A CNN-based self-supervised learning framework for small-sample near-infrared spectroscopy classification
Rongyue
Zhao
 a,
Wangsen
Li
a,
Wangsen
Li
 a,
Jinchai
Xu
a,
Linjie
Chen
a,
Xuan
Wei
*ab and
Xiangzeng
Kong
a,
Jinchai
Xu
a,
Linjie
Chen
a,
Xuan
Wei
*ab and
Xiangzeng
Kong
 *ab
*ab
aSchool of Future Technology, Fujian Agriculture and Forestry University, Fuzhou 350002, China. E-mail: xuanweixuan@126.com; xzkong@fafu.edu.cn
bCollege of Mechanical and Electrical Engineering, Fujian Agriculture and Forestry University, Fuzhou 350100, China
First published on 13th January 2025
Abstract
Near-infrared (NIR) spectroscopy, with its advantages of non-destructive analysis, simple operation, and fast detection speed, has been widely applied in various fields. However, the effectiveness of current spectral analysis techniques still relies on complex preprocessing and feature selection of spectral data. While data-driven deep learning can automatically extract features from raw spectral data, it typically requires large amounts of labeled data for training, limiting its application in spectral analysis. To address this issue, we propose a self-supervised learning (SSL) framework based on convolutional neural networks (CNN) to enhance spectral analysis performance with small sample sizes. The method comprises two learning stages: pre-training and fine-tuning. In the pre-training stage, a large amount of pseudo-labeled data is used to learn intrinsic spectral features, followed by fine-tuning with a smaller set of labeled data to complete the final model training. Applied to our own collected dataset of three tea varieties, the proposed model achieved a classification accuracy of 99.12%. Additionally, experiments on three public datasets demonstrated that the SSL model significantly outperforms traditional machine learning methods, achieving accuracies of 97.83%, 98.14%, and 99.89%, respectively. Comparative experiments further confirmed the effectiveness of the pre-training stage, with the highest accuracy improvement, reaching 10.41%. These results highlight the potential of the proposed method for handling small sample spectral data, providing a viable solution for improved spectral analysis.
1. Introduction
Near-infrared (NIR) spectroscopy is a versatile analytical technique that operates within the 780–2526 nm wavelength range. This range corresponds to the overtone and combination band absorption regions of hydrogen-containing functional groups, such as OH, CH, and NH, which are commonly found in organic molecules. When NIR light interacts with a sample, the chemical bonds of these functional groups absorb specific wavelengths, producing spectra that carry valuable information about the molecular structure.1 Given that different samples contain varying types and concentrations of these groups, the resulting spectra reflect unique characteristics such as peak positions, bandwidths, and shapes. These features enable both qualitative and quantitative analysis of various samples, making NIR spectroscopy a powerful, non-destructive, and cost-effective tool for a wide range of applications.2 However, the broad spectral range, low signal intensity, and overlapping spectral peaks often make direct interpretation challenging, necessitating advanced computational techniques for accurate data analysis.3In recent years, the rapid development of artificial intelligence technology, especially the widespread application of machine learning methods, has greatly promoted the development of chemometrics.4 By building models that map spectral data to sample characteristics, machine learning enables fast and accurate predictions of new sample components or properties. Traditionally, this process involves three key stages: spectral preprocessing, feature selection, and model construction. However, incorrect preprocessing often leads to distortion of the original signal, and feature selection, which often leads to partial loss of information, can reduce the accuracy of spectral data analyses.5 Therefore, there is a need for an integrated data-driven analysis method that works directly on spectral data and is capable of extracting data critical features and eliminating human interference.6 Recent studies have demonstrated that deep learning models, which can process raw, high-dimensional data end-to-end, excel in capturing complex patterns without requiring extensive preprocessing.7 These models, composed of multiple layers of nonlinear transformations, are capable of learning abstract representations from raw spectral data, thereby enhancing the accuracy of predictions. Despite their potential, training deep learning models often requires a large amount of labeled data, and labeling NIR spectral data typically relies on costly and time-consuming offline chemical analysis methods. This severely limits the scale of available labeled data and, consequently, constrains the optimization of model performance. Effectively training deep learning models with limited labeled data remains a key issue in the field of spectral data analysis.
The predominant methodologies employed for training models utilizing label information include supervised learning, unsupervised learning, semi-supervised learning, and transfer learning, each with its own strengths and limitations. A concise overview of these four learning paradigms is presented in Table 1. Supervised learning builds direct relationships between spectral data and their labels but is prone to overfitting, especially when labeled data is limited. Conversely, unsupervised learning does not depend on external labels, yet it is characterized by poor interpretability and presents challenges when applied directly to NIR spectroscopy analysis, which is often utilized for data dimensionality reduction prior to the implementation of supervised learning techniques.8 Semi-supervised learning combines the strengths of both supervised and unsupervised methods, but its performance can be limited by assumptions about data distribution. Transfer learning allows knowledge transfer from one domain to another, helping mitigate the challenge of limited labeled data, but differences between domains can lead to negative transfer effects. In summary, when dealing with limited sample sizes, each of these methods faces various degrees of challenge, complicating the effective training of models for spectral analysis.
| Methods | Definition | Labels requirements | Merits | Limitation | Models |
|---|---|---|---|---|---|
| Supervised learning | Training with labeled data to learn the mapping between inputs and outputs | High | High accuracy, effectively leverages existing knowledge | High cost of labeling data, limited generalization to new data, easily overfitted | VGGNet9 |
| Unsupervised learning | Training with unlabeled data to discover hidden structures and patterns | None | Label data not required | Poor interpretability, difficult to evaluate | PCA10 |
| HCA11 | |||||
| Semi-supervised learning | Training with a small amount of labeled data and a large amount of unlabeled data | Low | Improves model performance with less labeled data | Increased complexity of model training, reliance on data assumptions | S3VM12 |
| Transfer learning | Applying knowledge learned from one task to a different but related task | Medium | Uses knowledge from related tasks to boost performance on new, similar tasks | Requires correlation between source and target tasks, effectiveness depends on this correlation | CNN13,14 |
| MDEA15 |
SSL represents a novel paradigm within the field of machine learning, providing an innovative approach to the challenge posed by the scarcity of labeled samples.16 By designing learning tasks, SSL facilitates the automatic extraction of meaningful information from unlabeled data, a process referred to as pre-training, which establishes the initial parameters of the model. Following this, the model can be fine-tuned using a limited set of labeled data to optimize its performance for specific tasks. A significant advantage of SSL lies in its capacity to capture the intrinsic representations of data during the pre-training phase, which is essential for enhancing model performance. Firstly, since SSL does not depend on manual labeling, it can effectively leverage large volumes of unlabeled data for training, thereby facilitating the development of deeper and more complex neural network architectures that typically exhibit superior feature extraction and generalization capabilities. Secondly, by fine-tuning the model for diverse downstream tasks, SSL allows for rapid adaptation to the specific requirements of these tasks while preserving the model's generality. This not only improves network performance but also leads to substantial savings in computational resources and time.17 Given the high dimensionality and limited availability of labeled NIR spectral data, SSL is particularly well-suited to addressing these challenges, offering significant potential for improving spectral analysis.
Therefore, this study proposes a CNN-based SSL learning model designed to tackle the NIR spectral classification challenge. By leveraging the pre-training and fine-tuning mechanism inherent to self-supervised learning, the model offers a practical solution for small sample modeling. We first collected near-infrared spectral data from three different tea tree varieties and used these data to train and test the model, demonstrating its initial effectiveness. To thoroughly evaluate the model's generalization ability and robustness, we further introduced three publicly available NIR spectral datasets for training and testing. In addition, we emphasize the significant classification performance advantages of the proposed model by comparing it against traditional machine learning methods. Finally, a series of comparison and ablation experiments were conducted to optimize the model's framework parameters and validate its overall rationality.
2. Related work and background
2.1. NIR spectroscopy analysis based on deep learning
Deep learning represents an advanced machine learning paradigm that evolves from traditional shallow learning frameworks. The fundamental principle of deep learning is the abstract representation of data through a multi-layered neural network architecture, where the outputs of lower layers serve as inputs for higher layers, thereby establishing a bottom-up learning trajectory. This methodology facilitates the development of a mapping relationship between inputs and outputs, enabling the prediction, classification, or recognition of samples through multi-layer feature representation.6 In the domain of spectral analysis, numerous experiments and studies have demonstrated that deep learning techniques surpass conventional machine learning methods in terms of spectral analysis accuracy.18 Notably, CNN, characterized by their distinctive convolutional operations of and significantly reduce the number of parameters requiring optimization, thereby enhancing training efficiency and demonstrating robust feature extraction capabilities.13 Consequently, CNN and their variants have been extensively investigated and applied across various fields.In ref. 19, a CNN-based method for NIR data analysis was proposed, and classification experiments were conducted on four types of drugs from nine brands, achieving an accuracy of 97.3%. The results indicate that this method demonstrates superior recognition capabilities compared to traditional machine learning methods. Its advantages include the elimination of complex feature engineering, and the ability to handle high-dimensional data, thereby improving classification accuracy and reliability. In ref. 9, the authors utilized the VGGNet19, a variants of CNN, for classifying pine tree NIR spectroscopy data, achieving a classification accuracy of 98.41%. The performance is significantly higher than that of support vector machines (SVM) at 71.26% and backpropagation neural networks (BPNN) at 76.19%. Compared to traditional machine learning methods, deeper network architectures can effectively reduce noise, extract fine spectral features, address the issue of peak overlap in spectra, and enhance model generalization capabilities by incorporating dropout layers and batch normalization. Although deep models show great potential in spectral modeling, they typically require a substantial number of parameters usually rely on a large amount of labeled spectral data for training. In ref. 20, the authors argue that deep learning-based spectral modeling is most effective when the sample size exceeds 2000. In practice, obtaining labeled data is both time-consuming and costly, and acquiring data volumes in the thousands is even more challenging. To address the challenge of training deep learning models with strong generalization capabilities using limited labeled data, we designed an SSL solution, which provides an effective resolution to this problem.
2.2. Self-supervised learning
Self-supervised learning, a machine learning paradigm, leverages the intrinsic properties of data as supervisory signals, eliminating the dependence on large amounts of labeled data. This approach consists of two stages: model pre-training and model fine-tuning. In the pre-training stage, a series of prediction or classification tasks are constructed based on the intrinsic attributes of the data, such as partial image reconstruction, context prediction, and transformation recognition of signals.21 These tasks are designed to encourage the model to discover and extract deep patterns from the data while solving them. Subsequently, in the fine-tuning stage, a small amount of labeled data is used to make targeted adjustments to the pre-trained model, optimizing its performance for specific application scenarios and tasks. The applicability and efficiency of SSL have been fully validated across multiple fields. In computer vision,22 self-supervised learning models like SimCLR (a simple framework for contrastive learning of visual representations) have shown excellent performance in image classification tasks. In natural language processing,23,24 self-supervised models such as GPT (general pre-training) and BERT (bidirectional encoder representation from transformers) have been successfully applied to complex tasks like machine translation and language modeling. In time series,25 researchers have constructed positive and negative samples in the pretext task based on electroencephalogram (EEG) signals to achieve SSL, extracting characteristics of sleep EEG signals to facilitate emotion recognition tasks.Inspired by the successful application of SSL in various domains, we explored its potential for NIR spectral data processing. NIR spectral data presents challenges such as small sample sizes and significant complexity. SSL, with its unique advantages, emerges as a crucial solution to these challenges.
3. Materials and methods
3.1. Self-supervised learning framework
The proposed self-supervised framework comprises two distinct learning stages. In the initial stage, a transformation recognition network is developed to learn representations of NIR spectral data by identifying the transformations to the raw spectra. This stage emphasizes the acquisition of robust and generalized features from unlabeled NIR spectral data, with the task defined as the pretext task. In the subsequent stage, a spectral classification network is trained utilizing NIR spectral data that has been annotated with human-generated class labels to classify the types of samples; this task is referred to as the downstream task. The features derived from the NIR spectral data at the shared layer in the first stage are transferred to the qualitative analysis network in the second stage, thereby enhancing efficiency and performance through the reuse of network parameters. Fig. 1 illustrates the overall framework of our SSL approach, which consists of a two-part network: (A) the NIR spectroscopy transformation recognition network and (B) the NIR spectroscopy classification network. A detailed explanation of these two components will now be provided.The core of the network architecture consists of shared layers and task-specific layers, with the input layer serving as the data entry point. The shared layer plays a crucial role in extracting effective and generalizable features from the spectral data and transferring these learned features to the NIR spectral classification network. The specific structure and parameters of the shared layer are illustrated in Fig. 2(a). As illustrated, the shared layer is composed of three convolutional blocks. Each block begins with two consecutive one-dimensional convolutional layers for feature extraction, followed by batch normalization and ReLU activation functions to enhance the model's stability and nonlinear processing capabilities. A max pooling layer, with a pool size of 2, is then employed to diminish feature dimensions while retaining essential information. The convolutional kernel size is consistently set to 3, and the number of filters in the convolutional layers progressively increases from 32 to 64 and then to 128. This design is intended to extract universal features from the raw data that are effective across various transformation tasks. Following processing in the shared layers, the features are forwarded to the task-specific layers for further processing and classification. The task-specific layers consist of seven independent branches, each tasked with a binary classification task objective, determining whether the input spectrum corresponds to the raw data or one of the six predefined transformations. Each branch comprises three fully connected layers, with a ReLU activation function applied between them to facilitate nonlinear transformation, and 50% dropout strategy implemented after each fully connected layer to mitigate the risk of overfitting.
 | ||
| Fig. 2 Structure and parameters of (a) the shared layer in our proposed self-supervised learning framework and (b) the classifier in the NIR spectral classification network. | ||
3.1.1.1 Six NIR spectroscopy transformation. The primary objective of the NIR Spectral transformation recognition network is to extract features relevant to spectral classification through the execution of spectral transformation recognition tasks. Specifically, to effectively identify the various types of transformations in NIR spectroscopy, the recognition network must discern invariances within the transformed spectra. In this study, we implemented six distinct transformations on the raw spectral data. Following this, we labeled both the raw data and the transformed spectral data with their corresponding identifiers, with raw data labeled as 0 and the transformed data labeled from 1 to 6, corresponding to the six transformations. This combined dataset was utilized as the training dataset for the NIR spectroscopy transformation recognition network. A detailed description of the six spectral transformations is provided below.27
(1) Add noise: random noise with Gaussian distribution is added to the raw NIR spectroscopy.
(2) Offsets: offset refers to the movement of NIR spectroscopy along the longitudinal axis, where the offset is determined by a factor of 0.1 times the standard deviation of the training set. This means that the vertical axis value of each data point may increase the standard deviation of the training set by up to 10%.
(3) Multiplication: the amplitude of the raw NIR spectroscopy after multiplication is stretched or compressed, and the multiplication is performed at 1 ± 10.1 times the standard deviation of the training set.
(4) Horizontal flip: the raw NIR spectroscopy is flipped according to the horizontal line.
(5) Vertical flip: the raw NIR spectroscopy is flipped according to the vertical line.
(6) Permutation: the raw NIR spectroscopy is evenly divided into m segments and shuffled, randomly rearranging them.
3.1.1.2 Loss function. The entire NIR spectral transformation recognition network is performing a multi-task learning process, and each sub-task is a binary classification task. We use BCEWithLogitsLoss, a loss function commonly used for binary classification problems. This function combines sigmoid activation with binary cross-entropy loss, providing an efficient and stable gradient computation method for handling binary classification tasks. In the NIR spectroscopy transformation recognition task Tp, we define the input as a tuple table (Xj,Yj), where Xj is the NIR spectroscopy after jth transformation, Yj is the label generated after the jth transformation, and j ∈ (0,N) is the total number of NIR spectroscopy transformations, which is equal L × 7, where L is the total number of the samples. The loss function of each label is defined by the following equation, where the prediction probability of the jth task is defined as ψj:
Lj = [Yj![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ψj + (1 − Yj)ln(1 − ψj)] ψj + (1 − Yj)ln(1 − ψj)] | (1) |
In our multi-task learning approach, the model is trained by minimizing the total loss Ltotal, which is the weighted average of each individual loss Lj, from the NIR spectroscopy transformation recognition network. Here, αj is loss coefficient of the jth task:
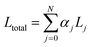 | (2) |
3.1.2.1 Loss function. In the NIR spectroscopy qualitative analysis task Tf, we define the input as a tuple table (Xi,yi), where Xi is the raw NIR spectral data, yi is the corresponding category label, and i ∈ (0,M), M is the total number of NIR spectroscopy input data. Accurate qualitative analysis of NIR spectroscopy is achieved by minimizing the cross-entropy loss function, which is defined by the following equation:
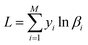 | (3) |
3.2. Datasets
3.2.1.1 Preparation of tea fresh leaf samples. In this experiment, three tea tree varieties cultivated in Anxi County, Quanzhou City, Fujian Province, China were selected: Benshan variety, Huangdan variety, and Tieguanyin variety. Fresh tea leaves were harvested on November 18, 2023, yielding a total of 855 leaves (285 leaves from each variety). The fresh tea leaves from different varieties are shown in Fig. 3, and different varieties of leaves cannot be effectively distinguished by the naked eye. After harvesting, the fresh tea leaves were placed in black sealed bags for subsequent spectral data collection.
 | ||
| Fig. 3 Fresh tea leaves from different tea plant varieties: (a) Benshan variety, (b) Huangdan variety, and (c) Tieguanying variety. (d) Handheld NIRez-G1 spectrometer. | ||
3.2.1.2 NIR spectral data acquisition. The instrument used in this study was a handheld long-wave near-infrared spectrometer (NIRez-G1, Isuzu Optical Corp., Taiwan, China), as shown in Fig. 3(d). The spectral wavelength range of the instrument is 950–1650 nm, and the spectral resolution is 10 nm. Prior to data collection, the spectrometer was preheated for 30 minutes to ensure the reliability of the initial spectral measurements of the fresh tea leaves. Before collecting the NIR spectral reflectance of tea leaves, spectral data of the dark current and reference board are first obtained for the calibration of the raw spectra prior to data processing. Initially, spectra from three different positions on a standard whiteboard (HIS-CT-250 × 280) are collected using the NIR spectrometer, and the average is recorded as W. Subsequently, a black opaque plastic disc is used to cover the sampling port, and spectra are collected three times, with the average recorded as B. To obtain as much spectral information as possible from the tea leaf surfaces, spectra are collected from three different locations on the leaf during the experiment, and the average is used as the raw spectral data for that sample. The reflectance is then calculated for the tea leaves using eqn (4) for calibration.
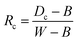 | (4) |
Spectral data were collected from 855 spectral data from fresh tea leaves of three varieties (3 varieties × 285 samples) following the method described above. In order to reduce noise and individual variability in the data and to improve representativeness, in each variety, three spectral data were averaged to generate new spectral data, resulting in a total of 285 average spectral reflectance data, which were varietally labelled for subsequent experiments.
| Datasets | Samples | Variables | Classes | Wavelength range | Available |
|---|---|---|---|---|---|
| Tea | 285 | 228 | 3 | 950–1650 nm | Data will be made available on request |
| Mango | 186 | 1557 | 4 | 1000–2500 nm | https://www.hub.uu2025.xyz/10.1016/j.dib.2019.104789 |
| Tablet | 310 | 404 | 4 | 700–2500 nm | http://www.models.life.ku.dk/Tablets |
| Coal | 5016 | 1499 | 12 | 1000–2500 nm | https://doi.org/10.5281/zenodo.11137126 |
3.2.2.1 Mango28. This dataset comprises NIR spectral data obtained from 186 whole mango samples representing four distinct varieties: Kweni, Cengkir, Palmer, and Kent. The spectral data, illustrated in Fig. 4(b), were collected using a benchtop Fourier transform infrared spectrometer (Thermo Nicolet Antaris II TM) over a wavelength range of 1000 to 2500 nm.
3.2.2.2 Tablet29. This publicly available dataset comprises 310 NIR spectroscopy samples of pharmaceutical drugs, which are categorized into four groups according to the concentration of active substances. The dataset encompasses a NIR spectroscopy wavelength range of 700 to 2500 nm. A representation of the NIR spectroscopy data is illustrated in Fig. 4(c).
3.2.2.3 Coal30. This publicly available dataset comprises NIR spectral data for 24 distinct types of coal and coal-measure rocks, with 12 types represented in each category. For each sample type, researchers collected approximately 418 NIR spectroscopy measurements, encompassing various levels of granularity and acquisition conditions. The wavelength range of the dataset extends from 1000 to 2500 nm, with selected spectral graphs illustrated in Fig. 4(d).
3.3. Implementation
To train the NIR spectroscopy transformation recognition network and the spectral classification network, we utilized the Adam optimizer with a learning rate of 0.0001 and a batch size of 32. The NIR spectroscopy transformation recognition network was trained for 100 epochs, while the spectral classification network underwent training for 250 epochs. Additionally, we implemented a 5-fold cross-validation method to assess model performance. Following the random shuffling of the dataset, 80% of the data was designated as the training set for training the model, with the remaining 20% allocated for testing model. This process was repeated five times; however, the data shuffling was conducted only once at the outset and was not repeated in subsequent iterations.The experiments were conducted on a system equipped with 16 GB of RAM and an Intel i7-13650HX CPU (4.9 GHz, 14 cores), paired with an NVIDIA RTX 4060 GPU with 8 GB of memory. The software environment consisted of Windows 11, Python 3.8.0, PyTorch 2.2.1, and CUDA 12.1. Both the proposed self-supervised learning model and the comparison models were trained and tested under this configuration. We will share the construction process and implementation details of the model for researchers in related areas at https://www.github.com/ryzhao0620/SSL-of-NIR-Spectroscopy-classification-based-on-CNN.git.
3.4. Comparison experiment algorithms and evaluation indicators
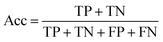 | (5) |
The formula for the F1 score is as follows:
 | (6) |
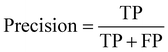 | (7) |
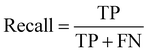 | (8) |
The F1 score represents the weighted harmonic means of precision and recall, considering the performance of both metrics, thus offering a more comprehensive and robust evaluation.
4. Results and discussion
4.1. NIR spectroscopy transformation recognition results
As shown in Table 3, our transformation recognition network achieved exceptionally high accuracy and F1 scores on the diverse datasets of Tea, Mango, Tablet, and Coal, with average accuracy exceeding 99% and F1 scores accuracy exceeding 97%. These results indicate that SSL enables the model to deeply explore the underlying features of spectral data, capturing both local details and global structures, thereby maintaining robust recognition capabilities across various transformations. Moreover, the network exhibited consistent performance and strong generalization across different datasets.| Transformation | Tea | Mango | Tablet | Coal | ||||
|---|---|---|---|---|---|---|---|---|
| Acc | F 1 | Acc | F 1 | Acc | F 1 | Acc | F 1 | |
| Raw | 0.9536 | 0.9324 | 1.0000 | 0.9189 | 1.0000 | 1.0000 | 0.9932 | 0.9899 |
| Adding noise | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Offsets | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Multiplication | 0.9879 | 0.97834 | 0.9432 | 0.9321 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Horizontal flipping | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.9901 |
| Vertical flipping | 1.0000 | 0.9741 | 0.9868 | 1.0000 | 0.9891 | 1.0000 | 0.9768 | 1.0000 |
| Permutation | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.9909 | 1.0000 | 1.0000 | 1.0000 |
| Average | 0.9916 | 0.9835 | 0.9900 | 0.9787 | 0.9971 | 1.0000 | 0.9957 | 0.9971 |
4.2. NIR spectroscopy classification results
Table 4 summarizes the classification accuracy and F1 score of each model across four classification tasks. The results show that in the presence of limited labeled data, SSL models significantly outperform all other traditional machine learning methods, the accuracy of the four classification tasks reached 0.9912, 0.9783, 0.9814 and 0.9989, respectively.| Datasets | Tea | Mango | Tablet | Coal | ||||
|---|---|---|---|---|---|---|---|---|
| Acc | F 1 | Acc | F 1 | Acc | F 1 | Acc | F 1 | |
| RF | 0.8421 | 0.8343 | 0.8872 | 0.8902 | 0.8516 | 0.8509 | 0.6637 | 0.6634 |
| SVM | 0.7614 | 0.7598 | 0.8550 | 0.8548 | 0.9129 | 0.9104 | 0.9334 | 0.9332 |
| kNN | 0.8281 | 0.8293 | 0.8390 | 0.8402 | 0.8548 | 0.8487 | 0.5981 | 0.5965 |
| ELM | 0.9509 | 0.9487 | 0.9622 | 0.9587 | 0.9426 | 0.9393 | 0.9234 | 0.9198 |
| BPNN | 0.8903 | 0.8897 | 0.9248 | 0.9231 | 0.8871 | 0.8823 | 0.9075 | 0.9026 |
| PLS-DA | 0.9018 | 0.9006 | 0.9356 | 0.9312 | 0.9581 | 0.9566 | 0.9181 | 0.9143 |
| Ours | 0.9912 | 0.9832 | 0.9783 | 0.9736 | 0.9814 | 0.9789 | 0.9989 | 0.9978 |
In this experiment, the sample sizes for the Tea, Mango and Tablet datasets were limited to 285, 186 and 310, respectively, which presents a challenge for deep learning models due to the small sample size.38 To assess the performance of the self-supervised model in the context of limited labeled data for coal classification, we conducted comparative experiments. These experiments employed a gradient-increase strategy, which involved gradually augmenting the amount of NIR spectral data from 5% to 50%. Through this methodology, we systematically evaluated the performance disparities between SSL algorithms and traditional machine learning methods across varying data quantities. Table 5 provides a visual representation of the accuracy achieved by various methods for each data quantity. The experimental results demonstrate that, under conditions of limited labeled data (specifically at 5% data quantity), the accuracy of SSL methods significantly surpasses that of the traditional machine learning methods. Furthermore, as the amount of labeled data increases, the accuracy of all algorithms improves; however, SSL methods consistently exhibit the highest accuracy throughout the evaluation.
| Sample size | Ours | RF | SVM | kNN | ELM | BPNN | PSL-DA |
|---|---|---|---|---|---|---|---|
| 50% | 0.9983 | 0.6057 | 0.8498 | 0.5609 | 0.8876 | 0.8744 | 0.8963 |
| 40% | 0.9976 | 0.5873 | 0.8478 | 0.5419 | 0.8648 | 0.8654 | 0.88.4 |
| 30% | 0.9951 | 0.5360 | 0.7967 | 0.5107 | 0.8167 | 0.8572 | 0.8616 |
| 20% | 0.9934 | 0.4570 | 0.7411 | 0.4288 | 0.6607 | 0.8266 | 0.8398 |
| 10% | 0.9916 | 0.3909 | 0.7143 | 0.3434 | 0.6019 | 0.6086 | 0.7114 |
| 5% | 0.9791 | 0.3750 | 0.6652 | 0.2583 | 0.5333 | 0.5587 | 0.6179 |
4.3. Effectiveness of self-supervised per-training
To evaluate the efficacy of self-supervised pre-training, a series of comparative experiments were conducted primarily varied in the self-supervised pre-training phase. Initially, the self-supervised model was pre-trained utilizing an unlabeled dataset, followed by fine-tuning the pre-trained model with labeled data, a process referred to as SSL fine-tuning. In contrast, we conducted an additional set of experiments in which the self-supervised pre-training phase was omitted, and the same architecture was trained from scratch using only the labeled data, a method termed fully supervised learning. It is worth noting that, apart from the use of unlabeled data for pretraining in the SSL fine-tuning group, all other experimental conditions were kept consistent between the SSL fine-tuning group and the fully supervised learning group, including data splitting, model hyperparameters, and classification evaluation metrics. Given the ample sample size of the Coal dataset, we established multiple gradients of data quantity to assess the effectiveness of SSL within the context of classification task involving the coal data. As illustrated in Fig. 5, the experimental results unequivocally demonstrate that SSL exhibits significant superiority across our tea dataset and the three public datasets, particularly in scenarios characterized by small sample sizes, where classification accuracies are significantly surpassed those of the supervised learning models employing the same architecture. On the coal dataset, as the volume of training data increased, the performance of both learning methodologies improved; however, SSL consistently maintained its advantage. This superiority can be attributed to the self-supervised pre-training process, during which the model effectively acquired multi-dimensional and general features by recognizing and differentiating various data transformations. This process positively influenced subsequent classification tasks, thereby thoroughly validating the effectiveness of self-supervised pre-training.4.4. The depth of the self-supervised model
In this section, we assess the quality of learned representations in relation to the depth of the self-supervised model. Representations obtained from each convolutional block were extracted and employed for downstream spectral classification of NIR spectral data, as depicted in Fig. 6(a). To validate the efficacy of our approach with small sample sizes, only 5% of the total Coal data volume was utilized as data for the experiments conducted in this section and in Section 4.5. During the experiments, Representations learned by convolutional blocks 1, 2, and 3 were extracted and applied to spectral classification. As illustrated in Fig. 6(b), the features extracted from convolutional block 3 demonstrated significant performance enhancements compared to those from Blocks 1 and 2. An attempted was made to stack three embedding vectors to generate a new embedding vector for the qualitative analysis task. However, the results indicated that the embedding derived from Convolutional Block 3 consistently produced the best performance. This find suggests that representations obtained from the final convolutional layer demonstrate superior generalization capabilities and improved performance in spectral classification tasks.4.5. The importance of multi-task learning
This section discusses the performance of spectral classification when performing multiple tasks to learn NIR spectroscopy representations. Multi-task networks may have an impact on the overall performance, which might be different from several individual tasks aggregated together. Fig. 7 illustrates a comparative spectral classification derived from learning NIR spectroscopy via multitasking versus a single-task approach. The findings indicate that the multitask network consistently surpasses the performance of the single-task network. This advantage can be attributed to the synergistic effects of various transformation recognition tasks, which enable the self-supervised model to acquire higher-level representations of spectral data more effectively. By adapting to these transformations, the neural network enhances its training performance.27 In particular, the addition of noise and offsets simulates diverse scattering and shift phenomena that may manifest in the spectra, while multiplication operations, along with horizontal and vertical flips, assist the model in capturing overall intensity or amplitude variations. Additionally, permutation operations augment the model's sensitivity to minor changes in wavelength positions. Collectively, these strategies facilitate a more profound understanding and representation of spectral data by the model. Furthermore, it is essential to investigate the impact of SSL of individual tasks on downstream tasks. Given that each transformation task contributes uniquely to the learning of NIR spectral representations, we optimize the overall model training process by assigning weights to the loss functions of different tasks during execution. This approach is conducive to the rapid convergence of the model.5. Conclusion
In this study, we proposed a novel SSL framework based on CNN to address the challenges of NIR spectroscopy classification with limited labeled data. Our method successfully combines the advantages of deep learning and SSL by employing a two-stage training process: pre-training with unlabeled data and fine-tuning with a small amount of labeled data. This approach enables the automatic extraction of intrinsic spectral features, reducing the need for complex preprocessing and feature engineering typically required in traditional machine learning techniques. The experimental results on both our own tea dataset and publicly available NIR spectroscopy datasets demonstrated that the proposed SSL method significantly outperforms traditional machine learning models. Our framework achieved classification accuracies of up to 99.12% on the tea dataset and showed superior generalization abilities on external datasets, with accuracies of 97.83%, 98.14%, and 99.89%, respectively. Moreover, the comparative and ablation experiments confirmed that the pre-training phase of SSL provided substantial performance improvements, particularly for small-sample spectral data. The highest observed accuracy improvement reached 10.41%, further underscoring the effectiveness of the SSL approach.In conclusion, the CNN-based SSL framework developed in this study provides a practical and effective tool for NIR spectroscopy analysis, enabling better performance in scenarios with limited data availability. This work opens new possibilities for integrating SSL into spectral analysis workflows, advancing the field toward more automated and reliable classification models.
Data availability
Data can be provided by the authors upon request.Author contributions
Rongyue Zhao: conceptualization, methodology, software, investigation, writing – original draft. Wangsen Li: software, data curation. Jingchai Xu: software, conceptualization. Linjie Chen: validation Xuan Wei: writing – review & editing. Xiangzeng Kong: supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the anonymous reviewers for their insightful comments and suggestions to significantly improve the quality of this article.References
- K. Martin, Appl. Spectrosc. Rev., 1992, 27, 325–383 CrossRef.
- J. Coates, Encyclopedia of Analytical Chemistry, 2000, vol. 12, pp. 10815–10837 Search PubMed.
- H. Heise and R. Winzen, Near-Infrared Spectroscopy: Principles, Instruments, Applications, 2001, pp. 125–162, DOI:10.1002/9783527612666.ch07.
- P. B. Joshi, Artif. Intell. Rev., 2023, 56, 9089–9114 CrossRef PubMed.
- W. Zhang, L. C. Kasun, Q. J. Wang, Y. Zheng and Z. Lin, Sensors, 2022, 22, 9764 CrossRef PubMed.
- J. Yang, J. Xu, X. Zhang, C. Wu, T. Lin and Y. Ying, Anal. Chim. Acta, 2019, 1081, 6–17 CrossRef PubMed.
- H. Zhou, H. Guan, X. Ma, B. Wei, Y. Zhang and Y. Lu, Microchem. J., 2024, 206, 111542 CrossRef.
- M. Usama, J. Qadir, A. Raza, H. Arif, K.-L. A. Yau, Y. Elkhatib, A. Hussain and A. Al-Fuqaha, IEEE Access, 2019, 7, 65579–65615 Search PubMed.
- Z. Wan, H. Yang, M. Gao, J. Xu, H. Mu, D. Qi and S. Han, IEEE Access, 2023, 11, 62721–62732 Search PubMed.
- M. El Maouardi, K. De Braekeleer, A. Bouklouze and Y. Vander Heyden, Food Control, 2024, 165, 110671 CrossRef.
- A. Gumieniczek, A. Berecka-Rycerz, H. Trębacz, A. Barzycka, E. Leyk and M. Wesolowski, Molecules, 2022, 27, 4283 CrossRef PubMed.
- L. Yang and Q. Sun, Chemometr. Intell. Lab. Syst., 2012, 114, 109–115 CrossRef.
- L. Li, X. Pan, W. Chen, M. Wei, Y. Feng, L. Yin, C. Hu and H. Yang, J. Innovative Opt. Health Sci., 2020, 13, 2050016 CrossRef.
- L. Pan, W. Wu, Z. Hu, H. Li, M. Zhang and J. Zhao, Biosyst. Eng., 2024, 245, 164–176 CrossRef.
- Z. Qiu, S. Zhao, X. Feng and Y. He, Sci. Total Environ., 2020, 740, 140118 CrossRef PubMed.
- X. Liu, F. Zhang, Z. Hou, L. Mian, Z. Wang, J. Zhang and J. Tang, IEEE Trans. Knowl. Data Eng., 2021, 35, 857–876 Search PubMed.
- P. Sarkar and A. Etemad, IEEE Trans. Affect. Comput., 2020, 13, 1541–1554 Search PubMed.
- X. Liu, H. An, W. Cai and X. Shao, TrAC, Trends Anal. Chem., 2024, 172, 117612 CrossRef.
- Z. An-Bing, Y. Hui-Hua, P. Xi-Peng, Y. Li-Hui and F. Yan-Chun, IEEE Access, 2021, 9, 3195–3206 Search PubMed.
- W. Ng, B. Minasny, W. d. S. Mendes and J. A. M. Demattê, Soil, 2020, 6, 565–578 CrossRef.
- K. Zhang, Q. Wen, C. Zhang, R. Cai, M. Jin, Y. Liu, J. Y. Zhang, Y. Liang, G. Pang and D. Song, IEEE Trans. Pattern Anal. Mach. Intell., 2024, 46(10), 6775–6794 Search PubMed.
- T. Chen, S. Kornblith, M. Norouzi and G. Hinton, A simple framework for contrastive learning of visual representations, International conference on machine learning, 2020, pp. 1597–1607 Search PubMed.
- J. Achiam, S. Adler, S. Agarwal, L. Ahmad, I. Akkaya, F. L. Aleman, D. Almeida, J. Altenschmidt, S. Altman and S. Anadkat, arXiv, 2023, preprint, arXiv:2303.08774, DOI: DOI:10.48550/arXiv.2303.08774.
- J. Devlin, M.-W. Chang, K. Lee and K. Toutanova, arXiv, 2018, preprint, arXiv:1810.04805, DOI: DOI:10.1109/tim.2024.3374300/mm1.
- X. Shen, X. Liu, X. Hu, D. Zhang and S. Song, IEEE Trans. Affect. Comput., 2023, 14(3), 2496–2511 Search PubMed.
- Y. Zhang and Q. Yang, Natl. Sci. Rev., 2018, 5, 30–43 CrossRef.
- E. J. Bjerrum, M. Glahder and T. Skov, arXiv, 2017, preprint, arXiv:1710.01927, DOI:10.48550/arXiv.1710.01927.
- A. A. Munawar and D. Wahyuni, Data Brief, 2019, 27, 104789 CrossRef PubMed.
- M. Dyrby, S. B. Engelsen, L. Nørgaard, M. Bruhn and L. Lundsberg-Nielsen, Appl. Spectrosc., 2002, 56, 579–585 CrossRef.
- Y. Lv, S. Wang, E. Yang and S. Ge, Sci. Data, 2024, 11, 628 CrossRef PubMed.
- R. Gao, J. Li, L. Dong, S. Wang, Y. Zhang, L. Zhang, Z. Ye, Z. Zhu, W. Yin and S. Jia, Microchem. J., 2024, 201, 110716 CrossRef.
- M. Y. Mohamed, M. I. Solihin, W. Astuti, C. K. Ang and W. Zailah, J. Phys.:Conf. Ser., 2019, 1367, 012029 CrossRef.
- R. M. Balabin, R. Z. Safieva and E. I. Lomakina, Anal. Chim. Acta, 2010, 671, 27–35 CrossRef PubMed.
- C. Tan, H. Chen and Z. Lin, Microchem. J., 2021, 160, 105691 CrossRef.
- Y. He, X. Li and X. Deng, J. Food Eng., 2007, 79, 1238–1242 CrossRef.
- F. S. Grasel and M. F. Ferrão, Anal. Methods, 2016, 8, 644–649 RSC.
- Z. Dong, J. Wang, P. Sun, W. Ran and Y. Li, J. Food Meas. Char., 2024, 18, 2237–2247 CrossRef.
- X. Li, J. Wu, T. Bai, C. Wu, Y. He, J. Huang, X. Li, Z. Shi and K. Hou, Comput. Electron. Agric., 2024, 223, 109122 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |





