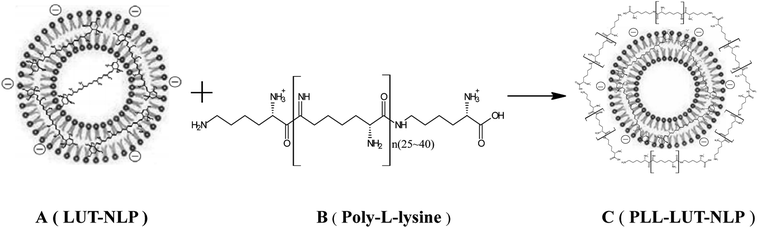
Open Access Article
Open Access ArticlePolypeptide – decorated nanoliposomes as novel delivery systems for lutein
Yan Jiao ab,
Dajing Li*a,
Chunquan Liua,
Ying Changb,
Jiangfeng Songa and
Yadong Xiaoa
ab,
Dajing Li*a,
Chunquan Liua,
Ying Changb,
Jiangfeng Songa and
Yadong Xiaoa
aInstitute of Farm Products Processing, Jiangsu Academy of Agricultural Sciences, Nanjing 210014, China. E-mail: jiaoyan_3000@126.com; lidajing@163.com
bCollege of Food and Biological Engineering, Qiqihar University, Qiqihar 161006, China
First published on 6th September 2018
Abstract
Lutein (LUT) is a bioactive food compound found in various vegetables and plays a critical role in the promotion of health and well-being. However, lutein is an unstable molecule which has a very low bioavailability caused by its poor solubility in aqueous media, and is poorly absorbed when administered orally. To enhance the stability, release and bioactivity of lutein, poly-L-lysine (PLL) decorated nanoliposomes (PLL–LUT-NLP) were developed as novel delivery systems for lutein. The mean particle size of PLL–LUT-NLP was found to be in the range 264–367 nm with a low polydispersity index (PDI < 0.4). The zeta potential changed from −38.6 mV in undecorated nanoliposomes to −27.9 mV in PLL-decorated nanoliposomes. Furthermore, the lutein entrapment efficiency (EE%) of PLL–LUT-NLP was found to be highest in nanoliposomes decorated with 0.06% (w/v) PLL. PLL could protect lutein in nanoliposomes from degradation and promote the lutein release from the nanoliposomes in gastrointestinal fluid conditions. Additionally, the PLL-decorated nanoliposomes maintained the antioxidant activity of the lutein, and the antiproliferative activity was more significant than that of undecorated nanoliposomes in inhibiting the proliferation of human tumor cells. These results suggest that PLL-decorated nanoliposomes have potential to be used for efficient delivery of lutein and further improve its bioavailability.
1. Introduction
Lutein is a hydroxycarotenoid (C40H56O2) present in dark green leafy vegetables.1 It is a natural pigment with a range of potential biological effects on health promotion through an array of putative bioactions, including antioxidation, anticancer, anti-inflammation, absorption of high energy blue light for protecting macula and skin by reducing UV-induced inflammation.2–5 Especially, lutein has strong antioxidant and anticancer activities which are due to its unique molecular structure (hydroxycarotenoid and unsaturated double bonds); therefore, the properties and structural stability of lutein are closely related to its antioxidant and anticancer activity.6 For instance, lutein could significantly induce breast cancer cell apoptosis under hypoxia in a dose-dependent manner. Hypoxia-induced production of ROS was also decreased by lutein.7 Lutein could inhibit sarcoma cell proliferation and tumor growth, and lutein–DOX combinatorial therapy significantly decreased the proliferation of S180 cells in vitro.8 In addition, natural carotenoids, α-carotene, β-carotene, lycopene and lutein could inhibit colonic aberrant crypt foci formation in rats, particularly lutein and lycopene in small doses may potentially prevent colon carcinogenesis.9Despite its numerous biological benefits, lutein is an unstable molecule of conjugated carbon–carbon double-bonds with a low absorption rate after administration and poor bioavailability due to its insolubility in the aqueous media. To enhance solubility, stability and bioavailability of lutein, extensive strategies have been developed for its encapsulation by suitable carriers, including nanoparticles, nanoencapsulation and nanoliposome.10,11 For example, lutein entrapped in chitosan/PLGA nanoparticles displayed a higher bioavailability than unmodified lutein in mice.12 The solubility of lutein could be improved by chitosan (CS)/poly-glutamic acid (PGA) nanoencapsulation.13
Nanoliposomes, spherical bilayer vesicles from dispersion of polar lipids in aqueous solvents, have been widely studied for many advantages. The technology of nanoliposomes can offer a novel approach for enhancing bioavailability of fat-soluble and unstable nutrients.14 They enhance bioactive agent performance by improving the solubility and bioavailability, in vitro and in vivo stability, as well as preventing the unwanted interactions with other molecules.15 In order to improve absorption and bioavailability of lutein, nanoliposomes are also one of the most extensively investigated delivery systems for encapsulation for protecting and controlling release of lutein.16,17
However, the practical application of nanoliposomes is limited by their low biological activity, due to the oxidative decomposition during long-term storage, digestion under low pH and enzymatic condition, and the weak release and absorption in the intestinal condition.18 Surface-decorated liposomes using functional cationic polymers to improve the stability and absorption of loaded active ingredients have been developed.19,20 Poly-L-lysine (PLL), a cationic polypeptide, has been shown to enhance the cellular uptake and bioactivity of drugs by increasing the permeability of various compounds as a modifier for active ingredients and drug nano-carriers.21
Microencapsules, based on decoration of poly-L-lysine (PLL) on the nanoparticles and liposomal surface, are an effective way for reducing the damage of liposomal membrane and the leakage of encapsulated compounds to improve encapsulation efficiency and bioactivity.22,23 Importantly, liposome surface modification with PLL significantly increased delivery of the marker coumarin-6, which had high biocompatibility and low toxicity in cells of the retina segment.24 PLL decoration could effectively penetrate and overcome biological barriers to cellular and tissue uptake and enhance the absorption effects in intestine.25,26
Hence, the present study focused primarily on the preparation of nanoliposomes and decoration with PLL for encapsulation of lutein. The physicochemical characteristics of the samples and their structural property were determined using dynamic light scattering (DLS), transmission electron microscopy (TEM) and fourier transform infrared (FTIR) spectroscopy. In addition, the encapsulation efficiency (EE), in vitro stability and release rate were studied. The antioxidant (DPPH, ABTS assay) and anticancer activity of free lutein, lutein nanoliposomes and PLL-decorated nanoliposomes were also investigated.
2. Materials and methods
2.1. Materials
Lutein (98%) and lecithin were purchased from Shanghai Yuanye Biotechnology Co., Ltd. (Shanghai, China) and Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China), respectively. Poly-L-lysine, cholesterol (MKBF1936 V), 1,1-diphenyl-2-picrylhydrazyl radical 2,2-diphenyl-1-(2,4,6-trinitrophenyl)hydrazyl (DPPH) and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) were from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). Ethanol, Tween 80, methanol, and other reagent chemicals were all of analytical grade.2.2. Preparation of PLL decorated nanoliposomes
Lutein nanoliposomes (LUT-NLP) were produced by an ethanol injection method, as described in a previous study.27 Lutein, cholesterol, Tween 80, and lecithin were mixed at a mass ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10. The mixture (1.0 mg mL−1 lutein, the lecithin concentration was kept at 10 mg mL−1) was well dissolved in absolute ethanol and quickly injected into the same volume of phosphate-buffered saline (PBS, pH 6.0, 0.05 M). It was stirred vigorously and kept for half an hour at 50 °C. The liposomal system was transferred to a rotary evaporator and incubated in a 50 °C water bath. The LUT-NLP (Fig. 1A) were obtained when ethanol was removed under a reduced pressure during the hydration process.
10. The mixture (1.0 mg mL−1 lutein, the lecithin concentration was kept at 10 mg mL−1) was well dissolved in absolute ethanol and quickly injected into the same volume of phosphate-buffered saline (PBS, pH 6.0, 0.05 M). It was stirred vigorously and kept for half an hour at 50 °C. The liposomal system was transferred to a rotary evaporator and incubated in a 50 °C water bath. The LUT-NLP (Fig. 1A) were obtained when ethanol was removed under a reduced pressure during the hydration process.
Surface decoration of prepared LUT-NLP was accomplished by mixing LUT-NLP suspension with an equal volume of the PLL (Fig. 1B) solution. An equal volume of PLL dispersion in PBS was added into the LUT-NLP suspension, the final concentration of PLL was adjusted from 0.04 to 0.08 mg mL−1. Then, the liposomal system was incubated at 50 °C for 120 min with subsequent cooling at room temperature to produce PLL–LUT-NLP (Fig. 1C). The final samples were filled into vials and stored at the same conditions as bare liposomes.28
2.3. Encapsulation efficiency of lutein
The encapsulation efficiency of lutein in the nanoliposomes was calculated by the difference between total amount of lutein used to prepare the loaded systems and the remaining amount of free lutein in aqueous medium.29 Free lutein was extracted to determine its amount as follows: 1.0 mL of nanoliposomes and 3 mL of petroleum ether were mixed by vortexing vigorously for 5 min at ambient temperature. The mixed sample was centrifuged at 2000 rpm for 5 min and the supernatant was collected in a tube. The operation procedure was repeated thrice.The supernatant was diluted to 10 mL with petroleum ether. Amount of free lutein was assayed for lutein content by HPLC at 445 nm (Agilent Technologies, Germany; column: YMC-C30) as described in our previous study.30 A calibration curve was made with solutions of lutein at concentrations from 5 to 100 μg mL−1. Each sample was assayed in triplicate. The entrapment efficiency (EE%) was calculated by the following formula:
| EE (%) = [1 − (Wfree/Wtotal)] × 100 | (1) |
2.4. Particle size and zeta potential analysis
Liposomal dispersions (1.0 mL) were suspended in 100 mL of phosphate buffer and taken into polystyrene cuvettes. Both z-average diameter (Dz) and zeta (ζ) potential were measured using a Nano-ZS90 particle size analyzer (Malvern Instruments Ltd., Malvern, UK).31 Each sample was analyzed thrice.2.5. Transmission electron microscopy (TEM)
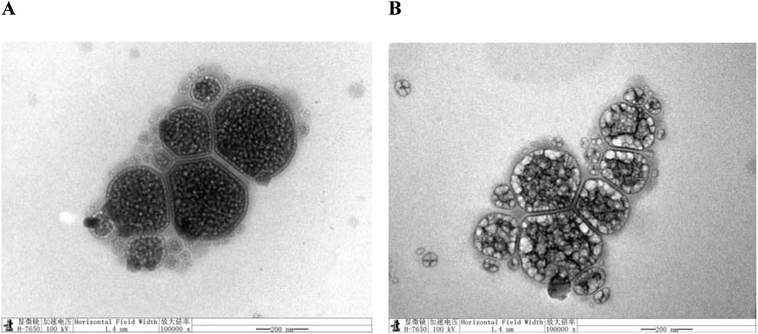
Morphological observation of nanoliposomes was performed by TEM (H-7000, Hitachi, Japan). TEM was performed to visualize the morphology of LUT-NLP and PLL–LUT-NLP. A small droplet of nanoliposomal suspension was deposited on a copper grid and allowed to air dry for 5 min. The grid was then negatively stained with 2% phosphotungstic acid for 1 min. Subsequently, the excess liquid was removed with a filter paper. After air drying at room temperature, the sample was observed under the TEM.322.6. FT-IR assay
An appropriate amount of KBr was dried under an infrared lamp, mixed with the freeze-dried LUT-NLP, PLL and PLL–LUT-NLP, and ground into a plate. The plate was scanned by an infrared spectrometer (Spectrum 100, Perkin Elmer, USA) from 4000 to 550 cm−1. The infrared spectra of PLL–LUT-NLP were recorded and analyzed in comparison with LUT-NLP. Infrared spectra of PLL and PLL–LUT-NLP were compared to further verify whether PLL was decorated in LUT-NLP.332.7. Stability evaluation of lutein in nanoliposome during in vitro digestion
In vitro digestion of LUT-NLP and PLL–LUT-NLP was carried out in simulated gastric fluid (SGF) and simulated intestinal fluid (SIF) separately according to the method of Davidov-Pardo et al. with a slight modification.34 By taking LUT as a control group, 5 mL of LUT, LUT-NLP and PLL–LUT-NLP solution was mixed with 25 mL SGF or SIF. The final concentration of LUT present in the suspension was 200 μg mL−1. The suspensions were incubated in a shaking water bath (100 rpm) at 37 °C and sampled after 0, 2, 4, 6, 8, 10 and 12 h, 1.0 mL suspensions were sonicated in the presence of 3 mL ethanol, the mixed sample was centrifuged at 2000 rpm for 5 min and the supernatant was collected. The supernatant was subsequently assayed for residual LUT content using HPLC assay as described above.SGF and SIF were prepared according to Frenzel et al. with a slight modification.35 SGF was produced by mixing 25 mL of distilled water with a pH value set to 2.0 with hydrochloric acid and 1.0 mL of 0.4% pepsin solution (800–2000 U mg−1 of protein). SIF contained sodium hydroxide (1.81 g L−1), potassium dihydrogen phosphate (8.09 g L−1), pancreatin (4.76 g L−1), and bile salts (5.16 g L−1).
Before in vitro digestion of LUT-NLP and PLL–LUT-NLP, the SGF and SIF were incubated at 37 °C for preheating in the water bath. HPLC assay of LUT before and after incubation in SGF and SIF was determined to further evaluate the detailed protective effect of nanoliposome decorated with PLL on the degradation rate of LUT. The degradation rate was calculated by the following equation.
| D (%) = [(C0 − Ct)/C0] × 100 | (2) |
2.8. In vitro release of lutein in nanoliposome
In vitro release of lutein from LUT-NLP and PLL–LUT-NLP was carried out in SGF and SIF separately according to the previous study, with a slight modification.36 10 mL of each nanoliposomal solution was taken in a dialysis bag with a cut off size of 10 kDa. The tube was immersed in 100 mL SGF and SIF respectively, thermostated at 37.0 ± 0.5 °C. The fluid outside the dialysis bag was stirred at 120 rpm and sampled after 0, 2, 4, 6, 8, 10, 12 and 20 h, respectively. Samples (1.0 mL) were taken to determine the lutein content at a certain time by HPLC method mentioned previously. In addition, equal volume of fresh dissolution medium was added into the tube to maintain the constant volume. The mean calculated values were obtained with 3 replicates. The release rate was calculated by the following equation.| Q (%) = (Mn/M) × 100 | (3) |
2.9. Antioxidant activities
| DPPH scavenging activity (%) = [(blank absorbance − sample absorbance)/blank absorbance] × 100 | (4) |
| ABTS scavenging activity (%) = [(Ablank − Asample)/Ablank] × 100 | (5) |
2.10. Cell culture
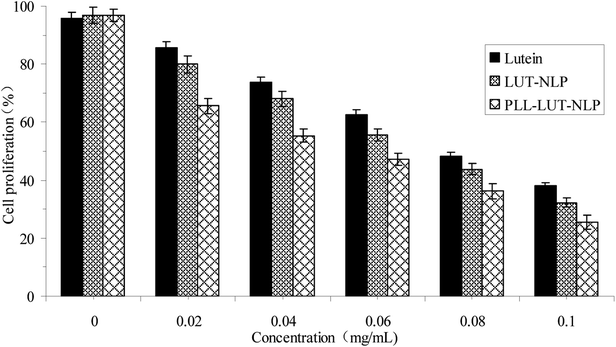
The Caco-2 cells (CBCAS, Shanghai, China) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Hyclone Laboratories, Inc., USA) and supplemented with 10% fatal bovine serum (FBS) (Gibco BRL Co., Ltd., USA), penicillin (100 kU L−1), and streptomycin (100 g L−1) at 37 °C in a 5% CO2 atmosphere in a humidified incubator. After reaching 70–80% confluence, the cells were sub-cultured and maintained with medium changes every 1–2 days.2.11. Cell proliferation
Cell proliferation was tested by the MTT assay.39 The Caco-2 cells were prepared and dispersed in 96-well cell culture plates at a cellular density of 1.0 × 105 cells per well. After incubating with different concentrations of lutein, LUT-NLP and PLL–LUT-NLP for 24 h, 0.020 mL of MTT solution (2 mg mL−1) in PBS was added to each well and incubated at 37 °C for 4 h, the supernatant was removed and replaced with 200 μL of DMSO. The absorbance of the MTT formazan was determined at 570 nm in an Multimode Plate Reader (Perkin Elmer EnSpire™, Perkin Elmer, Inc.). The relative cell viability was expressed as a percentage of the control that was not treated with free lutein or LUT-NLP and PLL–LUT-NLP.| Cell proliferation (%) = (ODsample/ODcontrol) × 100% | (6) |
2.12. Statistical analysis
All measurements were replicated thrice. The experimental results were statistically tested for significance (p ≤ 0.05) for analysis of variance using SPSS (statistical product and service solutions) software. All data were expressed as the mean ± standard deviation (SD).3. Results and discussion
3.1. Preparation and characterization of nanoliposomes
| Formulation | PLL (%) | EE (%) | z-Average diameter (nm) | Polydispersity index (PDI) | Zeta potential (mV) |
|---|---|---|---|---|---|
| NLP | 0 | — | 436.2 ± 7.34 | 0.707 ± 0.056 | −40.9 ± 1.28 |
| LUT-NLP | 0 | 90.20 ± 4.85 | 258.3 ± 7.45 | 0.451 ± 0.018 | −38.6 ± 1.06 |
| PLL–LUT-NLP 1 | 0.04 | 91.81 ± 5.21 | 264.8 ± 9.83 | 0.347 ± 0.027 | −34.3 ± 0.91 |
| PLL–LUT-NLP 2 | 0.06 | 92.49 ± 4.26 | 270.2 ± 8.04 | 0.355 ± 0.021 | −33.8 ± 0.88 |
| PLL–LUT-NLP 3 | 0.08 | 92.93 ± 5.39 | 367.1 ± 7.94 | 0.400 ± 0.036 | −27.9 ± 0.68 |
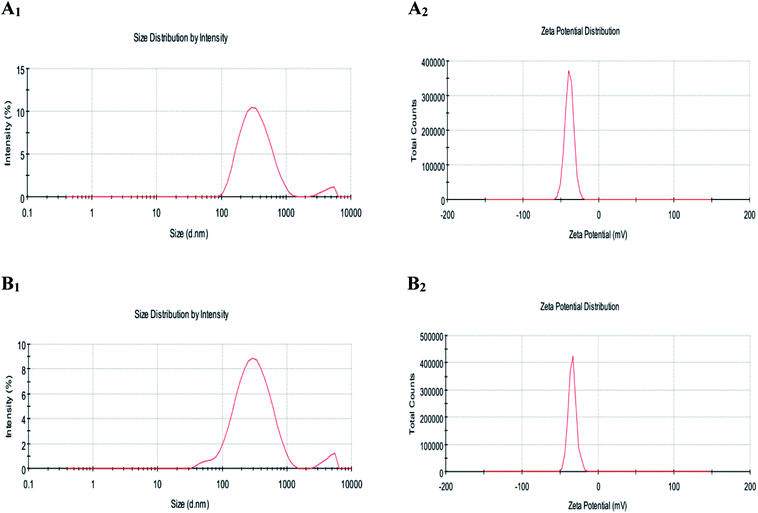 | ||
| Fig. 2 Figure showing the average particle size (A1 and B1) and zeta potential (A2 and B2) of LUT-NLP (A), PLL–LUT-NLP (B) at the PLL concentration of 0.06% (w/v) PLL. | ||
Zeta potential data showed that undecorated liposomes loading lutein possessed an lower average negative charge of −38.6 ± 1.06 mV (Fig. 2A2, Table 1). Small negative charge was attributed to the presence of phosphatidic acid in lecithin.42 After decorating with PLL at a concentration of 0.08 mg mL−1, there was a higher increase of LUT-NLP in zeta potential from −38.6 ± 1.06 mV to −27.9 ± 0.68 mV (Fig. 2B2, Table 1). The increased zeta potential of PLL–LUT-NLP was ascribed to the amino group of PLL and the increase in charge suggested that the electrostatic interaction of positively charged polymer chains with the negative regions of the liposome surface was a major contribution of energy in the formation of the charged PLL. With a more PLL addition, the potential values increased continuously by an enhanced density of positive charge.
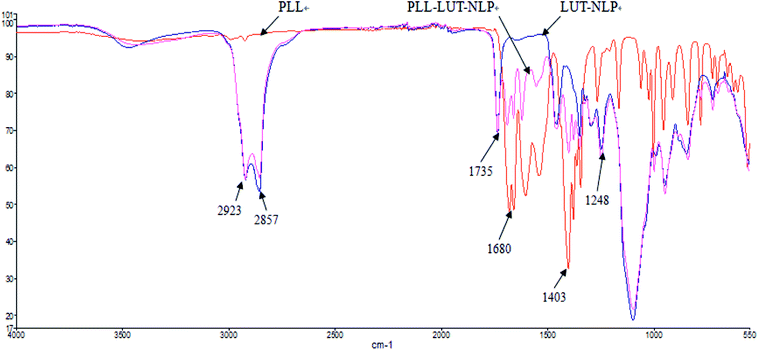
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (at 1735 cm−1) stretching bands and PO4 (at 1248 cm−1) antisymmetric stretching bands.45
O (at 1735 cm−1) stretching bands and PO4 (at 1248 cm−1) antisymmetric stretching bands.45
When PLL was incorporated in nanoliposome, at the wavenumbers of 2923 cm−1 and 2857 cm−1 (vibration absorption of CH2), 1735 cm−1 (symmetrical stretching vibration absorption of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 1248 cm−1 (PO4 antisymmetric stretching bands) region, the location of peaks and form of PLL–LUT-NLP and LUT-NLP nanoliposome were similar.
O) and 1248 cm−1 (PO4 antisymmetric stretching bands) region, the location of peaks and form of PLL–LUT-NLP and LUT-NLP nanoliposome were similar.
PLL–LUT-NLP exhibited peaks at 1680–1403 cm−1 region (stretching vibration absorption of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). They were characteristic peaks of PLL and indicated an existence of PLL in PLL–LUT-NLP. FT-IR spectra of the decorated nanoliposome confirmed that LUT-NLP had been successfully modified by PLL.
N). They were characteristic peaks of PLL and indicated an existence of PLL in PLL–LUT-NLP. FT-IR spectra of the decorated nanoliposome confirmed that LUT-NLP had been successfully modified by PLL.
3.2. In vitro digestion stability
Degradation rates of free lutein, LUT-NLP and PLL–LUT-NLP in SGF and in SIF were estimated respectively to investigate the digestion stability in vitro. The remaining content of lutein was used to evaluate degradation rates of lutein in nanoliposome by taking lutein as a control group. Fig. 5 shows that a rapid degradation of lutein occurred when incubated in SGF and SIF after 12 h. The free lutein degradation rates rose to 46.15 ± 1.743 and 37.20 ± 0.607 after 12 h, respectively.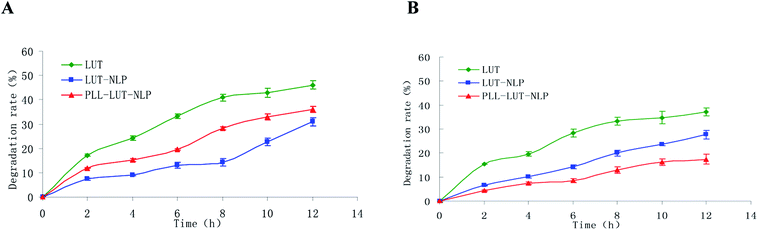 | ||
| Fig. 5 Lutein degradation rate of free lutein, LUT-NLP, PLL–LUT-NLP during digestion in SGF (A) and in SIF (B) for 12 h. | ||
Degradation of lutein was significantly reduced by encapsulating in NLP. The degradative lutein in LUT-NLP after incubating in SGF and SIF for 12 h were 30.95 ± 0.321% and 27.67 ± 0.80%, respectively.
After decoration with PLL, lutein degradation rate dropped to 17.47 ± 1.059% after 12 h incubation in SIF (Fig. 5B). But in SGF (Fig. 5A), it was 36.12 ± 0.613%, still higher than that in LUT-NLP. The results evidenced that hydrochloric acid in gastric juice and pepsase could weaken the interaction of PLL with the surface of LUT-NLP, and with the increased release of lutein from nanoliposome, the lutein degradation rates elevated slightly in SGF. When PLL aggregation was absorbed on the membrane surface, it could protect nanoliposome and thus reduce lutein degradation. The result verified that LUT-NLP could decrease the degradation of lutein in SGF and in SIF, with and without PLL decoration, and increase the stability of lutein. LUT-NLP decorated with PLL could not increase the stability of lutein in SGF. However, PLL–LUT-NLP decreased the degradation rates of lutein in SIF (Fig. 5B). Therefore, it can be concluded that the stability of lutein in SGF and SIF is improved by encapsulation in nanoliposome. Furthermore, the lutein protective capability of PLL–LUT-NLP was much higher than that of nanoliposome in absence of PLL.
3.3. In vitro release characteristics of LUT from nanoliposome
The differences in lutein release between LUT-NLP and PLL–LUT-NLP were performed for 20 h (longer than the intestinal digestion time) to verify the release characteristics. Sustained release of PLL–LUT-NLP was further evaluated by taking LUT-NLP as control groups. Fig. 6 showed that LUT in nanoliposomes was released faster in SIF than in SGF. After 20 h incubation in SGF, LUT release rates for two of the samples were 43.28 ± 2.16% and 51.26 ± 3.33%, respectively (Fig. 6A). However, after digestion in SIF, LUT release rate of LUT-NLP reached to 53.79 ± 1.56% and that of PLL–LUT-NLP to 70.32 ± 1.42% (Fig. 6B). These facts indicate that PLL–LUT-NLP had a good sustainable release profile. LUT release rate in SGF and in SIF were higher than that of undecorated nanoliposomes.46 Moreover, PLL–LUT-NLP released much more encapsulated lutein in SIF than in SGF and LUT release rate of PLL–LUT-NLP was much higher than LUT-NLP at 0–20 h in SIF. LUT-NLP released a similar proportion of lutein under both conditions.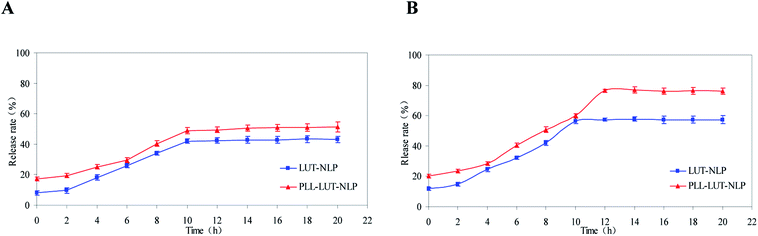 | ||
| Fig. 6 Lutein release rate of LUT-NLP, PLL–LUT-NLP during digestion in SGF (A) and in SIF (B) for 20 h. | ||
This experiment has verified that PLL–LUT-NLP has a strong sustainable capacity for LUT release. It is a suitable delivery system for LUT. The possible reasons are that (1) the membrane permeability of PLL–LUT-NLP was increased by PLL decoration, while leakage of LUT from PLL–LUT-NLP was caused by reassembly of the membrane; and (2) the hydrotropic PLL molecules were adsorbed on the surface of the NLP and LUT entrapped near the surface. As the dissolution rate of the PLL near the surface is high, the amount of LUT released will be high as well.
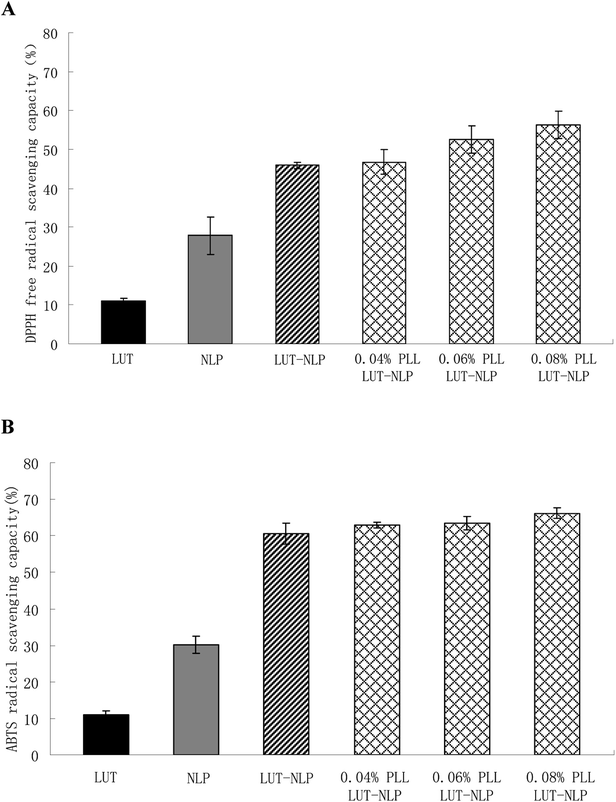
3.4. Determination of antioxidant activity
4. Conclusions
Our studies offer information about the physico-chemical properties of lutein nanoliposomes (LUT-NLP) and polypeptide-decorated lutein nanoliposomes (PLL–LUT-NLP). The increase in entrapment efficiency, size and zeta potential, as well as changes in morphology of PLL–LUT-NLP reflected several changes in the surface properties of nanoliposomes due to electrostatic attraction interactions. FTIR analysis showed that there were effective incorporated interactions between PLL and bilayer membrane of nanoliposomes. In addition, the performance of LUT was improved by NLP encapsulation during in vitro digestion. PLL–LUT-NLP decreased the degradation rates of lutein in SGF and in SIF, which showed a better stability in SIF than in SGF. Decoration of PLL on the surface of LUT-NLP efficiently promotes lutein release in vitro. Moreover, PLL-decorated nanoliposomes possessed the highest antioxidant activity compared with nanoliposomes and lutein, indicating that PLL-decorated liposomes protected lutein from oxidation and degradation in the external environment. The anticancer activity results showed that PLL-decorated nanoliposomes could inhibit proliferation of the Caco-2 cells in a dose dependent manner.These results suggest that PLL decorated nanoliposome system is a novel and effective delivery system for improving the physico-chemical properties (i.e. stability and release property) and bioactivity (i.e. antioxidant and anti-proliferative) of lutein and other hydrophobic functional compounds, which may be quite interesting for food and pharmaceutical industry. Further investigations should be performed to focus on the cell uptake and absorption mechanism of PLL-decorated lutein nanoliposome on molecular level.
Conflicts of interest
The authors declare that this article content has no conflict of interest.Acknowledgements
This research was financially supported by Jiangsu Planned Projects for Postdoctoral Research Funds (No. 1601028A) and by the University Nursing Program for Young Scholars with Creative Talents in Heilong jiang Province (No. UNPYSCT-2016091).References
- A. Kamil, D. E. Smith, J. B. Blumberg, C. Astete, C. Sabliov and C. Y. Oliver Chen, Food Chem., 2016, 192, 915–923 CrossRef PubMed.
- A. Sujak, J. Gabrielska, W. Grudziński, R. Borc, P. Mazurek and W. I. Gruszecki, Arch. Biochem. Biophys., 1999, 371, 301–307 CrossRef PubMed.
- E. S. M. Abdel-Aal and I. Rabalski, Am. J. Biomed. Sci., 2013, 5, 109–125 CrossRef.
- N. I. Krinsky, J. T. Landrum and R. A. Bone, Annu. Rev. Nutr., 2003, 23, 171–201 CrossRef PubMed.
- D. C. Musch, JAMA Ophthalmol., 2014, 132, 139–141 CrossRef PubMed.
- I. D. Nwachukwu, C. C. Udenigwe and R. E. Aluko, Trends Food Sci. Technol., 2016, 49, 74–84 CrossRef.
- Y. Li, Y. Zhang, X. Liu, M. Wang, P. Wang, J. Yang and S. Zhang, Int. J. Oncol., 2018, 52, 2119–2129 Search PubMed.
- R. L. Luan, P. C. Wang, M. X. Yan and J. Chen, Eur. Rev. Med. Pharmacol. Sci., 2018, 22, 1514–1520 Search PubMed.
- T. Narisawa, Y. Fukaura, M. Hasebe, M. Ito, R. Aizawa, M. Murakoshi, S. Uemurab, F. Khachik and H. Nishino, Cancer Lett., 1996, 107, 137–142 CrossRef PubMed.
- R. Arunkumar, K. V. H. Prashanth and V. Baskaran, Food Chem., 2013, 141, 327–337 CrossRef PubMed.
- S. Takagi, T. Yamagami, K. Takeda and T. Takagi, Agric. Biol. Chem., 1987, 51, 1567–1572 Search PubMed.
- A. Murugeshu, C. Astete, C. Leonardi, T. Morgan and C. M. Sabliov, Nanomedicine, 2011, 6, 1513–1528 CrossRef PubMed.
- D. Y. Hong, J. S. Lee and H. G. Lee, Int. J. Biol. Macromol., 2015, 85, 9–15 CrossRef PubMed.
- M. I. Adamczak, E. Hagesaether, G. Smistad and M. Hiorth, Int. J. Pharm., 2016, 498, 225–233 CrossRef PubMed.
- M. R. Mozafari, Methods Mol. Biol., 2010, 605, 29–50 CrossRef PubMed.
- C. Tan, B. Feng, X. Zhang, W. Xia and S. Xia, Food Hydrocolloids, 2016, 52, 774–784 CrossRef.
- A. Junghans, H. Sies and W. Stahl, Arch. Biochem. Biophys., 2001, 391, 160–164 CrossRef PubMed.
- W. Liu, M. Tian, Y. Kong, J. Lu, N. Li and J. Han, LWT–Food Sci. Technol., 2016, 75, 608–615 CrossRef.
- M. Murata, K. Nakano, K. Tahara, Y. Tozuka and H. Takeuchi, Eur. J. Pharm. Biopharm., 2012, 80, 340–346 CrossRef PubMed.
- A. Makhlof, S. Fujimoto, Y. Tozuka and H. Takeuchi, Eur. J. Pharm. Biopharm., 2011, 77, 216–224 CrossRef PubMed.
- K. Tahara, S. Furukawa, H. Yamamoto and Y. Kawashima, Int. J. Pharm., 2010, 392, 311–313 CrossRef PubMed.
- D. Volodkin, V. Ball, P. Schaaf, J. C. Voegel and H. Mohwald, BBA, Biochim. Biophys. Acta, Biomembr., 2007, 1768, 280–290 CrossRef PubMed.
- D. Volodkin, H. Mohwald, J. C. Voegel and V. Ball, J. Controlled Release, 2007, 117, 111–120 CrossRef PubMed.
- H. Sasaki, K. Karasawa, K. Hironaka, K. Tahara, Y. Tozuka and H. Takeuchi, Eur. J. Pharm. Biopharm., 2012, 83, 364–369 CrossRef PubMed.
- S. H. Kim, J. H. Jeong, K. W. Chun and T. G. Park, Langmuir, 2005, 21, 8852–8857 CrossRef PubMed.
- C. Lu, X. Li, W. Q. Xia, S. H. Lu, H. J. Luo, D. Y. Ye, Y. M. Zang and D. J. Liu, Acta Biomater., 2017, 49, 434–443 CrossRef PubMed.
- G. H. Shin, S. K. Chung, J. T. Kim, H. J. Joung and H. J. Park, J. Agric. Food Chem., 2013, 61, 11119–11126 CrossRef PubMed.
- C. Sebaaly, H. Greige-Gerges, S. Stainmesse, H. Fessi and C. Charcosset, Food Biosci., 2016, 15, 1–10 CrossRef.
- C. Tan, S. Xia, J. Xue, J. Xie, B. Feng and X. Zhang, J. Agric. Food Chem., 2013, 61, 8175–8184 CrossRef PubMed.
- D. J. Li, J. F. Song and C. Q. Liu, Int. J. Food Prop., 2015, 18, 178–185 CrossRef.
- F. S. Juveriya, I. Fathima, V. Abhishek and F. Khanum, Food Chem., 2016, 197, 562–570 CrossRef PubMed.
- C. Tan, J. Xue, X. Lou, X. W. Lou, S. Abbas, Y. Guan, B. Feng, X. M. Zhang and S. Q. Xia, Food Funct., 2014, 5, 1232–1240 RSC.
- L. Q. Zou, W. Liu, W. L. Liu, R. H. Liang, T. Li, C. M. Liu, Y. L. Cao, J. Niu and Z. Liu, J. Agric. Food Chem., 2014, 62, 934–941 CrossRef PubMed.
- G. Davidov-Pardo, C. E. Gumus and D. J. Mcclements, Food Chem., 2016, 196, 821–827 CrossRef PubMed.
- M. Frenzel and A. Steffen-Heins, Food Chem., 2015, 173, 1090–1099 CrossRef PubMed.
- M. Gibis, C. Ruedt and J. Weiss, Food Res. Int., 2016, 88, 105–113 CrossRef PubMed.
- K. Papoutsis, P. Pristijono, J. B. Golding, C. E. Stathopoulos, M. C. Bowyer, C. J. Scarlett and Q. V. Vuong, J. Food Process. Preserv., 2017, 41, 1–8 Search PubMed.
- L. Ramezanzade, S. F. Hosseini and M. Nikkhah, Food Chem., 2017, 234, 220–229 CrossRef PubMed.
- A. Sousa, P. Araújo, J. Azevedo, L. Cruz, I. Fernandes, N. Mateus and V. de Freitas, Food Chem., 2016, 192, 142–148 CrossRef PubMed.
- M. Reza Mozafari, C. Johnson, S. Hatziantoniou and C. Demetzos, J. Liposome Res., 2008, 18, 309–327 CrossRef PubMed.
- H. K. Park, J. K. Yoon and K. Kim, Langmuir., 2006, 22, 1626–1629 CrossRef PubMed.
- S. J. Choi, E. A. Decker, L. Henson, L. M. Popplewell, H. Xiao and D. J. Mcclements, Food Res. Int., 2011, 44, 3006–3012 CrossRef.
- Q. Lu, P. M. Lu, J. H. Piao, X. L. Xu, J. Chen, L. Zhu and J. G. Jiang, LWT–Food Sci. Technol., 2014, 57, 686–695 CrossRef.
- X. Wang, F. Wei, A. Liu, L. Wang, J. C. Wang, L. Ren, W. M. Liu, Q. Tu, L. Li and J. Y. Wang, Biomaterials, 2012, 33, 3719–3732 CrossRef PubMed.
- L. Q. Zou, S. F. Peng, W. Liu, L. Gan, W. L. Liu, R. H. Liang, C. M. Liu, J. Niu, Y. L. Cao, Z. Liu and X. Chen, Food Res. Int., 2014, 64, 492–499 CrossRef PubMed.
- T. Chuacharoen and C. M. Sabliov, Colloids Surf., A, 2016, 503, 11–18 CrossRef.
| This journal is © The Royal Society of Chemistry 2018 |