Open Access Article
Open Access ArticleStructure and mechanics of the human nuclear pore complex basket using correlative AFM-fluorescence superresolution microscopy†
Anthony
Vial
 a,
Luca
Costa
a,
Luca
Costa
 a,
Patrice
Dosset
a,
Pietro
Rosso
a,
Gaëlle
Boutières
a,
Orestis
Faklaris
a,
Patrice
Dosset
a,
Pietro
Rosso
a,
Gaëlle
Boutières
a,
Orestis
Faklaris
 b,
Heiko
Haschke
c,
Pierre-Emmanuel
Milhiet
b,
Heiko
Haschke
c,
Pierre-Emmanuel
Milhiet
 *a and
Christine M.
Doucet
*a and
Christine M.
Doucet
 *a
*a
aCBS (Centre de Biologie Structurale), Univ Montpellier, CNRS, INSERM, Montpellier, France. E-mail: pem@cbs.cnrs.fr; christine.doucet@cbs.cnrs.fr
bMRI, Biocampus, University of Montpellier, CNRS, INSERM, Montpellier, France
cBruker Nano GmbH, Berlin, Germany
First published on 6th February 2023
Abstract
Nuclear pore complexes (NPCs) are the only gateways between the nucleus and cytoplasm in eukaryotic cells. They restrict free diffusion to molecules below 5 nm while facilitating the active transport of selected cargoes, sometimes as large as the pore itself. This versatility implies an important pore plasticity. Recently, cryo-EM and AI-based protein modeling of human NPC revealed with acute precision how most constituents are arranged. But the basket, a fish trap-like structure capping the nucleoplasmic side of the pore, remains poorly resolved. Here by atomic force microscopy (AFM) coupled to single molecule localization microscopy (SMLM) we revealed that the basket is very soft and explores a large conformational landscape: apart from its canonical basket shape, it dives into the central pore channel or opens, with filaments reaching to the pore sides. Our observations highlight how this structure can adapt and let morphologically diverse cargoes shuttle through NPCs.
Introduction
In eukaryotic cells the nuclear envelope (NE) isolates the genome from the rest of the cell. This physical barrier is composed of two biological membranes: the outer nuclear membrane (ONM), that is continuous with the endoplasmic reticulum, and the inner nuclear membrane (INM), which harbors a unique integral protein composition.1 Nuclear pore complexes (NPCs) are the only gateways between the cytoplasm and nucleoplasm. Their primary function is to ensure the permeability barrier: they restrict passive diffusion to small molecules below 40 kDa, yet allowing regulated transport of larger protein assemblies, including the export of mRNAs.2 Very large MDa molecular assemblies can be transported through NPCs in their native state, such as proteasomes or viral capsids for instance.3–6 As a matter of fact, NPCs are the largest protein complexes found in eukaryotic cells, with a diameter around 100 nm in humans.7–10 They are composed of over 1000 polypeptides for a total size of 110 MDa in humans.7 With their 8-fold rotational symmetry, NPCs are composed of about 30 different proteins called nucleoporins (Nups), present in multiples of 8 copies per pore. The NPC scaffold is made of three stacked rings embedded in the NE, and a lumenal ring located in the perinuclear space. The central channel is filled with nucleoporins rich in FG-repeats whose physico-chemical properties rule the permeability barrier.11,12 This whole structure is capped by cytoplasmic filaments on one side, and a nuclear basket on the other (Fig. 1A).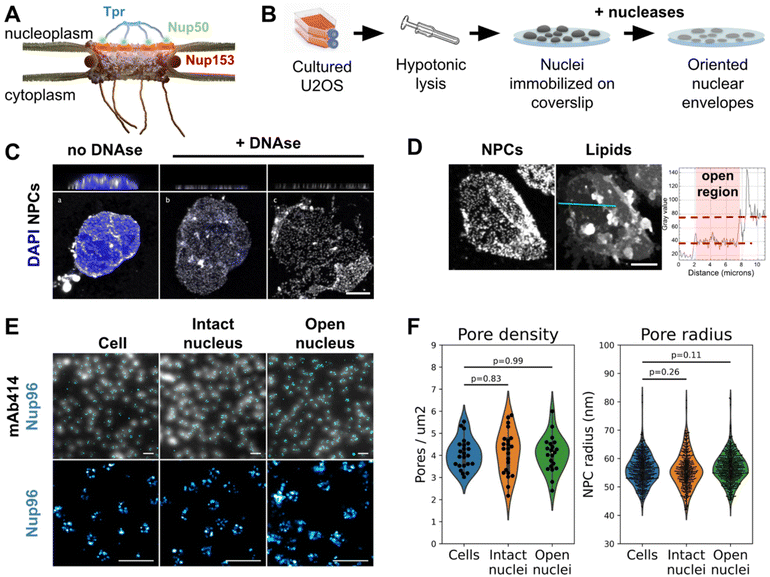 | ||
| Fig. 1 The nuclear membrane is preserved during NE preparation. (A) Schematics of the human nuclear pore complex, with emphasis on basket organization. (B) Schematic procedure for nuclear envelope preparation from cultured U2OS. (C) 3D-confocal imaging of nuclei isolated from U2OS and treated or not with nucleases. The chromatin was labeled with Hoechst (blue) and NPCs with WGA (white). Top panels show xz sections; bottom panels show the lower nuclear membrane. Scale bar is 5 μm. (D) Confocal imaging of an open nucleus. NPCs are labeled with WGA-AF594, lipids with DiOC6. Scale bar is 5 μm. The right panel shows the intensity profile of the lipid dye along the cyan section. (E) Nuclei and nuclear envelopes were extracted from U2OS/Nup96-SNAP. Intact cells, intact nuclei or open nuclei were labelled with mAb414-AF594 and AF647-SNAP ligand. Diffraction-limited (mAb414) and dSTORM images (Nup96-SNAP) were acquired in TIRF illumination; scale bars are 2 μm (top) and 500 nm (bottom). (F) Pore density was measured from confocal images (shown in Fig. S2C,†n > 20 nuclei); NPC radii were measured from dSTORM images as exemplified in E ( n ≥3 nuclei). | ||
Years of intense structural investigations have gradually revealed the molecular architecture of the human NPC scaffold. Cryo-electron microscopy (cryo-EM) performed on purified NPCs or in situ has depicted the arrangement of the luminal, inner, cytoplasmic and nucleoplasmic rings with respect to the NE membrane, at resolutions down to 2 nm.13–17 Integrative structural studies have positioned X-ray crystallographic and model structures of individual Nup fragments within the electron-dense volumes. We now visualize with subnanometer resolution how Nups are arranged within the scaffold.8,13,15–19 In striking contrast, the organization of cytoplasmic filaments and nuclear basket in human cells is not well solved in cryo-EM maps. One interpretation is that these domains are highly flexible, leading to a continuum of conformations that cannot be resolved by cryo-EM. While a near atomic composite model of NPC cytoplasmic face was recently published,13 the nuclear basket is notoriously missing. From scanning electron micrographs taken in the 90 s of Xenopus laevis nuclear envelopes,20 we know that the basket is made of eight filaments attached at their base to the NPC scaffold and joining together at their distal ends. The resulting structure appeared as a fish trap-like net, hence its name – the basket. In higher eukaryotes, this structure is mainly composed of three Nups: Nup153 and Nup50, located at the base of the basket, and Tpr which contains four predicted coiled-coil domains and is the major constituent of the filaments.21 During their nucleo-cytoplasmic shuttling, cargoes have to pass through the basket, which caps the nucleoplasmic opening of the channel. It is hard to envision how very large cargoes, such as intact viral capsids,5 can make their way in this intricate structure. Indeed, such large cargoes almost fill the entire space within the central pore channel, and their release in the nucleoplasm must involve some opening or disassembly of the basket. As a matter of fact, while most components of the scaffold are very stably associated, several Nups nevertheless can exchange from the NPC.22 It is in particular the case for Nups that connect core NPC components together23 and their high exchange rate was proposed to favor structural adaptation of the NPC during cargo translocation for instance. Likewise, components of the basket are highly dynamic: they exchange rapidly with the NPC, indicating a continuous turnover between NPC-bound and free Nups.22–25 Moreover, serial analysis of thousands of NPCs imaged by single molecule localization microscopy (SMLM) show that the main basket constituent, Tpr, exhibits a blurry spatial distribution while other components show a better-defined localization.26 However, most studies cited above rely on ensemble or averaged structural data. It is thus not clear how structural dynamics and/or variability of the basket translate at the single pore level. Therefore, to explore different basket conformations, we used atomic force microscopy (AFM), that does not rely on averaging.
Thanks to its very high signal to noise ratio, AFM provides sharp topography images of single particles and operates in liquid, making it an asset to study heterogeneous samples in physiological conditions.27 In AFM, samples are probed by a nanometric tip located at the end of a cantilever. Tip–sample interactions induce deflection of the cantilever, which can be detected and analyzed to reconstruct the topography of the sample surface and can also provide mechanical information. Furthermore, combined with super resolution fluorescence microscopy, molecular mapping can be correlated with AFM topography of biological membranes with lateral resolutions up to 10–30 nm (see a review in ref. 28 and a study carried out in our group29). AFM thus appears as a method of choice to observe and describe at high resolution the conformational states of NPCs. However, this technique requires direct accessibility to the probed surface. In the context of NPC study, it means to isolate nuclei and purify NEs. This technique has been developed on X. laevis stage-VI oocytes, since their nuclei are large enough (∼300 μm in diameter) to be manually teared open.30 High resolution images of the cytoplasmic and nucleoplasmic sides of Xenopus NPCs have revealed a certain degree of architectural plasticity – their morphology changes in response to different physiological stimuli or addition of transport receptors.25,31–38 However, these works did not provide an extensive description of the conformational landscape of NPC baskets. In other words, it is not clear whether baskets can adopt a finite number of conformations or freely explore a continuum of conformations. Considering the crucial role of NPCs in cellular homeostasis and the fact that they are associated with several diseases,39–42 there is a real interest in elucidating the structure of human nuclear pores and understanding their plasticity. Indeed, even though NPCs are highly conserved among eukaryotes, there are compositional and/or structural differences between species.14,43 Moreover, oocytes are in a particular metabolic state, and this translates into structural and functional specificities even in NPC organization: NPCs are ∼10 times denser than in somatic cells, and their structure and size evolves during gametogenesis.44 A few studies have been performed on biochemically extracted human NEs45,46,81,82 but none of them focused on NPC baskets.
We established a biochemical procedure to open nuclei isolated from cultured human U2OS cells and imaged by AFM the nucleoplasmic face of hundreds of NPCs in their physiological environment. Thorough statistical analysis of AFM images showed that the basket largely explores the pore central channel rather than systematically protruding into the nucleoplasm. We also located Tpr within the basket by correlating AFM with super-resolution microscopy and found pores with open baskets, i.e. with Tpr-containing filaments reaching towards the membrane around the pore. Comparing the topography of normal NPCs and NPCs devoid of basket demonstrated that the basket contributes to an important volume in the central channel of NPCs, and to a lesser extent at the surface of the scaffold rim. Finally, we found that the basket filaments are very soft, an undeniable asset for structural plasticity. Our findings explain why cryo-EM cannot currently decipher the basket structure but most importantly, they enlighten how it can adapt to cargoes of different morphologies and provide a flexible frame to support nucleo-cytoplasmic transport.
Results
Preparation of human nuclear envelopes (NEs) with preserved nuclear membrane organization
It appeared recently that biochemically purified NPCs are narrower than their counterparts imaged in situ.8,14 This is arguably due to a loss of tension in the NE. To avoid this artifact, we adapted a classical biochemical procedure to purify NEs,47 applying it to nuclei immobilized on glass in order to preserve their spatial organization, orientation and tension. Briefly, a hypotonic cell lysis is carried out using a Dounce homogenizer. The purified nuclei are immobilized on glass coverslips, then treated with a mixture of RNase and DNase in a hypotonic buffer (Fig. 1B, see Methods for more details). A fraction of nuclei spontaneously opens during this step (Fig. 1C). As nuclei are immobilized on glass prior to treatment, when they open, the top NE sheet is removed, leaving the bottom layer intact with its INM facing up. Samples were then fixed and fluorescently labeled for correlative AFM/fluorescence studies. Since nuclei are open, labeling can be performed without detergent, keeping the nuclear membranes intact. We demonstrated that the successive biochemical treatments indeed preserve the nuclear envelope and nuclear pores. While nuclease treatment completely digests chromatin in most nuclei (Fig. S1A†), it results in different configurations: some nuclei remain apparently intact (Fig. 1C-b) while a majority have a fragment of NE ripped off (Fig. 1C-c). The areas where one or two layers of NE are present are easily delineated thanks to the two-fold difference in NPC density (Fig. 1C and S1B†) or lipid staining (Fig. 1D). Some nuclei expose a well circumscribed central hole; in these cases the membrane organization is well preserved, based on the homogenous lamin-B1 staining (Fig. S1C-e and f†) and further confirmation by AFM imaging (Fig. 2A & B for instance). These nuclei, easily recognizable, were selected for further studies. On the contrary, some nuclei have large sections of NE ripped off; in those the organization of INM was disrupted, as shown by lamin staining (Fig. S1C-c and d†), and they were discarded.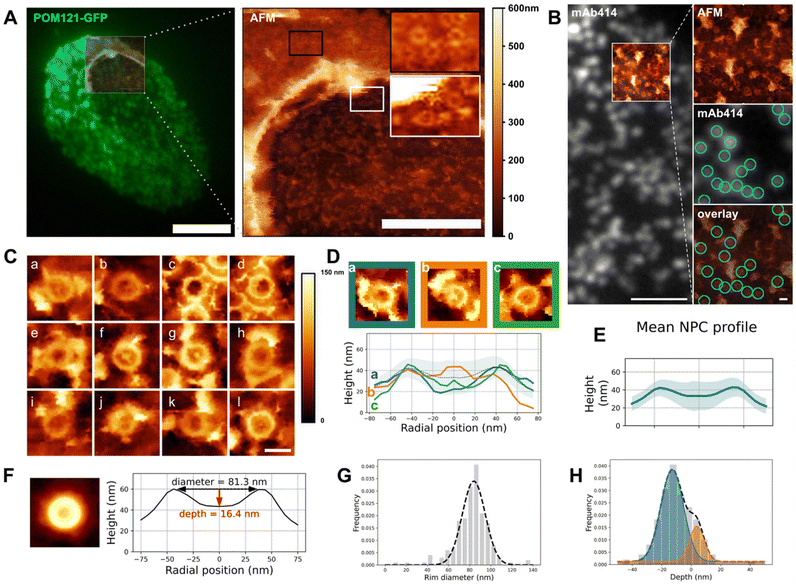 | ||
| Fig. 2 Structure of the NPC basket. (A) Correlative fluorescence/AFM image of an open nucleus isolated from U2OS overexpressing POM121-GFP. Left: TIRF image of the entire nucleus overlaid with the region scanned by AFM. Right: Height image acquired by AFM, encompassing the opening border. Scale bars are 5 μm (left), 2 μm (right). (B) Correlative TIRF/AFM image of the inner nuclear envelope of an open nucleus. NPCs are labelled with mAb414 and visualized as fluorescent dots that coincide with ring-like structures in the AFM image. Scale bars are 2 μm and 200 nm. (C) Representative samples of human nuclear pores (nucleoplasmic face) imaged by AFM. Scale bar is 100 nm. (D) Three configurations of NPC nucleoplasmic region and their respective height profile. “Empty” (blue), protruding (orange) and low basket (green). (E) Mean of rotationally averaged NPC height profiles (n = 210). The shaded area represents the standard deviation. (F) (Left) Average image of 210 NPC crops. AFM color scale as above. (Right) Height profile of the resulting image. (G and H) Diameter and depths distributions of 210 NPCs plotted as frequencies and fitted with one (respectively two) Gaussians. The dashed line represents the fit function. These graphs are also shown in Fig. S2D & E† to illustrate the analysis workflow. | ||
Regarding NPC organization, pore densities measured from cultured cells and purified nuclei – treated or not with nucleases – are similar (Fig. 1F & S1B†). Moreover, direct stochastic optical reconstruction microscopy (dSTORM) imaging on NPCs allowed us to measure their diameters (Fig. 1E and F). For this, we used a modified U2OS cell-line whose endogenous nucleoporin Nup96 is fused to a SNAP-tag.48 Nup96 is a main constituent of NPCs scaffold and its distribution is thus representative of the nuclear pore structure. We labeled intact cells and nuclei – intact or open – with an AlexaFluor 647 SNAP-ligand and performed dSTORM imaging. The diameter of Nup96-SNAP rings measured in the three situations shows no significant differences (Fig. 1F). Conservation of pore density and diameter suggests that adhesion of the NE prior to its rupture prevents NPC remodeling observed in biochemical NE fractions.8 In conclusion, we developed a method to prepare flat human NEs, free of chromatin, with areas of INM accessible to the AFM tip.
Topography of the NPC basket
The notorious absence of the pore basket in averaged structures requires assessing the nucleoplasmic topography of individual pores. Documenting the conformational landscape of the basket can reveal how rapid exchange of Tpr and Nup153 translates in the basket structure. Moreover, this will elucidate to which extent the pore basket can deviate from the canonical fishtrap-like structure into conformations compatible with transport of large cargoes. Fig. 2A shows an open nucleus prepared from U2OS cells expressing the transmembrane nucleoporin POM121 fused to GFP (POM121-GFP). Based on pore density, we assessed that this nucleus had a central, well-delineated hole as previously discussed. AFM imaging confirms the presence of this hole surrounded by a bulge of membrane (Fig. 2A, right panel). Nuclear pores are clearly delineated by the AFM tip on both the cytoplasmic (black inset) and nucleoplasmic faces (white inset). In order to probe the basket structure, we then focused on the INM. NEs were routinely prepared and labeled with a widely used fluorescent antibody against NPCs recognizing several Nups (mAb414 conjugated to Alexa Fluor 594). The fluorescent pore signal correlates with ring structures in the AFM image (Fig. 2B). 250 nm crops around each pore were extracted (examples shown in Fig. 2C) and a ring of about 82 nm diameter is visible in all NPCs, reminiscent of previous studies on Xenopus laevis NEs32–34,37 – this is the scaffold. In contrast, the central structure, that corresponds to the nuclear basket, is highly variable: in some cases, filaments are visible (Fig. 2C-a, b, d, f, g and j); they are rarely symmetric, but join at their distal ends into a structure reminiscent of the distal ring described in Xenopus.32–37 This point either protrudes above the scaffold ring, or stands below the ring level. In other cases, the basket is not clearly depicted and pores appear as an empty barrel. Fig. 2D exemplifies these three typical topologies and their radially averaged height profiles (methodological details in the methods section and Fig. S2A†). In the three examples shown in Fig. 2D, the ring diameter and height are very similar, while the central heights differ a lot. Averaging radial profiles of over 200 individual pores results in a mean profile with the central region lower than the ring (Fig. 2E & S2B†), far from the canonical protruding basket depicted in textbooks. Consistent with our qualitative description, the standard deviation is higher in the central region of the pore than at the ring level showing more structural variability in the basket than the scaffold. By curiosity we also built an averaged image from all NPC crops (Fig. 2F). No basket-like structure is visible in the resulting averaged NPC. This is coherent with our results presented in Fig. 2E and substantiates that systematic disappearance of basket electron density in high-resolution cryo-EM images is due to high heterogeneity.We then extracted the diameter and depth from each rotationally averaged profile (Fig. S2C† and Methods). Diameters are normally distributed, with a mean of 81.5 ± 16.3 nm (Fig. 2G). The depth distribution illustrates that the distal point of the basket can sweep across 60 nm in the axial direction (Fig. 2H), which is roughly the height of the human NPC scaffold itself.8 This suggests that the basket can explore a continuum of conformations. The height distribution is best fitted with two Gaussians corresponding to protruding (depth >0) and collapsed (depth <0) baskets. The relative Gaussian amplitudes confirm that protruding baskets are a minority.
Altogether our results show that, while the ring structure is rather conserved from pore to pore, the central region corresponding to the basket is extremely variable and a majority of basket structures do not protrude into the nucleoplasm but rather reside in the central pore channel. Considering that Tpr is a main component of the basket filaments,21 a quantitative evaluation of Tpr organization within pores, especially when an empty ring is observed, was performed.
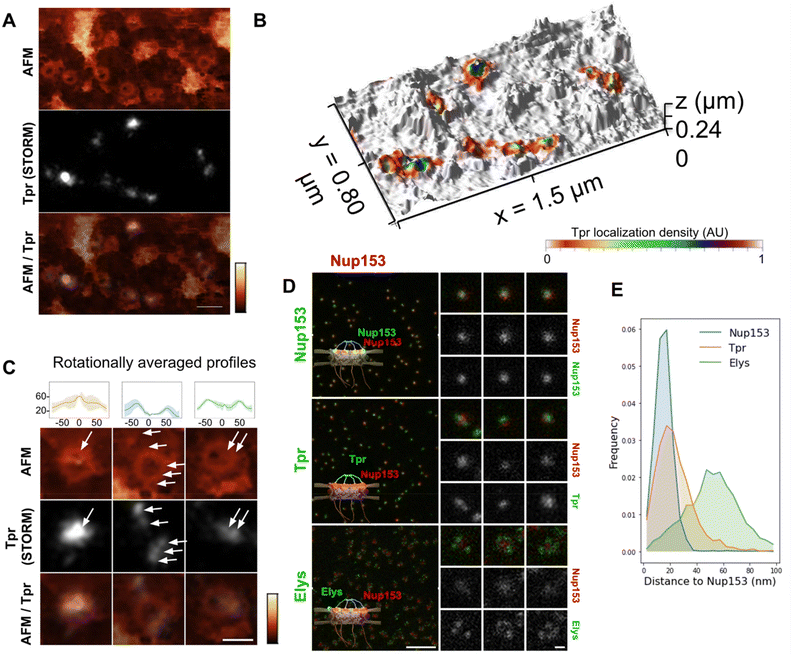
Tpr organization
Tpr is a 267 kDa protein that contains 4 coiled-coil regions in its N-terminal part while its C-terminus is mostly disordered. Coiled-coils are helical domains that can oligomerize and constitute rigid molecular rods.49,50 In the case of Tpr, the coiled-coils are interleaved with flexible linkers. Tpr thus contains the structural ingredients to provide filamentous yet articulated segments, which may be an asset for the basket to adapt its shape to cargoes.We labeled Tpr by indirect immuno-fluorescence on purified NEs and performed correlative dSTORM/AFM. As expected, Tpr signal correlates with NPCs in the TIRF and AFM images (Fig. 3A & S3A†). This is even more evident in the 3D representation (Fig. 3B), where Tpr density precisely coincides with height signal in the AFM images attributed to the basket domain. An ensemble view shows that Tpr localization densities are not homogenous (Fig. S3A†). Although this information cannot be quantitatively inferred to numbers of molecules, especially with immuno-fluorescence labeling,48,51 it nevertheless indicates a variability in the number of Tpr filaments bound to NPCs. This is most likely the result of Nup153 and Tpr turnover.22–24 At the single pore level, several configurations are observed (Fig. 3C): in the first example, a canonical basket is visible in the AFM image and a strong dSTORM signal correlates with the filaments and the distal ring. Importantly in this example, both height and Tpr signals are asymmetric, supporting that Tpr filaments can partially dissociate from the basket, leading to asymmetrical structures. Other NPCs, like the second example, look empty, with no apparent basket structure in the AFM image but Tpr signal is distributed in distinct localizations, located on top of the scaffold ring or even outside of it. Importantly, Tpr signal still colocalizes with filamentous structures visible in the AFM images. This suggests that the basket is in an “open” configuration, with nucleoplasmic filaments attached at their basis to the NPC scaffold, but their distal ends remaining free. Finally, a third example of NPC exhibits some internal density correlated with Tpr but lower than the NPC ring. This configuration looks like a “collapsed” basket, where the filaments may join by their distal ends but are directed inwards the NPC channel instead of protruding towards the nucleoplasm.
These images are reminiscent of the three NPC conformations described previously (Fig. 2D) and further document the fact that the NPC basket can adopt various configurations. Importantly, as dSTORM imaging was performed before AFM imaging, the asymmetry observed in Tpr organization in some NPCs is not due to deformation by the AFM tip. However, it could be due to deformations that arose during the biochemical preparation. We thus explored the basket structure in intact cells by 2-color STED (stimulated emission depletion) microscopy. This confocal-based super-resolution microscopy technique is highly convenient for multi-colour imaging and achieves a typical lateral resolution in fixed biological samples around 40 nm.52,53 We performed co-immuno-fluorescence in fixed cells against Tpr (coupled to a Star-635P fluorophore) and Nup153 (coupled to an Alexa Fluor 594) to visualize their relative organization. As a control, we used Nup153 co-labeled with Star-635P and Alexa Fluor 594; as expected, Nup153 imaged in both channels renders a very similar signal. Conversely, Elys, which is one of the outmost nucleoporins,26 is localized as a rim around Nup153 central dot, with some variability in the labeling efficiency (Fig. 3D). Regarding Tpr, it usually appears as one – sometimes two – dots that can be well aligned with Nup153 signal or slightly shifted (Fig. 3D, middle row). To characterize the relative positioning of these various proteins, we measured distances between Nup153 labeled with Alexa-594 and Nup153, Elys or Tpr labeled with Star-635P (Fig. 3E). The distribution shows that a fraction of Tpr is shifted away from Nup153, towards the nucleoplasmic ring where Elys resides. These results obtained on entire cells confirm the variability and asymmetry of the NPC basket observed in AFM images.
Altogether, our AFM and super-resolution microscopy data demonstrate important basket plasticity in the human NPC: Tpr-containing filaments can either protrude towards the nucleoplasm, collapse within the channel or partially disassemble at the level of the distal ring, with filaments pointing outwards from the pore channel axis.
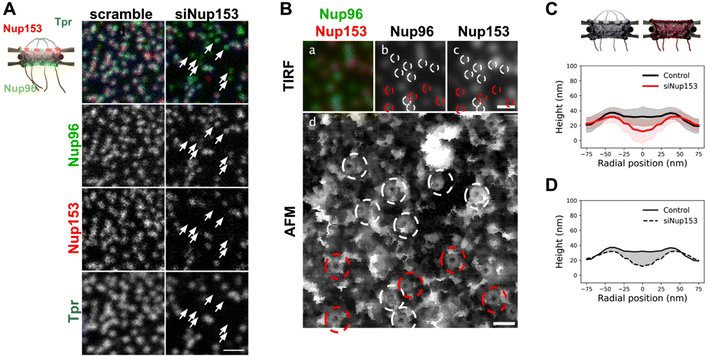
Contribution of the nuclear basket in the topography of NPCs
The presence of interleaved disordered linkers between its coiled-coil domains presumably allows Tpr to explore a large range of conformations, from a completely extended to a rather globular shape – as predicted by AlphaFold for instance.54 It is thus hard to predict how deep the basket could collapse into the pore channel. To understand the contribution of the basket in the overall NPC structure, we imaged pores in open nuclei depleted from Nup153. Indeed, as reported previously,55 siRNAs against Nup153 prevent Tpr binding to NPCs. Triple labeling of Nup153, Nup96, and Tpr (Fig. 4A) confirms that Nup153 depletion is associated with the absence of Tpr, while Nup96 remains visible (Fig. 4A, arrows point at pores without Nup153 or Tpr). It is important to note that siRNAs against Nups only achieve partial depletion.56 In the case of Nup153 depletion, it results in a mild decrease in pore density (Fig. S4A†) and importantly, only a fraction of the remaining pores are completely devoid of Nup153 (Fig. S4B†), which complicates the interpretation of topological differences observed between depleted and naive nuclei.In nuclei with co-labeled Nup96 and Nup153, we can distinguish in correlative AFM/fluorescence images pores that contain Nup153 (circled in white in Fig. 4B) or not (circled in red). In the corresponding AFM image, a central topography is more frequent in Nup153-associated NPCs. In contrast, a deep hole is more frequently observed in pores devoid of Nup153. Importantly, even when Nup153 is detected, its levels seem to vary from pore to pore but it is in all cases lower than in mock-depleted cells. To quantify how Nup153 depletion affects NPC topography, we generated average height profiles for pores imaged from Nup153-depleted NEs and compared to naive pores (Fig. 4C). Average height measured at the center is dramatically decreased when Nup153 is depleted. The height standard deviations (shaded areas in Fig. 4C) partly overlap, reflecting that some pores from depleted cells have similar topographies as compared to control pores. This overlap is likely due to remaining levels of Nup153. One can nevertheless note that very few Nup153-depleted NPCs were seen with protruding baskets (Fig. S4C†). This conformation may require high levels of Nup153 and Tpr, which is very unlikely to occur in depleted cells. Finally, the very low topographies observed in depleted nuclei probably reflect a total absence of basket. In this case, the AFM tip most likely probes the central FG-repeats. Variability of the central topography may reflect plasticity of the central channel FG-rich nucleoporins. Not to forget also that the central channel is partially occupied by shuttling cargoes, which may contribute to the overall topography and add up to the inherent variability.19 Anyway, this height difference between control and basket-depleted pores confirms that in most cases, even when the basket shape is not clearly depicted by AFM, it accounts for some molecular density occupying part of the central channel. This average contribution is visualized in Fig. 4D, as the shaded area. The height of the scaffold ring is also lower when Nup153 is depleted. This height difference reflects that the basket sits on top of the scaffold. Nup153 is probably present at this location since it interacts with the nucleoplasmic ring.24,57 Nup153 is mostly intrinsically disordered58 and its structure is likely more fuzzy than the highly ordered components of the nucleoplasmic ring.8 This could explain the important volume occupancy at the top of the scaffold ring.
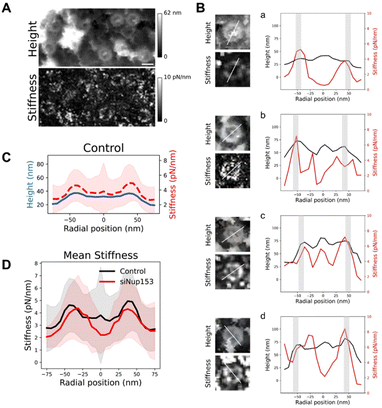
Mechanical properties of the basket
Altogether, our data show that the basket can adopt a large range of conformations. We wondered whether this important structural variability is related to mechanical flexibility. AFM is a very potent technique to measure mechanical properties of soft biological samples, down to stiffnesses of a few pN nm−1. Since the imaging mode that we used – quantitative imaging (QI, Bruker Nano GmbH, Germany) – is a force curve imaging mode allowing quantitative measurement of mechanical data at each pixel, stiffness maps were generated to understand the mechanical properties of NPCs. Fig. 5A shows an AFM height image and its corresponding stiffness map. Rings are seen in the stiffness maps and correspond to NPCs in the height images. Looking at individual pores revealed that stiffness at different regions of the basket varied greatly from pore to pore. To precisely correlate the stiffness to different parts of the NPC, we measured height and stiffness profiles in individual pores along a single line and plotted them on top of each other (Fig. 5B). The height profiles (black lines) reveal the scaffold region – the two peaks around ±42 nm, grey shaded areas – and the basket region – in between these two peaks. In most cases, the scaffold region is the stiffest (in the 4–8 pN nm−1 range) and the centermost region is the softest (2–5 pN nm−1). In a few cases though, the protrusion corresponding to the distal ring coincides with a point of high stiffness (Fig. 5B-c). Sometimes the region in between, where the basket filaments must stand, is stiffer (Fig. 5B-b & d). The origin of this variability is not clear but may reside in the transport status of pores: indeed, the density and size of cargoes located in the central channel may influence the stiffness measured in this area.To get a general view of NPCs mechanical properties, we generated rotationally averaged stiffness profiles, using the same methodology as for height profiles, and averaged them over 80 individual NPCs (Fig. 5C). This shows that on average, the basket region of NPCs is softer than the scaffold. Similarly, stiffness measurements of the NPC nucleoplasmic side in X. laevis oocytes obtained by AFM indicated a soft basket, relative to the nucleoplasmic ring.32 Interestingly, when the basket is absent in Nup153-depleted NPCs, the central region of pores is even softer (Fig. 5D), with a stiffness around 2 pN nm−1. In this configuration, the tip is likely directly probing the FG-repeats Nups that fill the central channel and are not shielded by the basket anymore. These proteins behave like long, flexible polymer molecules in vitro12,59 and their stiffness has already been measured by AFM, rendering values in the 5–10 pN nm−1 range.59–61
Since the basket, which is already very soft, appears suspended on top of an even softer medium, we wondered if it could be deflected by the AFM tip during imaging, which would affect the topographical images. Therefore we reconstructed the sample height with different forces applied (Fig. S5B†). Although a small indentation is visible with increasing forces, the overall conformation of the basket remains unchanged.
Discussion
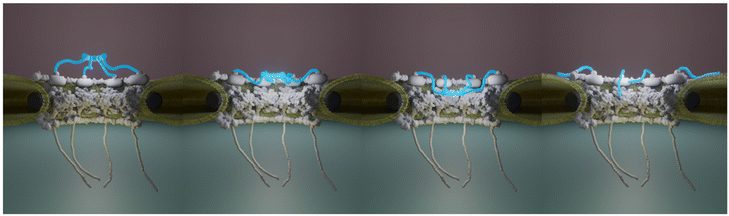
The huge size combined with their transmembrane nature make NPCs challenging objects to study and in particular at the structural level. Decades of intense integrative structural studies have provided a detailed view of the structure and molecular arrangement of NPC components in the scaffold domain. Yet, the central FG-Nups, cytoplasmic filaments and nuclear basket have been historically missing in these high resolution images obtained from purified NEs16,17,62 or in situ.8,14,63,64 Recently though, the cytoplasmic filaments structure and organization have been elucidated. But the detailed structure of the basket remains as a missing piece in the puzzle of NPC molecular organization. Here we show by AFM imaging of human inner nuclear membranes that the basket is highly plastic and explores a large volume of the central channel of the pore. In addition, the basket filaments can dissociate at their distal ends and open (Fig. 6). When reaching outwards the pore, they may scan around the pore and facilitate mRNP binding. Altogether, the conformational flexibility and the dissociation of the distal ends of the basket proteins allow NPC to adapt to cargoes of different sizes. They also explain why the basket in human cells is systematically missing in cryo-EM maps since it can adopt a continuum of conformations. In addition, the electron density in this region must be much lower than in the scaffold region. Indeed, a rapid calculation based on molecular weights of Nup153, Nup50 and Tpr, and their stoichiometry in human cells,47 indicates that the basket accounts for roughly 13% of the total NPC mass. Yet we show that it can explore a large volume (Fig. 4D). Consequently, the electron density from the basket might well fall below the detection limit in cryo-EM.These considerations underline the unique possibilities offered by AFM, which does not require averaging and provides mechanical information. As a matter of fact, NPCs have been widely studied by AFM in NEs extracted from X. laevis oocytes. These studies documented pore diameters around 81–85 nm for the cytoplasmic side25,32–34,65 and 80–88 nm at the nucleoplasmic side when measured at the ridge.32,33,65,66 The mean diameter that we measured here from human INM – 81.5 ± 16.3 nm – thus falls in the same range. Importantly, these studies also showed great variability of the basket conformation (protruding baskets and hollow pores). Moreover, some of this work showed that nuclear transport receptors65 or calcium36,37,67 induced conformational changes of the basket. Here, from unperturbed conditions, we observed various basket conformations, which may indeed reflect pores engaged in transport of different cargoes for instance. Fig. 2D illustrates three obviously different conformations, but it appeared rapidly that baskets can adopt many more conformations, suggesting it can explore a continuous conformational landscape. Quantification of the topography of hundreds of nuclear pore baskets performed in our study strongly suggests that the position of the highest region of the basket can protrude above the scaffold or collapse into the central channel with positions covering a continuum of distances of 60 nm (Fig. 2H), the basket exploring a continuous space within the central channel up to the nucleoplasm. Importantly, using correlative AFM/dSTORM microscopy appears here as an important asset to specifically assign Tpr fluorescence signal to structures delineated by the AFM tip. This let us unambiguously assign the basket structure, and was especially helpful in identifying unexpected conformations such as collapsed or open baskets (Fig. 3C).
Although it is tricky to compare data from different publications, we noticed that samples subjected to stronger fixation conditions (such as 1–2.5% glutaraldehyde) exhibit more protruding baskets,20,21 while mild (4% PFA for 5 minutes in this study) or absence of fixation32,37,65 leads to more hollow baskets. This suggests that chemical fixation, which is known to rigidify samples,68,69 may preclude pore plasticity. As a matter of fact, we and others19,32 showed that the basket is extremely soft and observed high standard deviation for stiffness values underlining the large mechanical variability in the central region of pores. The origin of this variability is not clear but may reside in the transport status of pores: indeed, the density and size of cargoes located in the central channel may influence the stiffness measured in this area. Of note, stiffness values given in previous work are higher than what we measured in human NPCs, which can be explained by the use of higher frequency (kHz) in PeakForce QNM by Stanley et al. This can overestimate the stiffness as compared to our quasi-static frequency range (typically 10–100 Hz for QI). As a matter of fact, supplementary control experiments performed by Stanley et al. using force volume, which probes the sample at lower speed, exhibit much lower stiffness values, closer to our results.32
Altogether these observations suggest that mechanical flexibility is the key parameter that allows the basket to explore a continuum of conformations. Being less restricted than other parts of the nuclear pore, the peripheral domains (cytoplasmic filaments and basket) require a lower entropic cost to change conformation. In this regard, they may be an asset for NPCs to adjust their shape. In addition to this deformation scenario, individual filaments may rapidly exchange from NPCs and reassemble in different configurations. This is supported by data showing the high exchange rate of Tpr and Nup153.22–24 In particular, Nup153 dynamics may destabilize Tpr association to the scaffold and loosen filament attachment. This is in agreement with many of our individual NPC images where the basket appears asymmetrical both in terms of topography and Tpr localization (Fig. 3). It was also shown recently in budding yeast that peripheral NPC components are loosely linked to the main scaffold and their interactions with the pore are labile.23 In this study the authors suggest that the dynamic nucleoporins could be envisioned as “a cloud of accessory factors surrounding and constantly exchanging with the NPC in vivo”. Such a model may explain how the basket filaments could rapidly change conformation while exchanging. This rapid switching could be a source of high variability in basket conformation and account at least in part for the heterogeneity we measure.
Importantly, the ability of the basket to explore a wide volume around its expected protruding position is functionally relevant. Indeed, the basket serves as a docking platform for mRNAs sorting and export.70–73 The capacity of the basket filaments to explore around the pore and their extended shape makes great assets to scan the pore vicinity for mRNA. As a matter of fact, Tpr depletion reduces the docking frequency of mRNAs to the nuclear pore and affects their diffusion through the central channel.73,74 Interestingly, the last steps of mRNA export and mRNP remodeling occur at the cytoplasmic filaments.75 These filaments, which are often represented as extended structures outreaching to the cytoplasm orthogonally from the NPC, also have the ability to bend towards the central channel.76 Our findings about the flexibility of the NPC basket and its ability to explore the central channel thus complete this picture and propose a continuous path for mRNPs during their journey across the NPC.
Taken together our results demonstrate that AFM/dSTORM correlative microscopy is an outstanding approach to decipher complex and heterogeneous biological structures. Thanks to a novel biochemical preparation of nuclear envelopes from human cells, we provided new structural details of the INM at nanometer resolution and revealed that the nuclear pore basket is very soft and explores a large conformational landscape. Our approach combining cellular content manipulation and high-resolution correlative AFM/fluorescence microscopy opens the way to new structure function characterization at the inner nuclear membrane of human cells.
Materials and methods
Cell strains, antibodies and siRNA
U2OS cells were obtained from ATCC. CRISPR-engineered Nup96-SNAP U2OS cell lines48 were obtained from CLS (clsgmbh.de). siRNAs were ordered from Eurogentec: siNup153 according to the sequence in ref. 77 (AAGGCAGACUCUACCAAAUGUUU dTdT) and scrambled siRNA (UAGAUACCAUGCACAAAUCC dTdT).All antibodies and fluorescent labels used in this study are commercially available. The purchase references and dilutions used are listed in Table 1.
| Reagent | Company | Reference | Dilution |
|---|---|---|---|
| mAb414 | Covance | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 2000 |
|
| mAb414 conjugated to AlexaFluor 594 | BioLegend | 682202 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
| Anti-lamin B1 | Thermo Scientific | PA5-19468 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
| Anti-Nup153 [SA1] | Abcam | ab96462 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 for STED 200 for STED |
| anti-Tpr | Abcam | ab84516 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 for STED and dSTORM 200 for STED and dSTORM |
| Anti-ELYS | Abcam | ab53540 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 for STED 200 for STED |
| WGA conjugated to Alexa Fluor 594 | Invitrogen | W11262 | 1 μg ml−1 from a stock solution prepared at 5 mg ml−1 in DMSO |
| DiOC6 | Invitrogen | D273 | 1–5 μM from a 5 mM stock solution prepared in DMSO |
| Alexa Fluor-647 SNAP ligand | New England BioLabs | S9136S | 1 μM for dSTORM |
| Alexa Fluor-647 anti-mouse | Molecular probes | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 2000 |
|
| Anti-rabbit or anti-mouse conjugated to Star580, StarRED or Star635P dyes | Abberior instruments | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 for STED 200 for STED |
Cell culture and transfection
Cells were cultured in DMEM/Glutamax (Gibco) supplemented with 10% fetal calf serum (Gibco) in a humid atmosphere at 37 °C, 5% CO2.For siRNA transfection, 1.5 millions of cells were seeded in 10 cm diameter plates. The day after they were transfected with 1.25 nmol of siRNA and 10 μL of Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendation. Medium was changed after 4–6 hours. Transfection was repeated 48 h later in identical conditions. Cells were lysed or imaged the day after.
Preparation of nuclear envelopes for AFM
All buffer compositions are indicated in Table 2. Nuclei were prepared from cultured cells as previously described.47 Briefly, around 40 millions cells (four 75 cm2 confluent flasks) were trypsinized. Cells were rinsed in PBS then hypotonic buffer (HB) by 5 minutes centrifugation at 300g. Cells were then resuspended in 1 mL HB, incubated on ice for 45 minutes and disrupted by mechanical shearing using a Dounce with a tight pestle (approximately 10 strokes are sufficient). Cell lysis was controlled by visual inspection using an inverted microscope (Primovert, ZEISS). The nuclei suspension was then loaded on top of 5 mL of S500 and spun for 15 minutes at 300g. The supernatant containing cellular debris was discarded, the pellet was resuspended in 300 μl of S500, transferred to a clean tube containing 5 mL of S500 and spun for 3 min at 300g. The pellet, containing mostly intact cells, was discarded. The supernatant was transferred to a clean tube and spun for 15 minutes at 300g. Nuclei were resuspended in S500 at a final density of 106 nuclei per mL.| Buffer | Composition | Ref. |
|---|---|---|
| Hypotonic buffer | 50 mM Tris pH 7.5, 1 mM DTT, Complete (Roche) | Ori et al., 2013![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 47 47 |
| S500 | 10 mM HEPES pH 7.5, 2.5 mM MgCl2, 50 mM KCl, 500 mM sucrose, 1 mM DTT, Complete | Doucet et al., 2010![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 56 56 |
| NE-A | 0.1 mM MgCl2, 1 mM DTT + 30 U DNaseI (Sigma, D5319) + 250 U RNase A | Ori et al., 2013![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 47 47 |
| NE-B | 20 mM Tris pH 8.5, 10% sucrose, 0.1 mM MgCl2, 1 mM DTT | Ori et al., 2013![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 47 47 |
| Nuclei buffer | 10 mM HEPES pH 7.5, 2.5 mM MgCl2, 83 mM KCl, 17 mM NaCl | |
| Immunofluorescence (IF) buffer | PBS wit 10 mg ml−1 BSA, 0.1% Triton X-100, 0.02% SDS | Doucet et al., 2010![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 56 56 |
| Permeabilization buffer (PB) | PBS with 0.4% Triton X-100 | Thevathasan et al., 2019![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 48 |
| Quenching solution (QS) | 100 mM NH4Cl in PBS | Thevathasan et al., 2019![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 48 |
| dSTORM buffer | PBS supplemented with 10% glucose, 0.5 mg mL−1 glucose oxidase (Sigma), 0.04 mg mL−1 catalase (Sigma, C3556), 35 mM mercaptoethylamine (MEA). | Thevathasan et al., 2019![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 48 |
25 mm high-precision type 1.5 glass coverslips of 0.17 mm thickness (Marienfeld) were coated with 200 μL of poly-L-lysine (Sigma) diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in water and filtered (0.01 mg ml−1 final concentration) for 2 minutes at room temperature. 250 μL nuclei were deposited on dried coverslips and incubated for 15 minutes at room temperature, the suspension was then removed and the coverslip was rinsed with S500.
10 in water and filtered (0.01 mg ml−1 final concentration) for 2 minutes at room temperature. 250 μL nuclei were deposited on dried coverslips and incubated for 15 minutes at room temperature, the suspension was then removed and the coverslip was rinsed with S500.
Meanwhile, the nuclear envelope buffers NE-A and NE-B (Table 2) were pre-warmed at 30 °C. Nuclei were treated with 200 μl of NE-A for 30 seconds. 800 μl of NE-B were added and the whole mixture was incubated for 30–40 minutes at room temperature as described in ref. 47. The nuclease mixture was removed and nuclei were rinsed several times with S500.
Labeling for confocal, STED and correlative TIRF/AFM microscopy
Cells and nuclei were labeled as previously described.56 Briefly, they were rinsed in PBS and fixed with 4% PFA (electron microscopy sciences) for 5 minutes, then rinsed twice in PBS. For intact cells labeling, an additional permeabilization step was performed for 10 minutes in IF buffer (Table 2). To keep the nuclear envelope intact, nuclei were simply incubated in blocking buffer (PBS supplemented with 3% goat serum) for 10 minutes. Primary antibodies were diluted in blocking buffer and incubated for 1 hour. Samples were rinsed 3 times in PBS. Secondary antibodies were incubated for 30 minutes; samples were then rinsed 3 times in PBS. All incubations were performed in the dark at room temperature. For confocal microscopy, coverslips were mounted on glass slides on a drop of Vectashield and sealed with nail polish. Slides were kept at 4 °C until imaging. For AFM microscopy, samples were kept at 4 °C in S500 until mounting in a coverslip holder designed to be adapted to JPK AFM and based on AttoFluor (Thermofisher) geometry.Labeling for dSTORM
25 mm high-precision type 1.5 glass coverslips of 0.17 mm thickness (Marienfeld) were sequentially washed in acetone, ethanol and water, then sonicated in 1 M KOH for 20 minutes. Coverslips were then extensively washed in milliQ water and air dried. For intact cells, the coverslips were UV-sterilized before seeding. 24 h later, cells were washed 2× with PBS and prefixed in 2.4% PFA for 30 s. Cells were then permeabilized with PB (Table 2) for 3 minutes, and fixation was completed in 2.4% PFA for 30 additional minutes. The fixing agent was quenched in QS buffer (Table 2) for 5 minutes, rinsed twice with PBS for 5 min and blocked for 10 min with PBS + 0.1% BSA.Staining with SNAP-ligand was performed for 2 h. Samples were then washed 3 times with PBS for 5 min, and post-fixed in PBS + 4% PFA for 5 minutes. All incubations were performed at room temperature. For imaging, the coverslips were set in an AttoFluor cell chamber and covered with 1.2 mL of dSTORM buffer (Table 2), covered by a glass coverslip to limit oxidation. The buffer was renewed after 2 hours.
Atomic force microscopy
Atomic force microscopy was performed on a JPK NanoWizard 4 combined with a CellHesion Z stage (Bruker Nano GmbH, Germany) and a homemade dSTORM setup.29 Experiments were performed in liquid in nuclei buffer, derived from Kramer et al.78 Bruker MSNL probes using cantilever E or Olympus OTR4 probes (with respective nominal stiffnesses 0.1 and 0.12 N m−1) were used for open nuclei. Both MSNL and OTR4 probes have pyramidal tips with a nominal tip radius of 2 nm for MSNL and 7 nm for OTR4. The spring constant of the cantilever was calibrated, prior to imaging, by acquisition of a force versus distance curve on a clean glass coverslip. Optical lever sensitivity was calibrated with a linear fit of the repulsive part of the force curve. Then, the spring constant was finally evaluated using the thermal noise method.79Images were recorded at resolution of 256 × 256 pixels in quantitative imaging (QI) mode. The force setpoint was below 300 pN to minimize the force applied while keeping the acquisition time short enough to avoid drift and facilitate the correlation with the super-resolution image. The length of the force curve was 200–300 nm to avoid tip–sample adhesion and facilitate tip detaching in each indentation cycle, with a tip approach-retract speed of typically 30 μm s−1.
Fluorescence microscopy
Confocal images were acquired on a Leica SP8 microscope with a 63×/1.4 NA Plan Apo objective. STED images were acquired on an Abberior Instrument Expert Line STED super-resolution microscope (Abberior Instruments GmbH, Göttingen, Germany), using 561 and 640 nm pulsed excitation laser sources and a pulsed STED laser operating at 775 nm and an 80 MHz repetition rate. The fluorescence excitation and collection were performed using a 100×/1.40 NA Plan Super Apochromat objective (Olympus, Hamburg). All acquisition operations were controlled by Imspector software (Abberior Instruments GmbH, Germany). Image series were recorded using Imspector software with the following parameters: pixel dwell time, 3.3 μs; 10 repetitions; pixel size, 15 nm, STED power (measured in the back focal plane of the objective), 250–300 mW. The gap delay between the excitation and STED laser pulses was 700 ps and the STED laser pulse duration was 8 ns.dSTORM experiments were performed on an AFM/dSTORM correlative setup29 consisting of a JPK NanoWizard 4 coupled with an inverted microscope equipped with an oil immersion Zeiss α-Plan-Apochromat 100×, 1.46 NA DIC objective. Proteins of interest in cells and nuclei were labeled with AlexaFluor 647.
An oxygen-scavenging dSTORM buffer containing 100 nm size Tetraspeck (Molecular Probes #T7279, 1/1000 dilution) fluorescent beads was loaded on the sample. A volume of 1.2 mL of this solution was used to fill the AttoFluor chamber. A glass coverslip was put on top of the AttoFluor to limit oxygen exchange with the ambient atmosphere.
dSTORM acquisition was performed by illuminating the sample with a 642 nm laser, set to 500 mW nominal power to obtain 3–7 kW cm−2 on the sample. 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames, with an acquisition time per frame of 20–30 ms, were recorded with an Andor iXon Ultra 897 EMCCD 512 × 512 pixels camera, with a pixel dimension of 16 μm. A 2× telescope was placed in front of the camera to obtain a final image magnification of 200x. 0–0.1 kW cm−2 of 405 nm was sometimes used for conversion from the dark state and to facilitate the dye blinking. The acquisition area was chosen so that a nucleus surrounded by several Tetraspeck fiducial markers was visible.
000 frames, with an acquisition time per frame of 20–30 ms, were recorded with an Andor iXon Ultra 897 EMCCD 512 × 512 pixels camera, with a pixel dimension of 16 μm. A 2× telescope was placed in front of the camera to obtain a final image magnification of 200x. 0–0.1 kW cm−2 of 405 nm was sometimes used for conversion from the dark state and to facilitate the dye blinking. The acquisition area was chosen so that a nucleus surrounded by several Tetraspeck fiducial markers was visible.
Data analysis
Images were treated with Fiji. dSTORM analysis was performed using the Fiji plug-in Thunderstorm.80 Drift was corrected thanks to the fiducial markers or in some cases by cross-correlation proposed by the plugin. To reconstruct the image, localizations were filtered to keep only intensity (number of photons) between 500 and 5000, localization precision below 45 nm and standard deviation of the fit of the point spread function (PSF) between 90 and 180 nm. For diameter measurements, analysis was performed using SMAP (Ries, 2020).AFM images were treated using Gwyddion software. Images were corrected by subtracting the mean plane, applying line correction and sometimes removing horizontal lines. Aberrant pixels (very high or very low) due to problems in the force curve were sometimes corrected by using a mask which interpolates a pixel with surrounding pixels. For crops around NPCs, data from images with scan sizes of 1 to 3 μm were used and crops of 300 nm × 300 nm were extracted. The Z scale was always ranging from 0 to 150 nm which is sufficient for the topography around NPCs. The offset and tilt of the nucleoplasmic ring was corrected using the Matlab algorithm from Stanley et al.32 To calculate the rotationally averaged height profiles, we measured the height profile along a 150 nm line crossing the pore center. We then rotated the line of 0.05π rad and repeated the process 20 times (Fig. S2A†), scanning the entire structure; the resulting radially averaged profiles are then plotted and fitted with three Gaussians corresponding to the scaffold rim and the basket distal ring (Fig. S2C†). The distance between the two extreme Gaussian peaks corresponds to pore diameter. Pore depth is calculated as the vertical distance between the average ring height and height at the pore center. This was automatically performed thanks to custom scripts written in Python.
For mechanical measurements from QI data, we used JPK data processing software. Experimentally calibrated spring constant was used for calculations, force curve baseline offset and tilt was subtracted before tip vertical position correction. Tip–sample contact point was determined using the Hertz/Sneddon model for a quadratic pyramid with tip pyramid angles modified to match MSNL probe geometry. The stiffness was measured by fitting the repulsive part of the force from the contact point over 20 nm. We did not generate Young modulus maps since its calculation is influenced by the tip–sample contact geometry that could vary when imaging very corrugated samples.
Author contributions
AV performed all AFM and AFM-correlative experiments; LC was involved in analysis of AFM force curves; PD was involved in AFM data analysis; PR and GB were involved in sample preparation and optimization; HH helped design and implement AFM experiments on nuclear envelope samples; OF helped design, optimize and analyze dSTORM data; CD performed confocal and STED experiments, wrote scripts for data analysis; PEM and CD designed and supervised the project. The manuscript was written by CD, AV and PEM and proofread by all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Jean-Bernard Fiche, Antoine Le Gall and Marcelo Nollmann for their help and advice regarding SMLM. We thank the members of the team ‘Integrative Biophysics of Membranes’ for their continuous support and feedback (https://integrativebiophysicsofmembranes.wordpress.com). The science-art model in Fig. 6 was designed and drawn by Zhanna Santybayeva (illustration4science.com).The CBS is a member of the France-BioImaging (FBI) and the French Infrastructure for Integrated Structural Biology (FRISBI), two national infrastructures supported by the French National Research Agency (ANR-10-INBS-04-01 and ANR-10-INBS-05, respectively). This work was supported by a grant from the French Research Agency (ANR-16-CE11-0004-01) and a Plan Cancer 2016 Equipment grant. Confocal and STED imaging were performed on the MRI imaging facility, member of the national France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04, “Investments for the future”). AV was supported by the Fondation ARC pour la recherche sur le cancer (grant no. DOC420190509114 and DOC42020070002524) and PR by the EpiGenMed Labex (ANR-10-LABX-12-01).
References
- E. C. Schirmer and L. Gerace, The nuclear membrane proteome: extending the envelope, Trends Biochem. Sci., 2005, 30, 551–558 CrossRef CAS PubMed.
- H. B. Schmidt and D. Görlich, Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles, Trends Biochem. Sci., 2016, 41, 46–61 CrossRef CAS PubMed.
- S. Albert, M. Schaffer, F. Beck, S. Mosalaganti, S. Asano and H. F. Thomas, et al., Proteasomes tether to two distinct sites at the nuclear pore complex, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 13726–13731 CrossRef CAS PubMed.
- R. Bayliss, A. H. Corbett and M. Stewart, The Molecular Mechanism of Transport of Macromolecules Through Nuclear Pore Complexes, Traffic, 2000, 1, 448–456 CrossRef CAS PubMed.
- V. Zila, E. Margiotta, B. Turoňová, T. G. Müller, C. E. Zimmerli and S. Mattei, et al., Cone-shaped HIV-1 capsids are transported through intact nuclear pores, Cell, 2021, 184(4), 1032–1046 CrossRef CAS PubMed.
- N. Panté and M. Kann, Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm, Mol. Biol. Cell, 2002, 13, 425–434 CrossRef PubMed.
- D. H. Lin and A. Hoelz, The Structure of the Nuclear Pore Complex (An Update), Annu. Rev. Biochem., 2019, 88, 725–783 CrossRef CAS PubMed.
- A. P. Schuller, M. Wojtynek, D. Mankus, M. Tatli, R. Kronenberg-Tenga and S. G. Regmi, et al., The cellular environment shapes the nuclear pore complex architecture, Nature, 2021, 598, 667–671 CrossRef CAS PubMed.
- B. Hampoelz, A. Andres-Pons, P. Kastritis and M. Beck, Structure and Assembly of the Nuclear Pore Complex, Annu. Rev. Biophys., 2019, 48, 515–536 CrossRef CAS PubMed.
- S. G. Brohawn, J. R. Partridge, J. R. R. Whittle and T. U. Schwartz, The nuclear pore complex has entered the atomic age, Structure, 2009, 17, 1156–1168 CrossRef CAS PubMed.
- S. Frey and D. Görlich, A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes, Cell, 2007, 130, 512–523 CrossRef CAS PubMed.
- B. W. Hoogenboom, L. E. Hough, E. A. Lemke, R. Y. H. Lim, P. R. Onck and A. Zilman, Physics of the Nuclear Pore Complex: Theory, Modeling and Experiment, Phys. Rep., 2021, 921, 1–53 CrossRef CAS PubMed.
- C. J. Bley, S. Nie, G. W. Mobbs, S. Petrovic, A. T. Gres and X. Liu, et al., Architecture of the cytoplasmic face of the nuclear pore, Science, 2022, 376, eabm9129 CrossRef CAS PubMed.
- S. Mosalaganti, A. Obarska-Kosinska, M. Siggel, R. Taniguchi, B. Turoňová and C. E. Zimmerli, et al., AI-based structure prediction empowers integrative structural analysis of human nuclear pores, Science, 2022, 376, eabm9506 CrossRef CAS PubMed.
- C. E. Zimmerli, M. Allegretti, V. Rantos, S. K. Goetz, A. Obarska-Kosinska and I. Zagoriy, et al., Nuclear pores dilate and constrict in cellulo, Science, 2021, 374, eabd9776 CrossRef CAS PubMed.
- J. Kosinski, S. Mosalaganti, A. von Appen, R. Teimer, A. L. DiGuilio and W. Wan, et al., Supp: Molecular architecture of the inner ring scaffold of the human nuclear pore complex, Science, 2016, 352, 363–365 CrossRef CAS PubMed.
- A. von Appen, J. Kosinski, L. Sparks, A. Ori, A. L. DiGuilio and B. Vollmer, et al., In situ structural analysis of the human nuclear pore complex, Nature, 2015, 526, 140–143 CrossRef CAS PubMed.
- P. Fontana, Y. Dong, X. Pi, A. B. Tong, C. W. Hecksel and L. Wang, et al., Structure of cytoplasmic ring of nuclear pore complex by integrative cryo-EM and AlphaFold, Science, 2022, 376, eabm9326 CrossRef CAS PubMed.
- S. Petrovic, D. Samanta, T. Perriches, C. J. Bley, K. Thierbach and B. Brown, et al., Architecture of the linker-scaffold in the nuclear pore, Science, 2022, 376, eabm9798 CrossRef CAS PubMed.
- M. Jarnik and U. Aebi, Toward a more complete 3-D structure of the nuclear pore complex, J. Struct. Biol., 1991, 107, 291–308 CrossRef CAS PubMed.
- S. Krull, J. Thyberg, B. Björkroth, H. R. Rackwitz and V. C. Cordes, Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket, Mol. Biol. Cell, 2004, 15(9), 4261–4277 CrossRef CAS PubMed.
- G. Rabut, V. Doye and J. Ellenberg, Mapping the dynamic organization of the nuclear pore complex inside single living cells, Nat. Cell Biol., 2004, 6, 1114–1121 CrossRef CAS PubMed.
- Z. Hakhverdyan, K. R. Molloy, S. Keegan, T. Herricks, D. M. Lepore and M. Munson, et al., Dissecting the Structural Dynamics of the Nuclear Pore Complex, Mol. Cell, 2021, 81(1), 153–165 CrossRef CAS PubMed.
- B. Souquet, E. Freed, A. Berto, V. Andric, N. Audugé and B. Reina-San-Martin, et al., Nup133 Is Required for Proper Nuclear Pore Basket Assembly and Dynamics in Embryonic Stem Cells, Cell Rep., 2018, 25, 1994 CrossRef CAS PubMed.
- I. Liashkovich, A. Meyring, A. Kramer and V. Shahin, Exceptional structural and mechanical flexibility of the nuclear pore complex, J. Cell Physiol., 2011, 226, 675–682 CrossRef CAS PubMed.
- V. J. Sabinina, M. J. Hossain, J.-K. Hériché, P. Hoess, B. Nijmeijer and S. Mosalaganti, et al., Three-dimensional superresolution fluorescence microscopy maps the variable molecular architecture of the nuclear pore complex, Mol. Biol. Cell, 2021, 32, 1523–1533 CrossRef CAS PubMed.
- Y. F. Dufrêne, T. Ando, R. Garcia, D. Alsteens, D. Martinez-Martin and A. Engel, et al., Imaging modes of atomic force microscopy for application in molecular and cell biology, Nat. Nanotechnol., 2017, 12, 295–307 CrossRef PubMed.
- A. Miranda, A. I. Gómez-Varela, A. Stylianou, L. M. Hirvonen, H. Sánchez and P. A. A. De Beule, How did correlative atomic force microscopy and super-resolution microscopy evolve in the quest for unravelling enigmas in biology?, Nanoscale, 2021, 13, 2082–2099 RSC.
- S. Dahmane, C. Doucet, A. Le Gall, C. Chamontin, P. Dosset and F. Murcy, et al., Nanoscale organization of tetraspanins during HIV-1 budding by correlative dSTORM/AFM, Nanoscale, 2019, 11, 6036–6044 RSC.
- T. D. Allen, S. A. Rutherford, S. Murray, H. S. Sanderson, F. Gardiner and E. Kiseleva, et al., Generation of cell-free extracts of Xenopus eggs and demembranated sperm chromatin for the assembly and isolation of in vitro-formed nuclei for Western blotting and scanning electron microscopy (SEM), Nat. Protoc., 2007, 2, 1173–1179 CrossRef CAS PubMed.
- V. Shahin, L. Albermann, H. Schillers, L. Kastrup, C. Schafer and Y. Ludwig, et al., Steroids dilate nuclear pores imaged with atomic force microscopy, J. Cell Physiol., 2005, 202, 591–601 CrossRef CAS PubMed.
- G. J. Stanley, A. Fassati and B. W. Hoogenboom, Atomic force microscopy reveals structural variability amongst nuclear pore complexes, Life Sci. Alliance, 2018, 1, e201800142 CrossRef PubMed.
- G. J. Stanley, B. Akpinar, Q. Shen, P. D. E. Fisher, C. P. Lusk and C. Lin, et al., Quantification of Biomolecular Dynamics Inside Real and Synthetic Nuclear Pore Complexes Using Time-Resolved Atomic Force Microscopy, ACS Nano, 2019, 13, 7949–7956 CrossRef CAS PubMed.
- Y. Sakiyama, A. Mazur, L. E. Kapinos and R. Y. H. Lim, Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy, Nat. Nanotechnol., 2016, 11, 719–723 CrossRef CAS PubMed.
- H. Wang and D. E. Clapham, Conformational changes of the in situ nuclear pore complex, Biophys. J., 1999, 77, 241–247 CrossRef CAS PubMed.
- O. L. Mooren, E. S. Erickson, D. Moore-Nichols and R. C. Dunn, Nuclear side conformational changes in the nuclear pore complex following calcium release from the nuclear membrane, Phys. Biol., 2004, 1, 125–134 CrossRef CAS PubMed.
- N.-P. Huang, M. Stubenrauch, J. Köser, N. Taschner, U. Aebi and M. Stolz, Towards monitoring transport of single cargos across individual nuclear pore complexes by time-lapse atomic force microscopy, J. Struct. Biol., 2010, 171, 154–162 CrossRef CAS PubMed.
- V. Shahin, T. Danker, K. Enss, R. Ossig and H. Oberleithner, Evidence for Ca2+- and ATP-sensitive peripheral channels in nuclear pore complexes, FASEB J., 2001, 15, 1895–1901 CrossRef CAS PubMed.
- L. Miorin, T. Kehrer, M. T. Sanchez-Aparicio, K. Zhang, P. Cohen and R. S. Patel, et al., SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 28344–28354 CrossRef CAS PubMed.
- F. Donnaloja, E. Jacchetti, M. Soncini and M. T. Raimondi, Mechanosensing at the Nuclear Envelope by Nuclear Pore Complex Stretch Activation and Its Effect in Physiology and Pathology, Front Physiol., 2019, 10, 896 CrossRef CAS PubMed.
- S. Sakuma and M. A. D'Angelo, The roles of the nuclear pore complex in cellular dysfunction, aging and disease, Semin. Cell Dev. Biol., 2017, 68, 72–84 CrossRef CAS PubMed.
- K. Zhang, L. Miorin, T. Makio, I. Dehghan, S. Gao and Y. Xie, et al., Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression, Sci. Adv., 2021, 7(6), eabe7386 CrossRef CAS PubMed.
- T. U. Schwartz, Solving the nuclear pore puzzle, Science, 2022, 376, 1158–1159 CrossRef CAS PubMed.
- J. Sellés, M. Penrad-Mobayed, C. Guillaume, A. Fuger, L. Auvray and O. Faklaris, et al., Nuclear pore complex plasticity during developmental process as revealed by super-resolution microscopy, Sci. Rep., 2017, 7, 14732 CrossRef PubMed.
- M. Asally, Y. Yasuda, M. Oka, S. Otsuka, S. H. Yoshimura and K. Takeyasu, et al., Nup358, a nucleoporin, functions as a key determinant of the nuclear pore complex structure remodeling during skeletal myogenesis, FEBS J., 2011, 278, 610–621 CrossRef CAS PubMed.
- K. T. Sapra, Z. Qin, A. Dubrovsky-Gaupp, U. Aebi, D. J. Müller and M. J. Buehler, et al., Nonlinear mechanics of lamin filaments and the meshwork topology build an emergent nuclear lamina, Nat. Commun., 2020, 11, 6205 CrossRef CAS PubMed.
- A. Ori, N. Banterle, M. Iskar, A. Andrés-Pons, C. Escher and K. H. Bui, et al., Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines, Mol. Syst. Biol., 2013, 9, 648 CrossRef CAS PubMed.
- J. V. Thevathasan, M. Kahnwald, K. Cieśliński, P. Hoess, S. K. Peneti and M. Reitberger, et al., Nuclear pores as versatile reference standards for quantitative superresolution microscopy, Nat. Methods, 2019, 16, 1045–1053 CrossRef CAS PubMed.
- A. Rose and I. Meier, Scaffolds, levers, rods and springs: diverse cellular functions of long coiled-coil proteins, Cell. Mol. Life Sci., 2004, 61, 1996–2009 CrossRef CAS PubMed.
- D. A. D. Parry, R. D. B. Fraser and J. M. Squire, Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure, J. Struct. Biol., 2008, 163, 258–269 CrossRef CAS PubMed.
- M. Bachmann, F. Fiederling and M. Bastmeyer, Practical limitations of superresolution imaging due to conventional sample preparation revealed by a direct comparison of CLSM, SIM and dSTORM, J. Microsc., 2016, 262, 306–315 CrossRef CAS PubMed.
- W. Hell, Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy, Opt. Lett., 1994, 19, 780 CrossRef PubMed.
- J. Bückers, L. Kastrup and S. W. Hell, Compact Multi-Color STED Microscopy, Biophys. J., 2011, 100, 354a–355a CrossRef.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov and O. Ronneberger, et al., Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- T. C. Walther, M. Fornerod, H. Pickersgill, M. Goldberg, T. D. Allen and I. W. Mattaj, The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins, EMBO J., 2001, 20, 5703–5714 CrossRef CAS PubMed.
- C. M. Doucet, J. A. Talamas and M. W. Hetzer, Cell cycle-dependent differences in nuclear pore complex assembly in metazoa, Cell, 2010, 141, 1030–1041 CrossRef CAS PubMed.
- B. Vollmer, M. Lorenz, D. Moreno-Andrés, M. Bodenhöfer, P. De Magistris and S. A. Astrinidis, et al., Nup153 Recruits the Nup107-160 Complex to the Inner Nuclear Membrane for Interphasic Nuclear Pore Complex Assembly, Dev. Cell, 2015, 1–12 Search PubMed.
- S. Milles and E. A. Lemke, Single molecule study of the intrinsically disordered FG-repeat nucleoporin 153, Biophys. J., 2011, 101, 1710–1719 CrossRef CAS PubMed.
- S. Frey, R. P. Richter and D. Gorlich, FG-Rich, Repeats of Nuclear Pore Proteins Form a Three-Dimensional Meshwork with Hydrogel-Like Properties, Science, 2006, 314, 815–817 CrossRef CAS PubMed.
- R. Y. H. Lim, N.-P. Huang, J. Köser, J. Deng, K. H. A. Lau and K. Schwarz-Herion, et al., Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9512–9517 CrossRef CAS PubMed.
- N. B. Eisele, S. Frey, J. Piehler, D. Görlich and R. P. Richter, Ultrathin nucleoporin phenylalanine-glycine repeat films and their interaction with nuclear transport receptors, EMBO Rep., 2010, 11, 366–372 CrossRef CAS PubMed.
- K. H. Bui, A. von Appen, A. L. DiGuilio, A. Ori, L. Sparks and M.-T. Mackmull, et al., Integrated Structural Analysis of the Human Nuclear Pore Complex Scaffold, Cell, 2013, 155, 1233–1243 CrossRef CAS PubMed.
- T. Maimon, N. Elad, I. Dahan and O. Medalia, The human nuclear pore complex as revealed by cryo-electron tomography, Structure, 2012, 20, 998–1006 CrossRef CAS PubMed.
- J. Mahamid, S. Pfeffer, M. Schaffer, E. Villa, R. Danev and L. K. Cuellar, et al., Visualizing the molecular sociology at the HeLa cell nuclear periphery, Science, 2016, 351, 969–972 CrossRef CAS PubMed.
- A. Bestembayeva, A. Kramer, A. A. Labokha, D. Osmanović, I. Liashkovich and E. V. Orlova, et al., Nanoscale stiffness topography reveals structure and mechanics of the transport barrier in intact nuclear pore complexes, Nat. Nanotechnol., 2015, 10, 60–64 CrossRef CAS PubMed.
- T. Danker, M. Mazzanti, R. Tonini, A. Rakowska and H. Oberleithner, Using atomic force microscopy to investigate patch-clamped nuclear membrane, Cell Biol. Int., 1997, 21, 747–757 CrossRef CAS PubMed.
- B. Maco, B. Fahrenkrog, N.-P. Huang and U. Aebi, Nuclear pore complex structure and plasticity revealed by electron and atomic force microscopy, Methods Mol. Biol., 2006, 322, 273–288 CrossRef CAS PubMed.
- E. Kozlova, A. Chernysh, E. Manchenko, V. Sergunova and V. Moroz, Nonlinear Biomechanical Characteristics of Deep Deformation of Native RBC Membranes in Normal State and under Modifier Action, Scanning, 2018, 2018, 1810585 CrossRef PubMed.
- K. J. Burkhart, T. E. Nowak, J. Blum, S. Kuhn, M. Welker and W. Sternstein, et al., Influence of formalin fixation on the biomechanical properties of human diaphyseal bone, Biomed. Tech., 2010, 55, 361–365 Search PubMed.
- J. Ma, Z. Liu, N. Michelotti, S. Pitchiaya, R. Veerapaneni and J. R. Androsavich, et al., High-resolution three-dimensional mapping of mRNA export through the nuclear pore, Nat. Commun., 2013, 4, 1–9 Search PubMed.
- V. Aksenova, A. Smith, H. Lee, P. Bhat, C. Esnault and S. Chen, et al., Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway, Nat. Commun., 2020, 11, 4577 CrossRef CAS PubMed.
- A. Ashkenazy-Titelman, Y. Shav-Tal and R. H. Kehlenbach, Into the basket and beyond: the journey of mRNA through the nuclear pore complex, Biochem. J., 2020, 477, 23–44 CrossRef CAS PubMed.
- Y. Li, V. Aksenova, M. Tingey, J. Yu, P. Ma and A. Arnaoutov, et al., Distinct roles of nuclear basket proteins in directing the passage of mRNA through the nuclear pore, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(37), e2015621118 CrossRef CAS PubMed.
- M. Tingey and W. Yang, Unraveling docking and initiation of mRNA export through the nuclear pore complex, Bioessays, 2022, e2200027 CrossRef PubMed.
- M. Oeffinger and D. Zenklusen, To the pore and through the pore: a story of mRNA export kinetics, Biochim. Biophys. Acta, 2012, 1819, 494–506 CrossRef CAS PubMed.
- J. Fernandez-Martinez, S. J. Kim, Y. Shi, P. Upla, R. Pellarin and M. Gagnon, et al., Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform, Cell, 2016, 167(5), 1215–1228 CrossRef CAS PubMed.
- J. Harborth, S. M. Elbashir, K. Bechert, T. Tuschl and K. Weber, Identification of essential genes in cultured mammalian cells using small interfering RNAs, J. Cell Sci., 2001, 114, 4557–4565 CrossRef CAS PubMed.
- A. Kramer, Y. Ludwig, V. Shahin and H. Oberleithner, A pathway separate from the central channel through the nuclear pore complex for inorganic ions and small macromolecules, J. Biol. Chem., 2007, 282, 31437–31443 CrossRef CAS PubMed.
- R. Proksch, T. E. Schäffer, J. P. Cleveland, R. C. Callahan and M. B. Viani, Finite optical spot size and position corrections in thermal spring constant calibration, Nanotechnology, 2004, 15, 1344 CrossRef.
- M. Ovesný, P. Křížek, J. Borkovec, Z. Svindrych and G. M. Hagen, ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging, Bioinformatics, 2014, 30, 2389–2390 CrossRef PubMed.
- M. S. Mohamed, A. Kobayashi, A. Taoka, T. Watanabe-Nakayama, Y. Kikuchi, M. Hazawa, T. Minamoto, Y. Fukumori, N. Kodera, T. Uchihashi, T. Ando and R. W. Wong, High-Speed Atomic Force Microscopy Reveals Loss of Nuclear Pore Resilience as a Dying Code in Colorectal Cancer Cells., ACS Nano, 2017, 11(6), 5567–5578 CrossRef CAS PubMed.
- M. S. Mohamed, M. Hazawa, A. Kobayashi, L. Guillaud, T. Watanabe-Nakayama, M. Nakayama, H. Wang, N. Kodera, M. Oshima, T. Ando and R. W. Wong, Spatiotemporally tracking of nano-biofilaments inside the nuclear pore complex core., Biomaterials, 2020, 256, 120198 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr06034e |
| This journal is © The Royal Society of Chemistry 2023 |