Open Access Article
Open Access ArticleMachine learning for screening active metabolites with metabolomics in environmental science
Hemi
Luan
 *
*
School of Medicine, Academy for Advanced Interdisciplinary Studies, Southern University of Science and Technology, 1088 Xueyuan Blvd, Xili, Nanshan District, Shenzhen, Guangdong, China. E-mail: luanhm@sustech.edu.cn; Fax: +86-0755-88010116; Tel: +86-0755-88010116
First published on 22nd August 2022
Abstract
Metabolites are substances produced during metabolism, playing roles in biological processes such as biochemical reactions, signaling and gene expression. Metabolomics is a study of all metabolites and metabolic patterns in the body in response to genetic and environmental stresses. With advanced analytical techniques, metabolomic analysis generates large-scale and complex datasets that must be interpreted for meaningful biological information. Machine learning is an emerging area and applied to reveal the data structure, achieve the predictability of trends, and discover the metabolic patterns of metabolomic data in environmental science. Here, we review the applicability of machine learning to screen active metabolites with metabolomics in environmental science, while presenting the use of machine learning for metabolomics data processing, toxic effects of environmental pollutants, and health outcomes of environmental exposure. We also discuss the potential of combining integrative metabolomics with novel machine learning algorithms for the challenges of complex relationships between active metabolites and environmental exposures.
Environmental significanceMetabolites are the substances produced during metabolism that occur naturally within cells and biological systems. Metabolomics has been used to analyze hundreds to thousands of metabolites, and enabled in-depth insight into metabolic changes of living organisms in response to environmental stresses, such as environmental pollution, diseases, and extreme climate. With emerging technologies leading to the continuous generation of highly accurate and dynamic environmental metabolomic data, there is a great need for more powerful machine learning algorithms to drive the widespread use of metabolomics for screening active metabolites associated with the toxic effects of environmental pollutants and health outcomes of environmental exposure. This paper will expand and contribute to that sphere of interest in machine learning-driven metabolomics research in environmental science. |
Introduction
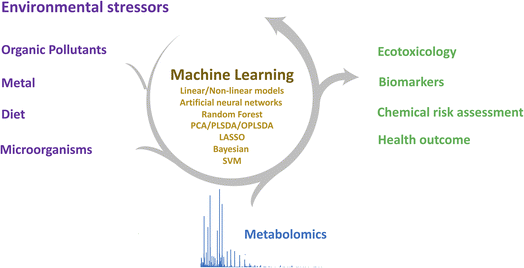
Metabolomics is the systematic study of the levels, function, and transformation of metabolites in a biological system. These endogenous metabolites are final products due to gene and protein activity and are directly related to biochemical activities, reflecting the changes in the physiology and pathology of living organisms, as well as the action of genetic and environmental stress.1 Metabolomics is increasingly used in environmental science, not only to understand the biological mechanisms of environmental stress on organisms but also to understand the interactions between active metabolites and functional molecules induced by environmental stress. Environmental stress may arise from abiotic stressors such as chemical pollutants,2 diet,3 and also biotic stressors such as pathogens4 and aging.5With the rapid development of a range of analytical instrumentals, such as liquid chromatography-mass spectrometry (LC-MS), gas chromatography-mass spectrometry (GC-MS), capillary electrophoresis-mass spectrometry (CE-MS), and nuclear magnetic resonance spectroscopy (NMR), metabolomics techniques enable the separation, detection, and quantitation of a wide range of metabolites and their associated biochemical metabolic pathways.6 Due to the diverse properties of metabolites, a combination of analytical techniques is required to detect all metabolites in biological samples, facilitating increased metabolite coverage. Advanced analytical techniques generate large volumes of highly sensitive and high-resolution metabolomics data, combined with new data analysis strategy approaches, allowing the identification of complex metabolites and the annotation of physiological and pathological phenomena arising from environmental stress7,8 (Fig. 1).
Machine learning is a data analytics technique that guides computers to learn data patterns, gain insight into the data structure, achieve the predictability of trends, and discover the scientific patterns in the metabolomic data. The use of classical multivariate statistical and machine learning methods, such as principal component analysis (PCA),9 partial least squares (PLS),10 support vector machines (SVMs),11 random forests (RFs),12 and least absolute shrinkage and selection operator (LASSO),13 has increased dramatically over the past decade, contributing to the identification of metabolites associated with environmental toxicology and human health. Several software tools help the implementation of machine learning algorithms, for example, TensorFlow Keras, mlr3, Scikit-Learn, or Pytorch. The purpose of this article is to summarize machine learning in metabolomic data processing and its application in environmental science and discuss the current challenges and opportunities for machine learning in the interdisciplinary fields of metabolomics and environmental science.
Metabolomics data processing for environmental research
The processing of raw data obtained from mass spectrometry-based metabolomics includes peak detection, mass spectra deconvolution, metabolite identification, data quality control, and active metabolite screening. A number of free and commercial software have been widely used for metabolomics data processing such as MZmine2,14 XCMS,15 and MS-DIAL.16 However, many challenges, such as false-positive feature extraction, unknown metabolite identification, and metabolic network annotation are still unresolved. Recently, it has been found that deep neural network-based algorithms for peak extraction, segmentation, and identification of metabolic data show higher accuracy than traditional peak extraction algorithms. A study by Melnikov et al. developed the “peakonly” algorithm for feature detection based on the use of a deep neural network for LC-MS chromatographic data.17 A series of machine learning based algorithms were furtherly proposed to evaluate and classify peaks based on peak quality metrics,17–20 such as deep neural network,21 adaBoost,22 SVMs, and RFs.23Metabolite identification remains one of the main challenges in metabolomics. In general, metabolites detected from mass spectrometry-based metabolomics are identified by comparing parameters of similarity degree between unknown compounds and reference compound data, such as retention time, exact mass, mass fragmentation patterns, collision cross-section value, and so on. However, this traditional approach is limited by the spectra quality and coverage of known compounds in the reference database. An effective strategy is to predict the spectra of known compounds, based on in silico methods of machine learning. Such software tools include CFM-ID,24 CSI:FingerID,25 and FingerID.26 It is noted that Chao et al. used CFM-ID to generate predicted spectra for compounds of the Distributed Structure Searchable Toxicity (DSSTox) database in the U.S. Environmental Protection Agency (EPA), with a total of approximately 765![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 substances.27
000 substances.27
The application of quality control in the field of metabolomics is essential to ensure the collection of high-quality data. Signal drift and batch effects are important factors affecting the data quality in large-scale metabolomics. A quality control-based machine learning algorithm: random forest signal correction (QC-RFSC) was developed in our previous work for the evaluation of data quality and removal of unwanted variations for large-scale metabolomic data.12,28 Alternatively, machine learning approaches such as SVMs and deep adversarial learning29 have been gradually introduced into the quality control procedures in the field of metabolomics.
While we have discussed the application of machine learning for challenges in data processing of metabolomics research, environmental metabolomics faces more specific problems in environmental science. Environmental contaminants are very complex and diverse, and their exposure is accompanied by the generation of transformed products and metabolites. For example, a recent study found that the exposure of mono (2-ethylhexyl) phthalate (MEHP) that is a transformed metabolite of a transformed product of Di (2-ethylhexyl) phthalate (DEHP) might lead to mutagenicity.30,31 The use of machine learning-based tools makes it possible to discover known or unknown transformed metabolites derived from environmental contaminants, such as BioTransformer.32 Kinds of model organisms are used in environmental metabolomics studies, such as earthworms33 and water flea.34 Machine learning has the potential to aid in the development of species-specific metabolic pathways and metabolic responses under environmental stress.35,36
Toxic effects of environmental pollutants
A large number of scientific papers have looked at the toxic effects of environmental pollutants, particularly metals,37 pesticides,38 and persistent organic pollutants (POPs).39 Previous studies on the toxicological effects of environmental pollutants have focused on physiological and biochemical indicators. Computer-assisted modeling methods such as quantitative structure–activity relationships (QSARs) are a key method to predict the chemical toxicity using structural and physicochemical environmental pollutants.40 Metabolomic approaches provide a new way to find active metabolites as biological indicators of the toxicity of environmental pollutants. Principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) in the form of multiple regression have been widely used for dimensionality reduction to a visual data structure and metabolic phenotype classification. PLS-DA is a supervised method, which allows identifying metabolites that contribute to classification as significantly changed metabolites. PLS-DA and orthogonal PLS-DA were the most widely used methods to screen the potential biomarkers of toxic effects of environmental pollutants, such as BDE-47,41 BDE-3,42 BDE-209,43 bisphenol-A,44 bisphenol S,45,46 and climbazole.47 In addition, Guo et al. applied PLS-DA in mass spectrometry imaging-based metabolomics for the discovery of kidney tissue-specific nephrotoxic biomarkers of cadmium exposure.48 Apart from the traditional multivariate statistical analysis methods, the backpropagation neural networks were recently reported for the first time and applied to metabolomic data for the investigation of metabolites and the disturbed metabolic pathways of nanotoxicity of engineered nanoparticles, allowing fast evaluation of environmental health risks induced by known and unknown engineered nanoparticles.49While we have discussed the application of machine learning for challenges in data processing of metabolomics research, environmental metabolomics faces more specific problems in environmental science. Environmental contaminants are very complex and diverse, and their exposure is accompanied by the generation of transformed products and metabolites. For example, a recent study found that the exposure of mono (2-ethylhexyl) phthalate (MEHP), a transformed metabolite of a transformed product of Di (2-ethylhexyl) phthalate (DEHP), might lead to mutagenicity.50,51 The use of machine learning-based tools makes it possible to identify known or unknown transformed metabolites derived from environmental contaminants, such as BioTransformer.52
Health outcome of environmental exposure
The human health risk is influenced by a combination of genetic and environmental exposure, such as diet,53 aging,54 and pollutants.55 Metabolomics provides insight into how environmental factors reshape the metabolic phenotype of living organisms and help to identify active metabolites associated with environmental exposures. Environmental pollutants may be one of the notable environmental factors that can significantly modify health outcomes. Machine learning-based approaches were applied to reveal the association between environmental contaminant exposure and metabolites. The linear model is one of the most popular methods to calculate the linear relationship between the levels of environmental pollutant exposure and metabolites. For example, Wang et al. performed a linear regression analysis to investigate the association of metals with the 18 endogenous metabolites among Chinese children and the elderly population.56 Yan et al. reported perturbations of the serum metabolome in response to pesticides and associations between groups of metabolites and multiple pesticides. However, linear models are not suitable to handle nonlinear data. Nonlinearity is a common phenomenon when assessing association among health outcomes, environmental exposure, and metabolites.Because of non-linear relationships between environmental pollutants and metabolites, nonlinear models are gradually being selected to study the interaction of environmental pollutants and metabolites. A trend of interest is that non-linear machine learning algorithms are being used for health outcome prediction. In addition, populations may be exposed to chemical mixtures, and the health effects of chemical mixtures can be revealed by using machine learning techniques, e.g. RFs, LASSO, logistic regression,57 and Bayesian kernel machine regression (BKMR).58 Luan et al. performed the quantitative analyses of multiple endocrine-disrupting chemicals and metabolites for a longitudinal cohort with 2317 pregnant women, and a random forest model with recursive feature elimination was successfully used to predict the gestational age with high accuracy, and interpret the mixture effect of endocrine-disrupting chemicals on pregnancy.59 BKMR based on the kernel machine framework was recently developed to estimate the health effects of multi-pollutant mixtures. Matta et al. studied the association between endometriosis and persistent organic pollutants, and BKMR was used to examine the joint effects of complex multi-pollutant mixtures and interactions between chemicals and metabolites.60 Compared to a whole population, personal exposure is more dynamic and spatiotemporal. Jiang et al. studied personal airborne biological and chemical exposure, and furtherly revealed associations among organisms, metabolites, and chemicals by using various models, such as sparse canonical correlation analysis, LASSO, and others.61
Conclusions and prospects
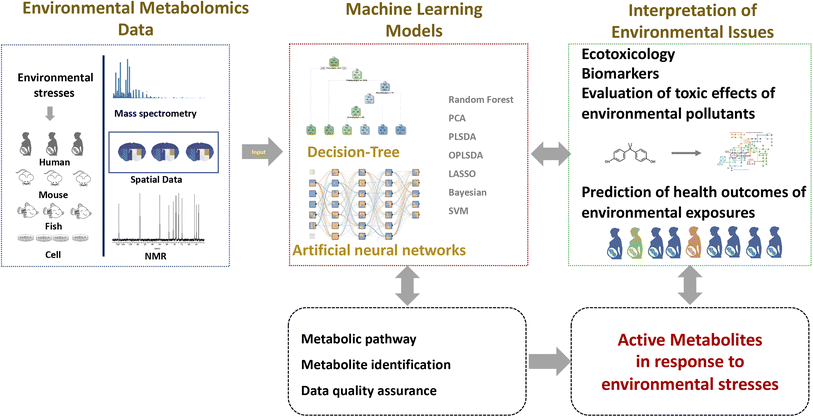
With growing interference from environmental stress, such as biotic and abiotic pollution, diseases, and extreme climate, the need to understand the effects of our environment would remain paramount. Metabolomics is an important tool for environmental research, capable of interpreting the biological effects of environmental stress through the measurement of a number of active metabolites. Machine learning has been used in the areas of metabolomics and environmental science due to recent advances in computing power. Notable machine learning achievements in environmental metabolomics, for example, have aided active metabolite screening, evaluation of toxic effects of environmental pollutants, and prediction of health outcomes of environmental exposures in the human population (Fig. 2). Outstanding challenges remain in the application and acceptance of machine learning for dynamic, heterogeneous, and multi-dimensional data from metabolomics and environmental exposure.Machine learning has been intensively used in metabolomics techniques, including data interpretation, identification of metabolic patterns and decision making, as well as metabolite identification. However, the application and acceptance of novel machine learning-based metabolomics for environmental science remain low. Most environmental metabolomic studies often use the traditional data analysis methods, such as PCA, PLS-DA, and OPLS-DA, ignoring their characteristics. Both of PCA and PLS-DA are typical linear methods to construct linear relationships between environmental stress and metabolic effects. However, non-linear relationships of environmental pollutants and toxic effects can often be observed. Some environmental contaminants' toxic effects may occur at low doses.62 Non-linear machine learning is likely to be the realistic approach to meeting requirements for the evaluation of toxic effects and the selection of key features.
As we know, the human body is exposed to multiple environmental stressors simultaneously, such as environmental pollutants, bacteria, and viruses. The joint effects of multiple exposures can be assessed by using metabolomics with the perturbation of metabolites or metabolic pathways in the human body.63 Moreover, there are tremendous amounts of dynamic, heterogeneous, and multi-dimensional data accumulated from exposome and metabolome studies. Artificial neural networks are an active subfield of machine learning. Compared to classical machine learning algorithms, artificial neural networks have better performance on big data sets because of the large number of hyperparameters that can be tuned.64 However, few studies were reported about the application of artificial neural networks in environmental metabolomic studies. It is critical to address these issues and provide novel computational methods to handle complex relationships between metabolite and multi-pollutant exposure. It is also conceivable that soon new technologies will lead to the continuous generation of highly accurate and dynamic data. There is a great need for more powerful machine learning algorithms to drive the widespread use of metabolomics in environmental science.
Author contributions
Hemi Luan: project administration, conceptualization, writing—review & editing and revising.Conflicts of interest
The authors declare that they have no conflicts of interest.Acknowledgements
The authors would like to acknowledge the financial support from the National Natural Science Foundation of China (grant no. 21904058).References
- J. G. Bundy, M. P. Davey and M. R. Viant, Environmental metabolomics: a critical review and future perspectives, Metabolomics, 2009, 5, 3–21 CrossRef CAS.
- C. Yang, J. Wei, G. Cao and Z. Cai, Lipid metabolism dysfunction and toxicity of BDE-47 exposure in white adipose tissue revealed by the integration of lipidomics and metabolomics, Sci. Total Environ., 2022, 806, 150350 CrossRef CAS PubMed.
- S. Andraos, K. L. Beck, M. B. Jones, T. L. Han, C. A. Conlon and J. V. de Seymour, Characterizing patterns of dietary exposure using metabolomic profiles of human biospecimens: a systematic review, Nutr. Rev., 2022, 80, 699–708 CrossRef PubMed.
- Y. Li, G. Hou, H. Zhou, Y. Wang, H. M. Tun, A. Zhu, J. Zhao, F. Xiao, S. Lin, D. Liu, D. Zhou, L. Mai, L. Zhang, Z. Zhang, L. Kuang, J. Guan, Q. Chen, L. Wen, Y. Zhang, J. Zhuo, F. Li, Z. Zhuang, Z. Chen, L. Luo, D. Liu, C. Chen, M. Gan, N. Zhong, J. Zhao, Y. Ren and Y. Xu, Multi-platform omics analysis reveals molecular signature for COVID-19 pathogenesis, prognosis and drug target discovery, Signal Transduction Targeted Ther., 2021, 6, 155 CrossRef CAS.
- J. L. Castro-Mejia, B. Khakimov, L. Krych, J. Bulow, R. L. Bechshoft, G. Hojfeldt, K. H. Mertz, E. S. Garne, S. R. Schacht, H. F. Ahmad, W. Kot, L. H. Hansen, F. J. A. Perez-Cueto, M. V. Lind, A. J. Lassen, I. Tetens, T. Jensen, S. Reitelseder, A. P. Jespersen, L. Holm, S. B. Engelsen and D. S. Nielsen, Physical fitness in community-dwelling older adults is linked to dietary intake, gut microbiota, and metabolomic signatures, Aging Cell, 2020, 19, e13105 CrossRef CAS.
- T. Hyotylainen, Analytical challenges in human exposome analysis with focus on environmental analysis combined with metabolomics, J. Sep. Sci., 2021, 44, 1769–1787 CrossRef PubMed.
- F. Bardanzellu and V. Fanos, Metabolomics, Microbiomics, Machine learning during the COVID-19 pandemic, Pediatr. Allergy Immunol., 2022, 33(Suppl 27), 86–88 CrossRef CAS PubMed.
- R. Schmid, D. Petras, L. F. Nothias, M. Wang, A. T. Aron, A. Jagels, H. Tsugawa, J. Rainer, M. Garcia-Aloy, K. Duhrkop, A. Korf, T. Pluskal, Z. Kamenik, A. K. Jarmusch, A. M. Caraballo-Rodriguez, K. C. Weldon, M. Nothias-Esposito, A. A. Aksenov, A. Bauermeister, A. Albarracin Orio, C. O. Grundmann, F. Vargas, I. Koester, J. M. Gauglitz, E. C. Gentry, Y. Hovelmann, S. A. Kalinina, M. A. Pendergraft, M. Panitchpakdi, R. Tehan, A. Le Gouellec, G. Aleti, H. Mannochio Russo, B. Arndt, F. Hubner, H. Hayen, H. Zhi, M. Raffatellu, K. A. Prather, L. I. Aluwihare, S. Bocker, K. L. McPhail, H. U. Humpf, U. Karst and P. C. Dorrestein, Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment, Nat. Commun., 2021, 12, 3832 CrossRef CAS PubMed.
- Z. He, H. Zhang, Y. Song, Z. Yang and Z. Cai, Exposure to ambient fine particulate matter impedes the function of spleen in the mouse metabolism of high-fat diet, J. Hazard. Mater., 2022, 423, 127129 CrossRef CAS PubMed.
- L. Liu, Q. Wu, X. Miao, T. Fan, Z. Meng, X. Chen and W. Zhu, Study on toxicity effects of environmental pollutants based on metabolomics: A review, Chemosphere, 2022, 286, 131815 CrossRef CAS PubMed.
- Z. Chen, S. Han, J. Zhang, P. Zheng, X. Liu, Y. Zhang and G. Jia, Metabolomics screening of serum biomarkers for occupational exposure of titanium dioxide nanoparticles, Nanotoxicology, 2021, 15, 832–849 CrossRef CAS PubMed.
- H. Luan, F. Ji, Y. Chen and Z. Cai, statTarget: A streamlined tool for signal drift correction and interpretations of quantitative mass spectrometry-based omics data, Anal. Chim. Acta, 2018, 1036, 66–72 CrossRef CAS.
- H. Wei, J. Sun, W. Shan, W. Xiao, B. Wang, X. Ma, W. Hu, X. Wang and Y. Xia, Environmental chemical exposure dynamics and machine learning-based prediction of diabetes mellitus, Sci. Total Environ., 2022, 806, 150674 CrossRef CAS PubMed.
- T. Pluskal, S. Castillo, A. Villar-Briones and M. Oresic, MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data, BMC Bioinf., 2010, 11, 395 CrossRef PubMed.
- N. G. Mahieu, J. L. Genenbacher and G. J. Patti, A roadmap for the XCMS family of software solutions in metabolomics, Curr. Opin. Chem. Biol., 2016, 30, 87–93 CrossRef CAS PubMed.
- H. Tsugawa, T. Cajka, T. Kind, Y. Ma, B. Higgins, K. Ikeda, M. Kanazawa, J. VanderGheynst, O. Fiehn and M. Arita, MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis, Nat. Methods, 2015, 12, 523–526 CrossRef CAS PubMed.
- A. D. Melnikov, Y. P. Tsentalovich and V. V. Yanshole, Deep Learning for the Precise Peak Detection in High-Resolution LC-MS Data, Anal. Chem., 2020, 92, 588–592 CrossRef CAS PubMed.
- H. Luan, X. Jiang, F. Ji, Z. Lan, Z. Cai and W. Zhang, CPVA: a web-based metabolomic tool for chromatographic peak visualization and annotation, Bioinformatics, 2020, 36, 3913–3915 CrossRef CAS PubMed.
- S. Toghi Eshghi, P. Auger and W. R. Mathews, Quality assessment and interference detection in targeted mass spectrometry data using machine learning, Clin. Proteomics, 2018, 15, 33 CrossRef PubMed.
- W. Zhang and P. X. Zhao, Quality evaluation of extracted ion chromatograms and chromatographic peaks in liquid chromatography/mass spectrometry-based metabolomics data, BMC Bioinf., 2014, 15(Suppl 11), S5 CrossRef PubMed.
- Y. Gloaguen, J. A. Kirwan and D. Beule, Deep Learning-Assisted Peak Curation for Large-Scale LC-MS Metabolomics, Anal. Chem., 2022, 94, 4930–4937 CrossRef CAS PubMed.
- K. Chetnik, L. Petrick and G. Pandey, MetaClean: a machine learning-based classifier for reduced false positive peak detection in untargeted LC-MS metabolomics data, Metabolomics, 2020, 16, 117 CrossRef CAS PubMed.
- T. Yu and D. P. Jones, Improving peak detection in high-resolution LC/MS metabolomics data using preexisting knowledge and machine learning approach, Bioinformatics, 2014, 30, 2941–2948 CrossRef CAS PubMed.
- F. Wang, J. Liigand, S. Tian, D. Arndt, R. Greiner and D. S. Wishart, CFM-ID 4.0: More Accurate ESI-MS/MS Spectral Prediction and Compound Identification, Anal. Chem., 2021, 93, 11692–11700 CrossRef CAS.
- M. A. Hoffmann, L. F. Nothias, M. Ludwig, M. Fleischauer, E. C. Gentry, M. Witting, P. C. Dorrestein, K. Duhrkop and S. Bocker, High-confidence structural annotation of metabolites absent from spectral libraries, Nat. Biotechnol., 2022, 40, 411–421 CrossRef CAS.
- M. Heinonen, H. Shen, N. Zamboni and J. Rousu, Metabolite identification and molecular fingerprint prediction through machine learning, Bioinformatics, 2012, 28, 2333–2341 CrossRef CAS.
- A. Chao, H. Al-Ghoul, A. D. McEachran, I. Balabin, T. Transue, T. Cathey, J. N. Grossman, R. R. Singh, E. M. Ulrich, A. J. Williams and J. R. Sobus, In silico MS/MS spectra for identifying unknowns: a critical examination using CFM-ID algorithms and ENTACT mixture samples, Anal. Bioanal. Chem., 2020, 412, 1303–1315 CrossRef CAS PubMed.
- H. Luan, W. Gu, H. Li, Z. Wang, L. Lu, M. Ke, J. Lu, W. Chen, Z. Lan, Y. Xiao, J. Xu, Y. Zhang, Z. Cai, S. Liu and W. Zhang, Serum metabolomic and lipidomic profiling identifies diagnostic biomarkers for seropositive and seronegative rheumatoid arthritis patients, J. Transl. Med., 2021, 19, 500 CrossRef CAS PubMed.
- Z. Rong, Q. Tan, L. Cao, L. Zhang, K. Deng, Y. Huang, Z. J. Zhu, Z. Li and K. Li, NormAE: Deep Adversarial Learning Model to Remove Batch Effects in Liquid Chromatography Mass Spectrometry-Based Metabolomics Data, Anal. Chem., 2020, 92, 5082–5090 CrossRef CAS.
- Y. J. Chang, C. Y. Tseng, P. Y. Lin, Y. C. Chuang and M. W. Chao, Acute exposure to DEHP metabolite, MEHP cause genotoxicity, mutagenesis and carcinogenicity in mammalian Chinese hamster ovary cells, Carcinogenesis, 2017, 38, 336–345 CrossRef CAS.
- H. Zhao, J. Li, Y. Zhou, L. Zhu, Y. Zheng, W. Xia, Y. Li, L. Xiang, W. Chen, S. Xu and Z. Cai, Investigation on Metabolism of Di(2-Ethylhexyl) Phthalate in Different Trimesters of Pregnant Women, Environ. Sci. Technol., 2018, 52, 12851–12858 CrossRef CAS PubMed.
- Y. Djoumbou-Feunang, J. Fiamoncini, A. Gil-de-la-Fuente, R. Greiner, C. Manach and D. S. Wishart, BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification, J. Cheminf., 2019, 11, 2 Search PubMed.
- M. J. Simpson and J. R. McKelvie, Environmental metabolomics: new insights into earthworm ecotoxicity and contaminant bioavailability in soil, Anal. Bioanal. Chem., 2009, 394, 137–149 CrossRef CAS PubMed.
- K. Toyota, A. Gavin, S. Miyagawa, M. R. Viant and T. Iguchi, Metabolomics reveals an involvement of pantothenate for male production responding to the short-day stimulus in the water flea, Daphnia pulex, Sci. Rep., 2016, 6, 25125 CrossRef CAS.
- C. M. Griffith, M. A. Morgan, M. M. Dinges, C. Mathon and C. K. Larive, Metabolic Profiling of Chloroacetanilide Herbicides in Earthworm Coelomic Fluid Using (1)H NMR and GC-MS, J. Proteome Res., 2018, 17, 2611–2622 CrossRef PubMed.
- L. Wang, X. Huang, A. K. C. Laserna and S. F. Y. Li, Untargeted metabolomics reveals transformation pathways and metabolic response of the earthworm Perionyx excavatus after exposure to triphenyl phosphate, Sci. Rep., 2018, 8, 16440 CrossRef.
- M. A. Garcia-Sevillano, T. Garcia-Barrera and J. L. Gomez-Ariza, Environmental metabolomics: Biological markers for metal toxicity, Electrophoresis, 2015, 36, 2348–2365 CrossRef CAS.
- Q. Yan, K. C. Paul, D. I. Walker, M. A. Furlong, I. Del Rosario, Y. Yu, K. Zhang, M. G. Cockburn, D. P. Jones and B. R. Ritz, High-Resolution Metabolomic Assessment of Pesticide Exposure in Central Valley, California, Chem. Res. Toxicol., 2021, 34, 1337–1347 Search PubMed.
- Y. Liang, Z. Tang, Y. Jiang, C. Ai, J. Peng, Y. Liu, J. Chen, X. Xin, B. Lei, J. Zhang and Z. Cai, Lipid metabolism disorders associated with dioxin exposure in a cohort of Chinese male workers revealed by a comprehensive lipidomics study, Environ. Int., 2021, 155, 106665 CrossRef CAS PubMed.
- G. Tsiliki, P. Nymark, P. Kohonen, R. Grafström and H. Sarimveis, Enriching Nanomaterials Omics Data: An Integration Technique to Generate Biological Descriptors, Small Methods, 2017, 1, 1700139 CrossRef.
- J. Wei, X. Li, L. Xiang, Y. Song, Y. Liu, Y. Jiang and Z. Cai, Metabolomics and lipidomics study unveils the impact of polybrominated diphenyl ether-47 on breast cancer mice, J. Hazard. Mater., 2020, 390, 121451 CrossRef CAS PubMed.
- Z. Wei, J. Xi, S. Gao, X. You, N. Li, Y. Cao, L. Wang, Y. Luan and X. Dong, Metabolomics coupled with pathway analysis characterizes metabolic changes in response to BDE-3 induced reproductive toxicity in mice, Sci. Rep., 2018, 8, 5423 CrossRef.
- Y. S. Jung, J. Lee, J. Seo and G. S. Hwang, Metabolite profiling study on the toxicological effects of polybrominated diphenyl ether in a rat model, Environ. Toxicol., 2017, 32, 1262–1272 CrossRef CAS PubMed.
- S. W. Lee, N. Chatterjee, J. E. Im, D. Yoon, S. Kim and J. Choi, Integrated approach of eco-epigenetics and eco-metabolomics on the stress response of bisphenol-A exposure in the aquatic midge Chironomus riparius, Ecotoxicol. Environ. Saf., 2018, 163, 111–116 CrossRef.
- P. Xie, X. Liang, Y. Song and Z. Cai, Mass Spectrometry Imaging Combined with Metabolomics Revealing the Proliferative Effect of Environmental Pollutants on Multicellular Tumor Spheroids, Anal. Chem., 2020, 92, 11341–11348 CrossRef CAS PubMed.
- C. Zhao, Z. Tang, J. Yan, J. Fang, H. Wang and Z. Cai, Bisphenol S exposure modulate macrophage phenotype as defined by cytokines profiling, global metabolomics and lipidomics analysis, Sci. Total Environ., 2017, 592, 357–365 CrossRef CAS.
- T. Zou, Y. Q. Liang, X. Liao, X. F. Chen, T. Wang, Y. Song, Z. C. Lin, Z. Qi, Z. F. Chen and Z. Cai, Metabolomics reveals the reproductive abnormality in female zebrafish exposed to environmentally relevant levels of climbazole, Environ. Pollut., 2021, 275, 116665 CrossRef CAS PubMed.
- T. Zeng, R. Zhang, Y. Chen, W. Guo, J. Wang and Z. Cai, In situ localization of lipids on mouse kidney tissues with acute cadmium toxicity using atmospheric pressure-MALDI mass spectrometry imaging, Talanta, 2022, 245, 123466 CrossRef CAS.
- T. Peng, C. Wei, F. Yu, J. Xu, Q. Zhou, T. Shi and X. Hu, Predicting nanotoxicity by an integrated machine learning and metabolomics approach, Environ. Pollut., 2020, 267, 115434 CrossRef CAS PubMed.
- Y.-J. Chang, C.-Y. Tseng, P.-Y. Lin, Y.-C. Chuang and M.-W. Chao, Acute exposure to DEHP metabolite, MEHP cause genotoxicity, mutagenesis and carcinogenicity in mammalian Chinese hamster ovary cells, Carcinogenesis, 2017, 38(3), 336–345 CrossRef CAS PubMed.
- H. Zhao, J. Li, Y. Zhou, L. Zhu, Y. Zheng, W. Xia, Y. Li, L. Xiang, W. Chen, S. Xu and Z. Cai, Investigation on Metabolism of Di(2-Ethylhexyl) Phthalate in Different Trimesters of Pregnant Women, Environ. Sci. Technol., 2018, 52(21), 12851–12858 CrossRef CAS PubMed.
- Y. Djoumbou-Feunang, J. Fiamoncini, A. Gil-de-la-Fuente, R. Greiner, C. Manach and D. S. Wishart, BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification, J. Cheminf., 2019, 11(1), 2 Search PubMed , 30612223.
- D. Thomas, Gene—environment-wide association studies: emerging approaches, Nat. Rev. Genet., 2010, 11, 259–272 CrossRef CAS.
- A. Peters, T. S. Nawrot and A. A. Baccarelli, Hallmarks of environmental insults, Cell, 2021, 184, 1455–1468 CrossRef CAS.
- B. S. Rathi, P. S. Kumar and D. N. Vo, Critical review on hazardous pollutants in water environment: Occurrence, monitoring, fate, removal technologies and risk assessment, Sci. Total Environ., 2021, 797, 149134 CrossRef CAS PubMed.
- Z. Wang, X. Xu, B. He, J. Guo, B. Zhao, Y. Zhang, Z. Zhou, X. Zhou, R. Zhang and Z. Abliz, The impact of chronic environmental metal and benzene exposure on human urinary metabolome among Chinese children and the elderly population, Ecotoxicol. Environ. Saf., 2019, 169, 232–239 CrossRef CAS.
- A. Jeong, G. Fiorito, P. Keski-Rahkonen, M. Imboden, A. Kiss, N. Robinot, H. Gmuender, J. Vlaanderen, R. Vermeulen, S. Kyrtopoulos, Z. Herceg, A. Ghantous, G. Lovison, C. Galassi, A. Ranzi, V. Krogh, S. Grioni, C. Agnoli, C. Sacerdote, N. Mostafavi, A. Naccarati, A. Scalbert, P. Vineis, N. Probst-Hensch and E. X. Consortium, Perturbation of metabolic pathways mediates the association of air pollutants with asthma and cardiovascular diseases, Environ. Int., 2018, 119, 334–345 CrossRef CAS PubMed.
- J. F. Bobb, L. Valeri, B. Claus Henn, D. C. Christiani, R. O. Wright, M. Mazumdar, J. J. Godleski and B. A. Coull, Bayesian kernel machine regression for estimating the health effects of multi-pollutant mixtures, Biostatistics, 2015, 16, 493–508 CrossRef PubMed.
- H. Luan, H. Zhao, J. Li, Y. Zhou, J. Fang, H. Liu, Y. Li, W. Xia, S. Xu and Z. Cai, Machine Learning for Investigation on Endocrine-Disrupting Chemicals with Gestational Age and Delivery Time in a Longitudinal Cohort, Research (Wash D C), 2021, 2021, 9873135 Search PubMed.
- K. Matta, T. Lefebvre, E. Vigneau, V. Cariou, P. Marchand, Y. Guitton, A. L. Royer, S. Ploteau, B. Le Bizec, J. P. Antignac and G. Cano-Sancho, Associations between persistent organic pollutants and endometriosis: A multiblock approach integrating metabolic and cytokine profiling, Environ. Int., 2022, 158, 106926 CrossRef CAS PubMed.
- C. Jiang, X. Wang, X. Li, J. Inlora, T. Wang, Q. Liu and M. Snyder, Dynamic Human Environmental Exposome Revealed by Longitudinal Personal Monitoring, Cell, 2018, 175, 277–291.e231 CrossRef CAS.
- C. Zhao, T. Yong, Y. Zhang, Y. Xiao, Y. Jin, C. Zheng, T. Nirasawa and Z. Cai, Breast cancer proliferation and deterioration-associated metabolic heterogeneity changes induced by exposure of bisphenol S, a widespread replacement of bisphenol A, J. Hazard. Mater., 2021, 414, 125391 CrossRef CAS.
- F. Wang, H. Zhang, N. Geng, X. Ren, B. Zhang, Y. Gong and J. Chen, A metabolomics strategy to assess the combined toxicity of polycyclic aromatic hydrocarbons (PAHs) and short-chain chlorinated paraffins (SCCPs), Environ. Pollut., 2018, 234, 572–580 CrossRef CAS.
- X. Liu, D. Lu, A. Zhang, Q. Liu and G. Jiang, Data-Driven Machine Learning in Environmental Pollution: Gains and Problems, Environ. Sci. Technol., 2022, 56, 2124–2133 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2022 |