Open Access Article
Open Access ArticleRecent advances in biological nanopores for nanopore sequencing, sensing and comparison of functional variations in MspA mutants
Huma Bhatti
a,
Rohil Jawed
b,
Irshad Alia,
Khurshid Iqbal
 a,
Yan Han
a,
Zuhong Lu
a and
Quanjun Liu
a,
Yan Han
a,
Zuhong Lu
a and
Quanjun Liu
 *a
*a
aState Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, No. 2 Sipailou, Nanjing 210096, People's Republic of China. E-mail: lqj@seu.edu.cn; humbhatti@yahoo.com; Fax: +86-25-83793283; Tel: +86-25-83793283
bSchool of Life Science and Technology, Southeast University, No. 2 Sipailou, Nanjing 210096, People's Republic of China
First published on 31st August 2021
Abstract
Biological nanopores are revolutionizing human health by the great myriad of detection and diagnostic skills. Their nano-confined area and ingenious shape are suitable to investigate a diverse range of molecules that were difficult to identify with the previous techniques. Additionally, high throughput and label-free detection of target analytes instigated the exploration of new bacterial channel proteins such as Fragaceatoxin C (FraC), Cytolysin A (ClyA), Ferric hydroxamate uptake component A (FhuA) and Curli specific gene G (CsgG) along with the former ones, like α-hemolysin (αHL), Mycobacterium smegmatis porin A (MspA), aerolysin, bacteriophage phi 29 and Outer membrane porin G (OmpG). Herein, we discuss some well-known biological nanopores but emphasize on MspA and compare the effects of site-directed mutagenesis on the detection ability of its mutants in view of the surface charge distribution, voltage threshold and pore–analyte interaction. We also discuss illustrious and latest advances in biological nanopores for past 2–3 years due to limited space. Last but not the least, we elucidate our perspective for selecting a biological nanopore and propose some future directions to design a customized nanopore that would be suitable for DNA sequencing and sensing of other nontrivial molecules in question.
1. Introduction
All living cells transport ions and molecules within and between the cells through protein channels located in their membranes, a prerequisite to maintain the integrity of a cell.1 DNA encodes all the genetic information in a cell by means of their four genetic letters and directs the cell to function accordingly. The predominant significance of DNA in human life necessitates the study of cellular functions at single-molecule level. In order to get insight at the single-molecule level, the scientists were zealous to invent such a device that can mimic the ubiquitous process of transportation (ions and/or molecules) via protein channels. The first manifestation of this kind of device was proposed by Wallace H. Coulter, named as Coulter counter a technique used for sizing and enumeration of microscopic particles hanging in a fluid.2 The journey of coulter counter from micrometer to nanometer scale was a great modernization in science to benefit human life. Therefore, nanopore sensing and sequencing is thought to be a part of this chain, in which a nanometer-scaled window is utilized to look over translocating ions and small molecules.Nanopore sensing has been gaining immense attention day by day and became a predominant technique for single-molecule detection over other techniques. One of the remarkable benefits of this technique is that it does not require complex labeling or laborious sample preparation3–5 and a small concentration of sample is enough to be identified.6 Besides rapid characterization of single-molecules,7–9 nanopore sensors are renowned for their high-throughput,10–12 ultra-sensitive,13–16 and being able to read long strands of DNA/RNA at single-molecule level.10,17,18 However, these advantages were difficult to achieve with conventional sequencing methods.19,20
Apart from discerning a variety of macromolecules, nanopores have also been employed for the detection of polymer,21 polypeptides22,23 and in the discrimination of protein sequences and their post-translational/chemical modifications.24 Moreover, molecules that are relatively smaller than the pore size, like heavy-metal ions,14,25–27 drugs28 and some health terrorist agents29 have also been investigated.
2. Principle of nanopore sensing
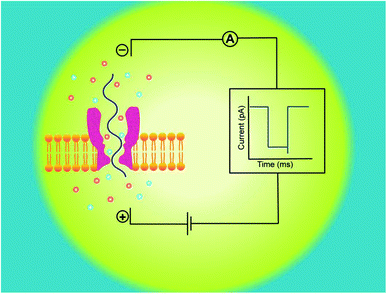
A nanopore device basically functions on the principle of Coulter Counter.2 It consists of two electrolyte-filled compartments that are set apart by means of an impermeable membrane and a nanometer-sized hole is established into the membrane.These two chambers are named as cis and trans. An Ag/AgCl electrode is dipped in each chamber. On applying electric potential to one of the chambers (usually trans), while the other is kept grounded (cis), a stable ionic current exists between these chambers. The steady-state of ions temporarily halts on addition of any charged analyte of interest, like DNA,13,30,31 RNA7,15 or protein32,33 to one of the chambers. The main forces involved in the capturing and translocation of a target-molecule are mainly the electric field gradient and electro-osmotic force across the membrane.12 During this process, the electric field force leads the macromolecule to pass through the nanopore; however, the direction of electro-osmotic force is opposite to that of electric field force. Moreover, other forces such as hydrodynamic force, entropic force and electrostatic (attraction/repulsion) force also affect the translocation of biomolecules but their effects are weak, comparatively.34
Hence, electrophoretic threading of single-molecule through a nanopore is evidenced with a transient current blockage imparted by simultaneous restriction to the ionic flow.35 The pattern of current blockage and the time consumed by a molecule in exploring the pore, depict the geometry, interactions and physicochemical features of an analyte that are subjected to nanopore sensing,36,37 as illustrated in Fig. 1.
3. Types of nanopore
Nanopores are classified into two main classes on the basis of the pore origin and the membrane embracing the nanopore: biological nanopore (organic)38,39 and solid-state nanopore (synthetic or inorganic),40–42 but more than a decade ago, a third type: hybrid nanopore (combination of biological and solid-state nanopore) have also been derived.43,44 In case of biological nanopores, when an exemplary lipid bilayer is installed on the orifice of the cup, a nanometer-sized porin protein like α-hemolysin or MspA is allowed to be incorporated in the membrane12 whereas in case of solid-state nanopore a nanometer-sized pore is fabricated in Si3N4,41,45 Al2O3,46 graphene,47 MoS2 (ref. 48) or TiO2 (ref. 49) synthetic membrane. Recently, the track-etched nanopores/membranes show promising applications in nanopore fabrication (with different geometries), easy functionalization, molecule sensing, energy transformation and detection of ion gating.50Here, in this review we will shed light on some commendable biological nanopores and their mutants that have been used so far for the detection of biological molecules and other nano-analytes. To avoid distractions, we will not discuss all findings, because the review mainly focuses on MspA nanopore, its engineered sub-types and effect of mutations on their sensing competence. Last but not the least; we will briefly account our perspective for selecting a biological nanopore according to the sample to be scrutinized, on behalf of research analytics.
4. Biological nanopores
Biological nanopores are the pore forming protein i.e. “porin” that is allowed to be inserted in a membrane of choice; mostly planar lipid bilayer membrane is used in biological nanopore sensing. The first nanopore ever to be known for the translocation of single-stranded DNA.12 Biological nanopores are more convenient owing to their natural interaction with bio-analyte and can highly be reproduced in bulk.38,39In addition to this, these pores are also advantageous over solid-state nanopore because biological nanopores can be engineered effectively and amino acids of choice can be induced to the specific location by site-directed mutagenesis.51 Although, other modes of nanopore engineering have also been adopted, such as incorporation of a tag (DNA or PEG) and/or enzyme to the nanopore,13,52,53 addition of adaptor54 or nontrivial truncation of amino acids to modulate the pore's physical and chemical features.55
As a rule, the translocation of a negatively charged biomolecule usually DNA causes the ionic flux perturbation which is commensurable to their interactions with constituent amino acids of channel protein.13,30,54 Therefore, a slight variation in the inherent composition or charge of a biological nanopore results in a striking change in their current–voltage response. A number of channel proteins have been known to date like α-hemolysin, MspA, phi 29 bacteriophage, aerolysin,56,57 OmpG,58,59 cytolysin A,60,61 FraC62 and so on. Interestingly, a new type of biological nanopore has been designed that makes use of multiple layers of DNA molecules to construct a nanometer-scaled channel with a prominent shape is termed as DNA origami nanopore.63,64 Moreover, a multitude of DNA nanotechnology is facilitating the nanopore sensing technique.65 The structures of some biological nanopores are shown in Fig. 2 while four well-known biological nanopores are briefly described below:
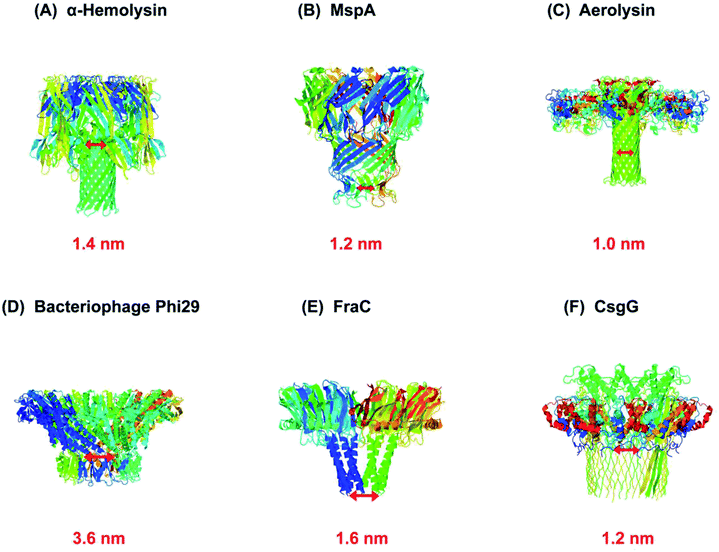 | ||
| Fig. 2 Biological nanopores. Structure of some customarily used biological nanopores with their marked constriction areas (red). (A) α-Hemolysin (PDB: 3ANZ), (B) MspA (PDB: 1UUN), (C) aerolysin (PDB: 5JZT), (D) bacteriophage Phi29 (PDB: 1H5W), (E) FraC (PDB: 4TSY) and (F) CsgG (PDB: 4UV3). | ||
4.1. α-Hemolysin
α-Hemolysin is a toxin secreted by human pathogen Staphylococcus aureus. It is a heptameric channel protein of about 232.4 kD. It is comprised of a mushroom-shaped cap (that is 10 nm both in length and in diameter) and a stem (5.2 nm long and 2.6 nm wide) that forms a transmembrane channel. The diameter of channel constriction is approximately 1.4 nm.38 The narrowest constriction of α-hemolysin channel is suitable enough to translocate a single stranded DNA or RNA as experimentally proved and published by J. J. Kasianowicz.12 If a nucleobase can vary the ionic current according to their geometry and chemical composition then nanopore sequencing could be possible by using this toxin.12Despite that, α-hemolysin has a long β-barrel which can accommodate approximately 10–12 nucleotides, consequently, the ionic flux modulated by a bio-analyte usually DNA, is the output of multiple recognition sites that come in contact with translocating bases.66 Firstly, it was found that the constriction is located in the middle of the channel's lumen around 111 residue.67 Later on, it was shown that the transmembrane region of α-hemolysin have three recognition sites that were found to be proficient in discriminating all four DNA nucleotides when a molecule of ssDNA was halted in the channel.68
At first, it was considered that two recognition sites (R1 and R2) in the transmembrane region might be favorable for sequencing, as the molecule transit through R1 would be proof-read by another constriction site R2.66 However, more than two sites would be inoperable because it was doubtful to evaluate the ionic current generated by three sites from electrical noise. To counteract this issue in reading signals, Stoddart et al., proposed to modify one recognition sites (R1) in α-hemolysin by voluminous mutagenesis to strengthen one sensing point for nucleotide detection. He explained that bulky groups in amino acid render hindrance to the flow of ions and signal recording would be more accurate in contrast to the lighter groups or side chains.69 Thereafter, Ervin et al., eliminated one or two sensing regions while concurrently boosting the third one by site-directed mutagenesis to form a single constriction for the identification of bases.70 Now, the channel's length of mutant α-hemolysin is ∼1.6 nm is closely similar to that of MspA (∼1.0 to 1.2 nm).70
4.2. Phi 29 and other bacteriophages
Bacteriophage phi 29 has also been remained an attractive biological nanopore owing to its sophisticated and enlarged pore size with an external wide end of 13.8 nm that tapers to give the connector a cone-shaped narrower end of 6.6 nm in diameter. The entire length of the channel from broad to narrow end is 7.5 nm.71 Wendell D. et al., is the first to publish data on the translocation of double-stranded DNA through a bacteriophage phi 29 DNA-packaging motor.72 The ingenious design of bacteriophage phi 29 is comprised of 12-subunits of gp10 connector protein to form a dodecamer channel,73 six copies of ATP-binding DNA packaging RNA (pRNA),74,75 and an ATPase protein76 which act as a linker and provide energy to translocate DNA. Essentially, it is more commonly employed in combination with other biological nanopore as a molecular motor.77 Currently, phi 29 has been found to act as a reverse transcriptase for FANA template (a xeno-nucleic acid).78 Therefore, this is thought to be the most energetic nano-motor hitherto.In addition to this, Wanunu et al. conducted a study in which a lipid-free hybrid nanopore has been designed. An engineered Thermus thermophilus G20c has electro-kinetically been inserted into the solid-state membrane, and then it was utilized for the identification and discrimination of several types of biological molecules.79
4.3. Aerolysin
Aerolysin is a β-pore-forming toxin from Aeromonas hydrophila that can be inserted into lipid bilayer membrane through its transmembrane region. Aerolysin is similar to α-hemolysin in shape, a broadly used protein nanopore, but lacks the vestibule. The constriction area of the aerolysin channel is ∼1.0–1.7 nm in diameter and the length of the transmembrane region is comparable to that of α-hemolysin.80 In 2006, aerolysin was first manifested as a biological nanopore and utilized for the detection of α-helical peptides.81 Afterwards, it was used to analyze the dynamics of unfolded protein,82 peptides,57 enzymatic degradations of polysaccharides83 and also served as a mass spectrometer for the discrimination of PEG polymers of various sizes.84 Subsequently, the characterization verge of 2–10 nucleotides of polydeoxyadenene (dAn) with well distinguishable current blockades is a breakthrough in nanopore sensing.56 Consequently, this nanopore gained much attention in nanopore sensing and later, used for the detection of short polynucleotides consists of four genetic letters of life.85 However, the main purpose of sequencing a genomic DNA is to read all the genetic letters continuously, in an error-free way. Therefore, to make aerolysin channel practical for longer ssDNA detection, scientists used KCl solution with asymmetric pH across the aerolysin nanopore and was able to detect ssDNA of ∼30 nucleotides length.86 Furthermore, altering the type of salt from KCl to LiCl showed interesting results for >100 nucleotides ssDNA detection.87 In order to increase the detection sensitivity, the key sensing zone of aerolysin was aimed to identify, which is K238.88 Besides, further mapping of aerolysin revealed that R220 and K238 are the most sensitive analytical points and R220 is accredited as the narrowest constriction of aerolysin. It is R220 that discriminates various short oligonucleotides, methylated cytosine and oxidized guanine in ssDNA.89 In addition, these two residues were replaced with glutamic acid to check the selectivity (R220E) and sensitivity (K238E) of aerolysin.90 Since 2016, Long Yi-Tao's group has been adding a great contribution in nanopore sensing platform by employing aerolysin nanopore.4.4. MspA
Mycobacterium smegmatis porin A (MspA) is a promising and an ideal nanopore for a variety of bio-nanotechnological applications for instance, sequencing DNA and RNA.91 MspA is the major porin of Mycobacterium smegmatis because it provides the main hydrophilic pathway via cell wall for the transport of nutrients required for the growth of mycobacterial cell.92,93 It is a homo-octameric channel of goblet-like shape and has a unique geometry that is 9.6 nm long and 8.8 nm wide. Interestingly, MspA forms a single, central-channel whose constriction is ∼1.0 nm long and ∼1.0 nm in diameter, relatively smaller and narrower than that of α-hemolysin,39 which is responsible for its higher spatial resolution. Besides this, MspA is a potent biosensor owing to its extreme stability towards high temperature up to 100 °C and over a wide range of pH from 0 to 14.94 MspA can be considered as a better candidate due to its highly pronounced signal-to-noise ratio and single-nucleotide discrimination sensitivity required for DNA sequencing.95,96 In addition to this, MspA nanopore can discriminate various abasic residues in a single-stranded DNA.97,985. MspA mutants
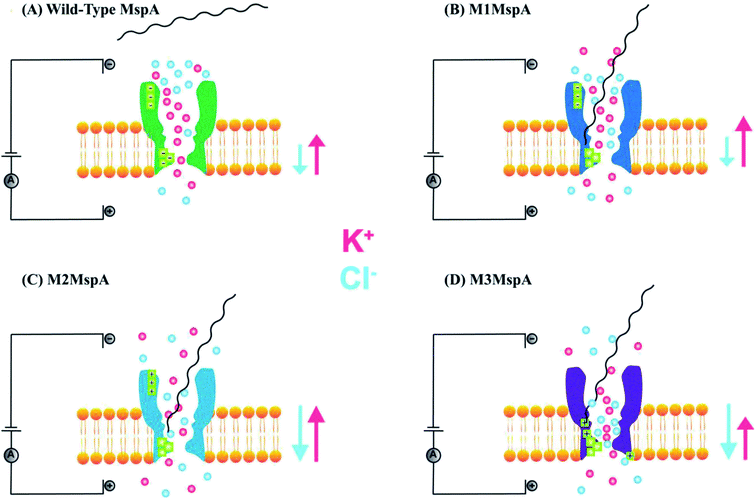
The promising features and perfect geometry of MspA make it a robust nanosensor and had led to the preparation of MspA mutants to meet the challenges of nanopore sensing. Further, the narrow constriction of MspA is found to be pretty ideal to characterize all four nucleotides in ssDNA, when a duplex is introduced between the nucleotides to be analyzed.91 Therefore, in this review, we are shedding light on MspA mutants to gain insights of their charge distributions and corresponding physicochemical status of the pore induced by site-directed mutagenesis. This will be advantageous in understanding the translocation dynamics, pore–analyte interactions of MspA mutants and to make it more skillful by altering some experimental protocols or to invent some new variants of MspA in future. On the basis of site-directed mutagenesis, three renowned MspA mutants have been known to date to the best of our knowledge as shown in Fig. 3 and are explained as follows: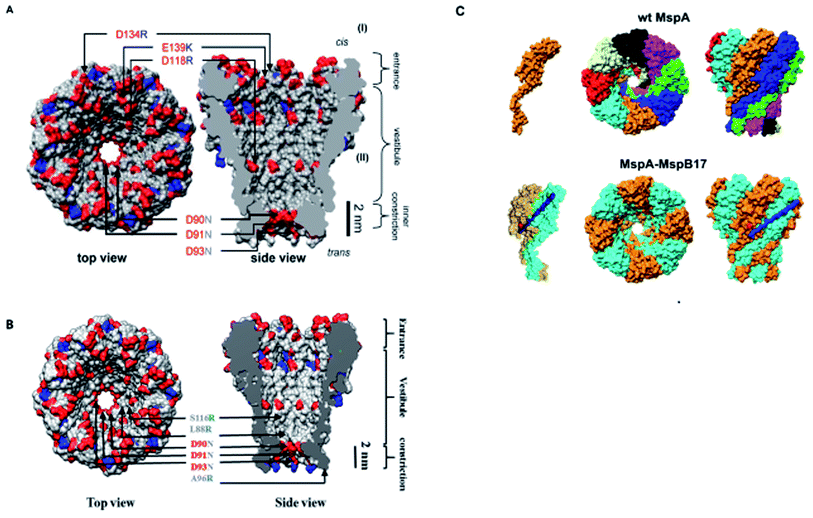 | ||
| Fig. 3 MspA mutants. Genetically engineered mutants of MspA nanopores (A) M1MspA and M2MspA,99 (B) M3MspA and (C) stoichiometric mutant of MspA, MspA–MspB17.100 Figures do not need permission from the references. | ||
5.1. M1MspA
This is the very first documented mutant of Mycobacterium smegmatis porin A (MspA) declared by Jens H. Gundlach's group in 2008. Initially, wild-type MspA (Wt-MspA) were examined as a nanopore for single-stranded DNA (ssDNA) detection. They found that if the voltage was exceeded above 60 mV the Wt-MspA exhibited incessant, spontaneous blockades of the ionic flow prior to the addition of ssDNA. They observed that some of the blockades were short lived and some needed voltage polarity to clear up the channel's cavity. On the contrary, they also found smooth and uninterrupted open pore current continuing for tens of seconds for electric potentials up to ∼100 mV. Furthermore, the introduction of 2–8 μM dC50 ssDNA to the cis side of Wt-MspA didn't manifest any signature blockades that could be ascribed to ssDNA translocation. Rather, the frequency of spontaneous blockades became so dominant that ssDNA detection was inoperable. They reasoned that overcrowding of negatively charged amino acids in the constriction zone of MspA would not allow the ssDNA (negatively charged) to thread through it. In order to settle down this matter, its constriction zone was engineered by the replacement of three negatively charged aspartic acid residues at positions 90, 91 and 93 with the neutral asparagine residues and this mutant is named as M1MspA (D90N/D91N/D93N). This mutant M1MspA pore has then been able to perform the ssDNA detection at voltages up to and above 180 mV, with a significant decrease in spontaneous blockades.99 Albeit, a matter of concern still left there that was needed to be addressed. This paved the way for further mutations in M1MspA.5.2. M2MspA
When a linear, ssDNA homopolymer of 50-mers was allowed to pass through M1MspA pore, a number of short-lived events ∼30 μs were found to be appeared. These events suggested the brief stay of ssDNA in the vestibule and finished with the exit of ssDNA back to the cis side. To overcome this issue, Butler et al., investigated the effect of charges in MspA pore and in this regard, they replaced three negatively charged residues; one in the vestibule and two at the mouth of the M1MspA, into positively charged residues at positions 118, 134 and 139. This second mutant of MspA is termed as M2MspA (D90N/D91N/D93N/D118R/D134R/E139K).99 The addition of positively charged moieties has dramatically found to increase the interaction of DNA with the MspA pore ∼20 times and this led to lengthen the stay of DNA in the vestibule approximately 100 times as compared to M1MspA. In addition, the voltage threshold for M2MspA had also been reduced to ∼80 mV that is almost half of the voltage required for M1MspA to initiate the capture and translocation of target ssDNA.995.3. M3MspA
With the aid of mutations brought in Wt-MspA, the mutant M2MspA is now able to allow ssDNA to translocate through and has reduced the escape of DNA back to the cis side and in turn capture rate has been increased. Nevertheless, DNA sequencing is still facing a key hurdle; the speed of translocation. To settle this problem, Hang et al. in 2016, designed and expressed a novel mutant of MspA, in which three negatively charged aspartic acid residues have been replaced by three neutral asparagine residues at positions 90, 91 and 93. Also, addition of three arginine amino acids has been done in the vicinity of constriction zone, at positions 88, 96 and 116 via site-directed mutagenesis and the resulting mutant is named as M3MspA (L88R/D90N/D91N/D93N/A96R/S116R). The addition of three arginine mutants have been shown to slow down the ssDNA translocation speed up to 10–30 folds and was also found to decrease the voltage threshold to initiate ssDNA translocation experiments.515.4. MspA subunit dimers
Another interesting modification of MspA is the combinatorial variation of MspA octamers that leads to the formation of single-chain subunit dimers by employing M1MspA. A number of peptide linkers ranging from 17 to 62 amino acids length were used to connect two monomers of MspA to generate a dimer. They found normal folding and insertion of these dimeric M1–M1MspA channels in their M. smegmatis membrane with an increased glucose uptake. Also, the voltage gating and current blockade of DNA hairpins through these M1–M1 dimers were appeared to be same as that of M1MspA. Altogether, this kind of stoichiometric alteration does not only produce functional channels of MspA but also provides flexibility in designing channels with customized constriction.1006. Comparative study of MspA
It has been discussed so far that α-hemolysin channel is widely used as a bio-nanopore but still it has some limitations, regarding its geometry with longer stem and its multiple recognition sites.66,68 Therefore, MspA is considered to be a promising candidate to overcome the above mentioned issues due to its ideal geometry with single, narrow and small constriction zone, dimensionally comparable with ssDNA.39 Nevertheless, another obstacle in sequencing DNA is the translocation speed of ssDNA which is too high for both M1MspA and M2MspA (∼2–10 bases per μs) as compared to α-hemolysin (∼0.5–1 base per μs).99This marked difference in speed have arisen ought to the geometrical difference in their transmembrane regions enclosing the restriction area of both types of porin protein, that is longer and wider for α-hemolysin. Additionally, the channel's composition is also of paramount importance in influencing the speed of ssDNA. As nucleotide bases and charged walls attract each other and this electrostatic attraction slows down the speed of ssDNA. Hence, ssDNA stays in α-hemolysin for longer period of time because it can accommodate 10–20 bases in its restricted and transmembrane area at a time.99 However, the constriction and transmembrane region of MspA is smaller and only 2–4 bases can accommodate. Also, lack of charged residues in channel constriction affects the speed of ssDNA.99
In the above section, we elaborated the specifications of each variant of MspA, now we will briefly analyze and compare the effects of site-directed mutations on their charge distribution, voltage gating/threshold voltage and sensing skills of the MspA mutants. Fig. 4 exhibits the genetically mutated parts of the MspA nanopores to better understand the significance of surface charge and positional effects.
6.1. M1MspA with wild-type MspA (Wt-MspA)
We see that in wild type MspA (Wt MspA) three negatively charged aspartic acid residues were not allowing negatively charged ssDNA to thread through, due to electrostatic repulsion, as shown in Fig. 4(A). Therefore, these residues were replaced by neutral asparagine residue. These changes didn't affect the channel forming ability of M1MspA, however the conductance of M1MspA was decreased by 2–3 parts.99 Since, the interior of Wt-MspA is negatively charged; therefore it was functioning as a cation-selective nanopore. We notice that the incorporation of neutral amino acids in the constriction of M1MspA was found to reduce the cation selectivity and hence, the conductance. Because of the change in charge distribution, M1MspA requires the voltage threshold of ∼+180 mV to detect ssDNA, indicated in Fig. 4(B). Moreover, the spontaneous blockades were found to be dropped significantly when a DNA hairpin was translocated through M1MspA,99 as depicted in Fig. 5(A).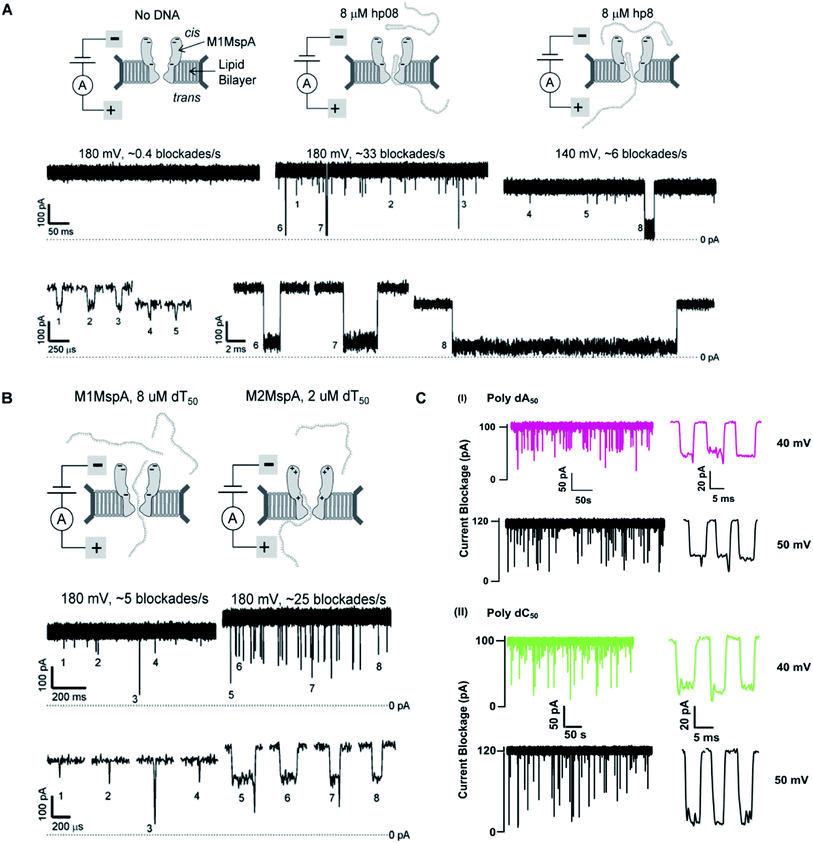 | ||
| Fig. 5 Comparison of DNA sensing through MspA mutants. (A) Translocation of DNA hairpin-08 through M1MspA at +180 and +140 mV. (B) The travelling of poly dT50 across M1MspA and M2MspA at +180 mV shows distinct difference in number of blockades, higher for M2MspA. Figures (A) and (B) have been reproduced from ref. 99 with permission from National Academy of Sciences, U.S.A, Copyright 2008. (C) Sensing of poly dA50 and poly dC50 through M3MspA shows signature events at +40 and +50 mV. The figure (C) does not need permission. | ||
On the other hand, the ionic current blockage for ssDNA traveling through M1MspA was found to be momentary <30 μs, which was too short to evaluate if it was real translocation event or merely skipping of ssDNA back to the cis side. Afterwards, M1MspA was employed in duplex interrupted (DI) DNA sequencing by making use of converted DNA structures and that was considered to be relatively simple in comparison with fluorescence and other perplexed labeling techniques.91
6.2. M2MspA with M1MspA
This scenario points towards an important aspect about the dependence of capturing and translocation of ssDNA on the charge distribution of a nanopore. We analyze that making the constriction neutral could allow ssDNA to traverse through M1MspA but the probability of successful events and returning of molecule back to the cis side were parallel. In this regard, there should be placement of some charged amino acids at the mouth of MspA to arrest ssDNA into the pore. This shortcoming was fulfilled by M2MspA, when the placement of three positively charged amino acids were done at the entrance of M1MspA, in place of three negatively charged residues. At that moment, the blockades were found to be increased from ∼5 blockades to ∼25 blockades per second,99 as shown in Fig. 5(B).We speculate that the positively charged arginine and lysine at positions 134 and 139, respectively, attract and capture ssDNA from cis side and further entry was accomplished by arginine at position 118, Fig. 4(C). On the contrary, the stay of ssDNA was relatively short as compared to α-hemolysin. This could be due to different geometry of transmembrane regions and/or availability of charged residues in or near around the constriction area. Although, we support the later for the short stay of ssDNA, because Wt-MspA and M1MspA bear the same transmembrane region and constriction area but different charged moieties in their vestibules. Furthermore, the minimum voltage threshold for M2MspA was reduced to ∼80 mV, whereas M1MspA did not give rise to any event below ∼140 mV.99 However, in our experiments, the minimum voltage threshold for M2MspA was found to be ∼120 mV, this might because of the different reagents used and/or atmospheric conditions (data not shown).
6.3. M3MspA with wild-type MspA (Wt-MspA)
In an effort to address the above said issue, Hang et al., expressed and purified a new mutant of MspA,51 which has previously been simulated.101 This mutant was found to reduce the speed of ssDNA ∼10–30 folds in real-time experiments. Recently, Bhatti, et al., explored this new mutant of MspA, termed as M3MspA through detection of ssDNA homopolymers and hetero–homopolymers with well-resolved signature events up to some milliseconds dwell time and corresponding signature events,102 as shown in Fig. 5(C). We scrutinize that, in M3MspA, the addition of three arginine amino acids in the vestibule and nearer to the constriction zone made this mutant anion-selective, noticeably, illustrated in Fig. 4(D). Consequently, crowding of anions (Cl− ions) in the pore's vestibule was anticipated in the absence of electric potential as well and accredited in lowering the threshold potential up to ∼± 40 mV. The attractive force from positively charged arginine in combination with small magnitude of applied voltage was favorable enough to initiate ssDNA translocation. This imbalance of ions was appeared to change the membrane's polarity but keep the pore functional up to ∼± 50 mV. Eventually, further increase in applied voltage ≥± 60 mV was found to halt the further flow of ions owing to the accumulation of Cl− ions at the trans side102 and K+ would also hang around trans to perpetuate the overall electrical neutrality of the system.103 Hence, M3MspA does not exhibit anion-selectivity at higher voltages. M3MspA shows transient or sometimes permanent (needs voltage polarity) spontaneous blockades beyond ∼± 60 mV.102If we compare M3MspA with Wt MspA, we would be able to understand the key role of charge distribution in nanopore sensing. Since, the gating potential of both Wt-MspA and M3MspA is comparable ∼± 60 mV, but it is the surface charge distribution inside the nanopore that decides the fate of ssDNA entry. Although, it is a very complex physicochemical mechanism to exactly comprehend at the nano-confined level but we can evaluate some research-based details. The ion-selectivity of both Wt-MspA and M3MspA is opposite. M3MspA is eminently selective for anions and therefore it can not only accommodate ssDNA but also generates well-resolved signals and remarkable discrimination between ssDNA homopolymers and hetero–homopolymers.102 We contemplate that the position and abundance of arginine amino acid are responsible for immense electrostatic attraction between ssDNA and M3MspA. Because placement of attractive sites (charged amino acids) near to the entry and exit found to have increased current blockage.104 Moreover, increased dwell time was observed in ssDNA translocation102 because arginine was found to be the top-most amino acid in making hydrogen bonds with DNA bases.105
7. Recent advances in biological nanopore
In past, many review articles have been published so far that take into account various aspects of biological nanopores,106,107 specifically, α-hemolysin.108,109 Therefore, in this section, we will elaborate some recent and remarkable detection carried out by employing wilt-type or engineered biological nanopores that can be beneficial in paving new methods for the prognosis and treatment of serious ailments and sequencing DNA.7.1. Recent advances in α-hemolysin
From the beginning of nanopore sequencing, α-hemolysin has been showing a great myriad of detection competence by employing its wild-type110,111 and various mutated or modified pores.112 As a matter of fact, α-hemolysin has been used for sensing a number of analytes that was needed to be known at the single-molecule level. Yet, the main challenge to nanopore DNA sequencing, that is the speed of DNA, is under way. In this regard, lithium salt gradient was established to retard the translocation speed at the level that even methylated cytosine can be detected,113 likewise, calcium flux is introduced at the trans site of α-hemolysin to control the mobility of DNA via strong Ca+–DNA interaction.114 However, another approach for increased capture rate (118.27 folds) and the decreased translocation speed (120 μs per base) of ssDNA was certainly achieved by introducing PEG molecules (PEG 4k) to the nanopore system.115Importantly, it is used to analyze and point out the position of the mutations in DNA via non-functionalized PNA,116 short fragments of DNA,117 sequence-specific ssDNA detection with gold nanoparticles,118 label-free detection of completely matched from mismatched DNA on the basis of enzymatic reaction4 and to quantify multiple cancer biomarkers in blood samples at picomolar level.119 Moreover, detection of microRNA120,121 and identification of methylated cytosine by employing lithium salt-gradient have also been achieved.113 Furthermore, identification of individual amino acids via their N-terminal derivatization,122,123 polypeptide–pore interaction124 and protein phosphorylation and dephosphorylation were well observed by utilizing the α-hemolysin nanopore.125
On the other hand, the detection skills of α-hemolysin are not limited to identify DNA or protein only; rather it has recently been used to scrutinize less-privileged but a significant class of macromolecules i.e. carbohydrates (glycosaminoglycan hyaluronic acid),126 Zn2+ ions detection,127 cancer drug–DNA interaction,128 in analyzing the isomers of guanine i.e. 5-guanidinohydantoin and iminoallantoin in a double-stranded DNA,129 isomers of glucose112 and analysis of enzymatic activity and inhibition of DNA methyltransferase130 and ADAM-17 cleavage reaction via salt-mediated nanopore.131 Additionally, identification of microcystin (MC) variants132 and discerning the MC variants on the basis of their molecular interaction with α-hemolysin nanopore have also been carried out.133
Currently, α-hemolysin nanopore has been used in exploring some new features of molecules, that is, the conformational heterogeneity of DNA assemblies,134 DNA compaction instigated by Na+ and K+ ions,135 determination of an intermediate in a catalyzed reaction cycle136 unzipping mechanism of free and poly arginine-attached DNA–PNA duplexes to unveil the differences in their unzipping kinetics in a nano-confined space137 and effect of glycerol and PEG molecules on the viscosity of aqueous solution together with their influence on molecular motion of hairpin DNA.138 Besides, early scanning of T790M drug resistance (infrequent mutation sequence) has become possible with a probe containing three locked nucleic acid (LNA) and α-hemolysin nanopore. The LOD for T790M is found to be 0.1 pM, ascertained as highly sensitive than any other available techniques.139 Surprisingly, this protein channel is so adaptable that it can discriminate aqueous sodium polysulfide chains (used in sulfur-based batteries) of various lengths differing by only single sulfur-atom, by introducing β-cyclodextrin into the channel, hence, batteries with improved electrochemical storage can be designed.140 Fig. 6 displays some significant advances in α-hemolysin nanopore, figures are reproduced with permission.
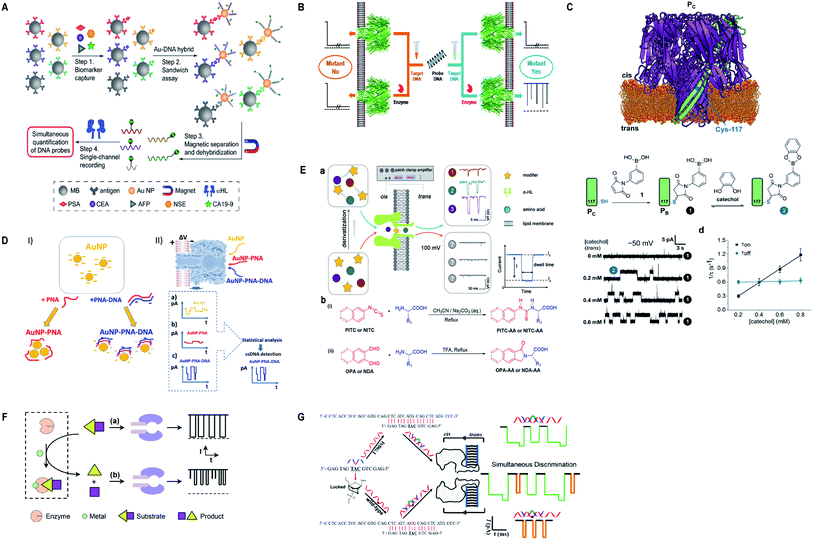 | ||
| Fig. 6 Recent advances in α-hemolysin. (A) Schematic representation of sandwich assay and signature events for corresponding barcode DNA (license number: 4938570920843). This figure has been reproduced from ref. 119 with permission from John Wiley and Sons, Copyright 2018. (B) Label-free detection of DNA mutations based on enzymatic reaction. This figure has been reproduced from ref. 4 with permission from American Chemical Society, Copyright 2018. (C) Determination of isomers of D-glucose and D-fructose by their bonding with boronic acid (license number: 4938581342453). This figure has been reproduced from ref. 112 with permission from John Wiley and Sons, Copyright 2018. (D) Detection of specific sequence by employing gold nanoparticles.118 This Figure does not need permission. (E) Identification of single amino acids by N-terminal derivatization. This figure has been reproduced from ref. 122 with permission from American Chemical Society, Copyright 2020. (F) Nanopore detection of zinc ions by enzymatic-reaction (license ID: 1074993-1). This figure has been reproduced from ref. 127 with permission from Royal Society of Chemistry, Copyright 2019. (G) Identification of drug-resistant mutations T790M at early stage. This figure has been reproduced from ref. 139 with permission from American Chemical Society, Copyright 2020. | ||
7.2. Recent advances in MspA
Almost, a huge part of analytes previously in question were well answered by α-hemolysin, but it needs to be more precise when it comes to DNA sequencing. Although, MspA is being employed by few researchers, but the information obtained from their studies are profound in overcoming many obstacles of DNA sequencing, which was duly covered by Jens H. Gundlach's group.141 Subsequently, MspA was subjected to SPRNT (Single-Molecule Picometer Resolution Nanopore Tweezers), a novel technique well-suited to enzymes that functions along DNA and RNA or other charged analytes.142 By employing SPRNT technique, temporal resolution of up to ∼40 pm and ten times more spatial resolution has been achieved for the first time to detect motion of enzymes.142 Later on, the dynamics of superfamily 2 helicase Hel308 was manifested at millisecond scale using MspA, in which the enzyme converts ATP hydrolysis into kinetic energy to move along the DNA.143,144 To improve signal to noise ratio, especially for motor enzymes that usually perform their functions at low ionic concentration, Gundlach's group suggested to maintain low cis [Cl−] provided, the trans [K+] at higher level.145 We notice that due to high speed of DNA through MspA nearly all detailed investigations were conducted by means of using molecular motors or adopters. Nevertheless, two basic errors hinder precision in sequencing: discontinuous steps of enzyme and indiscernible signals for similar sequences, which were greatly reduced by increasing the applied voltage across the membrane.146As in the above sections, we have already discussed about the ideal geometry of MspA that is a prerequisite for DNA sequencing and high speed of ssDNA through it that is a key hurdle.77,91,147 In order to figure out an ideal nanopore, a study is proposed by using molecular dynamic simulation,101 in which three variants of arginine amino acid have been designed and explored. To make it real time nanopore sensor, Hang et al., expressed and purified the arginine mutant of MspA (L88R/D90N/D91N/D93N/A96R/S116R) and verified it by successful insertion and poly (dT)100 translocation experiments.51 Furthermore, from the same group, Bhatti et al. explored this new arginine mutant of MspA (M3MspA) at different voltages and presented a remarkable discrimination between homopolymers and hetero–homopolymers with well resolved duration time (at a speed of ∼15–30 μs per base) and larger current amplitude. Apart from statistical analysis, the difference is quite apparent by just looking at the current trace, another key step towards nanopore DNA sequencing.102
Recently, MspA has been used in the detection of DNA lesions,148 identification and discrimination of small analytes (metal ions) up to ∼10 pA current amplitude.149 Besides, direct sequencing of xeno-nucleic acid (FANA),78 direct sequencing of naturally occurring microRNA for the first time and scrutiny of epigenetic modifications in microRNA150 were made possible by employing Nanopore-Induced Phase-Shift Sequencing (NIPSS).
Additionally, M2MspA was mutated to MspA-M (methionine at position 91) to detect chemistry events of a polyatomic ion, tetrachloroaurate(III) binding with methionine, a reversible Au(III)–thioether coordination reaction and further sensing of biothiols by the same Au-embedded (atomic-bridge) MspA-M nanopore.151 The maximum current blockage obtained through MspA-M was ∼55 pA, that has never been observed for any pore-ion interaction events to date.151 We really need such a sharp nanopore. Another hallmark from Gundlach's group is the utilization of MspA for sequencing unnatural base pairs (dTPT3-dNaM). Interestingly, this study manifests that unnatural base pairs in DNA can be replicated with high accuracy and precision similar to naturally occurring DNA and provides a significant platform to semisynthetic organisms that are known to produce proteins containing unnatural amino acids.152 Fig. 7 represents some impressive advances in MspA nanopore, figures are reproduced with permission.
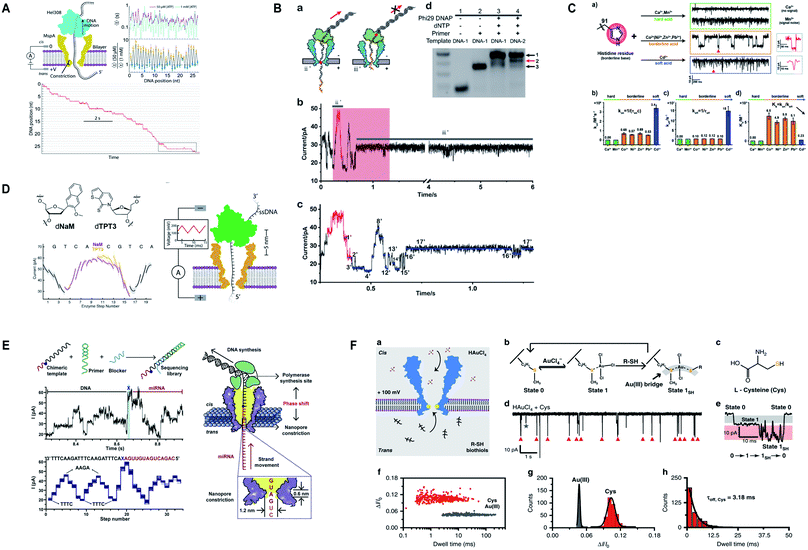 | ||
| Fig. 7 Recent advances in MspA. (A) Illustration for the SPRNT and Hel308 dynamics.143 This figure does not need permission. (B) Direct detection of O6-CMG DNA lesion (license ID: 5118040233795). This figure has been reproduced from ref. 148 with permission from John Wiley and Sons, Copyright 2019. (C) Representation of the interaction between histidine residues with various metal ions.149 This figure does not need permission. (D) Nanopore sequencing of unnatural DNA bases. This figure has been reproduced from ref. 152 with permission from American Chemical Society, Copyright 2020. (E) Direct sequencing of microRNA by employing NIPSS (license number: 5118050195676). This figure has been reproduced from ref. 150 with permission from Elsevier, Copyright 2020. (F) Engineering of M2MspA to MspA-M with the insertion of tetrachloroaurate (III) and detection of L-cysteine through it.151 This figure does not need permission. | ||
7.3. Recent advances in aerolysin
Aerolysin has been showing momentous advancements in nanopore sensing of not only protein but also some nucleic acids by genetically engineered nanopore.153 At first, we are taking into account the detection of peptides and proteins carried out by aerolysin nanopore, such that simultaneous screening of multiplex biomarkers of Alzheimer disease with some modified probes,154 pinpoints single amino acid difference among evenly charged homo-polypeptides,155 identification of single-molecule of cysteine and homocysteine in a mixture with the aid of 5′-benzaldehyde poly (dA)4 probe in the confinement of K238Q mutant aerolysin.156 Recently, the same mutant K238Q was employed in the first ever detection of single cysteine molecule without any modification or labeling with a longer blockade time ∼0.11 ± 0.02 ms.157 Moreover, protein kinase A activity has also been investigated by wild-type aerolysin nanopore.158Besides, Long Yi-Tao's group utilized aerolysin nanopore to investigate its sensing competence in the detection of nucleic acids as well. In this regard, direct sensing of single-molecule of RNA through aerolysin has been attained,159 ultrasensitive detection of cancer cells (as less as 5 Ramos cells) with the help of enzymatic signals amplified by aerolysin nanopore.160 By using some AdaBoost-based machine-learning model, it can accurately discriminate single nucleotide difference with the rise in accuracy from ∼0.293 to more than 0.991.161 Similarly, learning time-series (LTS) shapelets algorithm, the researchers were able to discern GA3 from AA3 in a same mixture of samples, without any laborious labeling or genetic mutations.162 The sensing skills of aerolysin mutant K238Q are highly appreciable as it can identify and quantify the different kinds of damaged nucleotides directly into a multi-analytes sample effortlessly.163 Recently, an impressive study is conducted that characterizes various states of phosphorylation and dephosphorylation of ssDNA in an orientation-dependent way, inside the aerolysin nanopore.164
Apart from protein and nucleic acids, aerolysin can detect diverse range of analytes and a variety of traits of a molecule that are restricted in its sensing region. A mutant K238C was utilized to scan the disulfide bond (S–S) forming and breaking between 5,5′-dithiobis-(2-nitrobenzoic acid) and the sulfhydryl group of cysteine at a single-molecule level, inside the lumen of the pore.165 Interestingly, the light was shed on the non-covalent attractions of the pore for analytes and it was concluded by scrutinizing few mutants of aerolysin that hydrogen bond is critical in inducing these interactions but various other factors also cannot be overshadowed.166 On top of that, the trans/cis conformation of azobenzene-ssDNA was ingeniously sensed by aerolysin nanopore on imparting alternating visible and UV light, with a translocation speed of 1.9 and 6.3 bases per s for the trans and cis Azo-ssDNA, respectively.167 In addition, a co-factor flavin adenine dinucleotide (FAD), pivotal for several metabolic reactions, was marvelously characterized into open, stack and four-quasi-stacked forms by employing aerolysin nanopore and further verified by conducting temperature-dependent and mutant aerolysin studies.168 Currently, an intriguing finding for the recognition of all twenty amino acids is proposed that predicts the bright future of nanopore protein sequencing.169 Fig. 8 illustrates some remarkable advances in aerolysin nanopore, figures are reproduced with permission.
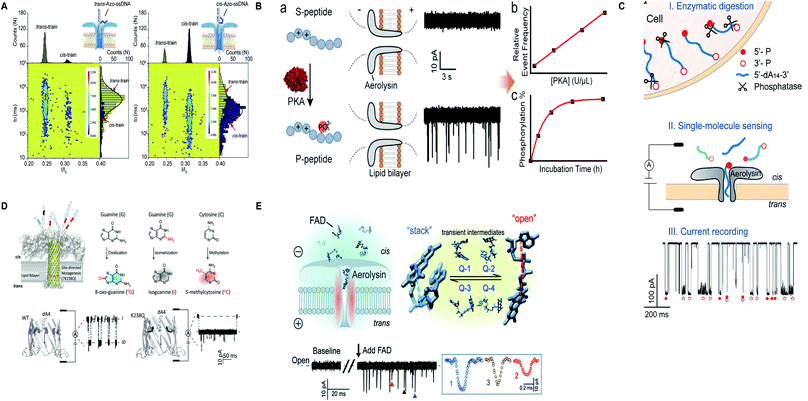 | ||
| Fig. 8 Recent advances in aerolysin. (A) Identification of trans-Azo-ssDNA and cis-Azo-ssDNA on applying visible light (left) and UV light (right) (license number: 5118541017008). This figure has been reproduced from ref. 167 with permission from Elsevier, Copyright 2018. (B) Schematic of the protein kinase activity that distinguishes unphosphorylated peptide from phosphorylated peptide though wild-type aerolysin. This figure has been reproduced from ref. 158 with permission from American Chemical Society, Copyright 2019. (C) Direct discrimination of DNA phosphorylation in an orientation-dependent manner. This figure has been reproduced from ref. 164 with permission from American Chemical Society, Copyright 2020. (D) Identification of damaged nucleotides by mutant K238Q aerolysin nanopore. This figure has been reproduced from ref. 163 (https://pubs.acs.org/doi/10.1021/acscentsci.9b01129) with permission from American Chemical Society, Copyright 2020, further permissions related to the material excerpted should be directed to the ACS. (E) Detection of possible conformations of FAD.168 This figure does not need permission. | ||
7.4. Recent advances in novel biological nanopores
The story of nanopore detection does not end on the legendary nanopores, but currently, many new biological nanopores have been manifesting excellent detection competence in their wild type and/or engineered form. Giovanni Maglia's group has been adding remarkable work by acquainting Fragaceatoxin C (FraC) nanopore as a single-molecule sensing platform. FraC nanopore can distinguish peptide and protein biomarkers from 25 kD to as low as 1.2 kD, even single amino acid dissimilarity in polypeptides was well differentiated.170 Later on, the same group tuned the size of FraC nanopore and utilized them in identifying diverse range of peptides differing in charge and mass with a ∼44 Da resolution and showed that at fixed applied voltage and pH = 3.8 sharp, it works as a mass spectrometer of peptides.171 Another momentous study is the formation of reversible photo-controlled FraC nanopore by attaching the photo-switchable azobenzene at various sites of insertion. The azobenzene can penetrate into the lipid membrane only in cis conformation. Hence, this method makes the FraC nanopore functional or non-functional depending upon irradiation to light.172 Besides, labeled or unlabeled peptides with single amino acid resolution can be attained by practicing molecular labels of diverse physicochemical peculiarities, mainly charge and mass.62 Significantly, phosphorylated and glycosylated peptides can be differentiated from their unmodified peptides by employing FraC nanopore. Also, a mixture containing phosphorylated and glycosylated peptides can also be discriminated profoundly.173Appreciably, Cytolysin A (ClyA) nanopore determined the exact quantity of glucose and asparagine directly from the blood and other bodily fluids.174 However, the engineering and molecular modeling of small proteins, dihydrofolate reductase (DHFR) and their conformational changes can be monitored by a system of forces to act upon it, like electrophoretic, electro-osmotic, electrostatic and steric forces, in a confined region of ClyA.175 On top of that, electrode-free nanopore spotting of small molecules (trimethyl-β-cyclodextrin), bio-macromolecules (dsDNA) and other macro-analytes (PEG) were carried out by diffusioptophysiology using ClyA nanopore.176
Oxford nanopore technologies (ONT) have executed direct sequencing of RNA and direct sensing of endogenous and exogenous modifications in RNA through CsgG nanopore, respectively.177,178 Recently, ONT constructed a double-constriction nanopore by joining CsgG and CsgF with an inter-constriction distance of ∼25 Å. This nanopore enhanced the single-nucleotide deciphering precision by 25–70% for homopolymers up to 9 nucleotides in length.179 We contemplate that this approach can also be practiced by employing other protein pores in combination with their functional variants to make the most of their distinct features together on a single nanopore platform.
The boost in nanopore sensing and sequencing is evident from the incorporation of several new biological nanopores for spotting diverse features of analytes in question. In this regard, FhuA nanopore was utilized for determining the protein–protein interaction outside the nanopore through reversible binding and unbinding of ligand (analyte) to the tethered-receptor of nanopore.180 Furthermore, introduction of positively charged amino acids into the PlyAB nanopore reduced the electro-osmotic forces by creating an electro-osmotic whirl inside the nanopore and eased the detection of larger proteins such as human serum albumin and human transferrin through a largest nanopore known to date.181 In addition, VDAC of the outer mitochondrial membrane was employed in parodying the effect of post-translational modifications on the translocation dynamics of α-Synuclein (α-Syn).182 A well-known OmpG channel was utilized to detect small molecules such as adenosine triphosphate and glutamate by erasing few residues around its loop 6 and modification of pore lumen.183 However, the kinetics and translocation dynamics of positively charged cyclic oligosaccharides were manifested distinctly by making use of CymA nanopore.184 On top of that, computational design of new protein channels opens up a broad way for producing customized pores that possess a better control over their ion selectivity, dimensions and chemical interactions for an analyte of interest.185 Fig. 9 shows some paramount advances in other novel biological nanopores, figures are reproduced with permission.
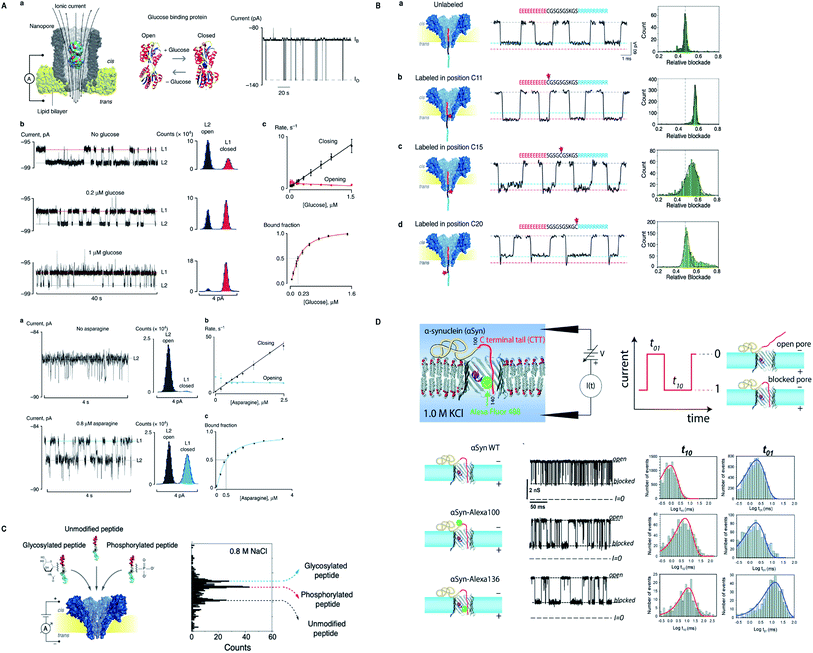 | ||
| Fig. 9 Recent advances in other novel biological nanopores. (A) Nanopore sensing of glucose and asparagine directly from bodily fluids by using cytolysin A.174 This figure does not need permission. (B) Identification of peptide labelled at different positions by FraC nanopore. This figure has been reproduced from ref. 62 (https://pubs.acs.org/doi/10.1021/acsnano.9b05156) with permission from American Chemical Society, Copyright 2019, further permissions related to the material excerpted should be directed to the ACS. (C) Label-free detection of post-translational modifications by FraC nanopore. This figure has been reproduced from ref. 173 (https://pubs.acs.org/doi/10.1021/acs.nanolett.9b03134) with permission from American Chemical Society, Copyright 2019, further permissions related to the material excerpted should be directed to the ACS. (D) α-Syn mimicking post-translational modifications by employing VDAC of the outer mitochondrial membrane (license ID: 1079204-1). This figure has been reproduced from ref. 182 with permission from Royal Society of Chemistry, Copyright 2020. | ||
8. Conclusion and perspective for selecting and designing a nanopore
Protein nanopores are convenient being economical, abundant in production and easy to engineer accurately according to the desired construct. Therefore, the past two decades show dramatic rise in the miscellaneous application of biological nanopores. Besides DNA sequencing, nanopores have revolutionized the prognosis and treatment of serious ailments at the disease onset by utilizing femto-molar (fM) concentration of target analytes.6Although, nanopore sensing has surpassed many present techniques but the choice for analyte detection is still limited on account of the desired features required for stochastic nanopore sensing that vary from analyte to analyte. Therefore, the confinement and physicochemical properties of a nanopore must be comparable with the analyte in question. In this regard, we have to consider the surface charge, geometry, physical measurements and hydrophobic/hydrophilic properties of a nanopore to make a better choice. Hence, to improve the sensitivity, selectivity and durability of a protein pore and to make it suitable for diagnostic purpose in hospitals, we should take into account the above described features before engineering a nanopore.
From the studies of well-known protein nanopores, we noticed that α-hemolysin is a versatile nanopore used for the detection of almost all domains of molecules mainly due to its suitably charged constriction region but dual constriction and long transmembrane region is inappropriate for DNA sequencing. However, MspA predominates in DNA sequencing owing to its single, sharp and small pore constriction but the translocation speed of ssDNA through it is faster than α-hemolysin. Contrarily, aerolysin nanopore cannot accommodate long strands of DNA at normal physiological conditions (see Section 4.3 for detail), but this nanopore is commendable for the detection of amino acids, peptides and proteins.
We ascertain that owing to the surface charge distribution and nanopore geometry, all kinds of protein pores behave in a different fashion even for the same analyte. In fact, we noticed that all the mutants of MspA discussed in preceding section showed peculiarity in their sensing skills, although all are MspA in real. The gist of this discussion points towards a pivotal fact that the choice of nanopore must not be dependent only on the dimension of the nanopore, besides, surface charge distribution should also be taken into consideration. Recently, a simulation-based study reasoned about different behaviors of biological nanopore and scrutinized out some newly engineered MspA mutants.186 We ascertain that incorporation of charged amino acids at the constriction zone of a nanopore has a pronounced effect on nanopore translocation pattern because of the deposition of oppositely charged ions in that confined area.
Certainly, the single, sharp constriction and stability of MspA nanopore suggest that it would be perfect for nanopore DNA sequencing. As this article mainly focuses on MspA and its variants, so we shed light on a fact that due to high speed of ssDNA through MspA, nearly all investigations were conducted by means of using molecular motors or adapters. Therefore, we suggest that by making use of MspA's ideal geometry, it would be more convenient to construct a mutant nanopore that can possibly meet the requirements of nanopore DNA sequencing together with eliminating laborious sample preparations. Currently, we are working on the new variant purified in our laboratory that is M3MspA by altering some experimental protocols to explore its hidden features. Concurrently, we anticipate that if we reduce some positively charged arginine amino acid from M3MspA then this newly fabricated mutant would be more convincing because it can reduce the counter-ions deposition in the nanopore. Additionally, the electric field inside the nanopore can possibly be managed by introducing some crowding molecules or metallic nanoparticles of desired sizes and charges outside of the nanopore. Thus, we envision that for any biological nanopore, proper positioning and suitable numbers of replaced residues (especially charged ones) are the keys to make an ideal and functional mutant nanopore, because it is observed that plenty of one kind of charge inside the nanopore creates ion selectivity in a nanopore.
Despite, nanopore has achieved many miraculous goals for stochastic sensing and significant steps of DNA sequencing but the speed of translocation, stability and single-read accuracy are the challenges left to be addressed for DNA/RNA sequencing, hitherto. However, these days Oxford Nanopore Technologies have been employing multiple variants of CsgG nanopore for direct RNA sequencing.177 By taking all the discrete details into account, we expect that computational designing and analysis of new protein channels and real time sophistication of the already present ones could pave a way towards a brighter future of nanopore DNA sequencing and sensing in the next few years.
Conflicts of interest
The authors declare no competing interest.Acknowledgements
We acknowledge the financial support by National Natural Science Foundation of China (61827814, 31872726) and the National Key R&D Program of China (2016YFA0501600).References
- E. Gouaux and R. MacKinnon, Science, 2005, 310, 1461 CrossRef CAS.
- W. H. Coulter, Means for Counting Particles Suspended in a Fluid, US Pat., 265650820, Chicago, Ill., 1953, Oct. , 1953 w, CQULTER 2,656,508. application August 27, 1949, Serial No. 112,819 17 Claims. (Cl. 324-ii) Search PubMed.
- M. Kukwikila and S. Howorka, Anal. Chem., 2015, 87, 9149–9154 CrossRef CAS PubMed.
- X. Chen, G. M. Roozbahani, Z. Ye, Y. Zhang, R. Ma, J. Xiang and X. Guan, ACS Appl. Mater. Interfaces, 2018, 10, 11519–11528 CrossRef CAS.
- S. Rauf, L. Zhang, A. Ali, Y. Liu and J. Li, ACS Sens., 2017, 2, 227–234 CrossRef CAS PubMed.
- H. Zhang, M. Hiratani, K. Nagaoka and R. Kawano, Nanoscale, 2017, 9, 16124–16127 RSC.
- M. Wanunu, T. Dadosh, V. Ray, J. Jin, L. McReynolds and M. Drndić, Nat. Nanotechnol., 2010, 5, 807–814 CrossRef CAS.
- R. Kawano, T. Osaki, H. Sasaki, M. Takinoue, S. Yoshizawa and S. Takeuchi, J. Am. Chem. Soc., 2011, 133, 8474–8477 CrossRef CAS PubMed.
- A. Meller, L. Nivon, E. Brandin, J. Golovchenko and D. Branton, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 1079 CrossRef CAS PubMed.
- A. H. Laszlo, I. M. Derrington, B. C. Ross, H. Brinkerhoff, A. Adey, I. C. Nova, J. M. Craig, K. W. Langford, J. M. Samson and R. Daza, Nat. Biotechnol., 2014, 32, 829–833 CrossRef CAS PubMed.
- Y. Wang, D. Zheng, Q. Tan, M. X. Wang and L.-Q. Gu, Nat. Nanotechnol., 2011, 6, 668–674 CrossRef CAS.
- J. J. Kasianowicz, E. Brandin, D. Branton and D. W. Deamer, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13770–13773 CrossRef CAS PubMed.
- S. Howorka, S. Cheley and H. Bayley, Nat. Biotechnol., 2001, 19, 636 CrossRef CAS PubMed.
- S. Wen, T. Zeng, L. Liu, K. Zhao, Y. Zhao, X. Liu and H.-C. Wu, J. Am. Chem. Soc., 2011, 133, 18312–18317 CrossRef CAS PubMed.
- M. Akeson, D. Branton, J. J. Kasianowicz, E. Brandin and D. W. Deamer, Biophys. J., 1999, 77, 3227–3233 CrossRef CAS PubMed.
- R. F. Purnell and J. J. Schmidt, ACS Nano, 2009, 3, 2533–2538 CrossRef CAS PubMed.
- J. Clarke, H.-C. Wu, L. Jayasinghe, A. Patel, S. Reid and H. Bayley, Nat. Nanotechnol., 2009, 4, 265–270 CrossRef CAS PubMed.
- J. A. Cracknell, D. Japrung and H. Bayley, Nano Lett., 2013, 13, 2500–2505 CrossRef CAS PubMed.
- J. Shendure and H. Ji, Nat. Biotechnol., 2008, 26, 1135 CrossRef CAS.
- F. Sanger, S. Nicklen and A. R. Coulson, Proc. Natl. Acad. Sci. U. S. A., 1977, 74, 5463 CrossRef CAS.
- C. Cao, Y.-L. Ying, Z. Gu and Y.-T. Long, Anal. Chem., 2014, 86, 11946–11950 CrossRef CAS.
- P. R. Singh, I. n. Bárcena-Uribarri, N. Modi, U. Kleinekathöfer, R. Benz, M. Winterhalter and K. R. Mahendran, ACS Nano, 2012, 6, 10699–10707 CrossRef CAS PubMed.
- Q. Zhao, R. S. S. D. Zoysa, D. Wang, D. A. Jayawardhana and X. Guan, J. Am. Chem. Soc., 2009, 131, 6324 CrossRef CAS PubMed.
- B. Cressiot, L. Bacri and J. Pelta, Small Methods, 2020, 4, 2000090 CrossRef CAS.
- O. Braha, L.-Q. Gu, L. Zhou, X. Lu, S. Cheley and H. Bayley, Nat. Biotechnol., 2000, 18, 1005 CrossRef CAS PubMed.
- M. T. Basel, R. K. Dani, M. Kang, M. Pavlenok, V. Chikan, P. E. Smith, M. Niederweis and S. H. Bossmann, ACS Nano, 2009, 3, 462–466 CrossRef CAS PubMed.
- Y. Wang, B.-Q. Luan, Z. Yang, X. Zhang, B. Ritzo, K. Gates and L.-Q. Gu, Sci. Rep., 2014, 4, 5883 CrossRef CAS PubMed.
- C. Shasha, R. Y. Henley, D. H. Stoloff, K. D. Rynearson, T. Hermann and M. Wanunu, ACS Nano, 2014, 8, 6425–6430 CrossRef CAS.
- H.-C. Wu and H. Bayley, J. Am. Chem. Soc., 2008, 130, 6813–6819 CrossRef CAS.
- A. Meller, L. Nivon, E. Brandin, J. Golovchenko and D. Branton, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 1079–1084 CrossRef CAS.
- A. Meller and D. Branton, Electrophoresis, 2002, 23, 2583–2591 CrossRef CAS.
- D. Rodriguez-Larrea and H. Bayley, Nat. Nanotechnol., 2013, 8, 288–295 CrossRef CAS PubMed.
- J. Nivala, D. B. Marks and M. Akeson, Nat. Biotechnol., 2013, 31, 247–250 CrossRef CAS PubMed.
- A. Aksimentiev, Nanoscale, 2010, 2, 468–483 RSC.
- D. K. Lubensky and D. R. Nelson, Biophys. J., 1999, 77, 1824–1838 CrossRef CAS PubMed.
- P. Yakovchuk, E. Protozanova and M. D. Frank-Kamenetskii, Nucleic Acids Res., 2006, 34, 564–574 CrossRef CAS PubMed.
- A. Meller, J. Phys.: Condens. Matter, 2003, 15, R581–R607 CrossRef CAS.
- L. Song, M. R. Hobaugh, C. Shustak, S. Cheley, H. Bayley and J. E. Gouaux, Science, 1996, 274, 1859–1865 CrossRef CAS PubMed.
- M. Faller, M. Niederweis and G. E. Schulz, Science, 2004, 303, 1189–1192 CrossRef CAS PubMed.
- J. Li, D. Stein, C. Mcmullan, D. Branton, M. J. Aziz and J. A. Golovchenko, Nature, 2001, 412, 166–169 CrossRef CAS PubMed.
- I. Yanagi, T. Ishida, K. Fujisaki and K. Takeda, Sci. Rep., 2015, 5, 14656 CrossRef CAS PubMed.
- L. Wu, H. Liu, W. Zhao, L. Wang, C. Hou, Q. Liu and Z. Lu, Nanoscale Res. Lett., 2014, 9, 140 CrossRef PubMed.
- A. R. Hall, A. Scott, D. Rotem, K. K. Mehta, H. Bayley and C. Dekker, Nat. Nanotechnol., 2010, 5, 874–877 CrossRef CAS PubMed.
- S. Cabello-Aguilar, S. Balme, A. A. Chaaya, M. Bechelany, E. Balanzat, J.-M. Janot, C. Pochat-Bohatier, P. Miele and P. Dejardin, Nanoscale, 2013, 5, 9582–9586 RSC.
- L. Zhu, Y. Xu, I. Ali, L. Liu, H. Wu, Z. Lu and Q. Liu, ACS Appl. Mater. Interfaces, 2018, 10, 26555–26565 CrossRef CAS.
- B. M. Venkatesan, A. B. Shah, J. M. Zuo and R. Bashir, Adv. Funct. Mater., 2010, 20, 1266–1275 CrossRef CAS.
- S. Garaj, S. Liu, J. A. Golovchenko and D. Branton, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12192 CrossRef CAS PubMed.
- K. Liu, C. Pan, A. Kuhn, A. P. Nievergelt, G. E. Fantner, O. Milenkovic and A. Radenovic, Nat. Commun., 2019, 10, 3 CrossRef CAS PubMed.
- R. Wang, T. Gilboa, J. Song, D. Huttner, M. W. Grinstaff and A. Meller, ACS Nano, 2018, 12, 11648–11656 CrossRef CAS PubMed.
- T. Ma, J.-M. Janot and S. Balme, Small Methods, 2020, 4, 2000366 CrossRef CAS.
- H. Liu, R. Wang, D. Gu, S. Tan, H. Wu and Q. Liu, J. Nanosci. Nanotechnol., 2017, 17, 9125–9129 CrossRef CAS.
- L. Movileanu, S. Howorka, O. Braha and H. Bayley, Nat. Biotechnol., 2000, 18, 1091–1095 CrossRef CAS PubMed.
- C. W. Fuller, S. Kumar, M. Porel, M. Chien, A. Bibillo, P. B. Stranges, M. Dorwart, C. Tao, Z. Li, W. Guo, S. Shi, D. Korenblum, A. Trans, A. Aguirre, E. Liu, E. T. Harada, J. Pollard, A. Bhat, C. Cech, A. Yang, C. Arnold, M. Palla, J. Hovis, R. Chen, I. Morozova, S. Kalachikov, J. J. Russo, J. J. Kasianowicz, R. Davis, S. Roever, G. M. Church and J. Ju, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 5233 CrossRef CAS PubMed.
- Y. Astier, A. Orit Braha and H. Bayley, J. Am. Chem. Soc., 2006, 128, 1705–1710 CrossRef CAS PubMed.
- D. Stoddart, M. Ayub, L. Höfler, P. Raychaudhuri, J. W. Klingelhoefer, G. Maglia, A. Heron and H. Bayley, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 2425 CrossRef CAS PubMed.
- C. Cao, Y. L. Ying, Z. L. Hu, D. F. Liao, H. Tian and Y. T. Long, Nat. Nanotechnol., 2016, 11, 713 CrossRef CAS PubMed.
- B. Cressiot, E. Braselmann, A. Oukhaled, A. H. Elcock, J. Pelta and P. L. Clark, ACS Nano, 2015, 9, 9050–9061 CrossRef CAS PubMed.
- D. A. Fajardo, J. Cheung, C. Ito, E. Sugawara, H. Nikaido and R. Misra, J. Bacteriol., 1998, 180, 4452–4459 CrossRef CAS PubMed.
- M. Fahie, C. Chisholm and M. Chen, ACS Nano, 2015, 9, 1089–1098 CrossRef CAS PubMed.
- M. Soskine, A. Biesemans, B. Moeyaert, S. Cheley, H. Bayley and G. Maglia, Nano Lett., 2012, 12, 4895–4900 CrossRef CAS PubMed.
- C. Wloka, V. V. Meervelt, D. V. Gelder, N. Danda, N. Jager, C. P. Williams and G. Maglia, ACS Nano, 2017, 11, 4387–4394 CrossRef CAS PubMed.
- L. Restrepo-Pérez, G. Huang, P. R. Bohländer, N. Worp, R. Eelkema, G. Maglia, C. Joo and C. Dekker, ACS Nano, 2019, 13, 13668–13676 CrossRef PubMed.
- A. Seifert, K. Göpfrich, J. R. Burns, N. Fertig, U. F. Keyser and S. Howorka, ACS Nano, 2015, 9, 1117–1126 CrossRef CAS PubMed.
- N. A. W. Bell, C. R. Engst, M. Ablay, G. Divitini, C. Ducati, T. Liedl and U. F. Keyser, Nano Lett., 2013, 12, 512–517 CrossRef.
- T. Ding, J. Yang, V. Pan, N. Zhao, Z. Lu, Y. Ke and C. Zhang, Nucleic Acids Res., 2020, 48, 2791–2806 CrossRef CAS PubMed.
- D. Stoddart, G. Maglia, E. Mikhailova, A. J. Heron and H. Bayley, Angew. Chem., Int. Ed., 2010, 49, 556–559 CrossRef CAS PubMed.
- L. Movileanu, S. Cheley, S. Howorka, O. Braha and H. Bayley, J. Gen. Physiol., 2001, 117, 239–252 CrossRef CAS.
- D. Stoddart, A. J. Heron, E. Mikhailova, G. Maglia and H. Bayley, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7702–7707 CrossRef CAS.
- D. Stoddart, A. J. Heron, J. Klingelhoefer, E. Mikhailova, G. Maglia and H. Bayley, Nano Lett., 2010, 10, 3633–3637 CrossRef CAS PubMed.
- E. N. Ervin, G. A. Barrall, P. Pal, M. K. Bean, A. E. P. Schibel and A. D. Hibbs, BioNanoScience, 2014, 4, 78–84 CrossRef PubMed.
- A. A. Simpson, Y. Tao, P. G. Leiman, M. O. Badasso, Y. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes and D. L. Anderson, Nature, 2000, 408, 745 CrossRef CAS PubMed.
- W. David, J. Peng, G. Jia, S. Varuni, L. T. Jin, M. Carlo and G. Peixuan, Nat. Nanotechnol., 2009, 4, 765 CrossRef PubMed.
- A. Guasch, J. Pous, B. Ibarra, F. X. Gomis-Rüth, J. M. Valpuesta, N. Sousa, J. L. Carrascosa and M. Coll, J. Mol. Biol., 2002, 315, 663–676 CrossRef CAS PubMed.
- P. X. Guo, S. Erickson and D. Anderson, Science, 1987, 236, 690–694 CrossRef CAS PubMed.
- P. Guo, C. Zhang, C. Chen, K. Garver and M. Trottier, Mol. Cell, 1998, 2, 149–155 CrossRef CAS PubMed.
- T. J. Lee and P. Guo, J. Mol. Biol., 2006, 356, 589–599 CrossRef CAS PubMed.
- E. A. Manrao, I. M. Derrington, A. H. Laszlo, K. W. Langford, M. K. Hopper, N. Gillgren, M. Pavlenok, M. Niederweis and J. H. Gundlach, Nat. Biotechnol., 2012, 30, 349–353 CrossRef CAS PubMed.
- S. Yan, X. Li, P. Zhang, Y. Wang, H.-Y. Chen, S. Huang and H. Yu, Chem. Sci., 2019, 10, 3110–3117 RSC.
- B. Cressiot, S. J. Greive, M. Mojtabavi, A. A. Antson and M. Wanunu, Nat. Commun., 2018, 9, 4652 CrossRef PubMed.
- M. W. Parker, J. T. Buckley, J. P. M. Postma, A. D. Tucker, K. Leonard, F. Pattus and D. Tsernoglou, Nature, 1994, 367, 292–295 CrossRef CAS PubMed.
- R. Stefureac, Y.-t. Long, H.-B. Kraatz, P. Howard and J. S. Lee, Biochemistry, 2006, 45, 9172–9179 CrossRef CAS PubMed.
- M. Pastoriza-Gallego, L. Rabah, G. Gibrat, B. Thiebot, F. G. van der Goot, L. Auvray, J.-M. Betton and J. Pelta, J. Am. Chem. Soc., 2011, 133, 2923–2931 CrossRef CAS PubMed.
- A. Fennouri, R. Daniel, M. Pastoriza-Gallego, L. Auvray, J. Pelta and L. Bacri, Anal. Chem., 2013, 85, 8488–8492 CrossRef CAS PubMed.
- G. Baaken, I. Halimeh, L. Bacri, J. Pelta, A. Oukhaled and J. C. Behrends, ACS Nano, 2015, 9, 6443–6449 CrossRef CAS.
- C. Cao, J. Yu, Y.-Q. Wang, Y.-L. Ying and Y.-T. Long, Anal. Chem., 2016, 88, 5046–5049 CrossRef CAS PubMed.
- Y. Wang, K. Tian, X. Du, R.-C. Shi and L.-Q. Gu, Anal. Chem., 2017, 89, 13039–13043 CrossRef CAS PubMed.
- Z.-L. Hu, M.-Y. Li, S.-C. Liu, Y.-L. Ying and Y.-T. Long, Chem. Sci., 2019, 10, 354–358 RSC.
- Y.-Q. Wang, M.-Y. Li, H. Qiu, C. Cao, M.-B. Wang, X.-Y. Wu, J. Huang, Y.-L. Ying and Y.-T. Long, Anal. Chem., 2018, 90, 7790–7794 CrossRef CAS PubMed.
- C. Cao, M.-Y. Li, N. Cirauqui, Y.-Q. Wang, M. Dal Peraro, H. Tian and Y.-T. Long, Nat. Commun., 2018, 9, 2823 CrossRef PubMed.
- Y.-Q. Wang, C. Cao, Y.-L. Ying, S. Li, M.-B. Wang, J. Huang and Y.-T. Long, ACS Sens., 2018, 3, 779–783 CrossRef CAS PubMed.
- I. M. Derrington, T. Z. Butler, M. D. Collins, E. Manrao, M. Pavlenok, M. Niederweis and J. H. Gundlach, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16060–16065 CrossRef.
- C. Stahl, S. Kubetzko, I. Kaps, S. Seeber, H. Engelhardt and M. Niederweis, Mol. Microbiol., 2001, 40, 451–464 CrossRef CAS.
- J. Stephan, J. Bender, F. Wolschendorf, C. Hoffmann, E. Roth, C. Mailänder, H. Engelhardt and M. Niederweis, Mol. Microbiol., 2005, 58, 714–730 CrossRef CAS PubMed.
- C. Heinz, H. Engelhardt and M. Niederweis, J. Biol. Chem., 2003, 278, 8678–8685 CrossRef CAS PubMed.
- E. A. Manrao, I. M. Derrington, M. Pavlenok, M. Niederweis and J. H. Gundlach, PLoS One, 2011, 6, e25723 CrossRef CAS.
- J. Schreiber, Z. L. Wescoe, R. Abu-Shumays, J. T. Vivian, B. Baatar, K. Karplus and M. Akeson, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 18910–18915 CrossRef CAS PubMed.
- Z. Wescoe, J. Schreiber and M. Akeson, J. Am. Chem. Soc., 2014, 136, 16582–16587 CrossRef CAS PubMed.
- A. H. Laszlo, I. M. Derrington, H. Brinkerhoff, K. W. Langford, I. C. Nova, J. M. Samson, J. J. Bartlett, M. Pavlenok and J. H. Gundlach, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 18904–18909 CrossRef CAS PubMed.
- T. Z. Butler, M. Pavlenok, I. M. Derrington, M. Niederweis and J. H. Gundlach, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20647–20652 CrossRef CAS PubMed.
- M. Pavlenok, I. M. Derrington, J. H. Gundlach and M. Niederweis, PLoS One, 2012, 7, e38726 CrossRef CAS PubMed.
- S. Bhattacharya, I. M. Derrington, M. Pavlenok, M. Niederweis, J. H. Gundlach and A. Aksimentiev, ACS Nano, 2012, 6, 6960–6968 CrossRef CAS PubMed.
- H. Bhatti, R. Jawed, H. Liu, I. Ali, L. Zhu and Q. Liu, Nanosci. Nanotechnol. Lett., 2019, 11, 1104–1115 CrossRef.
- E. B. Kalman, O. Sudre, I. Vlassiouk and Z. S. Siwy, Anal. Bioanal. Chem., 2009, 394, 413–419 CrossRef CAS PubMed.
- A. B. Kolomeisky and K. Uppulury, J. Stat. Phys., 2011, 142, 1268–1276 CrossRef CAS.
- N. M. Luscombe, R. A. Laskowski and J. M. Thornton, Nucleic Acids Res., 2001, 29, 2860–2874 CrossRef CAS PubMed.
- L. Liu and H. C. Wu, Angew. Chem., Int. Ed., 2016, 55, 15216–15222 CrossRef CAS PubMed.
- C. Cao and Y.-T. Long, Acc. Chem. Res., 2018, 51, 331–341 CrossRef CAS PubMed.
- H. Wang, J. Ettedgui, J. Forstater, J. W. F. Robertson, J. E. Reiner, H. Zhang, S. Chen and J. J. Kasianowicz, ACS Sens., 2018, 3, 251–263 CrossRef CAS PubMed.
- Y. Wang and L.-Q. Gu, AIMS Mater. Sci., 2015, 2, 448–472 Search PubMed.
- G. Liu, L. Zhang, D. Dong, Y. Liu and J. Li, Anal. Methods, 2016, 8, 7040–7046 RSC.
- R. T. Perera, A. M. Fleming, R. P. Johnson, C. J. Burrows and H. S. White, Nanotechnology, 2015, 26, 074002 CrossRef CAS PubMed.
- W. J. Ramsay and H. Bayley, Angew. Chem., Int. Ed., 2018, 57, 2841–2845 CrossRef CAS PubMed.
- T. Vu, J. Borgesi, J. Soyring, M. D'Alia, S.-L. Davidson and J. Shim, Nanoscale, 2019, 11, 10536–10545 RSC.
- S. Wang, Y. Wang, S. Yan, X. Du, P. Zhang, H.-Y. Chen and S. Huang, ACS Appl. Mater. Interfaces, 2020, 12, 26926–26935 CrossRef CAS.
- F. Yao, X. Peng, Z. Su, L. Tian, Y. Guo and X.-f. Kang, Anal. Chem., 2020, 92, 3827–3833 CrossRef CAS PubMed.
- A. Asandei, L. Mereuta, J. Park, C. H. Seo, Y. Park and T. Luchian, ACS Sens., 2019, 4, 1502–1507 CrossRef CAS PubMed.
- L. Mereuta, A. Asandei, I. Schiopu, Y. Park and T. Luchian, Anal. Chem., 2019, 91, 8630–8637 CrossRef CAS PubMed.
- L. Mereuta, A. Asandei, I. S. Dragomir, I. C. Bucataru, J. Park, C. H. Seo, Y. Park and T. Luchian, Sci. Rep., 2020, 10, 11323 CrossRef CAS.
- L. Liu, T. Li, S. Zhang, P. Song, B. Guo, Y. Zhao and H.-C. Wu, Angew. Chem., Int. Ed., 2018, 57, 11882–11887 CrossRef CAS PubMed.
- S. Fujii, K. Kamiya, T. Osaki, N. Misawa, M. Hayakawa and S. Takeuchi, Anal. Chem., 2018, 90, 10217–10222 CrossRef CAS.
- J. Ivica, P. T. F. Williamson and M. R. R. de Planque, Anal. Chem., 2017, 89, 8822–8829 CrossRef CAS.
- X. Wei, D. Ma, Z. Zhang, L. Y. Wang, J. L. Gray, L. Zhang, T. Zhu, X. Wang, B. J. Lenhart, Y. Yin, Q. Wang and C. Liu, ACS Sens., 2020, 5, 1707–1716 CrossRef CAS.
- X. Wei, D. Ma, L. Jing, L. Y. Wang, X. Wang, Z. Zhang, B. J. Lenhart, Y. Yin, Q. Wang and C. Liu, J. Mater. Chem. B, 2020, 8, 6792–6797 RSC.
- M. G. Larimi, L. A. Mayse and L. Movileanu, ACS Nano, 2019, 13, 4469–4477 CrossRef CAS PubMed.
- L. Harrington, L. T. Alexander, S. Knapp and H. Bayley, ACS Nano, 2019, 13, 633–641 CrossRef CAS PubMed.
- A. Fennouri, J. Ramiandrisoa, L. Bacri, J. Mathé and R. Daniel, Eur. Phys. J. E: Soft Matter Biol. Phys., 2018, 41, 127 CrossRef CAS.
- G. M. Roozbahani, Y. Zhang, X. Chen, M. H. Soflaee and X. Guan, Analyst, 2019, 144, 7432–7436 RSC.
- Y.-M. Liu, X.-Y. Fang, F. Fang and Z.-Y. Wu, Analyst, 2019, 144, 4081–4085 RSC.
- T. Zeng, A. M. Fleming, Y. Ding, H. Ren, H. S. White and C. J. Burrows, J. Org. Chem., 2018, 83, 3973–3978 CrossRef CAS.
- S. Rauf, L. Zhang, A. Ali, J. Ahmad, Y. Liu and J. Li, Anal. Chem., 2017, 89, 13252–13260 CrossRef CAS.
- X. Chen, Y. Zhang, G. Mohammadi Roozbahani and X. Guan, ACS Appl. Bio Mater., 2019, 2, 504–509 CrossRef CAS PubMed.
- J. J. S. Júnior, T. A. Soares, L. Pol-Fachin, D. C. Machado, V. H. Rusu, J. P. Aguiar and C. G. Rodrigues, RSC Adv., 2019, 9, 14683–14691 RSC.
- S. Zhou, H. Wang, X. Chen, Y. Wang, D. Zhou, L. Liang, L. Wang, D. Wang and X. Guan, ACS Appl. Bio Mater., 2020, 3, 554–560 CrossRef CAS PubMed.
- Y. Sheng, K. Zhou, Q. Liu, L. Liu and H.-C. Wu, Anal. Chem., 2020, 92, 7485–7492 CrossRef CAS PubMed.
- T. Vu, S.-L. Davidson and J. Shim, Analyst, 2018, 143, 906–913 RSC.
- W. J. Ramsay, N. A. W. Bell, Y. Qing and H. Bayley, J. Am. Chem. Soc., 2018, 140, 17538–17546 CrossRef CAS.
- I. S. Dragomir, I. C. Bucataru, I. Schiopu and T. Luchian, Anal. Chem., 2020, 92, 7800–7807 CrossRef CAS PubMed.
- M. Matsushita, K. Shoji, N. Takai and R. Kawano, J. Phys. Chem. B, 2020, 124, 2410–2416 CrossRef CAS PubMed.
- J. Lei, Y. Huang, W. Zhong, D. Xiao and C. Zhou, Anal. Chem., 2020, 92, 8867–8873 CrossRef CAS PubMed.
- F. Bétermier, B. Cressiot, G. Di Muccio, N. Jarroux, L. Bacri, B. Morozzo della Rocca, M. Chinappi, J. Pelta and J.-M. Tarascon, Commun. Mater., 2020, 1, 59 CrossRef.
- A. H. Laszlo, I. M. Derrington and J. H. Gundlach, Methods, 2016, 105, 75–89 CrossRef CAS PubMed.
- I. M. Derrington, J. M. Craig, E. Stava, A. H. Laszlo, B. C. Ross, H. Brinkerhoff, I. C. Nova, K. Doering, B. I. Tickman and M. Ronaghi, Nat. Biotechnol., 2015, 33, 1073–1075 CrossRef CAS PubMed.
- J. M. Craig, A. H. Laszlo, H. Brinkerhoff, I. M. Derrington, M. T. Noakes, I. C. Nova, B. I. Tickman, K. Doering, N. F. de Leeuw and J. H. Gundlach, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 11932 CrossRef CAS.
- C. C. Caldwell and M. Spies, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 11809 CrossRef CAS PubMed.
- I. C. Nova, I. M. Derrington, J. M. Craig, M. T. Noakes, B. I. Tickman, K. Doering, H. Higinbotham, A. H. Laszlo and J. H. Gundlach, PLoS One, 2017, 12, e0181599 CrossRef PubMed.
- M. T. Noakes, H. Brinkerhoff, A. H. Laszlo, I. M. Derrington, K. W. Langford, J. W. Mount, J. L. Bowman, K. S. Baker, K. M. Doering, B. I. Tickman and J. H. Gundlach, Nat. Biotechnol., 2019, 37, 651–656 CrossRef CAS PubMed.
- A. H. Laszlo, I. M. Derrington, B. C. Ross, H. Brinkerhoff, A. Adey, I. C. Nova, J. M. Craig, K. W. Langford, J. M. Samson, R. Daza, K. Doering, J. Shendure and J. H. Gundlach, Nat. Biotechnol., 2014, 32, 829–833 CrossRef CAS PubMed.
- Y. Wang, K. M. Patil, S. Yan, P. Zhang, W. Guo, Y. Wang, H.-Y. Chen, D. Gillingham and S. Huang, Angew. Chem., Int. Ed., 2019, 58, 8432–8436 CrossRef CAS PubMed.
- S. Wang, J. Cao, W. Jia, W. Guo, S. Yan, Y. Wang, P. Zhang, H.-Y. Chen and S. Huang, Chem. Sci., 2020, 11, 879–887 RSC.
- J. Zhang, S. Yan, L. Chang, W. Guo, Y. Wang, Y. Wang, P. Zhang, H.-Y. Chen and S. Huang, iScience, 2020, 23, 100916 CrossRef CAS.
- J. Cao, W. Jia, J. Zhang, X. Xu, S. Yan, Y. Wang, P. Zhang, H.-Y. Chen and S. Huang, Nat. Commun., 2019, 10, 5668 CrossRef PubMed.
- M. P. Ledbetter, J. M. Craig, R. J. Karadeema, M. T. Noakes, H. C. Kim, S. J. Abell, J. R. Huang, B. A. Anderson, R. Krishnamurthy, J. H. Gundlach and F. E. Romesberg, J. Am. Chem. Soc., 2020, 142, 2110–2114 CrossRef CAS PubMed.
- C. Cao, N. Cirauqui, M. J. Marcaida, E. Buglakova, A. Duperrex, A. Radenovic and M. Dal Peraro, Nat. Commun., 2019, 10, 4918 CrossRef PubMed.
- Z. Zou, H. Yang, Q. Yan, P. Qi, Z. Qing, J. Zheng, X. Xu, L. Zhang, F. Feng and R. Yang, Chem. Commun., 2019, 55, 6433–6436 RSC.
- F. Piguet, H. Ouldali, M. Pastoriza-Gallego, P. Manivet, J. Pelta and A. Oukhaled, Nat. Commun., 2018, 9, 966 CrossRef.
- Y. Lu, X.-Y. Wu, Y.-L. Ying and Y.-T. Long, Chem. Commun., 2019, 55, 9311–9314 RSC.
- B. Yuan, S. Li, Y.-L. Ying and Y.-T. Long, Analyst, 2020, 145, 1179–1183 RSC.
- F.-N. Meng, Y.-L. Ying, J. Yang and Y.-T. Long, Anal. Chem., 2019, 91, 9910–9915 CrossRef CAS.
- J. Yang, Y.-Q. Wang, M.-Y. Li, Y.-L. Ying and Y.-T. Long, Langmuir, 2018, 34, 14940–14945 CrossRef CAS.
- D. Xi, Z. Li, L. Liu, S. Ai and S. Zhang, Anal. Chem., 2018, 90, 1029–1034 CrossRef CAS PubMed.
- X.-J. Sui, M.-Y. Li, Y.-L. Ying, B.-Y. Yan, H.-F. Wang, J.-L. Zhou, Z. Gu and Y.-T. Long, J. Anal. Test., 2019, 3, 134–139 CrossRef.
- Z.-X. Wei, Y.-L. Ying, M.-Y. Li, J. Yang, J.-L. Zhou, H.-F. Wang, B.-Y. Yan and Y.-T. Long, Anal. Chem., 2019, 91, 10033–10039 CrossRef CAS.
- J. Wang, M.-Y. Li, J. Yang, Y.-Q. Wang, X.-Y. Wu, J. Huang, Y.-L. Ying and Y.-T. Long, ACS Cent. Sci., 2020, 6, 76–82 CrossRef CAS PubMed.
- M.-Y. Li, Y.-L. Ying, S. Li, Y.-Q. Wang, X.-Y. Wu and Y.-T. Long, ACS Nano, 2020, 14, 12571–12578 CrossRef CAS PubMed.
- B. Zhou, Y.-Q. Wang, C. Cao, D.-W. Li and Y.-T. Long, Sci. China: Chem., 2018, 61, 1385–1388 CrossRef CAS.
- M.-Y. Li, Y.-Q. Wang, Y. Lu, Y.-L. Ying and Y.-T. Long, Front. Chem., 2019, 7, 528 CrossRef CAS PubMed.
- Y.-L. Ying, Z.-Y. Li, Z.-L. Hu, J. Zhang, F.-N. Meng, C. Cao, Y.-T. Long and H. Tian, Chem, 2018, 4, 1893–1901 CAS.
- M.-Y. Li, Y.-Q. Wang, Y.-L. Ying and Y.-T. Long, Chem. Sci., 2019, 10, 10400–10404 RSC.
- H. Ouldali, K. Sarthak, T. Ensslen, F. Piguet, P. Manivet, J. Pelta, J. C. Behrends, A. Aksimentiev and A. Oukhaled, Nat. Biotechnol., 2020, 38, 176–181 CrossRef CAS PubMed.
- G. Huang, K. Willems, M. Soskine, C. Wloka and G. Maglia, Nat. Commun., 2017, 8, 935 CrossRef PubMed.
- G. Huang, A. Voet and G. Maglia, Nat. Commun., 2019, 10, 835 CrossRef.
- N. L. Mutter, J. Volarić, W. Szymanski, B. L. Feringa and G. Maglia, J. Am. Chem. Soc., 2019, 141, 14356–14363 CrossRef CAS PubMed.
- L. Restrepo-Pérez, C. H. Wong, G. Maglia, C. Dekker and C. Joo, Nano Lett., 2019, 19, 7957–7964 CrossRef.
- N. S. Galenkamp, M. Soskine, J. Hermans, C. Wloka and G. Maglia, Nat. Commun., 2018, 9, 4085 CrossRef PubMed.
- K. Willems, D. Ruić, A. Biesemans, N. S. Galenkamp, P. Van Dorpe and G. Maglia, ACS Nano, 2019, 13, 9980–9992 CrossRef CAS PubMed.
- Y. Wang, Y. Wang, X. Du, S. Yan, P. Zhang, H.-Y. Chen and S. Huang, Sci. Adv., 2019, 5, eaar3309 CrossRef PubMed.
- D. R. Garalde, E. A. Snell, D. Jachimowicz, B. Sipos, J. H. Lloyd, M. Bruce, N. Pantic, T. Admassu, P. James, A. Warland, M. Jordan, J. Ciccone, S. Serra, J. Keenan, S. Martin, L. McNeill, E. J. Wallace, L. Jayasinghe, C. Wright, J. Blasco, S. Young, D. Brocklebank, S. Juul, J. Clarke, A. J. Heron and D. J. Turner, Nat. Methods, 2018, 15, 201–206 CrossRef CAS PubMed.
- W. Stephenson, R. Razaghi, S. Busan, K. M. Weeks, W. Timp and P. Smibert, bioRxiv, 2020 DOI:10.1101/2020.05.31.126763.
- S. E. Van der Verren, N. Van Gerven, W. Jonckheere, R. Hambley, P. Singh, J. Kilgour, M. Jordan, E. J. Wallace, L. Jayasinghe and H. Remaut, Nat. Biotechnol., 2020, 38, 1415–1420 CrossRef CAS.
- A. K. Thakur and L. Movileanu, Nat. Biotechnol., 2019, 37, 96–101 CrossRef CAS PubMed.
- G. Huang, K. Willems, M. Bartelds, P. van Dorpe, M. Soskine and G. Maglia, Nano Lett., 2020, 20, 3819–3827 CrossRef CAS PubMed.
- D. P. Hoogerheide, P. A. Gurnev, T. K. Rostovtseva and S. M. Bezrukov, Nanoscale, 2020, 12, 11070–11078 RSC.
- R. R. Sanganna Gari, P. Seelheim, B. Liang and L. K. Tamm, ACS Sens., 2019, 4, 1230–1235 CrossRef CAS PubMed.
- D. Vikraman, R. Satheesan, K. S. Kumar and K. R. Mahendran, ACS Nano, 2020, 14, 2285–2295 CrossRef CAS PubMed.
- C. Xu, P. Lu, T. M. Gamal El-Din, X. Y. Pei, M. C. Johnson, A. Uyeda, M. J. Bick, Q. Xu, D. Jiang, H. Bai, G. Reggiano, Y. Hsia, T. J. Brunette, J. Dou, D. Ma, E. M. Lynch, S. E. Boyken, P.-S. Huang, L. Stewart, F. DiMaio, J. M. Kollman, B. F. Luisi, T. Matsuura, W. A. Catterall and D. Baker, Nature, 2020, 585, 129–134 CrossRef CAS.
- W. Zhou, H. Qiu, Y. Guo and W. Guo, J. Phys. Chem. B, 2020, 124, 1611–1618 CAS.
| This journal is © The Royal Society of Chemistry 2021 |