Open Access Article
Open Access ArticleAnalysis and application of volatile metabolic profiles of Escherichia coli: a preliminary GC-IMS-based study†
Yunwei Zheng‡
 a,
Fuxing Li‡a,
Chuwen Zhaoab,
Junqi Zhuab,
Youling Fangab,
Yaping Hang
a,
Fuxing Li‡a,
Chuwen Zhaoab,
Junqi Zhuab,
Youling Fangab,
Yaping Hang *a and
Longhua Hu*a
*a and
Longhua Hu*a
aDepartment of Clinical Laboratory, Jiangxi Province Key Laboratory of Immunology and Inflammation, Jiangxi Provincial Clinical Research Center for Laboratory Medicine, The Second Affiliated Hospital, Jiangxi Medical College, Nanchang University, Minde Road No. 1, Nanchang 330006, Jiangxi, China. E-mail: bingbinghang@163.com; longhuahu@163.com
bSchool of Public Health, Nanchang University, Nanchang, Jiangxi, China
First published on 13th August 2024
Abstract
Nosocomial infections caused by Escherichia coli (E. coli) may pose serious risks to patients, and early identification of pathogenic bacteria and drug sensitivity results can improve patient prognosis. In this study, we clarified the composition and relative content of volatile organic compounds (VOCs) generated by E. coli in tryptic soy broth (TSB) using gas chromatography-ion mobility spectrometry (GC-IMS). We explored whether imipenem (IPM) could be utilized to differentiate between carbapenem-sensitive E. coli (CSEC) and carbapenem-resistant E. coli (CREC). The results revealed that 36 VOCs (alcohols, aldehydes, acids, esters, ketones, pyrazines, heterocyclic compounds, and unknown compounds) were detected using GC-IMS. Besides, the results indicated that changes in the relative content of VOCs as well as changes in the signal intensity of fingerprints were able to assess the growth state of bacteria during bacterial growth and help identify E. coli. Lastly, under selective pressure of IPM, volatile fingerprints of E. coli could be employed as a model to distinguish CSEC from CREC strains.
1. Introduction
Escherichia coli (E. coli) is an important bacterium present in the intestinal microbiota of vertebrates1 and is one of the most common bacteria in the Enterobacteriaceae family. It possesses a wide range of virulence factors, such as toxins, adhesins, iron carriers, and other virulence factors, resulting in a wide range of pathogenic activities.2,3 In most cases, it is a common pathogen that causes urinary tract infections,4 diarrhea,5 bloodstream infections, and other illnesses.6 In clinical practice, empirical antibiotic therapy for various diseases caused by E. coli infections is often an effective therapeutic strategy until the results of drug sensitivity are clarified. However, inappropriate antibiotic application strategies can lead to multidrug-resistant E. coli. Therefore, shortening the reporting time for microbial identification and drug sensitivity testing has become an important challenge for microbiologists.The main resistance mechanism of carbapenem-resistant E. coli (CREC) includes the production of carbapenem-resistant enzymes with a predominance of New Delhi metalloenzyme (NDM).7 Especially in China, blaNDM accounted for 93% and 97.2% of adult and pediatric CREC, respectively.8 Data published by the China Bacterial Drug Resistance Monitoring Network in 2023 demonstrated that the resistance rates of E. coli to imipenem (IPM) and meropenem (MEM) were 1.9% and 2%, respectively, compared with 1.1% and 1.4% in 2005, which were at low levels of prevalence but have shown a slow rising trend. Notably, CREC infections increase patient mortality and prolong hospitalization compared to carbapenem-sensitive E. coli (CSEC) infections9,10 and are especially common in intensive care units (ICU).11
Traditional drug sensitivity tests are mainly based on paper diffusion and dilution (instrumental methods). However, these methods are cumbersome and time-consuming and increase the risk of delayed drug administration to patients. Accordingly, there is an urgent need for a rapid test to early identify CREC and related therapeutic measures in the clinic. Historically, microbiologists have revealed that bacteria have a powerful ability to produce large amounts of volatile substances12–14 and named them microbial volatile organic compounds (mVOCs). For example, Streptomyces can produce up to more than 80 volatile organic compounds (VOCs).15 In addition, using VOCs facilitates early identification of carbapenem-sensitive versus carbapenem-resistant Klebsiella pneumoniae,16 enabling appropriate measures to be taken to improve the prognosis of patients.
Gas chromatography-ion mobility spectrometry (GC-IMS) combines the excellent separation effect of gas chromatography (GC) with the high sensitivity of ion mobility spectrometry (IMS). GC-IMS can accurately analyze VOCs without cumbersome sample pre-treatment, greatly simplifying the analytical process and has already achieved remarkable results in the fields of food science17,18 and environmental monitoring.19 As an emerging detection technology, GC-IMS has been gradually applied in the medical field in recent years,20 especially in the rapid detection of pathogenic bacteria and identification of bloodstream-infected bacteria with certain advantages.21 Currently, few reports are found on the identification of E. coli strains and the determination of drug sensitivity results by GC-IMS. Consequently, the present study aimed to identify and analyze the volatile metabolic profiles produced by CREC and CSEC by GC-IMS and simultaneously rapidly identify and clinically validate the two, which will provide a reference for developing rational antibiotic treatment plans.
2. Materials and methods
2.1 Strain acquisition, identification, and sensitivity testing
The E. coli quality control strain (American Type Culture Collection, ATCC-25922) was purchased from the China General Microbial Strain Collection and Management Center. The rest of the strains were isolated from the Clinical Microbiology Unit, Department of Laboratory Medicine, the Second Affiliated Hospital of Nanchang University. Among them, CSEC and CREC were identified as E. coli by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) using ATCC-25922 as the control. The drug sensitivity results of VITEK® 2 Compact ASTGN16 (biomrieux) or Kirby–Bauer (KB) test suggested that they were sensitive or resistant to carbapenem antibiotics. The minimum inhibitory concentration (MIC) of IPM was determined according to the standards established by the Clinical Laboratory Standards Institute (CLSI). Modified carbapenem inactivation method (mCIM) and EDTA-carbapenem inactivation method (eCIM) were used to verify the carbapenemase type, and the drug resistance genes of the experimental strains were analyzed by next-generation sequencing technology. A total of 20 CSEC and 20 CREC strains isolated from clinical specimens were randomly selected as experimental strains, and all strains were preserved in glycerol broth (15% glycerol) (Solarbio, China) before experiments and stored in a refrigerator at −80 °C.2.2 Culture conditions and sample preparation
The enrichment medium used for experiments was Tryptic Soy Broth (TSB), whose main ingredients were soy peptone extract, peptone, glucose, and phosphate buffer. All experimental strains were inoculated on Columbia blood agar plates the day before and incubated in a 37 °C incubator for 18–22 h. The next day, single colonies of pure culture were picked and prepared into a 0.5 McFarland concentration bacterial suspension with sterile deionized water (ddH2O). The suspension was injected into a culture tube containing TSB to reach a final concentration of 107 colony-forming units (CFU) mL−1. The whole system had a total volume of 6 mL, with the blank group receiving an equal amount of ddH2O as the bacterial suspension. The samples were incubated at 37 °C with shaking at 200 rpm. Finally, about 500 μL of the bacterial culture was taken at different time points and placed in headspace flasks for GC-IMS analysis. To prevent the decomposition of IPM, the IPM solution was prepared 10 min before dosing; consequently, the final concentration of IPM added to the bacterial culture solution became 0.25 mg mL−1.222.3 Detection principle and equipment parameters
GC-IMS is mainly composed of several parts, such as a capillary column, ionization source, migration tube, and Faraday disk. When volatile compounds enter the equipment, they first enter the capillary column for pre-separation and then enter the migration tube as individual components. Sample molecules are ionized to molecules or ions by a tritium source in the ionization zone of the migration tube before entering the migration zone through the periodically opening ion gate. High-purity nitrogen gas enters the migration zone from the drift gas inlet and moves in the opposite direction to the ions, which, on the one hand, can blow some unionized molecules out of the outlet to avoid interference with the detection. On the other hand, it provides resistance to the movement of the ions, imparting their better separation. Finally, the ions reached the Faraday disk in the order of their migration rates from fast to slow. The time required to traverse the drift tube varied from short to long. Upon arrival, the ions were detected, and the peaks were sequentially recorded. This process yielded three-dimensional spectra, including the retention time (Rt), the drift time (Dt), and the signal intensity.The column model used in this study was MXT-WAX, with a length of 15 m, an inner diameter of 0.53 mm, a film thickness of 1 μm, and a migration tube length of 98 mm. Typically, 500 μL of the sample to be tested was placed in each headspace vial before the start of the experiment, closed with a magnetic screw cap and a septum, and then incubated at 60 °C with shaking at 500 rpm for 3 min, followed by the extraction of 1000 μL of headspace vials for 10 min. The gas was analyzed for 10 min. High-purity nitrogen (99.999%) was utilized as the carrier gas. The IMS drift gas flow rate was always maintained at 150 mL min−1, and the carrier gas gradient for the whole process was as follows: 0–1 min: 0–2 mL min−1; 1–3 min: 2–10 mL min−1; 3–10 min: 10–100 mL min−1. Other main parameters were as follows: T1 drift tube temperature was 45 °C, T2 gas chromatography column temperature was 80 °C, T3 inlet temperature was 80 °C, T4 and T5 transfer tube temperature was 80 °C, column temperature was 80 °C, injection needle temperature was 85 °C, and ionization source was tritium radioactive ionization source with an average radiation energy of 5.68 keV.
2.4 Data analysis
The analysis was conducted using the accompanying software VOCal version 0.1.3 in the FlavourSpec® flavor analyzer from G.A.S., Germany. After the samples were analyzed in a positive ionization mode, the signal peaks of the volatile components of the samples were collected, and visual two-dimensional maps were obtained from their color expression. Using the built-in library search NIST database of the GC-IMS instrument and the IMS database as a reference, volatile compounds were identified based on the retention index (RI) and drift time (Dt).23 The volatile compounds were identified using the VOCal software. The fingerprints were plotted using the VOCal plug-in Galerie and the Gallery Plot plug-in for intergroup comparison of fingerprints, Dynamic PCA 1.4.0 for principal component analysis (PCA), and nearest neighbor for similarity analysis. GraphPad Prism 8.3.0 was employed to plot the bacterial growth curves and the dynamic trends of VOCs using the online tools bioinformatics (https://www.bioinformatics.com.cn/), Microsoft Office PowerPoint 2010, and ChiPlot (https://www.chiplot.online/) for data visualization. The Mann–Whitney U test was utilized to compare VOCs between different groups, and VOCs were considered to be different between groups when p < 0.05.3. Results
3.1 Determination of drug sensitivity results of 20 CREC strains and time points for detecting VOCs
The results of 20 CREC strains to eight antibiotics are presented in Table 1. According to the drug sensitivity results, the resistance rates for these strains were as follows: cefepime (100%), piperacillin tazobactam (95%), ciprofloxacin (90%), levofloxacin (90%), and cotrimoxazole (85%). Additionally, all strains exhibited MIC values for IPM greater than 4 μg mL−1, indicating complete resistance to it.| Strain no. | MIC (μg mL−1) | mCIM | eCIM | Carbapenem gene | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ciprofloxacin | Levofloxacin | Aztreonam | Cefepime | Sulfamethoxazole | Tobramycin | Piperacillin tazobactam | Imipenem | ||||
| a Abbreviations: MIC, minimum inhibitory concentration; CREC, carbapenem-resistant Escherichia coli; S, susceptible; I, intermediate; R, resistant; mCIM, modified carbapenem inactivation method; eCIM, EDTA-carbapenem inactivation method. | |||||||||||
| CREC-1 | >2 (R) | 8 (R) | 16 (R) | >32 (R) | ≤2 (S) | ≤2 (S) | >128 (R) | 8 (R) | + | + | blaNDM-1 |
| CREC-2 | >2 (R) | >8 (R) | >16 (R) | >32 (R) | >4 (R) | >8 (R) | >64 (R) | 4 (R) | + | + | blaNDM-1 |
| CREC-3 | >2 (R) | >8 (R) | >16 (R) | 32 (R) | >4 (R) | ≤2 (S) | 64 (I) | >8 (R) | + | + | blaNDM-5 |
| CREC-4 | >2 (R) | 8 (R) | ≤1 (S) | >32 (R) | >4 (R) | >8 (R) | >128 (R) | 8 (R) | + | + | blaNDM-5 |
| CREC-5 | >2 (R) | 8 (R) | ≤1 (S) | >32 (R) | >4 (R) | >8 (R) | >128 (R) | 8 (R) | + | + | blaNDM-5 |
| CREC-6 | >4 (R) | 8 (R) | 16 (R) | >16 (R) | >4 (R) | ≤4 (S) | >128 (R) | >8 (R) | + | + | blaNDM-5 |
| CREC-7 | >4 (R) | >8 (R) | >32 (R) | >16 (R) | >4 (R) | 8 (I) | >128 (R) | 4 (R) | + | + | blaNDM-5 |
| CREC-8 | >4 (R) | >8 (R) | >32 (R) | >16 (R) | >4 (R) | 16 (R) | >128 (R) | 4 (R) | + | + | blaNDM-5 |
| CREC-9 | >4 (R) | >8 (R) | 8 (I) | >16 (R) | >4 (R) | ≤4 (S) | >128 (R) | 8 (R) | + | + | blaNDM-5 |
| CREC-10 | >4 (R) | >8 (R) | >32 (R) | >16 (R) | ≤2 (S) | >16 (R) | >128 (R) | >8 (R) | + | + | blaNDM-5 |
| CREC-11 | >4 (R) | >8 (R) | >32 (R) | >16 (R) | >4 (R) | ≤4 (S) | >128 (R) | 4 (R) | + | + | blaNDM-5 |
| CREC-12 | >4 (R) | >8 (R) | >32 (R) | >16 (R) | >4 (R) | >16 (R) | >128 (R) | >8 (R) | + | + | blaNDM-5 |
| CREC-13 | >4 (R) | >8 (R) | >32 (R) | >16 (R) | >4 (R) | >16 (R) | >128 (R) | 4 (R) | + | + | blaNDM-5 |
| CREC-14 | >4 (R) | >8 (R) | >32 (R) | >16 (R) | >4 (R) | 8 (I) | >128 (R) | >8 (R) | + | + | blaNDM-5 |
| CREC-15 | 0.5 (S) | >8 (R) | ≤4 (S) | >16 (R) | >4 (R) | >16 (R) | >128 (R) | 4 (R) | + | + | blaNDM-5 |
| CREC-16 | >4 (R) | 1 (S) | ≤4 (S) | 16 (R) | >4 (R) | >16 (R) | >128 (R) | 4 (R) | + | + | blaNDM-5 |
| CREC-17 | >4 (R) | 8 (R) | 16 (R) | >16 (R) | >4 (R) | ≤4 (S) | >128 (R) | >8 (R) | + | + | blaNDM-5 |
| CREC-18 | ≤0.25(S) | 8 (R) | 16 (R) | >16 (R) | ≤2 (S) | ≤4 (S) | >128 (R) | >8 (R) | + | + | blaNDM-5 |
| CREC-19 | >4 (R) | ≤0.5 (S) | ≤4 (S) | >16 (R) | >4 (R) | 8 (I) | >128 (R) | >8 (R) | + | + | blaNDM-5 |
| CREC-20 | >4 (R) | >8 (R) | 16 (R) | >16 (R) | >4 (R) | >16 (R) | >128 (R) | >8 (R) | + | + | blaNDM-5 |
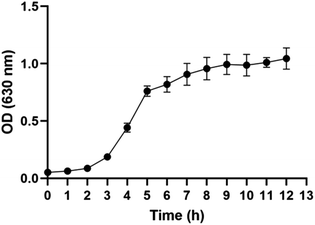
By determining the OD value of ATCC-25922 at different time points, the bacterial growth curve was plotted (Fig. 1). The observed growth patterns of the bacteria over time are as follows. In the first two hours, the bacteria growth is slow, indicating a growth stagnation period. From 2 to 5 hours, the rate of bacterial increase becomes visibly rapid, marking the exponential growth phase. After 5 hours, the rate of bacterial proliferation stabilizes as the rates of growth and death approach a dynamic equilibrium, leading to a stable bacterial population, known as the stationary phase. Consequently, it is appropriate to select five time points (3 h-T0, 4 h-T1, 5 h-T2, 6 h-T3, and 7 h-T4) for exploring VOCs between 3 and 7 hours for the present study and to compare the differences in VOCs among different groups at the end of the exponential growth period of the bacterial proliferation, specifically at the T2 time point.
3.2 Exploring the fingerprints of VOCs in blank bottles, CSEC, CREC and the relative content of each group at different time points
Under the established experimental conditions, one strain of CSEC and one strain of CREC were selected, and each bacterial strain repeated the same operation six times for the pre-exploratory experiments (hereinafter). In total, 4 alcohols, 6 aldehydes, 3 acids, 3 esters, 5 ketones, 3 pyrazines, 2 heterocyclic compounds, and 10 unknowns were detected using GC-IMS. The same substance was divided into monomer and dimer forms according to the drift time, resulting in the detection of 36 VOCs. The changes in the relative content of these 36 VOCs from T0 to T4 in the blank control group, the CSEC group, and the CREC group are detailed in ESI Table 1,† and detailed information on each substance is demonstrated in Table 2.| Chemical class | Compound | CAS# | Formula | MW | RI | Rt [s] | Dt [a.u.] |
|---|---|---|---|---|---|---|---|
| a Represents the substance that has two distinct peak positions in the GC-IMS system, with a shorter drift time corresponding to the monomer and a longer drift time corresponding to the dimer.b Abbreviations: VOCs, volatile organic compounds; GC-IMS, gas chromatography-ion mobility spectrometry; CAS#, chemical abstract service registry number; MW, molecular weight; RI, retention index; Rt, retention time; Dt, drift time. | |||||||
| Alcohols | Ethylene glycol | C107211 | C2H6O2 | 62.1 | 1668.2 | 492.458 | 1.10179 |
| 1-Butanol | C71363 | C4H10O | 74.1 | 1117.3 | 149.829 | 1.18152 | |
| 3-Methyl-1-butanola | C123513 | C5H12O | 88.1 | 1216 | 187.96 | 1.48891 | |
| 1218.9 | 189.125 | 1.24111 | |||||
| Aldehydes | Acetaldehyde | C75070 | C2H4O | 44.1 | 701.7 | 78.843 | 0.9626 |
| Propanal | C123386 | C3H6O | 58.1 | 774.8 | 87.347 | 1.04836 | |
| Nonanal | C124196 | C9H18O | 142.2 | 1404.3 | 280.573 | 1.48176 | |
| Benzaldehydea | C100527 | C7H6O | 106.1 | 1532.8 | 368.971 | 1.47003 | |
| 1531.2 | 367.72 | 1.15593 | |||||
| 3-Methyl-2-butenal | C107868 | C5H8O | 84.1 | 1217.3 | 188.489 | 1.09221 | |
| Acids | Acetic acid | C64197 | C2H4O2 | 60.1 | 1481.4 | 330.713 | 1.15909 |
| Propionic acida | C79094 | C3H6O2 | 74.1 | 1589.9 | 416.736 | 1.26668 | |
| 1580.6 | 408.609 | 1.11349 | |||||
| Esters | Ethyl acrylate | C140885 | C5H8O2 | 100.1 | 1015.3 | 123.315 | 1.42017 |
| Bornyl acetate | C76493 | C12H20O2 | 196.3 | 1581.3 | 409.215 | 1.20781 | |
| Methyl 2-methylbutanoate | C868575 | C6H12O2 | 116.2 | 1024.4 | 125.346 | 1.19505 | |
| Ketones | Acetone | C67641 | C3H6O | 58.1 | 823.2 | 93.49 | 1.11441 |
| Butan-2-one | C78933 | C4H8O | 72.1 | 905.8 | 104.961 | 1.25031 | |
| Cyclohexanone | C108941 | C6H10O | 98.1 | 1309.7 | 229.283 | 1.15745 | |
| 2,3-Pentanedione | C600146 | C5H8O2 | 100.1 | 1071.7 | 136.468 | 1.22922 | |
| 3-Hydroxybutan-2-one (acetoin) | C513860 | C4H8O2 | 88.1 | 1306.1 | 227.546 | 1.06994 | |
| Pyrazines | 2-Methylpyrazine | C109080 | C5H6N2 | 94.1 | 1286.8 | 218.389 | 1.08819 |
| 2-Ethyl-5-methylpyrazine | C13360640 | C7H10N2 | 122.2 | 1439.5 | 302.438 | 1.19545 | |
| 2,5-Dimethylpyrazine | C123320 | C6H8N2 | 108.1 | 1337 | 243.058 | 1.11647 | |
| Heterocyclic compound | Pyrrolidinea | C123751 | C4H9N | 71.1 | 1029.5 | 126.513 | 1.04138 |
| 1010.2 | 122.187 | 1.27592 | |||||
| Unidentified compounds | Unidentified-1 | Unidentified | * | 0 | 1312.1 | 230.476 | 1.22964 |
| Unidentified-2 | Unidentified | * | 0 | 1162.1 | 166.498 | 1.2147 | |
| Unidentified-3 | Unidentified | * | 0 | 1116.5 | 149.54 | 1.51926 | |
| Unidentified-4 | Unidentified | * | 0 | 1109.8 | 147.202 | 1.44227 | |
| Unidentified-5 | Unidentified | * | 0 | 964.4 | 113.957 | 1.16912 | |
| Unidentified-6 | Unidentified | * | 0 | 1148.4 | 161.222 | 1.07504 | |
| Unidentified-7 | Unidentified | * | 0 | 1145.5 | 160.126 | 1.33492 | |
| Unidentified-8 | Unidentified | * | 0 | 1166 | 168.052 | 1.03977 | |
| Unidentified-9 | Unidentified | * | 0 | 1180.5 | 173.87 | 1.12619 | |
| Unidentified-10 | Unidentified | * | 0 | 1217.3 | 188.507 | 1.31139 | |
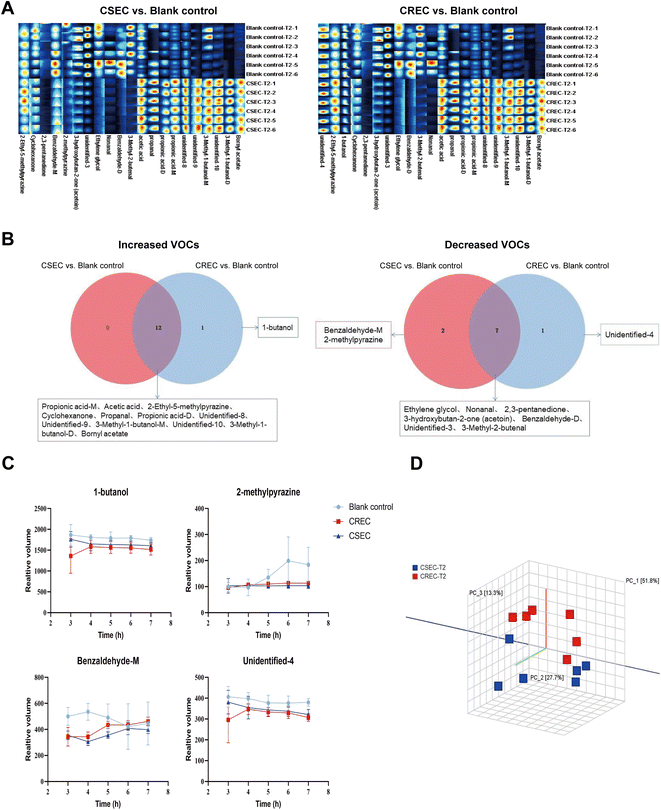
Based on the growth curves, we then focused on analyzing the differences in the relative content of each VOC among different groups at the T2 time point. By comparing the blank control group, there are 34 substances with the same trend of change in the CSEC and CREC groups, of which the contents of 16 increased and 18 decreased, and only two substances (acetaldehyde and unidentified-7) had the opposite trend of change. Notably, by comparing the CSEC group with the CREC group, only 11 VOCs differed, including alcohols: 3-methyl-1-butanol-M; aldehydes: acetaldehyde, benzaldehyde (monomer and dimer), 3-methyl-2-butenal; acids: acetic acid; esters: bornyl acetate, methyl 2-methylbutanoate; heterocyclic: pyrrolidine-M; unidentified (8 and 10). Detailed data are displayed in Table 3. The fingerprints of 36 VOCs among different groups at the T2 time point are illustrated in Fig. 2A. Obviously, although CSEC and CREC were divided into different groups because of different sensitivities to carbapenems, the fingerprints generated by the two groups showed almost no significant difference.
| Label | Blank control (T2) mean ± SD (n = 6) | CSEC (T2) mean ± SD (n = 6) | Up/down | CREC (T2) mean ± SD (n = 6) | Up/down | P1 | P2 | P3 |
|---|---|---|---|---|---|---|---|---|
| a P1: CSEC vs. blank control; P2: CREC vs. blank control; P3: CSEC vs. CREC.b Abbreviations: VOCs, volatile organic compounds; GC-IMS, gas chromatography-ion mobility spectrometry; SD, standard deviation; CSEC, carbapenem-sensitive Escherichia coli; CREC, carbapenem-resistant Escherichia coli; M, monomer; D, dimer. | ||||||||
| Ethylene glycol | 103.43 ± 27.69 | 54.9 ± 6.53 | Down | 57.44 ± 10.7 | Down | 0.0019 | 0.0035 | 0.6306 |
| 1-Butanol | 1788.99 ± 148.28 | 1635.3 ± 98.62 | Down | 1564.05 ± 142.13 | Down | 0.0606 | 0.0230 | 0.3368 |
| 3-Methyl-1-butanol-D | 118.3 ± 44.27 | 1217.46 ± 74.75 | Up | 1165.41 ± 74.34 | Up | 2.8722 × 10−11 | 4.4586 × 10−11 | 0.2544 |
| 3-Methyl-1-butanol-M | 242.46 ± 110.28 | 645.59 ± 17.37 | Up | 599.69 ± 35.08 | Up | 4.8344 × 10−6 | 1.9225 × 10−5 | 0.0166 |
| Acetaldehyde | 270.24 ± 98.29 | 307.96 ± 14.9 | Up | 268.48 ± 13.16 | Down | 0.3746 | 0.9661 | 0.0007 |
| Propanal | 79.5 ± 44.99 | 149.66 ± 6.51 | Up | 146.02 ± 8.31 | Up | 0.0036 | 0.0052 | 0.4182 |
| Nonanal | 246.83 ± 164.08 | 56.93 ± 17.85 | Down | 43.85 ± 12.96 | Down | 0.0182 | 0.0129 | 0.1772 |
| Benzaldehyde-D | 211.35 ± 83.33 | 86.75 ± 18.02 | Down | 109 ± 9.54 | Down | 0.0050 | 0.0136 | 0.0234 |
| Benzaldehyde-M | 490.92 ± 104.53 | 355.55 ± 23.81 | Down | 434.55 ± 28.2 | Down | 0.0114 | 0.2310 | 0.0004 |
| 3-Methyl-2-butenal | 197.67 ± 30.95 | 42.45 ± 3.32 | Down | 51.13 ± 4.46 | Down | 2.4722 × 10−7 | 4.4289 × 10−7 | 0.0033 |
| Acetic acid | 2472.66 ± 684.54 | 4870.41 ± 227.01 | Up | 5542.03 ± 211.99 | Up | 1.0071 × 10−5 | 1.0222 × 10−6 | 0.0003 |
| Propionic acid-D | 506.04 ± 107.29 | 969.72 ± 157.46 | Up | 1111.54 ± 159.08 | Up | 0.0001 | 1.5894 × 10−5 | 0.1517 |
| Propionic acid-M | 872.58 ± 74.15 | 1859.16 ± 114.56 | Up | 1898.72 ± 129.03 | Up | 7.0249 × 10−9 | 1.1121 × 10−8 | 0.5867 |
| Ethyl acrylate | 500.39 ± 429.04 | 176.7 ± 56.63 | Down | 181.89 ± 82.18 | Down | 0.0968 | 0.1044 | 0.9011 |
| Bornyl acetate | 123.23 ± 20.25 | 672.35 ± 43.01 | Up | 804.68 ± 44.9 | Up | 7.0665 × 10−11 | 1.1851 × 10−11 | 0.0004 |
| Methyl 2-methylbutanoate | 1111.6 ± 461.52 | 857.25 ± 32.21 | Down | 966.58 ± 105.78 | Down | 0.2078 | 0.4704 | 0.0360 |
| Acetone | 8141.33 ± 1285.66 | 8802.85 ± 819.94 | Up | 8819.36 ± 808.17 | Up | 0.3129 | 0.2997 | 0.9727 |
| Butan-2-one | 1229.63 ± 362.22 | 1314.57 ± 308.1 | Up | 1405.76 ± 309.14 | Up | 0.6711 | 0.3863 | 0.6199 |
| Cyclohexanone | 142.87 ± 25.34 | 184.76 ± 13.13 | Up | 180.47 ± 14.04 | Up | 0.0049 | 0.0098 | 0.5962 |
| 2,3-Pentanedione | 29.69 ± 9.32 | 17.14 ± 2.96 | Down | 15.43 ± 1.92 | Down | 0.0105 | 0.0043 | 0.2629 |
| 3-Hydroxybutan-2-one (acetoin) | 627.43 ± 116.83 | 437.08 ± 53.95 | Down | 388.02 ± 19.13 | Down | 0.0047 | 0.0006 | 0.0621 |
| 2-Methylpyrazine | 135.16 ± 31.02 | 102.81 ± 8.98 | Down | 109.87 ± 8.33 | Down | 0.0340 | 0.0825 | 0.1885 |
| 2-Ethyl-5-methylpyrazine | 65.48 ± 9.58 | 79.82 ± 12.44 | Up | 91.22 ± 10.14 | Up | 0.0493 | 0.0011 | 0.1122 |
| 2,5-Dimethylpyrazine | 478 ± 142.65 | 398.75 ± 91.67 | Down | 434.23 ± 75.49 | Down | 0.2789 | 0.5215 | 0.4811 |
| Pyrrolidine-D | 758.08 ± 266.7 | 677.02 ± 111.7 | Down | 679.6 ± 55.53 | Down | 0.5079 | 0.4965 | 0.9606 |
| Pyrrolidine-M | 611.36 ± 160.38 | 733.06 ± 32.91 | Up | 653.09 ± 79.44 | Up | 0.0987 | 0.5806 | 0.0459 |
| Unidentified-1 | 262.51 ± 113.53 | 246 ± 55.43 | Down | 249.74 ± 35.17 | Down | 0.7556 | 0.7978 | 0.8918 |
| Unidentified-2 | 746.99 ± 563.7 | 380.89 ± 111.58 | Down | 319.54 ± 70.47 | Down | 0.1497 | 0.0951 | 0.2814 |
| Unidentified-3 | 2733.67 ± 168.25 | 997.4 ± 163.68 | Down | 927.8 ± 236.18 | Down | 5.6261 × 10−9 | 2.9748 × 10−8 | 0.5662 |
| Unidentified-4 | 377.29 ± 36.87 | 343.66 ± 29.79 | Down | 333.54 ± 21.81 | Down | 0.1128 | 0.0313 | 0.5172 |
| Unidentified-5 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 264.39 ± 1673.03 264.39 ± 1673.03 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 787.63 ± 1969.04 787.63 ± 1969.04 |
Up | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 688.03 ± 2093.5 688.03 ± 2093.5 |
Up | 0.1793 | 0.2223 | 0.9340 |
| Unidentified-6 | 63.99 ± 10.7 | 59.12 ± 2.69 | Down | 62.32 ± 2.93 | Down | 0.3053 | 0.7209 | 0.0771 |
| Unidentified-7 | 27.53 ± 6.43 | 30.71 ± 5.96 | Up | 25.84 ± 4.79 | Down | 0.3954 | 0.6177 | 0.1502 |
| Unidentified-8 | 2402.09 ± 393.13 | 6006.3 ± 104.43 | Up | 6157.6 ± 125.46 | Up | 9.6343 × 10−10 | 7.4123 × 10−10 | 0.0465 |
| Unidentified-9 | 183.46 ± 15.34 | 962.88 ± 80.62 | Up | 1048.66 ± 53.25 | Up | 4.8704 × 10−10 | 3.5625 × 10−12 | 0.0547 |
| Unidentified-10 | 139.94 ± 32.98 | 741.83 ± 15.61 | Up | 787.02 ± 10.04 | Up | 2.0610 × 10−12 | 5.7045 × 10−13 | 0.0001 |
 | ||
| Fig. 2 (A) Fingerprints of 36 VOCs among different groups at the T2 time point. (B) Principal component analysis and similarity analysis using VOCs at T2 time point. | ||
3.3 Exploring whether VOCs can be used to distinguish CSEC from CREC at the T2 time point when IPM is not added
To investigate whether the VOCs detected at the T2 time point could be used to differentiate the different groups, the three groups of data were jointly imported into Dynamic PCA for analysis. The results disclosed that the 36 VOCs could differentiate the bacterial group (CSEC and CREC) from the blank control group. However, they were unable to further differentiate CSEC and CREC groups. Then, a similarity analysis was performed to verify the results of PCA analysis, which finally maintained the inability to effectively differentiate CSEC from CREC. Subsequently, a similarity analysis was performed on the above data to verify the results of PCA analysis, which finally maintained the inability to effectively differentiate between CSEC and CREC (Fig. 2B).Further comparison of the relative contents of VOCs in six sets of parallel samples from the CSEC group and the blank control group using the U-test reveals that of the 36 substances detected, 21 differed between the two groups, with 12 rising and 9 falling compared with the blank bottle. By comparing the relative contents of VOCs in six groups of parallel samples from CREC and blank bottles, there are also 21 substances with differences between the two groups, with 13 rising and 8 falling compared with the blank bottles. Fig. 3A displays the fingerprints of VOCs with differences in the comparison between different groups in the T2 time point, and Table 3 demonstrates the trends of all differences in the VOCs and their relative contents.
Finally, the Veen diagram (Fig. 3B) was employed to analyze the 12 content-increasing VOCs and 9 content-decreasing VOCs in the CSEC group compared to the blank control group versus the 13 content-increasing VOCs and 8 content-decreasing VOCs in the CREC group compared to the blank control group. The results indicated that, among all content-increasing VOCs, 1-butanol, which was unique to the CREC group, existed outside the 12 intersections. Among all content-decreasing VOCs, benzaldehyde-M and 2-methylpyrazine were unique to CSEC, and unidentified-4 was unique to CREC. The content change curves of the above four specific VOCs from T0 to T4 are presented in Fig. 3C. Finally, PCA was performed again using these four substances to explore their ability to differentiate between CSEC and CREC, and the results suggested that the two groups remained indistinguishable from each other (Fig. 3D).
3.4 Exploring whether IPM addition can distinguish CSEC from CREC using VOCs at the T2 time point
To explore the effect of adding IPM on the emission of VOCs from each group of bacteria, IPM was added during the T0 period (so that the final concentration of IPM in the whole system was 0.25 mg mL−1). ESI Table 2† demonstrates the changes in all detected VOCs for each group of samples in the T0–T4 time period after adding IPM. Although no new VOCs were generated when IPM was added, only the content of certain VOCs was changed. Table 4 demonstrates more intuitively the VOCs with differences between CSEC + IPM and CREC + IPM in the T2 period. In total, 27 differentiated VOCs were detected among 36 detected VOCs, imparting an increase of 16 differentiated VOCs compared to only 11 differentiated VOCs detected in the comparison of CSEC and CREC without IPM. Compared to the CSEC + IPM group, 10 of the 27 differential VOCs decreased (ethyl acrylate, cyclohexanone, methyl 2-methylbutanoate, 2,3-pentanedione, nonanal, pyrrolidine-D, ethylene glycol, butan-2-one, benzaldehyde-D, acetaldehyde) and 17 VOCs increased (unidentified-8, pyrrolidine-M, unidentified-10, 2-ethyl-5-methylpyrazine, unidentified-9, 3-methyl-1-butanol-D, unidentified-1, bornyl acetate, 1-butanol, acetic acid, 3-hydroxybutan-2-one (acetoin), unidentified-3, propanal, propionic acid-D, unidentified-4, unidentified-5, 3-methyl-2-butenal) in the CREC + IPM group.| Label | CSEC + IPM (T2) mean ± SD (n = 6) | CREC + IPM (T2) mean ± SD (n = 6) | Variation | P |
|---|---|---|---|---|
| a P: CSEC + IPM vs. CREC + IPM, variation: changes in VOCs in CREC + IPM using CSEC + IPM as a reference.b Abbreviations: VOCs, volatile organic compounds; IPM, imipenem; SD, standard deviation; CSEC, carbapenem-sensitive Escherichia coli; CREC, carbapenem-resistant Escherichia coli; M, monomer; D, dimer. | ||||
| Ethyl acrylate | 1417.25 ± 97.71 | 201.43 ± 69.79 | Down | 2.5943 × 10−10 |
| Unidentified-8 | 2493.5 ± 302.12 | 5863.61 ± 274.64 | Up | 1.9299 × 10−9 |
| Pyrrolidine-M | 399.24 ± 34.69 | 716.74 ± 17.06 | Up | 2.0255 × 10−9 |
| Unidentified-10 | 381.58 ± 30.86 | 730.34 ± 32.3 | Up | 3.3251 × 10−9 |
| 2-Ethyl-5-methylpyrazine | 17.09 ± 2.76 | 89.56 ± 8.96 | Up | 3.6544 × 10−9 |
| Cyclohexanone | 451.97 ± 39.32 | 169.56 ± 7.13 | Down | 8.7687 × 10−9 |
| Unidentified-9 | 392.07 ± 20.7 | 977.77 ± 97.87 | Up | 5.3767 × 10−8 |
| Methyl 2-methylbutanoate | 1171.33 ± 29.97 | 895.05 ± 41.47 | Down | 1.1631 × 10−7 |
| 3-Methyl-1-butanol-D | 777.74 ± 40.38 | 1129.12 ± 74.04 | Up | 1.3184 × 10−6 |
| Unidentified-1 | 83.26 ± 5.19 | 279.77 ± 53.22 | Up | 4.1343 × 10−6 |
| Bornyl acetate | 376.96 ± 100.43 | 765.26 ± 35.41 | Up | 4.4298 × 10−6 |
| 1-Butanol | 920.52 ± 30.03 | 1454.23 ± 153.1 | Up | 7.8301 × 10−6 |
| 2,3-Pentanedione | 32.88 ± 4.13 | 17.45 ± 2.33 | Down | 1.2124 × 10−5 |
| Acetic acid | 3793.23 ± 671.73 | 5684.85 ± 153.49 | Up | 5.2036 × 10−5 |
| Nonanal | 218.85 ± 67.62 | 37.12 ± 3.36 | Down | 6.2727 × 10−5 |
| 3-Hydroxybutan-2-one (acetoin) | 265.9 ± 33.7 | 424.49 ± 52.26 | Up | 9.5421 × 10−5 |
| Pyrrolidine-D | 815.98 ± 67.87 | 627.43 ± 38.91 | Down | 0.0002 |
| Unidentified-3 | 189.92 ± 20.9 | 776.88 ± 256.79 | Up | 0.0002 |
| Ethylene glycol | 85.41 ± 13.8 | 51.78 ± 5.88 | Down | 0.0003 |
| Propanal | 118.63 ± 5.93 | 141.43 ± 8.3 | Up | 0.0003 |
| Propionic acid-D | 585.02 ± 122.89 | 1021.79 ± 149.56 | Up | 0.0003 |
| Butan-2-one | 2015.94 ± 71.14 | 1365.89 ± 312.65 | Down | 0.0006 |
| Unidentified-4 | 240.1 ± 12.68 | 332.8 ± 45.25 | Up | 0.0007 |
| Unidentified-5 | 7499.96 ± 398.37 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 070.2 ± 2100.91 070.2 ± 2100.91 |
Up | 0.0022 |
| Benzaldehyde-D | 154.56 ± 29.98 | 110.22 ± 10.22 | Down | 0.0065 |
| 3-Methyl-2-butenal | 41.79 ± 2.6 | 48.3 ± 5.38 | Up | 0.0236 |
| Acetaldehyde | 294.95 ± 22.59 | 268.4 ± 12.58 | Down | 0.0306 |
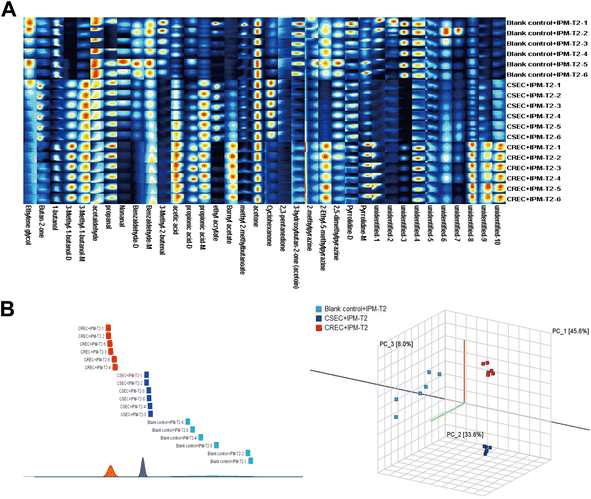
Fig. 4A further displays the fingerprints of the blank control, CSEC, and CREC groups after IPM addition at the T2 period. Remarkably, the most obvious change was in the CSEC group, while the least obvious change was in the CREC group, and the above trend continued until the end of the study. The reason for this is that after adding IPM at T0, the bacteria in the CSEC group were killed, while those in the CREC group were not killed because of their resistance to IPM. This is corroborated by the comparison between Fig. 4A and 2A in fingerprints.
Subsequently, PCA was again utilized to further explore whether IPM addition could differentiate CSEC from CREC. The results demonstrated that IPM addition could effectively differentiate between the blank control, CSEC, and CREC groups (Fig. 4B). The results of similarity analysis also confirmed that IPM addition could differentiate the groups, as detailed in Fig. 4B.
3.5 Expanded sample analyses of IPM addition distinguish CSEC from CREC at the T2 period
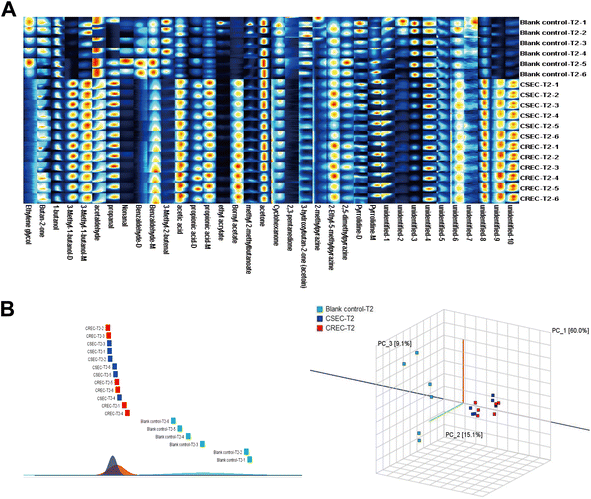
Twenty CSEC and CREC strains isolated from clinical patient specimens were collected, and three parallel experiments were conducted for all strains under the same experimental conditions. This results in the relative VOC contents of 60 CSEC and 60 CREC groups, and the abundance of VOCs for all clinical strains is given as a heatmap (Fig. 5A). Notably, the clinical strains also did not produce new VOCs. Compared to the 27 differential VOCs detected in the six parallel samples explored in the preliminary stage, the inter-group differences between the clinical strains in CSEC and CREC groups became more pronounced with IPM addition. This is attributed to the increase in the sample size, with the number of differential VOCs reaching 31. All the differential VOCs are presented in Table 5, of which the top 10 most differential substances are unidentified-3, unidentified-4, 2-ethyl-5-methylpyrazine, 1-butanol, unidentified-1, benzaldehyde-D, 3-hydroxybutan-2-one (acetoin), 3-methyl-1-butanol-M, pyrrolidine-D, 2,5-dimethylpyrazine. Finally, PCA was applied to verify the differentiation status of the clinical strains in the T2 period (Fig. 5B), and it can be observed that IPM addition can effectively differentiate CSEC strains from CREC strains.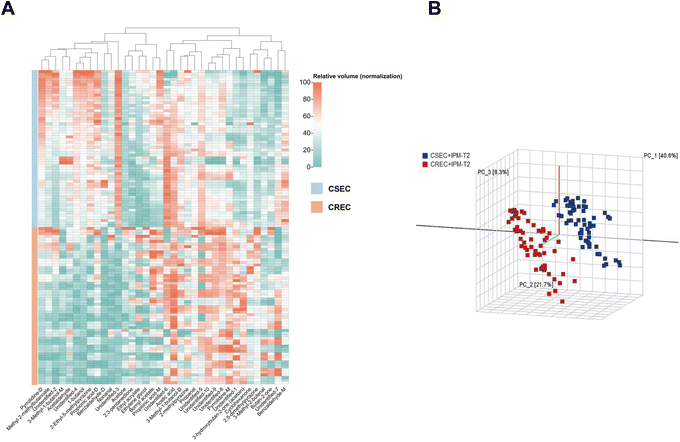 | ||
| Fig. 5 (A) Abundance of VOCs in clinical strains. (B) Principal component analysis validation to distinguish CSEC from CREC clinical strains. | ||
| Label | CSEC + IPM (T2) mean ± SD (n = 60) | CREC + IPM (T2) mean ± SD (n = 60) | Variation | P |
|---|---|---|---|---|
| a P: CSEC + IPM vs. CREC + IPM, variation: changes in VOCs in CREC + IPM using CSEC + IPM as a reference.b Abbreviations: VOCs, volatile organic compounds; IPM, imipenem; SD, standard deviation; CSEC, carbapenem-sensitive Escherichia coli; CREC, carbapenem-resistant Escherichia coli; M, monomer; D, dimer. | ||||
| Unidentified-3 | 247.7 ± 14.53 | 103.59 ± 14.53 | Down | 2.4537 × 10−85 |
| Unidentified-4 | 276.98 ± 30.73 | 146.92 ± 36.43 | Down | 6.1508 × 10−42 |
| 2-Ethyl-5-methylpyrazine | 91.72 ± 14.87 | 39.05 ± 23.32 | Down | 1.4973 × 10−28 |
| 1-Butanol | 1359.2 ± 195.29 | 923.04 ± 171.84 | Down | 1.7076 × 10−24 |
| Unidentified-1 | 136.6 ± 24.1 | 208.43 ± 35.65 | Up | 2.3258 × 10−24 |
| Benzaldehyde-D | 300.78 ± 129.69 | 91.29 ± 43.31 | Down | 7.3440 × 10−22 |
| 3-Hydroxybutan-2-one (acetoin) | 323.36 ± 40.05 | 415.06 ± 52.81 | Up | 3.9770 × 10−19 |
| 3-Methyl-1-butanol-M | 621.79 ± 43.92 | 542.45 ± 38.02 | Down | 8.4541 × 10−19 |
| Pyrrolidine-D | 1368.49 ± 167.57 | 1043.92 ± 182.01 | Down | 8.2665 × 10−18 |
| 2,5-Dimethylpyrazine | 356.32 ± 27.14 | 409.73 ± 34.76 | Up | 5.8263 × 10−16 |
| Unidentified-9 | 672.5 ± 136.84 | 884.46 ± 112.39 | Up | 1.0586 × 10−15 |
| Unidentified-10 | 599.35 ± 63.86 | 700 ± 61.7 | Up | 1.5105 × 10−14 |
| Pyrrolidine-M | 415.82 ± 73.63 | 515.25 ± 58.63 | Up | 3.6534 × 10−13 |
| Propionic acid-D | 625.25 ± 115.74 | 452.59 ± 158.77 | Down | 4.4050 × 10−10 |
| Nonanal | 135.76 ± 35.06 | 82.92 ± 51.67 | Down | 1.5385 × 10−9 |
| Acetaldehyde | 293.12 ± 27.69 | 264.31 ± 34.92 | Down | 1.9456 × 10−6 |
| Benzaldehyde-M | 545.88 ± 83.24 | 483.48 ± 62.94 | Down | 9.4168 × 10−6 |
| Butan-2-one | 960.75 ± 48.67 | 1024.58 ± 100.83 | Up | 2.2391 × 10−5 |
| Unidentified-8 | 4255.55 ± 435.6 | 4655.55 ± 580.52 | Up | 3.9900 × 10−5 |
| Acetic acid | 5529.08 ± 529.18 | 5913.92 ± 547.58 | Up | 0.0001 |
| Methyl 2-methylbutanoate | 1352.97 ± 134.6 | 1248.39 ± 155.82 | Down | 0.0001 |
| Acetone | 9675.73 ± 58.24 | 9781.19 ± 210.76 | Up | 0.0003 |
| Unidentified-6 | 410.63 ± 127.9 | 327.76 ± 134.64 | Down | 0.0008 |
| 2,3-Pentanedione | 22.55 ± 3.57 | 24.84 ± 4.13 | Up | 0.0016 |
| Unidentified-5 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 065.33 ± 364.08 065.33 ± 364.08 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 602.23 ± 1121.11 602.23 ± 1121.11 |
Down | 0.0029 |
| Unidentified-2 | 667.31 ± 236 | 543.1 ± 250.61 | Down | 0.0061 |
| Ethyl acrylate | 427.9 ± 145.81 | 541.76 ± 289.51 | Up | 0.0075 |
| 3-Methyl-2-butenal | 48.11 ± 7.6 | 53.35 ± 13.32 | Up | 0.0092 |
| 3-Methyl-1-butanol-D | 1206.69 ± 53.29 | 1164.28 ± 120.01 | Down | 0.0137 |
| Bornyl acetate | 429.63 ± 90.79 | 491.07 ± 169.18 | Up | 0.0146 |
| Cyclohexanone | 145.15 ± 15.34 | 155.76 ± 31.63 | Up | 0.0211 |
4. Discussion
Antibiotic therapy remains an effective way of treating bacterial infections, and to avoid resistance to inappropriate antibiotic treatment, accurate identification of the causative organisms and drug susceptibility testing are required. Existing microbial identification and drug susceptibility tests are still based on traditional methods, which increase the number of bacteria by enriching for a long period of time to achieve the minimum detection level. This prolonged process may miss the optimal time for patients to be treated, whereas the active development of rapid and effective tests can help doctors designate a rational drug plan and reduce the burden on patients.Recently, identifying bacteria using various emerging technologies for volatile metabolites produced by bacteria has become a new trend. Gas chromatography24 and high-performance liquid chromatography25 are unsuitable for clinical use because of their high operational requirements and relatively complex pre-treatment. Electronic nose methods26 are not highly sensitive and have no analytical capability for chemical compositions. GC-IMS technology—characterized by high sensitivity, selectivity, and rapid analysis, shows significant advantages in the detection of VOCs, and its unique ion mobility spectrometry analysis enables more accurate separation and identification of VOCs in complex mixtures. In addition, the non-destructive detection characteristic of GC-IMS technology ensures the integrity of the samples, thus avoiding, to a certain extent, the detection errors caused by chemical changes during sample processing. Although most of the studies proved that GC-IMS technology has made great contributions in the food27 and environmental28 fields, there are also studies confirming that GC-IMS can utilize specific mVOCs for the E. coli, Staphylococcus aureus, and Pseudomonas aeruginosa differentiation in mixed culture mode.29 The good performance of mVOCs in identifying bacterial species has successfully promoted their application in antibiotic susceptibility testing. In this study, we utilized the GC-IMS technique to side by side reflect the state of bacterial life activities and identify E. coli using VOCs produced during bacterial proliferation as a medium. With the addition of IPM and the application of PCA, similarity analysis, and other analytical methods, it is possible to differentiate between CSEC and CREC. To the best of our knowledge, this is the first work to report that the GC-IMS technique can effectively differentiate between carbapenem-sensitive and drug-resistant Escherichia coli based on VOCs. These findings not only provide new insights into understanding the metabolic adaptation mechanisms of Escherichia coli, but also lay the foundation for the development of rapid antibiotic susceptibility testing methods. More importantly, our findings are expected to enable faster diagnostic information for physicians and reduce unnecessary antibiotic use, thereby reducing healthcare costs.
Pseudomonas putida can utilize benzaldehyde dehydrogenase to reduce benzaldehyde to NADPH as an alternative energy source.30 Throughout the five aldehydes detected in this study, the concentrations of nonanal and benzaldehyde (monomer and dimer) were higher in the blank bottles than in the groups containing the other two. With IPM addition, the levels in the CREC group were lower than those in the CSEC group. Consequently, we conjecture that nonanal and benzaldehyde may be used as an energetic substance that is ingested in the proliferation phase of the bacterium. This seems to confirm that aldehydes can be utilized for survival during the metabolism of E. coli.
Acetone has been shown to be closely associated with bacterial growth and metabolism.31 Although acetone is considered to be a VOC-specific to E. coli14 and was successfully detected in this study, it did not greatly contribute to the pre-exploratory phase, either when distinguishing between E. coli and blank vials or when distinguishing between CSEC and CREC after IPM addition (Tables 3 and 4). Indole, which is converted from tryptophan by tryptophanase,32 is usually considered to be the specific VOC of E. coli.33,34 However, indole was undetected in this study, possibly due to the lack of tryptophan in the medium we used. However, it has been confirmed that Staphylococcus aureus,35 Klebsiella pneumoniae, and Acinetobacter baumannii36 release indole. Consequently, individual VOCs have limited ability to characterize different strains of bacteria. Besides, it is of great significance to explore the combined detection of multiple products to improve sensitivity and specificity of the VOCs determination method.37
When IPM was not added, there was little difference in the content of mVOCs between CSEC and CREC strains. In contrast, with IPM addition at the T0 period, the difference in the relative content of mVOCs between the groups became more pronounced. For the mVOCs with significantly decreased content, we hypothesized that it might be due to bacterial death, which terminated the whole metabolic process. For this hypothesis, we considered it by comparing the seven VOCs of 3-methyl-1-butanol-D, acetic acid, propionic acid-D, bornyl acetate, and unidentified-8, 9, and 10 in ESI Table 1 with ESI Table 2.† Firstly, these seven substances were found in the CSEC group without adding the IPM in the CSEC group, the relative content of their VOCs increased in the T0–T4 time period, but after the addition of IPM at the T0 time point, imipenem would bind to penicillin-binding proteins on the cell membrane of CSEC, preventing the normal function of transketolase, which led to the blockage of cell wall synthesis, which in turn caused the bacterial cell wall to become weak, and the bacterial cell eventually ruptured under the action of osmotic pressure which leads to bacterial death.38 Due to the termination of bacterial growth and metabolic processes at T0, the relative content of the above seven VOCs stagnated at T0, and remained unchanged or relatively decreased during the subsequent detection process. Among them, acetic acid39 and propionic acid40 were also shown to be VOCs closely related to the growth state of Escherichia coli. Therefore, for these substances, we believe that these mVOCs are closely linked to CSEC. Similarly, for mVOCs with significantly increased content, it might be due to bacterial death, which continuously accumulated nutrients in the TSB medium that were not consumed by the bacteria. As a result, the relative content of mVOCs was detected to be higher.
Notably, the monomer and dimer of four substances were detected in this study. Neither the 3-methyl-1-butanol monomer nor the dimer was affected by the selective pressure of IPM. The content of 3-methyl-1-butanol in both CSEC and CREC increased significantly compared with that of the blank control group (Tables 1 and 4). This indicates that they are more stable and are closely linked to E. coli. However, it was not possible to confirm these are specific VOCs for E. coli because it has been confirmed that Klebsiella pneumoniae can decompose leucine through the Ehrlich pathway to produce 3-methyl-1-butanol.41 Although the detection technology and the detection content of mVOCs differ, considering that Klebsiella pneumoniae and E. coli both belong to the Enterobacteriaceae family, and their metabolic pathways may not be very different, we are more inclined to consider 3-methyl 1-butanol as a volatile metabolite closely correlated with Enterobacteriaceae bacteria.
However, it has to be recognized that different growth environments can have important effects on the growth and metabolism of bacteria. In this study, we used TSB as the culture medium for Escherichia coli, mainly from the following considerations: firstly, TSB is a nutrient-rich medium containing a variety of nutrients required for the growth of microorganisms, such as proteins, carbohydrates, vitamins, and minerals, and so on. This comprehensiveness ensures the rapid growth of microorganisms in a suitable environment, which is conducive to the stable production and detection of VOCs. Secondly, TSB is one of the commonly used culture media in microbiology research, which is especially suitable for the culture of aerobic and partially anaerobic bacteria. Its wide application implies that its composition and performance have been widely verified as a basic medium for the study of VOCs, and a large number of studies have applied TSB medium to successfully identify different bacterial strains.42,43 In addition, this study is a preliminary exploratory experiment with a large number of unknowns, whereas TSB medium, as a widely used medium, is open and standardized in its formulation and preparation methods, which ensures reproducibility and relative certainty of the experiment. Whether comparing data within our lab or with other research organizations, the use of TSB ensures consistency in experimental conditions. Finally, compared to some special or customized media, TSB is relatively low cost and easy to obtain, which is especially important in large-scale experiments or long-term studies. This also gives us an insight into the future direction of our research: focusing on analyzing the variation of bacterial VOCs in different growth media to study the diversity and specificity of VOCs present in specific microbial species.
5. Limitations
Although we conducted a preliminary study on the identification of carbapenem-sensitive and carbapenem-resistant E. coli using GC-IMS, we have to admit that there are still some limitations. First, the number of experimental strains used was relatively small and originated from a single center. The enzyme types of the identified CREC were not further identified. In addition, due to the imperfection of the database, 10 substances could not be identified.6. Conclusions
In this study, the VOCs produced by bacteria were analyzed using the GC-IMS technique. Changes in the relative content of VOCs and the signal intensity of the fingerprints were used to assess the growth status of bacteria during the bacterial growth process and help identify E. coli. The advantages of this technique, such as rapid detection, real-time analysis, and no need for complex pre-treatment of the samples, allowed us to demonstrate for the first time that E. coli volatile fingerprinting can be employed as a model to distinguish CSEC from CREC strains. We intend to improve the experimental technique and collect more strains from multiple centers to increase the reliability of the results with the ultimate goal of rapidly identifying CSEC and CREC and even determining the enzyme type of CREC.Data availability
All datasets used during this study were obtained from the Clinical Laboratory of the Second Affiliated Hospital of Nanchang University; for access to datasets under reasonable conditions for the purpose, please contact the corresponding author.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was supported by the National Natural Science Foundation of China (82060391), the Natural Science Foundation of Jiangxi Province (20202BAB216021), the Medical Health Science and Technology Project of Jiangxi Provincial Health Commission (20201034), and the Youth Science Fund of the Science and Technology Program of Second affiliated Hospital, Jiangxi Medical College, Nanchang University (2019YNQN12005).References
- O. Tenaillon, D. Skurnik, B. Picard and E. Denamur, Nat. Rev. Microbiol., 2010, 8, 207–217 CrossRef CAS PubMed.
- B. Pakbin, W. M. Brück and J. W. A. Rossen, Int. J. Mol. Sci., 2021, 22, 9922 CrossRef CAS PubMed.
- C. D. Köhler and U. Dobrindt, Int. J. Med. Microbiol., 2011, 301, 642–647 CrossRef PubMed.
- M. R. Asadi Karam, M. Habibi and S. Bouzari, Mol. Immunol., 2019, 108, 56–67 CrossRef CAS PubMed.
- H. Chang, J. Guo, Z. Wei, Z. Huang, C. Wang, Y. Qiu, X. Xu and M. Zeng, PLoS One, 2021, 16, e0249888 CrossRef CAS PubMed.
- D. Z. Uslan, S. J. Crane, J. M. Steckelberg, F. R. Cockerill 3rd, J. L. St Sauver, W. R. Wilson and L. M. Baddour, Arch. Intern. Med., 2007, 167, 834–839 CrossRef.
- R. Zhang, L. Liu, H. Zhou, E. W. Chan, J. Li, Y. Fang, Y. Li, K. Liao and S. Chen, EBioMedicine, 2017, 19, 98–106 CrossRef.
- R. Han, Q. Shi, S. Wu, D. Yin, M. Peng, D. Dong, Y. Zheng, Y. Guo, R. Zhang and F. Hu, Front. Cell. Infect. Microbiol., 2020, 10, 314 CrossRef CAS PubMed.
- A. Martin, K. Fahrbach, Q. Zhao and T. Lodise, Open Forum Infect. Dis., 2018, 5, ofy150 CrossRef PubMed.
- M. D. Zilberberg, B. H. Nathanson, K. Sulham, W. Fan and A. F. Shorr, BMC Infect. Dis., 2017, 17, 279 CrossRef PubMed.
- S. Conlan, P. J. Thomas, C. Deming, M. Park, A. F. Lau, J. P. Dekker, E. S. Snitkin, T. A. Clark, K. Luong, Y. Song, Y. C. Tsai, M. Boitano, J. Dayal, S. Y. Brooks, B. Schmidt, A. C. Young, J. W. Thomas, G. G. Bouffard, R. W. Blakesley, J. C. Mullikin, J. Korlach, D. K. Henderson, K. M. Frank, T. N. Palmore and J. A. Segre, Sci. Transl. Med., 2014, 6, 254ra126 CrossRef PubMed.
- M. Kai, U. Effmert, G. Berg and B. Piechulla, Arch. Microbiol., 2007, 187, 351–360 CrossRef CAS PubMed.
- S. Schulz and J. S. Dickschat, Nat. Prod. Rep., 2007, 24, 814–842 RSC.
- M. Bunge, N. Araghipour, T. Mikoviny, J. Dunkl, R. Schnitzhofer, A. Hansel, F. Schinner, A. Wisthaler, R. Margesin and T. D. Märk, Appl. Environ. Microbiol., 2008, 74, 2179–2186 CrossRef CAS PubMed.
- J. S. Dickschat, T. Martens, T. Brinkhoff, M. Simon and S. Schulz, Chem. Biodiversity, 2005, 2, 837–865 CrossRef CAS.
- W. Filipiak, K. Żuchowska, M. Marszałek, D. Depka, T. Bogiel, N. Warmuzińska and B. Bojko, Front. Mol. Biosci., 2022, 9, 1019290 CrossRef CAS PubMed.
- Z. Xing, D. Zogona, T. Wu, S. Pan and X. Xu, Food Chem., 2023, 415, 135650 CrossRef CAS PubMed.
- S. P. Putri, M. M. M. Ikram, A. Sato, H. A. Dahlan, D. Rahmawati, Y. Ohto and E. Fukusaki, J. Biosci. Bioeng., 2022, 133, 425–435 CrossRef CAS PubMed.
- P. G. Tratnyek, E. Edwards, L. Carpenter and S. Blossom, Environ. Sci.: Processes Impacts, 2020, 22, 465–471 RSC.
- W. Vautz, J. Nolte, A. Bufe, J. I. Baumbach and M. Peters, J. Appl. Physiol., 2010, 108, 697–704 CrossRef PubMed.
- C. Drees, W. Vautz, S. Liedtke, C. Rosin, K. Althoff, M. Lippmann, S. Zimmermann, T. J. Legler, D. Yildiz, T. Perl and N. Kunze-Szikszay, Appl. Microbiol. Biotechnol., 2019, 103, 9091–9101 CrossRef CAS.
- H. Luo, Y. Hang, H. Zhu, Q. Zhong, S. Peng, S. Gu, X. Fang and L. Hu, Infect. Drug Resist., 2023, 16, 2601–2609 CrossRef CAS PubMed.
- X. Dou, L. Zhang, R. Yang, X. Wang, L. Yu, X. Yue, F. Ma, J. Mao, X. Wang and P. Li, Food Chem., 2022, 370, 131373 CrossRef CAS.
- W. Filipiak, A. Sponring, M. M. Baur, A. Filipiak, C. Ager, H. Wiesenhofer, M. Nagl, J. Troppmair and A. Amann, BMC Microbiol., 2012, 12, 113 CrossRef CAS PubMed.
- K. H. Kim, J. E. Szulejko, Y. H. Kim and M. H. Lee, Sci. World J., 2014, 2014, 308405 Search PubMed.
- M. V. Farraia, J. Cavaleiro Rufo, I. Paciência, F. Mendes, L. Delgado and A. Moreira, Porto Biomed. J., 2019, 4, e42 CrossRef PubMed.
- H. Zhu, D. Zhu and J. Sun, Front. Nutr., 2023, 10, 1247695 CrossRef PubMed.
- P. C. Moura and V. Vassilenko, Eur. J. Mass Spectrom., 2023, 29, 231–239 CrossRef CAS.
- Y. Lu, L. Zeng, M. Li, B. Yan, D. Gao, B. Zhou, W. Lu and Q. He, AMB Express, 2022, 12, 31 CrossRef CAS PubMed.
- M. P. D. Zahniser, S. Prasad, M. M. Kneen, C. A. Kreinbring, G. A. Petsko, D. Ringe and M. J. McLeish, Protein Eng., Des. Sel., 2017, 30, 271–278 CrossRef PubMed.
- S. Létoffé, B. Audrain, S. P. Bernier, M. Delepierre and J. M. Ghigo, mBio, 2014, 5, e00944 CrossRef PubMed.
- H. Devaraj, C. Pook, S. Swift, K. C. Aw and A. J. McDaid, J. Sep. Sci., 2018, 41, 4133–4141 CrossRef CAS PubMed.
- E. Tait, J. D. Perry, S. P. Stanforth and J. R. Dean, J. Chromatogr. Sci., 2014, 52, 363–373 CAS.
- Z. Chen, H. Sun, J. Huang, Y. Wu and D. Liu, PLoS One, 2015, 10, e0140508 CrossRef PubMed.
- N. Karami, F. Mirzajani, H. Rezadoost, A. Karimi, F. Fallah, A. Ghassempour and A. Aliahmadi, F1000Research, 2017, 6, 1415 Search PubMed.
- N. Karami, A. Karimi, A. Aliahmadi, F. Mirzajan, H. Rezadoost, A. Ghassempour and F. Fallah, Cell. Mol. Biol., 2017, 63, 112–121 CrossRef CAS PubMed.
- M. P. Romero-Gómez and J. Mingorance, J. Infect., 2011, 62, 251–253 CrossRef.
- S. Ranjitkar, F. Reck, X. Ke, Q. Zhu, G. McEnroe, S. L. Lopez and C. R. Dean, mSphere, 2019, 4, e00074 CrossRef CAS.
- S. Fitzgerald, L. Holland and A. Morrin, Front. Microbiol., 2021, 12, 693075 CrossRef PubMed.
- N. Drabińska, K. Hewett, P. White, M. B. Avison, R. Persad, N. M. Ratcliffe and B. de Lacy Costello, Adv. Med. Sci., 2022, 67, 1–9 CrossRef PubMed.
- A. Smart, B. de Lacy Costello, P. White, M. Avison, C. Batty, C. Turner, R. Persad and N. Ratcliffe, J. Pharm. Biomed. Anal., 2019, 167, 59–65 CrossRef CAS PubMed.
- W. Filipiak, A. Sponring, M. M. Baur, C. Ager, A. Filipiak, H. Wiesenhofer, M. Nagl, J. Troppmair and A. Amann, Microbiology, 2012, 158, 3044–3053 CrossRef CAS PubMed.
- C. A. Rees, A. Smolinska and J. E. Hill, J. Breath Res., 2016, 10, 027101 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03601h |
| ‡ Yunwei Zheng and Fuxing Li contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |