Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Collision cross sections of high-mannose N-glycans in commonly observed adduct states – identification of gas-phase conformers unique to [M − H]− ions†
W. B.
Struwe
*a,
J. L.
Benesch
a,
D. J.
Harvey
b and
K.
Pagel
*cd
aDepartment of Chemistry, University of Oxford, Oxford, UK OX1 3TA. E-mail: weston.struwe@chem.ox.ac.uk
bDepartment of Biochemistry, Oxford Glycobiology Institute, University of Oxford UK, OX1 3QU, UK
cFritz Haber Institute of the Max Planck Society, Faradayweg 4-6, 14195 Berlin, Germany
dInstitut für Chemie und Biochemie der Freien Universität Berlin, Takustr. 3, 14195 Berlin, Germany. E-mail: kevin.pagel@fu-berlin.de
First published on 7th July 2015
Abstract
We report collision cross sections (CCS) of high-mannose N-glycans as [M + Na]+, [M + K]+, [M + H]+, [M + Cl]−, [M + H2PO4]− and [M − H]− ions, measured by drift tube (DT) ion mobility-mass spectrometry (IM-MS) in helium and nitrogen gases. Further analysis using traveling wave (TW) IM-MS reveal the existence of distinct conformers exclusive to [M − H]− ions.
N-glycans represent the most common carbohydrate protein posttranslational modification and are attached to the protein backbone via asparagine residues. The extent of protein N-glycosylation in eukaryotes has been estimated up to 50%
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and the involved carbohydrates are important for protein structure, signalling, protein–protein recognition and protection from proteolysis. N-glycan structures contain a common core consisting of two N-acetylglucosamine (GlcNAc) residues at the reducing end (termed the chitobiose core) and three additional mannose (Man) residues. Individual structures are extended from the mannose residues and form elaborate branched structures; characterized as high-mannose, hybrid or complex type.
2 and the involved carbohydrates are important for protein structure, signalling, protein–protein recognition and protection from proteolysis. N-glycan structures contain a common core consisting of two N-acetylglucosamine (GlcNAc) residues at the reducing end (termed the chitobiose core) and three additional mannose (Man) residues. Individual structures are extended from the mannose residues and form elaborate branched structures; characterized as high-mannose, hybrid or complex type.
Due to the complexity inherent in their structure, the characterization of glycans remains challenging. Typically, their analysis relies on liquid chromatography techniques (with or without exoglycosidase digestions) for separation of complex mixtures and/or tandem mass spectrometry (MS) to produce fragment ions that facilitate structural assignments.3,4 A more recent approach is the application of ion mobility coupled to mass spectrometry (IM-MS) in which glycan mixtures are separated in the gas phase. The use of IM-MS in glycomics studies is still comparatively limited, yet several reports have demonstrated its potential using both travelling-wave (TW) and drift-tube (DT) IM-MS instruments.5–12 The emphasis of these studies has been on the separation of isomers from synthetically derived oligosaccharides or N-glycan mixtures purified from glycoprotein standards with alkali adduct ions being interrogated in the majority of cases.5,6,10–12 Over recent years, we have explored IM-MS of negative glycan ions for both deprotonated or phosphate adducts. Using glycans released from biologically derived material, we demonstrated the capability of negative polarity IM-MS to differentiate glycan isomers in complex mixtures6,8 and to extract glycan ions from non-carbohydrate material in the sample.7
IM separates ions based on the time required to traverse a region of inert neutral gas under the influence of a weak electric field. The separation is driven by differences in mass, charge, size and shape of each analyte and thereby provides information about the underlying three-dimensional structure. IM-MS data typically consists of mass-to-charge values (m/z) and arrival times which can be further converted into orientationally averaged collision cross sections (CCS).8,9,13 These CCSs are intrinsic to a particular glycan and are influenced by both the ionic state (i.e. positive/negative mode and adduction) as well as the particular drift gas (commonly helium or nitrogen). In a successful IM-MS glycomics experiment it can therefore be crucial to simultaneously consider multiple adduct states, instrument polarities and IM gases.
CCS values calculated from theoretical structures obtained by molecular dynamic simulations (MD) have been compared to experimental CCS data of native oligosaccharides11 and permethylated N-glycans10 as [M + Na]+ and [M + 2Na]2+ adducts, respectively. In both cases theoretical and experimental values were comparable providing insight into the gas-phase structure of the glycan adduct ions. These studies are valuable, but raise the questions as to whether, and how other adduct types alter the CCSs. In this context protonated and deprotonated ions are of particular interest, since the location of charge may vary, making MD simulations rather challenging. The simplest oligo-mannose structure Man3GlcNAc2 (or “Man3”, denoting the number of mannoses) for example carries 15 hydroxyl groups, all of which are potential sites for deprotonation. Although quantum chemical approaches may help to predict the location of the charge, and the three-dimensional structure, the computational resources required are very demanding and therefore often not practical for investigating a large variety of glycan structures. The separation power of IM rests solely in differences in glycan-adduct gas-phase structures and so a comprehensive study examining the CCSs of pure, known standards in all commonly observed ion states will aid the utility of the method and facilitate further development.
Here we report absolute DTCCS values of pure, synthetically-derived high-mannose N-glycans in six commonly observed ion adduct states ([M + H]+, [M + Na]+, [M + K]+, [M − H]−, [M + Cl]− and [M + H2PO4]−) using helium and nitrogen drift gases (Fig. 1). The investigated N-glycan standards were purchased from Dextra Laboratories (Reading, UK) and absolute DTCCS measurements were performed using a modified Synapt HDMS fitted with a linear drift tube as described previously.13 Glycan adducts were generated by the addition of respective salt solutions to promote adduct formation.
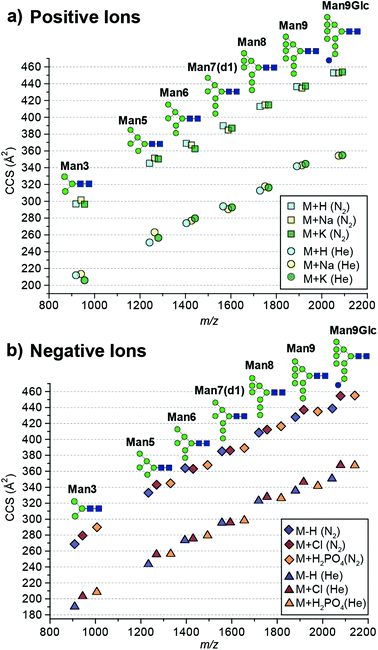 | ||
| Fig. 1 Absolute collision cross sections (DTCCS) of N-glycan standards measured in nitrogen and helium as [M + H]+, [M + Na]+, [M + K]+, [M − H]−, [M + Cl]− and [M + H2PO4]− adducts using drift tube ion mobility-mass spectrometry. For ATDs that exhibit multiple features the most abundant feature was used for CCS determination. Mannose residues are shown in green circles, N-acetylglucosamine residues are blue squares, and glucose residues are blue circles. Glycosidic linkages are based on the Oxford Glycan Nomenclature.1 | ||
The absolute DTCCS values of the investigated high-mannose structures ranged from 190 Å2 to 355 Å2 in helium and 269 Å2 to 455 Å2 in nitrogen drift gases. In general, the DTCCSs of proton, sodium and potassium adducts are rather similar especially so for the larger glycans Man8 (DTCCSN2 (H) 413 Å2, (Na) 415 Å2, (K) 415 Å2), Man9 (DTCCSN2 (H) 436 Å2, (Na) 435 Å2, (K) 437 Å2) and Man9Glc (DTCCSN2 (H) 453 Å2, (Na) 453 Å2, (K) 454 Å2). This is consistent with the specific cation having only minimal influence on the overall gas-phase conformation. Molecular dynamics simulations of [M + Na]+ ions have shown that glycans tend to “wrap” around the metal cation encouraging more compact gas-phase ions10,11 opposed to possible conformations where the different branching arms of the glycan are extended away from one another. Interestingly [M + H]+ ions had similar DTCCSs to sodium and potassium adduct ions yet the effect of protonation is unknown. It is likely that protons are located on nitrogens of N-acetylglucosamine residues. As these moieties are exclusively present in the chitobiose core, this would restrict charge effects in gas-phase structure formation. Overall, the CCS differences between protonated and cationated glycans are modest which suggests that they adopt rather similar gas-phase structures.
When examining positive ions we find that the similarity in CCS values of the different adducts/protomers contrasts considerably to negative ion DTCCS measurements where [M − H]− values were generally significantly smaller than those for chloride and phosphate anion adducts most notably for the smallest and largest N-glycans investigated. For example, the deprotonated Man3 DTCCSN2 is 269 Å2, the chloride adduct 279 Å2 (3.8% larger) and the phosphate adduct 290 Å2 (7.6% larger). Similarly, the Man9Glc DTCCSN2 values for [M − H]− (439 Å2) were significantly smaller compared to [M + H2PO4]− (455 Å2), [M + H]+ (453 Å2) and [M + K]+ (454 Å2) adducts. Unlike positive ions where the addition of a proton shows a minimal effect to the CCS (<1% change for higher mass glycans) compared to cationated glycans, where the removal of a proton significantly affects CCSs for high-mannose glycan structures.
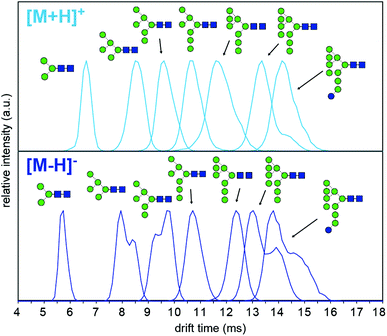
To understand why CCS values of deprotonated ions differ significantly to other negative as well as positive ions we investigated synthetic standards as [M + H]+, [M − H]−, [M + Cl]− and [M + H2PO4]− ions by TW IM-MS using a Synapt G2-Si instrument which has higher IM resolution than the DT IM-MS instrument used for absolute DTCCS measurements. The extracted arrival time distributions (ATDs) of [M − H]− ions of the synthetically-derived glycans show at least two distinct features for Man5, Man6, Man9 and Man9Glc structures that were absent in the respective [M + H]+ ions (Fig. 2). The shape and relative intensity of these distributions are not affected by activation until the threshold of dissociation is reached. In addition, the DTCCS and TWCCS values of [M − H]− ions are systematically lower than those of [M + H]+ ions (Fig. 1 and 2 and ESI Table 1†) for which the nearly Gaussian shape of the ATDs suggest the existence of only one species. Chloride and phosphate adducts ATDs also had single species (ESI Fig. 1†). Interestingly, the Man7 and Man8 [M − H]− ATD peaks are also nearly Gaussian but the estimated TWCCSs were lower (376 Å2 and 406 Å2) than those of protonated ions (392 Å2 and 421 Å2).
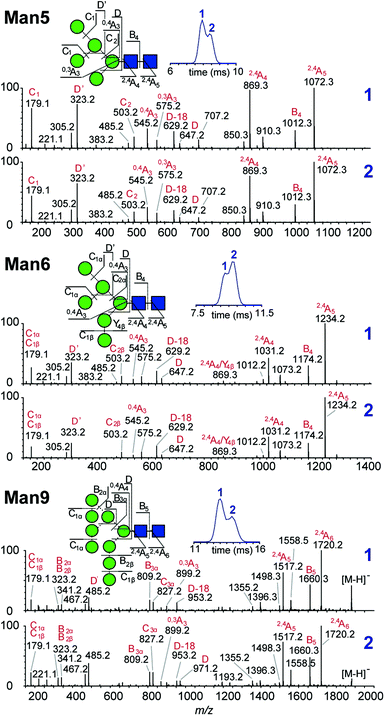
Multiple features in the ATDs could in principle arise from structural isomers or distinct gas-phase conformers that are only detectable as [M − H]− ions. It is well documented that CID of negative ions generates informative fragment ions specifically cross-ring (A-type) and D-type cleavages that can be used to differentiate high-mannose isomers.14 MS/MS of [M − H]− ions with collision-induced dissociation (CID) applied in the transfer region (i.e. MS/MS after IM separation) showed that the doublet peaks of Man5, Man6 and Man9 yield similar fragmentation patterns (Fig. 3). The CID spectra of the Man5 ATD peaks 1 and 2 match and the D′ (m/z 323), D (m/z 657) as well as cross ring A fragments (m/z 545 and 575) verify the existence of a single structure. Similarly, for Man6 we observed the same D-type fragment series, which indicates that the additional mannose residue is on the 3-arm. This is furthermore supported by the occurrence of characteristic 2,4A4/Y4β fragments m/z 869. The CID spectra of both Man9 ATDs also indicate a single isomer that is commonly observed on eukaryotic glycoproteins.15 Ions at m/z 971 (D), 827 (D-18), 809 (B3α) and 485 (D′) confirm this structure with five mannose residues on the 6-arm and three mannose residues substituted on the 3-arm. However, some ions (e.g. the parent ion and the m/z 971 D ion) are less abundant in ATD peak 2. It is unclear why the parent ions intensities differ here as equivalent CID voltages were applied. A possible explanation is the difference in size and CCS of both conformers, which could lead to considerably different internal energy deposition during CID.
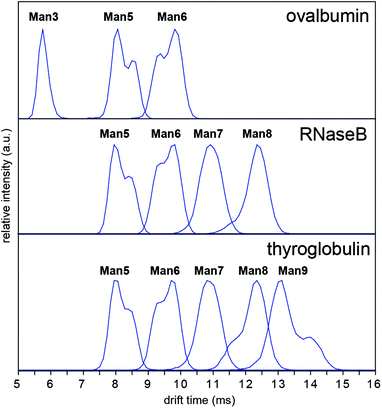
In order to investigate whether these conformations could be impurities deriving from the synthesis of the synthetic glycans, we examined N-glycans released from the glycoprotein standards porcine thyroglobulin, bovine ribonuclease B (RNase B) and chicken ovalbumin, which have been studied previously by IM-MS as [M + Na]+ and [M + H2PO4]− ions.8,9 The presence of multiple Man5, Man6 and Man9 [M − H]− conformers was observed in all cases (Fig. 4). However [M + Cl]− and [M + H2PO4]− ion ATDs indicated single conformers (ESI Fig. 2–4†) although the presence of structural isomers, particularly Man5 from RNase B16 may affect ATD peak widths. Interestingly the relative intensity of Man5, Man6 and Man9 [M − H]− conformers remained consistent between samples (i.e. ATD peak shapes). For example, the most abundant Man5 conformer had a lower DTCCS (thyroglobulin 321 Å2, RNase B 319 Å2, ovalbumin 322 Å2, synthetic 318 Å2) than the second species. This consistent ratio was also true for Man9 but the opposite was seen for Man6 where the most abundant conformer had a larger CCS (thyroglobulin 358 Å2, RNase B 359 Å2, ovalbumin 359 Å2, synthetic 357 Å2). MS/MS of thyroglobulin Man5 and Man6 confirmed the presence of single structures (ESI Fig. 5†). While it remains unclear what causes these N-glycans to adopt multiple distinct gas-phase conformations, it is clear that IM-MS provides an exquisitely sensitive means to separate, identify, and interrogate N-glycan structures.
Conclusions
We have reported the absolute DTCCSs of pure high-mannose N-glycans as commonly observed adducts by DT IM-MS in He and N2. These and other previously published values are deposited in the recently launched glyoconjugate CCS database GlycoMob (http://www.glycomob.org) housed within UniCarbKB, a glycomics mass spectrometry resource.17 CCS values deposited in GlycoMob were measured both by DT IM-MS and TW IM-MS and are well within instrument error of approximately 1%. We have found that DTCCS of [M − H]− were noticeably smaller than both [M + H]+ and anion/cationated glycans. Analysis by TW IM-MS furthermore revealed the presence of distinctive gas-phase conformers, specifically for Man5, Man6 and Man9 N-glycans among four sample sets. The presence of such conformers may lead to misinterpretation of IM-MS data unless careful attention is given to MS/MS data to discriminate between structural isomers and conformers. As IM-MS applications for glycomics studies are in their early stages further systematic studies are needed to understand how these biomolecules fold into their specific, adduct-dependent gas-phase structure.Acknowledgements
Financial support for this work was provided by the Biotechnology and Biological Sciences Research Council (BBSRC) [BB/L017733/1 to W.B.S., D.J.H. J.L.P.], the Free University Berlin and the Fritz Haber Institute of the Max Planck Society. Johanna Hofmann is gratefully acknowledged for reading the manuscript and fruitful comments.References
- D. J. Harvey, A. H. Merry, L. Royle, M. P. Campbell, R. A. Dwek and P. M. Rudd, Proteomics, 2009, 9, 3796–3801 CrossRef CAS PubMed.
- R. Apweiler, H. Hermjakob and N. Sharon, Biochim. Biophys. Acta, 1999, 1473, 4–8 CrossRef CAS.
- L. Royle, C. M. Radcliffe, R. A. Dwek and P. M. Rudd, Methods Mol. Biol., 2006, 347, 125–143 CAS.
- J. Zaia, OMICS, 2010, 14, 401–418 CrossRef CAS PubMed.
- L. S. Fenn and J. A. McLean, Phys. Chem. Chem. Phys., 2011, 13, 2196–2205 RSC.
- D. J. Harvey, C. A. Scarff, M. Edgeworth, M. Crispin, C. N. Scanlan, F. Sobott, S. Allman, K. Baruah, L. Pritchard and J. H. Scrivens, Electrophoresis, 2013, 34, 2368–2378 CrossRef CAS PubMed.
- D. J. Harvey, F. Sobott, M. Crispin, A. Wrobel, C. Bonomelli, S. Vasiljevic, C. N. Scanlan, C. A. Scarff, K. Thalassinos and J. H. Scrivens, J. Am. Soc. Mass Spectrom., 2011, 22, 568–581 CrossRef CAS PubMed.
- J. Hofmann, W. B. Struwe, C. A. Scarff, J. H. Scrivens, D. J. Harvey and K. Pagel, Anal. Chem., 2014, 86, 10789–10795 CrossRef CAS PubMed.
- K. Pagel and D. J. Harvey, Anal. Chem., 2013, 85, 5138–5145 CrossRef CAS PubMed.
- M. D. Plasencia, D. Isailovic, S. I. Merenbloom, Y. Mechref, M. V. Novotny and D. E. Clemmer, J. Am. Soc. Mass Spectrom., 2008, 19, 1706–1715 CrossRef CAS PubMed.
- J. P. Williams, M. Grabenauer, R. J. Holland, C. J. Carpenter, M. R. Wormald, K. Giles, D. J. Harvey, R. H. Bateman, J. H. Scrivens and M. T. Bowers, Int. J. Mass Spectrom., 2010, 298, 119–127 CrossRef CAS PubMed.
- F. Zhu, S. Lee, S. J. Valentine, J. P. Reilly and D. E. Clemmer, J. Am. Soc. Mass Spectrom., 2012, 23, 2158–2166 CrossRef CAS PubMed.
- M. F. Bush, Z. Hall, K. Giles, J. Hoyes, C. V. Robinson and B. T. Ruotolo, Anal. Chem., 2010, 82, 9557–9565 CrossRef CAS PubMed.
- D. J. Harvey, J. Am. Soc. Mass Spectrom., 2005, 16, 631–646 CrossRef CAS PubMed.
- H. Schachter, Glycoconjugate J., 2000, 17, 465–483 CrossRef CAS.
- J. M. Prien, D. J. Ashline, A. J. Lapadula, H. Zhang and V. N. Reinhold, J. Am. Soc. Mass Spectrom., 2009, 20, 539–556 CrossRef CAS PubMed.
- M. P. Campbell, R. Peterson, J. Mariethoz, E. Gasteiger, Y. Akune, K. F. Aoki-Kinoshita, F. Lisacek and N. H. Packer, Nucleic Acids Res., 2014, 42, D215–D221 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed methods including instrument acquisition settings and complete list of CCS values described in the main text. See DOI: 10.1039/c5an01092f |
| This journal is © The Royal Society of Chemistry 2015 |