low detection threshold of 307 nM. Upon binding to Cys residues on cancer cells, Ac triggers a turn-on response, resulting in increased fluorescence emission. This feature facilitates the clear visualization and identification of malignant cells. In cell line experiments, Ac successfully differentiated between healthy and cancerous cell lines, including HeLa, B16, RAW264.7, and NIH-3T3 cells. This finding underscores the potential of Ac as a valuable tool for early cancer detection and targeted therapeutic strategies. Developing such innovative probes paves the way for advancements in cancer diagnosis and treatment.173
5.3. Formaldehyde sensing
Formaldehyde (FA) is a compound that occurs naturally and is also a consequence of cellular metabolism.174 Reports have shown that occupational exposure to FA led to the occurrence of some cancers.175 Cancer detection and diagnosis require sensitive and selective FA sensors. Biotin-conjugated probes have been studied for FA sensing in cancer detection due to their high sensitivity and selectivity.176 Biotin can be conjugated to FA-reactive groups like hydrazines or amines to bind specifically to cancer cell FA. The FA-reactive group can be detected and quantified because biotin binds to SA or avidin. Biotin–hydrazine and biotin–amine probes have been developed for FA sensing in cancer detection. These tools have been used to detect cancer cell FA and assess chemotherapeutic response. Biotin-hydrazine probes are biotin-conjugated FA sensors for cancer detection. This probe forms a stable hydrazone bond with cancer cell FA, which SA-conjugated fluorescent or colorimetric probes can detect. The biotin-amine probe reacts with cancer cell FA to form a Schiff base detectable by SA-conjugated fluorescent or colorimetric probes.177Hydrazone formation and photo-induced electron transfer effect suppression were used to create a biotin-pendant-decorated naphthalimide-based FA sensor for cancer cells. After adding FA to a phosphate-buffered saline PBS-buffered aqueous sensor solution, absorbance at 428 nm increased slightly, and fluorescence enhancement at 541 nm increased 140-fold. In physiological conditions, the probe can detect FA with a dynamic range of 400 M FA in 1 M dye. After 20 minutes of pre-experiment FA (40 mM) incubation, biotin receptor-positive 4T-1 cells received probes 1 and 2. Probe 1 fluoresced more than probe 2 in one- and two-photon modes. Sensor 1's stronger fluorescence in biotin receptor-positive cells is crucial for tissue FA reporting. Probe 1 was tested for detecting endogenous FA concentrations in tumor tissues using two-photon excitation. Fluorescence intensity dropped dramatically after 20 minutes of incubation for probe 1. Probe 1 appears to only detect FA in tumor tissues. The researchers developed cancer-specific FA sensors to detect endogenous FA in cancer cells and tissues.178 These innovative platforms may help us understand FA's complex relationship with various diseases, particularly cancer development. Clinical use of these platforms may improve patient early detection, prognoses, and treatment options. Continued research and development of biotin-conjugated platforms is a crucial step toward better managing and preventing FA-related diseases using cutting-edge diagnostic tools and therapies.
5.4. Lysosome-specific biotin-conjugated probes
Lysosomes are vital for cell degradation and recycling. Dysregulated lysosomal function can cause cancer.179,180 Thus, lysosome-specific assays are desired to study cancer cell lysosomal function and identify therapeutic targets. Biotin-conjugated probes provide effective lysosomal function analysis in living cells. Targeting lysosomal proteins or lipids allows selective labeling and live imaging of cancer cell lysosomes.180 Targeting lysosomal pH with fluorescent probes has been widely used to study lysosomal function and physiology. These instruments usually use moderately basic lysosomotropic compounds that can bind to the lysosome and disrupt cell viability. These new probes aim to achieve similar sensitivity and accessibility while minimizing lysosomal dysfunction.181The first two-photon fluorescent probe with tumor-targeting and lysosome-specific capabilities, BN-lys, was created to assess live cell pH changes. In this work, malignancies were targeted with biotin, and fluorescence was controlled by morpholine as the pH site and lysosome-specific group using photoinduced electron transfer. Under the direction of the biotin group, BN-lys demonstrated robust fluorescence responses in cancer cells as opposed to modest fluorescence in normal cells. The instrument showed how well chloroquine changed the pH of lysosomes in natural cells. This probe comprises a tumor-targeting unit, a fluorophore with two photons, and a lysosome-specific group. The fluorophore serves as a fluorescence signal reporter. BN-lys has shown greater affinity to cancer cells as a two-photon fluorescence probe that can measure lysosomal pH in live cancer cells.182
5.5. Sensing vitamin-targeted polymers within cancers
Recently, sophisticated vitamin-targeted polymers have been created as sensors to find cancer cells. Based on cancer cells’ increased expression of vitamin receptors, these polymers have been designed to adhere to them alone. Including biotin molecules in these polymers is essential for enhancing the selective binding process.183 Biotin conjugates enable the targeted administration of medicinal or imaging substances to cancer cells by attaching biotin to the polymer and preferentially binding to cancer cells that need additional biotin.184 It has been reported that biotin-linked polymers could increase the contrast and selectivity of cancer imaging in vivo and the efficiency of medication administration to tumor locations. These polymers are excellent candidates for further clinical research because of their superior pharmacokinetics and safety characteristics.185Solid cancer cells overexpress biotin receptors, which are essential for tumor growth, metabolism, and metastasis. Researchers targeted solid cancer cells that overexpress biotin receptors with PG4.5 dendrimer NPs and biotin to improve chemotherapy efficacy and reduce side effects (Fig. 11). As determined by spectroscopy, the NPs have a spherical shape, 81.6 ± 6.08 nm size, and 0.47 ± 1.25 mV zeta potential. NPs had a maximum drug dosage of 10.84 ± 0.16% and an encapsulation efficiency of 47.01 ± 0.71. At pH values of 6.5 and 5, the NPs released 60.54 ± 1.99% and 73.96 ± 1.14% of gemcitabine (GEM). In vitro tests showed that the NPs selectively targeted HeLa cancer cells, reducing cell viability and apoptosis. This study found that biotin-coupled PG4.5-DETA nanovehicles selectively deliver GEM to tumor cells. The study suggests using vitamin-targeted NPs for tumor-specific drug delivery, with further research into an in vivo delivery system.186
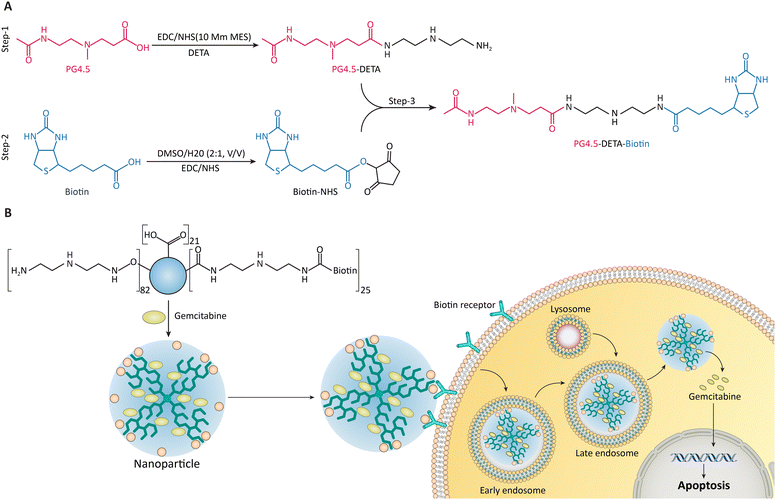 | ||
| Fig. 11 (A) Schematic representation of PG4.5-DETA and PG4.5-DETA-biotin synthesis. (B) This diagram demonstrates the delivery mechanism of GEMNPs via biotin receptor-mediated endocytosis, leading to the release of GEM inside HeLa cells, inducing apoptosis. Reproduced from ref. 186 under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/). | ||
5.6. Sensing huwentoxin-I
The peptide toxin Huwentoxin-I (HWTX-I) in spider venom is antimicrobial, neurotoxic, analgesic, cytotoxic, necrotizing, and hemagglutinating.187 Several peptide poisons in arachnid venom may treat cancer.188 These toxins control the cell cycle, activate caspase, and deactivate mitochondria, killing cancer cells. Spider venom peptides with an inhibitor cystine knot (ICK) motif can target ion channels and other pain-related targets, providing neuroprotective, antimicrobial, anticancer, and analgesic effects.189 Spider venom biotoxins have been shown to damage tumor cell membranes, inhibit proliferation, and induce apoptosis, suggesting antitumor potential. Psalmotoxin 1 (PcTx1), a spider peptide toxin, inhibits cation currents in malignant astroglioma cells and freshly removed glioblastoma (GBM) cells without affecting normal human astrocytes, making it a promising candidate for improving GBM patients’ prognoses with epileptic seizures.190 Other spider venom toxins, like those from the Macrothele raven, can induce apoptosis and necrosis in various tumor cells, inhibit DNA synthesis, affect cell viability, and stop the cell cycle. Spider peptides may inhibit cancer by regulating intracellular osmotic pressure and downstream signal molecules.191 Drug discovery and research use biotinylated HWTX-I, a peptidic neurotoxin. Biotinylation of HWTX-I isolates and identifies target proteins, which can help develop new drugs. It can also be used to study HWTX-I-receptor protein interactions and ion channel modulation, particularly voltage-gated calcium channels. Biotinylated HWTX-I can be used as a molecular probe to study its structure, function, and localization in cells and tissues, making it a useful neurobiology tool.61Yan et al.192 optimized a biotin labeling method for the spider toxin HWTX-I, achieving over 50% labeling without compromising its bioactivity. This enabled them to identify receptor proteins in nerve synapses using affinity purification. This method offers advantages over traditional techniques like voltage clamps in identifying toxin-binding proteins.
In another study,193 researchers employed biotin-tagged HWTX-IV to identify its interacting proteins. They confirmed that monobiotinylated HWTX-IV retained bioactivity and used various techniques to pinpoint interacting membrane proteins. This approach provides valuable insights into HWTX-IV's mechanism of action and a useful methodology for future research.
5.7. Detection of anti-human antibodies
Detecting anti-human antibodies is crucial in numerous diagnostic and research applications. A competitive immunoassay with a biotin probe is one method for detecting anti-human antibodies. In this procedure, the anti-human Ab sample is incubated with a biotinylated Ag and anti-biotin Ab solution. The MNPs conjugated with anti-biotin Ab are then separated using an external magnet, and the supernatant is removed. The particles are then rinsed twice with buffer 1 M PBS.194 Anti-biotin antibodies can enrich biotinylated peptides within complex peptide mixtures.195In situ hybridization can utilize biotinylated probes, whose signal can be amplified through the use of anti-biotin antibodies.196Using self-assembly techniques on gold, researchers created a high-throughput (HTP) protein chip platform to test the viability of protein sensors for detecting blood antibodies. Biotinylated single-layer structures were used to immobilize densely packed SA surfaces, which were then used to identify serum IgM antibodies in Lyme borreliosis patients. Using biotinylated peptide AAOspC8 test probes and blood samples as small as 1 L/spot and a high signal-to-noise ratio, the scientists were able to generate highly specific data on protein interactions. The biochip test requires only 1 L of reagent, compared to 2 L for ELISA. Self-assembled monolayers on gold made it possible to evaluate surface properties with optical, mechanical, and electrochemical instruments (Fig. 12).197 Using protein microarrays, it is anticipated that HTP protein interaction studies for disease diagnosis will be conducted frequently in the near future.
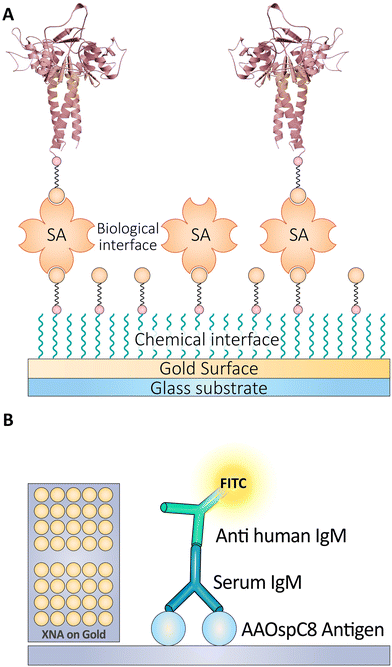 | ||
| Fig. 12 (A) This panel illustrates the immobilization of biotinylated probes on a sensor chip through biotin–SA binding for high-density detection. (B) Schematic depicting the use of FITC-labeled anti-human IgM antibodies in detecting the presence of Lyme disease Ag in a sandwich assay format. Reproduced from ref. 197 with permission from [American Society of Chemistry], copyright [2016]. | ||
Ultra-sensitive detection of molecules in biological samples can be made using nanohybrid materials. In a study, researchers used ZnO@Au nanocomposites and Biotin-SA as a molecular connector to bind antibodies and signal molecules. This resulted in forming a composite material consisting of gold nanorods (GNRs)-SA-biotinylated secondary antibodies (Biotin-Ab2), increasing the refractive index and permitting more signaling molecules. Thus, the surface plasmon resonance (SPR) signal intensity in biosensors was increased.198
Recent research employed the conventional sandwich technique with biotin-SA and a sensitive SPR biosensor based on ZnO@Au NPs to identify human immunoglobulin G (hIgG) (Fig. 13). Nano-zinc oxide (ZnO) was combined with gold-coated ZnO nanocrystals to construct a sensing substrate on a mercaptan-treated gold film. This enhanced the biocompatibility and optical properties of ZnO and its load capacity. Using the biotin–avidin technique, the SPR signal was improved. Under optimal experimental conditions, the secondary gold–SA–biotinylated antibodies (GSAB-Ab2)-conjugate SPR biosensor was able to detect hIgG in the range of 0.0375–40 g mL−1 with a minimum detection concentration approximately 67-fold lower than a conventional gold-plated SPR sensor. The sensitivity of SPR biosensors has increased within a specific range.199
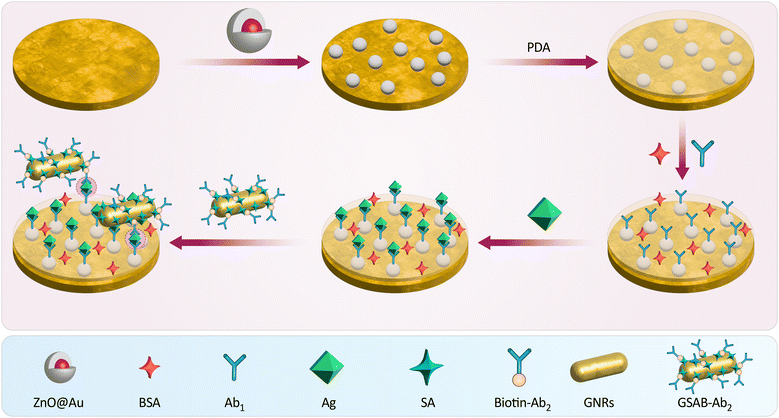 | ||
| Fig. 13 The scheme illustrates the assembly of a SPR biosensor, starting with a ZnO@Au nanocomposite base layer. Bovine serum albumin (BSA) and primary antibodies (Ab1) are applied to capture Ag, followed by SA and biotin–Ab2 to enhance the signal. GNRs and GSAB-Ab2 are then introduced for signal amplification, creating a sensitive, sandwich-type biosensor for detecting hIgG. Reproduced from ref. 199 with permission from [Elsevier], copyright [2022]. | ||
Although the gold standard measurement is an electrochemiluminescence (ECL) dot blot test based on a chicken anti-human thymidine kinase 1 (hTK1) IgY polyclonal Ab on a biotin-SA platform, measuring the amount of STK1p in serum enables accurate detection of early tumor progression. In addition to operator talent, the diversity of chicken antibodies can affect STK1p levels. Researchers have developed a fully automated sandwich-BSA technology to overcome these obstacles based on (hTK1)-IgY-rmAb#5, a stable recombinant chicken IgY monoclonal antibody (mAb). By immunizing hens with a 31-peptide sequence of hTK1, the researchers compiled a library of phage display SCFvs. Using hTK1 calibrators, this recombinant mAb demonstrated efficacy based on its high affinity and high sensitivity. As a result, it was highly accurate across multiple cohorts and extremely specific at low or elevated STK1p levels of 0.92 to 0.963 (Fig. 14).200
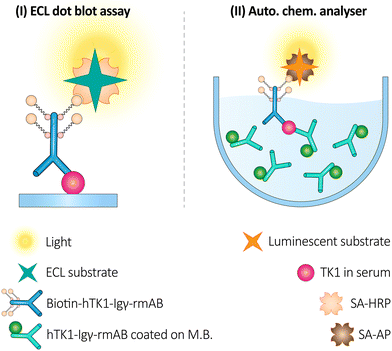 | ||
| Fig. 14 The scheme illustrates two assays for detecting hTK1 using recombinant monoclonal antibodies (rmAb). The first panel (I) shows an ECL dot blot assay for hTK1 identification using hTK1-IgY-rmAb coated on MBs. The second panel (II) depicts an automated chemiluminescence analyzer using the same antibodies for hTK1 detection in serum. These methods provide sensitive detection of hTK1, a potential biomarker for cell proliferation. Reproduced from ref. 200 under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/). | ||
5.8. Tumor marker detection
Multi-tumor marker monitoring is a promising method for detecting tumor Ag, which is crucial for cancer diagnosis and disease indicators. Several serum biomarkers have been linked to cancer diagnosis. These biomarkers are valuable for diagnosing and treating various cancers. Biomarkers such as alpha-fetoprotein (AFP), Epidermal growth factor receptor (EGFR), prostate-specific antigen (PSA), carcinoembryonic Ag (CEA), cancer Ag 125 (CA 125), cancer Ag 15-3 (CA 15-3), Matrix Metalloproteinase-1 (MMP-1), HER2, cytokeratin protein fragment 21-1 (CYFRA21-1), cancer Ag 19-9 (CA 19-9), beta-human chorionic gonadotropin (β-hCG), Serum pepsinogens (PGI/PGII), and Neuron-specific enolase (NSE) are notable. These markers indicate malignancy and progression. To provide quality care, medical staff must understand these markers and their role in cancer diagnosis.201 An electrochemical immunosensor is a sensitive and accurate tumor marker detection and analysis platform. Both unlabeled and labeled bedside immunosensors can analyze tumor Ag. Non-labeled immunosensors are used for quantitative Ag sensing, while labeled ones use sensitively detectable labels.202 An unlabeled immunosensor's electrochemical efficacy was improved by adding AuNPs to an electrode. Nanostructured electrode arrays and multi-label immunocomplexes improve unlabeled voltametric immunosensor sensitivity. Sandwich assay detects tumor Ag by sandwiching them between detection and tracer antibodies. Novel nanocomposites for tumor marker detection have been developed alongside NM research. Biomedical research uses antibodies to functionalize NPs. Electrostatic, direct covalent, or molecular interactions like SA-biotin make it possible to attach antibodies to NPs. Researchers used biotin–SA quantum-dot tagging to label extracellular vesicles for rigorous and repeatable immunophenotypic evaluation fluorescently. Indirect biotin labeling with NP allows regulated Ab bioconjugation, which may alter target binding site orientation and reduce Ab reactivity.203,204In addition, click chemistry, biotin–SA–biotin sandwich techniques, and Ag–Ab interactions created AFP-detecting fluorescent immunosensors. Three functionalized glasses were biotinylated with various click chemistry analogs after immobilizing anti-AFP antibodies with biotin, SA, and biotin sandwiched together. This study compared biotin functionalization and click-chemistry immobilization methods for six AFP microarray sensor fabrication methods. Two functionalization methods improved microarray sensor performance: epoxy–silane immobilization with biotin–amine and thiol–silane with biotin–maleimide. The array's 9.8 ± 2.9 g mL−1 sensitivity made it a fast and affordable screening sensor compared to more sensitive methods. Sandwiching a second biotinylated Ab with a fluorescently tagged SA allows label-free detection (Fig. 15). New antifouling and wettability surfaces should boost sensitivity. The study shows that binding chemistry can make sensitive protein biomarker sensors.210
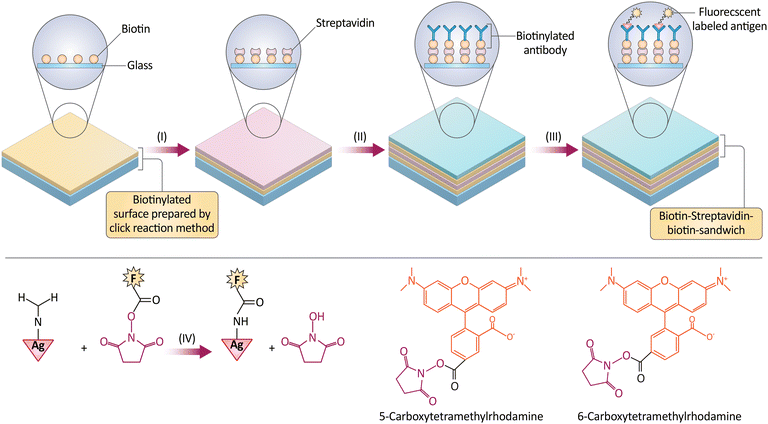 | ||
| Fig. 15 Representation of the process to create an AFP microarray. The method involves incubating SA with a biotinylated surface, attaching biotinylated anti-AFP antibodies, and reacting with NHS-rhodamine to produce fluorescently labeled AFP. Fluorescence intensity is measured to assess the microarray's sensitivity at various AFP concentrations. The microarrays are produced in a controlled humidity environment and imaged after incubation to confirm pattern integrity. Reproduced from ref. 210 under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/). | ||
A photoelectrochemical (PEC) sensor's anodic photocurrent is proportional to solution-solubilized electron donor concentration within a specific range. The multilayer of insulating protein blocks electrons from the solution's electron source from reaching the electrode surface, reducing photocurrent. Chen, Jiexia, and Guang-Chao Zhao developed encapsulated electron donors. Electrode electron donors can be created and released by enzymatic digestion using this method. Photocurrent and a signal-on PEC immunosensor improve AFP detection selectivity and sensitivity. Bio-APOAA and CdSe QDs were the amplification and photoactive units, respectively. Physicians can detect tumor markers like AFP with its linear range of 0.001 to 1000 ng mL−1 and low LOD of 0.31 pg mL−1. Clinical laboratories can use the immunosensor to detect AFP and other tumor markers before cancer screening or monitoring. The PEC immunosensor can detect novel tumor markers, and this research detects AFP sensitively and accurately. It also detects trypsin activity and inhibitors.211
Identifying alpha-fetoprotein-L3 (AFP-L3) is crucial for diagnosing HCC, but current techniques are hampered by low sensitivity and complex procedures. To resolve these issues, scientists have developed a straightforward and highly sensitive method for detecting AFP-L3, which is essential for diagnosing HCC. This novel technique employs biotinylated Lens culinaris agglutinin-linked silver NPs (B-LCA-AgNPs) to circumvent the insensitivity and complication of existing methods. The specific bond between Lens culinaris agglutinin and AFP-L3 enables direct AFP-L3 detection via the electrochemical signal output of AgNPs, circumventing the distinct processes typically required in clinical contexts. Following the recognition process between B-LCA-AgNPs and AFP-L3, avidin–biotin interactions accumulate many AgNPs at the binding site, amplifying the signal and enabling highly sensitive AFP-L3 detection. This novel method has a lower detection threshold (12 pg mL−1) and a stronger linear correlation (25–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pg mL−1) than previous techniques. In addition, it exhibits outstanding stability and consistency when analyzing AFP-L3 in human serum samples, making it a promising diagnostic instrument for clinical use.212
000 pg mL−1) than previous techniques. In addition, it exhibits outstanding stability and consistency when analyzing AFP-L3 in human serum samples, making it a promising diagnostic instrument for clinical use.212
The exceptionally malignant tumor known as HCC exhibits rapid growth.213 AFP is the most significant biomarker for HCC. Due to the unpredictability and instability of antibodies, Ab-based immunoassays have limitations. In recent research, aptamer was used instead of immunoassay to recognize AFP specifically. Aptamer-functionalized magnetic NPs (Ap-MNPs) were created by attaching the AFP-specific ssDNA aptamer to MNPs (Fe3O4@SiO2) via the avidin–biotin interaction. Ap-MNPs showed a high degree of targeting specificity. Ap-MNPs and HPLC were used to develop a label-free method for detecting AFP in blood. This technique demonstrates linearity between 1 g mL−1 and 50 g mL−1 with a correlation coefficient of 0.99999 and a LOD of 0.27 g mL−1. Ap-MNPs were revealed to be less effective than IgG, human serum albumin (HSA), and FIB.214
EGFR was detected by a sandwich electrochemical aptamer/Ab immunosensor with high sensitivity and specificity in a second experiment. Biotinylated anti-human EGFR Apts were immobilized on SA-coated MBs as capture probes and Abs as signaling probes. Apt-EGFR-Ab sandwiched between MBs was tested for EGFR complexation using AuNPs in HCl and differential pulse voltammetry (DPV). The immunosensor had a dynamic concentration range of 1 to 40 ng mL−1, a low LOD of 50 pg mL−1, and a relative standard deviation (RSD) of less than 4.2% under optimal conditions (Fig. 16). Magnetic particles separated samples quickly and precisely. This study shows immunosensors assess chemotherapy efficacy in breast cancer samples.218
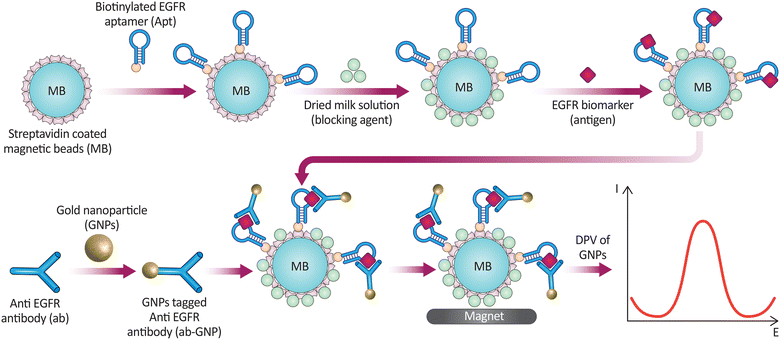 | ||
| Fig. 16 The main steps of the protocol for an assay to detect the EGFR biomarker using SA-coated MBs and AuNPs. The process involves attaching a biotinylated EGFR aptamer to the beads, blocking with dried milk, binding the EGFR Ag, and linking AuNP-tagged anti-EGFR antibodies to form a sandwich complex. This complex is isolated with a magnet, and the presence of EGFR is quantified using the DPV of the AuNPs. The method allows for sensitive detection of EGFR in varying concentrations, demonstrated through DPV measurements. Reproduced from ref. 218 with permission from [Elsevier], copyright [2015]. | ||
Scientists developed an automated DNA modification detection method. With the right treatments, this cutting-edge method can identify somatic EGFR gene mutations and treat NSCLC. BacMPs from Magnetospirillum magneticum AMB-1 were linked to SA to determine the minimum tumor cell count, speeding up the procedure and eliminating the laborious task of removing normal cells. The target PCR products from these BacMPs were biotin-labeled. The detection process integrated target PCR results with markers and fluorescent signals. An XYZ movable hand, 96-way automatic pipetter, solution dispenser, and fluorescent reader were used for novel automatic processing. Clinical lung cancer EGFR gene testing is possible with this versatile method. In less than 3.5 hours, it can detect in-frame deletions and point substitutions in the EGFR gene, even at low mutation rates.219
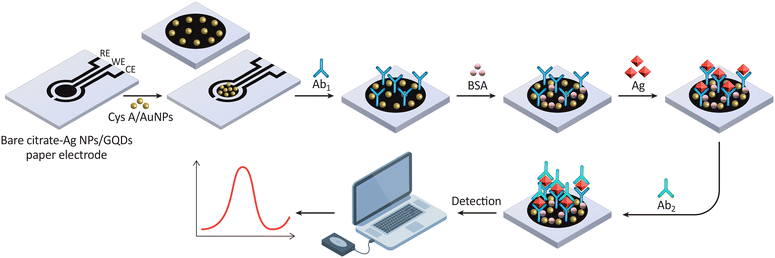 | ||
| Fig. 17 This illustration shows the steps for fabricating a paper-based electrochemical biosensor. It begins with a bare electrode modified with AgNPs and GQDs, followed by Ab immobilization, blocking with BSA to prevent non-specific binding, Ag capture, and finally, detection with a secondary antibody (Ab2), leading to signal generation for analysis. Reproduced from ref. 222. with permission from [Elsevier], copyright [2020]. | ||
Recent advancements in ECL have led to the development of more sensitive magnetic microbiosensors. Improvements in the distribution and enrichment of sandwich MBs have been achieved through double magnetic field actuation. Innovations include the use of circular-disc magnets, diamagnetic components, and external magnet actuation to preconcentrate MBs. These enhancements have significantly increased the sensitivity of ECL biosensors, demonstrating their potential to detect cancer biomarkers and exosomes with superior efficiency.223
Using redox probe tag identification technology, immunosensors were developed to detect four Ag, including CEA simultaneously. To immobilize capture antibodies on an electrode surface, a hybrid graphene/gold coating was co-deposited. Additional signal markers were added to the immunosensor in order to increase its sensitivity by detecting Ab bioconjugates via hybridization chain reaction (HCR) and biotin/SA methods. The signal was enhanced and amplified using AuNPs. The novel immunosensor detects AFP, CEA, CA 125, and PSA biomarkers within specific concentration ranges with greater sensitivity than comparable devices. The immunosensor demonstrated linear correlations between 0.2 and 800 pg mL−1 for AFP, CEA, CA 125, and PSA, with respective LODs of 62, 48, 77, and 60 fg M−1.225
Innovative techniques include a single-particle, inductively coupled plasma mass spectroscopy (ICP-MS)-based magnetic immunoassay for the simultaneous detection of CYFRA21-1, CEA, as well as CA 15-3. Antibodies were labeled with AuNPs, ZnSe QDs, and AgNPs so that single-particle ICP-MS would be sensitive enough to identify the target Ag. Biomarkers were effectively collected using MNPs coated with biotinylated antibodies (Fig. 18). With LODs of 0.02 ng mL−1, 0.006 ng mL−1, and 0.25 mU mL−1, CYFRA21-1, CEA, and CA 15-3 exhibited linear concentration ranges of 0.05–50 ng mL−1, 0.02–100 ng mL−1, and 0.6–250 mU mL−1, respectively. Blood samples from patients with lung cancer revealed simultaneous detection of all three biomarkers, with recovery rates ranging from 89.8 to 101.0%. The biomarkers’ RSDs were 3.2%, 4.4%, and 4.4%, respectively, indicating their precision.226
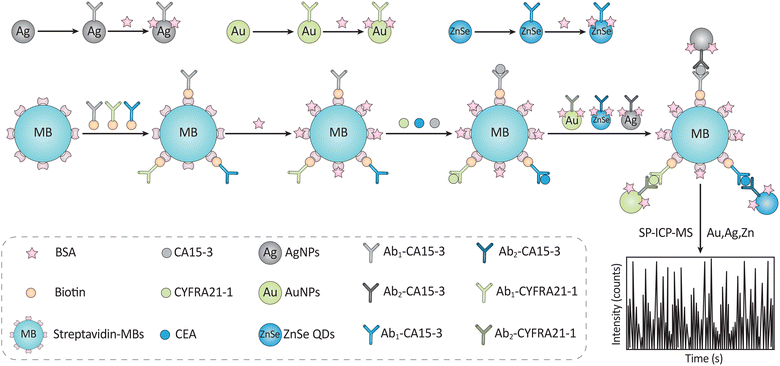 | ||
| Fig. 18 The serum of those with lung cancer may be analyzed using magnetic immunoassay and single molecule inductively coupled plasma mass spectrometry to identify three tumor biomarkers. To achieve this, a sandwich assay employing Ab-immobilized magnetic NP is used. Reproduced from ref. 226 with permission from [Elsevier], copyright [2020]. | ||
Recombinant DNA technology has combined the variable domain of a single-chain Ab with the core-SA domain in order to precisely deliver biotinylated therapeutic drugs to ovarian cancer cells. The bifunctional fusion protein (bfFp) was synthesized in Escherichia coli using the T7 expression method, then isolated and purified using an immobilized metal chelate affinity chromatography (IMAC) column. Confocal laser scanning microscopy (CLSM), ELISA, and western blotting all demonstrated that bfFp binds to biotin and anti-CA 125, respectively. CLSM was utilized to verify the specificity and bifunctional activity of bfFp's binding to OVCAR-3 cells, while ELISA and western blot analyses confirmed its specificity and bifunctional activity. bfFp increased the binding of biotinylated Ag and liposomes to OVCAR-3 cells as compared to control EMT6 cells lacking CA 125 expression. Using bfFp to target biotinylated medicinal medications, NPs, proteins, or liposomes may be feasible and effective for treating ovarian cancer patients. By administering the bfFp-therapeutic combination intraperitoneally, the most common site of metastasis may be effectively targeted.230,231
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106 U mL−1 was determined, with a linear range of CA 15-3 values from 50 to 15
106 U mL−1 was determined, with a linear range of CA 15-3 values from 50 to 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106 U mL−1. The designed immunosensor was more sensitive and stable than the industry-standard ELISA technique. As a result, it is advised to use the developed immunosensor for the clinical diagnosis of CA 15-3 and other cancer biomarkers to guarantee precise and fast patient quantification for efficient breast cancer treatment.234
106 U mL−1. The designed immunosensor was more sensitive and stable than the industry-standard ELISA technique. As a result, it is advised to use the developed immunosensor for the clinical diagnosis of CA 15-3 and other cancer biomarkers to guarantee precise and fast patient quantification for efficient breast cancer treatment.234
Different research used enzyme immunoassay with scanning electrochemical microscopy (SECM) to identify CA 15-3. First, two matching antibodies were used in a sandwich approach to concentrate and immobilize CA 15-3 on a flat substrate. While the second Ab was marked with HRP, the first Ab was collected on a substrate coated with SA. When H2Q and H2O2 were combined, HRP on the combination of CA 15-3 with the two antibodies catalyzed a process that changed H2Q into benzoquinone (BQ), an electroactive compound. The quantity of CA 15-3 that SECM measured and captured was in line with the reaction's produced reduction current of BQ. The researchers concentrated the Ab–Ag–Ab* combination on the substrate using a microcell to boost the method's sensitivity. The LOD for CA 15-3 determined by this approach was 2.5 U mL−1. In conclusion, CA 15-3 was concentrated and immobilized on a substrate, and its detection by SECM-enzyme immunoassay was accomplished by observing the reduction current of the electroactive product produced by the HRP-catalyzed reaction. The concentration of the Ab–Ag–Ab* combination in a microcell boosted the method's sensitivity.235
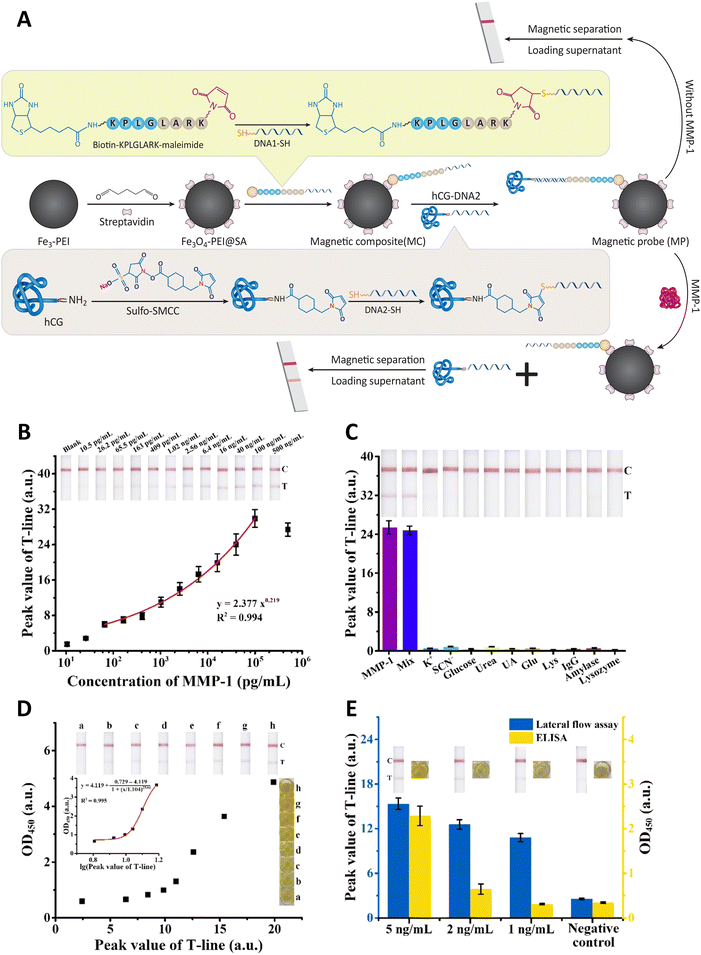 | ||
| Fig. 19 (A) To detect MMP-1, peptide-DNA, and hCG-DNA sensors are combined with pregnancy test strips. A diagram depicting the detection procedure is presented. (B) The relationship between the highest value of the T-line on a pregnancy test sheet and the MMP-1 levels is inverse. (C) The development of color findings to identify MMP-1 at an amount of 50 ng mL−1 and the outcomes for possible interfering substances at a concentration of 1 M are displayed. Using data points between 0 and 10 ng mL−1 MMP-1, the curve for the pregnancy strip test and ELISA detection signal is fitted. (D) The detection of MMP-1 in human saliva samples is reported using both pregnancy test strips and the ELISA method. Comparing points of data that range from 0 to 10 ng mL−1 MMP-1, the ELISA detecting signal and the fitting curve derived from pregnancy test strip results are examined. (E) The error bars in the MMP-1 detection results from human saliva specimens using the pregnancy test strip and ELISA techniques represent the standard deviation of trials conducted in triplicate. Reproduced from ref. 240 with permission from [American Chemical Society], copyright [2022]. | ||
Wang and colleagues have created a novel method for testing the HER2 tumor biomarker-binding capacity of peptides. Utilizing in situ single-bead sequencing on a microarray, this method proved successful in identifying two peptides with nanomolar affinity for HER2. Breast cancer, the most prevalent cancer among women, often exhibits overexpression of HER2. Peptides have significant advantages over antibodies as molecular probes for tumor detection and therapy monitoring owing to their small size, low immunogenicity, high penetration, biocompatibility, and simplicity of chemical modification. Two new octapeptides, H6 and H10, were found when the researchers used the OBOC combinatorial library technique to screen 7 × 105 candidates for affinity peptides. Several techniques, including cell culture, confocal fluorescence imaging, SPR imaging, and in vivo, fluorescence imaging, were used to establish the selectivity of these peptides towards HER2. These peptides showed HER2 with excellent affinity and specificity. In contrast to traditional QDs-only probes, these peptides might be turned into useful in vivo imaging probes when paired with NMs (Fig. 20). In conclusion, the effective in situ single-bead sequencing technique used in this work is a potential tool for peptide probe validation in live cancer cells and in vivo explant mouse models, and it may be helpful in breast cancer imaging and diagnostics.244
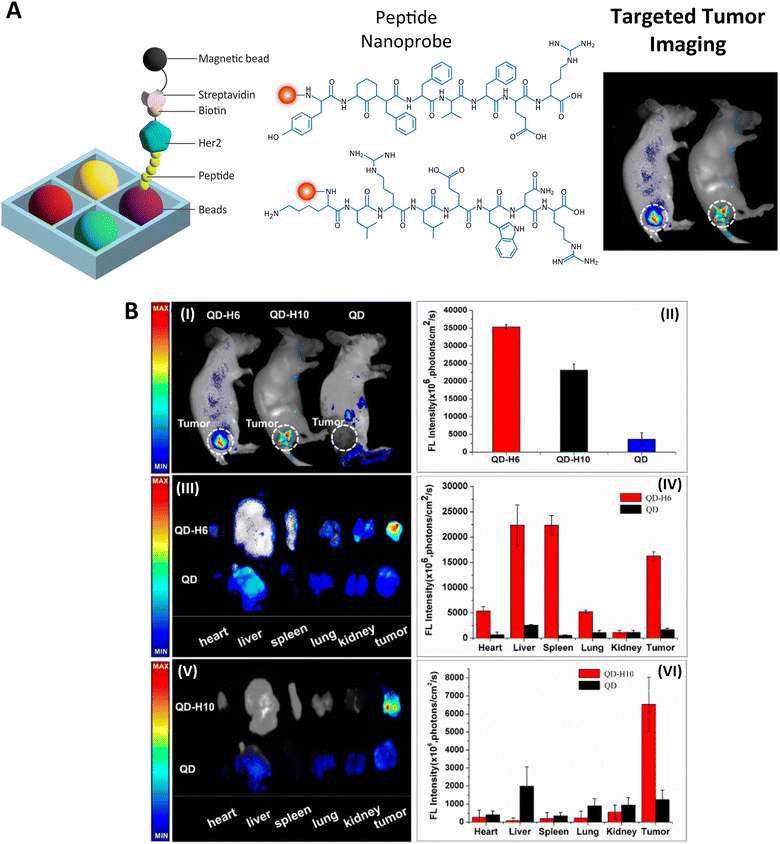 | ||
| Fig. 20 (A) A diagram showing two newly found peptides, H6 (YLFFVFER) and H10 (KLRLEWNR), which have a KD of 10–8 M and display a strong affinity for the HER2 tumor biomarker. These peptides were extracted using a screening approach from a library of 105 peptides. When conjugated to NPs in vivo, H6 and H10 probes predominantly accumulated in HER2-positive tumor tissues of xenografted mice. Using H6-QD and H10-QD probes, in vivo and ex vivo imaging of tumors in mice was performed. Imaging in vivo with fluorescence (I, II) revealed the accumulation of peptide probes in malignancies. Experiments involving biodistribution and ex vivo fluorescence imaging were conducted on tumor-bearing null mice that had been dissected. After quantifying the fluorescence signals from ex vivo experiments, the mean fluorescence intensity of three separate observations was calculated. Reproduced from ref. 244 with permission from [American Chemical Society], copyright [2015]. | ||
Another groundbreaking study proposes a biosensor that measures 12 tumor markers concurrently using a giant magnetoresistance (GMR) sensor chip (Fig. 21A), a microfluidic system, and magnetic nano-beads. This biosensor can rapidly and precisely identify tumor markers in blood samples using a double Ab sandwich immunoassay technique. It could be used for point-of-care testing (POCT) for the early identification of cancer. Additionally, a more effective method for biotin labeling is offered, expanding the clinical uses for the biosensor. These 12 tumor markers are AFP, CEA, CYFRA21-1, NSE, squamous cell carcinoma Ag (SCC), human pepsinogens I (PGI) and II (PGII), CA19-9, total PSA, free PSA, free-hCG, and thyroglobulin (Tg). Numerous cancer types, including lung, liver, digestive system, and prostate cancers, are associated with these indicators. The detecting platform is a GMR sensor chip comprising several GMR sensors that track changes in the magnetic field brought on by magnetic nano-beads. These nano-beads are covered with biotinylated detection antibodies that bind to Ag on the sensor surface to capture the antibodies that are caught. This creates a sandwich immunoassay configuration, resulting in magnified magnetic signals when the Ag is present. A microfluidic device controls the flow of samples and chemicals using a microfluidic chip with several channels and a syringe pump to control flow and rate. A biotin-PEG-NHS ester is used as the biotinylation reagent in the enhanced biotin labeling technique, which reacts with primary amine groups on antibodies. This compound has a lengthy PEG chain, which lowers steric resistance and boosts Ab solubility. The 12 tumor markers tested showed broad calibration curves, excellent specificity, and agreement with the traditional immunoradiometric assay (IRMA) technique for clinical samples in tests with standard solutions and clinical samples (Fig. 21B). The biosensor also offers the advantages of high throughput, small sample size, and quick assays.250
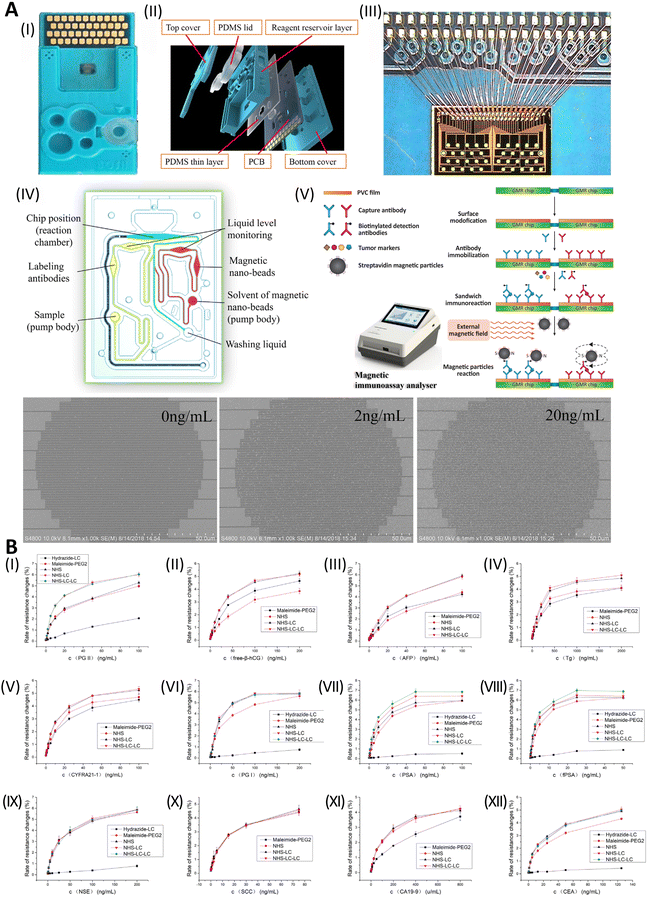 | ||
| Fig. 21 (A) Diagram showing the six parts of making a test card. The test card (II) is the first component's subject, followed by a description of its multilayered construction. (III) It also provides information on the GMR chip's connection to the PCB. In addition, it covers the microchannel system's construction in (IV). The GMR multi-biomarker immunoassay's reaction mechanism is covered in the following component (V). (VI) The last section contains SEM pictures of magnetic nano-beads attached to the GMR sensor surface after a protein assay. The three photos illustrate the results of three different doses of the CEA test. (B) Long-chain biotin compounds may increase the binding affinity of the protein avidin. In comparison to NHS-biotin, several biomarkers, including AFP, CYFRA21-1, PG II, Tg, free-hCG, PSA, and fPSA (I–V and VII–VIII), showed better reactivity with NHS-LC-biotin and NHS-LC-LC-biotin. The conjugating action between magnetic nano-beads and labeling antibodies may not be significantly affected by lengthening the spacer arm for certain antibodies because the binding sites may not be buried inside the surface plane. The reactivity of NHS-LC-biotin and NHS-LC-LC-biotin labeling was almost equal to that of NHS-biotin labeling in CEA, NSE, PG I, CA19-9, and SCC (VI and IX-II). The reactive group also influences the biotin–avidin system. In order of decreasing reactivity (highest to lowest), amine-reactive biotin had the most significant level of reactivity, followed by sulfhydryl-reactive and carbonyl-reactive biotin. Reprinted from ref. 250 with permission from [Elsevier], copyright [2018]. | ||
Table 2 provides a summary of biotin-based conjugates designed for detecting tumor markers. The conjugates consist of biotinylated antibodies, peptides, aptamers, NPs, and additional agents that target various tumor markers. The table also displays the detection techniques employed, including electrochemical-immunosensor, ELISA, immunohistochemistry, and others.
| Tumor marker | Biotinylated conjugate | Detection method | LOD | Advantages | Clinical significance | Ref. |
|---|---|---|---|---|---|---|
| a ng mL−1. b U mL−1, LOD. | ||||||
| AFP | A magnetic NP composite with biotin functionality (B-APTES@Fe3O4) | Electrochemical-immunosensor | 0.33 pg mL−1 | Reproducible and high-sensitivity | The immunosensor could potentially be used not only for detecting AFP but also for other purposes in the future | 209 |
| Ag–Ab interactions and the biotin-SA-biotin sandwich technique | Fluorescent immunosensor | 9.8 ± 2.9 g mL−1 | Quick and affordable | A sandwich strategy involving biotinylated antibodies and fluorescently labeled SA allows label-free detection | 210 | |
| The photoactive component is CdSe QDs, the detecting probe is a biotin-conjugated AFP Ab (bio-anti-AFP), the signal collection component is SA, and the amplification component is Bio-APOAA | PEC | 0.31 pg mL−1 | Selective and sensitive AFP detection | Promising for trypsin detection and inhibitor screening | 211 | |
| Biotinylated Lens culinaris agglutinin-linked silver NPs (B-LCA-AgNPs) | Electrochemical | 12 pg mL−1 | Exceptional stability and consistency | A simple and sensitive approach to detect AFP-L3, a crucial HCC marker | 212 | |
| Ap-MNPs (AFP-specific ssDNA aptamers avidin–biotin-attached to magnetic NPs (Fe3O4@SiO2)) | Label-free detection | 0.27 g mL−1 | Compared with IgG, HSA, and FIB, Ap-MNPs were more effective | An aptamer is demonstrated here to be a feasible and efficient tool for detecting biomarkers in clinical settings. | 214 | |
| ICP-MS SA-Au NPs tagged biotinylated tyramine | Dual amplification for sensitive ICP-MS-based immunoassay | 1.85 pg mL−1 | Simple, sensitive, and selective | The approach may detect biomarkers early in clinical settings and other proteins in human blood with excellent sensitivity | 251 | |
| EGFR | SA-conjugated gold NPs detect biotinylated capture probe sequences | PCR | 10−9 mol L−1 | Replicable and detects 5% gene mutation in combination samples | Highly expressed in various cancers such as lung, colorectal, pancreatic, and head and neck cancers. Used to identify patients who may benefit from targeted therapy | 217 |
| SA-coated MBs were immobilized with biotinylated anti-human EGFR Apt | Electrochemical | 50 pg mL−1 | Cost-effective and sensitive detection | The suggested method can potentially encourage the creation and advancement of a more accurate immunosensor that can precisely measure the EGFR biomarker in complicated biological samples | 218 | |
| Biotin-labeled target PCR products | PCR | 4.98% was LOQ | Detection of even extremely minute mutation rates | This technique may be used to find cancer cells mixed together with their non-cancerous counterparts | 219 | |
| PSA | Biotinylated PSA Ab | DPVs | 0.07 g L−1 | Detection of PSA glycoprotein in unprocessed human plasma | Used for screening and monitoring prostate cancer. It can also be used to monitor treatment effectiveness and detect recurrence | 222 |
| MB@SA·biotin-Ab1(PSA) | ELC | PSA at 0.28 ng mL−1 and exosomes at 4.9 × 102 Particle per mL | The enhanced sensibility of ECL MMbiosensors | A very sensitive ECL homogenous bioassay may be produced on standard ECL analyzers using novel approaches like as switching from a typical PMT to a diamagnetic PMT, switching from ring-disc magnets to circular-disc magnets for lain-in MGCE, and efficiently concentrating ECL MMbiosensors | 223 | |
| CEA | Biotin/SA-gold magnetic NPs and hybridization chain reaction | Electrochemical immunosensor | For CEA, CA 125, PSA, and 62, 48, 77, and 60 fg mL−1, respectively | Straightforward, rapid, and highly responsive | Used to monitor colorectal cancer progression and detect recurrence after treatment. It can also be elevated in lung, breast, pancreatic, and other cancers | 225 |
| MNPs coated with biotinylated antibodies | Magnetic immunoassay based on ICP-MS | 0.02 ng mL−1 | Improve the efficacy of testing procedures while minimizing the quantity of sample material used | In blood samples from lung cancer patients, the technique identified the biomarkers CYFRA21-1, CA 15-3, and CEA and has the potential to identify other biomarkers | 226 | |
| C125 | ZnS-encapsulated CdSe QDs coupled with biotin–SA | Fluorescence-imaging | — | QDs label and photograph dense samples. In sensitive immunoassays, their optical properties and photobleaching resistance may exceed organic fluorescent probes | Ovarian cancer's precise diagnosis | 252 |
| (Biotin)n-labeled BSA | ELISA and western blot | — | It may be feasible and convenient to administer effective adjuvant treatment to ovarian cancer patients using bfFp targeting to transport biotinylated therapeutic agents or utilizing pretargeting therapy with clinical relevance | Ovarian cancer's precise diagnosis | 230 | |
| CA 15-3 | biotinylated anti-CA 15-3 mAb | Immunosensor | 15 × 10−6 U mL−1 | high sensitivity, stability, and specificity | The created immunosensor may be utilized to diagnose CA 15-3 and other cancer biomarkers precisely and quickly | 234 |
| Biotinylated anti-CA 15-3 (Ab) | Enzyme immunoassay | 2.5 U mL−1 | Easy to use and requires less reagents | The SECM-enzyme immunoassay detects CA 15-3 and quantitatively quantifies Ag by examining the SECM scan curve at the location of the Ag complex formation using enzyme-labeled antibodies | 235 | |
| MMP-1 | A biotin-labeled, SA-modified magnetic NP-binding peptide containing peptide-DNA1 and hCG-DNA2 | The pregnancy test strip and magnetic separation technology | 65.5 pg mL−1 | High level of accuracy in detecting the target molecule or substance without interference from other substances or factors | It may be made easily and essentially without the need for sophisticated ingredients, making it a good option for investigating oral cancer in various situations | 240 |
| HER2 | Biotin-binding protein NeutrAvidin™-NPs composed of gelatin and human serum albumin (HAS) | Utilizing the unique interaction between Her2 receptors on breast carcinoma cells, the sandwich immunoassay design | — | Incorporating antibodies into NPs allows targeted drug delivery to specific tumor cells that overexpress HER2 | Aggressive breast cancer is linked to HER2 overexpression. Used to determine whether individuals may benefit from targeted treatment | 101 |
| Biotinylated-HER2 | Lab-on-chip system | — | Improved affinity and selectivity | HER2 peptide nanoprobes have been validated on live cancer cells and in vivo explant mouse models using in situ single-bead sequencing | 244 | |
| CYFRA21 | Biotinylated anti-CYFRA21 Ab | PEC immunosensor | 3.5 fg mL−1 | High sensitivity, stability, selectivity, and excellent repeatability | PEC identification of CYFRA21-1 in non-small cell carcinoma samples may enable an early diagnosis of lung cancer. | 248 |
| Other tumor markers (CA19-9, β-hCG, etc.) | Goat anti-mouse IgG biotinylated (Fc-specific) | Microimmunoassay | Less than 12 g l−1 for the hormones AFP, CEA, and hCG, or less than 17 kU l−1 for the hormones CA 125, CA 19-9, and CA 15–3 | Shortened assay time, low sample volume, and high throughput | It may be used to measure several tumor markers in oncology and other areas of medicine, as well as in various scientific applications | 249 |
| Biotin-PEG-NHS ester | — | AFP (0.52)a, CEA (0.27)a, CYFRA21–1 (0.25)a, NSE (0.5)a, SCC (0.3)a, PGI (1)a, PG II (0.5)a, CA19–9 (2)b, total PSA (0.02)a, free PSA (0.07)a, free-β-hCG (0.3)a, and Tg (1)a | — | — | 250 | |
6. Limitations and future directions
6.1 Biotin as a versatile molecular tag for cancer detection
The use of biotin as a molecular tool for cancer diagnosis has gained significant attention in the scientific community. Biotin possesses a strong binding affinity for avidin or SA, which has paved the way for its incorporation into various diagnostic platforms.253 In the context of cancer diagnosis, biotin has been extensively utilized as a versatile molecular tag for the specific and sensitive detection of cancer biomarkers. One prominent application of biotin in cancer detection is through the functionalization of NPs. Biotinylated NPs offer several advantages, including augmented target specificity, signal amplification, and increased imaging contrast. Furthermore, these NPs can be tailored to selectively identify and adhere to biotin receptors, which are frequently overexpressed on cancer cell surfaces.254 Through the conjugation of biotin to NP surface, researchers can achieve precise tumor targeting and visualization using imaging modalities such as magnetic resonance imaging (MRI), PET scanning, and optical imaging. This conjugation can be accomplished through various strategies, such as covalent attachment or surface modification techniques. For instance, tethering biotin molecules to the NP surface via chemically stable covalent bonds enhances long-term stability and target specificity.255 Alternatively, biotin can be incorporated through surface coating or functionalization, enabling reversible interactions with avidin or SA for efficient detection and imaging.In diagnostic assays, biotin serves as an integral component in immunoassays and biosensors utilized for detecting cancer biomarkers.250 Biotinylated antibodies or aptamers, which are specific to the target biomarkers, are used to sequester the analyte of interest. The subsequent addition of avidin or SA conjugated with fluorescent dyes, enzymes, or NPs, along with robust interaction between biotin and avidin/SA, allows signal amplification and ultrasensitive detection of.
Despite the numerous advantages of using biotin in cancer diagnosis, several limitations must be considered for the practical application of this approach. One major concern is the potential for non-specific binding of biotin to endogenous biotin-binding proteins in biological samples.256 This nonspecific binding can result in false-positive signals, compromising cancer biomarker detection accuracy. Thus, careful optimization of experimental conditions, such as the biotin density on the diagnostic platform and the use of blocking agents, is crucial to minimize non-specific interactions and enhance the specificity of biotin-based assays.257 Moreover, elevated levels of biotin in blood samples due to supplementation can interfere with certain biotin-based diagnostic tests. This biotin interference is a critical factor that could result in inaccurate test outcomes, thereby impacting clinical decision-making.258
In addition to NP-based applications, biotin has also proven to be instrumental in developing biotin electrosensors, which have shown promise in cancer diagnosis. These sensors exploit the specific binding between biotin and avidin/SA to enable sensitive and selective detection of cancer biomarkers. These electrosensors typically consist of a biotinylated capture probe immobilized on an electrode surface which is specifically designed to recognize and bind the target biomarker. Upon binding, the sensor generates a measurable and quantifiable electrochemical signal. Biotin electrosensors are advantageous as they provide a label-free detection method, eliminating the need for additional labeling steps and reducing assay complexity. This simplifies the detection process and improves the overall efficiency of the diagnostic assay. Furthermore, biotin electrosensors can achieve high sensitivity and selectivity due to the strong and specific interaction between biotin and avidin/SA. The binding event between the target biomarker and the biotinylated capture probe leads to a measurable change in the electrochemical signal, enabling the detection of cancer biomarkers even at low concentrations.
Biotin electrosensors offer exciting possibilities for cancer diagnosis, but some hurdles need to be cleared for reliable detection. A key challenge is optimizing the sensor's performance. This involves selecting a high attraction and specificity capture probe for the target cancer biomarker. Fine-tuning the electrode surface's chemistry and the detection conditions are also crucial for maximizing sensitivity and achieving a wider range of detectable concentrations with a low minimum detection level. Researchers are further exploring various electrode materials and designs to improve sensitivity and stability. Another significant hurdle is the non-specific binding of biomolecules in complex samples, leading to false-positive signals. To address this, scientists are investigating appropriate blocking agents to prevent unwanted interactions with the electrode surface. Additionally, control experiments are essential to differentiate specific binding from non-specific interactions. By overcoming these challenges, biotin electrosensors can potentially become a valuable tool for cancer diagnosis.
Several strategies have been employed to improve the performance of biotin electrosensors. One approach entails the incorporation of NMs into sensor design. NMs, such as carbon nanotubes, graphene, and metal NPs, have unique properties that can enhance the electrochemical signal and improve the sensor's performance. These NMs can serve as highly conductive platforms, providing an expansive surface area for immobilizing the biotinylated capture probe and facilitating electron transfer during the detection procedure. The integration of NMs into biotin electrosensors has demonstrated enhanced sensitivity, selectivity, and stability. Additionally, signal amplification techniques have been employed to improve the detection limits of biotin electrosensors, particularly when targeting low-abundance cancer biomarkers. One commonly used amplification method involves enzymatic amplification, wherein enzymes catalyze the conversion of a substrate into a detectable product. For instance, SA-conjugated HRP can be employed after binding the target biomarker to the biotinylated capture probe. The HRP catalyzes the oxidation of a chromogenic substrate, producing a colored or fluorescent product that can be easily detected and quantified. This enzymatic amplification step significantly enhances the sensitivity of the biotin electrosensor. Moreover, integrating microfluidics technology with biotin electrosensors has shown great potential in improving the efficiency and reliability of cancer diagnosis. Microfluidic devices provide precise control over sample flow, enabling rapid and accurate analysis of biological samples. By integrating biotin-functionalized surfaces within microfluidic channels, cancer biomarkers can be efficiently captured and detected, with the added benefits of reduced sample volume, faster analysis time, and multiplexed detection capabilities.
In summary, integrating biotin as a molecular tag in cancer diagnosis represents a versatile and robust approach for the specific and sensitive detection of cancer biomarkers. Its application in NP-based platforms and the development of biotin electrosensors are particularly promising. Biotin electrosensors afford label-free detection with high sensitivity and selectivity. However, challenges remain, such as optimizing sensor performance and addressing non-specific binding. Ongoing advancements in electrode materials, NM integration, signal amplification strategies, and microfluidics technology are expected to enhance the capabilities of biotin electrosensors in cancer diagnosis, paving the way for early and accurate detection of cancer biomarkers.
6.2 Regulatory and manufacturing challenges
The successful translation of biotin-functionalized NPs from bench to clinical practice necessitates tackling regulatory and manufacturing challenges. The regulatory pathway for nanomedicines is rigorous and mandates exhaustive evaluation of safety, efficacy, and quality assessments.259 Additionally, scaling up the production of biotin-functionalized NPs while maintaining batch-to-batch consistency and quality control poses significant manufacturing challenges. Future efforts should be channeled toward establishing standardized protocols and guidelines pertinent to the production, characterization, and quality control of biotin-functionalized NPs. Multidisciplinary collaboration between researchers, clinicians, regulatory agencies, and pharmaceutical manufacturers is crucial to streamlining the translation pipeline and ascertaining the reproducibility, safety, and quality of these NPs for widespread clinical use.260Overcoming the challenges associated with the diagnosis by biotin conjugation on nanosensors requires careful consideration and implementation of several strategies. One of the primary challenges in utilizing biotin conjugation on nanosensors for diagnosis is the potential for non-specific binding, leading to false-positive signals and reduced specificity. To overcome this challenge, researchers can employ several techniques. Firstly, optimizing the density of biotin molecules on the nano carrier's surface is crucial. By controlling the density, researchers can minimize the chances of non-specific interactions while ensuring sufficient binding sites for the target biomarkers.261 Additionally, blocking agents, such as BSA or casein, mitigate non-specific binding by occluding unoccupied sites on the nanocarrier's surface.
Achieving high target specificity is vital for accurate cancer diagnosis. One strategy to enhance specificity is to carefully select the targeting ligands in addition to biotin.262 By incorporating ligands specific to cancer cells or tumor biomarkers, the nanosensors can be designed to specifically recognize and bind to cancer cells, reducing off-target interactions. Moreover, utilizing a combination of targeting ligands can further enhance the specificity of the nanovehicles. For example, antibodies or aptamers specific to cancer biomarkers can be conjugated along with biotin, allowing for dual targeting and improved specificity.263
Stability is crucial for successfully implementing biotin conjugation on nanosensors for diagnostic purposes. Stable and long-lasting conjugation is essential to ensure the integrity of the NPs during storage, transportation, and analysis. The covalent attachment of biotin to the nano carrier's surface provides stable conjugation and long-term stability. Additionally, proper surface modification techniques, such as functionalizing the NPs with hydrophilic polymers or introducing a protective coating, can enhance stability and prevent degradation or detachment of the biotin conjugates.264
The development and optimization of diagnostic assays play a vital role in overcoming challenges associated with biotin conjugation on NPs.265 Optimization involves determining the optimal assay conditions, including incubation time, temperature, and buffer composition, to maximize the binding efficiency and minimize non-specific interactions. Additionally, utilizing appropriate controls, such as negative controls and isotype controls, helps validate the specificity of the biotin-conjugated NPs in the diagnostic assays.266 Employing a multimodal detection approach by integrating multiple techniques can further enhance the accuracy and sensitivity of the diagnosis by biotin conjugation on nanosensors.
For instance, incorporating imaging modalities, such as MRI, PET, or optical imaging, alongside the biotin-conjugated nano-biosensors can provide multimodal detection and visualization of cancer cells or tumors.267 This integration allows for complementary information from different detection modalities, improving the overall diagnostic performance.
By implementing these strategies, researchers can overcome the challenges associated with diagnosis by biotin conjugation on nanosensors. These approaches contribute to developing more reliable and accurate diagnostic platforms, ultimately improving patient outcomes through early and precise detection of cancer biomarkers.
7. Conclusion
Biotin-functionalized NPs represent a significant breakthrough in the field of oncological diagnostics, showcasing exceptional specificity and efficiency through targeted delivery and innovative surface modifications. These NPs utilize the strong affinity between biotin and avidin or streptavidin, facilitating precise tumor targeting and enhanced imaging contrast that are crucial for early detection and accurate cancer diagnosis. This review systematically explores the application of biotin in various diagnostic platforms, where its conjugation to NPs significantly improves signal sensitivity and diagnostic accuracy while also enabling the detailed visualization of tumor sites. Despite their numerous advantages, challenges such as non-specific binding remain a concern. This issue can be effectively managed by optimizing the biotin density on the NPs and employing robust blocking agents to refine assay conditions, thereby minimizing false positives and enhancing reliability. Looking to the future, the potential integration of biotin-functionalized NPs with cutting-edge developments in nanosensor technology, microfluidics, and advanced signal amplification techniques is expected to revolutionize cancer diagnostics. Such advancements will not only enhance the capacity for early tumor detection but also significantly improve the precision of diagnostic processes. This progress promises to pave the way for highly personalized therapeutic strategies, ultimately improving patient management outcomes and reducing the overall burden on healthcare systems. By pushing the boundaries of current technologies and continuing to integrate these innovative tools into clinical practice, biotin-functionalized NPs are composed to transform the landscape of cancer diagnostics and therapy, offering new hope for patients and clinicians alike.Author contributions
Conceptualization, S.S.; writing – original draft preparation, S.F.-K., A.K., M.C., S.S., and B.F.-F.; writing – review and editing, S.S., S.M., M.C. A.Z., A.K; supervision, final draft, S.S., S.G. All authors have read and agreed to the published version of the manuscript.Abbreviations
| Ag | Antigen |
| CTC | Circulating cancer cell |
| CA 125 | Cancer Ag 125 |
| CA 15-3 | Cancer Ag 15-3 |
| NPs | Nanoparticles |
| NP | Nanoparticle |
| MRS | Magnetic resonance spectroscopy |
| CT | X-ray computed tomography |
| PET | Positron emission tomography |
| ELISAs | Enzyme-linked immunosorbent assays |
| NMs | Nanomaterials |
| NM | Nanomaterial |
| UNPs | Up-conversion nanoparticles |
| MNPs | Magnetic nanoparticles |
| QDs | Quantum dots |
| CNTs | Carbon nanotubes |
| SA | Streptavidin |
| Tyr | Tyrosine |
| PEG | Polyethylene glycol |
| HER2 | Human epidermal growth factor receptor 2 |
| SMVT | Sodium-dependent multivitamin transporter |
| HRP | Poly-horseradish peroxidase |
| T3 | Triiodothyronine |
| T4 | Thyroxine |
| Ab | Antibody |
| hCG | Human chorionic gonadotropin |
| EGFR | Epidermal growth factor receptors |
| MBs | Magnetic beads |
| EpCAM | Epithelial cell adhesion molecule |
| HNPs | Hybrid nanoparticles |
| NSCLC | Non-small cell lung cancer |
| HCC | Hepatocellular carcinoma |
| ROS | Reactive oxygen species |
| SPR | Surface plasmon resonance |
| GSH | Glutathion |
| Cys | Cysteine |
| NOXs | NADPH oxidases |
| O2˙ | Superoxide |
| H2O2 | Hydrogen peroxide |
| SODs | Superoxide dismutases |
| TRX | Thioredoxins |
| PRX | Peroxiredoxins |
| GPX | Glutathione peroxidases |
| ER | Endoplasmic reticulum |
| Hcy | Homocysteine |
| MQ | Mono-quenching |
| ICT | Intramolecular charge transfer |
| LOD | Limit of detection |
| AuNP | Gold nanoparticle |
| FA | Formaldehyde |
| GEM | Gemcitabine |
| HWTX-I | Huwentoxin-I |
| ICK | Inhibitor cystine knot |
| PcTx1 | Psalmotoxin 1 |
| GBM | Glioblastoma |
| PBS | Phosphate-buffered saline |
| HTTP | High-throughput |
| ZnO | Zinc oxide |
| SPR | Surface plasmon resonance |
| BSA | Bovine serum albumin |
| Ab1 | Primary antibodies |
| Biotin-Ab2 | Biotinylated secondary antibodies |
| GNRs | Gold nanorods |
| GSAB-Ab2 | Secondary gold-SA-biotinylated antibodies |
| hIgG | Human immunoglobulin G |
| ECL | Electrochemiluminescence |
| hTK1 | Human thymidine kinase 1 |
| mAb | Monoclonal antibody |
| rmAb | Recombinant monoclonal antibodies |
| AFP | Alpha-fetoprotein |
| CEA | Carcinoembryonic Ag |
| CYFRA21-1 | Cytokeratin protein fragment 21-1 |
| CA 19-9 | Cancer Ag 19-9 |
| β-hCG | Beta-human chorionic gonadotropin |
| PGI/PGII | Serum pepsinogens |
| NSE | Neuron-specific enolase |
| PEC | Photoelectrochemical |
| AFP-L3 | Alpha-fetoprotein-L3 |
| Ap-MNPs | Aptamer-functionalized magnetic NPs |
| HAS | Human serum albumin |
| PCR | Polymerase chain reaction |
| DPV | Differential pulse voltammetry |
| RSD | Relative standard deviations |
| Cys-AuNPs | AuNPs functionalized with cysteamine |
| LLOQ | lower limit of quantitation |
| Ab2 | Secondary antibody |
| ICP-MS | Inductively coupled plasma mass spectroscopy |
| bfFp | Bifunctional fusion protein |
| IMAC | Immobilized metal chelate affinity chromatography |
| CLSM | Confocal laser scanning microscopy |
| SECM | Scanning electrochemical microscopy |
| BQ | Benzoquinone |
| MMP-1 | Matrix Metalloproteinase-1 |
| PDAC | Pancreatic ductal adenocarcinoma |
| GMR | Giant magnetoresistance |
| POCT | Point-of-care testing |
| SCC | Squamous cell carcinoma |
| PGI | Human pepsinogens I |
| Tg | Thyroglobulin |
| IRMA | Immunoradiometric assay |
| MRI | Magnetic resonance imaging |
Data availability
This article's data sharing is not applicable as no new data were created or analyzed in this study.Conflicts of interest
The authors declare that there is no conflict of interest regarding the publication of this article.Acknowledgements
Marco Cordani was supported by grant RYC2021-031003I funded by MICIU/AEI/10.13039/501100011033 and by European Union NextGenerationEU/PRTR.References
- B. Cao, F. Bray, A. Ilbawi and I. Soerjomataram, Effect on longevity of one-third reduction in premature mortality from non-communicable diseases by 2030: a global analysis of the Sustainable Development Goal health target, Lancet Global Health, 2018, 6(12), 1288–1296 CrossRef PubMed.
- J. Alizadeh, M. Kavoosi, N. Singh, S. Lorzadeh, A. Ravandi and B. Kidane, et al., Regulation of Autophagy via Carbohydrate and Lipid Metabolism in Cancer, Cancers, 2023, 15(8), 2195 CrossRef CAS PubMed.
- A. Martelli, M. Omrani, M. Zarghooni, V. Citi, S. Brogi and V. Calderone, et al., New Visions on Natural Products and Cancer Therapy: Autophagy and Related Regulatory Pathways, Cancers, 2022, 14(23), 5839 CrossRef CAS PubMed.
- S. Sargazi, R. Arshad, R. Ghamari, A. Rahdar, A. Bakhshi and S. F. Karkan, et al., siRNA–based nanotherapeutics as emerging modalities for immune–mediated diseases: A preliminary review, Cell Biol. Int., 2022, 46(9), 1320–1344 CrossRef CAS PubMed.
- R. L. Siegel, K. D. Miller, N. S. Wagle and A. Jemal, Cancer statistics, 2023, CA Cancer J. Clin., 2023, 73(1), 17–48 CrossRef PubMed.
- C. Allemani, H. K. Weir, H. Carreira, R. Harewood, D. Spika and X.-S. Wang, et al., Global surveillance of cancer survival 1995–2009: analysis of individual data for 25 676 887 patients from 279 population-based registries in 67 countries (CONCORD-2), Lancet, 2015, 385(9972), 977–1010 CrossRef PubMed.
- R. De Angelis, E. Demuru, P. Baili, X. Troussard, A. Katalinic and M. D. C. Lopez, et al., Complete cancer prevalence in Europe in 2020 by disease duration and country (EUROCARE-6): a population-based study, Lancet Oncol., 2024, 25(3), 293–307 CrossRef PubMed.
- M. A. Benderra, A. G. Serrano, E. Paillaud, C. M. Tapia, T. Cudennec and C. ChouaÏd, et al., Prognostic value of comorbidities in older patients with cancer: the ELCAPA cohort study, ESMO Open, 2023, 8(5), 101831 CrossRef PubMed.
- K. F. Brown, H. Rumgay, C. Dunlop, M. Ryan, F. Quartly and A. Cox, et al., The fraction of cancer attributable to modifiable risk factors in England, Wales, Scotland, Northern Ireland, and the United Kingdom in 2015, Br. J. Cancer, 2018, 118(8), 1130–1141 CrossRef PubMed.
- A. Pulumati, A. Pulumati, B. S. Dwarakanath, A. Verma and R. V. Papineni, Technological advancements in cancer diagnostics: Improvements and limitations, Cancer Rep., 2023, e1764 CrossRef PubMed.
- M. Razlansari, S. Jafarinejad, A. Rahdar, M. Shirvaliloo, R. Arshad and S. Fathi-Karkan, et al., Development and classification of RNA aptamers for therapeutic purposes: an updated review with emphasis on cancer, Mol. Cell. Biochem., 2022, 1–26 Search PubMed.
- J. Alizadeh, S. C. da Silva Rosa, X. Weng, J. Jacobs, S. Lorzadeh and A. Ravandi, et al., Ceramides and ceramide synthases in cancer: Focus on apoptosis and autophagy, Eur. J. Cell Biol., 2023, 102(3), 151337 CrossRef CAS PubMed.
- B. Behrooz A, H. Latifi-Navid, S. C. da Silva Rosa, M. Swiat, E. Wiechec and C. Vitorino, et al., Integrating Multi-Omics Analysis for Enhanced Diagnosis and Treatment of Glioblastoma: A Comprehensive Data-Driven Approach, Cancers, 2023, 15(12), 3158 CrossRef PubMed.
- L. Jiang, X. Lin, F. Chen, X. Qin, Y. Yan and L. Ren, et al., Current research status of tumor cell biomarker detection, Microsyst. Nanoeng., 2023, 9, 123 CrossRef CAS PubMed.
- U. Laraib, S. Sargazi, A. Rahdar, M. Khatami and S. Pandey, Nanotechnology-based approaches for effective detection of tumor markers: A comprehensive state-of-the-art review, Int. J. Biol. Macromol., 2022, 195, 356–383 CrossRef CAS PubMed.
- A. Seyfoori, M. Shokrollahi Barough, P. Mokarram, M. Ahmadi, P. Mehrbod and A. Sheidary, et al., Emerging Advances of Nanotechnology in Drug and Vaccine Delivery against Viral Associated Respiratory Infectious Diseases (VARID), Int. J. Mol. Sci., 2021, 22(13), 6937 CrossRef CAS PubMed.
- R. Geevarghese, S. S. Sajjadi, A. Hudecki, S. Sajjadi, N. R. Jalal and T. Madrakian, et al., Biodegradable and Non-Biodegradable Biomaterials and Their Effect on Cell Differentiation, Int. J. Mol. Sci., 2022, 23(24), 16185 CrossRef CAS PubMed.
- F. Farshchi and M. Hasanzadeh, Nanomaterial based aptasensing of prostate specific antigen (PSA): recent progress and challenges in efficient diagnosis of prostate cancer using biomedicine, Biomed. Pharmacother., 2020, 132, 110878 CrossRef CAS PubMed.
- M. Pourmadadi, S. Ostovar, G. Ruiz-Pulido, D. Hassan, M. Souri and A.-L. E. Manicum, et al., Novel epirubicin-loaded nanoformulations: advancements in polymeric nanocarriers for efficient targeted cellular and subcellular anticancer drug delivery, Inorg. Chem. Commun., 2023, 110999 CrossRef CAS.
- A. Sani, M. Pourmadadi, M. Shaghaghi, M. M. Eshaghi, S. Shahmollaghamsary and R. Arshad, et al., Revolutionizing anticancer drug delivery: Exploring the potential of tamoxifen-loaded nanoformulations, J. Drug Delivery Sci. Technol., 2023, 104642 CrossRef CAS.
- F. Davodabadi, S. Mirinejad, S. Fathi–Karkan, M. Majidpour, N. Ajalli and R. Sheervalilou, et al., Aptamer–functionalized quantum dots as theranostic nanotools against cancer and bacterial infections: a comprehensive overview of recent trends, Biotechnol. Prog., 2023, 39(5), e3366 CrossRef CAS PubMed.
- M. Barani, A. Rahdar, S. Sargazi, M. S. Amiri, P. K. Sharma and N. Bhalla, Nanotechnology for inflammatory bowel disease management: Detection, imaging and treatment, Sens. Bio-Sens. Res., 2021, 32, 100417 CrossRef.
- S. Sargazi, I. Fatima, M. H. Kiani, V. Mohammadzadeh, R. Arshad and M. Bilal, et al., Fluorescent-based nanosensors for selective detection of a wide range of biological macromolecules: A comprehensive review, Int. J. Biol. Macromol., 2022, 206, 115–147 CrossRef CAS PubMed.
- M. Barani, M. Mukhtar, A. Rahdar, S. Sargazi, S. Pandey and M. Kang, Recent advances in nanotechnology-based diagnosis and treatments of human osteosarcoma, Biosensors, 2021, 11(2), 55 CrossRef CAS PubMed.
- S. Sargazi, Z. Ahmadi, M. Barani, A. Rahdar, S. Amani and M. F. Desimone, et al., Can nanomaterials support the diagnosis and treatment of human infertility? A preliminary review, Life Sci., 2022, 120539 CrossRef CAS PubMed.
- H. Ghaznavi, M. Shirvaliloo, A. Zarebkohan, Z. Shams, F. Radnia and Z. Bahmanpour, et al., An updated review on implications of autophagy and apoptosis in tumorigenesis: possible alterations in autophagy through engineered nanomaterials and their importance in Cancer therapy, Mol. Pharmacol., 2021, 100(2), 119–143 CrossRef CAS PubMed.
- M. Qindeel, F. Sabir, S. Sargazi, V. Mohammadzadeh and S. I. Mulla, New insights into the application of nanoghosts as theranostic tools with an emphasis on cardiovascular diseases, J. Nanopart. Res., 2021, 23, 1–27 CrossRef.
- R. Arshad, S. Sargazi, I. Fatima, A. Mobashar, A. Rahdar and N. Ajalli, et al., Nanotechnology for Therapy of Zoonotic Diseases: A Comprehensive Overview, ChemistrySelect, 2022, 7(21), e202201271 CrossRef CAS.
- M. Ahmadi, T. Madrakian and S. Ghavami, Preparation and Characterization of Simvastatin Nanocapsules: Encapsulation of Hydrophobic Drugs in Calcium Alginate, Methods Mol. Biol., 2020, 2125, 47–56 CrossRef CAS PubMed.
- L. Sun, H. Liu, Y. Ye, Y. Lei, R. Islam and S. Tan, et al., Smart nanoparticles for cancer therapy, Signal Transduction Targeted Ther., 2023, 8(1), 418 CrossRef CAS PubMed.
- C. J. Bashor, I. B. Hilton, H. Bandukwala, D. M. Smith and O. Veiseh, Engineering the next generation of cell-based therapeutics, Nat. Rev. Drug Discovery, 2022, 21(9), 655–675 CrossRef CAS PubMed.
- F. Böttger, A. Vallés-Martí, L. Cahn and C. R. Jimenez, High-dose intravenous vitamin C, a promising multi-targeting agent in the treatment of cancer, J. Exp. Clin. Cancer Res., 2021, 40(1), 343 CrossRef PubMed.
- M. A. Malik, A. A. Hashmi, A. S. Al-Bogami and M. Y. Wani, Harnessing the Power of Gold: Advancements in Anticancer Gold Complexes and functionalized Nanoparticles, J. Mater. Chem. B, 2024, 552–576 RSC.
- I. Fatima, A. Rahdar, S. Sargazi, M. Barani, M. Hassanisaadi and V. K. Thakur, Quantum dots: synthesis, antibody conjugation, and HER2-receptor targeting for breast cancer therapy, J. Funct. Biomater., 2021, 12(4), 75 CrossRef CAS PubMed.
- D. A. Alromi, S. Y. Madani and A. Seifalian, Emerging application of magnetic nanoparticles for diagnosis and treatment of cancer, Polymers, 2021, 13(23), 4146 CrossRef CAS PubMed.
- T. A. Tabish, P. Dey, S. Mosca, M. Salimi, F. Palombo and P. Matousek, et al., Smart gold nanostructures for light mediated cancer theranostics: combining optical diagnostics with photothermal therapy, Adv. Sci., 2020, 7(15), 1903441 CrossRef CAS PubMed.
- P. Kesharwani, R. Ma, L. Sang, M. Fatima, A. Sheikh and M. A. Abourehab, et al., Gold nanoparticles and gold nanorods in the landscape of cancer therapy, Mol. Cancer, 2023, 22(1), 98 CrossRef CAS PubMed.
- Y. Wu, M. R. Ali, K. Chen, N. Fang and M. A. El-Sayed, Gold nanoparticles in biological optical imaging, Nano Today, 2019, 24, 120–140 CrossRef CAS.
- R. Mohammadi, H. Naderi-Manesh, L. Farzin, Z. Vaezi, N. Ayarri and L. Samandari, et al., Fluorescence sensing and imaging with carbon-based quantum dots for early diagnosis of cancer: A review, J. Pharm. Biomed. Anal., 2022, 212, 114628 CrossRef CAS PubMed.
- G. Pittiglio, J. H. Chandler, T. da Veiga, Z. Koszowska, M. Brockdorff and P. Lloyd, et al., Personalized magnetic tentacles for targeted photothermal cancer therapy in peripheral lungs, Commun. Eng., 2023, 2(1), 50 CrossRef.
- E. M. Materón, C. M. Miyazaki, O. Carr, N. Joshi, P. H. Picciani and C. J. Dalmaschio, et al., Magnetic nanoparticles in biomedical applications: A review, Appl. Surf. Sci. Adv., 2021, 6, 100163 CrossRef.
- M. Chehelgerdi, M. Chehelgerdi, O. Q. B. Allela, R. D. C. Pecho, N. Jayasankar and D. P. Rao, et al., Progressing nanotechnology to improve targeted cancer treatment: overcoming hurdles in its clinical implementation, Mol. Cancer, 2023, 22(1), 169 CrossRef PubMed.
- L. Chen, W. Hong, W. Ren, T. Xu, Z. Qian and Z. He, Recent progress in targeted delivery vectors based on biomimetic nanoparticles, Signal Transduction Targeted Ther., 2021, 6(1), 225 CrossRef PubMed.
- G. Russell-Jones, K. McTavish, J. McEwan, J. Rice and D. Nowotnik, Vitamin-mediated targeting as a potential mechanism to increase drug uptake by tumours, J. Inorg. Biochem., 2004, 98(10), 1625–1633 CrossRef CAS PubMed.
- L. C.-C. Lee and K. K.-W. Lo, Chapter Two - Strategic design of photofunctional transition metal complexes for cancer diagnosis and therapy, in Advances in Inorganic Chemistry. 80, ed. P. C Ford and R. van Eldik, Academic Press, 2022, pp. 35–94 Search PubMed.
- S. H. Helppolainen, K. P. Nurminen, J. A. Määttä, K. K. Halling, J. P. Slotte and T. Huhtala, et al., Rhizavidin from Rhizobium etli: the first natural dimer in the avidin protein family, Biochem. J., 2007, 405(3), 397–405 CrossRef CAS PubMed.
- M. Kamel, S. Atta, S. Maher, H. Abd Elaziz and Z. Demerdash, Modified streptavidin-biotin based lateral flow test strip for rapid detection of SARS-CoV-2 S1 antigen in saliva samples, Sci. Rep., 2024, 14(1), 7319 CrossRef CAS PubMed.
- E. K. Wagner, A. Vedadghavami, T. D. Jacobsen, S. A. Goel, N. O. Chahine and A. G. Bajpayee, Avidin grafted dextran nanostructure enables a month-long intra-discal retention, Sci. Rep., 2020, 10(1), 12017 CrossRef CAS PubMed.
- S. Wang, M. Z. Hossain, T. Han, K. Shinozuka, T. Suzuki and A. Kuwana, et al., Avidin–Biotin Technology in Gold Nanoparticle-Decorated Graphene Field Effect Transistors for Detection of Biotinylated Macromolecules with Ultrahigh Sensitivity and Specificity, ACS Omega, 2020, 5(46), 30037–30046 CrossRef CAS PubMed.
- C. E. Chivers, A. L. Koner, E. D. Lowe and M. Howarth, How the biotin–streptavidin interaction was made even stronger: investigation via crystallography and a chimaeric tetramer, Biochem. J., 2011, 435(1), 55–63 CrossRef CAS PubMed.
- A. Jain and K. Cheng, The principles and applications of avidin-based nanoparticles in drug delivery and diagnosis, J. Controlled Release, 2017, 245, 27–40 CrossRef CAS PubMed.
- M.-M. Chen, X. Tang, J.-J. Li, F.-Y. Chen, Z.-T. Jiang and R. Fu, et al., Active targeting tumor therapy using host-guest drug delivery system based on biotin functionalized azocalix [4] arene, J. Controlled Release, 2024, 368, 691–702 CrossRef CAS PubMed.
- S. Maiti, N. Park, J. H. Han, H. M. Jeon, J. H. Lee and S. Bhuniya, et al., Gemcitabine–coumarin–biotin conjugates: a target specific theranostic anticancer prodrug, J. Am. Chem. Soc., 2013, 135(11), 4567–4572 CrossRef CAS PubMed.
- R. Tripathi, A. Guglani, R. Ghorpade and B. Wang, Biotin conjugates in targeted drug delivery: is it mediated by a biotin transporter, a yet to be identified receptor, or (an)other unknown mechanism(s)?, J. Hyg. Res., 2023, 38(1), 185–189 Search PubMed.
- H. Li, G. Bruce, N. Childerhouse, G. Keegan, G. Mantovani and S. Stolnik, Biotin receptor-mediated intracellular delivery of synthetic polypeptide-protein complexes, J. Controlled Release, 2023, 357, 333–341 CrossRef CAS PubMed.
- A. Gharehzadehshirazi, M. Zarejousheghani, S. Falahi, Y. Joseph and P. Rahimi, Biomarkers and Corresponding Biosensors for Childhood Cancer Diagnostics, Sensors, 2023, 23(3), 1482 CrossRef CAS PubMed.
- Y. Yildizhan, K. Driessens, H. S. K. Tsao, R. Boiy, D. Thomas and N. Geukens, et al., Detection of Breast Cancer-Specific Extracellular Vesicles with Fiber-Optic SPR Biosensor, Int. J. Mol. Sci., 2023, 24(4), 3764 CrossRef CAS PubMed.
- D. Panjwani, S. Patel, D. Mishra, V. Patel, M. Yadav and A. Dharamsi, et al., Avidin-Biotin functionalized self-assembled protein nanoparticles as EGFR targeted therapeutics for the treatment of lung cancer: characterization and cell viability, J. Dispersion Sci. Technol., 2023, 44(13), 2483–2496 CrossRef CAS.
- D. Duijvesz, C. Y. L. Versluis, C. A. Van Der Fels, M. S. Vredenbregt-van den Berg, J. Leivo and M. T. Peltola, et al., Immuno–based detection of extracellular vesicles in urine as diagnostic marker for prostate cancer, Int. J. Cancer, 2015, 137(12), 2869–2878 CrossRef CAS PubMed.
- Q. Lv, Y. Wang, C. Su, T. Lakshmipriya, S. C. Gopinath and K. Pandian, et al., Human papilloma virus DNA-biomarker analysis for cervical cancer: signal enhancement by gold nanoparticle-coupled tetravalent streptavidin-biotin strategy, Int. J. Biol. Macromol., 2019, 134, 354–360 CrossRef CAS PubMed.
- S. Maiti and P. Paira, Biotin conjugated organic molecules and proteins for cancer therapy: A review, Eur. J. Med. Chem., 2018, 145, 206–223 CrossRef CAS PubMed.
- M. Jurczyk, K. Jelonek, M. Musiał-Kulik, A. Beberok, D. Wrześniok and J. Kasperczyk, Single- versus Dual-Targeted Nanoparticles with Folic Acid and Biotin for Anticancer Drug Delivery, Pharmaceutics, 2021, 13(3), 326 CrossRef CAS PubMed.
- D. J. Lanska, The Discovery of Niacin, Biotin, and Pantothenic Acid, Ann. Nutr. Metab., 2012, 61(3), 246–253 CrossRef CAS PubMed.
- G. T. DeTitta, J. W. Edmonds, W. Stallings and J. Donohue, Molecular structure of biotin. Results of two independent crystal structure investigations, J. Am. Chem. Soc., 1976, 98(7), 1920–1926 CrossRef CAS PubMed.
- S. Fanali, P. R. Haddad, C. Poole and M.-L. Riekkola, Liquid chromatography: applications, Elsevier, 2017 Search PubMed.
- R. S. Carling and C. Turner, Methods for assessment of biotin (Vitamin B7). Laboratory assessment of vitamin status, Elsevier, 2019, pp. 193–217 Search PubMed.
- K. L. Levert, The role of cysteine 230 and lysine 238 of biotin carboxylase in the deprotonation of biotin and synthesis of a bisubstrate analog inhibitor of carboxyltransferase, Louisiana State University and Agricultural & Mechanical College, 2002 Search PubMed.
- T. C. Bruice and A. Hegarty, Biotin-bound CO2 and the mechanism of enzymatic carboxylation reactions, Proc. Natl. Acad. Sci. U. S. A., 1970, 65(4), 805–809 CrossRef CAS PubMed.
- G. L. Waldrop, H. M. Holden and M. S. Maurice, The enzymes of biotin dependent CO2 metabolism: what structures reveal about their reaction mechanisms, Protein Sci., 2012, 21(11), 1597–1619 CrossRef CAS PubMed.
- R. B. Guchhait, S. E. Polakis, D. Hollis, C. Fenselau and M. D. Lane, Acetyl coenzyme A carboxylase system of Escherichia coli: site of carboxylation of biotin and enzymatic reactivity of 1′-N-(ureido)-carboxybiotin derivatives, J. Biol. Chem., 1974, 249(20), 6646–6656 CrossRef CAS PubMed.
- H. M. Said, Cell and molecular aspects of human intestinal biotin absorption, J. Nutr., 2009, 139(1), 158–162 CrossRef CAS PubMed.
- X. Xiang, Y. Liu, X. Zhang, W. Zhang and Z. Wang, Effects of biotin on blood glucose regulation in type 2 diabetes rat model, J. Hyg. Res., 2015, 44(2), 185–189 CAS.
- A. T. Tankeu, G. Van Winckel, J. Elmers, E. Jaccard, A. Superti-Furga and B. Wolf, et al., Biotinidase deficiency: What have we learned in forty years?, Mol. Genet. Metab., 2023, 138(4), 107560 CrossRef CAS PubMed.
- S. B. van der Meer, T. Knuschke, A. Frede, N. Schulze, A. M. Westendorf and M. Epple, Avidin-conjugated calcium phosphate nanoparticles as a modular targeting system for the attachment of biotinylated molecules in vitro and in vivo, Acta Biomater., 2017, 57, 414–425 CrossRef CAS PubMed.
- S. Dhakal, H. Shafaat and V. Balasubramaniam, Thermal and high–pressure treatment stability of egg–white avidin in aqueous solution, J. Food Process Eng., 2020, 43(9), e13481 CrossRef CAS.
- S. H. Helppolainen, K. P. Nurminen, J. A. E. Määttä, K. K. Halling, J. P. Slotte and T. Huhtala, et al., Rhizavidin from Rhizobium etli: the first natural dimer in the avidin protein family, Biochem. J., 2007, 405(3), 397–405 CrossRef CAS PubMed.
- J. M. Marangoni, S. C. Wu, D. Fogen, S. L. Wong and K. K. S. Ng, Engineering a disulfide-gated switch in streptavidin enables reversible binding without sacrificing binding affinity, Sci. Rep., 2020, 10(1), 12483 CrossRef CAS PubMed.
- J. H. T. Luong and S. K. Vashist, Chemistry of Biotin-Streptavidin and the Growing Concern of an Emerging Biotin Interference in Clinical Immunoassays, ACS Omega, 2020, 5(1), 10–18 CrossRef CAS PubMed.
- N. Kresge, R. Simoni and R. Hill, The Discovery of Avidin by Esmond E. Snell, J. Biol. Chem., 2004, 279, e5 CrossRef CAS.
- O. Livnah, E. A. Bayer, M. Wilchek and J. L. Sussman, The structure of the complex between avidin and the dye, 2-(4′-hydroxyazobenzene) benzoic acid (HABA), FEBS Lett., 1993, 328(1–2), 165–168 CrossRef CAS PubMed.
- O. Livnah, E. A. Bayer, M. Wilchek and J. L. Sussman, Three-dimensional structures of avidin and the avidin-biotin complex, Proc. Natl. Acad. Sci. U. S. A., 1993, 90(11), 5076–5080 CrossRef CAS PubMed.
- P. Lehtolainen, T. Wirth, A. K. Taskinen, P. Lehenkari, O. Leppänen and M. Lappalainen, et al., Targeting of biotinylated compounds to its target tissue using a low-density lipoprotein receptor–avidin fusion protein, Gene Ther., 2003, 10(25), 2090–2097 CrossRef CAS PubMed.
- G. Poletto, D. Cecchin, P. Bartoletti, F. Venturini, N. Realdon and L. Evangelista, Radionuclide Delivery Strategies in Tumor Treatment: A Systematic Review, Curr. Issues Mol. Biol., 2022, 44(8), 3267–3282 CrossRef CAS PubMed.
- S.-Y. Wu, F.-G. Wu and X. Chen, Antibody-Incorporated Nanomedicines for Cancer Therapy, Adv. Mater., 2022, 34(24), 2109210 CrossRef CAS PubMed.
- Z. Guo, L. Zhu, W. Xu, X. Luo, H. Chen and X. Li, et al., PD-L1 ImmunoPET on the basis of Avidin/Biotin pre-targeted cancer imaging, Biochem. Biophys. Res. Commun., 2023, 673, 23–28 CrossRef CAS PubMed.
- S. Wang, M. Z. Hossain, T. Han, K. Shinozuka, T. Suzuki and A. Kuwana, et al., Avidin-Biotin Technology in Gold Nanoparticle-Decorated Graphene Field Effect Transistors for Detection of Biotinylated Macromolecules with Ultrahigh Sensitivity and Specificity, ACS Omega, 2020, 5(46), 30037–30046 CrossRef CAS PubMed.
- X. Zeng, Y.-X. Sun, W. Qu, X.-Z. Zhang and R.-X. Zhuo, Biotinylated transferrin/avidin/biotinylated disulfide containing PEI bioconjugates mediated p53 gene delivery system for tumor targeted transfection, Biomaterials, 2010, 31(17), 4771–4780 CrossRef CAS PubMed.
- M. Ogawa, N. Kosaka, P. L. Choyke and H. Kobayashi, Tumor-specific detection of an optically targeted antibody combined with a quencher-conjugated neutravidin “quencher-chaser”: a dual “quench and chase” strategy to improve target to nontarget ratios for molecular imaging of cancer, Bioconjugate Chem., 2009, 20(1), 147–154 CrossRef CAS PubMed.
- R. P. Chauhan, G. Singh, S. Singh, N. Bag, M. Patra and S. R. Vadera, et al., Biotinylated magnetic nanoparticles for pretargeting: synthesis and characterization study, Cancer Nanotechnol., 2011, 2(1–6), 111–120 CrossRef CAS PubMed.
- D.-J. Chung, K.-C. Kim and S.-H. Choi, Electrochemical DNA biosensor based on avidin–biotin conjugation for influenza virus (type A) detection, Appl. Surf. Sci., 2011, 257(22), 9390–9396 CrossRef CAS.
- H. P. Lesch, M. U. Kaikkonen, J. T. Pikkarainen and S. Ylä-Herttuala, Avidin-biotin technology in targeted therapy, Expert Opin. Drug Delivery, 2010, 7(5), 551–564 CrossRef CAS PubMed.
- W. X. Ren, J. Han, S. Uhm, Y. J. Jang, C. Kang and J.-H. Kim, et al., Recent development of biotin conjugation in biological imaging, sensing, and target delivery, Chem. Commun., 2015, 51(52), 10403–10418 RSC.
- I. Santos-Barriopedro, G. van Mierlo and M. Vermeulen, Off-the-shelf proximity biotinylation using ProtA-TurboID, Nat. Protoc., 2023, 18(1), 36–57 CrossRef CAS PubMed.
- D. P. Minde, M. Ramakrishna and K. S. Lilley, Biotin proximity tagging favours unfolded proteins and enables the study of intrinsically disordered regions, Commun. Biol., 2020, 3(1), 38 CrossRef CAS PubMed.
- S. Yamanaka, Y. Horiuchi, S. Matsuoka, K. Kido, K. Nishino and M. Maeno, et al., A proximity biotinylation-based approach to identify protein-E3 ligase interactions induced by PROTACs and molecular glues, Nat. Commun., 2022, 13(1), 183 CrossRef CAS PubMed.
- C. Sirithanakorn and J. E. Cronan, Biotin, a universal and essential cofactor: synthesis, ligation and regulation, FEMS Microbiol. Rev., 2021, 45(4), 1–18 Search PubMed.
- W. W. Hu, W. J. Syu, W. Y. Chen, R. C. Ruaan, Y. C. Cheng and C. C. Chien, et al., Use of biotinylated chitosan for substrate-mediated gene delivery, Bioconjugate Chem., 2012, 23(8), 1587–1599 CrossRef CAS PubMed.
- A. Nieto-Orellana, H. Li, R. Rosiere, N. Wauthoz, H. Williams, C. J. Monteiro, C. Bosquillon, N. Childerhouse, G. Keegan, D. Coghlan, G. Mantovani and S. Stolnik, Targeted PEG-poly(glutamic acid) complexes for inhalation protein delivery to the lung, J. Controlled Release, 2019, 316, 250–262 CrossRef CAS PubMed.
- L. Wang and F. J. Sigworth, Liposomes on a streptavidin crystal: a system to study membrane proteins by cryo-EM, Methods Enzymol., 2010, 481, 147–164 CAS.
- B. Gautam, C.-H. Luo, H.-D. Gao, J.-C. Hsiao, H.-R. Tseng and H.-M. Lee, et al., Nano-On-Nano: Responsive Nanosubstrate-Mediated Liposome Delivery with High Cellular Uptake Efficiency, ACS Appl. Bio Mater., 2023, 6(4), 1611–1620 CrossRef CAS PubMed.
- H. Wartlick, K. Michaelis, S. Balthasar, K. Strebhardt, J. Kreuter and K. Langer, Highly Specific HER2-mediated Cellular Uptake of Antibody-modified Nanoparticles in Tumour Cells, J. Drug Targeting, 2004, 12(7), 461–471 CrossRef CAS PubMed.
- P. M. Ahmad and F. Ahmad, Mammalian pyruvate carboxylase: effect of biotin on the synthesis and translocation of apo-enzyme into 3T3-L adipocyte mitochondria, FASEB J., 1991, 5(10), 2482–2485 CrossRef CAS PubMed.
- Z. Fatima, X. Jin, Y. Zou, H. Y. Kaw, M. Quinto and D. Li, Recent trends in analytical methods for water-soluble vitamins, J. Chromatogr. A, 2019, 1606, 360245 CrossRef CAS PubMed.
- J. A. Chmiel, G. A. Stuivenberg, K. F. Al, P. P. Akouris, H. Razvi and J. P. Burton, et al., Vitamins as regulators of calcium-containing kidney stones - new perspectives on the role of the gut microbiome, Nat. Rev. Urol., 2023, 20(10), 615–637 CrossRef PubMed.
- A. Dasgupta, Chapter 2-Biotin: Pharmacology, Pathophysiology, and Assessment of Biotin Status, in Biotin and Other Interferences in Immunoassays, Elsevier, Amsterdam, The Netherlands, 2019, pp. 17–35 Search PubMed.
- F. Grafe, M. Brandsch, W. Wohlrab and R. H. Neubert, Transport of biotin in human keratinocytes, J. Invest. Dermatol., 2003, 120(3), 428–433 CrossRef CAS PubMed.
- L. C. Phillips, Sodium-dependent multivitamin transporter: a potential novel cause of cardiomyopathy, Newcastle University, 2021 Search PubMed.
- C. Neophytou and C. Pitsouli, Biotin controls intestinal stem cell mitosis and host-microbiome interactions, Cell Rep., 2022, 38(10), 110505 CrossRef CAS PubMed.
- M. Montomoli, A. Vetro, F. Tubili, M. A. Donati, M. Daniotti and F. Pochiero, et al., A novel SLC5A6 homozygous variant in a family with multivitamin-dependent neurometabolic disorder: Phenotype expansion and long-term follow-up, Eur. J. Med. Genet., 2023, 66(8), 104808 CrossRef CAS PubMed.
- C. H. Hsieh, J. Lee, H. H. Sung, Y. F. Huang, Y. S. Ding and C. Y. Li, et al., Novel SLC5A6 mutations lead to B lymphocyte maturation defects with metabolic abnormality rescuable by biotin replenishment, Clin. Immunol., 2023, 257, 109855 CrossRef CAS PubMed.
- T. Holling, S. Nampoothiri, B. Tarhan, P. E. Schneeberger, K. P. Vinayan and D. Yesodharan, et al., Novel biallelic variants expand the SLC5A6-related phenotypic spectrum, Eur. J. Hum. Genet., 2022, 30(4), 439–449 CAS.
- M. Patel, R. K. Vadlapatla, S. Shah and A. K. Mitra, Molecular expression and functional activity of sodium dependent multivitamin transporter in human prostate cancer cells, Int. J. Pharm., 2012, 436(1–2), 324–331 CrossRef CAS PubMed.
- K. T. Fam, M. Collot and A. S. Klymchenko, Probing biotin receptors in cancer cells with rationally designed fluorogenic squaraine dimers, Chem. Sci., 2020, 11(31), 8240–8248 RSC.
- L. Kou, Q. Yao, H. Zhang, M. Chu, Y. D. Bhutia and R. Chen, et al., Transporter-targeted nano-sized vehicles for enhanced and site-specific drug delivery, Cancers, 2020, 12(10), 2837 CrossRef CAS PubMed.
- I. Ojima, E. S. Zuniga, W. T. Berger and J. D. Seitz, Tumor-targeting drug delivery of new-generation taxoids, Future Med. Chem., 2012, 4(1), 33–50 CrossRef CAS PubMed.
- J. Liu, Y. Y. Liu, C. S. Li, A. Cao and H. Wang, Exocytosis of Nanoparticles: A Comprehensive Review, Nanomaterials, 2023, 13(15), 2215 CrossRef CAS PubMed.
- R. Augustine, A. Hasan, R. Primavera, R. J. Wilson, A. S. Thakor and B. D. Kevadiya, Cellular uptake and retention of nanoparticles: Insights on particle properties and interaction with cellular components, Mater. Today Commun., 2020, 25, 101692 CrossRef CAS.
- V. Balan, G. Dodi, C. T. Mihai, A. M. Serban and V. C. Ursachi, Biotinylated chitosan macromolecule based nanosystems: A review from chemical design to biological targets, Int. J. Biol. Macromol., 2021, 188, 82–93 CrossRef CAS PubMed.
- S. Chen, X. Zhao, J. Chen, J. Chen, L. Kuznetsova and S. S. Wong, et al., Mechanism-Based Tumor-Targeting Drug Delivery System. Validation of Efficient Vitamin Receptor-Mediated Endocytosis and Drug Release, Bioconjugate Chem., 2010, 21(5), 979–987 CrossRef CAS PubMed.
- R. Al-Obaidy, A. J. Haider, S. Al-Musawi and N. Arsad, Targeted delivery of paclitaxel drug using polymer-coated magnetic nanoparticles for fibrosarcoma therapy: in vitro and in vivo studies, Sci. Rep., 2023, 13(1), 3180 CrossRef CAS PubMed.
- A. A. Hariri, S. S. Newman, S. Tan, D. Mamerow, A. M. Adams and N. Maganzini, et al., Improved immunoassay sensitivity and specificity using single-molecule colocalization, Nat. Commun., 2022, 13(1), 5359 CrossRef CAS PubMed.
- R. Gupta, P. Gupta, S. Wang, A. Melnykov, Q. Jiang and A. Seth, et al., Ultrasensitive lateral-flow assays via plasmonically active antibody-conjugated fluorescent nanoparticles, Nat. Biomed. Eng., 2023, 7(12), 1556–1570 CrossRef CAS PubMed.
- M. Haun and S. Wasi, Biotinylated antibodies bound to streptavidin beads: a versatile solid matrix for immunoassays, Anal. Biochem., 1990, 191(2), 337–342 CrossRef CAS PubMed.
- M. J. Khosravi and R. C. Morton, Novel application of streptavidin-hapten derivatives as protein-tracer conjugate in competitive-type immunoassays involving biotinylated detection probes, Clin. Chem., 1991, 37(1), 58–63 CrossRef CAS.
- M. Mishra, S. Tiwari, A. Gunaseelan, D. Li, B. D. Hammock and A. V. Gomes, Improving the sensitivity of traditional Western blotting via Streptavidin containing Poly-horseradish peroxidase (PolyHRP), Electrophoresis, 2019, 40(12–13), 1731–1739 CrossRef CAS PubMed.
- T. Lakshmipriya, S. C. Gopinath and T. H. Tang, Biotin-Streptavidin Competition Mediates Sensitive Detection of Biomolecules in Enzyme Linked Immunosorbent Assay, PLoS One, 2016, 11(3), e0151153 CrossRef PubMed.
- M. K. O. Grant, S. L. Shapiro, K. H. Ashe, P. Liu and K. R. Zahs, A Cautionary Tale: Endogenous Biotinylated Proteins and Exogenously-Introduced Protein A Cause Antibody-Independent Artefacts in Western Blot Studies of Brain-Derived Proteins, Biol. Proced. Online, 2019, 21(1), 6 CrossRef PubMed.
- N. Wang, C. H. Gibbons and R. Freeman, Novel immunohistochemical techniques using discrete signal amplification systems for human cutaneous peripheral nerve fiber imaging, J. Histochem. Cytochem., 2011, 59(4), 382–390 CrossRef CAS PubMed.
- I. A. Darwish, Immunoassay Methods and their Applications in Pharmaceutical Analysis: Basic Methodology and Recent Advances, Int. J. Biomed. Sci., 2006, 2(3), 217–235 CrossRef CAS PubMed.
- J. H. T. Luong, K. B. Male and J. D. Glennon, Biotin interference in immunoassays based on biotin-strept(avidin) chemistry: An emerging threat, Biotechnol. Adv., 2019, 37(5), 634–641 CrossRef CAS PubMed.
- S. K. Grebe, Laboratory testing in thyroid disorders, in The Thyroid and Its Diseases: A Comprehensive Guide for the Clinician, 2019, pp. 129–159 Search PubMed.
- M. R. Kosorok and E. B. Laber, Precision medicine, Annu. Rev. Stat. Appl., 2019, 6, 263–286 CrossRef PubMed.
- J. Li, L. Jin, Z. Wang, Q. Peng, Y. Wang and J. Luo, et al., Towards precision medicine based on a continuous deep learning optimization and ensemble approach, NPJ Digit. Med., 2023, 6(1), 18 CrossRef PubMed.
- C. Y. Lu, V. Terry and D. M. Thomas, Precision medicine: affording the successes of science, NPJ Precis. Oncol., 2023, 7(1), 3 CrossRef PubMed.
- E. M. Czekanska, J. Geng, M. Glinka, K. White, J. Kanczler and N. D. Evans, et al., Combinatorial delivery of bioactive molecules by a nanoparticle-decorated and functionalized biodegradable scaffold, J. Mater. Chem. B, 2018, 6(27), 4437–4445 RSC.
- I. Venugopal, A. I. Mehta and A. A. Linninger, Drug Delivery Applications of Nanoparticles in the Spine, Methods Mol. Biol., 2020, 2059, 121–143 CrossRef CAS PubMed.
- Y. Wang, Z. Li, F. Mo, T.-J. Chen-Mayfield, A. Saini and A. M. LaMere, et al., Chemically engineering cells for precision medicine, Chem. Soc. Rev., 2023, 1068–1102 RSC.
- H. Liu, J. Cao and S.-N. Ding, Simultaneous detection of two ovarian cancer biomarkers in human serums with biotin-enriched dendritic mesoporous silica nanoparticles-labeled multiplex lateral flow immunoassay, Sens. Actuators, B, 2022, 371, 132597 CrossRef CAS.
- A. Mobed, F. Kohansal, S. Dolati and M. Hasanzadeh, Label-free immunosensing of telomerase using bio-conjugation of biotinylated antibody to poly(chitosan) gold nanoparticles, Bioanalysis, 2023, 567–580 CrossRef CAS PubMed.
- T. Yudhistira, E. C. Da Silva, A. Combes, M. Lehmann, A. Reisch and A. S. Klymchenko, Biotinylated Fluorescent Polymeric Nanoparticles for Enhanced Immunostaining, Small Methods, 2023, 7(4), 2201452 CrossRef CAS PubMed.
- S. Aniu Lincy, Y. Allwin Richard, T. Vinitha, K. Balamurugan and V. Dharuman, Streptavidin Fe2O3-gold nanoparticles functionalized theranostic liposome for antibiotic resistant bacteria and biotin sensing, Biosens. Bioelectron., 2023, 219, 114849 CrossRef CAS PubMed.
- A. C. da Silva, C. F. de Freitas, C. A. Cardinali, T. L. Braga, W. Caetano and M. Ida Bonini Ravanelli, et al., Biotin-functionalized silica nanoparticles loaded with Erythrosine B asselective photodynamic treatment for Glioblastoma Multiforme, J. Mol. Liq., 2022, 345, 117898 CrossRef.
- D. Panjwani, S. Patel, D. Mishra, V. Patel, M. Yadav and A. Dharamsi, et al., Avidin-Biotin functionalized self-assembled protein nanoparticles as EGFR targeted therapeutics for the treatment of lung cancer: characterization and cell viability, J. Dispersion Sci. Technol., 2022, 1–14 Search PubMed.
- K. Xiong, C. Ouyang, J. Liu, J. Karges, X. Lin and X. Chen, et al., Chiral RuII-PtII Complexes Inducing Telomere Dysfunction against Cisplatin-Resistant Cancer Cells, Angew. Chem., Int. Ed., 2022, 61(33), e202204866 CrossRef CAS PubMed.
- G. Moku, V. R. Gopalsamuthiram, T. R. Hoye and J. Panyam, Surface modification of nanoparticles: methods and applications, in Surface modification of polymers: Methods and applications, 2019, pp. 317–346 Search PubMed.
- M. Azizi, M. Shahgolzari, S. Fathi-Karkan, M. Ghasemi and H. Samadian, Multifunctional plant virus nanoparticles: An emerging strategy for therapy of cancer, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, e1872 Search PubMed.
- M. T. Manzari, Y. Shamay, H. Kiguchi, N. Rosen, M. Scaltriti and D. A. Heller, Targeted drug delivery strategies for precision medicines, Nat. Rev. Mater., 2021, 6(4), 351–370 CrossRef CAS PubMed.
- R. Lawrence, M. Watters, C. R. Davies, K. Pantel and Y. J. Lu, Circulating tumour cells for early detection of clinically relevant cancer, Nat. Rev. Clin Oncol., 2023, 20(7), 487–500 CrossRef PubMed.
- M. P. Ward, L. E. Kane, L. A. Norris, B. M. Mohamed, T. Kelly and M. Bates, et al., Platelets, immune cells and the coagulation cascade; friend or foe of the circulating tumour cell?, Mol. Cancer, 2021, 20(1), 1–17 CrossRef PubMed.
- C. Mazzitelli, D. Santini, A. G. Corradini, C. Zamagni, D. Trerè and L. Montanaro, et al., Liquid Biopsy in the Management of Breast Cancer Patients: Where Are We Now and Where Are We Going, Diagnostics, 2023, 13(7), 1241 CrossRef CAS PubMed.
- A. Ring, B. D. Nguyen-Sträuli, A. Wicki and N. Aceto, Biology, vulnerabilities and clinical applications of circulating tumour cells, Nat. Rev. Cancer, 2023, 23(2), 95–111 CrossRef CAS PubMed.
- S. Wang, J. Cui, Q. Fan, J. Gan, C. Liu and Y. Wang, et al., Reversible and Highly Ordered Biointerfaces for Efficient Capture and Nondestructive Release of Circulating Tumor Cells, Anal. Chem., 2022, 94(26), 9450–9458 CrossRef CAS PubMed.
- N.-N. Lu, M. Xie, J. Wang, S.-W. Lv, J.-S. Yi and W.-G. Dong, et al., Biotin-triggered decomposable immunomagnetic beads for capture and release of circulating tumor cells, ACS Appl. Mater. Interfaces, 2015, 7(16), 8817–8826 CrossRef CAS PubMed.
- H. J. Lee, H.-Y. Cho, J. H. Oh, K. Namkoong, J. G. Lee and J.-M. Park, et al., Simultaneous capture and in situ analysis of circulating tumor cells using multiple hybrid nanoparticles, Biosens. Bioelectron., 2013, 47, 508–514 CrossRef CAS PubMed.
- C. Pipatwatcharadate, P. R. Iyer and D. Pissuwan, Recent Update Roles of Magnetic Nanoparticles in Circulating Tumor Cell (CTC)/Non-CTC Separation, Pharmaceutics, 2023, 15(10), 2482 CrossRef CAS PubMed.
- N. Liang, L. Liu, P. Li, Y. Xu, Y. Hou and J. Peng, et al., Efficient isolation and quantification of circulating tumor cells in non-small cell lung cancer patients using peptide-functionalized magnetic nanoparticles, J. Thorac. Dis., 2020, 12(8), 4262 CrossRef PubMed.
- F.-B. Wang, Y. Rong, M. Fang, J.-P. Yuan, C.-W. Peng and S.-P. Liu, et al., Recognition and capture of metastatic hepatocellular carcinoma cells using aptamer-conjugated quantum dots and magnetic particles, Biomaterials, 2013, 34(15), 3816–3827 CrossRef CAS PubMed.
- J. Zuo, Z. Zhang, M. Luo, L. Zhou, E. C. Nice and W. Zhang, et al., Redox signaling at the crossroads of human health and disease, MedComm, 2022, 3(2), e127 CrossRef CAS PubMed.
- P. V. Oliveira and F. R. Laurindo, Implications of plasma thiol redox in disease, Clin. Sci., 2018, 132(12), 1257–1280 CrossRef CAS PubMed.
- E. L. Lieu, T. Nguyen, S. Rhyne and J. Kim, Amino acids in cancer, Exp. Mol. Med., 2020, 52(1), 15–30 CrossRef CAS PubMed.
- N. P. Ward and G. M. DeNicola, Sulfur metabolism and its contribution to malignancy, Int. Rev. Cell Mol. Biol., 2019, 347, 39–103 CAS.
- M. Chen, D. Liu, F. Liu, Y. Wu, X. Peng and F. Song, Recent advances of redox-responsive nanoplatforms for tumor theranostics, J. Controlled Release, 2021, 332, 269–284 CrossRef CAS PubMed.
- H. Kong and N. S. Chandel, Regulation of redox balance in cancer and T cells, J. Biol. Chem., 2018, 293(20), 7499–7507 CrossRef CAS PubMed.
- D. Jung, S. Maiti, J. H. Lee, J. H. Lee and J. S. Kim, Rational design of biotin–disulfide–coumarin conjugates: A cancer targeted thiol probe and bioimaging, Chem. Commun., 2014, 50(23), 3044–3047 RSC.
- H. Zhou, D. Tang, Y. Yu, L. Zhang, B. Wang and J. Karges, et al., Theranostic imaging and multimodal photodynamic therapy and immunotherapy using the mTOR signaling pathway, Nat. Commun., 2023, 14(1), 5350 CrossRef CAS PubMed.
- Y. Yang, B. Sun, S. Zuo, X. Li, S. Zhou and L. Li, et al., Trisulfide bond–mediated doxorubicin dimeric prodrug nanoassemblies with high drug loading, high self-assembly stability, and high tumor selectivity, Sci. Adv., 2020, 6(45), eabc1725 CrossRef CAS PubMed.
- S. Bhuniya, S. Maiti, E. J. Kim, H. Lee, J. L. Sessler and K. S. Hong, et al., An activatable theranostic for targeted cancer therapy and imaging, Angew. Chem., 2014, 126(17), 4558–4563 CrossRef.
- J. He, C. Li, L. Ding, Y. Huang, X. Yin and J. Zhang, et al., Tumor targeting strategies of smart fluorescent nanoparticles and their applications in cancer diagnosis and treatment, Adv. Mater., 2019, 31(40), 1902409 CrossRef CAS PubMed.
- S. Chakravorty, A. Malvi, A. Chaturvedi, K. Sonkusare and N. Dave, Glutathione–the master antioxidant, Int. J. Med. Res. Pharm. Sci., 2020, 7(2), 1–11 Search PubMed.
- L. Kennedy, J. K. Sandhu, M.-E. Harper and M. Cuperlovic-Culf, Role of glutathione in cancer: From mechanisms to therapies, Biomolecules, 2020, 10(10), 1429 CrossRef CAS PubMed.
- Y. Yuan, R. T. Kwok, G. Feng, J. Liang, J. Geng and B. Z. Tang, et al., Rational design of fluorescent light-up probes based on an AIE luminogen for targeted intracellular thiol imaging, Chem. Commun., 2014, 50(3), 295–297 RSC.
- A. W. Scott, V. Garimella, C. M. Calabrese and C. A. Mirkin, Universal Biotin–PEG-linked gold nanoparticle probes for the simultaneous detection of nucleic acids and proteins, Bioconjugate Chem., 2017, 28(1), 203–211 CrossRef CAS PubMed.
- X. Pei, F. Huo, Y. Yue, T. Chen and C. Yin, Cancer cell recognition by a Cys-reactive turn-on significant enhanced fluorescent emission targeting biotin receptors, Sens. Actuators, B, 2020, 304, 127431 CrossRef CAS.
- M. Pietzke, G. Burgos-Barragan, N. Wit, J. Tait-Mulder, D. Sumpton and G. M. Mackay, et al., Amino acid dependent formaldehyde metabolism in mammals, Commun. Chem., 2020, 3(1), 78 CrossRef CAS PubMed.
- D. Adamović, Z. Čepić, S. Adamović, M. Stošić, B. Obrovski and S. Morača, et al., Occupational exposure to formaldehyde and cancer risk assessment in an anatomy laboratory, Int. J. Environ. Res. Public Health, 2021, 18(21), 11198 CrossRef PubMed.
- E. Agathokleous and E. J. Calabrese, Formaldehyde: Another hormesis-inducing chemical, Environ. Res., 2021, 199, 111395 CrossRef CAS PubMed.
- L. D. Adair, N. K. Tan and E. J. New, Activity–based Fluorescent Sensors and Their Applications in Biological Studies, Mol. Fluoresc. Sens. Cell. Stud., 2022, 129–172 CAS.
- Y. H. Lee, Y. Tang, P. Verwilst, W. Lin and J. S. Kim, A biotin-guided formaldehyde sensor selectively detecting endogenous concentrations in cancerous cells and tissues, Chem. Commun., 2016, 52(75), 11247–11250 RSC.
- S. Kumar, M. Sanchez-Alvarez, F. N. Lolo, F. Trionfetti, R. Strippoli and M. Cordani, Autophagy and the Lysosomal System in Cancer, Cells, 2021, 10(10), 2752 CrossRef CAS PubMed.
- M. Cao, X. Luo, K. Wu and X. He, Targeting lysosomes in human disease: from basic research to clinical applications, Signal Transduction Targeted Ther., 2021, 6(1), 379 CrossRef CAS PubMed.
- N.-E. Choi, J.-Y. Lee, E.-C. Park, J.-H. Lee and J. Lee, Recent advances in organelle-targeted fluorescent probes, Molecules, 2021, 26(1), 217 CrossRef CAS PubMed.
- B. Dong, X. Song, X. Kong, C. Wang, N. Zhang and W. Lin, A tumor-targeting and lysosome-specific two-photon fluorescent probe for imaging pH changes in living cells, J. Mater. Chem. B, 2017, 5(5), 988–995 RSC.
- R. P. Bhole, S. Jadhav, Y. B. Zambare, R. V. Chikhale and C. G. Bonde, Vitamin-anticancer drug conjugates: a new era for cancer therapy, İstanbul J. Pharm., 2020, 50(3), 312–322 Search PubMed.
- X. Cai, Z. Zhang, Y. Dong, T. Hao, L. Yi and X. Yang, A biotin-guided near-infrared fluorescent probe for imaging hydrogen sulfide and differentiating cancer cells, Org. Biomol. Chem., 2023, 332–338 RSC.
- Y. Dai, Y. Zhang, T. Ye and Y. Chen, Synthesis and Antitumor Evaluation of Biotin-SN38-Valproic Acid Conjugates, Molecules, 2023, 28(9), 3936 CrossRef CAS PubMed.
- E. Y. Hanurry, T. W. Mekonnen, A. T. Andrgie, H. F. Darge, Y. S. Birhan and W.-H. Hsu, et al., Biotin-Decorated PAMAM G4. 5 dendrimer nanoparticles to enhance the delivery, anti-proliferative, and apoptotic effects of chemotherapeutic drug in cancer cells, Pharmaceutics, 2020, 12(5), 443 CrossRef CAS PubMed.
- G. Li, Z. Yang, C. Zhang, C. Li, Y. Zheng and X. Wu, et al., Pharmacological research progress and prospects of spiders toxins especially on ion channels, Int. J. Adv. Pharm., Biol. Chem., 2015, 4(1), 171–195 CAS.
- S. Mirzaei, H. S. Fekri, F. Hashemi, K. Hushmandi, R. Mohammadinejad and M. Ashrafizadeh, et al., Venom peptides in cancer therapy: An updated review on cellular and molecular aspects, Pharmacol. Res., 2021, 164, 105327 CrossRef CAS PubMed.
- J. Montnach, L. A. Blömer, L. Lopez, L. Filipis, H. Meudal and A. Lafoux, et al., In vivo spatiotemporal control of voltage-gated ion channels by using photoactivatable peptidic toxins, Nat. Commun., 2022, 13(1), 417 CrossRef CAS PubMed.
- F. Yang, X. Sun, Y. Ding, H. Ma, T. O. Yang and Y. Ma, et al., Astrocytic acid-sensing ion channel 1a contributes to the development of chronic epileptogenesis, Sci. Rep., 2016, 6(1), 31581 CrossRef CAS PubMed.
- S. Atd, G. S. Cruz and G. R. Baptista, Anti-inflammatory activities of arthropod peptides: a systematic review, J. Venomous Anim. Toxins Incl. Trop. Dis., 2021, 27 DOI:10.1590/1678-9199-JVATITD-2020-0152.
- Y.-Z. Yan, H. Liu, R. Lv, S. Huang, J.-R. Wang and T.-Y. Guo, et al., Biotin labeling and receptor analysis of huwentoxin-I, Anal. Methods, 2013, 5(16), 3888–3894 RSC.
- H. Yu, H. Liu, Y. Yan, Z. Duan and X. Wang, Detection and identification of huwentoxin-IV interacting proteins by biotin-avidin chemistry combined with mass spectrometry, J. Venomous Anim. Toxins Incl. Trop. Dis., 2014, 20, 1–9 CrossRef PubMed.
- W.-Z. Lin, Y.-H. Chen, C.-K. Liang, C.-C. Liu and S.-Y. Hou, A competitive immunoassay for biotin detection using magnetic beads and gold nanoparticle probes, Food Chem., 2019, 271, 440–444 CrossRef CAS PubMed.
- N. D. Udeshi, K. Pedram, T. Svinkina, S. Fereshetian, S. A. Myers and O. Aygun, et al., Antibodies to biotin enable large-scale detection of biotinylation sites on proteins, Nat. Methods, 2017, 14(12), 1167–1170 CrossRef CAS PubMed.
- G. Niedobitek, T. Finn, H. Herbst and H. Stein, In situ hybridization using biotinylated probes: an evaluation of different detection systems, Pathol., Res. Pract., 1989, 184(3), 343–348 CrossRef CAS PubMed.
- P. Pavlickova, N. M. Jensen, H. Paul, M. Schaeferling, C. Giammasi and M. Kruschina, et al., Antibody detection in human serum using a versatile protein chip platform constructed by applying nanoscale self-assembled architectures on gold, J. Proteome Res., 2002, 1(3), 227–231 CrossRef CAS PubMed.
- M. Mansouri, F. Fathi, R. Jalili, S. Shoeibie, S. Dastmalchi and A. Khataee, et al., SPR enhanced DNA biosensor for sensitive detection of donkey meat adulteration, Food Chem., 2020, 331, 127163 CrossRef CAS PubMed.
- H. Yang, X. Zhao, Z. Zhang, P. Ma, X. Wang and D. Song, et al., Biotin-streptavidin sandwich integrated PDA-ZnO@ Au nanocomposite based SPR sensor for hIgG detection, Talanta, 2022, 246, 123496 CrossRef CAS PubMed.
- X. Wang, S. Li, R. Zhao, H. Li, P. Gao and C. Jin, et al., Development of a Novel Recombinant Full-Length IgY Monoclonal Antibody against Human Thymidine Kinase 1 for Automatic Chemiluminescence Analysis on a Sandwich Biotin-Streptavidin Platform for Early Tumour Discovery, J. Immunol. Res., 2023, 2023, 7612566 Search PubMed.
- K. K. Reddy, H. Bandal, M. Satyanarayana, K. Y. Goud, K. V. Gobi and T. Jayaramudu, et al., Recent trends in electrochemical sensors for vital biomedical markers using hybrid nanostructured materials, Adv. Sci., 2020, 7(13), 1902980 CrossRef CAS PubMed.
- M. Li, F. Jiang, L. Xue, C. Peng, Z. Shi and Z. Zhang, et al., Recent Progress in Biosensors for Detection of Tumor Biomarkers, Molecules, 2022, 27(21), 7327 CrossRef CAS PubMed.
- K. M. Al-Qaoud, Y. M. Obeidat, T. Al-Omari, M. Okour, M. M. Al-Omari and M. I. Ahmad, et al., The development of an electrochemical immunosensor utilizing chicken IgY anti-spike antibody for the detection of SARS-CoV-2, Sci. Rep., 2024, 14(1), 748 CrossRef CAS PubMed.
- H. Abdulkarim and M. Siaj, Label-free multiplex electrochemical immunosensor for early diagnosis of lysosomal storage disorders, Sci. Rep., 2022, 12(1), 9334 CrossRef CAS PubMed.
- A. G. Chaudhuri, S. Samanta, M. Dey, N. Raviraja and S. Dey, Role of Alpha-Fetoprotein in the Pathogenesis of Cancer, J. Environ. Pathol., Toxicol. Oncol., 2024, 43, 57–76 CrossRef PubMed.
- A. X. Zhu, R. S. Finn, Y. K. Kang, C. J. Yen, P. R. Galle and J. M. Llovet, et al., Serum alpha-fetoprotein and clinical outcomes in patients with advanced hepatocellular carcinoma treated with ramucirumab, Br. J. Cancer, 2021, 124(8), 1388–1397 CrossRef CAS PubMed.
- T. Chen, X. Dai, J. Dai, C. Ding, Z. Zhang and Z. Lin, et al., AFP promotes HCC progression by suppressing the HuR-mediated Fas/FADD apoptotic pathway, Cell Death Dis., 2020, 11(10), 822 CrossRef CAS PubMed.
- A. Mohammadinejad, R. K. Oskuee, R. Eivazzadeh-Keihan, M. Rezayi, B. Baradaran and A. Maleki, et al., Development of biosensors for detection of alpha-fetoprotein: As a major biomarker for hepatocellular carcinoma, TrAC, Trends Anal. Chem., 2020, 130, 115961 CrossRef CAS.
- L. Jiang, F. Li, J. Feng, P. Wang, Q. Liu and Y. Li, et al., An optionality further amplification of an sandwich-type electrochemical immunosensor based on biotin–streptavidin–biotin strategy for detection of alpha fetoprotein, RSC Adv., 2016, 6(29), 24373–24380 RSC.
- S. M. M. Dadfar, S. Sekula-Neuner, V. Trouillet, H.-Y. Liu, R. Kumar and A. K. Powell, et al., Evaluation of click chemistry microarrays for immunosensing of alpha-fetoprotein (AFP), Beilstein J. Nanotechnol., 2019, 10(1), 2505–2515 CrossRef CAS PubMed.
- J. Chen and G.-C. Zhao, A novel signal-on photoelectrochemical immunosensor for detection of alpha-fetoprotein by in situ releasing electron donor, Biosens. Bioelectron., 2017, 98, 155–160 CrossRef CAS PubMed.
- J. Li, T. Gao, S. Gu, J. Zhi, J. Yang and G. Li, An electrochemical biosensor for the assay of alpha-fetoprotein-L3 with practical applications, Biosens. Bioelectron., 2017, 87, 352–357 CrossRef CAS PubMed.
- N. E. Rich, B. V. John, N. D. Parikh, I. Rowe, N. Mehta and G. Khatri, et al., Hepatocellular Carcinoma Demonstrates Heterogeneous Growth Patterns in a Multicenter Cohort of Patients With Cirrhosis, Hepatology, 2020, 72(5), 1654–1665 CrossRef CAS PubMed.
- Y. Su, T. Xue, L. Wu, Y. Hu, J. Wang and Q. Xu, et al., Label-free detection of biomarker alpha fetoprotein in serum by ssDNA aptamer functionalized magnetic nanoparticles, Nanotechnology, 2019, 31(9), 095104 CrossRef PubMed.
- V. Subramaniyan, S. Fuloria, G. Gupta, D. H. Kumar, M. Sekar and K. V. Sathasivam, et al., A review on epidermal growth factor receptor's role in breast and non-small cell lung cancer, Chem.-Biol. Interact., 2022, 351, 109735 CrossRef CAS PubMed.
- P. Krawczyk, D. M. Kowalski, R. Ramlau, E. Kalinka-Warzocha, K. Winiarczyk and K. Stencel, et al., Comparison of the effectiveness of erlotinib, gefitinib, and afatinib for treatment of non-small cell lung cancer in patients with common and rare EGFR gene mutations, Oncol. Lett., 2017, 13(6), 4433–4444 CrossRef CAS PubMed.
- L. Xue, J.-J. Fei, Y. Song, R.-H. Xu and Y.-J. Bai, Visual DNA microarray for detection of epidermal growth factor receptor (EGFR) gene mutations, Scand. J. Clin. Lab. Invest., 2014, 74(8), 693–699 CrossRef CAS PubMed.
- H. Ilkhani, M. Sarparast, A. Noori, S. Z. Bathaie and M. F. Mousavi, Electrochemical aptamer/antibody based sandwich immunosensor for the detection of EGFR, a cancer biomarker, using gold nanoparticles as a signaling probe, Biosens. Bioelectron., 2015, 74, 491–497 CrossRef CAS PubMed.
- K. Maruyama, H. Takeyama, T. Mori, K. Ohshima, S.-I. Ogura and T. Mochizuki, et al., Detection of epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) using a fully automated system with a nano-scale engineered biomagnetite, Biosens. Bioelectron., 2007, 22(9–10), 2282–2288 CrossRef CAS PubMed.
- P. Suttipasit and S. Wongwittayapanich, Detection of prostate specific antigen and semenogelin in specimens from female rape victims, J. Forensic Leg. Med., 2018, 54, 102–108 CrossRef PubMed.
- S. Rasheed, T. Kanwal, N. Ahmad, B. Fatima, M. Najam-ul-Haq and D. Hussain, Advances and challenges in portable optical biosensors for onsite detection and point-of-care diagnostics, TrAC, Trends Anal. Chem., 2024, 117640 CrossRef CAS.
- F. Farshchi, A. Saadati and M. Hasanzadeh, A novel immunosensor for the monitoring of PSA using binding of biotinylated antibody to the prostate specific antigen based on nano-ink modified flexible paper substrate: efficient method for diagnosis of cancer using biosensing technology, Heliyon, 2020, 6(7), e04327 CrossRef PubMed.
- Y. Wei, J. Zhang, X. Yang, Z. Wang, J. Wang and H. Qi, et al., Performance enhancement of electrochemiluminescence magnetic microbiosensors by using double magnetic field actuation for cancer biomarkers and exosomes, Talanta, 2023, 259, 124485 CrossRef CAS PubMed.
- C. Hall, L. Clarke, A. Pal, P. Buchwald, T. Eglinton and C. Wakeman, et al., A review of the role of carcinoembryonic antigen in clinical practice, Ann. Coloproctol., 2019, 35(6), 294 CrossRef PubMed.
- Q. Zhu, Y. Chai, Y. Zhuo and R. Yuan, Ultrasensitive simultaneous detection of four biomarkers based on hybridization chain reaction and biotin–streptavidin signal amplification strategy, Biosens. Bioelectron., 2015, 68, 42–48 CrossRef CAS PubMed.
- Y. Cao, J. Feng, L. Tang, G. Mo, W. Mo and B. Deng, Detection of three tumor biomarkers in human lung cancer serum using single particle inductively coupled plasma mass spectrometry combined with magnetic immunoassay, Spectrochim. Acta, Part B, 2020, 166, 105797 CrossRef CAS.
- P. Charkhchi, C. Cybulski, J. Gronwald, F. O. Wong, S. A. Narod and M. R. Akbari, CA125 and Ovarian Cancer: A Comprehensive Review, Cancers, 2020, 12(12), 3730 CrossRef PubMed.
- J. Bailes, Photostability of Semiconductor Quantum Dots in Response to UV Exposure, Methods Mol. Biol., 2020, 2118, 343–349 CrossRef CAS PubMed.
- W. E. Omer, M. F. Abdelbar, N. M. El-Kemary, N. Fukata and M. A. El-Kemary, Cancer antigen 125 assessment using carbon quantum dots for optical biosensing for the early diagnosis of ovarian cancer, RSC Adv., 2021, 11(49), 31047–31057 RSC.
- W. W.-S. Wang, D. Das, S. A. McQuarrie and M. R. Suresh, Design of a bifunctional fusion protein for ovarian cancer drug delivery: single-chain anti-CA125 core-streptavidin fusion protein, Eur. J. Pharm. Biopharm., 2007, 65(3), 398–405 CrossRef CAS PubMed.
- J. M. Ryu, D. Kang, J. Cho, J. E. Lee, S. W. Kim and S. J. Nam, et al., Prognostic Impact of Elevation of Cancer Antigen 15–3 (CA15–3) in Patients With Early Breast Cancer With Normal Serum CA15–3 Level, J. Breast Cancer, 2023, 26(2), 126–135 CrossRef PubMed.
- M. J. Salih, L. Al-Mansouri and H. M. Hasson, Exploring the Prognostic Value of CS15–3 Tumor Marker in Breast Cancer Recurrence, Asian Pac. J. Cancer Prev., 2024, 25(1), 229–232 CrossRef CAS PubMed.
- S. Saharti and M. Qari Elevated CA15-3 Levels in Myeloid Disorders: Clinicopathological Correlation. 2022.
- S. A. Nakhjavani, B. Khalilzadeh, P. S. Pakchin, R. Saber, M. H. Ghahremani and Y. Omidi, A highly sensitive and reliable detection of CA15–3 in patient plasma with electrochemical biosensor labeled with magnetic beads, Biosens. Bioelectron., 2018, 122, 8–15 CrossRef PubMed.
- X. Zhang, X. Peng and W. Jin, Scanning electrochemical microscopy with enzyme immunoassay of the cancer-related antigen CA15–3, Anal. Chim. Acta, 2006, 558(1–2), 110–114 CrossRef CAS.
- G. Gonzalez-Avila, B. Sommer, D. A. Mendoza-Posada, C. Ramos, A. A. Garcia-Hernandez and R. Falfan-Valencia, Matrix metalloproteinases participation in the metastatic process and their diagnostic and therapeutic applications in cancer, Crit. Rev. Oncol. Hematol., 2019, 137, 57–83 CrossRef PubMed.
- G. I. Murray, M. E. Duncan, P. O'Neil, J. A. McKay, W. T. Melvin and J. E. Fothergill, Matrix metalloproteinase-1 is associated with poor prognosis in oesophageal cancer, J. Pathol., 1998, 185(3), 256–261 CrossRef CAS PubMed.
- G. Zhang, T. Li, G. Tan, Y. Song, Q. Liu and K. Wang, et al., Identity of MMP1 and its effects on tumor progression in head and neck squamous cell carcinoma, Cancer Med., 2022, 11(12), 2516–2530 CrossRef CAS PubMed.
- L. Dai, J. Mugaanyi, X. Cai, M. Dong, C. Lu and C. Lu, Comprehensive bioinformatic analysis of MMP1 in hepatocellular carcinoma and establishment of relevant prognostic model, Sci. Rep., 2022, 12(1), 13639 CrossRef CAS PubMed.
- Z.-T. Zhong, G. Ashraf, W. Chen, B. Liu, G.-P. Wang and Y.-D. Zhao, Detection of Matrix Metalloproteinase-1 in Human Saliva Based on a Pregnancy Test Strip Platform, Anal. Chem., 2022, 94(47), 16384–16392 CrossRef CAS PubMed.
- S. M. Swain, M. Shastry and E. Hamilton, Targeting HER2-positive breast cancer: advances and future directions, Nat. Rev. Drug Discovery, 2023, 22(2), 101–126 CrossRef CAS PubMed.
- M. Gamez-Chiachio, A. Molina-Crespo, C. Ramos-Nebot, J. Martinez-Val, L. Martinez and K. Gassner, et al., Gasdermin B over-expression modulates HER2-targeted therapy resistance by inducing protective autophagy through Rab7 activation, J. Exp. Clin. Cancer Res., 2022, 41(1), 285 CrossRef CAS PubMed.
- S. Chupradit, S. A. Jasim, D. Bokov, M. Z. Mahmoud, A. B. Roomi and K. Hachem, et al., Recent advances in biosensor devices for HER-2 cancer biomarker detection, Anal. Methods, 2022, 14(13), 1301–1310 RSC.
- Z. Wang, W. Wang, X. Bu, Z. Wei, L. Geng and Y. Wu, et al., Microarray based screening of peptide nano probes for HER2 positive tumor, Anal. Chem., 2015, 87(16), 8367–8372 CrossRef CAS PubMed.
- T. Li, Q. Xie, Y. Y. Fang, Y. Sun, X. M. Wang and Z. Luo, et al., Prognostic value of CYFRA 21-1 and Ki67 in advanced NSCLC patients with wild-type EGFR, BMC Cancer, 2023, 23(1), 295 CrossRef CAS PubMed.
- M. Ju, X. Ge, X. Di, Y. Zhang, L. Liang and Y. Shi, Diagnostic, prognostic, and recurrence monitoring value of plasma CYFRA21–1 and NSE levels in patients with esophageal squamous cell carcinoma, Front. Oncol., 2022, 11, 789312 CrossRef PubMed.
- H. Huang, Y. Yang, Y. Zhu, H. Chen, Y. Yang and L. Zhang, et al., Blood protein biomarkers in lung cancer, Cancer Lett., 2022, 215886 CrossRef CAS PubMed.
- J. Zhang, X. Xue, Y. Du, J. Zhao, H. Ma and X. Ren, et al., Antigen-Down PEC Immunosensor for CYFRA21–1 Detection Based on Photocurrent Polarity Switching Strategy, Anal. Chem., 2022, 94(36), 12368–12373 CrossRef CAS PubMed.
- S.-P. Song, B. Li, J. Hu and M.-Q. Li, Simultaneous multianalysis for tumor markers by antibody fragments microarray system, Anal. Chim. Acta, 2004, 510(2), 147–152 CrossRef CAS.
- Y. Gao, W. Huo, L. Zhang, J. Lian, W. Tao and C. Song, et al., Multiplex measurement of twelve tumor markers using a GMR multi-biomarker immunoassay biosensor, Biosens. Bioelectron., 2019, 123, 204–210 CrossRef CAS PubMed.
- X. Li, B. Chen, M. He, G. Xiao and B. Hu, Gold nanoparticle labeling with tyramide signal amplification for highly sensitive detection of alpha fetoprotein in human serum by ICP-MS, Talanta, 2018, 176, 40–46 CrossRef CAS PubMed.
- H.-Z. Wang, H.-Y. Wang, R.-Q. Liang and K.-C. Ruan, Detection of tumor marker CA125 in ovarian carcinoma using quantum dots, Acta Biochim. Biophys. Sin., 2004, 36(10), 681–686 CrossRef CAS PubMed.
- H. Alawieh, T. El Chemaly, S. Alam and M. Khraiche, Towards point-of-care heart failure diagnostic platforms: BNP and NT-proBNP biosensors, Sensors, 2019, 19(22), 5003 CrossRef CAS PubMed.
- Y. Zhong, F. Meng, C. Deng and Z. Zhong, Ligand-directed active tumor-targeting polymeric nanoparticles for cancer chemotherapy, Biomacromolecules, 2014, 15(6), 1955–1969 CrossRef CAS PubMed.
- R. Bilan, F. Fleury, I. Nabiev and A. Sukhanova, Quantum dot surface chemistry and functionalization for cell targeting and imaging, Bioconjugate Chem., 2015, 26(4), 609–624 CrossRef CAS PubMed.
- J. H. Luong, K. B. Male and J. D. Glennon, Biotin interference in immunoassays based on biotin-strept (avidin) chemistry: an emerging threat, Biotechnol. Adv., 2019, 37(5), 634–641 CrossRef CAS PubMed.
- O. Tokel, F. Inci and U. Demirci, Advances in plasmonic technologies for point of care applications, Chem. Rev., 2014, 114(11), 5728–5752 CrossRef CAS PubMed.
- P. Kabiri, R. Weiskirchen and J. van Helden, The biotin interference within interference suppressed immunoassays, J. Clin. Lab. Anal., 2021, 35(9), e23940 CrossRef CAS PubMed.
- R. K. Thapa and J. O. Kim, Nanomedicine-based commercial formulations: Current developments and future prospects, J. Pharm. Invest., 2023, 53(1), 19–33 CrossRef CAS PubMed.
- J. R. Yu, J. Navarro, J. C. Coburn, B. Mahadik, J. Molnar and J. H. Holmes IV, et al., Current and future perspectives on skin tissue engineering: key features of biomedical research, translational assessment, and clinical application, Adv. Healthcare Mater., 2019, 8(5), 1801471 CrossRef PubMed.
- S. Nakamura, H. Mitomo, M. Aizawa, T. Tani, Y. Matsuo and K. Niikura, et al., Dna brush-directed vertical alignment of extensive gold nanorod arrays with controlled density, ACS Omega, 2017, 2(5), 2208–2213 CrossRef CAS PubMed.
- Y. J. Ko, W. J. Kim, K. Kim and I. C. Kwon, Advances in the strategies for designing receptor-targeted molecular imaging probes for cancer research, J. Controlled Release, 2019, 305, 1–17 CrossRef CAS PubMed.
- Y. Wen, C. L. Schreiber and B. D. Smith, Dual-targeted phototherapeutic agents as magic bullets for cancer, Bioconjugate Chem., 2020, 31(3), 474–482 CrossRef CAS PubMed.
- A. Anwar, S.-Y. Teow and Y. S. Wu, Nanogel-based drug delivery system as a treatment modality for diverse diseases: Are we there yet?, J. Drug Delivery Sci. Technol., 2023, 105224 Search PubMed.
- L. Muthukrishnan, Multidrug resistant tuberculosis–Diagnostic challenges and its conquering by nanotechnology approach–An overview, Chem.-Biol. Interact., 2021, 337, 109397 CrossRef CAS PubMed.
- C. Ehlinger, N. Spear, R. Doddareddy, G. Shankar and A. Schantz, A generic method for the detection of polyethylene glycol specific IgG and IgM antibodies in human serum, J. Immunol. Methods, 2019, 474, 112669 CrossRef CAS PubMed.
- C. Gao, Y. Wang, Z. Ye, Z. Lin, X. Ma and Q. He, Biomedical micro–/nanomotors: from overcoming biological barriers to in vivo imaging, Adv. Mater., 2021, 33(6), 2000512 CrossRef CAS PubMed.
Footnote |
| † These authors have senior co-authorship. |
| This journal is © The Royal Society of Chemistry 2024 |