Open Access Article
Open Access ArticleA silicon-rhodamine chemical-genetic hybrid for far red voltage imaging from defined neurons in brain slice†
Gloria
Ortiz
a,
Pei
Liu
a,
Parker E.
Deal
 a,
Ashley K.
Nensel
a,
Kayli N.
Martinez
a,
Kiarash
Shamardani
a,
Ashley K.
Nensel
a,
Kayli N.
Martinez
a,
Kiarash
Shamardani
 b,
Hillel
Adesnik
bc and
Evan W.
Miller
b,
Hillel
Adesnik
bc and
Evan W.
Miller
 *abc
*abc
aDepartment of Chemistry, University of California, Berkeley, California 94720-1460, USA. E-mail: evanwmiller@berkeley.edu
bDepartment of Molecular & Cell Biology, University of California, Berkeley, California 94720-1460, USA
cHelen Wills Neuroscience Institute, University of California, Berkeley, California 94720-1460, USA
First published on 29th September 2021
Abstract
We describe the design, synthesis, and application of voltage-sensitive silicon rhodamines. Based on the Berkeley Red Sensor of Transmembrane potential, or BeRST, scaffold, the new dyes possess an isomeric molecular wire for improved alignment in the plasma membrane and 2′ carboxylic acids for ready functionalization. The new isoBeRST dyes have a voltage sensitivity of 24% ΔF/F per 100 mV. Combined with a flexible polyethyleneglycol (PEG) linker and a chloroalkane HaloTag ligand, isoBeRST dyes enable voltage imaging from genetically defined cells and neurons and provide improved labeling over previous, rhodamine-based hybrid strategies. isoBeRST-Halo hybrid indicators achieve single-trial voltage imaging of membrane potential dynamics from cultured hippocampal neurons or cortical neurons in brain slices. With far-red/near infrared excitation and emission, turn-on response to action potentials, and effective cell labeling in thick tissue, the new isoBeRST-Halo derivatives provide an important complement to voltage imaging in neurobiology.
Introduction
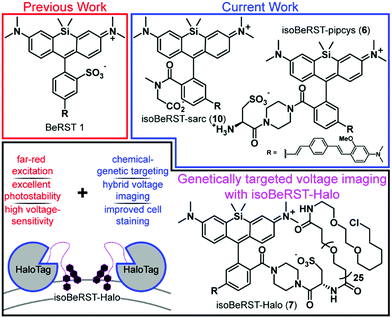
Voltage imaging in the central nervous system promises to transform the ways in which we observe brain systems.1,2 Recently, a number of approaches to voltage imaging have emerged, including methods that rely solely on synthetic dyes3–9 or genetically encoded proteins.10–17 Alternatively, hybrid methodologies can combine the unique properties of synthetic dyes—high molecular brightness, wide availability of colors, or fast response kinetics—with the cellular specificity of genetically encoded methods.18–24 Our group recently reported the development of a completely synthetic voltage-sensitive fluorophore, Berkeley Red Sensor of Transmembrane potential 1, or BeRST 1, a silicon-rhodamine-based indicator that operates via voltage-sensitive photoinduced electron transfer (PeT)25 and provides a faithful readout of fast voltage changes, outperforming completely genetically encoded voltage indicators.26 The high sensitivity (24% ΔF/F per 100 mV), fast response kinetics, photostability, and far red/near infra-red excitation and emission profile enable the use of BeRST 1 in a number of voltage imaging applications.27–29However, the use of BeRST 1 has been largely restricted to in vitro systems of homogeneous cell types because, unlike genetically encoded indicators, BeRST cannot be targeted to specific cells. Usage in more complex settings, like thick brain tissue, remains a challenge because of a lack of methods to genetically target BeRST 1 to defined cells (Scheme 1). Here we report two new synthetic BeRST dyes and show that this new class of indicator can be combined with a genetically-encoded protein tether to enable voltage imaging from defined cells in mouse brain slice.
Results
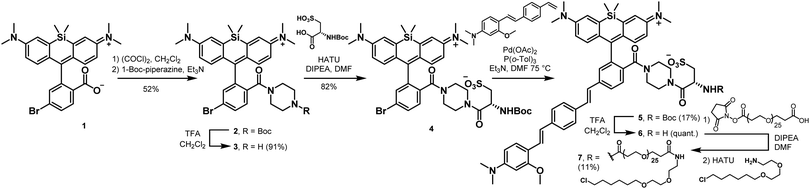
To enable genetic targeting of BeRST-style dyes, we redesigned the synthesis of BeRST. We replaced the 2′-sulfonate of BeRST with a carboxylate: this allows for addition of covalent tethers and mimics our previous design success with Rhodamine-based Voltage Reporters (RhoVRs).30,31 We also used the 5′ version of molecular wire, since the 5′, or isomeric, version showed improved voltage sensitivity compared to the 4′ RhoVR.30 Additionally, the commercial availability of the precursors to the aldehyde starting material substantially simplified the synthetic route (Scheme S1, ESI†). The optimized synthesis of isoBeRST-sarc 10 begins with a Heck reaction between fluorophore 134,35 and (E)-3-methoxy-N,N-dimethyl-4-(4-vinylstyryl)aniline36 to obtain carboxy silicon rhodamine 8 (Scheme S1, ESI†). Dye 8 is coupled to sarcosine tert-butyl ester using oxalyl chloride, followed by a TFA-catalyzed deprotection of the tert-butyl ester to give the voltage-sensitive fluorophore isoBeRST-sarc 10, which is the Si-rhodamine analog of RhoVR 1.30We also synthesized the piperazine-cysteic acid conjugate of isoBeRST, or isoBeRST-pipcys 6, since this configuration allowed us to target RhoVR dyes to specific cells using HaloTag (Scheme 2).31 The synthesis of isoBeRST-pipcys 6 and isoBeRST-Halo 7 follows a sequential amide-coupling/Heck coupling sequence. This provided higher overall yields than amide coupling with the assembled molecular wire/fluorophore 8. The cyclic, piperazine-derived tertiary amide of 3 appears more stable than the amide formed from sarcosine, based on its ability to undergo successful Pd-catalyzed synthesis of isoBeRST-pipcys 6. The route begins with oxalyl chloride mediated coupling of reported silicon rhodamine 1 with 1-Boc-piperazine, followed by TFA deprotection to yield silicon rhodamine 2 (Scheme 2). A second coupling mediated by HATU installed Boc-L-cysteic acid, affording 3. Compound 3 was then submitted to a Heck reaction with (E)-3-methoxy-N,N-dimethyl-4-(4-vinylstyryl)aniline to provide 5. Subsequent TFA deprotection gives voltage-sensitive fluorophore isoBeRST-pipcys 6 in 17% yield. In a one-pot sequence, isoBeRST-pipcys 6 is reacted with acid-dPEG25-NHS ester, followed by addition of HaloTag amine37 (Scheme 2) and HATU. The complete reaction was purified via preparative-scale HPLC to yield the genetically-targetable isoBeRST-Halo 7 in 11% yield. We selected a PEG25 linker because, for RhoVR-Halo, a PEGn linker with n = 25 monomer units almost completely recovered the full voltage sensitivity of the untethered, parent indicator (34% ΔF/F vs. 38% ΔF/F).31 Shorter PEG linkers (n = 13, 9, or 5) had lower voltage sensitivity, and PEG0 had no voltage sensitivity.
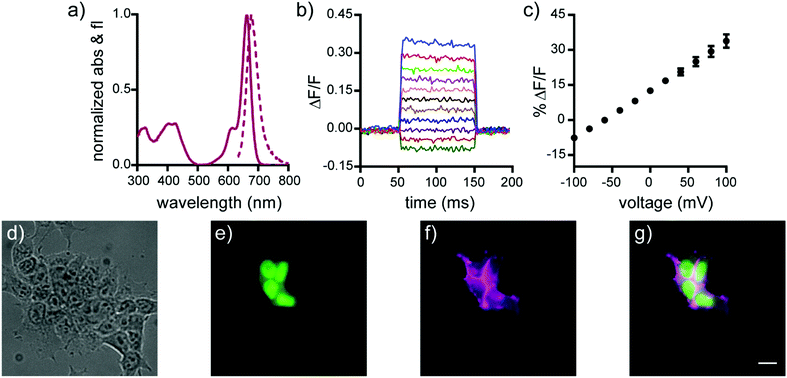
Spectroscopic characterization of isoBeRST-sarc 10, isoBeRST-pipcys 6, and isoBeRST-Halo 7 reveals that all three voltage indicators possess similar photophysical properties (Table 1, Fig. 1, Fig. S1 and S2, ESI†). IsoBeRST-sarc 10 displays a λmax at 661 nm, similar to BeRST 1 (λmax = 658 nm) and identical to isoBeRST-pipcys 6 and isoBeRST-Halo 7. IsoBeRST-sarc 10 possesses an emission maximum of 681 nm and a quantum yield (Φ) of 8.0%, while isoBeRST-pipcys 6 has an emission maximum of 681 nm and Φ of 5.0%. IsoBeRST-Halo 7 has an emission maximum of 677 nm and Φ of 3.4%.
| Compound | λ max nm−1 | λ em nm−1 | ε | Φ | % ΔF/F/100 mVc | Relative brightnessd (%) |
|---|---|---|---|---|---|---|
| a In PBS, pH 7.4, 0.1% SDS. b Referenced to Cy5.5-carboxylic acid in PBS, (Φ = 0.23).32,33 c Voltage-clamped HEK cells. Error is ± S.D. for n = 5–6 cells. d In HEK cells. Error is ± S.E.M for n = 4 coverslips (>100 cells per coverslip for relative brightness). | ||||||
| isoBeRST-pipcys 6 | 662 | 681 | 172![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.050 | 24 ± 1.9 | 100 |
| isoBeRST-Halo 7 | 662 | 677 | — | 0.034 | 21 ± 1.2 | 30 |
| isoBeRST-sarc 10 | 661 | 681 | 107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
0.080 | 24 ± 2.6 | — |
All of the new Si-rhodamine indicators are voltage-sensitive. In human embryonic kidney (HEK) cells untargeted dyes isoBeRST-sarc 10 (Fig. S1, ESI†) and isoBeRST-pipcys 6 (Fig. S2, ESI†) localize to the plasma membrane and are voltage sensitive. IsoBeRST-pipcys 6 has a voltage sensitivity of 24% ± 2% ΔF/F per 100 mV (SNR = 110 ± 15), identical to BeRST 1 (24% ± 5% ΔF/F per 100 mV)25 and to isoBeRST-sarc 10 (Table 1). We selected isoBeRST-pipcys 6 to evaluate in neurons because of the higher yielding synthesis and stability compared to isoBeRST-sarc 10. In cultured rat hippocampal neurons, isoBeRST-pipcys 6 (500 nM) provided clear resolution of action potentials (Fig. S3, ESI†).
The genetically-targetable isoBeRST-Halo 7 selectively labels HEK cells expressing cell-surface HaloTag (Fig. 1 and Fig. S4, ESI†). We expressed HaloTag on the surface of mammalian cells using a fusion with a single-pass transmembrane domain.31 At 500 nM isoBeRST-Halo 7, cells expressing cell-surface HaloTag are approximately 14 fold brighter than un-transfected control cells (Fig. S4f, ESI†). At lower concentrations (50 nM), fluorescence intensity in HaloTag-expressing cells increases to approximately 30 fold over non-HaloTag expressing cells (Fig. S4f, ESI†). This is three times better contrast than RhoVR-Halo labeling (10–15 fold).31 Although expression levels of HaloTag vary with transient transfection, a screen of isoBeRST-Halo 7 concentrations reveals that HaloTag binding sites saturate at around 50 to 100 nM (Fig. S4f, ESI†). The drop in contrast ratio, from ∼30 fold at 50 nM to 14 fold at 500 nM comes from a small increase in background staining in control cells (an increase of about 4 percentage points, from 5% to 9%). Importantly, isoBeRST-Halo 7 (50 nM) is voltage-sensitive, with a voltage sensitivity of 21% ± 1% ΔF/F per 100 mV and an SNR of 42 ± 7 (Fig. 1 and Table 1). IsoBeRST-Halo 7 maintains about the same voltage sensitivity as isoBeRST-pipcys 6 (500 nM), indicating that the covalently tethered dye remains properly oriented in the plasma membrane (Table 1 and Fig. 1).
Covalently-tethered isoBeRST-Halo 7 visualizes voltage changes in genetically-defined neurons. Dissociated, cultured rat hippocampal neurons transfected with HaloTag under control of the synapsin promoter were labeled with 50 nM isoBeRST-Halo. Neurons expressing HaloTag show excellent selectivity, revealing good localization of the dye to the outer membrane (Fig. 2 and Fig. S5, ESI†). The best contrast between HaloTag-expressing and control cells is achieved using 50 nM isoBeRST-Halo (50× brighter than untransfected cells). However, similarly high levels of contrast can be achieved, even when using higher isoBeRST-Halo concentrations (18× to 30× brighter when using 100 to 500 nM isoBeRST-Halo). By comparison, RhoVR-Halo,31 under identical conditions, can only achieve a selectivity ratio of 9.5× (50 nM RhoVR-Halo), which drops to 5× at higher concentrations (200 to 500 nM, Fig. S5f, ESI†). High isoBeRST-Halo fluorescence correlates with high levels of HaloTag/GFP (Fig. S5e–g, ESI†). Using these optimized loading conditions, we demonstrated the ability to record spontaneous and evoked activity in neurons (Fig. 2e and Fig. S6, ESI†). IsoBeRST-Halo responded to field stimulated evoked action potentials with a 10% ± 0.3% ΔF/F and SNR of 15 ± 1 (19 cells).
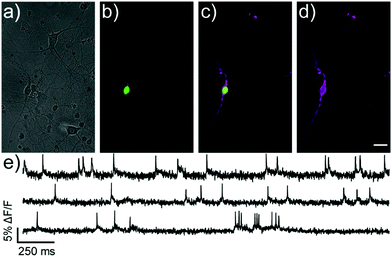
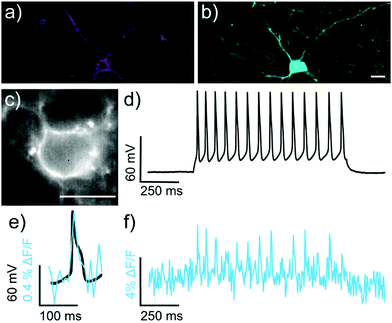
We next evaluated the ability of isoBeRST-Halo 7 to monitor voltage dynamics from neurons in brain slice. We introduced genes for HaloTag and a co-expression marker, blue fluorescent protein, or BFP, on separate plasmids via in utero electroporation in mouse embryos.38 We prepared tissue slices from the brains of these mice and stained the slices with isoBeRST-Halo 7 (250 to 500 nM, 15 min). Confocal fluorescence microscopy reveals localization of isoBeRST-Halo fluorescence in the cell membranes of neurons that express BFP and HaloTag (Fig. 3a and b). Both cell bodies and more distal processes like axonal and dendritic membranes appear fluorescent (Fig. 3a and b and Fig. S7, ESI†), mirroring results in dissociated rat neurons (Fig. 2). Unlabeled cell bodies appear as dark spots, indicating that labeling of neurons requires HaloTag expression (Fig. S7, ESI†). In mouse brain slices, isoBeRST-Halo is voltage-sensitive. Simultaneous patch clamp electrophysiology and fluorescence imaging establishes that isoBeRST-Halo tracks action potentials in a single trial (Fig. 3c–f). The voltage-sensitive fluorescence of isoBeRST-Halo corresponds well with the electrode-based recording of action potentials (Fig. 3e). IsoBeRST-Halo 7 detects action potentials (Fig. 3f) with a SNR of 4.9 ± 1.3 (S.D., n = 10 spikes) and a ΔF/F of 3.3% ± 0.6% (S.D., n = 10 spikes). The sensitivity of isoBeRST-Halo 7 in brain slices compares favorably to RhoVR-Halo, which has a higher voltage sensitivity (34%) than isoBeRST-Halo (21%) in HEK cells and has an SNR of 3.3 and a ΔF/F of 4.3% in brain slice (Table S1, ESI†).31 IsoBeRST-Halo 7 shows photostability comparable to RhoVR-Halo in brain slice, (Fig. S8 and Table S1, ESI†). This photostability tracks with the photostability measured for the untargeted dyes (isoBeRST-pipcys 6) in HEK293T cells (Fig. S8, ESI†).
Conclusion
In summary, we describe the design, synthesis, and application of new silicon-rhodamine-based voltage-sensitive fluorophores. The new BeRST derivatives rely on a 2′ carboxylate, rather than sulfonate, and can be combined with secondary amines to generate tertiary amides that function well as voltage indicators in their own right, with ΔF/F values matching the sulfonate-based BeRST 1.25 Unlike BeRST 1, however, the new carboxy-containing isoBeRST derivatives reported here can be readily incorporated into a hybrid genetic targeting framework. When combined in this way, isoBeRST-Halo 7 enables selective labeling of cells expressing cell-surface HaloTag, including HEK293T cells, dissociated rat hippocampal neurons, and cortical neurons in mouse brain slices. Labeling with isoBeRST-Halo provides improved contrast between HaloTag-expressing and non-expressing cells, compared to RhoVR1-Halo (Fig. S5f, ESI†).31 In all of these contexts, isoBeRST-Halo is voltage-sensitive, with ΔF/F values comparable to the parent BeRST 1 indicator.Voltage imaging with isoBeRST-Halo 7 provides an important complement to voltage imaging efforts. It offers a turn-on indicator for action potentials, possesses an excitation spectrum aligned with common excitation sources, operates in the far-red/near infrared some 90 nm red-shifted relative to previous RhoVR1-Halo strategies,31 and takes advantage of the high photostability of silicon-rhodamines (Table S1, ESI†).25,39,40 In the future, we will maximize expression of cell-surface HaloTag, since one limitation of the covalent tethering approach is that the stoichiometric labeling limits the number of indicators that can be added to a cell membrane. Finally, we envision that isoBeRST-Halo can pair with optically31 and enzymatically orthogonal hybrid genetic labeling strategies41–43 to provide multi-color voltage imaging in complex tissues.
Ethical statement
All animal procedures were approved by the UC Berkeley Animal Care and Use Committees and conformed to the NIH Guide for the Care and Use and Laboratory Animals and the Public Health Policy.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Research in the Miller lab is supported by grants from the NIH (R01NS098088) and Klingenstein-Simon Foundations (40746). E.W.M and H. A. acknowledge support from NSF Neuronex (1707350). Research in the Adesnik lab is supported by NIH (UF1NS107574 and RF1MH120680). H. A. is a NYSCF fellow. This work (H. A.) was supported by the New York Stem Cell Foundation. G. O. was supported by a Gilliam Research Fellowship from Howard Hughes Medical Institute. P. L. was supported by a graduate fellowship from A*STAR. K. N. M. was supported in part by a training grant from the NIH (T32GM066698). Confocal imaging experiments were performed at the CRL Molecular Imaging Center, supported by the Helen Wills Neuroscience Institute. HRMS data were collected at the QB3/Chemistry Mass Spectrometry Facility (UC Berkeley) with the assistance of Dr Ulla N. Andersen.Notes and references
- D. S. Peterka, H. Takahashi and R. Yuste, Neuron, 2011, 69, 9–21 CrossRef CAS PubMed.
- M. Scanziani and M. Häusser, Nature, 2009, 461, 930–939 CrossRef CAS PubMed.
- P. Liu and E. W. Miller, Acc. Chem. Res., 2020, 53, 11–19 CrossRef CAS PubMed.
- N. A. Sayresmith, A. Saminathan, J. K. Sailer, S. M. Patberg, K. Sandor, Y. Krishnan and M. G. Walter, J. Am. Chem. Soc., 2019, 141, 18780–18790 CrossRef CAS PubMed.
- J. E. Reeve, A. D. Corbett, I. Boczarow, W. Kaluza, W. Barford, H. Bayley, T. Wilson and H. L. Anderson, Angew. Chem., Int. Ed., 2013, 52, 9044–9048 CrossRef CAS PubMed.
- C. E. Rowland, K. Susumu, M. H. Stewart, E. Oh, A. J. Mäkinen, T. J. O’Shaughnessy, G. Kushto, M. A. Wolak, J. S. Erickson, A. L. Efros, A. L. Huston and J. B. Delehanty, Nano Lett., 2015, 15, 6848–6854 CrossRef CAS PubMed.
- P. Yan, C. D. Acker, W. L. Zhou, P. Lee, C. Bollensdorff, A. Negrean, J. Lotti, L. Sacconi, S. D. Antic, P. Kohl, H. D. Mansvelder, F. S. Pavone and L. M. Loew, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20443–20448 CrossRef CAS PubMed.
- E. W. Miller, J. Y. Lin, E. P. Frady, P. A. Steinbach, W. B. Kristan and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2114–2119 CrossRef CAS PubMed.
- J. S. Treger, M. F. Priest, R. Iezzi and F. Bezanilla, Biophys. J., 2014, 107, L09–L12 CrossRef CAS PubMed.
- K. D. Piatkevich, E. E. Jung, C. Straub, C. Linghu, D. Park, H. J. Suk, D. R. Hochbaum, D. Goodwin, E. Pnevmatikakis, N. Pak, T. Kawashima, C. T. Yang, J. L. Rhoades, O. Shemesh, S. Asano, Y. G. Yoon, L. Freifeld, J. L. Saulnier, C. Riegler, F. Engert, T. Hughes, M. Drobizhev, B. Szabo, M. B. Ahrens, S. W. Flavell, B. L. Sabatini and E. S. Boyden, Nat. Chem. Biol., 2018, 14, 352–360 CrossRef CAS PubMed.
- D. R. Hochbaum, Y. Zhao, S. L. Farhi, N. Klapoetke, C. A. Werley, V. Kapoor, P. Zou, J. M. Kralj, D. Maclaurin, N. Smedemark-Margulies, J. L. Saulnier, G. L. Boulting, C. Straub, Y. K. Cho, M. Melkonian, G. K. Wong, D. J. Harrison, V. N. Murthy, B. L. Sabatini, E. S. Boyden, R. E. Campbell and A. E. Cohen, Nat. Methods, 2014, 11, 825–833 CrossRef CAS PubMed.
- M. Kannan, G. Vasan, C. Huang, S. Haziza, J. Z. Li, H. Inan, M. J. Schnitzer and V. A. Pieribone, Nat. Methods, 2018, 15, 1108–1116 CrossRef CAS PubMed.
- A. S. Abdelfattah, S. L. Farhi, Y. Zhao, D. Brinks, P. Zou, A. Ruangkittisakul, J. Platisa, V. A. Pieribone, K. Ballanyi, A. E. Cohen and R. E. Campbell, J. Neurosci., 2016, 36, 2458–2472 CrossRef CAS PubMed.
- L. Jin, Z. Han, J. Platisa, J. R. A. Wooltorton, L. B. Cohen and V. A. Pieribone, Neuron, 2012, 75, 779–785 CrossRef CAS PubMed.
- Y. Gong, C. Huang, J. Z. Li, B. F. Grewe, Y. Zhang, S. Eismann and M. J. Schnitzer, Science, 2015, 350, 1361–1366 CrossRef CAS PubMed.
- M. Z. Lin and M. J. Schnitzer, Nat. Neurosci., 2016, 19, 1142–1153 CrossRef PubMed.
- J. Wu, Y. Liang, S. Chen, C.-L. Hsu, M. Chavarha, S. W. Evans, D. Shi, M. Z. Lin, K. K. Tsia and N. Ji, Nat. Methods, 2020, 17, 287–290 CrossRef CAS PubMed.
- B. Chanda, R. Blunck, L. C. Faria, F. E. Schweizer, I. Mody and F. Bezanilla, Nat. Neurosci., 2005, 8, 1619–1626 CrossRef CAS PubMed.
- N. Ghitani, P. O. Bayguinov, Y. Ma and M. B. Jackson, J. Neurophysiol., 2015, 113, 1249–1259 CrossRef PubMed.
- D. N. Ng and P. Fromherz, ACS Chem. Biol., 2011, 6, 444–451 CrossRef CAS PubMed.
- M. Sundukova, E. Prifti, A. Bucci, K. Kirillova, J. Serrao, L. Reymond, M. Umebayashi, R. Hovius, H. Riezman, K. Johnsson and P. A. Heppenstall, Angew. Chem., Int. Ed., 2019, 58, 2341–2344 CrossRef CAS PubMed.
- T. Fiala, J. Wang, M. Dunn, P. Šebej, S. J. Choi, E. C. Nwadibia, E. Fialova, D. M. Martinez, C. E. Cheetham, K. J. Fogle, M. J. Palladino, Z. Freyberg, D. Sulzer and D. Sames, J. Am. Chem. Soc., 2020, 142, 9285–9301 CrossRef CAS PubMed.
- A. S. Abdelfattah, T. Kawashima, A. Singh, O. Novak, H. Liu, Y. Shuai, Y.-C. Huang, L. Campagnola, S. C. Seeman, J. Yu, J. Zheng, J. B. Grimm, R. Patel, J. Friedrich, B. D. Mensh, L. Paninski, J. J. Macklin, G. J. Murphy, K. Podgorski, B.-J. Lin, T.-W. Chen, G. C. Turner, Z. Liu, M. Koyama, K. Svoboda, M. B. Ahrens, L. D. Lavis and E. R. Schreiter, Science, 2019, 365, 699–704 CrossRef CAS PubMed.
- S. Liu, C. Lin, Y. Xu, H. Luo, L. Peng, X. Zeng, H. Zheng, P. R. Chen and P. Zou, Nat. Chem., 2021, 13, 472–479 CrossRef CAS PubMed.
- Y.-L. Huang, A. S. Walker and E. W. Miller, J. Am. Chem. Soc., 2015, 137, 10767–10776 CrossRef CAS PubMed.
- M. M. Milosevic, J. Jang, E. J. McKimm, M. H. Zhu and S. D. Antic, eNeuro, 2020, 7, ENEURO.0060–0020.2020 CrossRef CAS PubMed.
- S. P. Ginebaugh, E. D. Cyphers, V. Lanka, G. Ortiz, E. W. Miller, R. Laghaei and S. D. Meriney, J. Neurosci., 2020, 40, 3504–3516 CrossRef CAS PubMed.
- A. Klimas, G. Ortiz, S. C. Boggess, E. W. Miller and E. Entcheva, Prog. Biophys. Mol. Biol., 2020, 154, 62–70 CrossRef PubMed.
- H. M. McNamara, R. Salegame, Z. Al Tanoury, H. T. Xu, S. Begum, G. Ortiz, O. Pourquie and A. E. Cohen, Nat. Phys., 2020, 16, 357 Search PubMed.
- P. E. Deal, R. U. Kulkarni, S. H. Al-Abdullatif and E. W. Miller, J. Am. Chem. Soc., 2016, 138, 9085–9088 CrossRef CAS PubMed.
- P. E. Deal, P. Liu, S. H. Al-Abdullatif, V. R. Muller, K. Shamardani, H. Adesnik and E. W. Miller, J. Am. Chem. Soc., 2020, 142, 614–622 CrossRef CAS PubMed.
- S. R. Mujumdar, R. B. Mujumdar, C. M. Grant and A. S. Waggoner, Bioconjugate Chem., 1996, 7, 356–362 CrossRef CAS PubMed.
- Y. Koide, Y. Urano, K. Hanaoka, W. Piao, M. Kusakabe, N. Saito, T. Terai, T. Okabe and T. Nagano, J. Am. Chem. Soc., 2012, 134, 5029–5031 CrossRef CAS PubMed.
- B. Wang, X. Chai, W. Zhu, T. Wang and Q. Wu, Chem. Commun., 2014, 50, 14374–14377 RSC.
- E. Kozma, G. Estrada Girona, G. Paci, E. A. Lemke and P. Kele, Chem. Commun., 2017, 53, 6696–6699 RSC.
- C. R. Woodford, E. P. Frady, R. S. Smith, B. Morey, G. Canzi, S. F. Palida, R. C. Araneda, W. B. Kristan, C. P. Kubiak, E. W. Miller and R. Y. Tsien, J. Am. Chem. Soc., 2015, 137, 1817–1824 CrossRef CAS PubMed.
- T. K. Neklesa, H. S. Tae, A. R. Schneekloth, M. J. Stulberg, T. W. Corson, T. B. Sundberg, K. Raina, S. A. Holley and C. M. Crews, Nat. Chem. Biol., 2011, 7, 538–543 CrossRef CAS PubMed.
- H. Tabata and K. Nakajima, Neuroscience, 2001, 103, 865–872 CrossRef CAS PubMed.
- Y. Koide, Y. Urano, K. Hanaoka, T. Terai and T. Nagano, ACS Chem. Biol., 2011, 6, 600–608 CrossRef CAS PubMed.
- J. B. Grimm, T. A. Brown, A. N. Tkachuk and L. D. Lavis, ACS Cent. Sci., 2017, 3, 975–985 CrossRef CAS PubMed.
- P. Liu, V. Grenier, W. Hong, V. R. Muller and E. W. Miller, J. Am. Chem. Soc., 2017, 139, 17334–17340 CrossRef CAS PubMed.
- G. Ortiz, P. Liu, S. H. H. Naing, V. R. Muller and E. W. Miller, J. Am. Chem. Soc., 2019, 141, 6631–6638 CrossRef CAS PubMed.
- V. Grenier, B. R. Daws, P. Liu and E. W. Miller, J. Am. Chem. Soc., 2019, 141, 1349–1358 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Spectra, supporting figures, experimental details. See DOI: 10.1039/d1cb00156f |
| This journal is © The Royal Society of Chemistry 2021 |