Open Access Article
Open Access ArticleMass spectrometry imaging in plants, microbes, and food: a review†
Mudita
Vats
a,
Berta
Cillero-Pastor
 ab,
Eva
Cuypers
ab,
Eva
Cuypers
 a and
Ron M. A.
Heeren
a and
Ron M. A.
Heeren
 *a
*a
aMaastricht MultiModal Molecular Imaging Institute (M4i), Division of Imaging Mass Spectrometry, Maastricht University, Maastricht, the Netherlands. E-mail: r.heeren@maastrichtuniversity.nl; Fax: +31433884154; Tel: +31433881499
bMERLN Institute for Technology-inspired Regenerative Medicine, Department of Cell Biology-Inspired Tissue Engineering (cBITE), Maastricht University, Maastricht, the Netherlands
First published on 28th August 2024
Abstract
Plant health, which affects the nutritional quality and safety of derivative food products, is influenced by symbiotic interactions with microorganisms. These interactions influence the local molecular profile at the tissue level. Therefore, studying the distribution of molecules within plants, microbes, and plant-based food is crucial to assess plant health, ensure the safety and quality of the agricultural products that become part of our food supply, and plan agricultural management practices. Within this framework, the molecular distribution within plant-based samples can be visualized with mass spectrometry imaging (MSI). This review describes key MSI methodologies, highlighting the role they play in unraveling the localization of metabolites, lipids, proteins, pigments, and elemental components across plants, microbes, and food products. Furthermore, investigations that involve multimodal molecular imaging approaches combining MSI with other imaging techniques are described. The advantages and limitations of the different MSI techniques that influence their applicability in diverse agro-food studies are described to enable informed choices for tailored analyses. For example, some MSI technologies involve meticulous sample preparation while others compromise spatial resolution to gain throughput. Key parameters such as sensitivity, ionization bias and fragmentation, reference database and compound class specificity are described and discussed in this review. With the ongoing refinements in instrumentation, data analysis, and integration of complementary techniques, MSI deepens our insight into the molecular biology of the agricultural ecosystem. This in turn empowers the quest for sustainable and productive agricultural practices.
1. Introduction
Plants are the primary source of food for both humans and animals. The quality and safety of food depend on multiple factors, such as plant growth, agricultural practices and storage. Plants grow in close proximity with microbes, including those in the rhizosphere (soil around the roots) and phyllosphere (aerial plant parts). This symbiotic relationship significantly influences plant nutrition, growth, defense mechanisms2,3 and the quality of the food product. The most common techniques for quality assessment of agro-food are liquid chromatography and gas chromatography coupled to mass spectrometry (LC-MS and GC-MS, respectively), and nuclear magnetic resonance (NMR), which provides precise chemical information but does not give spatial information.4–11 Microscopic techniques such as atomic force microscopy (AFM) have the ability to characterize the structural arrangements of polysaccharides such as starch in banana and potato. Moreover, it was applied to study the plant cell wall molecules such as cellulose, hemicellulose and pectin. The arrangement of these polysaccharides in part determines the quality of agricultural produce. However, it can only detect a limited range of molecules.12,13 Mass spectrometry imaging (MSI) techniques offer spatially resolved molecular information directly on intact tissue sections without the need for homogenization. MSI techniques have been used to visualize molecular distributions during the nitrogen cycle, fruit ripening, seed development and plant growth stages (ESI Table 1†). The information about plant physiology helps in understanding nutrient uptake, optimizing plant nutrition, plant–microbe interactions, and planning sustainable farming practices. In addition, MSI has enabled study of the mechanisms of plant–pathogen interactions and localization of heavy metals, pesticides and alkaloids in agricultural samples. This knowledge is helpful in ensuring safety in the food supply chain. It is important to note MSI techniques are affected by ion suppression in complex plant environments. This is a consequence of the lack of chromatographic separation prior to ionization. Consequently, the number of metabolites detected in MSI techniques is typically lower when compared with LC-MS analysis of plant extracts. Continuous advancements in MSI sensitivity combined with image-guided laser capture microdissection enhance the coverage of compounds. This review offers insight into the basic principles and performance parameters of MSI for plant-based molecular imaging studies. A selected set of plant-based MSI studies is discussed to demonstrate the advances made in this thriving field.1.1 Fundamentals of MSI
The fundamental principle behind MSI is the combination of two essential aspects: mass spectrometry and spatial localization. Mass spectrometry identifies and quantifies molecules based on their mass-to-charge ratio (m/z). In MSI, this principle is utilized to detect and spatially resolve the molecules present in a sample. MSI creates a two-dimensional map in which each pixel illustrates molecular information specific to its corresponding location within the sample. MSI typically follows a basic workflow that involves several key steps. First, the sample is prepared, often as thin (plant) tissue sections, to facilitate the analysis. Next, a local desorption and ionization strategy is employed to convert the sample's molecules into ions. This conversion can be achieved using various ionization techniques such as MALDI (matrix-assisted laser desorption ionization), ESI (electrospray ionization) and other methods described further in this review. These ions are subsequently introduced into a mass analyzer, where they are separated based on their m/z. The most common mass analyzers are TOF (time-of-flight), Q-TOF (Quadrupole-TOF), FT-ICR (Fourier transform-ion cyclotron resonance) and FT-MS (Orbitrap). Following the acquisition of a mass spectrum from a specific location on the tissue, the sample plate is repositioned to commence the spectral acquisition on the next location. The procedure is recurrently executed to gather a multitude of spectra by systematically scanning various x and y positions. The collected spectra are compiled and processed after the completion of the acquisition process. Chemical images corresponding to each observed ion of interest are subsequently constructed using dedicated software tools. Finally, registration with other imaging modalities is combined with statistical image and data analysis and provides valuable insights into the sample's molecular composition and localization. The quality of the results depends on each of these steps; therefore, it is essential to optimize both the sample preparation and performance parameters to make the best use of this state-of-the-art technology.1.2 Sample preparation and challenges
Effective sample handling and preparation are critical steps in preserving the original spatial distribution of compounds, such as small metabolites. Several studies have previously documented sample preparation methodologies for plant tissues.14 The choice of sample preparation method largely depends on the nature of the sample and specific MSI technique used. The three most important considerations for sample preparation for MSI are: (1) is the sample vacuum compatible? (2) does the sample need to be perfectly flat? and (3) does the surface need to be modified prior to MSI analysis? Several examples that address these issues are discussed below.Some solutions have been developed for the abovementioned challenges. A recent study addressed agar flaking, aiming to enhance the attachment of cultivable Bacillus subtilis colonies to the MALDI-target plate.27 DHB was used as adhesive agent to improve the bonding of agar samples to MALDI targets. B. subtilis was cultured on four different agar media, and a region of agar including the bacterial colony was laid upon a target that was sprayed with DHB matrix (plate coat). Subsequently, a pre-coat and final matrix layer were applied on top of the agar using the sprayer. This method prevented agar flaking, which in turn saved the MALDI source from the pollution damage caused by agar. It also enabled the visualization of metabolites in B. subtilis colonies. The implementation of this method widens the scope of MALDI MSI, aiding in high-throughput screening and optimization of ionization conditions for targeted metabolites in agar-based microbial cultures. Another study reported removing the trichomes before MSI analysis by a surface treatment.28 However, in studies where trichomes are important, DESI-MSI is an advantageous technique. A study was conducted on Cannabis sativa leaves to visualize the distribution of cannabinoids and flavonoids on trichomes using DESI-MSI. Additionally, cross-sections of the sugar leaves of C. sativa L. were analyzed using MALDI-MSI.29 In this investigation, the trichomes were analyzed by direct DESI, where leaves are attached to a glass slide, and indirect DESI using imprints of the leaf surface. While cannabinoids were detected by both the methods, the direct DESI images were smudged whereas imprinting maintained the spatial distribution. The cross-sections were analyzed using MALDI-MSI as it provides higher spatial resolution compared with DESI. Freeze-drying samples can ameliorate the shrinking of fresh plant samples during vacuum-based MSI. The samples should be freeze-dried before obtaining an optical image30 to prevent a discrepancy between the optical and MS images. Freeze-drying also prevents the delocalization of compounds.
1.3 Performance parameters
In addition to sample preparation considerations, performance parameters are also important considerations in selecting the most suitable technique for a specific investigation. These parameters, including sensitivity, spatial resolution, acquisition speed and mass resolution, collectively influence the quality of MSI results.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–15
000–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 pixels per s. However, these measurements were acquired on mammalian samples such as fingerprints and rat tissues, so acquisition speed using plant-based samples remains to be determined.
500 pixels per s. However, these measurements were acquired on mammalian samples such as fingerprints and rat tissues, so acquisition speed using plant-based samples remains to be determined.
2. Applications of MSI techniques analyzing food plants and microbes
2.1 Matrix-assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI)
The workflow of MALDI-MSI analysis is explained in the previous section. The application of MALDI-MSI to localize a large range of biomolecules in tissues is becoming an integral part of spatial biology in various domains such as biomedicine, forensics, environment, food and agriculture. Commercially available instruments offer ∼5–200 μm spatial resolution.44 MALDI-MSI is a soft ionization technique that can detect the spatial distributions of metabolites, lipids, peptides, proteins and other small molecules. It predominantly yields singly charged ionic species from analyte molecules. MALDI-TOF-MSI has found valuable applications in the study of agricultural samples, specifically to study plant–microbe relationships, food safety and nutrition, and plant-based cosmetics.MALDI-MSI has been employed to investigate the symbiotic relationship between Medicago truncatula root nodules and the bacteria Sinorhizobium meliloti during nitrogen metabolism.46 This study applied DHB as a matrix for positive ion mode and DMAN (1,8-bis (dimethyl-amino) naphthalene) for negative ion mode, to enhance the breadth of analyte detection. This combination of matrices aided in detecting organic acids, amino acids, sugars, lipids and flavonoids (Fig. 1). The metabolic differences of wild-type plant and rhizobia were compared with mutants that were defective in nitrogen fixation. The spatial distribution of metabolites in negative ion modes such as maleic acid (m/z 115.0), citric acid (m/z 191.0), and deprotonated sucrose (m/z 341.07) was obtained by MALDI-MSI in a medicago root and nodules section with a spot diameter of 50 μm. Additionally, sucrose was visualized in sodiated and potassiated forms (m/z 365.11 and m/z 381.08) in positive ion mode. ClinProTools facilitated a comparative statistical analysis of metabolite abundance between the root and nodules of medicago. In this particular context, the nodules of medicago exhibited notably higher intensities of sucrose and glutamine. This observation aligns with known metabolic processes, as sucrose serves as an essential energy source which is subsequently utilized in glycolysis, leading to the release of metabolites such as phosphoenolpyruvate or malate. The bacteria consume these substrates to perform nitrogen fixation, after which the fixed ammonia is assimilated into glutamine. Moreover, the authors were able visualize the heme moiety (m/z 616.18) in wild-type root nodules that is absent in mutant plants and bacteria. This study helped in understanding the role played by the aforementioned metabolites in nitrogen fixation and other metabolic pathways, thereby providing insights into plant health.
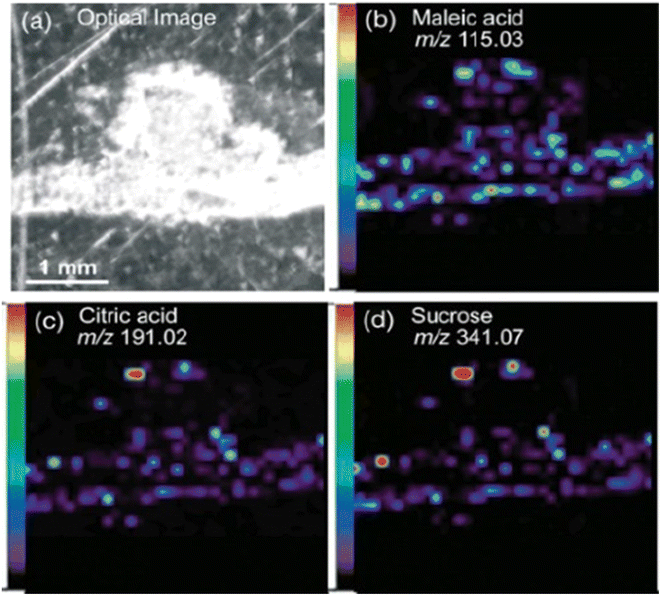 | ||
| Fig. 1 Spatial distribution of metabolites in medicago nodules. Optical image of a medicago section (a) and representative negative-mode MS images of deprotonated metabolite peaks, including (b) maleic acid (m/z 115.03), (c) citric acid (m/z 191.02) and (d) sucrose (m/z 341.07). Figure reprinted from ref. 46 with permission from John Wiley and Sons, copyright 2013. | ||
Another study utilized MALDI-MSI and SIMS imaging to understand the distribution, translocation, and metabolism of nitrogen in radish plants.30 To illustrate nutrient cycles in plants, radish seeds were planted and enriched with 15N-KNO3 constituting the first-generation group. Subsequently, upon harvesting, the plants were homogenized to create a tea, which served as the sole nitrogen source for the second-generation 15N group. Unenriched control plants were also cultivated and prepared for MSI experiments. Multiple MSI experiments were performed on fresh frozen radish bulbs and freeze-dried leaf samples. A MALDI-MSI experiment was performed at a spatial resolution of 150 μm within a mass range of m/z 100–500. The MSI images of leaves and bulbs showed the distribution of choline (m/z 104.1) in control samples, while labeled samples had an integration of 15N, resulting in the formation of an ion at (m/z 105.1). Similarly phosphocholine (m/z 184) and 15N phosphocholine (m/z 185) were detected and imaged. The intensity pattern of the unlabeled and labeled compounds in the MSI images demonstrated that 15N was first metabolized by the plant and integrated into its structure, followed by transfer of 15N to the next generation. For SIMS-MSI, the radish bulb sections were gold-coated and measurements were performed with an Au liquid metal gun. Each 125 μm square consisted of 256 × 256 pixels. A central portion of the bulb was measured with SIMS, which showed the choline distribution in control, first- and second-generation groups. Unlabeled and labeled choline was observed in the string-like network of the bulb section. The isotopic ratio of m/z 104/105 in control, first-generation, and second-generation samples was 2.5322, 0.1963, and 0.4689, respectively. The authors observed a variation in the MSI images and spectrum of MALDI and SIMS, which might be because of the small portion of a large sample measured in SIMS. MALDI was able to acquire the distribution of choline in the entire radish bulb section. This combination of MALDI and SIMS MSI could be useful in monitoring the distribution and transport of compounds in other plants. Moreover, it can assist in improved strategies for plant nutrition cycling and schedules. This knowledge can also contribute to minimizing excess nutrient runoff into water bodies, which can cause pollution and ecosystem damage, thus help reduce the environmental impact of agriculture.
A combination of MALDI-MSI and NALDI-MSI (nanoparticle-assisted laser desorption ionization) has been applied to investigate rice–bacteria interactions on the plant leaves using TiO2 and Fe3O4 nanoparticles.47 Furthermore, imprinting and fracturing, as a sample preparation method, were applied to mitigate the challenges posed by variations in sample height. Imprinting squeezed out cytoplasmic hydrophilic compounds that were subsequently visualized. Imprinting can be applied to a wide range of leaf types regardless of size or surface composition but cannot capture membrane-bound molecules such as lipids. Fracturing sandwiches the sample between adhesive sides of packing tape and passes it through a rolling mill, after which matrix is applied to the exposed insides and then analyzed. Fracturing is complementary to imprinting because it allows the detection of compounds present in fracture-opened areas, namely phospholipids and chlorophylls. The limitations of this method are the lack of control over which layer will fracture open. Fox example, leaves of the rice plant exhibit greater fragility because the mesophyll adjacent to the veins is most susceptible to fracturing. NALDI was helpful in visualizing the plant metabolites that are otherwise not detected, but potentially compromises the spatial resolution. The authors visualized the compounds that are released by a host-defense mechanism at a spatial resolution of 10 μm. Metabolites in the range of m/z 100–1200 such as phytoalexins, momilactones, and phytocassanes were visualized on the sites of leaves infected with bacteria. Additional observations included the detection of nucleobases, phosphocholine, and amino acids that corresponded to the dark regions on the soybean leaf surface. A future direction could be an additional investigation to understand the nitrogen metabolism of aphids corresponding to the detection of molecules in the dark spots of soybean leaf. The development of NALDI has presented new avenues for assessing the efficiency of nanoparticles in the detection of plant metabolites, thereby enhancing plant pathology research and applications.
Similarly, blueberries were also analyzed using MALDI-FT-ICR-MSI, to visualize the spatial distribution of primary and secondary metabolites during fruit development.49 The blueberries were collected at different stages of ripening and 50 μm cross-sections were obtained. MSI measurements were acquired with 60 μm raster step size and laser beam width of 30 μm, within a mass range of m/z 150–2000 in both positive and negative ion mode. Different species of anthocyanins, polyphenols, sucrose, hexose sugars and amino acids were detected and localized. Sucrose was distributed only in the placenta region during the early stage of blueberry ripening but throughout the entire fruit at the later ripening stage. A signal from a potassiated hexose sugar [M + K]+ (m/z 219.0271) was visualized in the mesocarp. Chlorogenic acids were distributed in the outer mesocarp at an early stage and in the exocarp and mesocarp regions in later stages. Image segmentation using SCiLS lab facilitated the visualization of the metabolite distribution in different stages of berry development (Fig. 2). For example, the composition of anthocyanin in bilberry tissues remains consistent and the accumulation displays a gradient of skin to the flesh of fruit. This investigation improved the understanding of the link between primary and secondary metabolism during berry development. The results will help the food industry to identify the molecular fruit-quality traits.
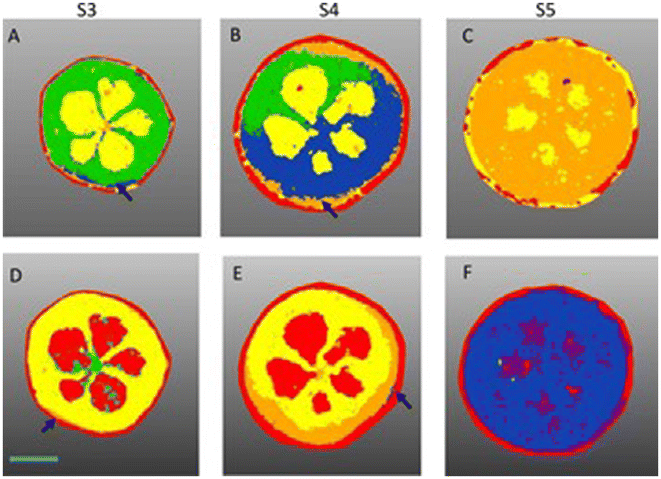 | ||
| Fig. 2 Segmentation analysis of blueberry development imaged in negative (A to C) and positive ion mode (D to F). (A) S3 bilberry fruit primarily comprising one chemical zone (green). Arrow marks the beginning of the S4 zone (blue). (B) S4 fruit consists of S3 (green) S4 (blue) and the proceeding S5 zone (orange) beginning to appear around the exocarp. (C) S5 bilberry (orange) S5 zone encompasses the entire fruit flesh (D) S3 bilberry comprising two chemical zones yellow and green. Arrow marks the beginning of the S4 zone in orange. (E) S4 bilberry S4 region (orange) shown in outer mesocarp. Arrow marks the beginning of the S5 zone (blue). (F) S5 bilberry S5 region (blue) encompasses the entire fruit flesh. Red regions correspond to chemical background signals derived from the MALDI matrix. Scale bar = 2 mm (for interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article). Reprinted from ref. 49, with permission from Elsevier copyright (2022). | ||
MALDI-MSI has been utilized to visualize the metabolic response to tissue-specific stress such as wounds or pest attacks at micro-regional levels in tomatoes.50 The local responses occurring in the vicinity of the wound region have remained elusive due to the limitations of conventional methods in analyzing the distribution of various biomolecules in the anatomical structures of samples such as fruits. 10 μm-thick fresh frozen tomato sections were obtained using a cryo-microtome. The sections were analyzed at 50 μm spatial resolution, with one experiment using DHB as a matrix and another using 9-AA (9-aminoacridine) as a matrix. Primary and secondary metabolites were detected and localized in the epicarp, mesocarp, locule, and seeds of the fruit. Identification was conducted using MS/MS and compared with the MS/MS profiles of standard compounds. This comparison resulted in the identification of sour and umami compounds among the primary metabolites and caffeate as a secondary metabolite. Unripened (green) tomato sections were compared with ripe tomato sections for the differences in the intensities of metabolites. For instance, sour compound malate (m/z 133.02) was found to be more intense in green fruit compared with ripened fruit, while caffeate (m/z 179.02) and glutamine (m/z 145.05) had similar distributions in both unripe and ripe sections. Since signal intensity data only represent the relative abundance of biomolecules, the authors conducted LC-MS to measure the compounds quantitatively. However, it is worth noting that in the LC-MS experiments, it was challenging to achieve a complete separation of the epicarp from the mesocarp using fresh fruit. Therefore, these two techniques complement each other effectively. Furthermore, this study effectively investigated the spatially resolved metabolism of a glycoalkaloid, tomatine, by examining local responses to the ripening process and wounding stress using MALDI-MSI.
Another study on tomatoes unveiled the function of the GORKY transporter in preventing tomato bitterness, revealing an intriguing aspect of tomato physiology and flavor modulation.51 GORKY, derived from a Russian word meaning “bitter”, belongs to the NPF transporter family and prevents the accumulation of high levels of α-tomatine in ripe tomato fruit. The objective of this investigation was to unravel the molecular mechanism(s) responsible for the maintenance of low α-tomatine levels and the non-bitter taste in tomatoes. This study encompassed gene expression examinations utilizing PCR (polymerase chain reaction), and metabolic profiling conducted through LC-MS and MALDI-MSI techniques. For the MALDI-MSI experiment, fresh frozen tomato samples were embedded in M1 medium and sectioned at 45 μm thickness. The measurements were performed in positive ion mode within the mass range of m/z 150–3000 at a pixel size of 50 μm. MALDI-MSI measurements were performed on ripe transgenic Del/Ros1 tomato sections, which are known for their purple, anthocyanin-rich fruit. Interestingly, when Del and Ros1 were silenced, the natural red color of the tomatoes was restored. The unaffected areas of the fruit were visualized using delphinidin and petunidin. In the areas affected by GORKY silencing, there was an accumulation of α-tomatine and acetoxytomatine, while the levels of esculeoside A declined. The GORKY gene appears to be involved in transferring α-tomatine and other steroidal alkaloids from the vacuole to the cytosolic region as tomatoes ripen. This movement aids in converting the entire α-tomatine pool into non-bitter forms, enhancing the fruit's appeal. Consequently, it will enhance the palatability of tomatoes for consumers.
MALDI-MSI has been used to study the metabolites and lipids corresponding to the flavor profile of white button mushrooms.52 In this study, fresh frozen (35 μm sections) and heat-treated (17 μm sections) mushrooms were analyzed to visualize the distribution of taste compounds. The sections were sprayed with DHB for detecting lipids in positive ion mode, norharmane for lipid in negative ion mode, and NEDC for metabolites in negative ion mode. In addition, on-tissue chemical derivatization was performed using TAHS (p-N,N,N-trimethylamonioanilyl N-hydroxysuccinimidylcarbamate iodide) to enhance the ionization efficiency of amino acids which are important compounds in the flavor profile. MALDI-MSI results revealed the localization of sweet, sour, bitter and umami taste compounds in cap, gills and stipe of the mushrooms. Amino acids such as glycine (Gly-TAHS m/z 252.13), valine (Val-TAHS m/z 294.18), malic acid (m/z 133.01), citric acid (m/z 191.01), histidine (His-TAHS m/z 332.17), phenylalanine (Phe-TAHS m/z 342.18), asparagine (Asp-TAHS m/z 310.14), and glutamine (Gln-TAHS m/z 323.17), and lipids such as PC 18:2 (m/z 520.33), PE 36:4 (m/z 778.48), PI 18:2 (m/z 595.28), and PA 34:2 (m/z 671.46) among others were visualized. This study demonstrated the sample preparation and molecular imaging in high water-containing samples using MALDI-MSI. This information is helpful for the food industry to decide which part of the mushroom should be used to extract specific flavor compounds. Moreover, the flavor compounds extracted from such sources are natural and healthy.
Expanding the scope of application, MALDI-MSI has been used to detect coffee adulteration, a concerning practice often involving mixing low and high-quality beans from various geographical sources. Coffee beans consist of endogenous molecules such as sugars, chlorogenic acid, caffeine and fatty acids. These compounds are pivotal for molecular-level coffee bean grading, geographical origin determination, and optimizing storage conditions. MALDI-MSI can spatially resolve these compounds to compare the differences between different geographical origins. In a study, AP (atmospheric pressure) MALDI-MSI was utilized to identify and visualize the distribution of endogenous molecules in coffee bean sections from eight different geographical regions.54 The measurements were performed in both positive and negative ion mode within a mass range of m/z 100–900 and at a spatial resolution of 75 μm. In positive ion mode, several compounds, including caffeine (m/z 195.09, [M + H]+), theophylline (m/z 181.06, [M + H]+), theobromine (m/z 181.06, [M + H]+), sucrose (m/z 381.08, [M + K]+), caffeoylquinic acid (m/z 393.06, [M + K]+), and dicaffeoylquinic acid (m/z 517.09, [M + H]+), were successfully identified and their spatial distribution patterns were investigated. In the negative ion mode, caffeoylquinic acid (m/z 353.08, [M − H]−) and dicaffeoylquinic acid (m/z 515.09) were identified and visualized. The validity of these identifications was further confirmed through MALDI-MS/MS experiments and comparison with existing literature. Protonated caffeine molecules (m/z 195.09, [M + H]+) were observed to be widely dispersed within sections of coffee beans from different coffee-producing regions. Their distribution, while prevalent throughout the entire beans, showed variation in relative abundance among beans from diverse sources. Notably, coffee beans originating from Colombia and Tanzania exhibited notably higher relative caffeine levels compared with beans from other regions. Additionally, the study explored the presence of caffeoylquinic acid (m/z 393.06, [M + K]+). Interestingly, ions with m/z values of 393.06, 186.96, and 220.12 were predominantly situated in the central part of the endosperm, while ions with m/z 296.07 and m/z 222.03 were concentrated towards the periphery of the endosperm. Furthermore, principal component analysis (PCA) was conducted on the samples originating from various geographical regions and revealed distinct geographical separations among coffee beans sourced from Uganda, Guatemala, Ethiopia, and China. These findings underscore significant disparities in chemical species and composition across these regions.
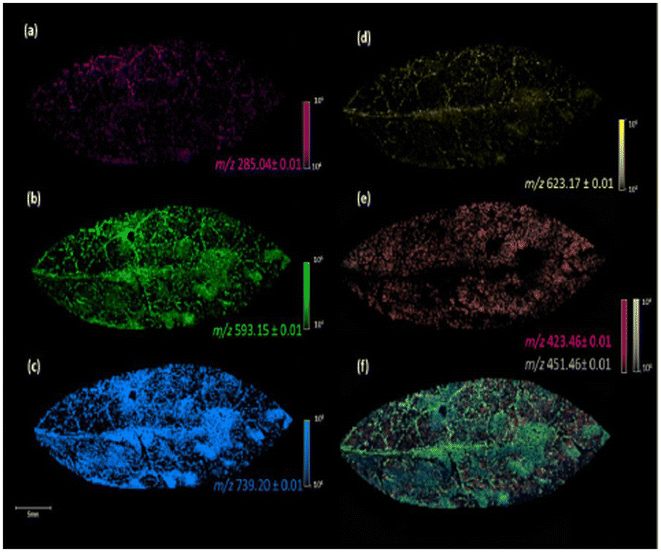 | ||
| Fig. 3 Image of the fixed leaves of G. sepium in MALDI(−) MSI analysis, using DHB as a matrix, with the different m/z values obtained: kaempferol, m/z 285.04 (a), kaempferol 3-O-a-rhamnopyranosyl(1–6)-β-galactopyranoside, m/z 593.15 (b), kaempferol 3-O-α rhamnopyranosyl(1–2)[α-rhamnopyranosyl(1–6)]-β-galactopyranoside, m/z 739.20 (c), iorhamnetin-3-O-rutinoside, m/z 623.17 (d), fatty acids, m/z 423.46 and 451.46 (e), and colocalization of the ions at m/z 285.04, 423.46, 451.46, 593.15, 623.17, and 739.20 (f). Reprinted from ref. 55 with permission from American Chemical Society, copyright (2022). | ||
Despite these advantages, MALDI-MSI also presents certain limitations. Ions with widely divergent desorption and ionization efficiencies within the same sampling area cause an ionization bias (or suppression) effect. This phenomenon results in the preferential detection of certain, often abundant, ions, thus suppressing the detection of low-abundance molecules or those with low ionization efficiencies. Moreover, the generation of high-abundance, low-molecular-weight ions from the matrix compounds causes the presence of interfering signals when using low-resolution mass analyzers.31
2.2 Secondary ion mass spectrometry (SIMS)
SIMS is based on ion beam sputtering of a sample surface and subsequent analysis and detection of secondary ions produced. There are many types of primary ion guns, such as Ar+, C60+, Bi, Bi3+, Au+, Aun+, Csn+ and Arn+ used in combination with a mass analyzer, usually TOF or quadrupole. SIMS can be operated in two modes, static and dynamic, defined by the total ion dose delivered to the sample. Static SIMS has a lower primary ion dose, and removes a minimal amount of material from the sample surface. Conversely, dynamic SIMS removes a larger amount of sample surface and it helps in the detection of small molecules, elemental ions, lipids, and small peptides. It offers a spatial resolution of 100 nm using this technique.57 In addition, nano-SIMS offers the capability of acquisition at nanoscale spatial resolution, achieving <50 nm resolution with Cs+ and <200 nm with O−.58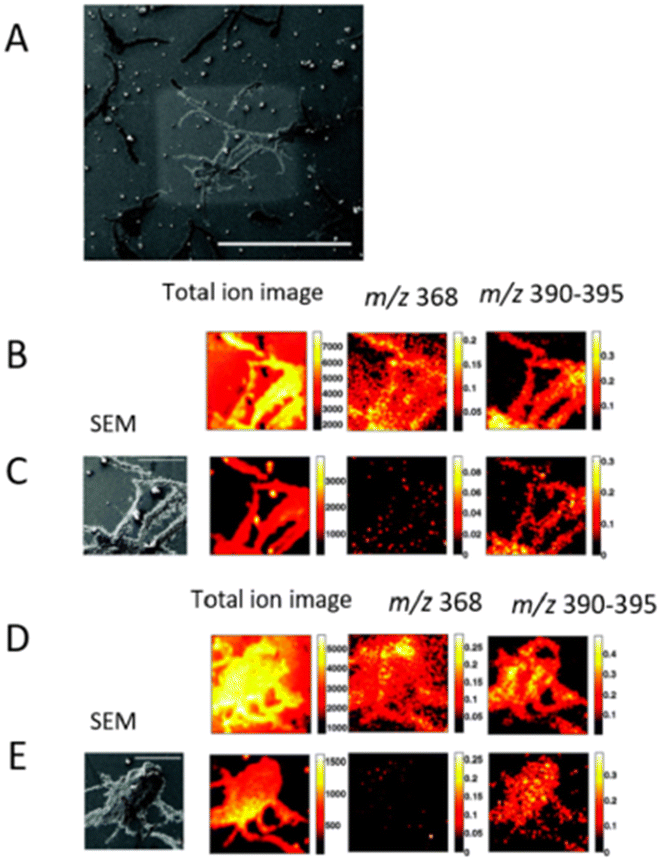 | ||
| Fig. 4 Subsurface imaging of antibiotic distribution demonstrated on a salt-stressed bacterial population. A SEM image of one of the sample areas analyzed (bar = 500 μm) is shown with the etch crater visible (A). Two different areas are imaged before (B and D) and after (C and E) sputter etching the surface with a primary ion dose density of 6 × 1013 and 2 × 1014 ions per cm2, respectively. The total ion spectrum for the four cases is shown on the left along with the corresponding total ion images (256 × 256 pixels) and the normalized images showing the distribution of the antibiotic peaks at m/z 368 and m/z 390–395 (128 × 128 pixels). An SEM image of the analyzed area (bar = 100 μm) is also shown for the etched surfaces for reference. Adapted and reprinted from ref. 59 with permission from American Chemical Society, copyright (2008). | ||
Another study aimed at detecting carbon (C) and nitrogen (N) assimilation by individual bacterial cells and fungal hyphae in soil using TOF-SIMS and epifluorescence microscopy.60 The authors investigated the spatial distribution of isotopically labeled carbon and nitrogen within a soil sample, focusing on a high-resolution scale of 200 nm. This analysis was achieved through the use of a fundamental model system utilizing kaolin clay (0.25 g), suspended in water, filtered onto a glass fiber filter, and subsequently labeled with 1 mM, 99 atom% 15NO3−. The experimental setup involved adding straw and fresh dairy manure to the clay, followed by covering the assembly with a Si contact slide, resulting in a cylindrical structure of approximately 15 mm diameter. The achieved spatial resolution proved to be highly effective in discerning the distinct activities of individual bacterial cells engaged in carbon and nitrogen assimilation. Moreover, the utilization of secondary ion images offered clear visualization and differentiation of fungal hyphae. The researchers were further able to precisely identify, map, and elucidate the specific locations and functions of both bacteria and fungal hyphae involved in the crucial process of nutrient uptake. Notably, the application of TOF-SIMS allowed for the discrimination of relative variations in 15N content (atoms percent) across microsite boundaries, providing valuable insights into isotopic composition dynamics. The study also integrated epifluorescence microscopy images to enhance the analysis, seamlessly superimposing them onto the TOF-SIMS data. This research elucidated the nutrient dynamics in soil ecosystems and the role played by soil microbial communities in nutrient cycling.
The combination of TOF-SIMS with other analytical techniques allowed researchers to obtain valuable insights into agriculturally relevant phenomena such as plant–microbe interactions. In this context, TOF-SIMS combined with ESI-MS has been used to study the in situ production of cyclic lipopeptides (a class of antibiotics) by Bacillus amyloliquefaciens S499 in the roots of tomato plants.61 The lipopeptides secreted by bacillus colonies were recovered, and extracted material was analyzed using LC-ESI-MS. Lipopeptide production was compared under in vivo and in vitro conditions, which revealed that the lipopeptide signature varies in different conditions. TOF-SIMS facilitated the imaging of lipopeptides secreted by S499 cells with developing roots of tomato plantlets cultivated from sterilized seeds on gelified medium. This information can be used to identify the most effective biocontrol agents for protecting crops against diseases, thus reducing the need for pesticides.
Furthermore, nano-SIMS was used in combination with a transmission electron microscope (TEM) to measure and visualize the distribution of 15N/14N in situ in symbiotic relationships, which cannot be accomplished with conventional mass spectrometry imaging techniques.62 Spatially resolved detection of 15N assimilation by individual microorganisms in the ectorhizosphere, endorhizosphere and rhizoplane was obtained at 100 nm spatial resolution (Fig. 5). The variability of 15N assimilation between active and inactive cells was directly verified. TEM was used to provide high-resolution imaging of the root and rhizosphere, allowing researchers to visualize the structural and spatial organization of the plant root and the microorganisms present in the rhizosphere. TEM imaging of extracted roots revealed the presence of individual microorganisms in direct proximity to the root surface. Subsequent nano-SIMS analyses of these regions indicated that certain microorganisms were enriched with 15N, although not all microorganisms showed the same level of enrichment. It was shown that following a 0.5-hour exposure to 15N, the microorganisms exhibited a nearly 50-fold increase in the 15N/14N ratio, a pattern that persisted after 24 hours of continuous 15N exposure. Remarkably, even in microorganisms highly enriched with 15N, where a substantial 50% of the N originated from the introduced 15N, these results underscore the rapidity of N uptake and subsequent assimilation. Gaining insights into rapid N uptake and assimilation by microorganisms has significant implications for the comprehension of nutrient and carbon cycling within the plant–soil interface. Such knowledge is pivotal for understanding metal toxicity, as the root exudates can increase the bioavailability and uptake of metals/metalloids in the rhizosphere by chelation and solubilization.63,64 Furthermore, it aids in the strengthening of defense mechanisms against potential pathogen attacks, and optimizing nutrient uptake efficiency, all to mitigate risks for plant health.
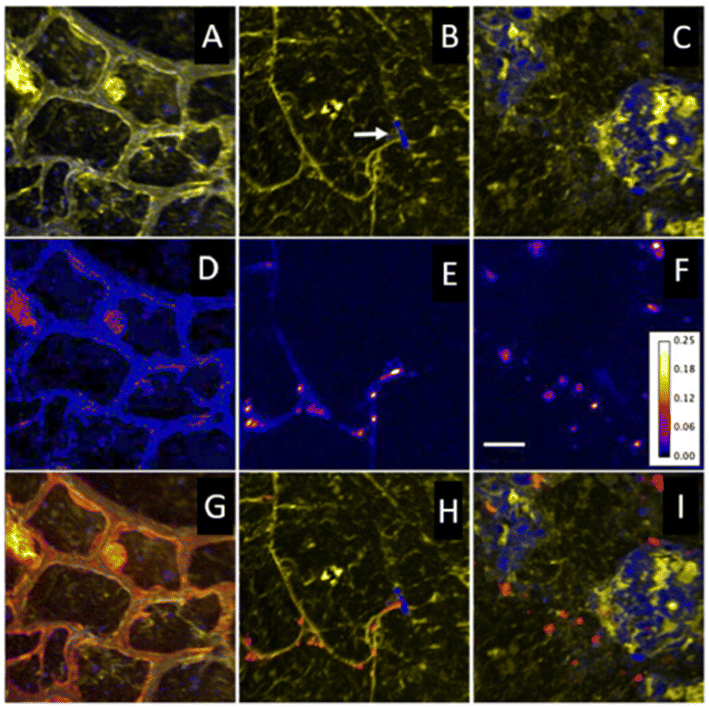 | ||
| Fig. 5 Nano-SIMS ion images highlighting in situ spatial relationships, coupled with levels of 15N enrichment. (A to C) Overlays of 12C14N (yellow) and 28Si (blue) ion images representative of key regions: plant root cells (A), rhizosphere (B), and soil matrix (C). A root hair that has encountered a soil particle and diverged is seen in B (arrow). (D to F) 15N/14N ratio images from same regions, showing areas of 15N enrichment. The color depicts the 15N/14N ratio, with the ratio scale shown in (F). Bar = 5 μm. (G to I) Overlay of 12C14N (yellow), 28Si (blue), and 12C15N/12C14N (red) images from each region. Figure adapted and reprinted from ref. 62 with permission from Oxford University press, copyright (2009). | ||
Nano-SIMS can also facilitate research on the individual cellular responses to environmental changes and enable a quantitative description of cellular behavior.65 This effort was demonstrated by integrating EL-FISH (element labeling fluorescence in situ hybridization) and nano-SIMS imaging to study the mutualistic interaction of filamentous Anabaena and Rhizobium. Upon co-culturing the Rhizobium species, the presence of isotopically labeled carbon and nitrogen was detected at 150 nm spatial resolution. This detection implied that the assimilation of carbon and nitrogen compounds by the epibiont was fixed by the cyanobacterium. The utilization of EL-FISH introduced fluorine labeling to specifically visualize one member of a dual-species consortium for nano-SIMS analysis, which localized fluorine relative to carbon and the distribution of 13C and 15N enrichment. In this approach, Anabaena cells, which were not targeted by the alpha proteobacteria probe, exhibited minimal fluorine levels. As a result, rhizobium cells lacking 13C enrichment were also visible, which was facilitated by EL-FISH due to heightened fluorine content. These observations highlight the capacity of EL-FISH to enable accurate cell detection and phylogenetic identification, irrespective of the cell's metabolic status. The investigation on the nitrifying and denitrifying bacteria presented in this research aids in developing strategies to improve soil health, plant growth, and more sustainable agricultural practices.
Similarly, the presence of heavy metals in the soil affects the health of crops and ultimately determines the safety of food products. Contaminated soil containing heavy metals can result in metal uptake by crops, potentially posing health hazards to consumers, including the risk of cancer.67 A combined approach of TOF-SIMS and scanning electron microscope (SEM) was utilized to study the distributions of heavy metals in the rhizospheric region of Populous tremuloides. Through this analysis, the distribution of elements, namely calcium (Ca), magnesium (Mg), sodium (Na), potassium (K), silicon (Si) and aluminium (Al) was examined in different structures of the plant roots.68 SEM results indicated that Al and Si are found in close proximity within a region. TOF-SIMS was advantageous as it provided the detailed distribution of elements within the cell walls of plant root. Ca and Mg were detected and imaged on similar regions of the cell walls and were actively transported into the root structure. By comparison, the movement of Al into the root was restricted, likely due to the presence of specific root exudates. In this investigation, the results of SEM provided a comprehensive interpretation of the SIMS data. This research deepened the understanding of distribution and uptake of metals between soil and root cells.
2.3 Liquid extraction surface analysis–mass spectrometry imaging (LESA-MSI)
Liquid extraction surface analysis–mass spectrometry imaging (LESA-MSI) is an atmospheric ionization technique that consists of a liquid microjunction coupled to mass spectrometry. In LESA-MSI, a droplet of solvent is deposited on the sample surface. This droplet forms a liquid microjunction with the sample and facilitates the diffusion of analytes from the sample into the solvent. The droplet is then robotically taken back into the electro-conductive pipette tip, which subsequently goes to the mass spectrometer, by nano-ESI through a dedicated spray tip. The spatial resolution of LESA-MSI depends on the angle of contact made by the extraction solvent, as well as the type and volume of the extraction solvent used. A spatial resolution of 125 μm has been reported.71 LESA can efficiently detect protein complexes, individual intact proteins and metabolites.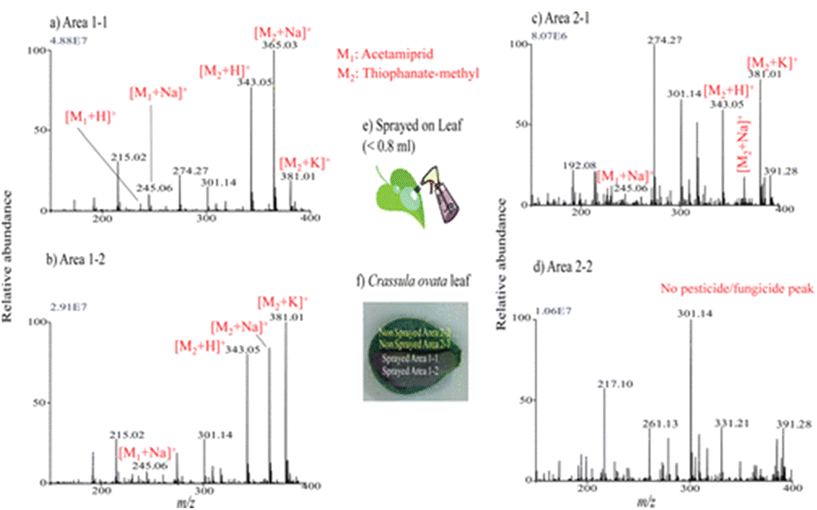 | ||
| Fig. 6 Mass spectra measured by LESA-MS in different regions of Crassula ovata leaf for monitoring of the presence of sprayed pesticide and fungicide after a week. (a–d) Mass spectra measured at the different regions: area 1-1 (a), area 1-2 (b), area 2-1(c), and area 2-2 (d), shown in panel (f). (e) Schematic for the spraying procedures of pesticide and fungicide mixture solution on leaf. Reprinted with permission from ref. 72 with permission from American Chemical Society, copyright (2013). | ||
In another study, LESA-HRMS (LESA–high resolution mass spectrometry) and Raman microscopy were employed to study the metabolic interactions between two wood-rot fungi, Schizophyllum commune and Hypholoma fasciculare.74 The spatial distribution of secondary metabolites such as indigo, indirubin and isatin produced at the interaction zone of the fungi was studied. This analysis provided a better understanding of the biochemical processes that occur between wood-degrading microbial species. The exclusive detection of indigo in the co-cultures implies that it may serve as a stress or defense response against antagonistic microorganisms. To ensure the preservation of fungal diversity in ecosystems, a comprehensive understanding of the intricate mechanisms driving fungal community ecology is essential. The complementary use of LESA-HRMS and Raman proved beneficial for imaging delicate and three-dimensional colonies, eliminating the need for matrix application that can potentially disturb their structures during analysis. This strategic approach provided a non-invasive means of visualizing and studying microbial communities and their interactions within diverse ecosystems.
2.4 Laser ablation inductively coupled plasma mass spectrometry imaging (LA-ICP-MSI)
LA-ICP-MSI works by irradiating the sample surface using UV beam (213 nm) which volatilizes or ablates the sample. The aerosols produced go into the ICP-MS where there is a hot carrier plasma (argon) that achieves complete atomization and ionization, followed by transport to the mass spectrometer for analysis. It is mostly coupled to a quadrupole mass analyzer. The spatial resolution of LA-ICP-MSI relies on the spot size of the laser, and a spatial resolution of under 10 μm can be achieved. It is a hard ionization technique, and therefore it can detect elements and isotopes.LA-ICP-MSI has been applied to analyze the elemental composition in plants.
Furthermore, the compositional balance of micro- and macroelements plays a pivotal role in determining the nutritional profile of plant products. In this context, the application of LA-ICP-MS has been instrumental in the analysis of sunflower leaves.77 It was found that certain elements are specifically distributed in the veins and tip of the leaves, while others were homogeneously distributed throughout the leaf. The leaf was ablated orthogonal to the main leaf vein with a spot size of 180 μm. The elements were categorized into four main groups based on their distribution patterns. The first group included elements that displayed a uniform distribution across the leaf surface, namely Ca, Cr, and P. In the second group, an enrichment in the tip leaf tissue was observed for Fe and S. The third group comprised elements like Cd, Ce, Cu, La, Mn, and Zn, which were primarily found to accumulate within the leaf's veins. Finally, K and Ni, forming the fourth group, exhibited a tendency to preferentially accumulate within the mesophyll. Importantly, these distinct groups were similarly identified in other sunflower leaves cultivated under the same conditions within the same pot. Certain elements, such as Cd, Ce, Cr, and La, can be taken up by plants even without metabolic function and can be toxic at low levels. Elemental mapping aids in comprehending distribution patterns of elements and their impact on biochemical processes.
Shifting the focus to medicinal plants, the significance of detecting (heavy) metal contamination becomes even more pronounced. This importance is underscored by the dual role these plants play, serving as both sources of valuable therapeutic compounds and components of human consumption. Coptis chinensis Franch, which is an important medicinal plant in China, was analyzed for spatial mapping of metals using LA-ICP-MSI and synchrotron radiation microscopic X-ray fluorescence (SR-μXRF) at a spot diameter of 44 μm.78 LA-ICP-MSI offered a lower detection limit with its superior sensitivity (0.01 μg g−1) compared with XRF (0.1–1 μg g−1); however, this integration provided a non-destructive spatial visualization of multiple elements concurrently. The root cross-sections studied using XRF showed significant Cr accumulation, along with Fe, Mn, Ca, and Zn. Interestingly, the distribution pattern of Cr in rhizome cross-sections differed, with higher intensity signals detected in the external layer, including the periderm and cortex. Given the relatively low chromium concentrations in the leaves, LA-ICP-MS was employed for precise spatial micromapping. The images indicate that chromium is primarily concentrated in the cortex and within certain vascular bundles. In addition, the images indicated that Ca preferentially accumulates in the cortex and specific vascular bundles, resembling the pattern observed for Cr. Similarly, Mn distribution is similar to that of Ca. Moreover, there was an even distribution of P and Cu throughout cross-sections of the entire petiole (Fig. 7). This study elucidated the mechanisms underlying the accumulation of Cr in medicinal plants, offering insights into the high Cr levels found in C. chinensis Franch within agricultural settings. These findings contribute to the development of strategies that mitigate the potential transfer of Cr to humans.
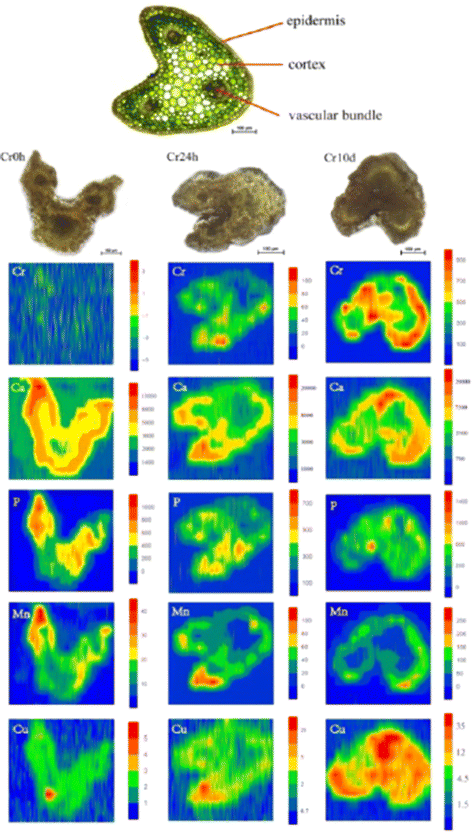 | ||
| Fig. 7 LA-ICP-MS elemental images of Cr, Ca, P, Mn, and Cu in petiole cross-sections of C. chinensis Franch. The upper image show the thin cross-sections (10 μm thick) for indicating the tissue structure of petioles, and the three images below it are the scanned sample cross-sections (300 μm thick) images of petioles from the (left to right) with Cr exposure of 0 hours (Cr0h), 24 hours (Cr24h), and 10 days (Cr10d groups). Each elemental image indicates the relative distribution of a specific element. The color scale (red to blue; highest to lowest) denotes element concentrations. Figure reprinted from ref. 78 with permission from Springer Nature, copyright (2018). | ||
2.5 Laser desorption ionization mass spectrometry imaging (LDI-MSI)
In LDI-MSI, the laser ionizes the molecules directly on the sample surface without the need for a matrix. The analyte ions which are formed are then analyzed by the mass spectrometer.82 In plant MSI, LDI aids in the detection of flavonoids and low molecular weight compounds that have a strong absorbance at the laser wavelength used. LDI has been demonstrated to be able to detect metabolites, glycans, lipids and small proteins.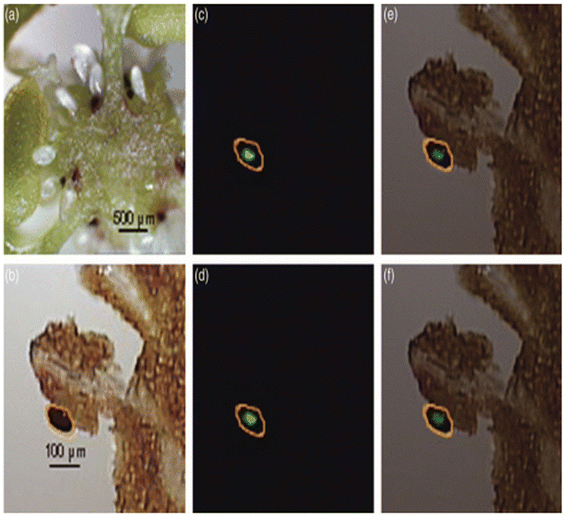 | ||
| Fig. 8 LDI-MSI detection of hypericins in appendices of the placenta of H. perforatum. (a) Cryosection of the ovary of H. perforatum showing the placenta (ovary wall), the funiculi, and ovules. (b) Optical image of the measured region of placenta. The area circled in orange shows expected localization of secondary metabolites. (c and d) Molecular images of (b) for m/z 503 (blue) and m/z 519 (green), respectively. (e and f) Overlays of optical image (b) and molecular images for m/z 503 (blue) and m/z 519 (green), respectively. Figure reprinted from ref. 83 with permission from John Wiley and Sons, copyright (2009). | ||
LDI-MSI has been coupled with other analytical techniques to provide accurate information. For instance, study of the production of phytoalexins during biotic and abiotic stress was to understand the defense mechanisms of grapevines against environmental stress. The spatial distributions of viniferins imaged at a spatial resolution of 50 μm in UV-stressed and Plasmopara viticola-infected Vitis vinifera leaves have been studied using MSI and fluorescence microscopy.84 The high spatial resolution fluorescence microscopy images showed different structures of the leaves and that stilbene phytoalexins were produced in the stomata and protected the leaves from UV radiation. LDI-MSI enabled the identification of specific phytoalexins in the stomata.
2.6 Laser ablation electrospray ionization (LAESI)
LAESI consist of a source that utilizes an infrared laser (2940 nm) to irradiate the sample, followed by analyte extraction and electrospray ionization (ESI). The laser photons are typically absorbed by the water present in the sample surface and result in aerosolization prior to ionization by ESI. The resulting ions are subsequently directed to the mass analyzer.85 It provides a spot size of 15–350 μm depending on source instrumentation.86,87 Studies have shown that LAESI allows the analysis of a wide range of molecules such as sugars, lipids, peptides, proteins and metabolites under ambient conditions, and as such has shown potential in plant and food science applications.87–89 Specifically, LAESI has been applied to study plant metabolism and pesticide accumulation.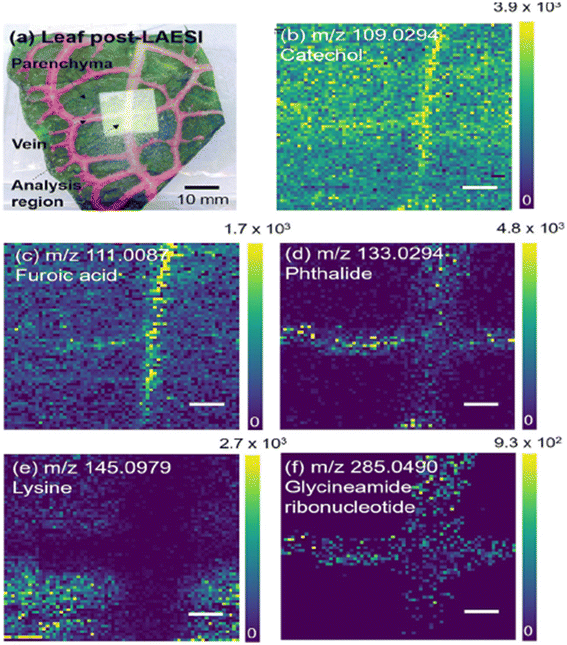 | ||
| Fig. 9 LAESI-MSI of F. argyroneura leaf in high spatial resolution (40 μm) in negative ion mode. (a) Optical image of leaf post-LAESI imaging showing analysis region (lighter region). Single ion images (b–f): (b) catechol (vein), (c) furoic acid (vein), (d) phthalide (vein), (e) lysine (parenchyma), (f) glycineamideribonucleotide (vein). Color bars shown as signal/noise, and scale bars are 200 μm. Reprinted (adapted) from ref. 1 with permission from American Chemical Society, copyright.1 | ||
LAESI enabled the in situ cell-by-cell imaging in plant tissues by taking each cell as a voxel. This chemical imaging approach distinguished between pigmented and non-pigmented cells of onion (A. cepa) at a spatial resolution of 30 μm.90 The molecular image obtained through cell-by-cell analysis revealed the presence of the metabolite cyanidin (m/z 287), responsible for the purple pigmentation in A. cepa epidermal cells. Orthogonal partial least squares discriminant analysis (OPLS-DA) was conducted to investigate the cell-to-cell variations within and between tissues. In the purple A. cepa, the epidermal tissue, non-pigmented cells were abundant with some pigmented cells. The pigmented cells were detected with the presence of flavonoids, anthocyanidins and the associated glucosides. That study introduced an innovative atmospheric pressure MS imaging technique for single-cell analysis. This approach also offers advantages in avoiding the topographical changes that often occur in plant samples under vacuum conditions.
LAESI has also been effectively employed as a validation method to confirm the presence of glucosinolate in the leaves of Arabidopsis thaliana, which had initially been analyzed using MALDI-TOF.26 The A. thaliana leaves exhibited a highly similar set of ions to those obtained from MALDI measurements of a matrix-sublimed leaf containing abundant 4MTB, 4MSOB, I3M, and 8MSOO. These glucosinolates can act as a primary defense against pathogens, especially for A. thaliana rosette leaves, which are closer to the ground and more susceptible to colonization by soil-borne pathogens, necessitating effective defenses. Moreover, adequate concentrations of glucosinolates on A. thaliana leaf surfaces were observed, which proved to be attractive to specialist feeding lepidopterans Plutella xylostella and Pieris rapae for oviposition. The combination of these two techniques has shown remarkable capabilities in understanding plant–pollinator interactions, a crucial aspect of ecological studies.
LAESI-MSI offers another significant application, enabling the imaging of pesticide residues on plant surfaces to ensure the quality and safety of food products.17 This technique has been utilized to study the localization of pesticides in various crops, including lemon, rose, rye, cherry tomato, and maize at a spatial resolution of 150 μm. For instance, in citrus fruit peels, LAESI-MSI identified imazalil and thiabendazole, while in tomato samples, lycoperoside and esculeoside were detected (Fig. 10). This technique facilitated the imaging and detection of pesticides without the need for intricate sample sectioning at ambient ionization conditions.
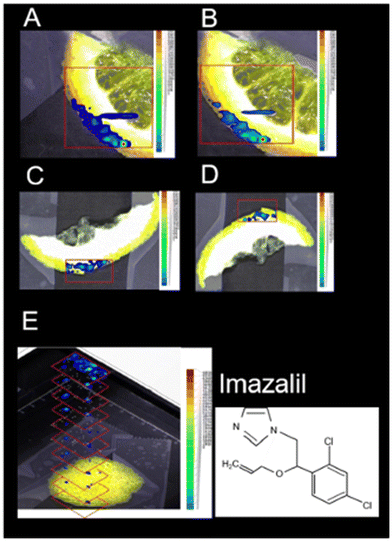 | ||
| Fig. 10 Positive-ion LAESI-TOF-MSI accurate ion maps of (A, C) the 35Cl35Cl isotope [M + H]+ ion at m/z 297.056 (±5 mDa), and (B, D) the 35Cl37Cl isotope [M + H]+ ion at m/z 299.056 (±5 mDa), on (A, B) orange slice and (C, D) lemon slice, showing the spatial distributions of the post-harvest fungicide imazalil. (E) 3D profiling of imazalil in a lemon-peel slice, represented by a stack of 2D ion maps of m/z 297.055. Figure adapted and reprinted from ref. 17 with permission from Springer Nature, copyright (2014 ). | ||
2.7 Desorption electrospray ionization (DESI-MSI)
DESI is an ambient ionization mass spectrometry imaging technique that combines electrospray ionization (ESI) with desorption ionization. The method involves supersonic spraying of electrically charged solvent over the sample surface, which is affixed to a moving stage. The ionized analytes are transported by the high-speed solvent droplets into the mass spectrometer at ambient pressure. Spatial resolution typically varies between 35–200 μm, depending on the instrumentation used.41 An alternate ambient approach, nano-DESI, utilizes a flow probe system that continuously samples a flowing solvent through a solvent micro junction on the sample and channels the extracted analytes into the mass spectrometer through a capillary for nano-ESI analysis. This offers a spatial resolution of 10–200 μm.92–94 DESI exhibits the capability to detect a wide range of molecules, including small metabolites, lipids, peptides, and proteins. DESI has been applied for investigating metabolites, pesticide distribution, and microbial interactions in plant and food samples.The versatility of DESI-MSI in detecting diverse molecules has been effectively employed to study the distribution of metabolites in grapevine stems.14 The grapevine stem was sectioned using a razor blade, and DESI imaging was subsequently conducted with a step size of 200 μm. This analysis revealed the localization of endogenous and exogenous organic acids including malic acid, tartaric acid, citric acid, glutaric acid, and adipic acid in the mass range of m/z 50–200. The organic acids were distributed in the pith, secondary xylem, and secondary phloem of the stem cross-section. The metabolic composition of grapevines plays a crucial role in determining the taste and overall quality of wine, making DESI-MSI a valuable tool in viticulture and enology research.
Continuing in plant metabolomics with DESI-MSI, the spatial distribution and molecular mechanisms of terpenoid biosynthesis in Salvia miltiorrhiza and S. grandifolia were investigated.96 This study focused on the tanshinone and carnosol biosynthetic pathways, two significant sources of diterpenoid components. Molecular imaging research revealed distinct structural features and spatial distributions of these intermediates within the pathways. A total of 33 metabolites involved in the diterpenoid biosynthetic pathway were successfully localized. Despite some similarities, distinct distribution patterns of these target metabolites were observed between the two species. Notably, three upstream precursors (GGPP, CPP, and miltiradiene) were detected only in trace amounts, limiting their identification using the metabolomics approach. Within the tanshinone synthetic pathway, tricyclic diterpenes like 11-hydroxy-sugiol, 11-hydroxy-ferruginol, 11,20-dihydroxy-sugiol, and 11,20-dihydroxy-ferruginol were widely distributed in the xylem, phloem, and periderm of S. grandifolia, while they were specifically localized in the pericardium of S. miltiorrhiza, albeit at lower relative concentrations compared with S. grandifolia. Additionally, the distribution of the metabolites carnosol, ferulic acid, and rosmarinic acid was observed in S. miltiorrhiza and S. grandifolia leaves. This investigation involved a multi-omics approach and utilized LC-MS/MS, PCR and microscopy in addition to DESI-MSI. The combination of multi-omics techniques and DESI-MSI offers a powerful and efficient approach for visualizing the accumulation patterns of natural products and gaining deeper insights into secondary metabolism.
Nano-DESI has also been applied in wine profiling to detect the presence of anthocyanin in dried wine stains and grapes from different cultivars. Moreover, a wine stain on cotton fabric was investigated for the presence of anthocyanin.97 Anthocyanin represents the predominant group of pigmented flavonoids found in nature. They are glycosides of anthocyanidins, where a sugar moiety is primarily attached to position 3 and/or 5 of the molecule. The anthocyanin profile present in fruits and vegetables holds practical value, enabling differentiation among species or varieties and facilitating quality control and authentication of related food products. This application is of significance due to the reported illegal practice of coloring wines using anthocyanins sourced from other fruits. In addition to its use in DESI-MSI for the analysis of stained cotton fabric, this study successfully differentiated between mixtures of different wines and grape slices. This methodology highlights the potential of nano-DESI in exploring compounds within complex matrices like cotton fabric, with potential applications in forensic science for identifying unknown stains. Its capacity to distinguish between wines from distinct cultivars also positions it as a valuable tool in combating fraud within the beverage industry.
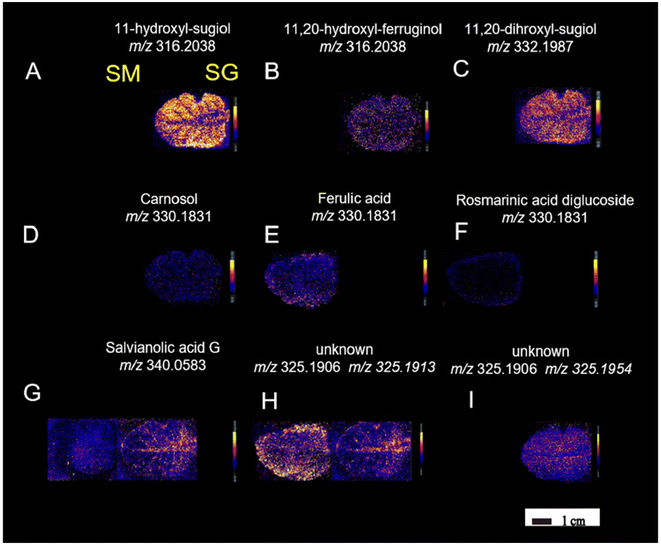 | ||
| Fig. 11 DESI-MSI of metabolites (A–I) in S. miltiorrhiza and S. grandifolia leaves. SM, S. miltiorrhiza; SG, S. grandifolia. Figure adapted and reprinted from ref. 96 with permission from Oxford University Press, copyright (2023). | ||
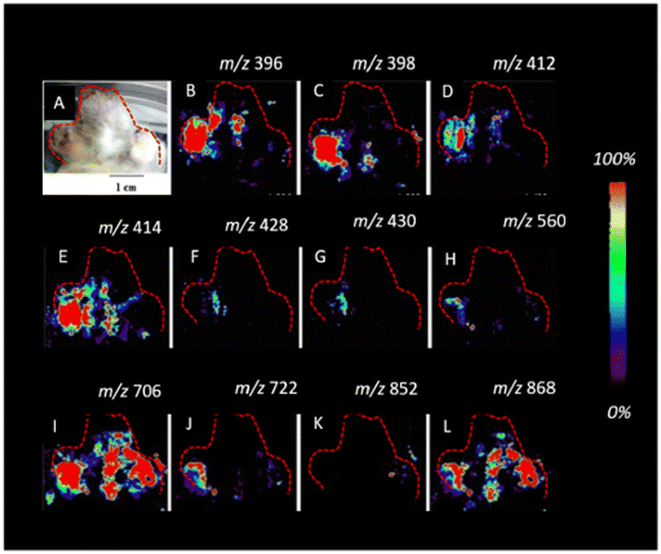 | ||
| Fig. 12 Imaging DESI-MS of an infected potato sprout imprinted on tape, 8 d after inoculation. Localization of the new metabolites (A–L) biosynthesized after fungus invasion can be observed. The infected potato sprout is imprinted directly on tape without further manipulation. The red outline indicates the portion of the sprout and the tuber imaged. Figure adapted and reprinted with permission from ref. 101 with permission from American Chemical Society, copyright (2015). | ||
Elucidation of the spatial distribution of plant secondary metabolites during infection enables the identification of distinct metabolic pathways localized in the vicinity of the infection site. DESI-MSI has also been employed to investigate the interactions between endophytic bacteria and cacao pathogen monocultures.102 In this study, the spatial metabolic distributions of Burkholderia seminalis, the fungus Moniliophthora perniciosa and the oomycetes Phytophthora citrophthora, P. capsici, and P. palmivora were studied. The DESI-MSI measurements were carried out at 200 μm spatial resolution. The samples were cultured, dehydrated, and sprayed with methanol to obtain a flat surface prior to DESI-MSI analysis. In the B. seminalis monocultures, the spatial localization of metabolites and phospholipids such as pyrrolnitrin glycerophosphoethanolamines, glycerophosphatidic acid, and glycerophosphoglycerols were localized. Pyrrolnitrin is known for its potent antifungal properties and the phospholipids act as a permeability barrier. Moreover, rhamnolipids, with their multifaceted functions in motility, cell signaling, and biofilm formation, were also identified in the bacterial colonies. DESI-MSI analysis was further extended to include the fungal and oomycete cultures, leading to the detection and localization of glycerophosphoethanolamines, glycerophosphoglycerols, phosphocholines and glycerophosphoinositols. Additionally, B. seminalis was co-cultured with the pathogenic fungi and oomycetes of cacao to observe its defense mechanisms in action. The defense mechanism of B. seminalis relies on the production of antimicrobial metabolites. Notably, the partially diffuse rhamnolipids (m/z 799 and 815) identified appear to play a significant role in its antifungal properties. The antifungal pyrrolnitrin was localized within the B. seminalis colony (m/z 254). However, its involvement in the inhibitory activity under the specific experimental conditions and incubation times used in this study seems to be limited. This analysis offered valuable insights into the chemical defense mechanisms utilized to combat devastating fungal and oomycete diseases affecting cacao crops worldwide.
2.8 3-D imaging with laser-assisted rapid evaporative ionization mass spectrometry (LA-REIMS)
REIMS operates by directing a laser onto the sample, generating vapors that are then transported to the mass spectrometer through tubing. This tubing, connected to the laser probe on one end and the top of a venturi incorporated in the REIMS source on the other end, facilitated the ionization of aerosols and vapors upon contact with a heated impactor surface in the venturi. Vapor transport is realized by an aspirator pump, and a Q-TOF MS is used to analyze the ions produced. Notably, various (phospho) lipids have been reported as the most abundantly detected species using this ambient approach.105 LA-REIMS can be combined with an automated 3D MS scanner featuring various axes, a sample holder, a distance sensor, clamps to secure the laser probe, and a control box (Fig. 13). Positioned beneath the laser probe, the sample is systematically moved for measurements, maintaining a consistent distance and angle between the probe and the surface.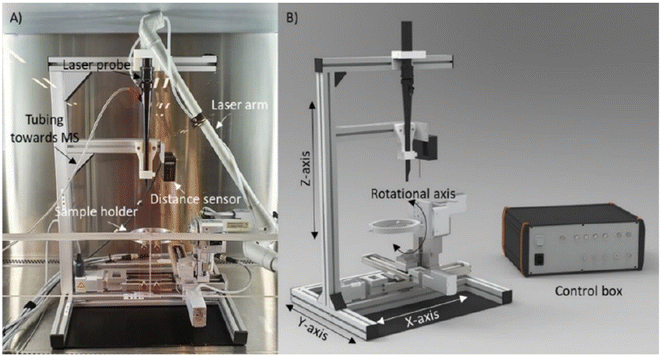 | ||
| Fig. 13 Images of the 3D MS scanner. (A) Picture of the 3D MS scanner setup in the biosafety cabinet with the CO2 laser probe inserted. Besides the laser probe, the following elements are indicated: the tubing through which the created smoke is aspirated toward the mass spectrometer (MS), the distance sensor that is used for the topographical measurement, and the sample holder. (B) Design image of the 3D MS scanner with indication of the three translational axes, the rotational axis, and the control box. Figure reprinted from ref. 16 with permission from American Chemical Society, copyright (2021). | ||
REIMS has demonstrated remarkable potential in acquiring the lipidomic profile of bacteria and fungi, enabling species-level identification.106 It has been successfully employed to analyze various meat products, including beef, pork, fish, and chicken, for the detection and identification of fatty acids, phospholipids, and diacylglycerol.106–110 Moreover, the intelligent knife (i-knife) based on REIMS showcased its potential in determining the geographical origin of pistachios, highlighting its significance in ensuring food authenticity.111
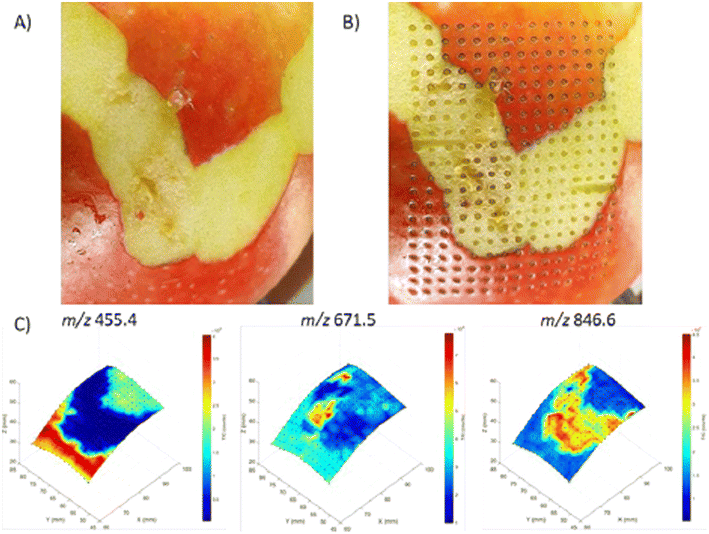 | ||
| Fig. 14 3D visualization of molecular distributions obtained from an apple with the 3D MS scanner. (A) Apple before analysis with LA-REIMS using the 3D MS scanner. Parts of the peel were removed to show different molecular patterns can be created. (B) Apple after analysis with LA-REIMS using the 3D MS scanner. The total number of measurement points acquired was 300 with a spatial resolution of 2 mm, thus covering an area of 28 × 38 mm (15 × 20 measurement points). (C) 3D visualization of the molecular distributions (absolute intensities) of the imaging experiment on an apple. Three m/z values were selected based on their different distributions: 455.4, 671.5, and 846.6. Figure reprinted from ref. 16 with permission from American Chemical Society, copyright (2021). | ||
3. Conclusion
The health of the agricultural ecosystem directly influences the quality and safety of the food we consume, as plants are an essential element in our food chain. This review has described various MSI techniques used to study the localization of molecules in various plant parts, microbial species, and food products. MSI analysis provides spatial information with promising potential to optimize agricultural practices for ensuring quality plant-based food products. Some challenges persist in terms of sample preparation, spatial resolution, lack of plant-focused molecular databases and analysis of complex datasets, that are ameliorated with innovative approaches. The current state-of-the-art and developments in sample preparation protocols and the multimodal imaging approaches have opened up molecular profiling possibilities for complex plant-based samples. For example, studying the spatial distribution of molecules on surface and internal structures of a plant-leaf is possible with a combination of DESI and MALDI-MSI. In this case, DESI can ionize small metabolites on the surface while MALDI can provide the higher spatial resolution required to differentiate, for instance, xylem and phloem. Similarly, a combined LDI and MALDI-MSI analytical approach can solve the problem of detecting small metabolites within the same sample, that are often suppressed by MALDI-matrix signals, and compounds that necessitate matrices for ionization. In addition, analyzing complex datasets necessitates robust and standardized data-processing pipelines to ensure comparability of MSI data across different studies and laboratories. Ongoing and future developments in instrumentation and database availability will make MSI complementary to chromatography-based techniques to confirm the detection and identities. Despite the challenges, MSI techniques continue to revolutionize our understanding of plant and microbial metabolism, leading to safer and more authentic food. By bridging the gap between MSI methodologies and biological processes, these innovations provide insights into plant biology, thereby contributing to a more sustainable and productive agricultural future.Author contributions
R. M. A. H., B. C. P. and E. C. conceived the idea for the review and outlined the scope and structure. M. V. conducted the literature search and drafted the initial manuscript. R. M. A. H., B. C. P. and E. C. provided critical feedback and revisions to the draft, figures and tables, and edited the final version of the manuscript. All authors reviewed and approved the final version of the review before submission.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors thank Hang Nguyen (Maastricht University) for assistance in improving the English language and readability of this article. This research was supported by FoodTraNet Project funded by European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 956265.References
- M. J. Taylor, A. Liyu, A. Vertes and C. R. Anderton, Ambient Single-Cell Analysis and Native Tissue Imaging Using Laser-Ablation Electrospray Ionization Mass Spectrometry with Increased Spatial Resolution, J. Am. Soc. Mass Spectrom., 2021, 32(9), 2490–2494 CrossRef CAS PubMed.
- T. F. C. Chin-A-Woeng, G. V. Bloemberg, A. J. van der Bij, K. M. G. M. van der Drift, J. Schripsema, B. Kroon, R. J. Scheffer, C. Keel, P. A. H. M. Bakker, H.-V. Tichy, F. J. de Bruijn, J. E. Thomas-Oates and B. J. J. Lugtenberg, Biocontrol by Phenazine-1-carboxamide-Producing Pseudomonas chlororaphis PCL1391 of Tomato Root Rot Caused by Fusarium oxysporum f. sp. radicis-lycopersici, Mol. Plant-Microbe Interact., 1998, 11(11), 1069–1077 CrossRef CAS.
- S. C. Jung, A. Martinez-Medina, J. A. Lopez-Raez and M. J. Pozo, Mycorrhiza-Induced Resistance and Priming of Plant Defenses, J. Chem. Ecol., 2012, 38(6), 651–664 CrossRef CAS PubMed.
- V. Brighenti, M. Licata, T. Pedrazzi, D. Maran, D. Bertelli, F. Pellati and S. Benvenuti, Development of a new method for the analysis of cannabinoids in honey by means of high-performance liquid chromatography coupled with electrospray ionisation-tandem mass spectrometry detection, J. Chromatogr. A, 2019, 1597, 179–186 CrossRef CAS PubMed.
- J. M. Montiel-León, S. V. Duy, G. Munoz, M.-A. Verner, M. Y. Hendawi, H. Moya, M. Amyot and S. Sauvé, Occurrence of pesticides in fruits and vegetables from organic and conventional agriculture by QuEChERS extraction liquid chromatography tandem mass spectrometry, Food Control, 2019, 104, 74–82 CrossRef.
- M. Esteki, Z. Shahsavari and J. Simal-Gandara, Food identification by high performance liquid chromatography fingerprinting and mathematical processing, Food Res. Int., 2019, 122, 303–317 CrossRef CAS PubMed.
- R. Weng, S. Lou, X. Pang, Y. Song, X. Su, Z. Xiao and J. Qiu, Multi-residue analysis of 126 pesticides in chicken muscle by ultra-high-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry, Food Chem., 2020, 309, 125503 CrossRef CAS PubMed.
- E. Schievano, M. Stocchero, V. Zuccato, I. Conti and L. Piana, NMR assessment of European acacia honey origin and composition of EU-blend based on geographical floral markers, Food Chem., 2019, 288, 96–101 CrossRef CAS PubMed.
- J. Pinto, A. S. Oliveira, J. Azevedo, V. De Freitas, P. Lopes, I. Roseira, M. Cabral and P. Guedes de Pinho, Assessment of oxidation compounds in oaked Chardonnay wines: A GC-MS and (1)H NMR metabolomics approach, Food Chem., 2018, 257, 120–127 CrossRef CAS PubMed.
- A. Steele, D. T. Goddard and I. B. Beech, An atomic force microscopy study of the biodeterioration of stainless steel in the presence of bacterial biofilms, Int. Biodeterior. Biodegrad., 1994, 34(1), 35–46 CrossRef CAS.
- T. Yamada, H. Arakawa, T. Okajima, T. Shimada and A. Ikai, Use of AFM for imaging and measurement of the mechanical properties of light-convertible organelles in plants, Ultramicroscopy, 2002, 91(1), 261–268 CrossRef CAS PubMed.
- E. Lesniewska, M. Adrian, A. Klinguer and A. Pugin, Cell wall modification in grapevine cells in response to UV stress investigated by atomic force microscopy, Ultramicroscopy, 2004, 100(3), 171–178 CrossRef CAS PubMed.
- Y. Wen, Z. Xu, Y. Liu, H. Corke and Z. Sui, Investigation of food microstructure and texture using atomic force microscopy: A review, Compr. Rev. Food Sci. Food Saf., 2020, 19(5), 2357–2379 CrossRef CAS PubMed.
- Y. Dong, B. Li, S. Malitsky, I. Rogachev, A. Aharoni, F. Kaftan, A. Svatoš and P. Franceschi, Sample Preparation for Mass Spectrometry Imaging of Plant Tissues: A Review, Front. Plant Sci., 2016, 7, 60 Search PubMed.
- C. Li, X. Kang, J. Nie, A. Li, M. A. Farag, C. Liu, K. M. Rogers, J. Xiao and Y. Yuan, Recent advances in Chinese food authentication and origin verification using isotope ratio mass spectrometry, Food Chem., 2023, 398, 133896 CrossRef CAS PubMed.
- S. P. Nauta, P. Huysmans, G. J. M. Tuijthof, G. B. Eijkel, M. Poeze, T. P. Siegel and R. M. A. Heeren, Automated 3D Sampling and Imaging of Uneven Sample Surfaces with LA-REIMS, J. Am. Soc. Mass Spectrom., 2022, 33(1), 111–122 CrossRef CAS PubMed.
- M. W. F. Nielen and T. A. van Beek, Macroscopic and microscopic spatially-resolved analysis of food contaminants and constituents using laser-ablation electrospray ionization mass spectrometry imaging, Anal. Bioanal. Chem., 2014, 406(27), 6805–6815 CrossRef CAS PubMed.
- C. J. Shih, P. Y. Chen, C. C. Liaw, Y. M. Lai and Y. L. Yang, Bringing microbial interactions to light using imaging mass spectrometry, Nat. Prod. Rep., 2014, 31(6), 739–755 RSC.
- J. Y. Yang, V. V. Phelan, R. Simkovsky, J. D. Watrous, R. M. Trial, T. C. Fleming, R. Wenter, B. S. Moore, S. S. Golden, K. Pogliano and P. C. Dorrestein, Primer on Agar-Based Microbial Imaging Mass Spectrometry, J. Bacteriol., 2012, 194(22), 6023–6028 CrossRef CAS PubMed.
- L. A. McDonnell and R. M. A. Heeren, Imaging mass spectrometry, Mass Spectrom. Rev., 2007, 26(4), 606–643 CrossRef CAS PubMed.
- B. Bartels, P. Kulkarni, N. Danz, S. Böcker, H. P. Saluz and A. Svatoš, Mapping metabolites from rough terrain: laser ablation electrospray ionization on non-flat samples, RSC Adv., 2017, 7(15), 9045–9050 RSC.
- M. Maia, A. McCann, C. Malherbe, J. Far, J. Cunha, J. Eiras-Dias, C. Cordeiro, G. Eppe, L. Quinton, A. Figueiredo, E. D. Pauw and M. S. Silva, Grapevine leaf MALDI-MS imaging reveals the localisation of a putatively identified sucrose metabolite associated to Plasmopara viticola development, bioRxiv, 2022, preprint, DOI:10.1101/2022.08.01.502001.
- L. A. McDonnell, T. H. Mize, S. L. Luxembourg, S. Koster, G. B. Eijkel, E. Verpoorte, N. F. de Rooij and R. M. Heeren, Using matrix peaks to map topography: increased mass resolution and enhanced sensitivity in chemical imaging, Anal. Chem., 2003, 75(17), 4373–4381 CrossRef CAS PubMed.
- V. K. Balasubramanian, D. Veličković, M. D. M. Rubio Wilhelmi, C. R. Anderton, C. N. Stewart, S. DiFazio, E. Blumwald and A. H. Ahkami, Spatiotemporal metabolic responses to water deficit stress in distinct leaf cell-types of poplar, Front. Plant Sci., 2024, 15, 1346853 CrossRef PubMed.
- T. Hoffmann and P. C. Dorrestein, Homogeneous Matrix Deposition on Dried Agar for MALDI Imaging Mass Spectrometry of Microbial Cultures, J. Am. Soc. Mass Spectrom., 2015, 26(11), 1959–1962 CrossRef CAS PubMed.
- R. Shroff, K. Schramm, V. Jeschke, P. Nemes, A. Vertes, J. Gershenzon and A. Svatoš, Quantification of plant surface metabolites by matrix-assisted laser desorption-ionization mass spectrometry imaging: glucosinolates on Arabidopsis thaliana leaves, Plant J., 2015, 81(6), 961–972 CrossRef CAS PubMed.
- J. N. Jens, D. J. Breiner and V. V. Phelan, Spray-Based Application of Matrix to Agar-Based Microbial Samples for Reproducible Sample Adherence in MALDI MSI, J. Am. Soc. Mass Spectrom., 2022, 33(4), 731–734 CrossRef CAS PubMed.
- M. Maia, A. McCann, C. Malherbe, J. Far, J. Cunha, J. Eiras-Dias, C. Cordeiro, G. Eppe, L. Quinton, A. Figueiredo, E. De Pauw and M. Sousa Silva, Grapevine leaf MALDI-MS imaging reveals the localisation of a putatively identified sucrose metabolite associated to Plasmopara viticola development, Front. Plant Sci., 2022, 13, 1012636 CrossRef PubMed.
- M. D. B. B. Lorensen, S. Y. Hayat, N. Wellner, N. Bjarnholt and C. Janfelt, Leaves of Cannabis sativa and their trichomes studied by DESI and MALDI mass spectrometry imaging for their contents of cannabinoids and flavonoids, Phytochem. Anal., 2023, 34(3), 269–279 CrossRef CAS PubMed.
- C. Seaman, B. Flinders, G. Eijkel, R. M. A. Heeren, N. Bricklebank and M. R. Clench, “Afterlife Experiment”: Use of MALDI-MS and SIMS Imaging for the Study of the Nitrogen Cycle within Plants, Anal. Chem., 2014, 86(20), 10071–10077 CrossRef CAS PubMed.
- B. A. Boughton, D. Thinagaran, D. Sarabia, A. Bacic and U. Roessner, Mass spectrometry imaging for plant biology: a review, Phytochem. Rev., 2016, 15(3), 445–488 CrossRef CAS PubMed.
- L. Í. L. Maciel, R. A. Bernardo, R. O. Martins, A. C. Batista Junior, J. V. A. Oliveira, A. R. Chaves and B. G. Vaz, Desorption electrospray ionization and matrix-assisted laser desorption/ionization as imaging approaches for biological samples analysis, Anal. Bioanal. Chem., 2023, 415(18), 4125–4145 CrossRef PubMed.
- K. M. Engel, P. Prabutzki, J. Leopold, A. Nimptsch, K. Lemmnitzer, D. R. N. Vos, C. Hopf and J. Schiller, A new update of MALDI-TOF mass spectrometry in lipid research, Prog. Lipid Res., 2022, 86, 101145 CrossRef CAS PubMed.
- R. Sun, Y. Zhang, W. Tang and B. Li, Submicron 3,4-dihydroxybenzoic acid–TiO2 composite particles for enhanced MALDI MS imaging of secondary metabolites in the root of differently aged baical skullcap, Analyst, 2022, 147(13), 3017–3024 RSC.
- M. Noun, R. Akoumeh and I. Abbas, Cell and Tissue Imaging by TOF-SIMS and MALDI-TOF: An Overview for Biological and Pharmaceutical Analysis, Microsc. Microanal., 2022, 28(1), 1–26 CrossRef CAS PubMed.
- F. P. Y. Barré, M. R. L. Paine, B. Flinders, A. J. Trevitt, P. D. Kelly, R. Ait-Belkacem, J. P. Garcia, L. B. Creemers, J. Stauber, R. J. Vreeken, B. Cillero-Pastor, S. R. Ellis and R. M. A. Heeren, Enhanced Sensitivity Using MALDI Imaging Coupled with Laser Postionization (MALDI-2) for Pharmaceutical Research, Anal. Chem., 2019, 91(16), 10840–10848 CrossRef PubMed.
- L. Molina-Millán, A. Körber, B. Flinders, B. Cillero-Pastor, E. Cuypers and R. M. A. Heeren, MALDI-2 Mass Spectrometry for Synthetic Polymer Analysis, Macromolecules, 2023, 56(19), 7729–7736 CrossRef PubMed.
- R. M. A. Heeren, L. A. McDonnell, E. Amstalden, S. L. Luxembourg, A. F. M. Altelaar and S. R. Piersma, Why don't biologists use SIMS?: A critical evaluation of imaging MS, Appl. Surf. Sci., 2006, 252(19), 6827–6835 CrossRef CAS.
- A. R. Buchberger, K. DeLaney, J. Johnson and L. Li, Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights, Anal. Chem., 2018, 90(1), 240–265 CrossRef CAS PubMed.
- P. Agüi-Gonzalez, S. Jähne and N. T. N. Phan, SIMS imaging in neurobiology and cell biology, J. Anal. At. Spectrom., 2019, 34(7), 1355–1368 RSC.
- D. I. Campbell, C. R. Ferreira, L. S. Eberlin and R. G. Cooks, Improved spatial resolution in the imaging of biological tissue using desorption electrospray ionization, Anal. Bioanal. Chem., 2012, 404(2), 389–398 CrossRef CAS PubMed.
- Y. J. Lee, D. C. Perdian, Z. Song, E. S. Yeung and B. J. Nikolau, Use of mass spectrometry for imaging metabolites in plants, Plant J., 2012, 70(1), 81–95 CrossRef CAS PubMed.
- A. Körber, J. D. Keelor, B. S. R. Claes, R. M. A. Heeren and I. G. M. Anthony, Fast Mass Microscopy: Mass Spectrometry Imaging of a Gigapixel Image in 34 Minutes, Anal. Chem., 2022, 94(42), 14652–14658 CrossRef PubMed.
- J. L. Norris and R. M. Caprioli, Analysis of Tissue Specimens by Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry in Biological and Clinical Research, Chem. Rev., 2013, 113(4), 2309–2342 CrossRef CAS PubMed.
- M. Maia, A. Aziz, P. Jeandet and V. Carré, Profiling and Localization of Stilbene Phytoalexins Revealed by MALDI-MSI during the Grapevine–Botrytis cinerea Interaction, J. Agric. Food Chem., 2023, 71(42), 15569–15581 CrossRef PubMed.
- H. Ye, E. Gemperline, M. Venkateshwaran, R. Chen, P. M. Delaux, M. Howes-Podoll, J. M. Ané and L. Li, MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula-Sinorhizobium meliloti symbiosis, Plant J., 2013, 75(1), 130–145 CrossRef CAS PubMed.
- A. T. Klein, G. B. Yagnik, J. D. Hohenstein, Z. Ji, J. Zi, M. D. Reichert, G. C. MacIntosh, B. Yang, R. J. Peters, J. Vela and Y. J. Lee, Investigation of the Chemical Interface in the Soybean–Aphid and Rice–Bacteria Interactions Using MALDI-Mass Spectrometry Imaging, Anal. Chem., 2015, 87(10), 5294–5301 CrossRef CAS PubMed.
- H. Enomoto, T. Sensu, E. Yumoto, T. Yokota and H. Yamane, Derivatization for detection of abscisic acid and 12-oxo-phytodienoic acid using matrix-assisted laser desorption/ionization imaging mass spectrometry, Rapid Commun. Mass Spectrom., 2018, 32(17), 1565–1572 CrossRef CAS PubMed.
- A. P. Dare, C. S. Günther, A. C. Grey, G. Guo, N. J. Demarais, S. Cordiner, T. K. McGhie, H. Boldingh, M. Hunt, C. Deng, K. Karppinen, L. Jaakola and R. V. Espley, Resolving the developmental distribution patterns of polyphenols and related primary metabolites in bilberry (Vaccinium myrtillus) fruit, Food Chem., 2022, 374, 131703 CrossRef CAS PubMed.
- J. Nakamura, T. Morikawa-Ichinose, Y. Fujimura, E. Hayakawa, K. Takahashi, T. Ishii, D. Miura and H. Wariishi, Spatially resolved metabolic distribution for unraveling the physiological change and responses in tomato fruit using matrix-assisted laser desorption/ionization–mass spectrometry imaging (MALDI–MSI), Anal. Bioanal. Chem., 2017, 409(6), 1697–1706 CrossRef CAS PubMed.
- Y. Kazachkova, I. Zemach, S. Panda, S. Bocobza, A. Vainer, I. Rogachev, Y. Dong, S. Ben-Dor, D. Veres, C. Kanstrup, S. K. Lambertz, C. Crocoll, Y. Hu, E. Shani, S. Michaeli, H. H. Nour-Eldin, D. Zamir and A. Aharoni, The GORKY glycoalkaloid transporter is indispensable for preventing tomato bitterness, Nat. Plants, 2021, 7(4), 468–480 CrossRef CAS PubMed.
- M. Vats, B. Cillero-Pastor, B. Flinders, E. Cuypers and R. M. A. Heeren, Mass spectrometry imaging reveals flavor distribution in edible mushrooms, J. Food Sci. Technol., 2023, 61, 888–896 CrossRef PubMed.
- N. A. dos Santos, C. M. de Almeida, F. F. Gonçalves, R. S. Ortiz, R. M. Kuster, D. Saquetto and W. Romão, Analysis of Erythroxylum coca Leaves by Imaging Mass Spectrometry (MALDI–FT–ICR IMS), J. Am. Soc. Mass Spectrom., 2021, 32(4), 946–955 CrossRef CAS PubMed.
- N. Li, J. Dong, C. Dong, Y. Han, H. Liu, F. Du and H. Nie, Spatial Distribution of Endogenous Molecules in Coffee Beans by Atmospheric Pressure Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging, J. Am. Soc. Mass Spectrom., 2020, 31(12), 2503–2510 CrossRef CAS PubMed.
- A. C. Hertel Pereira, A. C. Auer, L. Biedel, C. M. de Almeida, W. Romão and D. C. Endringer, Analysis of Gliricidia sepium Leaves by MALDI Mass Spectrometry Imaging, J. Am. Soc. Mass Spectrom., 2022, 33(3), 573–583 CrossRef CAS PubMed.
- M. Arts, Z. Soons, S. R. Ellis, K. A. Pierzchalski, B. Balluff, G. B. Eijkel, L. J. Dubois, N. G. Lieuwes, S. M. Agten and T. M. Hackeng, Detection of localized hepatocellular amino acid kinetics by using mass spectrometry imaging of stable isotopes, Angew. Chem., Int. Ed., 2017, 56(25), 7146–7150 CrossRef CAS PubMed.
- T. Hoshi and M. Kudo, High resolution static SIMS imaging by time of flight SIMS, Appl. Surf. Sci., 2003, 203–204, 818–824 CrossRef CAS.
- E. A. Bonnin and S. O. Rizzoli, Novel Secondary Ion Mass Spectrometry Methods for the Examination of Metabolic Effects at the Cellular and Subcellular Levels, Front. Behav. Neurosci., 2020, 14, 124 CrossRef CAS PubMed.
- S. Vaidyanathan, J. S. Fletcher, R. Goodacre, N. P. Lockyer, J. Micklefield and J. C. Vickerman, Subsurface Biomolecular Imaging of Streptomyces coelicolor Using Secondary Ion Mass Spectrometry, Anal. Chem., 2008, 80(6), 1942–1951 CrossRef CAS PubMed.
- J. B. Cliff, D. J. Gaspar, P. J. Bottomley and D. D. Myrold, Exploration of Inorganic C and N Assimilation by Soil Microbes with Time-of-Flight Secondary Ion Mass Spectrometry, Appl. Environ. Microbiol., 2002, 68(8), 4067–4073 CrossRef PubMed.
- V. Nihorimbere, H. Cawoy, A. Seyer, A. Brunelle, P. Thonart and M. Ongena, Impact of rhizosphere factors on cyclic lipopeptide signature from the plant beneficial strain Bacillus amyloliquefaciens S499, FEMS Microbiol. Ecol., 2012, 79(1), 176–191 CrossRef CAS PubMed.
- P. L. Clode, M. R. Kilburn, D. L. Jones, E. A. Stockdale, J. B. Cliff III, A. M. Herrmann and D. V. Murphy, In Situ Mapping of Nutrient Uptake in the Rhizosphere Using Nanoscale Secondary Ion Mass Spectrometry, Plant Physiol., 2009, 151(4), 1751–1757 CrossRef CAS PubMed.
- A. Cuypers, T. Remans, N. Weyens, J. Colpaert, A. Vassilev and J. Vangronsveld, Soil-Plant Relationships of Heavy Metals and Metalloids, 2013, pp. 161–193 Search PubMed.
- P. Hinsinger and F. Courchesne, Mobility and bioavailability of heavy metals and metalloids at the soil-root interface, in Biophysico-chemical Processes of Heavy Metals and Metalloids in Soil Environments, 2008, pp. 267–311 Search PubMed.
- S. Behrens, T. Lösekann, J. Pett-Ridge, P. K. Weber, W. O. Ng, B. S. Stevenson, I. D. Hutcheon, D. A. Relman and A. M. Spormann, Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS, Appl. Environ. Microbiol., 2008, 74(10), 3143–3150 CrossRef CAS PubMed.
- M. C. Perkins, G. Bell, D. Briggs, M. C. Davies, A. Friedman, C. A. Hart, C. J. Roberts and F. J. M. Rutten, The application of ToF-SIMS to the analysis of herbicide formulation penetration into and through leaf cuticles, Colloids Surf., B, 2008, 67(1), 1–13 CrossRef CAS PubMed.
- S. Yang, M. He, Y. Zhi, S. X. Chang, B. Gu, X. Liu and J. Xu, An integrated analysis on source-exposure risk of heavy metals in agricultural soils near intense electronic waste recycling activities, Environ Int, 2019, 133, 105239 CrossRef CAS PubMed.
- R. R. Martin, S. J. Naftel, S. Macfie, W. Skinner, F. Courchesne and V. Séguin, Time of flight secondary ion mass spectrometry studies of the distribution of metals between the soil, rhizosphere and roots of Populus tremuloides Minchx growing in forest soil, Chemosphere, 2004, 54(8), 1121–1125 CrossRef CAS PubMed.
- P. Massonnet and R. M. A. Heeren, A concise tutorial review of TOF-SIMS based molecular and cellular imaging, J. Anal. At. Spectrom., 2019, 34(11), 2217–2228 RSC.
- L. Wu, X. Lu, K. S. Kulp, M. G. Knize, E. S. F. Berman, E. J. Nelson, J. S. Felton and K. J. J. Wu, Imaging and differentiation of mouse embryo tissues by ToF-SIMS, Int. J. Mass Spectrom., 2007, 260(2), 137–145 CrossRef CAS.
- J. G. Swales, N. Strittmatter, J. W. Tucker, M. R. Clench, P. J. H. Webborn and R. J. A. Goodwin, Spatial Quantitation of Drugs in tissues using Liquid Extraction Surface Analysis Mass Spectrometry Imaging, Sci. Rep., 2016, 6(1), 37648 CrossRef CAS PubMed.
- M. K. Mandal, T. Ozawa, S. Saha, M. M. Rahman, M. Iwasa, Y. Shida, H. Nonami and K. Hiraoka, Development of Sheath-Flow Probe Electrospray Ionization Mass Spectrometry and Its Application to Real Time Pesticide Analysis, J. Agric. Food Chem., 2013, 61(33), 7889–7895 CrossRef PubMed.
- K. I. Kocurek, L. Stones, J. Bunch, R. C. May and H. J. Cooper, Top-Down LESA Mass Spectrometry Protein Analysis of Gram-positive and Gram-negative Bacteria, J. Am. Soc. Mass Spectrom., 2017, 28(10), 2066–2077 CrossRef CAS PubMed.
- R. C. Menezes, M. Kai, K. Krause, C. Matthäus, A. Svatoš, J. Popp and E. Kothe, Monitoring metabolites from Schizophyllum commune interacting with Hypholoma fasciculare combining LESA-HR mass spectrometry and Raman microscopy, Anal. Bioanal. Chem., 2015, 407(8), 2273–2282 CrossRef PubMed.
- X. Liu and A. B. Hummon, Mass Spectrometry Imaging of Therapeutics from Animal Models to Three-Dimensional Cell Cultures, Anal. Chem., 2015, 87(19), 9508–9519 CrossRef CAS PubMed.
- N. Yamaji and J. F. Ma, Bioimaging of multiple elements by high-resolution LA-ICP-MS reveals altered distribution of mineral elements in the nodes of rice mutants, Plant J., 2019, 99(6), 1254–1263 CrossRef PubMed.
- A. Kötschau, G. Büchel, J. W. Einax, C. Fischer, W. von Tümpling and D. Merten, Mapping of macro and micro elements in the leaves of sunflower (Helianthus annuus) by Laser Ablation–ICP–MS, Microchem. J., 2013, 110, 783–789 CrossRef.
- W. Huang, J. Jiao, M. Ru, Z. Bai, H. Yuan, Z. Bao and Z. Liang, Localization and Speciation of Chromium in Coptis chinensis Franch. using Synchrotron Radiation X-ray Technology and Laser Ablation ICP-MS, Sci. Rep., 2018, 8(1), 8603 CrossRef PubMed.
- A. Pedrosa Diniz, A. Rodrigues Kozovits, C. de Carvalho Lana, A. Trópia de Abreu and M. Garcia Praça Leite, Quantitative analysis of plant leaf elements using the LA-ICP-MS technique, Int. J. Mass Spectrom., 2019, 435, 251–258 CrossRef CAS.
- D. Günther and B. Hattendorf, Solid sample analysis using laser ablation inductively coupled plasma mass spectrometry, TrAC, Trends Anal. Chem., 2005, 24(3), 255–265 CrossRef.
- N. Miliszkiewicz, S. Walas and A. Tobiasz, Current approaches to calibration of LA-ICP-MS analysis, J. Anal. At. Spectrom., 2015, 30(2), 327–338 RSC.
- K. Downard, Mass Spectrometry: A Foundation Course, The Royal Society of Chemistry, 2004 Search PubMed.
- D. Hölscher, R. Shroff, K. Knop, M. Gottschaldt, A. Crecelius, B. Schneider, D. G. Heckel, U. S. Schubert and A. Svatos, Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species, Plant J., 2009, 60(5), 907–918 CrossRef PubMed.
- L. Becker, S. Bellow, V. Carré, G. Latouche, A. Poutaraud, D. Merdinoglu, S. C. Brown, Z. G. Cerovic and P. Chaimbault, Correlative Analysis of Fluorescent Phytoalexins by Mass Spectrometry Imaging and Fluorescence Microscopy in Grapevine Leaves, Anal. Chem., 2017, 89(13), 7099–7106 CrossRef CAS PubMed.
- B. Bartels and A. Svatoš, Spatially resolved in vivo plant metabolomics by laser ablation-based mass spectrometry imaging (MSI) techniques: LDI-MSI and LAESI, Front. Plant Sci., 2015, 6, 471 Search PubMed.
- B. Shrestha and A. Vertes, In Situ Metabolic Profiling of Single Cells by Laser Ablation Electrospray Ionization Mass Spectrometry, Anal. Chem., 2009, 81(20), 8265–8271 CrossRef CAS PubMed.
- P. Nemes and A. Vertes, Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry, Anal. Chem., 2007, 79(21), 8098–8106 CrossRef PubMed.
- P. Nemes, A. A. Barton, Y. Li and A. Vertes, Ambient Molecular Imaging and Depth Profiling of Live Tissue by Infrared Laser Ablation Electrospray Ionization Mass Spectrometry, Anal. Chem., 2008, 80(12), 4575–4582 CrossRef CAS PubMed.
- B. Shrestha, P. Nemes, J. Nazarian, Y. Hathout, E. P. Hoffman and A. Vertes, Direct analysis of lipids and small metabolites in mouse brain tissue by AP IR-MALDI and reactive LAESI mass spectrometry, Analyst, 2010, 135(4), 751–758 RSC.
- B. Shrestha, J. M. Patt and A. Vertes, In Situ Cell-by-Cell Imaging and Analysis of Small Cell Populations by Mass Spectrometry, Anal. Chem., 2011, 83(8), 2947–2955 CrossRef CAS PubMed.
- S. P. Annangudi, E. Gemperline and J. R. Gilbert, Spatial and Depth Profiling of Agricultural Formulations in Leaf Tissue Using LAESI Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2024, 35(5), 1007–1011 CrossRef CAS PubMed.
- R. Yin, J. Kyle, K. Burnum-Johnson, K. J. Bloodsworth, L. Sussel, C. Ansong and J. Laskin, High Spatial Resolution Imaging of Mouse Pancreatic Islets Using Nanospray Desorption Electrospray Ionization Mass Spectrometry, Anal. Chem., 2018, 90(11), 6548–6555 CrossRef CAS PubMed.
- J. Laskin, B. S. Heath, P. J. Roach, L. Cazares and O. J. Semmes, Tissue Imaging Using Nanospray Desorption Electrospray Ionization Mass Spectrometry, Anal. Chem., 2012, 84(1), 141–148 CrossRef CAS PubMed.
- C.-C. Hsu, P.-T. Chou and R. N. Zare, Imaging of Proteins in Tissue Samples Using Nanospray Desorption Electrospray Ionization Mass Spectrometry, Anal. Chem., 2015, 87(22), 11171–11175 CrossRef CAS PubMed.
- Y. Zhao, M. Jiang, M. Liu, H. Wang, W. Wang, T. Zhang, X. Tian, L. Hong, F. Yang, Y. Wang, Y. Zou, H. Yu, Z. Li and W. Yang, Spatial Distribution and Characterization of the Small-Molecule Metabolites and In Situ Hydrolyzed Oligosaccharides in the Rhizome of Glycyrrhiza uralensis by Desorption Electrospray Ionization-Mass Spectrometry Imaging and High-Resolution Liquid Chromatography–Mass Spectrometry, J. Agric. Food Chem., 2023, 71(50), 20372–20385 CrossRef CAS PubMed.
- J. Xia, G. Lou, L. Zhang, Y. Huang, J. Yang, J. Guo, Z. Qi, Z. Li, G. Zhang, S. Xu, X. Song, X. Zhang, Y. Wei, Z. Liang and D. Yang, Unveiling the spatial distribution and molecular mechanisms of terpenoid biosynthesis in Salvia miltiorrhiza and S. grandifolia using multi-omics and DESI–MSI, Hortic. Res., 2023, 10(7), uhad109 CrossRef CAS PubMed.
- L. Hartmanova, V. Ranc, B. Papouskova, P. Bednar, V. Havlicek and K. Lemr, Fast profiling of anthocyanins in wine by desorption nano-electrospray ionization mass spectrometry, J. Chromatogr., A, 2010, 1217(25), 4223–4228 CrossRef CAS PubMed.
- S. Gerbig, H. E. Brunn, B. Spengler and S. Schulz, Spatially resolved investigation of systemic and contact pesticides in plant material by desorption electrospray ionization mass spectrometry imaging (DESI-MSI), Anal. Bioanal. Chem., 2015, 407(24), 7379–7389 CrossRef PubMed.
- A. Tata, C. J. Perez, T. S. Hamid, M. A. Bayfield and D. R. Ifa, Analysis of metabolic changes in plant pathosystems by imprint imaging DESI-MS, J. Am. Soc. Mass Spectrom., 2015, 26(4), 641–648 CrossRef CAS PubMed.
- K. A. Aliferis and S. Jabaji, FT-ICR/MS and GC-EI/MS Metabolomics Networking Unravels Global Potato Sprout's Responses to Rhizoctonia solani Infection, PLoS One, 2012, 7(8), e42576 CrossRef CAS PubMed.
- A. Tata, C. J. Perez, T. S. Hamid, M. A. Bayfield and D. R. Ifa, Analysis of Metabolic Changes in Plant Pathosystems by Imprint Imaging DESI-MS, J. Am. Soc. Mass Spectrom., 2015, 26(4), 641–648 CrossRef CAS PubMed.
- F. D. S. Araújo, R. L. Vieira, E. P. L. Molano, H. J. Máximo, R. J. D. Dalio, P. H. Vendramini, W. L. Araújo and M. N. Eberlin, Desorption electrospray ionization mass spectrometry imaging reveals chemical defense of Burkholderia seminalis against cacao pathogens, RSC Adv., 2017, 7(48), 29953–29958 RSC.
- M. W. Towers, T. Karancsi, E. A. Jones, S. D. Pringle and E. Claude, Optimised Desorption Electrospray Ionisation Mass Spectrometry Imaging (DESI-MSI) for the Analysis of Proteins/Peptides Directly from Tissue Sections on a Travelling Wave Ion Mobility Q-ToF, J. Am. Soc. Mass Spectrom., 2018, 29(12), 2456–2466 CrossRef CAS PubMed.
- A. Bodzon-Kulakowska, T. Cichon, A. Golec, A. Drabik, J. Ner and P. Suder, DESI-MS as a tool for direct lipid analysis in cultured cells, Cytotechnology, 2015, 67(6), 1085–1091 CrossRef CAS PubMed.
- T. Paxton, Chapter 8 - Rapid evaporative ionization mass spectrometry, in Ambient Ionization Mass Spectrometry in Life Sciences, ed. K. Zaitsu, Elsevier, 2020, pp. 241–270 Search PubMed.
- F. Bolt, S. J. S. Cameron, T. Karancsi, D. Simon, R. Schaffer, T. Rickards, K. Hardiman, A. Burke, Z. Bodai, A. Perdones-Montero, M. Rebec, J. Balog and Z. Takats, Automated High-Throughput Identification and Characterization of Clinically Important Bacteria and Fungi using Rapid Evaporative Ionization Mass Spectrometry, Anal. Chem., 2016, 88(19), 9419–9426 CrossRef CAS PubMed.
- M. Rienth, J. Crovadore, S. Ghaffari and F. Lefort, Oregano essential oil vapour prevents Plasmopara viticola infection in grapevine (Vitis Vinifera) and primes plant immunity mechanisms, PLoS One, 2019, 14(9), e0222854 CrossRef CAS PubMed.
- Y. Guitton, G. Dervilly-Pinel, R. Jandova, S. Stead, Z. Takats and B. Le Bizec, Rapid evaporative ionisation mass spectrometry and chemometrics for high-throughput screening of growth promoters in meat producing animals, Food Addit. Contam., Part A: Chem., Anal., Control, Exposure Risk Assess, 2018, 35(5), 900–910 CrossRef CAS PubMed.
- C. Black, O. P. Chevallier, S. A. Haughey, J. Balog, S. Stead, S. D. Pringle, M. V. Riina, F. Martucci, P. L. Acutis, M. Morris, D. S. Nikolopoulos, Z. Takats and C. T. Elliott, A real time metabolomic profiling approach to detecting fish fraud using rapid evaporative ionisation mass spectrometry, Metabolomics, 2017, 13(12), 153 CrossRef PubMed.
- R. S. Barlow, A. G. Fitzgerald, J. M. Hughes, K. E. McMillan, S. C. Moore, A. L. Sikes, A. B. Tobin and P. J. Watkins, Rapid Evaporative Ionization Mass Spectrometry: A Review on Its Application to the Red Meat Industry with an Australian Context, Metabolites, 2021, 11(3), 171 CrossRef CAS PubMed.
- F. Rigano, S. Stead, D. Mangraviti, R. Jandova, D. Petit, N. Marino and L. Mondello, Use of an “Intelligent Knife” (iknife), Based on the Rapid Evaporative Ionization Mass Spectrometry Technology, for Authenticity Assessment of Pistachio Samples, Food Anal. Methods, 2019, 12(2), 558–568 CrossRef.
- N. Ogrinc, A. Kruszewski, P. Chaillou, P. Saudemont, C. Lagadec, M. Salzet, C. Duriez and I. Fournier, Robot-Assisted SpiderMass for In Vivo Real-Time Topography Mass Spectrometry Imaging, Anal. Chem., 2021, 93(43), 14383–14391 CrossRef PubMed.
- V. Plekhova, L. Van Meulebroek, M. De Graeve, A. Perdones-Montero, M. De Spiegeleer, E. De Paepe, E. Van de Walle, Z. Takats, S. J. S. Cameron and L. Vanhaecke, Rapid ex vivo molecular fingerprinting of biofluids using laser-assisted rapid evaporative ionization mass spectrometry, Nat. Protoc., 2021, 16(9), 4327–4354 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an00644e |
| This journal is © The Royal Society of Chemistry 2024 |