Advances in electrochemiluminescence for single-cell analysis
Qian
Yang
ab,
Xiaoyu
Huang
a,
Beibei
Gao
a,
Lu
Gao
a,
Feng
Yu
 *b and
Fu
Wang
*b and
Fu
Wang
 *a
*a
aSchool of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai 200240, China. E-mail: wangfu@sjtu.edu.cn
bKey Laboratory for Green Processing of Chemical Engineering of Xinjiang Bingtuan, School of Chemistry and Chemical Engineering, Shihezi University, Shihezi 832003, China. E-mail: yufeng05@mail.ipc.ac.cn
First published on 7th December 2022
Abstract
Recent years have witnessed the emergence of innovative analytical methods with high sensitivity and spatiotemporal resolution that allowed qualitative and quantitative analysis to be carried out at single-cell and subcellular levels. Electrochemiluminescence (ECL) is a unique chemiluminescence of high-energy electron transfer triggered by electrical excitation. The ingenious combination of electrochemistry and chemiluminescence results in the distinct advantages of high sensitivity, a wide dynamic range and good reproducibility. Specifically, single-cell ECL (SCECL) analysis with excellent spatiotemporal resolution has emerged as a promising toolbox in bioanalysis for revealing individual cells’ heterogeneity and stochastic processes. This review focuses on advances in SCECL analysis and bioimaging. The history and recent advances in ECL probes and strategies for system design are briefly reviewed. Subsequently, the latest advances in representative SCECL analysis techniques for bioassays, bioimaging and therapeutics are also highlighted. Then, the current challenges and future perspectives are discussed.
1. Introduction
The cell is the basic unit of structure and function of an organism, and many critical areas of life sciences require research to be addressed at the single-cell level (e.g., stem cell differentiation and disease progression). Intrinsically, single-cell analysis plays a vital role in biology and medicine, presenting morphological changes in individual cells during physiological and pathological processes and promoting further study of the biological functions of cells in complex cellular environments.1–3 Therefore, developments in analytical techniques are a driving force in the single-cell field.4–6 The observation of cell surface-active substances versus intracellular substance transport and the highly sensitive detection of molecules produced by single-cell metabolic processes are essential for studying cellular heterogeneity and disease states.Cell imaging analysis has become a core technology and cutting-edge science in life sciences since the 21st century. It has attracted enormous interest from researchers in a wide range of disciplines.7–10 With continuous efforts, a range of innovative analytical methods with high sensitivity and spatiotemporal resolution (e.g., confocal laser fluorescence microscopy,11 confocal Raman microscopy,12 super-resolution microscopy,13etc.) have emerged to allow both qualitative and quantitative analyses at the single-cell levels for morphological characterization and quantitative chemical analysis of single cells. However, the conventional single-cell fluorescence analysis techniques suffer from photobleaching and autofluorescence caused by real-time excitation light, limiting the signal-to-noise ratio (SNR) and sensitivity in biosensing and bioimaging.14 Therefore, it is necessary to explore alternative methods with low fluorescence backgrounds to analyse the function of individual cells efficiently. Electro-chemiluminescence (ECL), also referred to as electrogenerated chemiluminescence, is a type of chemiluminescence generated by electrochemical reactions with the conversion from a light signal to an electrical signal by optical readout devices, exhibiting excellent potential applications in single-cell analysis.15–17 The measurement of ECL luminescence spectra and intensities by photomultiplier tubes (PMT) or other optical components enables qualitative and quantitative analyses of cells and their organelles at the single-cell level.18–20 As an electrochemical-triggered luminescence imaging technique, single-cell ECL (SCECL) analysis is a very powerful sensing and imaging platform with excellent spatiotemporal control of light emission, low background and good reproducibility.
Generally, ECL can be generated by the annihilation pathway and the co-reactant pathway.21–24 The annihilation pathway is a positive and negative double-step pulse potential applied to the electrode where luminophores undergo reduction and oxidation reactions at the cathode and anode, respectively, producing anionic and cationic radicals. Subsequently, the anionic radical combines with the cationic radical to undergo an annihilation reaction near the electrode surface to produce the excited species, which leaps back to the ground state to produce optical radiation.25 Different from the annihilation pathway, which requires the application of positive and negative bi-directional voltages, co-reactant ECL only requires a single voltage to be applied to the electrode in either the positive or negative direction. During oxidation or reduction, the co-reactants are often converted into certain intermediates with strong reducing or oxidizing properties, which in turn can react with ECL probes or their intermediate states to form an excited state of ions or molecules, and then achieve ECL luminescence.26 Several recent reviews have summarized the detailed mechanism of ECL, highlighting the advantages of varying ECL routes for ECL imaging systems.27–30 For example, Wu et al. systematically described the mechanism of ECL cell analysis based on the quantum dot (QD) system.31 Zhang et al. introduced the mechanism of the ECL reaction in ECL imaging based on the co-reactant pathway.32 In contrast, SCECL analysis encounters two major dilemmas due to cellular characteristics and ECL response properties. On the one hand, cells adhering to the electrode surface hinder the transfer of electrons and the diffusion of electroactive molecules (luminophores and co-reactants) to the electrode surface, resulting in a weak ECL signal. On the other hand, there is a technical limitation of insufficient spatiotemporal resolution due to the weak luminescence signal produced by the ECL molecules. Thereby, improving the sensitivity and spatiotemporal resolution of SCECL analysis is a significant challenge for this technology in life science research.
In recent years, SCECL analysis has developed in leaps and bounds with the advances in ECL analysis. While many review articles have highlighted ECL sensing or imaging based on the ECL reaction principles, SCECL analysis techniques for novel sensing and imaging strategies have not been systematically reviewed. Herein, we present the ECL detection and imaging systems and the analysing principles to characterize single-cell behaviour, visualize cells and organelles, and analyze cellular dynamic behaviour. Compared to previous research results on SCECL analysis, this review classifies SCECL analysis strategies to better understand the relationship between single-cell level analysis and ECL systems.
2. Development of new ECL probes
The emergence of single-cell analysis techniques has opened up a new area for exploring the molecular basis of life. ECL probes are emitters that generate luminescent signals during electrochemical processes which play the most important role in the ECL procedure. The performance of the ECL probe directly affects the ECL efficiency, and the emergence of novel ECL probes with high emission efficiency accelerates the ECL response, enabling high-throughput analysis of micro-targets. ECL probes can generally be classified into three categories: organic compounds, metal complexes, and nanomaterials.33,342.1. Organic ECL probes
Organic compounds were the first reported ECL probe. In the 1920s, Dufford et al. observed electrolytic luminescence during the electrolysis of Grignard's reagent, opening a gate to the exploration of the field of ECL.35 Organic compound probes mainly include hydrazides such as luminol,36,37 and polycyclic aromatic hydrocarbons such as porphyrins,38–40 rubrenes,41,42 anthracenes,43etc. The luminescence of organic probes usually requires the involvement of hydrogen peroxide (H2O2) and is very sensitive to H2O2. Therefore, some bioactive substances capable of producing H2O2 can be indirectly determined by organic ECL probes based on this principle.44–46 Most organic ECL probes have high ECL yields, but only produce strong ECL signals in alkaline solutions, making them difficult to apply in biological systems.2.2. Metal complex ECL probes
In the 1970s, Bard's group discovered that tris-bipyridyl-ruthenium(II) (Ru(bpy)32+) had high luminescence efficiency and stable intensity in aqueous solutions.47 The Ru(bpy)32+-tripropylamine (TPrA) system is a typical example of ECL technology in practical analytical applications, which has been successfully used in commercial ECL immunoassays and clinical diagnostics.48–51 Meanwhile, several novel metal complexes with photochemical properties and electrochemical activity, such as Ir(III) with a probe coordination pattern similar to Ru(II), have attracted great interest.52–54 However, research on the ECL of Ir(III) is mainly restricted to organic solution systems, which is not conducive to its further application in biochemical analysis and other fields.55 Several new two-dimensional structures such as rare earth probes and near-infrared (NIR) lanthanide complex probes with inherently excellent luminescence properties and biocompatibility are currently attracting a lot of attention.56–58Metal complex ECL probes are commonly expensive and the introduction of heavy elements makes them potentially toxic to cells. However in combination with advanced nanotechnology, Ru(bpy)32+ remains of significant research value.59–61 For example, mixing nanomaterials with Ru(bpy)32+ (or its derivatives) can emit enhanced ECL when reacted with co-reactants, resulting in better analytical performance.62
2.3. Nanomaterial ECL probes
In 2002, Ding et al. observed the ECL of silicon nanocrystals, which sparked the interest of researchers in the study of ECL probes in nanomaterials.63 With advances in the preparation of high-performance nanomaterials, various semiconductors are characterized for highly efficient ECL nanoprobes.64 For example, metal QDs,65,66 carbon dots (CDs),67–69 and metal nanoclusters (NCs)70–72 are widely used in ECL research.Metal QDs are usually made of Group II–VIB (e.g., CdSe, CdTe, CdS, ZnSe) or Group III–VA (e.g., InP, InAs) elements or their alloyed semiconductor materials.73–75 With homogeneous and core–shell structures for easy surface modification and confined sizes at the nanoscale (1–10 nm) for better quantum confinement effects, metal QDs are widely used in bioanalyses such as molecular detection and cell imaging.76–78 CDs (including graphene dots) are promising ECL luminophores used in bioanalysis due to their good stability, ease of modification and biocompatibility.79–81 However, conventional CDs exhibit weak ECL emission and require high excitation potentials, which limit their sensitivity and safety. A strategy of embedding zero-dimensional CDs into the skeletal network of metal nanosheets could provide an ingenious solution to this problem.82,83 Metal NCs are usually composed of tens of metal atoms, are ultra-small in size (<2 nm), and act as a bridge between organometallic compounds and crystalline metal NPs, and thus have potential applications in the field of single-cell analysis.84–86
Most nanomaterials have excellent optical properties but suffer from poor biocompatibility or electrochemical activity, and their luminescence properties are easily affected during modification. Therefore, the development of nanoprobes with good water solubility, stability, optoelectronic properties and low toxicity is of great value in the field of SCECL analysis.
3. Single-cell ECL systems
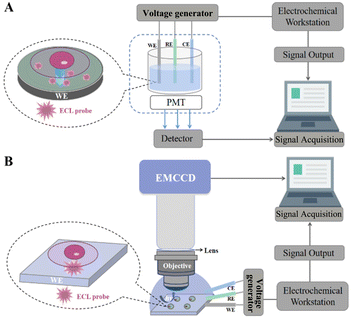
In life analytical chemistry, the detection of chemical components involved in cell activities has become the focus of life sciences to explore new detection principles and establish the corresponding analytical methods.21 Many new cross-cutting areas have been formed in recent years. SCECL analysis is an imaging technique developed in recent years which closely integrates electrochemistry and photophysics to provide both optical images and electrochemical signals at the single-cell level. SCECL analysis enables the development of analytical sciences that require high throughput, visualization, and information diversification.87,88 Although the construction of single-cell electrochemical analysis instruments and test conditions are different, the principle of ECL testing is consistent, using the intensity of the ECL signal, image greyscale, spectral peaks and other relationships between the content of the components to be measured to achieve quantitative analysis.89–91 Currently, most laboratory ECL test platforms are self-built according to experimental requirements. The basic ECL analysis system consists of three core components: an electrochemical signal excitation system, an optical signal detection system and a three-electrode cell system.92 Electrical signal excitation devices are generally used for common electrochemical workstations. Optical detectors include PMT, avalanche photodiodes (APD), charge-coupled devices (CCD), electron-multiplying charge-coupled devices (EMCCD), etc. The three-electrode cell system is composed of the working electrode (WE), the counter electrode (CE) and the reference electrode (RE), where the WE of the sensing system is usually a round glassy carbon electrode (GCE) and the imaging system is usually composed of square fluorine tin oxide (FTO) conductive glass or indium tin oxide (ITO) conductive glass.93–953.1. ECL sensing system
Cell sensing involves cell capture, cell adhesion, signal transduction and signal output, and the accuracy can be assured using an ECL cell sensor with high sensitivity and selectivity.96 As shown in Fig. 1A, the SCECL sensing system includes an electrochemical workstation, a three-electrode cell system, a detector, and a computer. SCECL sensing technology mainly uses optical receptors such as PMT to collect the light intensity signals of photons generated on the surface of the electrode, convert the light signal into an electrical signal through the PMT, and output the corresponding ECL signal after amplification processing.97,98 Depending on the sensitivity of the ECL sensor assembly, the ECL probe and the PMT determine the sensitivity of its analytical performance. The ECL active material can modify the WE as an ECL probe to provide a signal output, which the PMT converts into an electrical signal for final signal acquisition.The principles of antigen–antibody-specific binding, hybridisation between DNA or aptamers, and enzyme-based catalysis of biomolecules are often used to construct ECL biosensors.99–101 Based on the high bio-affinity of the antigen and antibody reaction, immunosensors are most widely used in the detection of clinical samples. Using TPrA as an electron donor, ECL is produced by electron transfer under electrical excitation by labelling the antibody or antigen with tris-bipyridyl-ruthenium. Compared to ECL immunosensors, DNA and aptamer-based ECL sensors offer many advantages, such as easy labelling and good stability. DNA and aptamer-based ECL sensors are based on the principle of complementary base pairing which relies on efficient hybridisation between nucleic acids to convert the information sensed by the probe into a detectable signal. In enzyme-based ECL sensors, many of the test substrates can be broken down by the action of the corresponding biological enzymes and converted into easily detectable small biological molecules. ECL analysis based on the luminol enzyme is the most common, as many substrates can be broken down to H2O2 in the presence of this enzyme.
3.2. ECL imaging system
The signals collected by ECL are divided into two main types: intensity and image.102 Although higher sensitivity and lower detection limits can be obtained with the intensity signal acquisition approach, the imaging devices show many advantages that cannot be replaced by the former, such as rapid characterisation of electrochemically active sites and high throughput array detection. For example, PMT-based ECL detection can only obtain the intensity over the entire electrode but does not have spatial resolution. To allow more direct observation of specific localised areas on the electrodes, optical detection devices are used in microscopy imaging systems to acquire and output the signal in the form of an image.As optical imaging instruments and techniques continue to improve, ultra-high sensitivity single-photon level optical detection devices (e.g., EMCCD) are being used in microscopy imaging systems.103–105 Single-cell ECL microscopy (ECLM) holds significant potential for ultra-sensitive microscopy imaging that combines the advantages of chemiluminescence in detectability with improved resolution. A single-cell ECLM system consists of an electrochemical workstation, an electrochemical cell, a microscope with EMCCD and other optical components (e.g., objective, lens, etc.), and a computer (Fig. 1B).106–108 The electrochemical workstation is the electrochemical signal driving device and EMCCD is the optical signal acquisition device. The process of acquiring ECL images starts with the application of a suitable potential to the electrode to initiate the ECL reaction. Subsequently, the light signal generated by the electrochemical reaction is first focused by an optical lens, then captured and converted into a digital signal by an EMCCD, and finally transferred to a computer for analysing and processing. Various high luminescence efficiency ECL probes have been designed to accelerate ECL luminescence, along with novel electrode designs and imaging strategies (e.g., array electrode ECL imaging, bipolar electrode ECL imaging, etc.). ECL imaging can achieve nanoscale spatial resolution and millisecond temporal resolution, respectively. SCECL imaging and super-resolved ECL imaging at the single-cell level are the future research hotspots.109,110
4. Application of single-cell ECL analysis in bioassays
Chemical analysis of individual living cells with high throughput and spatiotemporal resolution can provide valuable cellular characterization information related to cellular heterogeneity. An in-depth exploration into the processes of cellular life activities (e.g., elucidation of individual cell structure–function relationships and study of extracellular release from each region of the cell surface) will significantly facilitate the study of biological phenomena and disease development.88 With the up-to-date advances, SCECL analysis studies have focused on two main areas: one is on the visualization and quantification of intracellular molecules, and another is on the visualization of cell surface structures.1114.1. Single-cell detection
Cell heterogeneity is a crucial area of research in SCECL analysis which could allow more accurate and higher resolution studies on the heterogeneous expression of cells, revealing extra information about cellular life activities. Cytosensing uses the cell as the object of study to monitor cellular life processes by converting cells, cell surface-active substances, and cellular metabolites into analysable detection signals for qualitative or quantitative resolution of certain functional information of the cell. Through the design and study of ECL cell sensors, information-rich electrochemical and optical signals can be obtained simultaneously. Until now, various strategies have been developed for analysing single cells and their intracellular molecules, such as determining DNA,112 miRNA,113 proteins,114 cancer cell receptor expression,115etc.Various low abundance but information-rich proteins exist on the cell surface. They are often used as markers for early diagnosis, therapeutic efficacy or as targets for treatment in disease therapy.118 Qiu et al. (2017) prepared solid-state zinc-coadsorbed CQD nanocomposite probes by attaching zinc-coated CQDs to gold nanoparticles (AuNPs), with 120-fold higher ECL strength than bare CQDs (Fig. 2A).119 Meanwhile, a graphene oxide (GO)/AuNP interface was formed by modifying GO and AuNPs on ITO electrodes to improve electrical conductivity. They successfully assessed CD44 expression on the surface of single cells through the changes in the ECL signal intensity, and could detect MDA-MB-231 cells and MCF-7 cells from 1 to 18 cells and 1 to 12 cells. By increasing the distance between the cell membrane and the electrode to accommodate more luminescent molecules, the sensitivity of cell analysis can be enhanced. Long et al. (2018) used 11-mercaptodecanoic acid (MUA) to provide the optimum gap on electrodeposited PANI/AuNP substrates (Fig. 2B).120 They synthesized a bifunctional Au@Cu–PbCQD nanoprobe in which the AuNP core is connected to a lead bead-adsorbed CQD via a copper(II) ion bridge. The ECL intensity of the resulting sensor with a spatial structure modified by MUA and distinctive ECL probe design was increased by 37.5 ± 3.9%. Thus, the sensor can achieve high sensitivity in specifically identifying CD44.
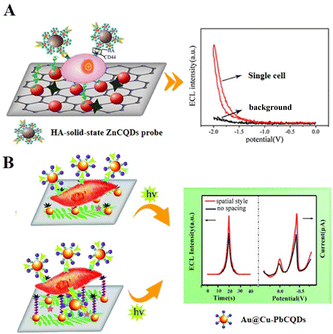 | ||
| Fig. 2 (A) Single-cell analysis platform based on a solid-state ZnCQD nanocomposite ECL probe. Reprinted with permission from ref. 119. Copyright 2017 American Chemical Society. (B) A single-cell sensor with MUA-modified electrodes based on an Au@Cu–PbCQD ECL nanoprobe. Reproduced with permission from ref. 120. Copyright 2018 The Royal Society of Chemistry. | ||
Liu et al. (2019) proposed a strategy for ECL detection using graphene hydrogel (GH) electrodes modified with glucose transporter protein 4 (GLUT4).121 They labelled HSMC cells with GLUT4-Ab-functionalized CDs (CDs-GLUT4-Ab), which were then immobilized on the permeable and conductive GH electrodes. Based on the specific recognition of GLUT4 by GLUT4-Ab, CDs-GLUT4-Ab can specifically capture GLUT1 on HSMC cell membranes. This ECL assay strategy without cellular steric hindrance improved the accuracy of the assessment of GLUT4 on the cell surface and the sensitivity of the analytical performance of HSMC cells, which could detect from 500 to 1.0 × 106 cells per mL, with a limit of detection (LOD) of 200 cells per mL.
An important indicator for assessing disease severity and predicting recurrence is tumour markers, which are biomolecules associated with cancer cyclin D1 (CCND1), which is overexpressed in human MCF-7 breast cancer cells and can be used as a tumour marker to reflect the malignancy of the cancer cells. Zhao's group (2019) prepared Au@Cu–Bi2S3 QD ECL nanoprobes by linking Bi2S3 QDs with AuNPs via copper(II) ion bridges, further constructing an ECL immunosensor to detect CCND1.122 Based on the conductivity of Cu2+ and the surface plasmon resonance (SPR) effect of AuNPs, the ECL signal was enhanced more than 20 fold. The ECL immunosensor has good stability and presented a relatively wide detection range of 10 fg mL−1–1 μg mL−1.
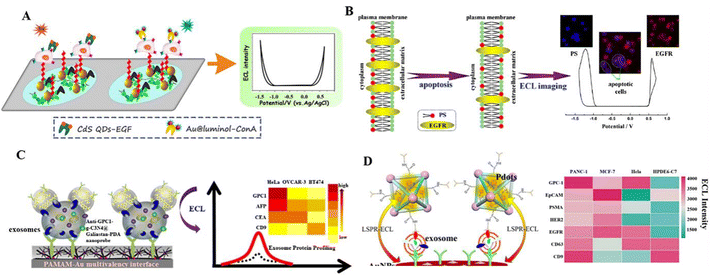 | ||
| Fig. 3 (A) Au@luminol and CdS QDs as ECL nanoprobes to detect the expression levels of multiple cell–surface receptors, mannose and epidermal growth factor receptor (EGFR). Reprinted with permission from ref. 123. Copyright 2017 American Chemical Society. (B) Identifying epidermal growth factor receptor (EGFR) and phosphatidylserine (PS) based on the principle of complementary base pairing. Reprinted with permission from ref. 124. Copyright 2019 American Chemical Society. (C) An ECL biosensor for exosome protein using an anti-GPC1–g-C3N4@Galinstan–PDA nanoprobe. Reprinted with permission from ref. 127. Copyright 2019 American Chemical Society. (D) The ECL immunosensor for pancreatic exosome detection is based on the LSPR interaction between MOF@Pdots and AuNPs. Reprinted with permission from ref. 129. Copyright 2022 American Chemical Society. | ||
Abnormally expressed tumour markers in biological samples (e.g., living cancer cells or specific RNA/DNA fragments) could be biomarkers for clinical diagnosis and disease treatment. Liu et al. (2020) successfully synthesized two potential-resolved ECL nanoprobes and used them to directly analyse the apoptosis diagnosis of living cancer cells (Fig. 3B).124 In this research, Au@L012 and g-C3N4 were modified by EGF and peptide (PSBP) to recognize EGFR and phosphatidylserine (PS) on the cell surface, respectively, which correlated with the degree of cell apoptosis. Furthermore, using their self-built ECLM, EGFR and PS expressions in many individual cells could be assessed simultaneously, thus enabling visual analysis of apoptosis rates in normal and cancerous cell samples. An aberrant expression of microRNAs (miRNAs) is associated with an extensive array of cancer types that could be the analytes for cancer diagnosis. Chai et al. (2022) successfully synthesized bimetallic cluster-doped MnS:CdS@ZnS core–shell QDs as ECL emitters, which were applied to cancer cell (MCF7) lysates for miRNA let-7a analysis.125 Due to the surface defect passivation of the MnS:CdS@ZnS QDs, the ECL efficiency was as high as 84%. Based on the miRNA-powered strand displacement amplification as a signal amplification strategy, they constructed an ECL “signal-off” biosensor for ultra-sensitive detection of miRNA let-7a, with a LOD of 4.1 aM.
Exosomes secreted by living cells have a phospholipid bilayer structure carrying various biologically active substances (proteins, lipids, nucleic acids, etc.). When the exosomes deliver these substances to the recipient cells, they will alter the physiological or pathological function of these cells. Detecting genetic material and proteins in exosomes will help in the early diagnosis of diseases. Liu's group (2019) designed a sensitive ECL biosensor by combining a g-C3N4 conjugated polydopamine coated Galinstan liquid metal shell–core (g-C3N4@Galinstan–PDA) nanoprobe with a multivalent PAMAM–AuNP electrode interface (Fig. 3C).127 A highly sensitive diagnosis of exosomes from HeLa cells can be achieved, with a LOD of 31 particles per μL obtained. Based on a similar strategy, they used in situ formed AuNP decorated Ti3C2 MXene hybrid nanoprobes with aptamer modification (AuNPs–MXenes–Apt) in 2021.128 The AuNPs–MXenes–Apt hybrid nanoprobes not only present an efficient and specific recognition of exosomes, but also provide a bare catalytic surface with high electrocatalytic activity. In the ECL biosensor for highly sensitive ECL detection of exosomes by AuNPs–MXenes–Apt hybrid nanoprobes, the detection limit for exosomes from HeLa cell lines was 30 particles per μL. Distinguishing subtle changes in exosomal surface protein markers in different cell lines helps to understand exosome heterogeneity and gather phenotypic information about cancer. Xiong et al. (2022) prepared a sandwich-type ECL immunosensor to assess the exosome surface protein expression levels of different cell lines.129 They designed an ECL probe using polymer dots (Pdots) loaded on a metal–organic backbone (MOF@Pdots) (Fig. 3D). Based on the localized surface plasmon resonance (LSPR) interaction between MOF@Pdots and AuNPs, they offered a universal platform for pancreatic cancer exosome detection. The ECL immunosensor showed high sensitivity, with a wide calibration range of 1.0 × 103 to 1.0 × 106 particles per mL.
H2O2 is an essential chemical in the human body, and moderate concentrations of H2O2 have potential therapeutic implications for many diseases. Using a luminol-based ECL system, H2O2 in single cells can be detected. Chen et al. (2020) designed novel nitrogen-doped hydrazide conjugated CDs (NHCDs) as an ECL nanoprobe, which had a strong anodic ECL at low excitation potentials.130 The ECL quantum efficiency of NHCDs increased to 2.5 times as the nitrogen atoms were doped into the graphite core of the CDs. Using NHCDs modified the electrode surface to prepare ECL biosensors that can detect the cell-secreted H2O2, enabling rapid differentiation between cancer cells and normal cells.
With a large surface area, electrical conductivity, biocompatibility, and easy mixing with other nanomaterials, CDs are widely used in ECL cell sensors. Wang et al. (2020) synthesized Au@CD nano-hybridized probes with 12-fold higher Au@CD ECL emission efficiency than pure CDs (Fig. 4A).132 Based on the high ECL efficiency of Au@CDs, they prepared a sensitive cell sensor to detect cells directly, by immobilizing human mucin 1 (MUC1) on the surface of Au@CDs on an electrode. The Au@CD-based cell sensor could detect the MCF-7 cells ranging from 100 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL, and with a LOD of 34 cells per mL. Liu et al. (2021) designed a CoCu–ZIF@CD cell sensor for detection of B16eF10 cells (Fig. 4B).133 As zero-dimensional (0D) CDs were effectively embedded into two-dimensional (2D) Co–Cu–ZIF nanosheets, the 0D/2D heterogeneous nanostructures exhibited better electrical chemical activity and excellent fluorescence properties. The CoCu–ZIF@CD-based cell sensor exhibited superior stability and sensitivity for detecting B16–F10 cells, ranging from 1 × 102–1 × 105 cells per mL.
000 cells per mL, and with a LOD of 34 cells per mL. Liu et al. (2021) designed a CoCu–ZIF@CD cell sensor for detection of B16eF10 cells (Fig. 4B).133 As zero-dimensional (0D) CDs were effectively embedded into two-dimensional (2D) Co–Cu–ZIF nanosheets, the 0D/2D heterogeneous nanostructures exhibited better electrical chemical activity and excellent fluorescence properties. The CoCu–ZIF@CD-based cell sensor exhibited superior stability and sensitivity for detecting B16–F10 cells, ranging from 1 × 102–1 × 105 cells per mL.
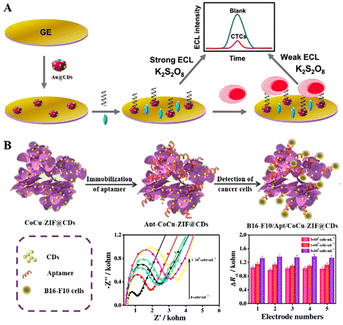 | ||
| Fig. 4 (A) An ECL biosensor for MCF-7 cell detection by an Au@CD nanoprobe. Reprinted with permission from ref. 132. Copyright 2020 Elsevier. (B) An ECL biosensor for B16–F10 cell detection by a CoCu–ZIF@CD nanoprobe. Reprinted with permission from ref. 133. Copyright 2021 Springer Nature BV. | ||
Apart from the strategy of compounding various nanomaterials to enhance ECL, a surface vacancy is also important to improve the luminescence efficiency of ECL. Gao et al. (2021) developed a rare earth nanosheet of ultrathin Lu2O3–S enriched with oxygen vacancies as a novel cell sensing ECL probe, which was combined with a DNA device cyclic-amplification technology to construct a cross-enhanced ECL cell-sensing platform.56 Lu2O3–S nanosheets are rich in oxygen vacancies and exhibit a special 2D structural morphology, resulting in stronger and more stable ECL emissions. This ECL cell sensor demonstrates excellent analytical performance, with a proportional response of the ECL signal to the CEM cell concentration over a wide range of 50 to 1 × 106 cells per mL, and an extremely low LOD of 10 cells per mL (Table 1).
| ECL probes | Recognition elements | Analyte | Cell line | LOD | Linear range | Ref. |
|---|---|---|---|---|---|---|
| N/A = not available; CQDs: carbon quantum dots; CDs: carbon dots; QDs: quantum dots; HA: hyaluronic acid; GLUT4: glucose transporter protein 4; MUC1: mucin 1; GPC1: glypican 1 antibody; recombinant glypican 1; AFP: alpha-fetoprotein prostate; CEA: carcinoembryonic antigen; ConA: concanavalin A; EGFR: epidermal growth factor receptor; CTCs: circulating tumour cells; MDA-MB-231: human breast cancer cell line; MCF-7: breast cancer cell line; H9c2: cardiomyocyte cell line; HSMCs: human skin mast cells; OVCAR-3: human ovarian adenocarcinoma cells; BT474: human breast cancer cells; HPDE6-C7: human normal pancreatic duct epithelial cells; B16–F10: B16–F10 mouse cells. | ||||||
| ZnCQDs | HA | CD44 | MDA-MB-231 | 1 cell per mL | 1–18 cells per mL | 119 |
| Au@Cu–PbCQDs | HA | CD44 | MCF-7 | 20 cells | 20 cells | 120 |
| CDs | GLUT4 antibody | GLUT4, cell detection | HSMCs | 200 cells per mL | 500–106 cells per mL | 121 |
| Au@Cu–Bi2S3 QDs | Cyclin D1 | Protein | BSA | 6.34 fg mL−1 | 10 fg mL−1–1 μg mL−1 | 122 |
| Au@luminol, CdS QDs | MUC1 | Cell detection, mannose, EGFR | MCF-7 cells | 20 cells per mL | 102–106 cells per mL | 123 |
| g-C3N4, Au@L012 | Complementary base pairing | EGFR, PS | HSM, H9C2, MCF-7 cells | N/A | More than 100 cells | 124 |
| MnS:CdS@ZnS | Complementary base pairing | miRNA | N/A | 4.1 aM | 10 aM–1 nM | 125 |
| g-C3N4@Galinstan–PDA | Exosome; AFP, CEA | Exosome, protein | HeLa, OVCAR-3, BT474 cells | 31 particles per μL | 50–105 particles per μL | 127 |
| AuNPs–Ti3C MXenes | Exosome; AFP, CEA | Exosome, protein | HeLa, OVCAR-3, BT474 cells | 30 particles per μL | 102–105 particles per μL | 128 |
| MOF@Pdots | Protein | Exosome, protein | PANC-01, HeLa, MCF-7, HPDE6–C7 cells | 400 | 103–106 particles per mL | 129 |
| NHCDs | H2O2 | H2O2 | HeLa, MCF-7 cells | N/A | 10–104 cells per mL | 130 |
| CdTe QDs/luminol | ConA; EGF | Cell detection | MCF-7 cells | 80 cells per mL | 100–6500 cells per mL | 131 |
| Au@CDs | CTCs | Cell detection | MCF-7 cells | 34 cells per mL | 102–104 cells per mL | 132 |
| CoCu–ZIF@CDs | B16eF10 cells | Cell detection | B16–F10 cells | 34 cells per mL | 102–105 cells per mL | 133 |
| Lu2O3–S nanosheets | Complementary base pairing | Cell detection | CEM cells | 10 cells per mL | 50–106 cells per mL | 56 |
4.2. Single-cell bioimaging
Microelectrodes based on electrochemical methods can offer excellent local sensitivity, allowing complex biological processes to be studied at the single-cell level.136,137 Based on the luminol ECL system, H2O2 and glucose can be detected in single cells. Jiang's group (2016) applied luminol ECL to analyse intracellular glucose.138 They retained individual cells in cell-sized microwells on a gold-coated ITO slide. Subsequently, they introduced Triton X-100 to disrupt the plasma membrane of individual cells in the microwells, enabling analysis of intracellular glucose. In combination with this ECL imaging platform, 64 individual cells can be analysed in 60 s, speeding up single-cell analysis significantly (Fig. 5C). Quantitative measurement of H2O2 concentrations in the extracellular environment can provide a better understanding of its role in the physiological processes of organisms. He et al. (2016) prepared an Au–luminol-microelectrode to induce ECL with H2O2, enabling the electrochemical visualization of H2O2 in a single cell (Fig. 5A).139 They reported an ECL imaging assay for detecting intracellular H2O2 with a LOD of 1 mM. The higher detection limit of H2O2 in cells will limit its practical application. The sensitivity for H2O2 detection can be improved by optimizing the composition and surface structure of the electrode. Li et al. (2017) prepared an AuNP-modified FTO electrode by applying a luminol-based ECL system to determine the release of H2O2 from living cells.140 The AuNP–FTO electrode exhibits significant electrocatalytic activity for the luminol-based ECL system, which covers the H2O2 concentration of 10 nM to 1 μM.
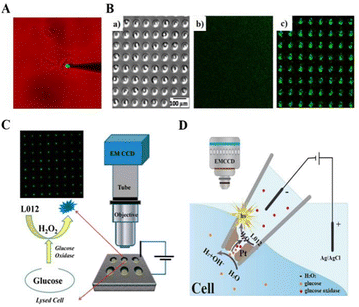 | ||
| Fig. 5 Schematic diagram of intracellular molecular measurement using SCECL imaging analysis: (A) the schematic system is used for the electrochemical imaging of intracellular hydrogen peroxide. Reprinted with permission from ref. 139. Copyright 2016 American Chemical Society. (B) Schematic diagram of gold-coated PDMS microwells loaded with individual cells: (a) the bright field image, (b) the background luminescence image, (c) the ECL image. Reprinted with permission from ref. 145. Copyright 2018 Springer Nature BV. (C) Schematic ECL imaging setup and the detection strategy for fast analysis of intracellular glucose in single cells. Reprinted with permission from ref. 138. Copyright 2016 American Chemical Society. (D) Representation of bipolar ECL detection on the porous Pt deposit inside the nanopipette, which is inserted into the cytosol for intracellular wireless electroanalysis. Reprinted with permission from ref. 149. Copyright 2020 Wiley-VCH. | ||
Array electrodes have attracted a lot of attention for their advantages of integration, miniaturization and high throughput. In combination with ECL imaging systems, they can achieve high throughput analysis of a wide range of target analytes.141–143 Xu et al. (2018) achieved enhanced ECL using microporous arrays modified with g-C3N4 nanosheets at low concentrations of luminol–H2O2 at 500 nM and analysed total cholesterol using ECL imaging analysis.144 This strategy was designed to increase the sensitivity of the assay, and the ECL visualization detection limit for H2O2 in the microtiter array was increased to 500 nM, ensuring the parallel detection of low concentrations of cholesterol in single cells. Xia et al. (2018) prepared gold-plated polydimethylsiloxane (PDMS) chips with cell-sized microwells by a stamping and spraying process, which had cell-retained microwells with higher luminescence intensity than that in the planar region among the microwells (Fig. 5B).145 They successfully monitored intracellular glucose by applying the PDMS chips.
A bipolar electrode (BPE) is a conductor between the cathode and anode of an electrolytic cell in a certain electrolyte, usually isolated in a microchannel filled with solution.146–148 Under the action of an applied electric field, the potential difference formed at the two ends of the conductor will be polarised as the anode and cathode for oxidation and reduction reactions respectively. Wang et al. (2020) established a bipolar ECL detection scheme to perform a wireless single-cell electroanalysis (Fig. 5D).149 They prepared a Pt deposit at the nanopipette tip and applied a very low potential to the nanopipette tip which can produce ECL emission of luminol at lower voltages in contrast to classical bipolar ECL. The porous structure of the Pt deposit permitted the transfer of target molecules into the nanopipette, thus enabling the measurement of intracellular glucose or H2O2 concentrations in vivo.
In addition to imaging analysis of the overall distribution of molecules within a single cell, single nanoparticle ECL technology offers a promising high throughput and sub-micron spatial resolution for local electrochemistry, enabling cellular focal molecular assays. Cui et al. (2019) used individual TiO2 NPs to visualize the local ECL of H2O2 efflux (Fig. 6A).150 The oxygen vacancies on the surface of rutile TiO2 NPs achieve steady-state adsorption of H2O2, which allowed the continuous production of superoxide radicals (O2˙−) and hydroxyl radicals (OH˙−) on the TiO2 NPs. Subsequently, aqueous luminol could be oxidized to a luminol derivative on the ITO and TiO2 surfaces, thus reacting with free radicals to produce ECL continuously. This strategy did not require complex microprocessing techniques and provided information on different cell regions. To achieve the ECL enhancement effect, Zhang et al. (2020) applied single lithium iron phosphate (LiFePO4, LFP) NPs to the surface of ITO, resulting in enhanced ECL illumination at low voltages due to the interaction between LFP and ITO.151 In this system, oxidation of L012 was promoted at a low voltage of 0.5 V. At the same time, the conversion of LiFePO4 to FePO4 accelerated the generation of oxygen intermediates from H2O2, allowing direct visualization of the H2O2 efflux from single living cells.
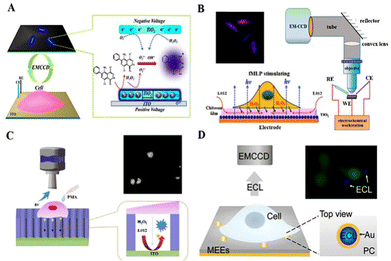 | ||
| Fig. 6 Schematic diagram of SCECL imaging: (A) schematic process of the ECL sensing of single TiO2 nanoparticles for visualizing local H2O2 efflux from single cells. Reprinted with permission from ref. 150. Copyright 2019 American Chemical Society. (B) Direct ECL imaging of a single cell on the chitosan-modified electrodes. Reprinted with permission from ref. 134. Copyright 2018 American Chemical Society. (C) Scheme of confined luminol ECL for mapping the H2O2 efflux from single living cells. The simplified reaction scheme of L012 is shown inside the SMCs. Reprinted with permission from ref. 135. Copyright 2019 Elsevier. (D) Illustration of confined ECL on gold MEEs for detecting H2O2 efflux from single cells. Reprinted with permission from ref. 152. Copyright 2021 Wiley-VCH. | ||
Due to the short lifespan and limited diffusion distances of the intermediates involved in the electrochemical reaction, the ECL signal is restricted to the marginal region of the cell membrane immediately adjacent to the electrode surface. Liu et al. (2018) prepared chitosan and nano-TiO2 modified FTO (FTO/TiO2/CS) (Fig. 6B).152 A permeable chitosan film avoided direct cell-to-electrode contact, storing more ECL reagents and ensuring high sensitivity for quantitative analysis. Their strategy demonstrated that expanding the space between the cells and the electrode was beneficial for ECL visualization studies of cells. Chen et al. (2019) developed an ECL device for the generation of luminol and H2O2 from vertically ordered silica microchannels (SMCs) (Fig. 6C).153 The vertical alignment of SMCs on ITO slides restricted the lateral diffusion of species produced during the ECL process, allowing the H2O2 efflux from cells to be observed at submicron spatial resolution. Using the ECL technique for cell analysis or cell imaging, the ECL response is difficult to occur in the cell-covered area due to the spatial resistance of the cells on the common electrode. Based on a similar strategy to limit the lateral spread of ECL, Ding et al. (2021) designed a vertically aligned combination of gold microtubule electrode ensembles (MEEs) for local sensing of single cells (Fig. 6D).154 The closed vertical structure of the MEEs limits ECL production to only a single microtubule on the PC membrane template. Local changes in the H2O2 concentration could be shown by changes in the ECL signal on individual MEEs. MEEs with sufficient spatial resolution allow parallel single-cell sensing, making them a promising platform for the analysis of intercellular information exchange.
SCECL analysis can distinguish between cellular variability and selectively measure membrane proteins overexpressed on tumour cells, essential for diagnosis and dedicated disease treatment. A significant breakthrough is the imaging of ruthenium complex dye-labelled cell membranes and proteins under surface-constrained ECLM. Sojic's group (2017) developed the first surface-constrained ECLM, which is a co-reactant-based surface-constrained ECLM system that allows imaging of living cells and their membrane proteins (Fig. 7A).155 Inkjet-printed disposable electrodes based on carbon nanotubes (CNT) were chosen for direct ECL imaging of tagged plasma receptors overexpressed on tumor cells. Then, Liu et al. (2021) developed Ru(bpy)32+-doped silica/AuNPs (RuDSNs/AuNPs) as ECL nano-emitters, using surface-constrained ECLM to image membrane proteins on a single fixed cell, enabling the ECL imaging of individual proteins on cell membranes at an electrode.156 RuDSN/AuNP nano-emitters were labelled with antibodies attached to cell membranes, and since ECL emission was restricted to the local surface of RuDSNs, the local surface limitation effect led to a significant enhancement of ECL from RuDSN/AuNP nano-emitters. Recently, Chen et al. (2021) designed a Hi-AuNF@G-ssDNA–Apt nanoprobe to achieve the ECL imaging of carcinoembryonic antigen (CEA) on fixed cell membranes (Fig. 7B).157 ECL imaging of CEA was achieved using the highly selective Hi-AuNF@G-ssDNA–Apt nanoprobe. The ssDNA–Apt nanoprobe can distinguish MCF-7 cells from normal human skeletal muscle cells at the single-cell level by ECLM.
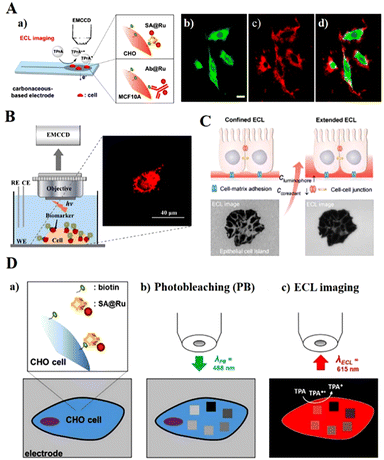 | ||
| Fig. 7 Schematic diagram of the imaging strategy for SCECL systems. (A) Surface-confined microscopy: spatially resolved ECL imaging of single cells (a), photoluminescence (b), ECL (c), and the overlaying (d) images of Chinese hamster ovary cells grown on a glassy carbon electrode. Reprinted with permission from ref. 155. Copyright 2017 American Chemical Society. (B) CEA imaging on the cytomembrane. Reprinted with permission from ref. 157. Copyright 2021 American Chemical Society. (C) Imaging cell–matrix adhesions by ECLM. Reprinted with permission from ref. 159. Copyright 2021 Wiley-VCH. (D) ECL microscopy of labelled cells with photobleached ROIs. Reprinted with permission from ref. 160. Copyright 2021 Wiley-VCH. | ||
The high concentration of co-reactants in ECL imaging systems can cause severe damage to cells. Wang et al. (2020) developed a dual intramolecular electron transfer strategy and tertiary amine conjugated Pdots (TEA–Pdots), enabling ECL imaging detection of individual living cell membrane proteins without the need for an additional co-reactant.158 They solved the QDs’ coalescence problem with a simple approach for achieving ECL in an aqueous phase system. The ECL intensity of TEA–Pdot is 132 times stronger than bare Pdots due to its superstructure and intramolecular electron transfer. Ding et al. (2021) used ECLM for spatially selective imaging of cell–matrix adhesion by varying the co-reactant and ECL probe concentrations (Fig. 7C).159 With the adjustment of the co-reactant and ECL probe concentrations, the thickness of the luminescent layer matched the spatial location of different cell adhesions, achieving spatially selective localization recognition of cell–matrix adhesion and cell–cell interconnection in fixed cells.
Photobleaching is the irreversible extinction of fluorophores under continuous light excitation, resulting in periodic fluorescence fading while observing single cells. Han et al. (2021) investigated the effect of photobleaching on ECLM. They attached streptavidin-modified Ru(bpy)32+ tags (SA@Ru) to biotinylated proteins in the cytoplasmic membrane of mammalian Chinese hamster ovary (CHO) fixed cells (Fig. 7D).160 The markers immobilized on CHO membranes were photobleached sequentially with different laser powers, photobleached areas showed low ECL emission as observed by confocal microscopy, and a linear relationship between ECL reduction and photobleaching was found in the study.
In recent work, Zhang et al. (2021) established ECL-based capacitance ECLM for label-free detection at the single-cell level, enabling the imaging of cell membrane surface proteins (Fig. 8A).161 The specific capacitance (Cs) of the surface region decreases when the species binds to the electrode surface or cell membrane while inducing a large potential drop (Vdl) across the bilayer in this region. So it was possible to image antigens on a single cell by the change in capacitance following the formation of antigen–antibody complexes directly. Cell morphology can be identified by observing the brighter ECL spots on the electrode surface. In another work, Qin et al. (2021) proposed a rapid label-free method for single-cell anodic ECL imaging using a closed BPE array (Fig. 8B).162 The closed BPE system avoided interference between anodic and cathodic reactions. Since the attached cell electrodes block oxygen diffusion, the high oxygen concentration produced a significant oxygen reduction current on the cathode side and a higher ECL intensity on the anode side, resulting in excellent contrast ECL imaging of the cell-adherent region. The system successfully achieves higher image acquisition of cell morphology and adhesion intensity by separating ECL responses.
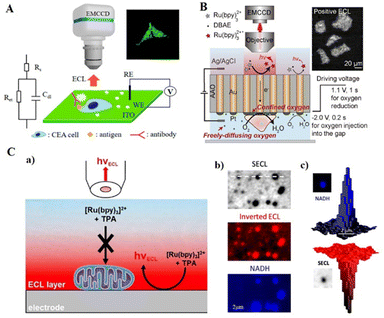 | ||
| Fig. 8 Schematic diagram of SCECL label-free imaging: (A) schematic illustration of ECL-based capacitance ECLM. Reprinted with permission from ref. 161. Copyright 2021 American Chemical Society. (B) Schematic illustration for label-free ECL imaging of single cells using a closed bipolar electrode array. Reprinted with permission from ref. 162. Copyright 2021 Wiley-VCH. (C) (a) Schematic diagram of the label-free shadow ECL microscopy (SECLM) of single mitochondria based on the spatial confinement of the ECL-emitting layer. (b) Typical SECLM and fluorescence images recorded at a high magnification of the same ROI showing single mitochondria. (c) An example of 3D imaging of a single mitochondrion recorded by NADH fluorescence (top) and SECLM (bottom). Reprinted with permission from ref. 163. Copyright 2021 Wiley-VCH. | ||
Mitochondria play a central role in critical cellular processes such as metabolism and apoptosis. The label-free method is well suited for imaging living mitochondria without labelling active mitochondria that affect their metabolic and respiratory processes. Ma et al. (2021) reported a method to image the individual living mitochondria that were deposited on the electrode surface with excellent spatial resolution (Fig. 8C).163 By adjusting the oxidative reduction of Ru(bpy)32+/TPrA to constrain the ECL emission reaction layer space, a similar thickness between the luminescent layer and the mitochondria was ensured. Therefore, the mitochondria effectively impede the diffusion of the ECL reagent and appear as a shadow around the ECL. Correlation fluorescence staining experiments showed that this method did not affect mitochondrial metabolism, while co-localization fluorescence analysis verified the feasibility of the proposed in vivo mitochondria-shadowed ECLM.
In 2018, Sojic's group reported surface-constrained ECLM that could provide information on cell membrane transport properties.164 They imaged the entire basement membrane of individual fixed cells by labelling the cell membrane with ECL probes. However, the ECL probe required a direct electrode surface and the generated ECL remains confined to the basement membrane of the cell, making it challenging to image intracellular structures and biomolecules. Ma et al. (2021) reported bio-coreactant-enhanced ECLM and revealed intracellular structures and dynamic transport (Fig. 9A).165 They used intracellular amine-rich biomolecules as the co-reactants in ECL reactions via a ‘catalytic pathway’, thus enabling the imaging of intracellular hierarchical structures such as the nucleolus, nucleus and endoplasmic reticulum without the use of multiple labels. Ru(bpy)32+ was generated on the surface of an electrode imposed with a constant anodic potential, driven by intracellular amine-rich biomolecules. On the one hand, the emitted ECL could be extended to a diffuse thickness of Ru(bpy)33+ that can cover the entire cell height, thus enabling the imaging of intracellular structures. On the other hand, the transmembrane and intracellular transport of Ru(bpy)33+ could be traced by time-resolved ECL signals.
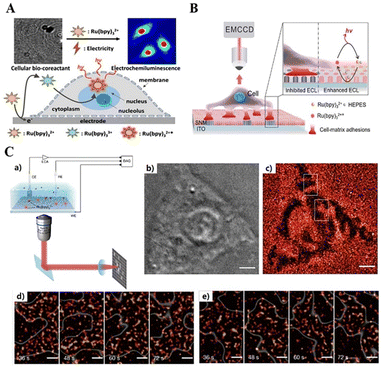 | ||
| Fig. 9 Schematic of the combined wide-field optical imaging and electrochemical recording setup(a). Bright-field image (b) and super-resolved ECL image (c) of a live HEK293 cell. Dynamic visualization of the cell adhesions at different moments (d) and (e). Reprinted with permission from ref. 167. Copyright 2021 Springer Nature BV. | ||
Imaging cell–matrix adhesion dynamics can reveal more precisely the orientation of cells in collective migration. Ding et al. (2019) reported a method for label-free imaging of cell–matrix adhesion in living cells, which led to a spatial distribution and dynamic changes in cell–matrix adhesion and adhesion strength at the subcellular level, visualizing the collective migration of living cells (Fig. 9B).166 An ITO electrode modified with a silica nanochannel membrane (SNM/ITO) was used as the substrate electrode. Cell–matrix adhesion of single live cells was performed on the surface of the nano-porous SNM/ITO electrode, followed by imaging of the cell–matrix adhesion of live cells using ECLM. Cell–matrix adhesion formed a tight contact with the electrode surface, thereby locally blocking the ECL response and the diffusion of luminophores, highlighted by the ECL as a dark map of cell–matrix adhesion. This method provided a spatial distribution and dynamics of cell adhesion kinetics, and could also be recorded with super-resolution. Feng's group (2021) achieved the wide-field spatial imaging of single-molecule ECL signals and developed super-resolution ECLM for living cell adhesion super-resolution imaging (Fig. 9C).167 To verify the feasibility of this super-resolution ECL imaging, they compared the super-resolution ECLM of the cell adhesion region with the bright-field images recorded by optical microscopy and found that the cell adhesion region in the figure was consistent. The ECLM imaged the adhesion dynamics of living cells with a high spatiotemporal resolution, and was able to observe the motion of the adhesion region with a temporal resolution of 12 s and a spatial resolution of 150 nm, allowing clearer microstructure and cell images to be seen.
4.3. ECL imaging-guided therapeutic approach
In cancer treatment, light therapy is gaining increasing interest due to its non-invasive nature, simple maneuverability, flexible controllability, high spatiotemporal resolution and high biocompatibility.168 To overcome the barriers of external light sources, therapeutic applications of internal self-illuminating light sources such as chemiluminescence, bioluminescence, and ECL have been developed recently. However, the intensity and area of most self-illuminated treatments are difficult to control, limiting the effectiveness of the treatment. Therefore, developing efficient, self-illuminating photodynamic therapy with good spatial and temporal controllability is important. In situ emitted photons can replace conventional excitation light to construct chemically excited photodynamic therapy or drug release systems to monitor and treat deep-seated diseases or tumours. As a light source that emits photons in situ, ECL is the ideal light source for phototherapy with the advantages of high temporal and spatial controllability, stable luminescence and high luminous flux. Dai's group (2018) reported the use of ECL as a light source in photodynamic therapy (ECL-therapeutics), constructing an ECL-based electrically driven luminescence system for killing pathogenic bacteria.169Zhang et al. (2019) successfully obtained parallel imaging of miRNA-21 in single cancer cells using the excellent performance of ECLM and AuNCs@PMA probes to construct a strategy for combining tumour diagnosis and therapy (Fig. 10A).170 After in situ labelling of miRNA-21 with a gold nanocage loaded phorbol-12-myristate-13-acetate (PMA) probe (AuNCs@PMA probe), PMA is released intracellularly thereby CL inducing ROS production, as revealed by ECL imaging. Recently, Xu's group (2022) combined ECL as an external light source for photodynamic therapy (PDT) with real-time imaging to report an ECL-driven PDT (Fig. 10B).171 The photosensitizer Ce6 absorbs ECL emission and sensitizes the surrounding oxygen to generate singlet oxygen (1O2) which can kill cancer cells. Furthermore, under ECLM with high spatiotemporal resolution, it is possible to monitor the dynamic process of progressive changes in the cell morphology at the single-cell level, changes in cell–matrix adhesion and increased cell membrane permeability during apoptosis.
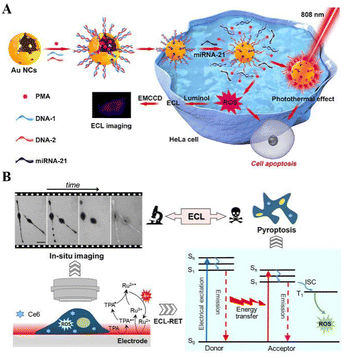 | ||
| Fig. 10 Schematic diagram of ECL imaging-guided therapeutic approach: (A) schematic diagram of ECL-imaging and combined therapy. Reprinted with permission from ref. 170. Copyright 2019 American Chemical Society. (B) Left: Schematic illustration of in situ imaging of the process of ECL–PDT by ECLM at a single living cell level. Right: Mechanism of ECL–PDT. Reprinted with permission from ref. 171. Copyright 2022 Wiley-VCH. | ||
5. Conclusions
SCECL analysis is a powerful analytical method that enables high sensitivity, low detection limit, high spatial and temporal resolution imaging of single-cell level bioassays, and even super-resolution dynamic imaging of cells. Based on the single-cell analysis, life science research can reveal the nature and laws of activities at a deeper level, providing a more reliable scientific basis for investigating the causes, development and treatment of major diseases. This review summarizes the recent advances in SCECL analysis based on various novel designs of ECL probes with high luminescence efficiency and the development of advanced ECL analysis systems (sensing systems and imaging systems) for biosensing, imaging, and ECL imaging-guided therapy, respectively. Although the rapid development of ECL probes and optics has facilitated SCECL analysis, spatial resolution and temporal resolution are the main challenges in the development of single-cell analysis methods, in addition to the highly sensitive detection of weak signals in tiny spaces extracted during rapid processes.For high spatiotemporal imaging resolution and dynamic analysis of complex cell systems, firstly, development of future ECL probes should continue to focus on the design of efficient and stable luminophores, and on the strategy of surface modification and nanomaterial combination to obtain ECL probes with good optical properties, stability, and biocompatibility. Secondly, high-throughput detection systems based on microplates, bipolar electrochemistry and wireless systems can be further developed, along with the design of new ECL imaging techniques to cater more sensitive and stable detection of small biological molecules. Subsequently, the design of high-resolution microscopy with increased spatiotemporal resolution and reduced background noise is also necessary to enable precise imaging of single cells, organelles and their surface-active substances, and real-time monitoring of dynamic molecular activities and intracellular processes.
Author contributions
All authors contributed to the article and approved the final manuscript. Qian Yang mainly contributed to the writing of the original draft and summarized related literature reports. Xiaoyu Huang, Beibei Gao, Lu Gao collected the related literature reports and polished the manuscript. Feng Yu contributed to the supervision, revision and polished the manuscript. Fu Wang completed the frame of this article, revised and polished the manuscript, and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is financially supported by the Shenzhen Science and Technology Program (JCYJ20210324123610030), the Shenzhen Science and Technology Innovation grants (JCYJ20200109115633343) and the Guangdong Provincial Key Laboratory of Biomedical Optical Imaging (2020B121201010).References
- T. Stuart and R. Satija, Nat. Rev. Genet., 2019, 20, 257–272 CrossRef.
- L. Keller and K. Pantel, Nat. Rev. Cancer, 2019, 19, 553–567 CrossRef.
- M. Labib and S. O. Kelley, Nat. Rev. Chem., 2020, 4, 143–158 CrossRef.
- Y. Fu and Q. Ma, Nanoscale, 2020, 12, 13879–13898 RSC.
- Y. Zhou, H. Yin, W.-W. Zhao and S. Ai, Coord. Chem. Rev., 2020, 424, 213519 CrossRef.
- B. Mohan, S. Kumar, V. Kumar, T. Jiao, H. K. Sharma and Q. Chen, TrAC, Trends Anal. Chem., 2022, 157, 116735 CrossRef CAS.
- Y. T. Chong, J. L. Y. Koh, H. Friesen, S. K. Duffy, M. J. Cox, A. Moses, J. Moffat, C. Boone and B. J. Andrews, Cell, 2015, 162, 221 CrossRef CAS.
- A. Görgens, Science, 2016, 352, 1479–1479 Search PubMed.
- A. M. Valm, S. Cohen, W. R. Legant, J. Melunis, U. Hershberg, E. Wait, A. R. Cohen, M. W. Davidson, E. Betzig and J. Lippincott-Schwartz, Nature, 2017, 546, 162–167 CrossRef CAS PubMed.
- H. Mikami, C. Lei, N. Nitta, T. Sugimura, T. Ito, Y. Ozeki and K. Goda, Chem, 2018, 4, 2278–2300 CAS.
- A. M. John, A. Haroon, G. Francisco and B. K. Rao, J. Clin. Oncol., 2018, 36, e21614–e21614 Search PubMed.
- N. Gierlinger, T. Keplinger and M. Harrington, Nat. Protoc., 2012, 7, 1694–1708 CrossRef PubMed.
- G. Vicidomini, P. Bianchini and A. Diaspro, Nat. Methods, 2018, 15, 173–182 CrossRef PubMed.
- R. A. Hoebe, C. H. Van Oven, T. W. J. Gadella, P. B. Dhonukshe, C. J. F. Van Noorden and E. M. M. Manders, Nat. Biotechnol., 2007, 25, 249–253 CrossRef PubMed.
- M. M. Richter, Chem. Rev., 2004, 104, 3003–3036 CrossRef PubMed.
- L. Hu and G. Xu, Chem. Soc. Rev., 2010, 39, 3275–3304 RSC.
- Z. Liu, W. Qi and G. Xu, Chem. Soc. Rev., 2015, 44, 3117–3142 RSC.
- H. Qi and C. Zhang, Anal. Chem., 2020, 92, 524–534 CrossRef CAS PubMed.
- W. Zhao, H.-Y. Chen and J.-J. Xu, Chem. Sci., 2021, 12, 5720–5736 RSC.
- H. Ding, B. Su and D. Jiang, ChemistryOpen, 2022, e202200113 Search PubMed.
- Y. Chen, S. Zhou, L. Li and J.-J. Zhu, Nano Today, 2017, 12, 98–115 CrossRef CAS.
- Y.-M. Long, L. Bao, Y. Peng, Z.-L. Zhang and D.-W. Pang, Carbon, 2018, 129, 168–174 CrossRef.
- A. Zanut, A. Fiorani, S. Canola, T. Saito, N. Ziebart, S. Rapino, S. Rebeccani, A. Barbon, T. Irie, H.-P. Josel, F. Negri, M. Marcaccio, M. Windfuhr, K. Imai, G. Valenti and F. Paolucci, Nat. Commun., 2020, 11, 2668 CrossRef.
- H. Wang, Anal. Bioanal. Chem., 2021, 413, 4119–4135 CrossRef PubMed.
- J. R. Adsetts, K. Chu, M. Hesari, J. Ma and Z. Ding, Anal. Chem., 2021, 93, 11626–11633 CrossRef.
- L. Li, W. Zhao, L. Luo, X. Liu, X. Bi, J. Li, P. Jiang and T. You, Electroanalysis, 2022, 34, 608–622 CrossRef.
- L. Chen, D. J. Hayne, E. H. Doeven, J. Agugiaro, D. J. D. Wilson, L. C. Henderson, T. U. Connell, Y. H. Nai, R. Alexander, S. Carrara, C. F. Hogan, P. S. Donnelly and P. S. Francis, Chem. Sci., 2019, 10, 8654–8667 RSC.
- H. Peng, W. Wu, Z. Huang, L. Xu, Y. Sheng, H. Deng, X. Xia and W. Chen, Electrochem. Commun., 2020, 111, 106667 CrossRef CAS.
- K.-M. Kim, J. Kim, J. Kim and J.-I. Hong, Chem. Commun., 2022, 58, 7542–7545 RSC.
- K. Chu, J. R. Adsetts, J. Ma, C. Zhang, M. Hesari, L. Yang and Z. Ding, J. Phys. Chem. C, 2021, 125, 22274–22282 CrossRef CAS.
- R. Wu, Z. Feng, J. Zhang, L. Jiang and J.-J. Zhu, TrAC, Trends Anal. Chem., 2022, 148, 116531 CrossRef CAS.
- J. Zhang, S. Arbault, N. Sojic and D. Jiang, Annu. Rev. Anal. Chem., 2019, 12, 275–295 CrossRef CAS.
- Z. Cao, Y. Shu, H. Qin, B. Su and X. Peng, ACS Cent. Sci., 2020, 6, 1129–1137 CrossRef CAS.
- A. Abdussalam and G. Xu, Anal. Bioanal. Chem., 2022, 414, 131–146 CrossRef CAS.
- R. T. Dufford, D. Nightingale and L. W. Gaddum, J. Am. Chem. Soc., 1927, 49, 1858–1864 CrossRef.
- C. Fang, H. Li, J. Yan, H. Guo and T. Yifeng, ChemElectroChem, 2017, 4, 1587–1593 CrossRef.
- M. Mayer, S. Takegami, M. Neumeier, S. Rink, A. Jacobi von Wangelin, S. Schulte, M. Vollmer, A. G. Griesbeck, A. Duerkop and A. J. Baeumner, Angew. Chem., Int. Ed., 2018, 57, 408–411 CrossRef.
- G. Liu, J. Hong, K. Ma, Y. Wan, X. Zhang, Y. Huang, K. Kang, M. Yang, J. Chen and S. Deng, Biosens. Bioelectron., 2020, 150, 111963 CrossRef PubMed.
- Q. Han, C. Wang, P. Liu, G. Zhang, L. Song and Y. Fu, Chem. Eng. J., 2021, 421, 129761 CrossRef.
- Z. Han, J. Wang, P. Du, J. Chen, S. Huo, H. Guo and X. Lu, Anal. Chem., 2022, 94, 8426–8432 CrossRef CAS PubMed.
- M. Zhao, L. Bai, W. Cheng, X. Duan, H. Wu and S. Ding, Biosens. Bioelectron., 2019, 127, 126–134 CrossRef.
- J. Wu, P. Ran, S. Zhu, F. Mo, C. Wang and Y. Fu, Sens. Actuators, B, 2019, 278, 97–102 CrossRef.
- D. Zhu, Y. Zhang, S. Bao, N. Wang, S. Yu, R. Luo, J. Ma, H. Ju and J. Lei, J. Am. Chem. Soc., 2021, 143, 3049–3053 CrossRef.
- R. Liu, C. Liu, H. Li, M. Liu, D. Wang and C. Zhang, Biosens. Bioelectron., 2017, 94, 335–343 CrossRef.
- M. Wu, N. Xu, J. Qiao, J. Chen and L. Jin, Analyst, 2019, 144, 4633–4638 RSC.
- X. Zhang, P. Wang, Y. Nie and Q. Ma, TrAC, Trends Anal. Chem., 2021, 143, 116410 CrossRef.
- N. E. Tokel and A. J. Bard, J. Am. Chem. Soc., 1972, 94, 2862–2863 CrossRef.
- K. K.-W. Lo, A. W.-T. Choi and W. H.-T. Law, Dalton Trans., 2012, 41, 6021–6047 RSC.
- T. Chen, Y. Xu, Z. Peng, A. Li and J. Liu, Polym. Chem., 2016, 7, 5880–5887 RSC.
- J. Cai, T. Chen, Y. Xu, S. Wei, W. Huang, R. Liu and J. Liu, Biosens. Bioelectron., 2019, 124–125, 15–24 CrossRef PubMed.
- J. Berrones Reyes, M. K. Kuimova and R. Vilar, Curr. Opin. Chem. Biol., 2021, 61, 179–190 CrossRef PubMed.
- K. N. Swanick, S. Ladouceur, E. Zysman-Colman and Z. Ding, Chem. Commun., 2012, 48, 3179–3181 RSC.
- M. A. Haghighatbin, S. E. Laird and C. F. Hogan, Curr. Opin. Electrochem., 2018, 7, 216–223 CrossRef.
- M. A. Haghighatbin, S. E. Laird and C. F. Hogan, Curr. Opin. Electrochem., 2018, 8, R1 CrossRef.
- P. Chen, W. Han, M. Zhao, J. Su, Z. Li, D. Li, L. Pi, X. Zhou and T. Zhai, Adv. Funct. Mater., 2021, 31, 2008790 CrossRef.
- H. Gao, J. Zhang, X. Wei, Q. Zhu and T. Wei, Talanta, 2021, 228, 122230 CrossRef PubMed.
- L. Zhao, X. Song, X. Ren, D. Fan, Q. Wei and D. Wu, Anal. Chem., 2021, 93, 8613–8621 CrossRef PubMed.
- Y. Zhu, M. Zhao, X. Hu, X. Wang and L. Wang, Electrochim. Acta, 2017, 228, 1–8 CrossRef.
- G. Valenti, E. Rampazzo, S. Bonacchi, L. Petrizza, M. Marcaccio, M. Montalti, L. Prodi and F. Paolucci, J. Am. Chem. Soc., 2016, 138, 15935–15942 CrossRef PubMed.
- F.-K. Du, H. Zhang, X.-C. Tan, J. Yan, M. Liu, X. Chen, Y.-Y. Wu, D.-F. Feng, Q.-Y. Chen, J.-M. Cen, S.-G. Liu, Y.-Q. Qiu and H.-Y. Han, Biosens. Bioelectron., 2018, 106, 50–56 CrossRef.
- J.-Y. Wang, Y.-K. Li, X. Jing, P. Luo, X.-Y. Dong and S.-Q. Zang, ACS Mater. Lett., 2022, 4, 960–966 CrossRef.
- Y. Wang, Q. Liu, J. Wei, Z. Dai, L. Ding, R. Yuan, Z. Wen and K. Wang, Biosens. Bioelectron., 2021, 173, 112771 CrossRef.
- Z. Ding, B. M. Quinn, S. K. Haram, L. E. Pell, B. A. Korgel and A. J. Bard, Science, 2002, 296, 1293–1297 CrossRef.
- Y. Zhao, L. Bouffier, G. Xu, G. Loget and N. Sojic, Chem. Sci., 2022, 13, 2528–2550 RSC.
- G. Jie, J. Zhang, G. Jie and L. Wang, Biosens. Bioelectron., 2014, 52, 69–75 CrossRef.
- A. Khojastehnezhad, F. Taghavi, E. Yaghoobi, M. Ramezani, M. Alibolandi, K. Abnous and S. M. Taghdisi, Talanta, 2021, 235, 122753 CrossRef PubMed.
- E. Yang, Z. Ning, F. Yin, Z. Fang, M. Chen, M. Zhang, W. Xu, Y. Zhang and Y. Shen, Sens. Actuators, B, 2022, 369, 132288 CrossRef.
- F. Arcudi, L. Đorđević, S. Rebeccani, M. Cacioppo, A. Zanut, G. Valenti, F. Paolucci and M. Prato, Adv. Sci., 2021, 8, 2100125 CrossRef.
- T. Guerrero-Esteban, C. Gutiérrez-Sánchez, T. García-Mendiola, M. Revenga-Parra, F. Pariente and E. Lorenzo, Sens. Actuators, B, 2021, 343, 130096 CrossRef.
- G. Valenti, E. Rampazzo, S. Kesarkar, D. Genovese, A. Fiorani, A. Zanut, F. Palomba, M. Marcaccio, F. Paolucci and L. Prodi, Coord. Chem. Rev., 2018, 367, 65–81 CrossRef.
- H. Peng, Z. Huang, H. Deng, W. Wu, K. Huang, Z. Li, W. Chen and J. Liu, Angew. Chem., Int. Ed., 2020, 59, 9982–9985 CrossRef PubMed.
- Y. Cao, J.-L. Zhou, Y. Ma, Y. Zhou and J.-J. Zhu, Dalton Trans., 2022, 51, 8927–8937 RSC.
- J. Yao, L. Li, P. Li and M. Yang, Nanoscale, 2017, 9, 13364–13383 RSC.
- M. J. Lim, N. N. M. Shahri, H. Taha, A. H. Mahadi, E. Kusrini, J. W. Lim and A. Usman, Carbohydr. Polym., 2021, 260, 117806 CrossRef CAS.
- E. Yang, Y. Zhang and Y. Shen, Anal. Chim. Acta, 2022, 1209, 339140 CrossRef CAS.
- S. Zhang, H. Zheng, Y. Chen, H. Yi, H. Dai, Z. Hong and Y. Lin, ACS Appl. Nano Mater., 2019, 2, 7061–7066 CrossRef.
- S. Qi, J. Chen, X. Bai, Y. Miao, S. Yang, C. Qian, B. Wu, Y. Li and B. Xin, RSC Adv., 2021, 11, 21813–21823 RSC.
- K. Yan, P. Wang, M. Wang, M. Zhang, W. Liu and Q. Ma, Microchem. J., 2022, 179, 107589 CrossRef.
- M. Wang, R. Sun, Q. Wang, L. Chen, L. Hou, Y. Chi, C. H. Lu, F. Fu and Y. Dong, Chem. – Eur. J., 2018, 24, 4250–4254 CrossRef PubMed.
- F. Li, D. Yang and H. Xu, Chem. – Eur. J., 2019, 25, 1165–1176 CrossRef PubMed.
- X.-H. Wei, X. Qiao, J. Fan, Y.-Q. Hao, Y.-T. Zhang, Y.-L. Zhou and M.-T. Xu, Microchem. J., 2022, 173, 106910 CrossRef CAS.
- G.-X. Liang, K.-R. Zhao, Y.-S. He, Z.-J. Liu, S.-Y. Ye and L. Wang, Microchem. J., 2021, 171, 106787 CrossRef CAS.
- D. Bahari, B. Babamiri, K. Moradi, A. Salimi and R. Hallaj, Biosens. Bioelectron., 2022, 195, 113657 CrossRef CAS PubMed.
- S. Han, Z. Zhang, S. Li, L. Qi and G. Xu, Sci. China: Chem., 2016, 59, 794–801 CrossRef CAS.
- M. Hesari and Z. Ding, Acc. Chem. Res., 2017, 50, 218–230 CrossRef CAS.
- Y. Zhou, Y. Zheng, X. Zhu, Y. Chai and R. Yuan, Biosens. Bioelectron., 2021, 190, 113449 CrossRef CAS PubMed.
- C. Ma, Y. Cao, X. Gou and J.-J. Zhu, Anal. Chem., 2020, 92, 431–454 CrossRef CAS.
- K. Hiramoto, E. Villani, T. Iwama, K. Komatsu, S. Inagi, K. Y. Inoue, Y. Nashimoto, K. Ino and H. Shiku, Micromachines, 2020, 11, 530 CrossRef.
- M. Hesari and Z. Ding, Nat. Protoc., 2021, 16, 2109–2130 CrossRef CAS.
- M. Bhaiyya, P. K. Pattnaik and S. Goel, Curr. Opin. Electrochem., 2021, 30, 100800 CrossRef.
- Z. Zhang, P. Du, G. Pu, L. Wei, Y. Wu, J. Guo and X. Lu, Mater. Chem. Front., 2019, 3, 2246–2257 RSC.
- X. Ma, W. Gao, F. Du, F. Yuan, J. Yu, Y. Guan, N. Sojic and G. Xu, Acc. Chem. Res., 2021, 54, 2936–2945 CrossRef.
- Q. Li, J. Y. Zheng, Y. Yan, Y. S. Zhao and J. Yao, Adv. Mater., 2012, 24, 4745–4749 CrossRef.
- S. Li, Y. Liu and Q. Ma, TrAC, Trends Anal. Chem., 2019, 110, 277–292 CrossRef.
- J. Xu, Y. Hu, S. Wang, X. Ma and J. Guo, Analyst, 2020, 145, 2058–2069 RSC.
- X. Chen, Y. Liu and Q. Ma, J. Mater. Chem. C, 2018, 6, 942–959 RSC.
- W. Gao, M. Saqib, L. Qi, W. Zhang and G. Xu, Curr. Opin. Electrochem., 2017, 3, 4–10 CrossRef CAS.
- P. Nikolaou, G. Valenti and F. Paolucci, Electrochim. Acta, 2021, 388, 138586 CrossRef CAS.
- Y. Zhang, R. Zhang, X. Yang, H. Qi and C. Zhang, J. Pharm. Anal., 2019, 9, 9–19 CrossRef PubMed.
- R. A. Husain, S. R. Barman, S. Chatterjee, I. Khan and Z.-H. Lin, J. Mater. Chem. B, 2020, 8, 3192–3212 RSC.
- B. Babamiri, D. Bahari and A. Salimi, Biosens. Bioelectron., 2019, 142, 111530 CrossRef CAS.
- H. Ding, W. Guo and B. Su, ChemPlusChem, 2020, 85, 725–733 CrossRef PubMed.
- Z. Zhang, C. Ma, Q. Xu and J.-J. Zhu, Analyst, 2022, 147, 2884–2894 RSC.
- W. Zhao and J.-J. Xu, Chin. J. Chem., 2022, 40, 1975–1986 CrossRef.
- W. Guo, Y. Liu, Z. Cao and B. Su, J. Anal. Test., 2017, 1, 14 CrossRef.
- C. Ma, W. Wu, L. Li, S. Wu, J. Zhang, Z. Chen and J.-J. Zhu, Chem. Sci., 2018, 9, 6167–6175 RSC.
- C. Ma, H.-F. Wei, M.-X. Wang, S. Wu, Y.-C. Chang, J. Zhang, L.-P. Jiang, W. Zhu, Z. Chen and Y. Lin, Nano Lett., 2020, 20, 5008–5016 CrossRef CAS.
- S. Knezevic, L. Bouffier, B. Liu, D. Jiang and N. Sojic, Curr. Opin. Electrochem., 2022, 35, 101096 CrossRef CAS.
- M.-M. Chen, C.-H. Xu, W. Zhao, H.-Y. Chen and J.-J. Xu, J. Am. Chem. Soc., 2021, 143, 18511–18518 CrossRef CAS PubMed.
- H. Zhu, D. Jiang and J.-J. Zhu, Chem. Sci., 2021, 12, 4794–4799 RSC.
- A. Zanut, A. Fiorani, S. Rebeccani, S. Kesarkar and G. Valenti, Anal. Bioanal. Chem., 2019, 411, 4375–4382 CrossRef CAS.
- Y. Wu, J. Yang, Z. Zheng, Z. Li, F. Lu, Y. Chen and W. Gao, Talanta, 2018, 188, 58–65 CrossRef CAS.
- L. Peng, Y. Yuan, X. Fu, A. Fu, P. Zhang, Y. Chai, X. Gan and R. Yuan, Anal. Chem., 2019, 91, 3239–3245 CrossRef PubMed.
- H. Sha, Y. Wang, Y. Zhang, H. Ke, X. Xiong and N. Jia, Sens. Actuators, B, 2018, 277, 157–163 CrossRef.
- M.-S. Wu, Z. Liu, J.-J. Xu and H.-Y. Chen, ChemElectroChem, 2016, 3, 429–435 CrossRef.
- M. Poudineh, E. H. Sargent, K. Pantel and S. O. Kelley, Nat. Biomed. Eng., 2018, 2, 72–84 CrossRef.
- Y.-G. Feng, J.-H. Zhu, X.-Y. Wang, A.-J. Wang, L.-P. Mei, P.-X. Yuan and J.-J. Feng, J. Electroanal. Chem., 2021, 882, 115010 CrossRef.
- G. Zheng, F. Patolsky, Y. Cui, W. U. Wang and C. M. Lieber, Nat. Biotechnol., 2005, 23, 1294–1301 CrossRef CAS.
- Y. Qiu, B. Zhou, X. Yang, D. Long, Y. Hao and P. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 16848–16856 CrossRef CAS PubMed.
- D. Long, C. Chen, C. Cui, Z. Yao and P. Yang, Nanoscale, 2018, 10, 18597–18605 RSC.
- G. Liu, C. Ma, B.-K. Jin, Z. Chen, F.-L. Cheng and J.-J. Zhu, Anal. Chem., 2019, 91, 3021–3026 CrossRef CAS PubMed.
- L. Liu, A. Yang, W. Luo, H. Liu, X. Liu and W. Zhao, Analyst, 2021, 146, 2057–2064 RSC.
- B. Zhou, Y. Qiu, Q. Wen, M. Zhu and P. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 2074–2082 CrossRef CAS.
- G. Liu, B.-K. Jin, C. Ma, Z. Chen and J.-J. Zhu, Anal. Chem., 2019, 91, 6363–6370 CrossRef CAS PubMed.
- Y.-T. Yang, J.-L. Liu, M.-F. Sun, R. Yuan and Y.-Q. Chai, Anal. Chem., 2022, 94, 6874–6881 CrossRef CAS PubMed.
- R. Strack, Nat. Methods, 2018, 15, 481–481 CrossRef CAS PubMed.
- Y. Zhang, F. Wang, H. Zhang, H. Wang and Y. Liu, Anal. Chem., 2019, 91, 12100–12107 CrossRef CAS.
- H. Zhang, Z. Wang, F. Wang, Y. Zhang, H. Wang and Y. Liu, Anal. Chem., 2020, 92, 5546–5553 CrossRef CAS PubMed.
- H. Xiong, Z. Huang, Q. Lin, B. Yang, F. Yan, B. Liu, H. Chen and J. Kong, Anal. Chem., 2022, 94, 837–846 CrossRef CAS PubMed.
- A. Chen, W. Liang, H. Wang, Y. Zhuo, Y. Chai and R. Yuan, Anal. Chem., 2020, 92, 1379–1385 CrossRef CAS.
- C. Ding, Y. Li, L. Wang and X. Luo, Anal. Chem., 2019, 91, 983–989 CrossRef CAS PubMed.
- P. Liu, L. Wang, K. Zhao, Z. Liu, H. Cao, S. Ye and G. Liang, Sens. Actuators, B, 2020, 316, 128131 CrossRef CAS.
- Y. Liu, S. Huang, J. Li, M. Wang, C. Wang, B. Hu, N. Zhou and Z. Zhang, Microchim. Acta, 2021, 188, 69 CrossRef CAS PubMed.
- C. Amatore, C. Pebay, L. Servant, N. Sojic, S. Szunerits and L. Thouin, ChemPhysChem, 2006, 7, 1322–1327 CrossRef CAS.
- L. S. Dolci, S. Zanarini, L. D. Ciana, F. Paolucci and A. Roda, Anal. Chem., 2009, 81, 6234–6241 CrossRef CAS.
- W. Shen, S. Das, F. Vitale, A. Richardson, A. Ananthakrishnan, L. A. Struzyna, D. P. Brown, N. Song, M. Ramkumar, T. Lucas, D. K. Cullen, B. Litt and M. G. Allen, Microsyst. Nanoeng., 2018, 4, 30 CrossRef.
- J. E. Dick, Chem. Commun., 2016, 52, 10906–10909 RSC.
- J. Xu, P. Huang, Y. Qin, D. Jiang and H.-Y. Chen, Anal. Chem., 2016, 88, 4609–4612 CrossRef CAS PubMed.
- R. He, H. Tang, D. Jiang and H.-Y. Chen, Anal. Chem., 2016, 88, 2006–2009 CrossRef CAS.
- M. Li, H. Gao, X. Wang, Y. Wang, H. Qi and C. Zhang, Microchim. Acta, 2017, 184, 603–610 CrossRef CAS.
- Z.-M. Lyu, X.-L. Zhou, X.-N. Wang, P. Li, L. Xu and E. H. Liu, Sens. Actuators, B, 2019, 284, 437–443 CrossRef CAS.
- N. Wang, H. Ao, W. Xiao, W. Chen, G. Li, J. Wu and H. Ju, Biosens. Bioelectron., 2022, 201, 113959 CrossRef CAS PubMed.
- P. Bertoncello and P. Ugo, ChemElectroChem, 2017, 4, 1663–1676 CrossRef CAS.
- J. Xu, D. Jiang, Y. Qin, J. Xia, D. Jiang and H.-Y. Chen, Anal. Chem., 2017, 89, 2216–2220 CrossRef CAS PubMed.
- J. Xia, J. Zhou, R. Zhang, D. Jiang and D. Jiang, Anal. Bioanal. Chem., 2018, 410, 4787–4792 CrossRef CAS PubMed.
- Z.-Y. Che, X.-Y. Wang, X. Ma and S.-N. Ding, Coord. Chem. Rev., 2021, 446, 214116 CrossRef CAS.
- S. E. Fosdick, K. N. Knust, K. Scida and R. M. Crooks, Angew. Chem., Int. Ed., 2013, 52, 10438–10456 CrossRef CAS PubMed.
- T. J. Anderson, P. A. Defnet and B. Zhang, Anal. Chem., 2020, 92, 6748–6755 CrossRef CAS PubMed.
- Y. Wang, R. Jin, N. Sojic, D. Jiang and H.-Y. Chen, Angew. Chem., Int. Ed., 2020, 59, 10416–10420 CrossRef CAS PubMed.
- C. Cui, Y. Chen, D. Jiang, H.-Y. Chen, J. Zhang and J.-J. Zhu, Anal. Chem., 2019, 91, 1121–1125 CrossRef CAS PubMed.
- J. Zhang, R. Jin, Y. Chen, D. Fang and D. Jiang, Sens. Actuators, B, 2021, 329, 129208 CrossRef CAS.
- G. Liu, C. Ma, B.-K. Jin, Z. Chen and J.-J. Zhu, Anal. Chem., 2018, 90, 4801–4806 CrossRef CAS PubMed.
- J. Zhang, H. Ding, S. Zhao, D. Jiang and H.-Y. Chen, Electrochem. Commun., 2019, 98, 38–42 CrossRef CAS.
- H. Ding, W. Guo, L. Ding and B. Su, Chin. J. Chem., 2021, 39, 2911–2916 CrossRef CAS.
- G. Valenti, S. Scarabino, B. Goudeau, A. Lesch, M. Jović, E. Villani, M. Sentic, S. Rapino, S. Arbault, F. Paolucci and N. Sojic, J. Am. Chem. Soc., 2017, 139, 16830–16837 CrossRef CAS PubMed.
- Y. Liu, H. Zhang, B. Li, J. Liu, D. Jiang, B. Liu and N. Sojic, J. Am. Chem. Soc., 2021, 143, 17910–17914 CrossRef PubMed.
- Y. Chen, X. Gou, C. Ma, D. Jiang and J.-J. Zhu, Anal. Chem., 2021, 93, 7682–7689 CrossRef PubMed.
- N. Wang, H. Gao, Y. Li, G. Li, W. Chen, Z. Jin, J. Lei, Q. Wei and H. Ju, Angew. Chem., Int. Ed., 2021, 60, 197–201 CrossRef PubMed.
- H. Ding, P. Zhou, W. Fu, L. Ding, W. Guo and B. Su, Angew. Chem., Int. Ed., 2021, 60, 11769–11773 CrossRef PubMed.
- D. Han, B. Goudeau, D. Manojlovic, D. Jiang, D. Fang and N. Sojic, Angew. Chem., Int. Ed., 2021, 60, 7686–7690 CrossRef PubMed.
- J. Zhang, R. Jin, D. Jiang and H.-Y. Chen, J. Am. Chem. Soc., 2019, 141, 10294–10299 CrossRef CAS PubMed.
- X. Qin, H.-J. Jin, X. Li, J. Li, J.-B. Pan, K. Wang, S. Liu, J.-J. Xu and X.-H. Xia, Chem. – Eur. J., 2022, 28, e202103964 CAS.
- Y. Ma, C. Colin, J. Descamps, S. Arbault and N. Sojic, Angew. Chem., Int. Ed., 2021, 60, 18742–18749 CrossRef CAS PubMed.
- S. Voci, B. Goudeau, G. Valenti, A. Lesch, M. Jović, S. Rapino, F. Paolucci, S. Arbault and N. Sojic, J. Am. Chem. Soc., 2018, 140, 14753–14760 CrossRef CAS PubMed.
- C. Ma, S. Wu, Y. Zhou, H.-F. Wei, J. Zhang, Z. Chen, J.-J. Zhu, Y. Lin and W. Zhu, Angew. Chem., Int. Ed., 2021, 60, 4907–4914 CrossRef CAS PubMed.
- H. Ding, W. Guo and B. Su, Angew. Chem., Int. Ed., 2020, 59, 449–456 CrossRef CAS PubMed.
- J. Dong, Y. Lu, Y. Xu, F. Chen, J. Yang, Y. Chen and J. Feng, Nature, 2021, 596, 244–249 CrossRef CAS PubMed.
- S. H. Yun and S. J. J. Kwok, Nat. Biomed. Eng., 2017, 1, 0008 CrossRef CAS PubMed.
- S. Liu, H. Yuan, H. Bai, P. Zhang, F. Lv, L. Liu, Z. Dai, J. Bao and S. Wang, J. Am. Chem. Soc., 2018, 140, 2284–2291 CrossRef PubMed.
- H. Zhang, W. Gao, Y. Liu, Y. Sun, Y. Jiang and S. Zhang, Anal. Chem., 2019, 91, 12581–12586 CrossRef PubMed.
- M.-M. Chen, C.-H. Xu, W. Zhao, H.-Y. Chen and J.-J. Xu, Angew. Chem., Int. Ed., 2022, 61, e202117401 Search PubMed.
| This journal is © The Royal Society of Chemistry 2023 |