Open Access Article
Open Access ArticleLipid- and gut microbiota-modulating effects of graphene oxide nanoparticles in high-fat diet-induced hyperlipidemic mice†
Juan Li a,
Shengmei Yanga,
Jiaqi Yuc,
Rongli Cuia,
Ru Liua,
Runhong Leia,
Yanan Chang
a,
Shengmei Yanga,
Jiaqi Yuc,
Rongli Cuia,
Ru Liua,
Runhong Leia,
Yanan Chang a,
Huan Genga,
Yanxia Qina,
Weihong Gu
a,
Huan Genga,
Yanxia Qina,
Weihong Gu a,
Shibo Xiaa,
Kui Chena,
Jianglong Konga,
Guogang Chen*b,
Chongming Wu*c and
Gengmei Xing*a
a,
Shibo Xiaa,
Kui Chena,
Jianglong Konga,
Guogang Chen*b,
Chongming Wu*c and
Gengmei Xing*a
aCAS Key Laboratory for Biomedical Effects of Nanomaterial & Nanosafety, Institute of High Energy Physics, Chinese Academy of Science (CAS), Beijing 100049, China. E-mail: xinggm@ihep.ac.cn
bCollege of Food Science, Shihezi University, Shihezi 832000, China. E-mail: cgg611@shzu.edu.cn
cPharmacology and Toxicology Research Center, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100193, China. E-mail: cmwu@implad.ac.cn
First published on 6th September 2018
Abstract
Graphene oxide (GO) suspensions can act as a good dispersant and drug delivery system for effective dispersion and drug sustained release. In this study, we investigated the impact of GO on blood/liver lipids and gut microbiota structure in high-fat diet (HFD)-induced hyperlipidemic mice. Oral administration of GO for 28 days remarkably decreased the lipid levels in blood and liver. GO did not decrease the total number of gut bacteria but increased the relative abundance of short-chain fatty acid (SCFA)-producing bacteria such as Clostridium clusters IV and Allobaculum spp. GO also enhanced the copying of bacterial butyryl coenzyme A transferase (BcoA), a key butyrate-producing gene. Although further pharmacological studies are still needed, these results provided an interesting hint that GO may exert beneficial effects on the host's metabolism via selective modulation of SCFA-producing gut microbes.
1 Introduction
Graphene oxide (GO) is the most widely used water-soluble derivative of graphene in biological and medical fields such as DNA detection,1 cell imaging2 and drug loading/delivery3 due to their good biocompatibility and low cytotoxicity.4 Previous studies have shown that GO exhibited adequate antimicrobial effects in vitro.5–11 GO was reported to significantly suppress the reproduction of bacterial and fungal pathogens at a concentration of 500 μg mL−1.6 The antibacterial action of GO can be achieved via directly contacting microbes with the extremely sharp edges of the nanowalls.5 It has been reported that the bactericidal activity of GO is size-dependent.12 A recent study has shown that GO is easily biodegraded via bacterial respiration at rapid time scales.13 Accordingly, GO sheets could also act as biocompatible sites for adsorption of bacteria but the bacterially-reduced GO sheets potently inhibit bacteria proliferation on their surfaces.14 Up to now, the antimicrobial properties of GO have been successfully used to stainless steel,5 cotton fabric,15 polymer films,16–20 and water treatment membranes.6,21–27 However, whether GO can modulate microbial community in vivo still remains unknown.More than 100 trillion bacteria reside in our digestive tracts which were 10 times the number of human body cells. The gut microbiota is believed to involve in the energy storage and metabolism, playing key roles in maintenance of human health and mediating physiological and pharmacological actions of foreign matters including nanomaterials.28–30 Previously, we have demonstrated that the fullerenol nanoparticles could change the overall structure of the gut microbiota and exert antihyperlipidemic effects in vivo.31 GO could be easily obtained through a chemical oxidation and exfoliation route upon natural graphite and is rich in oxidative groups such as epoxy, hydroxyl, carbonyl, and carboxyl groups.32,33 The atomic 2D graphene layer is considered as a basic building block for graphitic carbons of other dimensionalities; it can either be wrapped up into 0D fullerenes or be rolled into 1D nanotubes.32,34,35 For 2D graphene platelets, their planar surface makes them much easier to be densely packed in a face-to-face manner on the basis of stronger van der Waal's interactions than 0D fullerenes and 1D nanotubes, as well as the contribution originated from the π-conjugated domains.32,36 Therefore, it is interesting to see what will happen when GO meets gut microbes after oral administration.
Hyperlipidemia, a disorder of lipid metabolism, is becoming an epidemic throughout the world, which dramatically increases the incidence of cardiovascular diseases37 and nonalcoholic fatty liver disease.38 Recent studies have demonstrated that hyperlipidemia is closely related to the dysregulation of the gut microbiota.39 Some antihyperlipidemic agents such as bebeerine and fullerenols have been reported to exert their pharmacological actions through modulating the gut microbiota structure.31,40
In the present study, we investigated the impact of GO on blood and liver lipids as well as on the gut microbiota in high-fat diet (HFD)-induced hyperlipidemic mice. The influence of GO on the relative abundance of SCFA-producing bacteria and key enzyme in SFCA synthesis were particularly focused. Our findings provided insights for the modulating effects of GO on gut microbes in vivo and made a potential utility of graphene-based nanomaterials in the management of dyslipidemia.
2 Materials and methods
2.1 Physicochemical characterization of GO
GO was prepared and purified by a previously reported method in our laboratory.41,42 Dry samples were analyzed on a Nicolet Magna-IR750 FTIR spectrophotometer equipped with a Nic-plan IR microscope. Atomic force microscopy (AFM) images were obtained by using a scanning probe microscope (Bruker, Dimension Edge), operated in tapping mode.41,42 The hydrodynamic sizes of GO were detected by dynamic light scattering (DLS, Bruker, USA).2.2 Animal experiments
All the animal experiments were performed in accordance with the National Institutes of Health regulations for the care and use of animals in research and were approved by the Medical Ethics Committee of Peking Union Medical College. All efforts were made to minimize animal suffering.Sixteen male specific pathogen-free (SPF) C57BL/6 mice (8 week-old) mice were obtained from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Animals were kept in a humidity-controlled room on a 12 h light–dark cycle with food and water available ad libitum for 1 week. Afterward, the mice were divided randomly into 2 groups (8 animals per group) and fed with high-fat diet (HFD) which contained 60% fat, 14% protein, and 26% carbohydrate and provided a total energy content of 21.0 kJ g−1. The GO groups were given GO (200 mg kg−1 per day, suspended in distilled water) via gavage for 28 day, while the control group (NC) was given an equal volume of distilled water. Fresh feces were collected from each mouse on 0, 3, 7, 14 and 28 day after GO treatment and the body weights were recorded. The fecal samples were snap-frozen in liquid nitrogen and stored at −80 °C for subsequential analysis. At the end of the experiment, animals were fasted overnight then anesthetized in chambers saturated with isoflurane and sacrificed by cardiac puncture. Blood and liver tissue were taken for the measure of serum and liver levels of triglycerides (TG) and total cholesterol (TC) by respective kits (BioSino Co., Ltd, Beijing, China).
2.3 Analysis of gut microbes by realtime PCR
Total bacteria number and the relative abundance of SCFA-producing bacteria (Allobaculum spp., Clostridium cluster IV and XIVa) and key genes involved in butyrate production (butyryl coenzyme A transferase (BcoA)) in the feces were quantified by realtime PCR as previously reported.31 The gene-specific primers designed to target the total and specific bacteria and butyrate-producing genes (BcoA) were provided in Table S1.† Cloned 16S rRNA genes of E. coli were used to construct standard curve for total bacteria copies. The total bacteria were expressed as log10 copies (16 S DNA gene)/g feces. The relative abundance of specific bacteria or functional genes was normalized to the total bacteria.2.4 Histological analysis
After animals were euthanized, liver and gastrointestinal tissues were obtained from each mice at a similar position. The tissue samples were fixed in 4% formaldehyde, embedded in paraffin and cut into slices about 3 μm thick. The sections were stained with hematoxylin and eosin (H&E) and their morphological changes were observed by microscopy.2.5 Statistics
Data are presented as the means ± SEM. SPSS 17.0 software was used for the statistical analysis. The significance of group differences for normally distributed data was assessed by one-way ANOVA followed by Tukey post hoc tests. P < 0.05 was considered statistically significant.3 Results
3.1 Physicochemical properties of GO
AFM analysis revealed that the surface morphology of GO and showed an irregular sheet morphology (Fig. 1a). The FTIR spectra indicated that there are abundant –OH groups on GO nanoparticles (Fig. 1b). The size of GO was between 200–300 nm (Fig. 1a) and the hydrodynamic size of GO was 431 ± 81 nm (Fig. 1c). The hydrodynamic size of GO was much bigger than that estimated by AFM and the distribution of GO NPs were wide.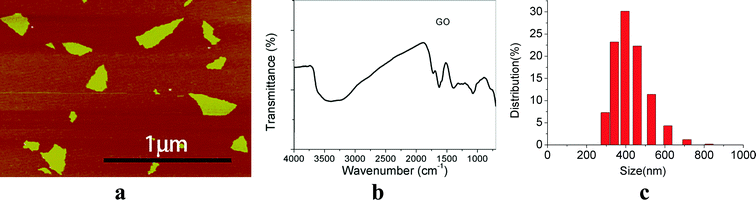 | ||
| Fig. 1 Physicochemical characterization of GO. (a) AFM images; (b) FTIR spectra; (c) GO size distribution. | ||
3.2 GO NPs decrease TC and TG levels in blood and liver
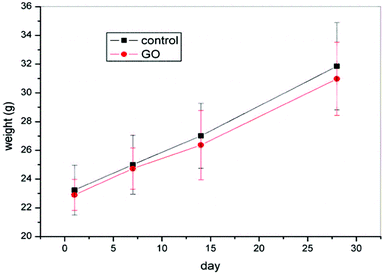
Gavage of GO NPs showed no obvious influence on the body weight of mice (Fig. 2). In contrast, treatment with GO NPs for 28 days significantly decreased the serum levels of total cholesterol (TC) and triglycerides (TG) (Fig. 3). GO NPs showed more pronounced impact on TG level than on TC content. At the same time, the liver contents of TC and TG were dramatically decreased by GO NPs, suggesting marked ameliorating effect of GO on HFD-induced liver steatosis. In accordance, HFD-induced fatty degeneration (ballooning) of hepatocytes was attenuated by GO NPs (Fig. 4). We also checked the impact of GO NPs on the histology of intestine, i.e. duodenal, jejunum, ileum and cecum. The results from hematoxylin–eosin (H&E) staining of mice intestinal tissue showed that gavage of GO NPs for 28 days had no obvious harmful effects on intestinal tissue including duodenal, jejunum, ileum and cecum (Fig. S1†).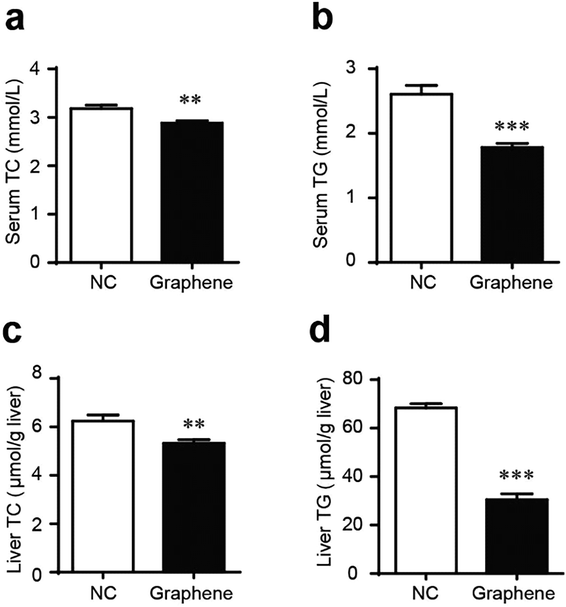 | ||
| Fig. 3 GO NPs decrease blood and liver TC and TG in mice. Blood levels of TC (a) and TG (b), liver levels of TC (c) and TG (d). *P < 0.05, **P < 0.01, ***P < 0.001 vs. NC. | ||
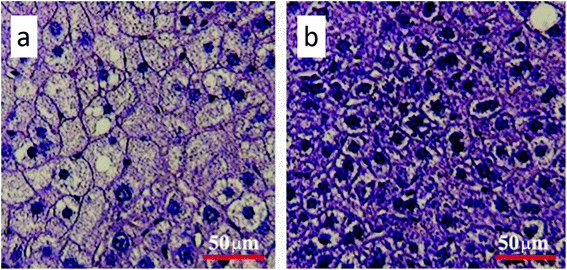 | ||
| Fig. 4 Hematoxylin and eosin staining of the liver (a) for control and (b) for GO-treated mice. The scale bar is 50 μm. | ||
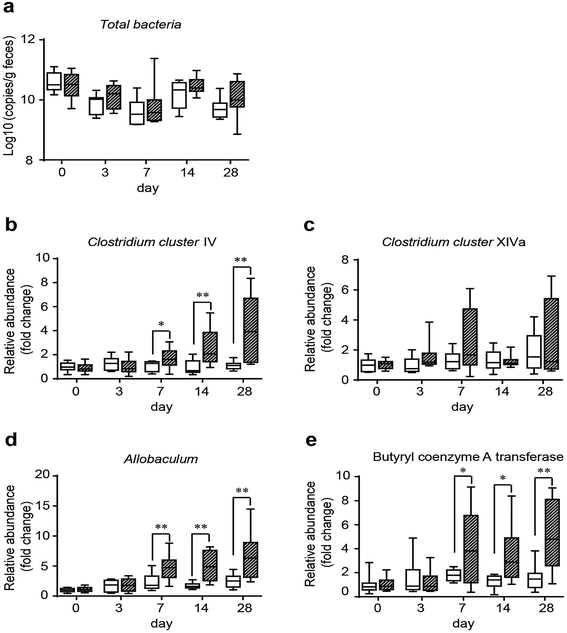
3.3 GO NPs dynamically enhance SCFA-producing bacteria in vivo
To assess the influence of GO on gut microbes and provide a potential way through which GO NPs may exert its beneficial effect on blood and liver lipids, we evaluated the modulating effect of GO NPs on SCFA-producing bacteria, an important gut microbial community playing key roles in improving host metabolism. Clinical studies have shown that SCFAs can substantially ameliorate HFD-induced hyperlipidemia especially on TG level.43 Fecal samples were obtained from each mouse after treatment with GO NPs for 0, 3, 7, 14 and 28 days. Realtime PCR analysis displayed that GO NPs dynamically modulated gut microbial composition. Oral administration of GO NPs did not remarkably decrease the total numbers of gut bacteria (Fig. 5a) but the relative abundance of SCFA-producing genera such as Clostridium clusters IV and Allobaculum spp. were significantly increased after treatment for 7 days (Fig. 5b and d). GO NPs also enhanced the abundance of Clostridium clusters XIVa but did not reach significance (Fig. 5c). As Clostridium clusters IV and XIVa are main members to produce butyrate, the most beneficial SCFA for human health, we assessed the influence of GO NPs on the relative abundance of butyryl coenzyme A transferase (BcoA), a key gene in bacterial butyrate production. The results showed that oral administration of GO NPs significantly enhanced the relative abundance of BcoA after treatment for 7 days (Fig. 5e). These results indicated that GO NPs are adequate to promote SCFA-producing gut microbes and key gene.4 Discussions
Graphene oxidized (GO) is widely used in biological and medical fields. Although the antimicrobial activity of GO has been confirmed in vitro,5–11 the in vivo modulating effect of GO on gut microbiota is remained to be elucidated. In this study, we investigated the impact of GO NPs on gut microbiota and on blood lipids in HFD-induced hyperlipidemic mice.Our results showed that oral administration GO NPs for 28 days had no obvious effects on the body weight and the histomorphology of intestinal tissues including duodenal, jejunum, ileum and cecum, but significantly alleviated HFD-induced hyperlipidemia as manifested by the decrease in serum TC and TG. Accordingly, the HFD-elicited liver steatosis as indicated by elevated liver TC, TG levels and fatty degeneration (ballooning) of hepatocytes were dramatically decreased by GO NPs, suggesting marked ameliorating effect of GO on HFD-induced liver steatosis. To the best of our knowledge, this may be the first observation of the anti-hyperlipidemic and hepatic steatosis-alleviating effects of GO.
As hyperlipidemia is closed related to the dysregulation of gut microbiota, especially to gut microbes-derived SCFA,44 we assessed the in vivo influence of GO NPs on the relative abundance of SCFA-producing bacteria, namely Clostridium clusters IV/XIVa and Allobaculum spp., and on BcoA, a key enzyme in butyrate synthesis. Our results showed that oral administration of GO NPs for 0, 3, 7, 14 and 28 days did not remarkably decrease the total numbers of gut bacteria, but dynamically increased the relative abundance of main SCFA-producing genera such as Clostridium clusters IV and Allobaculum spp. after treatment for 7 days. At the same time, the relative abundance of BcoA, a key gene in bacterial butyrate production, was also significantly increased by GO NPs treatment. These data not only proved that oral administration of GO NPs can modulate gut microbiota in vivo but indicated that enhancing the relative abundance of SCFA-producing gut microbes and key gene may play a key in the hyperlipidemia-alleviating effect of GO NPs.
It's worth mentioning that the chemical structures of GO and fullerenols both are rich in –OH groups in complex status which involved peroxo groups to hydroxy groups and peroxo groups to epoxy groups. Like the hypothesis in the previously published paper,45 it may be considered that there are furan-like and pyran-like structures on stereostructures of the fullerenol or GO as deduced by the different status of hydroxyl groups on the carbon cages of fullerene and the fullerenol or GO were degrade by the gut microbacteria. And then promote SCFAs producing bacteria survival and the TC and TG levels of mice were reduced.45 Otherwise, the underlying mechanism of GO NPs to lipid- and gut microbiota-modulating effects remains to be further investigated and many details was still not clear.
Authors' contributions
Juan Li: wrote the manuscript, analyzed all data, prepared all tables and figures, and conducted most of the experimental work. Shengmei Yang and Yanan Chang: assisted in conducting animal experiments and the manuscript revision. Rongli Cui and Ru Liu: characterized the GO nanoparticles. Runhong Lei: assisted in conducting animal experiments and the manuscript revision. Huan Geng: prepared and observed the HE tissue slides. Yanxia Qin, Weihong Gu, Shibo Xia, Kui Chen and Jianglong Kong: assisted in conducting animal experiments. Jiaqi Yu and Chongming Wu: analyzed realtime PCR and gut microbiota. Gengmei Xing, Chongming Wu and Guogang Chen: designed experiments, contributed to the writing of manuscript and supported data interpretation. All authors read and approved the final manuscript.Ethics approval and consent to participate
All the animal experiments were performed in accordance with the National Institutes of Health regulations for the care and use of animals in research and were approved by the Medical Ethics Committee of Peking Union Medical College (No. YZS201709004).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported financially by the National Basic Research Program of China (973 Program) (2015CB930104), National Natural Science Foundation of China (31571028, 51571185) and the Beijing Natural Science Foundation (2182089).References
- X. Liu, R. Aizen, R. Freeman, O. Yehezkeli and I. Willner, ACS Nano, 2012, 6, 3553–3563 CrossRef PubMed.
- L. Qinghai, W. Li, W. Jinyong, P. Fei, L. Da, C. Rongli, N. Yang, Q. Xiaojun, L. Yu, S. Hao, Y. Juan and L. Yan, Nanoscale, 2012, 4, 7084–7089 RSC.
- B. Hongqian, P. Yongzheng, P. Yuan, N. G. Sahoo, W. Tongfei, L. Lin, L. Jun and G. Leong Huat, Small, 2011, 7, 1569–1578 CrossRef PubMed.
- X. X. Chen, B. Cheng, Y. X. Yang, A. Cao, J. H. Liu, L. J. Du, Y. Liu, Y. Zhao and H. Wang, Small, 2013, 9, 1765–1774 CrossRef PubMed.
- O. Akhavan and E. Ghaderi, ACS Nano, 2010, 4, 5731–5736 CrossRef PubMed.
- J. N. Chen, H. Peng, X. P. Wang, F. Shao, Z. D. Yuan and H. Y. Han, Nanoscale, 2014, 6, 1879–1889 RSC.
- Z. Elham, A. Shahram and S. Omid, Med. Hypotheses, 2015, 85, 1019–1020 CrossRef PubMed.
- D. L. Huang, X. Wang, C. Zhang, G. M. Zeng, Z. W. Peng, J. Zhou, M. Cheng, R. Z. Wang, Z. X. Hu and X. Qin, Chemosphere, 2017, 186, 414–421 CrossRef PubMed.
- Y. S. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. R. Liu, Q. Huang, C. H. Fan, H. P. Fang and R. H. Zhou, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef PubMed.
- F. Wang, S. Ma, Y. Si, L. F. Dong, X. L. Wang, J. Yao, H. L. Chen, Z. J. Yi, W. C. Yao and B. S. Xing, Carbon, 2017, 114, 671–678 CrossRef.
- W. C. Yu, S. H. Zhan, Z. Q. Shen, Q. X. Zhou and D. Yang, Chem. Eng. J., 2017, 313, 836–846 CrossRef.
- F. Perreault, A. F. de Faria, S. Nejati and M. Elimelech, ACS Nano, 2015, 9, 7226–7236 CrossRef PubMed.
- E. C. Salas, Z. Z. Sun, A. Luttge and J. M. Tour, ACS Nano, 2010, 4, 4852–4856 CrossRef PubMed.
- O. Akhavan and E. Ghaderi, Carbon, 2012, 50, 1853–1860 CrossRef.
- L. Karimi, M. E. Yazdanshenas, R. Khajavi, A. Rashidi and M. Mirjalili, Cellulose, 2014, 21, 3813–3827 CrossRef.
- M. Dhanasekar, V. Jenefer, R. B. Nambiar, S. G. Babu, S. P. Selvam, B. Neppolian and S. V. Bhat, Mater. Res. Bull., 2018, 97, 238–243 CrossRef.
- R. Kurapati, M. Vaidyanathan and A. M. Raichur, RSC Adv., 2016, 6, 39852–39860 RSC.
- H. N. Nguyen, E. T. Nadres, B. G. Alamani and D. F. Rodrigues, J. Mater. Chem. B, 2017, 5, 6616–6628 RSC.
- C. M. Santos, M. C. R. Tria, R. A. M. V. Vergara, F. Ahmed, R. C. Advincula and D. F. Rodrigues, Chem. Commun., 2011, 47, 8892–8894 RSC.
- P. C. Wu, H. H. Chen, S. Y. Chen, W. L. Wang, K. L. Yang, C. H. Huang, H. F. Kao, J. C. Chang, C. L. L. Hsu, J. Y. Wang, T. M. Chou and W. S. Kuo, J. Nanobiotechnol., 2018, 16, 349–380 Search PubMed.
- R. Kanchanapally, B. P. V. Nellore, S. S. Sinha, F. Pedraza, S. J. Jones, A. Pramanik, S. R. Chavva, C. Tchounwou, Y. Shi, A. Vangara, D. Sardar and P. C. Ray, RSC Adv., 2015, 5, 18881–18887 RSC.
- H. J. Kim, Y. S. Choi, M. Y. Lim, K. H. Jung, D. G. Kim, J. J. Kim, H. Kang and J. C. Lee, J. Membr. Sci., 2016, 514, 25–34 CrossRef.
- H. Liu, H. Wang and X. Zhang, Adv. Mater., 2015, 27, 249–254 CrossRef PubMed.
- F. Perreault, M. E. Tousley and M. Elimelech, Environ. Sci. Technol. Lett., 2014, 1(1), 71–76 CrossRef.
- E. Shams, H. Yeganeh, H. Naderi-Manesh, R. Gharibi and Z. M. Hassan, J. Mater. Sci.: Mater. Med., 2017, 28, 75–88 CrossRef PubMed.
- J. Wang, Y. M. Wang, Y. T. Zhang, A. Uliana, J. Y. Zhu, J. D. Liu and B. Van der Bruggen, ACS Appl. Mater. Interfaces, 2016, 8, 25508–25519 CrossRef PubMed.
- J. Y. Zhu, J. Wang, J. W. Hou, Y. T. Zhang, J. D. Liu and B. Van der Bruggen, J. Mater. Chem. A, 2017, 5, 6776–6793 RSC.
- J. C. Clemente, L. K. Ursell, L. W. Parfrey and R. Knight, Cell, 2012, 148, 1258–1270 CrossRef PubMed.
- N. R. Shin, J. C. Lee, H. Y. Lee, M. S. Kim, T. W. Whon, M. S. Lee and J. W. Bae, Gut, 2014, 63, 727–735 CrossRef PubMed.
- J. Li, S. Yang, R. Lei, W. Gu, Y. Qin, S. Ma, K. Chen, Y. Chang, X. Bai, S. Xia, C. Wu and G. Xing, Nanoscale, 2018, 10, 7736–7745 RSC.
- J. Li, R. Lei, X. Li, F. Xiong, Q. Zhang, Y. Zhou, S. Yang, Y. Chang, K. Chen, W. Gu, C. Wu and G. Xing, Part. Fibre Toxicol., 2018, 15, 5 CrossRef PubMed.
- Z. Li, Z. Liu, H. Y. Sun and C. Gao, Chem. Rev., 2015, 115, 7046–7117 CrossRef PubMed.
- D. R. Dreyer, S. Park, C. W. Bielawski and R. S. Ruoff, Chem. Soc. Rev., 2010, 39, 228–240 RSC.
- A. K. Geim and K. S. Novoselov, Nat. Mater., 2007, 6, 183–191 CrossRef PubMed.
- Nat. Nanotechnol., 2010, 5, 755 Search PubMed.
- L. J. Cote, F. Kim and J. X. Huang, J. Am. Chem. Soc., 2009, 131, 1043–1049 CrossRef PubMed.
- T. A. Pearson, Med. Care, 2017, 55, 1–3 CrossRef PubMed.
- M. Tomizawa, Y. Kawanabe, F. Shinozaki, S. Sato, Y. Motoyoshi, T. Sugiyama, S. Yamamoto and M. Sueishi, Biomed. Rep., 2014, 2, 633–636 CrossRef PubMed.
- X. Tong, J. Xu, F. Lian, X. Yu, Y. Zhao, L. Xu, M. Zhang, X. Zhao, J. Shen, S. Wu, X. Pang, J. Tian, C. Zhang, Q. Zhou, L. Wang, B. Pang, F. Chen, Z. Peng, J. Wang, Z. Zhen, C. Fang, M. Li, L. Chen and L. Zhao, MBio, 2018, 9, 1–12 CrossRef PubMed.
- Y. Wang, Q. Tong, J. W. Shou, Z. X. Zhao, X. Y. Li, X. F. Zhang, S. R. Ma, C. Y. He, Y. Lin, B. Y. Wen, F. Guo, J. Fu and J. D. Jiang, Theranostics, 2017, 7, 2443–2451 CrossRef PubMed.
- R. L. Cui, J. Li, H. Huang, M. Y. Zhang, X. H. Guo, Y. A. Chang, M. Li, J. Q. Dong, B. Y. Sun and G. M. Xing, Nano Res., 2015, 8, 1259–1268 CrossRef.
- J. Li, R. L. Cui, Y. N. Chang, X. H. Guo, W. H. Gu, H. Huang, K. Chen, G. M. Lin, J. Q. Dong, G. M. Xing and B. Y. Sun, RSC Adv., 2016, 6, 58028–58033 RSC.
- S. Sharma and S. Puri, Altern. Ther. Health Med., 2015, 21, 34–42 Search PubMed.
- L. R. Hoving, S. Katiraei, M. Heijink, A. Pronk, L. van der Wee-Pals, T. Streefland, M. Giera, K. Willems van Dijk and V. van Harmelen, Mol. Nutr. Food Res., 2018, 62, e1700942 CrossRef PubMed.
- J. Li, R. H. Lei, X. Li, F. X. Xiong, Q. Y. Zhang, Y. Zhou, S. M. Yang, Y. A. Chang, K. Chen, W. H. Gu, C. M. Wu and G. M. Xing, Part. Fibre Toxicol., 2018, 15, 5, DOI:10.1186/s12989-018-0241-9.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06058d |
| This journal is © The Royal Society of Chemistry 2018 |