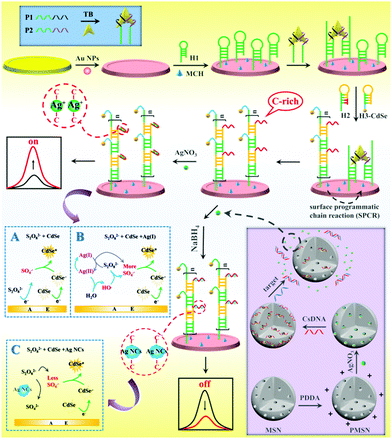
Versatile “on–off” biosensing of thrombin and miRNA based on Ag(I) ion-enhanced or Ag nanocluster-quenched electrochemiluminescence coupled with hybridization chain reaction amplification†
Junjun
Ge
,
Chunli
Li
,
Yu
Zhao
,
Xijuan
Yu
and
Guifen
Jie
 *
*
Key Laboratory of Optic-electric Sensing and Analytical Chemistry for Life Science, MOE, Shandong Key Laboratory of Biochemical Analysis, Key Laboratory of Analytical Chemistry for Life Science in Universities of Shandong, College of Chemistry and Molecular Engineering, Qingdao University of Science and Technology, Qingdao 266042, P. R. China. E-mail: guifenjie@126.com
First published on 6th June 2019
Abstract
A novel biosensing platform based on the Ag(I) ion-enhanced or Ag nanocluster (NC)-quenched electrochemiluminescence (ECL) of CdSe quantum dots (QDs) was designed for versatile “on–off” assays of thrombin (TB) and miRNA, in which bipedal molecular machine (BMM)-triggered surface programmatic chain reaction (SPCR) coupled with mesoporous silica nanoparticle (MSN) multiple amplification is used to introduce plentiful QDs and Ag+ ions to significantly improve the ECL signal.
Thrombin (TB) plays a vital role in some pathological and physiological processes, including inflammation, wound healing, and blood coagulation.1,2 MicroRNAs (miRNAs) play significant functions in various biological processes, such as cell differentiation, apoptosis, and cancer development.3 Therefore, sensitive and selective detection of TB and miRNAs is highly significant for clinical diagnosis and treatment.
Recently, DNA molecular machines have attracted intense attention because of their highly programmable nature, high precision, and ease of preparation; they have been well developed in the biosensing4,5 and intracellular molecular imaging6 fields. Of particular interest is that these molecular machines can be cyclically reused, and have been demonstrated to be a powerful signal amplification technique for assays of different targets.7
MSNs have attracted substantial attention due to their high porosity, good tunability, ease of functionalization, and ordered pore volume for a wide range of guests, from small molecules to proteins.8,9 They have been widely used as efficient drug or protein delivery carriers due to their controlled pore sizes.10,11
Electrochemiluminescence has attracted increasing attention in various bioanalytical applications due to its simplicity, high sensitivity, wide dynamic range, and low background.12,13 QDs have gradually become widely used ECL emitters due to their stable physical and chemical properties, strong anti-interference capacity and easy film-forming ability.14,15 Promotion of QDs’ ECL efficiency is of great importance for the widespread application of ECL biosensors. The use of S2O82− to catalyze QDs’ ECL was a classical ECL-coreactant system.16 ECL resonance energy transfer (ECL-RET) has been extensively used for biosensing due to its high sensitivity and wide range of analytes.17,18 Yuan's group reported an ECL-RET system from tris(4,4′-dicarboxylic acid-2,2′-bipyridyl) ruthenium(II) dichloride (Ru(dcbpy)32+) to CdSe@ZnS QDs for miRNA-141 detection.19 Xu et al. employed ECL-RET of gold-nanoparticle-luminol for dual-wavelength ratiometric miRNA detection.20 Although these methods can change the ECL efficiency of QDs, improvement of ECL intensities is still necessary for sensitive ECL bioassay.
In this study, a novel molecular machine-powered chain reaction amplification strategy was combined with Ag(I) ion-accelerated and Ag NC-quenched QDs’ ECL to develop a highly sensitive ECL biosensing platform for dual target assay. Large amounts of QDs and Ag+ were first assembled into an electrode by SPCR to much improve the ECL signal. Ag(I) ions were utilized as coreaction accelerators in the CdSe–S2O82− system to remarkably enhance the ECL intensity for TB detection. CsDNA-capped MSNs were developed as nanocarriers for controlled release of numerous Ag+ ions to achieve amplified ECL detection of miRNA-21 based on Ag NC quenching of QDs’ ECL. This method can detect two targets in one ECL biosensing platform, showing wide linear ranges and very low detection limits, which is expected to be applied in early disease diagnosis.
As shown in Scheme 1, the versatile “on–off” ECL biosensing platform was designed for sensitive detection of thrombin and miRNA-21. The BMM was first formed through specific binding of one target protein with two DNA aptamers, which then hybridized with hairpin DNA 1 (H1) on the electrode to open its prelocked domain and triggered subsequent hybridization chain reaction (HCR) with C-rich hairpin DNA 2 (H2) and CdSe-labeled hairpin DNA 3 (H3-CdSe), resulting in the attachment of large amounts of QDs and C-rich DNA on the electrode. Then, numerous Ag(I) ions were captured by C-rich DNA via C–C mismatches to form stable Ag(I)-embedded DNA complexes; the Ag(I) ions as efficient coreaction accelerators can react with S2O82− to produce massive amounts of SO4−˙ with the recycling of the Ag(I) and Ag(II) ions, and SO4−˙ can react with CdSe QDs to achieve extremely enhanced ECL for sensitive “signal on” detection of TB.
Moreover, MSNs were functionalized with poly(diallyldimethylammonium chloride) (PDDA) to carry positive charges; then AgNO3 molecules were entrapped in the pores of the MSNs and capped by the biogate CsDNA. The presence of target miRNA-21 triggered release of AgNO3 due to DNA–RNA hybridization, which resulted in a relationship between miRNA-21 and AgNO3 concentration. The released AgNO3 molecules were introduced into the SPCR-produced C-rich DNA to form stable cytosine–Ag+–cytosine; thus Ag NCs were in situ generated by NaBH4 reduction. Due to the ECL resonance energy transfer (RET) between Ag NC acceptors and CdSe QD donors, the ECL signal of CdSe QDs was quenched.21 In addition, Ag NCs competitively reacted with S2O82− to reduce the production of ECL co-reactive free radicals (SO4−˙), which further decreased the ECL signal; thus a “signal off” ECL biosensor for ultrasensitive detection of miRNA-21 was developed.
Fig. S2A (ESI†) shows a transmission electron microscopy (TEM) image of CdSe QDs in aqueous solution; the QDs with an average diameter of about 3 ± 0.5 nm were uniformly distributed. The size of the QDs estimated from the absorption peak in UV-vis spectra (Fig. S2B, ESI†) according to the empirical equation22 (D = (1.6122 × 10−9)λ4 − (2.6575 × 10−6)λ3 + (1.6242 × 10−3)λ2 − (0.4277)λ + 41.57 = 2.173) was consistent with that in TEM. Fig. S2C (ESI†) displays the PL spectra of the CdSe QDs, and the emission peak was at 526 nm.
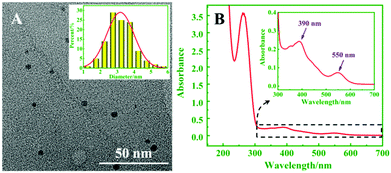
The TEM image in Fig. 1A revealed that the Ag NCs were monodispersed, and the average size of the Ag NCs was about 3.0 nm. Fig. 1B shows two peaks of UV-vis absorption at 390 and 550 nm, respectively, which were consistent with different particle sizes of Ag NCs. Among them, the absorption peak at 550 nm matched the fluorescence emission peak of the CdSe QDs, which proves that ECL resonance energy transfer between the QDs and Ag NCs can be performed.
In order to confirm the feasibility of the proposed ECL biosensor for TB detection based on BMM-triggered SPCR amplification, gel electrophoresis (GE) was employed to characterize the process. As shown in Fig. 2, lanes m, a, b, and c show the GE results for the marker, H1, H2, and H3, respectively. The thiol group in H1 easily combines into a disulfide bond to increase the molecular weight of DNA, producing two distinct bright bands (lane a). Lane d exhibits several distinct bands of the products by SPCR at the top of the strip, suggesting that H1, H2, and H3 were hybridized in the presence of TB to achieve SPCR amplification.
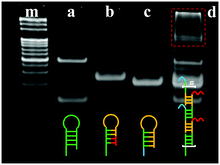 | ||
| Fig. 2 Polyacrylamide gel electrophoretogram (PAGE) analysis of SPCR: (m) marker, (a) H1, (b) H2, (c) H3, and (d) product of SPCR. | ||
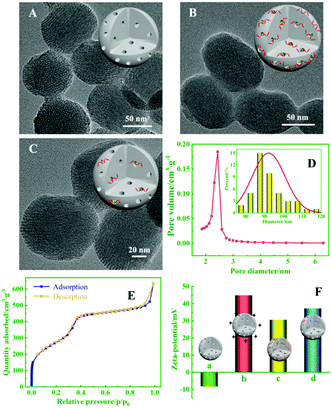
To demonstrate that the MSNs can load AgNO3 molecules to quantify target miRNA using the biogate, the morphological properties of MSNs, CsDNA capped PMSNs, and PMSN-CsDNA in the presence of miRNA-21 were characterized with TEM images. As shown in Fig. 3A–C, the average diameter of the spherical MSNs was about 90 nm, and the CsDNA-capped PMSNs showed a much rougher surface and fewer pores (B) than MSNs (A), indicating that CsDNA successfully adhered onto the PMSNs. After being incubated with the target miRNA, the pore outlines of MSNs became clearer because the rigid CsDNA–RNA duplex structure left the surface of the PMSN and the AgNO3 molecules were released (C). Fig. 3D also shows that the average diameter of the MSNs is about 90 nm, while the pore size with narrow distribution is about 2.44 nm, which guaranteed efficient loading of AgNO3 molecules.23 As shown in Fig. 3E, the MSN samples displayed an excellent N2 adsorption–desorption isotherm curve with a sharp increase in adsorbed volume between 0.4 and 0.5 of P/P0, which is characteristic of a well defined mesopore structure.
Moreover, the analysis process for miRNA using PMSN-AgNO3/CsDNA was characterized in terms of the zeta potential. As shown in Fig. 3F, after the positively charged PDDA was attached on MSNs, the zeta potential changed from −8.26 mV (MSNs, column a) to +44.98 mV (PMSNs, column b). Furthermore, as the DNA phosphate backbone has negative charge, the zeta potential of CsDNA-capped PMSNs decreased to +30.72 mV (column c). After further reacting with the miRNA, PMSNs showed a slight increase in potential (+37.37 mV, column d), so the results verified the successful fabrication and ability of the CsDNA-PMSNs for miRNA detection.
The atomic force microscopy (AFM) technique was also employed to confirm the fabrication process of the biosensor. Fig. S3A and B (ESI†) show the images of Au NPs on the electrode, which possessed a uniformly distributed surface structure. The thickness of the pure gold film was ∼15 nm, and the gold NPs can also be observed. After the CdSe QDs were immobilized on the Au NP-electrode (Fig. S3C, ESI†), the surface became more dense due to more nanoparticles. After the Ag NCs were in situ generated on the C-rich ssDNA via reduction on the electrode (Fig. S3D, ESI†), the surface was covered with abundant smaller nanoparticles in appearance, suggesting that the biosensor based on CdSe QDs and Ag NCs was successfully fabricated for dual target detection.
In order to confirm the feasibility analysis of the ECL biosensor for TB and miRNA-21 detection based on the Ag(I) ion enhanced or Ag NC quenched ECL of the CdSe–S2O82− system. The ECL signals under different conditions were employed to characterize the process. As shown in Fig. S5A (ESI†), almost no ECL was observed on the bare electrode (curve a). In the absence of TB, the ECL signal is similar to that of the bare electrode (curve b). When TB was present, an evident ECL peak with high intensity (curve c, 6798 a.u.) was observed, as the TB–aptamer complex first opened H1 and then initiated HCR to assemble numerous H3-CdSe QDs onto the electrode. Moreover, the HCR introduced abundant C-rich hairpin DNA 2 (H2) to capture large amounts of Ag(I) ions on the electrode, the Ag(I) ions as coreaction accelerators can react with S2O82− to supply more SO4−˙ to significantly increase the ECL intensity, the improved ECL signal is about 2.56 times (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 448 a.u., curve d) that of curve c, so the “signal on” ECL biosensor was developed for sensitive TB detection. The possible ECL mechanism is as follows:24
448 a.u., curve d) that of curve c, so the “signal on” ECL biosensor was developed for sensitive TB detection. The possible ECL mechanism is as follows:24
| Ag(II) + H2O → Ag(I) + AgO+ + 2H+ | (1) |
| AgO+ + H2O → Ag(I) + H2O2 | (2) |
| H2O2 + e− → OH− + OH˙ | (3) |
| OH˙ + S2O82− → SO4˙− + HSO4− + O2 | (4) |
| O2 + H2O + e− → OH− + H2O2 | (5) |
| CdSe + e− → CdSe˙− | (6) |
| CdSe˙− + SO4˙− → CdSe* + SO42− | (7) |
| CdSe* → CdSe + hν | (8) |
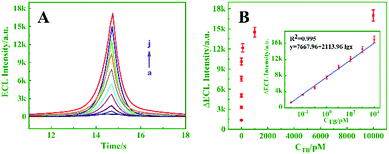
To investigate the performance of the proposed ECL biosensor, ECL responses to TB with various concentrations were recorded under optimal conditions. Fig. 4A displays the ECL intensity–time curves, wherein the ECL response signals increase gradually with the increasing concentration of TB from 0 to 1000 pM. The relationship between ΔECL and the TB concentration is shown in Fig. 4B. It can be found that the ΔECL is logarithmically related to the concentration of TB in the range from 0.001 to 1000 pM (R2 = 0.995), and the detection limit is 0.165 fM (S/N =3). A comparison of the reported TB detection methods indicated that the prepared biosensor exhibited better sensitivity (Table S1, ESI†).
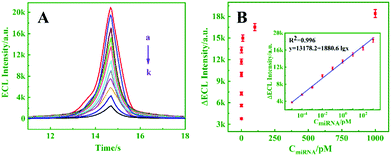
Under optimal conditions, the proposed biosensor was successively used to detect target miRNA-21. As exhibited in Fig. 5A, the ECL intensity decreased upon increasing the concentration of miRNA-21 from 0 to 103 pM (curves a–k), and an excellent linear relationship between ΔECL and the logarithm of the miRNA-21 concentration (10−5–103 pM) is shown in Fig. 5B (inset, R2 = 0.996). Moreover, the detection limit was calculated to be 4.97 aM (S/N = 3). By comparison, the prepared biosensor exhibited better performance and higher sensitivity than the other analysis methods for miRNA detection (Table S2, ESI†).
The selectivity of the proposed sensor for TB detection was verified; AFP, CEA, PSA, TB and a mixture of them were detected, respectively. The results are shown in Fig. S9A (ESI†). No remarkable changes in the ECL signals were observed for detecting the interferents compared with that of TB (102 pM) and its mixture, indicating the excellent selectivity of the proposed ECL biosensor for TB assay.
In conclusion, based on a novel molecular machine-powered chain reaction amplification strategy combined with the Ag(I) ion-accelerated and Ag NC-quenched ECL of the CdSe–S2O82− system, a highly sensitive ECL-biosensing platform was designed for versatile “on–off” detection of dual targets. Target TB–aptamer binding first initiated SPCR not only to bring a large number of QD signal probes on the electrode, but also to generate abundant C-rich DNA chains. The C-rich DNA was fully utilized to capture large amounts of Ag(I) ions for enhancement of QD ECL to develop a novel “signal on” ECL biosensor for sensitive TB assay, and also in situ electrochemically generate massive amounts of Ag NCs for ECL quenching to achieve “signal off” ECL detection of miRNA-21. It is for the first time that the molecular machine powered-SPCR amplification was combined with Ag+-induced “on off” ECL in the same biosensing system for versatile assays of dual targets. The proposed sensing platform showed very wide linear ranges and lower detection limits compared to previously reported methods. The biosensor can be applied to human serum samples with good precision, which has promising applications in bioanalysis and clinical diagnosis.
This work was supported by the National Natural Science Foundation of China (No. 21575072).
Conflicts of interest
There are no conflicts of interest to declare.Notes and references
- G. Shen, H. Zhang, C. Yang, Q. Yang and Y. Tang, Anal. Chem., 2016, 89, 548–551 CrossRef PubMed.
- C. X. Zhu, M. Y. Liu, X. Y. Li, X. H. Zhang and J. H. Chen, Chem. Commun., 2018, 54, 10359–10362 RSC.
- P. Zhang, J. Jiang, R. Yuan, Y. Zhuo and Y. Q. Chai, J. Am. Chem. Soc., 2018, 140, 9361–9364 CrossRef CAS PubMed.
- X. L. Yang, Y. N. Tang, S. D. Mason, J. B. Chen and F. Li, ACS Nano, 2016, 10, 2324–2330 CrossRef CAS PubMed.
- C. Feng, Z. H. Wang, T. S. Chen, X. X. Chen, D. S. Mao, J. Zhao and G. X. Li, Anal. Chem., 2018, 90, 2810–2815 CrossRef CAS PubMed.
- H. Y. Peng, X. F. Li, H. Q. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378–14390 CrossRef PubMed.
- J. Zhu, H. Gan, J. Wu and H. X. Ju, Anal. Chem., 2018, 90, 5503–5508 CrossRef CAS PubMed.
- F. Tang, L. Li and D. Chen, Adv. Mater., 2012, 24, 1504–1534 CrossRef CAS PubMed.
- A. Popat, S. B. Hartono, F. Stahr, J. Liu, S. Z. Qiao and G. Q. Lu, Nanoscale, 2011, 3, 2801–2818 RSC.
- Q. Zhang, F. Liu, K. T. Nguyen, X. Ma, X. Wang, B. Xing and Y. Zhao, Adv. Funct. Mater., 2012, 22, 5144–5156 CrossRef CAS.
- J. Lee, J. Park, K. Singha and W. J. Kim, Chem. Commun., 2013, 49, 1545–1547 RSC.
- G. F. Jie, L. Tan, Y. Zhao and X. C. Wang, Biosens. Bioelectron., 2017, 94, 243–249 CrossRef CAS PubMed.
- L. L. Li, Y. Chen and J. J. Zhu, Anal. Chem., 2017, 89, 358–371 CrossRef CAS PubMed.
- Y. Q. Dong, H. C. Pang, H. B. Yang, C. X. Guo, J. W. Shao, Y. W. Chi, C. M. Li and T. Yu, Angew. Chem., Int. Ed., 2013, 52, 7800–7804 CrossRef CAS PubMed.
- M. Y. Daniel, C. R. Estefania and C. G. Agustin, Anal. Chem., 2016, 88, 3739–3746 CrossRef PubMed.
- R. Zhang, J. R. Adsetts, Y. Nie, X. Sun and Z. Ding, Carbon, 2018, 129, 45–53 CrossRef CAS.
- J. Wang, X. Jiang and H. Han, Biosens. Bioelectron., 2016, 82, 26–31 CrossRef CAS PubMed.
- S. C. Phung, J. M. Cabot, M. Macka, S. M. Powell, R. M. Guijt and M. Breadmore, Anal. Chem., 2017, 89, 6513–6520 CrossRef CAS PubMed.
- Z. Li, Z. Lin, X. Wu, H. Chen, Y. Q. Chai and R. Yuan, Anal. Chem., 2017, 89, 6029–6035 CrossRef CAS PubMed.
- X. L. Huo, N. Zhang, H. Yang, J. J. Xu and H. Y. Chen, Anal. Chem., 2018, 90, 13723–13728 CrossRef CAS PubMed.
- Y. Y. Zhang, Q. M. Feng, J. J. Xu and H. Y. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 26307–26314 CrossRef CAS PubMed.
- T. Osinga, W. Lipinski, E. Guillot, G. Olalde and A. Steinfeld, Exp. Heat Transfer, 2006, 19, 69–79 CrossRef CAS.
- P. P. Gai, C. C. Gu, T. Hou and F. Li, ACS Appl. Mater. Interfaces, 2018, 10, 9325–9331 CrossRef CAS PubMed.
- Y. M. Lei, R. X. Wen, J. Zhou, Y. Q. Chai, R. Yuan and Y. Zhuo, Anal. Chem., 2018, 90, 6851–6858 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section and supporting figures and tables. See DOI: 10.1039/c9cc03235e |
| This journal is © The Royal Society of Chemistry 2019 |