Open Access Article
Open Access ArticleEnhanced antibacterial activity of a novel silver-based metal organic framework towards multidrug-resistant Klebsiella pneumonia†
Payam B.
Hassan
a,
Sameera Sh.
Mohammed Ameen
b,
Lana
Mohammed
c,
Sirwan M.
Muhammed Ameen
a and
Khalid M.
Omer
 *d
*d
aDepartment of Biology, College of Science, University of Sulaimani, Sulaymaniyah, 46002, Kurdistan Region, Iraq
bDepartment of Chemistry, College of Science, University of Zakho, Zakho, Kurdistan Region, Iraq
cDepartment of Medical Laboratory, College of Health and Medical Technology, Sulaimani Polytechnic University, Sulaymaniyah, Iraq
dDepartment of Chemistry, College of Science, University of Sulaimani, Qliasan St. 46002, Sulaymaniyah, Kurdistan Region, Iraq. E-mail: khalid.omer@univsul.edu.iq
First published on 12th June 2024
Abstract
The growth and spread of multidrug-resistant bacterial species, such as Klebsiella pneumoniae, pose a serious threat to human health and require the development of innovative antibacterial agents. The search for an acceptable, safe, and efficient antibacterial is a matter of significant concern. In the present work, silver-based metal–organic frameworks (Ag-MOFs) showed efficient antibacterial activity against multidrug-resistant K. pneumoniae (KBP 11) with a minimum inhibitory concentration and minimum bactericidal concentration of 10 μg mL−1. Moreover, the Ag-MOF showed enhanced antibacterial activity compared to silver ions and silver nanoparticles. Our experimental investigation showed that the antibacterial efficacy is attributed to the production of reactive oxygen species and the release of cellular constituents, such as K+ ions and proteins. The MOF scaffold enhances the stability and controlled release of silver ions, enabling sustained antibacterial activity and minimizing the risk of bacterial resistance development. Additionally, the MOF class, due to the high surface area and porous nature, enhances the transfer of bacteria into and on the surface of the MOF.
Introduction
Multidrug resistant bacteria (MDRB) have emerged as a major human health concern, offering a serious challenge to traditional antibiotic therapy.1,2 The advent of these resistant strains requires the development of innovative materials for fighting bacterial infections.3,4K. pneumoniae is a non-motile Gram-negative bacterium, which belongs to the Enterobacteriaceae family, commonly present in the respiratory and gastrointestinal tracts of humans.5,6 It causes many infections, including pneumonia, urinary tract infections, septicemia, wound infections, and nosocomial infections within healthcare settings. Significantly, the capacity of this organism to acquire resistance to several antibiotics has resulted in the formation of strains that are resistant to numerous drugs, posing considerable difficulties in medical settings. Due to its clinical impact and potential for antibiotic resistance, effective detection, infection control strategies, and appropriate antibiotic usage are crucial in managing K. pneumoniae infections.7,8
Metal–organic frameworks (MOFs), which consist of crystalline microporous materials made from metal nodes connected by organic linkers, have gained significant interest owing to their wide-ranging properties and potential uses.9–14
MOFs have emerged as interesting possibilities for combating MDRB in recent years due to their unique antibacterial properties.15–17 The growing problem of multidrug resistance has encouraged researchers to design and explore alternative strategies beyond conventional antibiotics. MOFs, with their tunable composition, catalytic activity, high surface area, relative biocompatibility, and porous structure, offer a range of distinctive characteristics that can be harnessed to combat bacterial infections, and they can also be used in bioimaging, biosensing for tumor therapy, and as cargo delivery agents for cancer treatment.18–22
Few reports of pure silver-based MOFs with no doping and functionalization have been reported as antimicrobial agents. Lu et al.23 prepared two biocompatible silver carboxylate MOFs, 1 and 2. Both MOFs showed antimicrobial activities towards Gram-negative bacteria, Escherichia coli has MIC values of 5–10 and 10–15 ppm. While for a Gram-positive bacterium such as Staphylococcus aureus, MIC values were 10–15 and 15–20 ppm. Jaros et al.24 prepared a novel bioactive pure Ag-based MOF (bioMOF 1) from silver(I) oxide, 1,3,5-triaza-7-phosphaadamantane (PTA), and pyromellitic acid (H4pma). The bioMOF 1 showed diverse bioactivity as a robust antimicrobial agent against pathogenic strains of typical Gram-negative bacteria (Escherichia coli and Pseudomonas aeruginosa), Gram-positive bacteria (Staphylococcus aureus), and yeast (Candida albicans). The minimum inhibitory concentration (MIC) value for Gram-negative bacteria was 5 μg mL−1.
The principal reason behind the antibacterial activity of pure Ag-MOFs stems from the release of silver ions. The primary objective revolves around liberating these silver ions from the MOF framework, whether through the weakened bond between the ligand and silver metal or the cooperative antibacterial impact with silver. This process facilitates a cascade of reactions involving cell membrane denaturation due to ROS generation or DNA intercalation, hampering cell transcription and replication, and ultimately culminating in bacterial demise.16,25,26 Many other articles were reports on using pure Ag-MOF as an antimicrobial agent.27–29 However, the majority of the antimicrobial-based Ag-MOFs suffer from the complex structure of the linker and high MIC. In addition, the cytotoxicity of MOFs is of enormous importance. Available toxicity data are rare, most of which are associated with the inorganic and organic precursors rather than the MOFs directly. On comparing MOF cytotoxicity to other commercialized nanosystems, it is lower and strongly associated with its composition.30
Zhang et al.17 developed a Ag2[HBTC][im]–polylactic acid (PA) fibrous mat, an antibacterial wound dressing, by utilizing silver ion-MOFs, which have the ability to progressively release powerful antibacterial Ag+ ions to replace antibiotics as the antibacterial agent. The in vivo experimental results demonstrated that when used as an antibacterial wound dressing, the wound healing ratio could reach 99.9%, which was significantly higher than that in other comparison groups. This fabricated fibrous mat serves as a valuable guide for the subsequent creation of antibiotic-free wound dressing. In addition, the characterization results showed thay the prepared fibrous mat possesses excellent water and thermal stability, up to 300 °C.
He et al.31 synthesized a novel Ag@MOF@PDA by functionalizing cyclodextrin metal–organic frameworks (CD-MOFs) with polydopamine (PDA) and ultrafine silver nanoparticles (Ag NPs). Under a NIR laser, the constructed Ag@MOF@PDA enhanced the release rate of Ag+ ions in a controllable way and also efficiently damaged the integrity of biofilms via photothermal therapy (PTT). The results demonstrated that the Ag@MOF@PDA nanosystem is effective in healing bacterial and biofilm infections with minimal biotoxicity, and providing a potential antibiotic-free alternative in the “post-antibiotic era.”
In the present work, submicron sized Ag-based MOFs without functionalization and/or encapsulation were prepared from common organic linkers, terephthalic acid, and silver ions. In an effort to broaden the narrow spectrum of antibiotics active against K. pneumonia, here, we examined Ag-MOF's in vitro susceptibility to clinical isolates of MDR K. pneumonia, which are commonly seen in clinical microbiology labs. Moreover, the mechanisms of antibacterial action are also investigated.
Experimental section
Materials and methods
All the information on chemicals and instrumentation are briefly provided in the ESI.†Synthesis of the Ag-MOF
Briefly, 2 mmol of AgNO3, 4 mmol of TP and 8 mmol of TEA were dissolved in 100 mL solution of methanol (50%). Then, the resulting solution was mixed and stirred for 30 minutes and left for 12 hours. The white powder of Ag-MOF was produced and washed with methanol and water many times, respectively. Then, the produced Ag-MOF was dried at 70 °C in a vacuum oven for 24 h.Bacterial strain used in this study
For the antibacterial experiment, K. pneumonia was selected as a model. The multidrug-resistant K. pneumonia was isolated from wound swab specimens in Sulaymaniyah Shar Hospital. Molecular identification was applied for further confirmation of the isolate by PCR amplification of 16S rDNA using the primers Lee et al.,32 16S rRNA (sense, 5-TGGCTCAGATTGAACGCTGGCGGC; antisense, 5 TACCTTGTTACGACTTCACCCCA). Using a PCR thermocycler (Techne, USA), the PCR was carried out with 35 cycles at 94 °C for 30 seconds, 62 °C for 40 seconds, and 72 °C for 40 seconds, with a final extension of 5 min at 72 °C and a 4 °C hold. Following the PCR, agarose gel electrophoresis was used to examine the reaction result (1500 bp). Then, the purification of PCR products was applied according to the manufacturer's instructions on the genomic DNA purification kit (Geneaid, Taiwan). DNA sequencing was performed by Macrogen (Seoul, South Korea) using a 3500xL genetic analyzer (Applied Biosystems). Later, sequences were modified manually using the BioEdit software (7.2.5 version). The consensus sequence was BLASTed against the GenBank NCBI's contents after it was submitted to GenBank (National Center for Biotechnology Information, Bethesda, MD, USA) for an entry number. The phylogenetic tree was constructed (Fig. S1, ESI†) by the MEGA 11 program with the neighbor joining method, to represent the phylogenetic position of strain.33 The antibiotic susceptibility test was carried out for the isolated K. pneumonia using automated system VITEK2 with cards AST-N417 for the following antibiotics: piperacillin/tazobactam, cefazolin, cefuroxime, gentamicin, ciprofloxacin, amoxicillin/clavulanic acid, cefuroxime axeti, ceftazidime, ceftriaxone, cefepime, ertapenem, imipenem, meropenem, amikacin, fosfomycin, nitrofurantoin, and trimethoprim/sulfamethoxazole. Additionally, the disc diffusion method according to the Kirby–Bauer method34 was also used for the determination of extra antibiotic sensitivity profiles, using the following antibacterial discs: CFM (cefixime 5 μg), CD (cefdinir 5 μg), CPC (cefpodoxime–clavulanate 40 μg), CPD (cefpodoxime 10 μg), CT (colistin 10 μg), S (streptomycin 10 μg), MXF (moxifloxacin 5 μg), LEV (levofloxacin 5 μg), DO (doxycycline 30 μg), TE (tetracycline 30 μg), TGC (tigecycline 15 μg), FO (fosfomycin 200 μg), AZM (azithromycin 15 μg), and CLR (clarithromycin 15 μg).Antibacterial activity determination
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 minutes at 4 °C, and subsequently, the supernatant was discarded. The bacteria were washed and then resuspended in 1.8 mL of PBS buffer. Afterwards, the bacterial suspensions were treated with 200 μL of the prepared solution and incubated at a temperature of 37 °C for 30 minutes in the dark. Then, the cells were subjected to thermal degradation at a temperature of 95 °C for 15 minutes. An ELISA reader was used to detect the absorbance of the resultant lysate at a wavelength of 520 nm. The experiments were performed in triplicate.
000 g for 10 minutes at 4 °C, and subsequently, the supernatant was discarded. The bacteria were washed and then resuspended in 1.8 mL of PBS buffer. Afterwards, the bacterial suspensions were treated with 200 μL of the prepared solution and incubated at a temperature of 37 °C for 30 minutes in the dark. Then, the cells were subjected to thermal degradation at a temperature of 95 °C for 15 minutes. An ELISA reader was used to detect the absorbance of the resultant lysate at a wavelength of 520 nm. The experiments were performed in triplicate.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 minutes at 4 °C; subsequently, the supernatant was collected. A volume of 500 μL of the supernatant was mixed with 250 μL of a solution containing 20% trichloroacetic acid. The resulting mixture was subjected to centrifugation at 10
000 g for 10 minutes at 4 °C; subsequently, the supernatant was collected. A volume of 500 μL of the supernatant was mixed with 250 μL of a solution containing 20% trichloroacetic acid. The resulting mixture was subjected to centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 15 minutes, while maintaining a temperature of 4 °C. Afterwards, a volume of 300 μL of the supernatant was mixed with 200 μL of a TBA buffer solution containing 0.8% TBA in a 2 M NaOH solution. The solution was heated at a temperature of 95 °C for one hour. Subsequently, it was allowed to cool down to ambient temperature. At this point, the absorbance was measured (at a wavelength of 532 nm) using a microtiter ELISA reader. The experiments were conducted in triplicate.
000 g for 15 minutes, while maintaining a temperature of 4 °C. Afterwards, a volume of 300 μL of the supernatant was mixed with 200 μL of a TBA buffer solution containing 0.8% TBA in a 2 M NaOH solution. The solution was heated at a temperature of 95 °C for one hour. Subsequently, it was allowed to cool down to ambient temperature. At this point, the absorbance was measured (at a wavelength of 532 nm) using a microtiter ELISA reader. The experiments were conducted in triplicate.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 30 minutes. The supernatant was used to determine the quantity of proteins that had leaked from the treated cells using the precise technique described in ref. 39. In addition, the determination of potassium ion leakage in the bacterial suspensions was conducted at various time intervals (0, 30, 60, 90, and 120 minutes) following treatment. This was accomplished using the Na-tetraphenylborate (Na-TPB) manual kit, with the measurement being dependent on the turbidity of the suspensions. In summary, a 25 μL supernatant was blended with 1 mL of Na-TPB reagent and subjected to incubation at ambient temperature for 5 minutes. Concurrently, standard potassium was employed as a positive control. Subsequently, the turbidity was quantified through photometric analysis at a wavelength of 578 nm using a spectrophotometer. The experiments were conducted in triplicate.
000 g for 30 minutes. The supernatant was used to determine the quantity of proteins that had leaked from the treated cells using the precise technique described in ref. 39. In addition, the determination of potassium ion leakage in the bacterial suspensions was conducted at various time intervals (0, 30, 60, 90, and 120 minutes) following treatment. This was accomplished using the Na-tetraphenylborate (Na-TPB) manual kit, with the measurement being dependent on the turbidity of the suspensions. In summary, a 25 μL supernatant was blended with 1 mL of Na-TPB reagent and subjected to incubation at ambient temperature for 5 minutes. Concurrently, standard potassium was employed as a positive control. Subsequently, the turbidity was quantified through photometric analysis at a wavelength of 578 nm using a spectrophotometer. The experiments were conducted in triplicate.
Results and discussion
Characterization of Ag-MOF
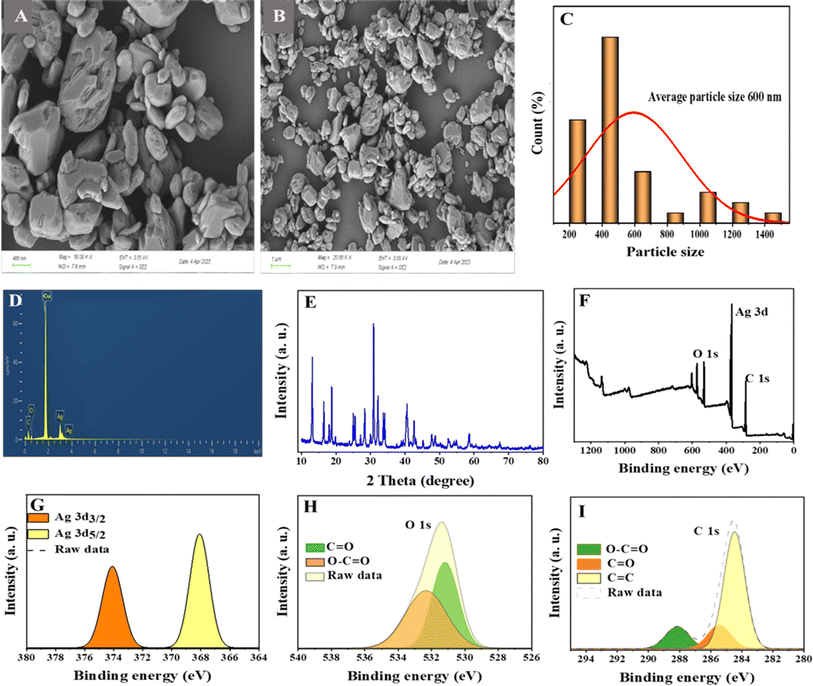
Various characterization techniques were carried out to investigate the morphology, shape and size of the as prepared Ag-MOF. Fig. 1A and B show the FE-SEM images of the Ag-MOFs at two different magnifications. Fig. 1C shows the histogram of particle size diameter of Ag-MOF, with the average particle size between 300 and 600 nm. Elemental analysis using the EDS technique was used to confirm the presence of elements of the Ag-MOF. Fig. 1D shows the presence of Ag, C, and O atoms which confirms the formation of the Ag-MOF. Fig. 1E shows the XRD spectrum of the Ag-MOF. The XPS spectra were obtained to examine the chemical composition of the Ag-MOFs. The survey spectrum (Fig. 1F) shows the peaks of O, C, and Ag. The resolved spectra of Ag in Fig. 1G display two well-resolved peaks of Ag 3d3/2 (373.8 eV) and Ag 3d5/2 (367.9 eV), respectively.40,41 The O 1s spectra as shown in Fig. 1H have two main peaks at 531.8 and 532.44 eV which are attributed to the O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds in the Ag-MOF.42Fig. 1I shows the C 1s spectrum which reveals three peaks at 288.52, 285.86 and 284.56 eV which are attributed to O–C
O bonds in the Ag-MOF.42Fig. 1I shows the C 1s spectrum which reveals three peaks at 288.52, 285.86 and 284.56 eV which are attributed to O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C.42–44
C.42–44
Identification of the bacterial strain
The multidrug-resistant K. pneumonia was isolated from clinical samples at Shar Hospital in Sulaymaniyah, Iraq. The resulting 16S rRNA sequences for the isolate were compared to the closely comparable sequences present in the NCBI database. The strain exhibited a sequence similarity of 99% to K. pneumoniae. The 16S rRNA sequence was submitted to the GenBank (NCBI) with an accession number of OR500977. A phylogenetic tree was created using the 16S rRNA gene sequencing of the isolate. Based on the neighbor joining phylogenetic data, our isolate of K. pneumonia KPB11 showed a cluster with Klebsiella sp., as seen in Fig. S1 (ESI†). In addition, the result of antibiotic sensitivity testing by using a VITEK 2 system and the Kirby–Bauer method showed that the strain has resistance to the following antibiotics, including cefepime, ertapenem, piperacillin/tazobactam, cefixime, cefdinir, cefazolin, cefuroxime, amoxicillin/clavulanic acid, cefuroxime axeti, ceftazidime, ceftriaxone, imipenem, meropenem, ciprofloxacin, nitrofurantoin, trimethoprim/sulfamethoxazole, cefpodoxime–clavulanate, cefpodoxime, moxifloxacin, levofloxacin, doxycycline, tetracycline, fosfomycin, azithromycin, and clarithromycin as shown in Table S1 (ESI†).Antibacterial activity
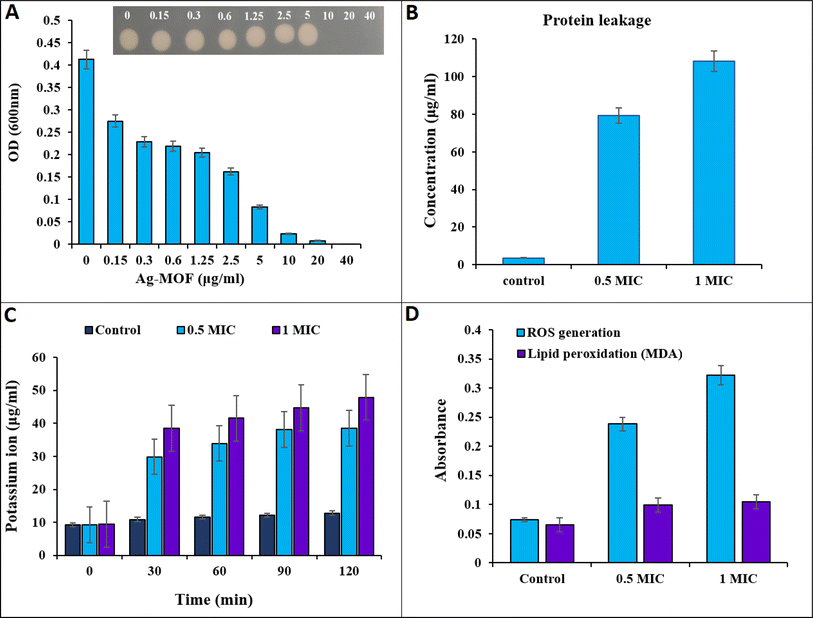
MOFs are new fascinating microporous biomaterials that found many uses in pharmaceutical and biological applications because they provide an alternative to existing antibacterial agents and have the potential to be an innovative solution to the issue of microbial drug resistance.30 At different concentrations, the antibacterial activity of the synthesized Ag-MOF was investigated using MIC and MBC evaluations. MBC is the lowest concentration of an antibiotic substance necessary to kill 99.9% of the bacterial population.45 MIC is the lowest concentration of an antimicrobial agent that inhibits bacterial growth without killing the bacteria throughout the incubation. The results demonstrated significant dose-dependent antimicrobial activity against the examined bacterium. The MIC and MBC values of Ag-MOF were 10 μg mL−1 (Fig. 2A). Furthermore, bacterial proliferation was shown to decrease while the concentration of Ag-MOF increased. According to the findings, Ag MOF exhibited strong antibacterial action against multidrug-resistant K. pneumonia KPB11. Multiple investigations have shown that Ag-MOFs have antibacterial properties against a variety of bacteria, such as E. coli, B. subtilis, and S. aureus.23,46Table 1 displays previous studies examining antibacterial agents and their antibacterial activity, in contrast to our samples. The findings indicate that Ag-MOF has potential for being used as an antibacterial agent in biological applications. The antibacterial efficacy of Ag-based MOFs is not solely determined by the ligand, but also relies on the liberation of the core metal ion. The Ag-MOF solution releases a greater quantity of Ag+ ions compared to the Ag-NP solution. Consequently, the antibacterial activity of Ag-based MOFs can be attributed to the release of Ag+ ions.23| Antibacterial agent | Bacterial strains | MIC (μg mL−1) | References |
|---|---|---|---|
| [(AgL)NO3]·2H2O | Escherichia coli | 300 | 61 |
| Staphylococcus aureus | 297 | ||
| Ag-dop γ-Fe2O3@SiO2@ZIF-8-Ag (FSZ-Ag) | Escherichia coli | 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
62 |
| Staphylococcus aureus | 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
||
| Cu(I)MOF-AgCl/Ag | Escherichia coli | 7.8 | 63 |
| Staphylococcus aureus | 16 | ||
| Ni-gallate MOF | Pseudomonas aeruginosa | 225 | 64 |
| Staphylococcus aureus | 35 | ||
| ZIF-8@SnO2@CoFe2O4 | Escherichia coli | 1250 | 65 |
| Staphylococcus aureus | 2500 | ||
| Ag NPs | Staphylococcus aureus | 625 | 66 |
| Ag NPs | Klebsiella pneumonia | 12.5 | 67 |
| Salmonella enteritidis | 25 | ||
| Na-Alg-s-AgNPs@SiO2 | Klebsiella pneumonia | 95 | 68 |
| Enterobacter cloacae | 60 | ||
| Bacillus cereus | 80 | ||
| Ag-MOF | K. pneumonia KPB11 | 10 | This study |
Antibacterial mechanism
The measurement of proteins and potassium ion leakage from the cell membrane of K. pneumonia KPB11 was conducted to investigate the bactericidal mechanism of action of Ag-MOF (Fig. 2). The data presented in Fig. 2B illustrate a substantial increase in protein leakage simultaneously with an increase in the concentration of Ag-MOF. At the outset, a protein leakage of 3.66 μg mL−1 was observed in untreated cells. However, cells treated with Ag-MOF at concentrations of 0.5 MIC and 1 MIC showed protein leakages of 79.3 μg mL−1 and 108.3 μg mL−1, respectively.Fig. 2C illustrates the amount of potassium ions that were leaked from cells when subjected to varying concentrations of Ag-MOF. The cells exposed to 1 MIC of Ag-MOF exhibited the highest diffusion of potassium ions. The potassium ion leakage levels increased from less than 9.41 μg mL−1 in the control cells to 38.4, 41.5, 44.7, and 47.8 μg mL−1 at time intervals of 30, 60, 90, and 120 minutes, respectively. Furthermore, the release of potassium ions from cells that were exposed to a concentration of 0.5 MIC of Ag-MOF was measured at four different time intervals: 30, 60, 90, and 120 minutes. The recorded values for potassium ion leakage were 29.8, 33.9, 38.1, and 38.5 μg mL−1, respectively.
The exact mechanism underlying the antibacterial activity of Ag-MOF remains unclear, and only a small group of researchers have conducted studies on this topic, although the antibacterial mechanisms of Ag nanoparticles have been extensively investigated for a long time. It is worth noting that the antibacterial activities exhibited by MOFs containing Ag might vary from those observed in commercial Ag nanoparticles. However, several strategies have been proposed to address this issue. The primary mechanism of action may be attributed to the elevated electrical conductivity exhibited by metallic silver.47 This results in the generation of static electricity that exhibits a pronounced affinity for sulphur proteins, thereby causing silver ions to adhere to the cellular membrane. The adhesion of silver ions might increase the permeability of the cytoplasmic membrane, ultimately resulting in the disruption of the cell membrane. Furthermore, upon entry into the cells, silver ions induce enzyme inactivation and the generation of ROS48,49 ROS have been found to enhance the occurrence of cell membrane rupture and disrupt processes including DNA replication, transcription and translation.50–53
The current study clearly demonstrates that Ag-MOF exhibits significant capability in disrupting bacterial cell membranes, leading to the release of cellular contents (Fig. 2). This phenomenon has been quantified through the measurement of membrane proteins and the leakage of potassium ions from the cells. Prior research has shown that the bactericidal impact of Ag-MOF on both types of bacteria, Gram-negative E.coli and Gram-positive S. aureus, is due to the disturbance of cell membrane permeability.23
To better understand the bactericidal mechanism of Ag-MOF against K. pneumonia KPB11 ROS and lipid peroxidation production were measured in cells treated with Ag-MOF. The results presented in Fig. 2D demonstrate a significant increase in ROS production in K. pneumoniae KPB11. Specifically, untreated cells displayed a ROS production level of 0.07 μg mL−1, whereas cells treated with a 0.5 MIC of Ag-MOF showed a notably higher ROS production level of 0.23 μg mL−1. This observed increase represents a more than threefold elevation in ROS production. Simultaneously, the production of ROS increased considerably to 0.47 μg mL−1 when exposed to a concentration of 1 MIC of Ag-MOF, resulting in a fourfold enhancement. The untreated cells exhibited a lipid peroxidation level of 0.065 μg mL−1, whereas the cells treated with a 0.5 MIC of MOF-Ag demonstrated an increase to 0.099 μg mL−1, indicating a 1.5-fold elevation. Nevertheless, the lipid peroxidation levels exhibited a subsequent elevation in cells subjected to a concentration of 1 MIC of MOF-Ag, reaching a value of 0.104 μg mL−1. This increment corresponds to an approximate 1.6-fold augmentation. Similarly, Mohammed and Hamzah54 found that treating pandrug resistant K. pneumoniae with AgNPs led to a notable increase in ROS production. Moreover, in a study conducted by Hamida et al.,55 it was discovered that the exposure of K. pneumoniae to AgNPs resulted in a significant increase in the generation of ROS. This increase in ROS levels coincided with a reduction in the functioning of antioxidant enzymes.
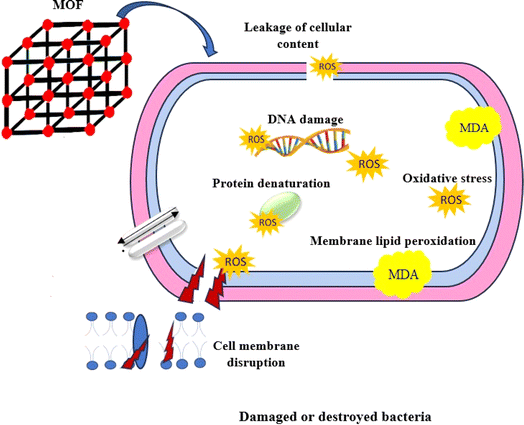
The overproduction of ROS within bacterial cells has been documented as a significant mechanism by which Ag nanoparticles induce oxidative stress and subsequent microbial mortality (Fig. 2).56,57 Moreover, lipid peroxidation, as evidenced by the presence of malondialdehyde (MDA) as a by-product, is considered a detrimental outcome of increased ROS generation.58–60 Based on our findings, it was observed that the production of ROS in K. pneumoniae KPB11 exhibited a dose-dependent enhancement. This observation was further shown by the measurement of MDA concentrations, which displayed a similar pattern of escalation. The aforementioned results suggest that the increased production of ROS and subsequent elevation of MDA levels have an indirect impact on the functionality of K. pneumonia KPB11. Fig. 3 shows the schematic diagram of the mechanism of antibacterial action.
Conclusions
Ag-MOFs were fabricated at the nanoscale and utilized as antibacterial agents against MDR K. pneumoniae strain KPB11. The results of the study showed that the MIC and MBC values were both 10 μg mL−1. Furthermore, Ag-MOF may induce the generation of ROS in cells, leading to cell wall damage and the release of cellular biomolecules including K+ ions and proteins, potentially contributing to its antibacterial effect. The MIC and MBC of the Ag-MOF were lower compared to those of most Ag nanoparticles and Ag+ ions evaluated against different pathogenic bacteria. This could be due to the greater surface area and porosity of MOFs. Further study is required to determine the antibacterial effectiveness of Ag-MOFs against different pathogenic bacterial strains and to comprehend their precise mechanisms of action.Conflicts of interest
The authors declare that there are no financial conflicts of interest.References
- C. M. Courtney, S. M. Goodman, J. A. McDaniel, N. E. Madinger, A. Chatterjee and P. Nagpal, Nat. Mater., 2016, 15, 529–534 CrossRef CAS PubMed.
- A. Parmanik, S. Das, B. Kar, A. Bose, G. R. Dwivedi and M. M. Pandey, Curr. Microbiol., 2022, 79, 388 CrossRef CAS PubMed.
- R. J. Melander and C. Melander, ACS Infect. Dis., 2017, 3, 559–563 CrossRef CAS PubMed.
- M. Khosravi, A. Mirzaie, A. B. Kashtali and H. Noorbazargan, Arch. Microbiol., 2020, 202, 2105–2115 CrossRef CAS PubMed.
- F. Foroohimanjili, A. Mirzaie, S. M. M. Hamdi, H. Noorbazargan, M. Hedayati Ch, A. Dolatabadi, H. Rezaie and F. M. Bishak, J. Basic Microbiol., 2020, 60, 216–230 CrossRef CAS PubMed.
- G. Wang, G. Zhao, X. Chao, L. Xie and H. Wang, Int. J. Environ. Res. Public Health, 2020, 17, 6278 CrossRef CAS PubMed.
- T. Le, L. Wang, C. Zeng, L. Fu, Z. Liu and J. Hu, Antimicrob. Resist. Infect. Control, 2021, 10, 41 CrossRef PubMed.
- A. Mirzaie and R. Ranjbar, AMB Express, 2021, 11, 122 CrossRef CAS PubMed.
- R. Freund, O. Zaremba, G. Arnauts, R. Ameloot, G. Skorupskii, M. Dincă, A. Bavykina, J. Gascon, A. Ejsmont, J. Goscianska, M. Kalmutzki, U. Lächelt, E. Ploetz, C. S. Diercks and S. Wuttke, Angew. Chem., Int. Ed., 2021, 60, 23975–24001 CrossRef CAS PubMed.
- Q. Wang and D. Astruc, Chem. Rev., 2020, 120, 1438–1511 CrossRef CAS PubMed.
- S. S. Mohammed Ameen, N. M. Sher Mohammed and K. M. Omer, Talanta, 2023, 254, 124178 CrossRef CAS PubMed.
- S. Sh. Mohammed Ameen, N. M. Sher Mohammed and K. M. Omer, Microchem. J., 2022, 181, 107721 CrossRef.
- A. H. Alshatteri, G. K. Ali and K. M. Omer, ACS Appl. Mater. Interfaces, 2023, 15, 21239–21251 CrossRef CAS PubMed.
- S. S. Mohammed Ameen, F. O. Qasim, H. S. Alhasan, K. H. Hama Aziz and K. M. Omer, ACS Appl. Mater. Interfaces, 2023, 15, 46098–46107 CrossRef CAS PubMed.
- L. Yan, A. Gopal, S. Kashif, P. Hazelton, M. Lan, W. Zhang and X. Chen, Chem. Eng. J., 2022, 435, 134975 CrossRef CAS.
- R. Karimi Alavijeh, S. Beheshti, K. Akhbari and A. Morsali, Polyhedron, 2018, 156, 257–278 CrossRef CAS.
- S. Zhang, J. Ye, Y. Sun, J. Kang, J. Liu, Y. Wang, Y. Li, L. Zhang and G. Ning, Chem. Eng. J., 2020, 390, 124523 CrossRef CAS.
- R. Li, T. Chen and X. Pan, ACS Nano, 2021, 15, 3808–3848 CrossRef CAS PubMed.
- K. Chattopadhyay, M. Mandal and D. K. Maiti, Mater. Adv., 2024, 5, 51–67 RSC.
- S. S. Mohammed Ameen and K. M. Omer, Food Chem., 2024, 139170 CrossRef CAS PubMed.
- S. S. M. Ameen, I. B. Qader, H. A. Qader, F. K. Algethami, B. Y. Abdulkhair and K. M. Omer, Microchim. Acta, 2023, 191, 62 CrossRef PubMed.
- S. H. Al-Jaf, S. S. Mohammed Ameen and K. M. Omer, Lab on a Chip, 2024, 24, 2306–2316 RSC.
- X. Lu, J. Ye, D. Zhang, R. Xie, R. F. Bogale, Y. Sun, L. Zhao, Q. Zhao and G. Ning, J. Inorg. Biochem., 2014, 138, 114–121 CrossRef CAS PubMed.
- S. W. Jaros, J. Król, B. Bażanów, D. Poradowski, A. Chrószcz, D. S. Nesterov, A. M. Kirillov and P. Smoleński, Molecules, 2020, 25, 2119 CrossRef CAS PubMed.
- A. Mirzaie, F. Badmasti, H. Dibah, S. Hajrasouliha, F. Yousefi, R. Andalibi, A. B. Kashtali, A. H. Rezaei and R. Bakhtiatri, Iran. J. Public Health, 2022, 51, 1097–1106 Search PubMed.
- V. Paratore, D. Franco, S. Guglielmino, F. Lo Presti, F. Traina, S. Conoci and G. G. Condorelli, Mater. Adv., 2024, 5, 1033–1044 RSC.
- S. W. Jaros, M. F. C. Guedes da Silva, J. Król, M. Conceição Oliveira, P. Smoleński, A. J. L. Pombeiro and A. M. Kirillov, Inorg. Chem., 2016, 55, 1486–1496 CrossRef CAS PubMed.
- P. Cao, X. Wu, W. Zhang, L. Zhao, W. Sun and Z. Tang, Ind. Eng. Chem. Res., 2020, 59, 1559–1567 CrossRef CAS.
- B.-P. Xie, J.-W. Chai, C. Fan, J.-H. Ouyang, W.-J. Duan, B. Sun, J. Chen, L.-X. Yuan, X.-Q. Xu and J.-X. Chen, ACS Appl. Bio Mater., 2020, 3, 8525–8531 CrossRef CAS PubMed.
- G. Wyszogrodzka, B. Marszałek, B. Gil and P. Dorożyński, Drug Discovery Today, 2016, 21, 1009–1018 CrossRef CAS PubMed.
- Y. He, X. Wang, C. Zhang, J. Sun, J. Xu and D. Li, Small, 2023, 19, 2300199 CrossRef CAS PubMed.
- H. W. Lee, Y. M. Koh, J. Kim, J. C. Lee, Y. C. Lee, S. Y. Seol, D. T. Cho and J. Kim, Clin. Microbiol. Infect., 2008, 14, 49–54 CrossRef CAS PubMed.
- K. Tamura, G. Stecher, D. Peterson, A. Filipski and S. Kumar, Mol. Biol. Evol., 2013, 30, 2725–2729 CrossRef CAS PubMed.
- A. W. Bauer, W. M. Kirby, J. C. Sherris and M. Turck, Am. J. Clin. Pathol., 1966, 45, 493–496 CrossRef CAS.
- S. Agnihotri, S. Mukherji and S. Mukherji, RSC Adv., 2014, 4, 3974–3983 RSC.
- B. Das, S. K. Dash, D. Mandal, T. Ghosh, S. Chattopadhyay, S. Tripathy, S. Das, S. K. Dey, D. Das and S. Roy, Arabian J. Chem., 2017, 10, 862–876 CrossRef CAS.
- A. Haddadian, F. F. Robattorki, H. Dibah, A. Soheili, E. Ghanbarzadeh, N. Sartipnia, S. Hajrasouliha, K. Pasban, R. Andalibi, M. H. Ch, A. Azari, A. Chitgarzadeh, A. B. Kashtali, F. Mastali, H. Noorbazargan and A. Mirzaie, Sci. Rep., 2022, 12, 21938 CrossRef CAS PubMed.
- S. Liao, Y. Zhang, X. Pan, F. Zhu, C. Jiang, Q. Liu, Z. Cheng, G. Dai, G. Wu, L. Wang and L. Chen, Int. J. Nanomed., 2019, 14, 1469–1487 CrossRef CAS PubMed.
- V. Tiwari, N. Mishra, K. Gadani, P. S. Solanki, N. A. Shah and M. Tiwari, Front. Microbiol., 2018, 9, 1218 CrossRef PubMed.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and R. J. Randall, J. Biol. Chem., 1951, 193, 265–275 CrossRef CAS PubMed.
- W. S. A. El-Yazeed and A. I. Ahmed, RSC Adv., 2019, 9, 18803–18813 RSC.
- A. Kushwaha, G. Singh and M. Sharma, RSC Adv., 2020, 10, 13050–13065 RSC.
- G. Kumar and D. T. Masram, ACS Omega, 2021, 6, 9587–9599 CrossRef CAS PubMed.
- H. D. Chul, R. Vinodh, C. V. V. Muralee Gopi, C. Deviprasath, H.-J. Kim and M. Yi, Dalton Trans., 2019, 48, 14808–14819 RSC.
- P. Mitra, D. Dutta, S. Das, T. Basu, A. Pramanik and A. Patra, ACS Omega, 2018, 3, 7962–7970 CrossRef CAS PubMed.
- M. D. Firouzjaei, A. A. Shamsabadi, M. Sharifian Gh, A. Rahimpour and M. Soroush, Adv. Mater. Interfaces, 2018, 5, 1701365 CrossRef.
- P. R. More, S. Pandit, A. Filippis, G. Franci, I. Mijakovic and M. Galdiero, Microorganisms, 2023, 11, 369 CrossRef CAS PubMed.
- T. Ameh, M. Gibb, D. Stevens, S. H. Pradhan, E. Braswell and C. M. Sayes, Nanomaterials, 2022, 12, 2402 CrossRef CAS PubMed.
- M. Mehrabi, M. F. Ghasemi, B. Rasti, M. Falahati, A. Mirzaie and A. Hasan, J. Biomol. Struct. Dyn., 2021, 39, 2595–2606 CrossRef CAS PubMed.
- H. M. Abd El Salam, H. N. Nassar, A. S. A. Khidr and T. Zaki, J. Inorg. Organomet. Polym. Mater., 2018, 28, 2791–2798 CrossRef CAS.
- S. Eckhardt, P. S. Brunetto, J. Gagnon, M. Priebe, B. Giese and K. M. Fromm, Chem. Rev., 2013, 113, 4708–4754 CrossRef CAS PubMed.
- S. W. Jaros, M. F. C. Guedes da Silva, M. Florek, M. C. Oliveira, P. Smoleński, A. J. L. Pombeiro and A. M. Kirillov, Cryst. Growth Des., 2014, 14, 5408–5417 CrossRef CAS.
- R. Behdad, M. Pargol, A. Mirzaie, S. Z. Karizi, H. Noorbazargan and I. Akbarzadeh, J. Basic Microbiol., 2020, 60, 494–507 CrossRef CAS PubMed.
- L. Mohammed and H. Hamzah, J. Bionanosci., 2024 DOI:10.1007/s12668-024-01334-y.
- R. S. Hamida, M. A. Ali, D. A. Goda, M. I. Khalil and M. I. Al-Zaban, Front. bioeng. biotechnol., 2020, 8, 433 CrossRef PubMed.
- K. Muthukumar, S. Rajakumar, M. N. Sarkar and V. Nachiappan, Antonie van Leeuwenhoek, 2011, 99, 761–771 CrossRef CAS PubMed.
- Y. Dong, H. Zhu, Y. Shen, W. Zhang and L. Zhang, PLoS One, 2019, 14, e0222322 CrossRef CAS PubMed.
- M. A. Quinteros, V. Cano Aristizábal, P. R. Dalmasso, M. G. Paraje and P. L. Páez, Toxicol. in Vitro, 2016, 36, 216–223 CrossRef CAS PubMed.
- Z. Yu, Q. Li, J. Wang, Y. Yu, Y. Wang, Q. Zhou and P. Li, Nanoscale Res. Lett., 2020, 15, 115 CrossRef CAS PubMed.
- M. J. Hajipour, K. M. Fromm, A. Akbar Ashkarran, D. Jimenez de Aberasturi, I. R. d. Larramendi, T. Rojo, V. Serpooshan, W. J. Parak and M. Mahmoudi, Trends Biotechnol., 2012, 30, 499–511 CrossRef CAS PubMed.
- Y. Liu, X. Xu, Q. Xia, G. Yuan, Q. He and Y. Cui, Chem. Commun., 2010, 46, 2608–2610 RSC.
- Z. Rahmati, J. Abdi, M. Vossoughi and I. Alemzadeh, Environ. Res., 2020, 188, 109555 CrossRef CAS PubMed.
- F. Mao, Y. Su, X. Sun, B. Li and P. F. Liu, ACS Omega, 2023, 8, 2733–2739 CrossRef CAS PubMed.
- A. A. G. El-Shahawy, E. M. Dief, S. I. El-Dek, A. A. Farghali and F. I. Abo El-Ela, Cancer Nanotechnol., 2023, 14, 60 CrossRef CAS.
- R. Roudbari, N. Keramati and M. Ghorbani, J. Mol. Liq., 2021, 322, 114524 CrossRef CAS.
- P. Parvekar, J. Palaskar, S. Metgud, R. Maria and S. Dutta, Biomater. Invest. Dent., 2020, 7, 105–109 CAS.
- M. A. Huq and S. Akter, Molecules, 2021, 26, 5996 CrossRef CAS PubMed.
- S. Pandey and J. Ramontja, Int. J. Biol. Macromol., 2016, 93, 712–723 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00037d |
| This journal is © The Royal Society of Chemistry 2024 |