Open Access Article
Open Access ArticleTackling breast cancer with gold nanoparticles: twinning synthesis and particle engineering with efficacy
Suvadeep
Mal
 a,
Subhasis
Chakraborty
b,
Monalisa
Mahapatra
a,
Subhasis
Chakraborty
b,
Monalisa
Mahapatra
 a,
Kakarla
Pakeeraiah
a,
Kakarla
Pakeeraiah
 a,
Suvadra
Das
c,
Sudhir Kumar
Paidesetty
a,
Suvadra
Das
c,
Sudhir Kumar
Paidesetty
 *a and
Partha
Roy
*a and
Partha
Roy
 *d
*d
aMedicinal Chemistry Research Laboratory, School of Pharmaceutical Sciences, Siksha ‘O’ Anusandhan (Deemed to be University), Campus-2, Ghatikia, Kalinga Nagar, Bhubaneswar, Odisha 751003, India. E-mail: sairampaidesetty@gmail.com
bDmbH Institute of Medical Science, Dadpur, Hooghly, West Bengal 712305, India
cBasic Science and Humanities Department, University of Engineering and Management, Action Area III, B/5, Newtown, Kolkata, West Bengal 700160, India
dGITAM School of Pharmacy, GITAM (Deemed to be University), Vishakhapatnam, 530045, India. E-mail: partharoy2502@gmail.com
First published on 17th April 2024
Abstract
The World Health Organization identifies breast cancer as the most prevalent cancer despite predominantly affecting women. Surgery, hormonal therapy, chemotherapy, and radiation therapy are the current treatment modalities. Site-directed nanotherapeutics, engineered with multidimensional functionality are now the frontrunners in breast cancer diagnosis and treatment. Gold nanoparticles with their unique colloidal, optical, quantum, magnetic, mechanical, and electrical properties have become the most valuable weapon in this arsenal. Their advantages include facile modulation of shape and size, a high degree of reproducibility and stability, biocompatibility, and ease of particle engineering to induce multifunctionality. Additionally, the surface plasmon oscillation and high atomic number of gold provide distinct advantages for tailor-made diagnosis, therapy or theranostic applications in breast cancer such as photothermal therapy, radiotherapy, molecular labeling, imaging, and sensing. Although pre-clinical and clinical data are promising for nano-dimensional gold, their clinical translation is hampered by toxicity signs in major organs like the liver, kidneys and spleen. This has instigated global scientific brainstorming to explore feasible particle synthesis and engineering techniques to simultaneously improve the efficacy and versatility and widen the safety window of gold nanoparticles. The present work marks the first study on gold nanoparticle design and maneuvering techniques, elucidating their impact on the pharmacodynamics character and providing a clear-cut scientific roadmap for their fast-track entry into clinical practice.
1. Introduction
Cancer ranked as the leading global mortality cause in 2020, characterized by the typical presence of uncontrolled cell growth, angiogenesis and metastasis. According to the WHO factsheet, breast cancer (BC) ranks as the most commonly diagnosed with 2.26 million cases, surpassing all other cancers such as lung (2.21 million), colorectal (1.93 million), prostate (1.41 million), skin (1.20 million) and stomach (1.09 million) cancer, irrespective of gender specificity. The fatality of this variant makes it the second leading cause of cancer death (685![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths globally in 2020) in women after lung cancer.1 Most BCs originate as benign fibrocysts in different parts of the breast that become malignant and start to metastasize.2,3
000 deaths globally in 2020) in women after lung cancer.1 Most BCs originate as benign fibrocysts in different parts of the breast that become malignant and start to metastasize.2,3
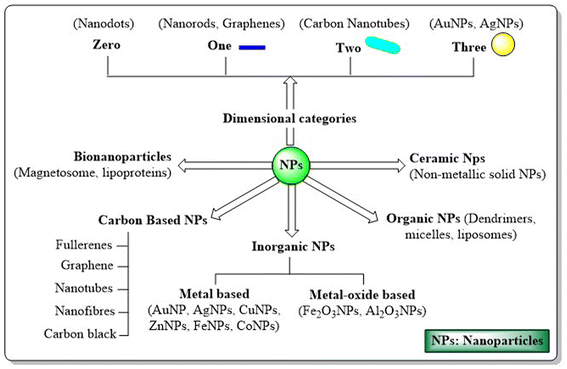
Therapeutic nanoparticles (NPs) display unique colloidal, optical, quantum, magnetic, mechanical and electrical properties depending on their size, shape, surface area and surface charge, resulting in major breakthroughs in therapeutic interventions.4 The spotlight on cancer management reveals the advantages of NPs, including transporting and shielding high drug payloads, ease of engineering with various targeting ligands, a facile route to induce multi-functionality, accommodation of drug cocktails, tuning drug release to map the intended mechanism or pharmacodynamics and their unique ability to evade clinical challenges like multidrug resistance.5,6 Based on dimensions, NPs can be divided into zero, one, two or three dimensional frameworks. Based on their morphology and chemical properties, NPs are classified as metal nanoparticles, bio-nanoparticles, organic nanoparticles, polymeric nanoparticles, solid lipid NPs, ceramic NPs, carbon-base NPs, quantum dots etc.7 (Fig. 1).
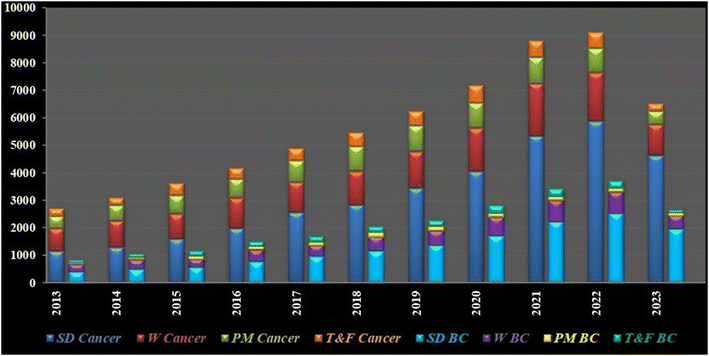
Granulations of gold or gold nuggets can be traced back to as early as 3000 BC and thereafter its increasing applications led to its reputation as the “elixir of life,” believed to bestow eternal life. In ancient times, gold was the remedy for a myriad of clinical dysfunctions. Later, in the year 1848, the “California Gold Rush” began when James W. Marshall reported the discovery of gold and its true value at Colona, California.8 Gold nanoparticles (AuNPs) in clinical applications can be traced back over 150 years with similar increasing trends of scientific focus (Fig. 2) due to its inertness along with facile fabrication and functionalization and specific optical or physico-chemical attributes like localized surface plasmon resonance.9 However, tackling breast cancer with AuNPs captured scientific curiosity a little later compared to their intervention in other cancer variants.
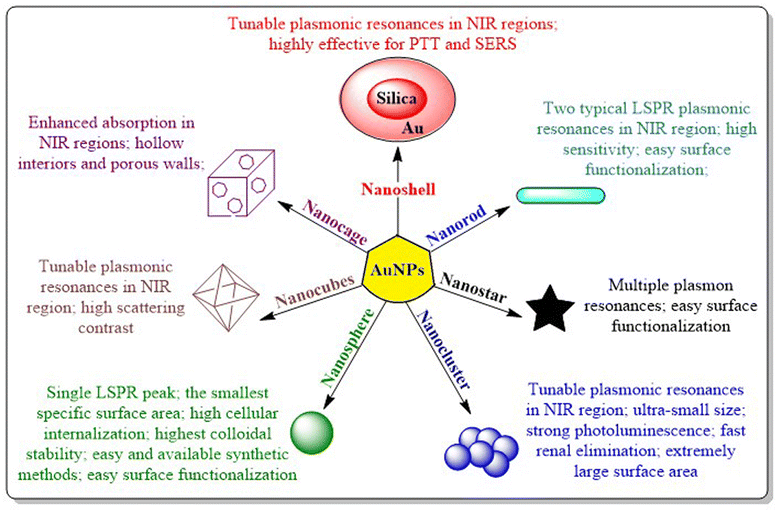
AuNPs consist of a gold atom (Au) in the inner core decorated by a negative charge at the surface. Particle engineering of AuNPs with different biomolecules (proteins, enzymes or DNA) assures the site-specific localization of NPs with programmed delivery of drug cargo.10 AuNPs exist in diverse morphologies viz. nanospheres, nanoclusters, nanorods, nanocubes, nanocages, nanostars or nanoshells (Fig. 3).11 Some morphologies are more prominent in a specific pharmacological response such as targeted delivery,10 photodermal and photothermal therapy,12 SERS based imaging,13 photo-electronics, clinical management of cancer,14 microbial infections,15 contrast agents and field enhancers,16 chemical and biochemical sensors17 and as radiosensitizers.18
AuNPs allow easy association of site-directed ligands, imaging probes, therapeutic bioactive materials and other functional moieties to facilitate diagnostic, therapeutic or theranostic applications in cancer at the molecular level.19 Engineering of AuNPs has become a mandatory art to favor the intended clinical application. Various mechanisms for the synthesis of AuNPs containing functional moieties are being explored either to increase their bonding with biological molecules or to make them better drug-carriers with improved specificity. Modern trends in the tuning of AuNPs involve the use of one or a combination of the functional moieties like polyethylene glycol (PEG), bovine serum albumin (BSA), amino acids and polypeptides, oligonucleotides, antibodies or biomarkers, which are discussed in this review for management of BC.
AuNPs have distinct advantages depending on their properties and intended use. The most important property of AuNPs in clinical translation is biocompatibility as they are less likely to induce immune responses or toxicity as compared to other nanoparticles viz. copper oxide, zinc or cobalt NPs. The facile feasibility of AuNPs for convenient surface modification is another major advantage in applications like targeted drug delivery and molecular sensing. While other nanomaterials such as silver, copper, or iron oxide nanoparticles also have surface modification capabilities, they typically require additional ligands or modifiers for surface applications. Without the need for surface engineering, AuNPs also possess distinctive optical characteristics, notably localized surface plasmon resonance (LSPR), which can be explored for diverse applications including biosensing, imaging, and photothermal therapy, making AuNPs superior to other nanoparticles.20 With their higher atomic number and robust scattering and absorption properties, AuNPs offer various bioimaging modalities such as optical, photoacoustic, and computed tomography (CT) imaging which adds to their clinical value.
Beside all these, AuNPs have excellent chemical and thermal stability, making them suitable for applications requiring high temperatures (photothermal or radiotherapy) with high drug loading capacity. AuNPs offer similar electrical conductivity to metal oxide nanoparticles (NPs), while possessing the additional advantage of being resistant to oxidation.21 This property ensures their efficacy and performance in harsh environments, contrasting with iron oxide or copper oxide NPs which are prone to oxidation, potentially compromising their effectiveness and durability over time.22
Diverse strategies exist in combating BC using AuNPs. AuNPs can be engineered to act as targeted drug delivery vehicles to deliver therapeutics to BC cells safeguarding the healthy cells. AuNPs can be used as sensors for probing and imaging BC cells using surface enhanced Raman spectroscopy (SERS). AuNPs can tackle BC through plasmonic photothermal therapy (PPTT), radiotherapy or by preventing angiogenesis and metastasis.23 AuNPs have proved effective in critical clinical challenges like triple negative breast cancer (TNBC) by interacting with over-expressed cell-surface receptors which are not connected with cell proliferation or survival.24 Engineered AuNPs successfully delivered drugs and induced apoptosis even in multi-drug resistant BC cells.25 The acceptance of AuNPs in clinical applications is further intensified due to their safety towards normal cells.26
However, although bulk gold acts as a noble metal, AuNPs do exhibit toxicities in different systems depending on their size and shape, particle engineering, dosage, route of administration, exposure time etc. Increased cellular uptake and low bio-distribution often lead to long term toxicity, which is considered to be a major hurdle for nano-gold formulations. Although several AuNPs have translated into pre-clinical and clinical stages, none of them have yet progressed to the point of entering the market. As such, researchers are coming up with newer strategic manoeuvres to address their drawbacks. This review summarizes the entire portfolio of nano-scale gold, covering synthesis routes, stabilization techniques, particle engineering, toxicity, and pre-clinical and clinical developments to establish their role in BC management either as therapeutics, diagnostics or theranostics. Although numerous reports have successfully confirmed the clinical backdrop of exploring gold nanoparticles in BC, scientific insights into their formulation perspectives to arrive at tailor-made solutions remain unexplored. The present work addresses this niche area to provide a comprehensive scientific databank to assist fast-track clinical translation of nano-scale gold.
2. AuNP synthesis and stabilization
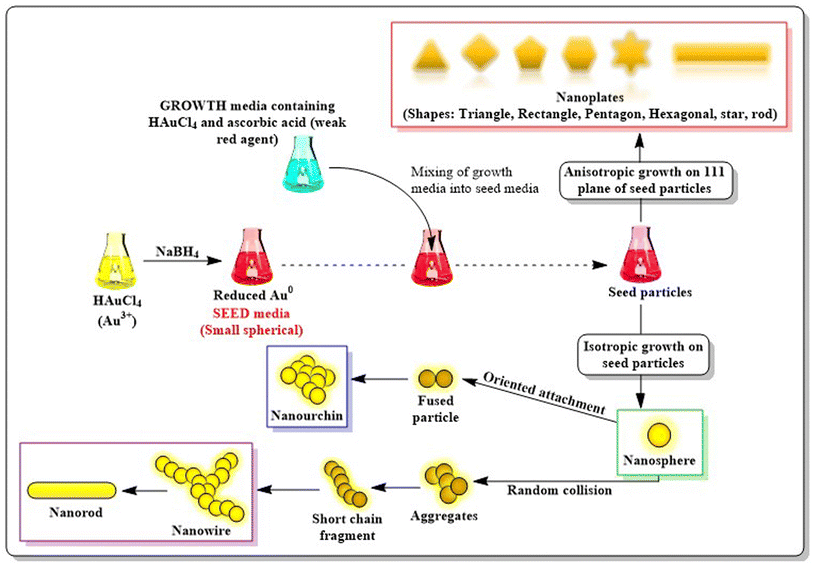
Basic strategies for nanoparticles synthesis revolve around two pivotal pillars: the “top-down” method and the “bottom-up” method. Top-down methods involve destruction, in which bulk materials are broken into smaller particles to produce nanoparticles by thermal decomposition,27 ball milling,28 laser ablation,29 sputtering,30 UV and IR irradiation31 and aerosol technology.32 On the contrary, bottom-up synthesis is constructive i.e. nanoparticles are synthesised starting from the atomic level using different techniques such as the sol–gel approach,33 spinning,34 chemical vapour deposition35 and green biological synthesis using plant materials, extracts, and microorganisms.36,37 The formation of AuNPs generally follows a bottom-up synthesis route consisting of two steps: first the reduction of bulk gold precursor (Au3+) is carried out with the help of reducing agents, followed by stabilization of the AuNPs formed with specified capping agents, which helps in restricting further agglomeration of the nanoparticles. Several synthetic approaches are available to synthesize AuNPs, which are discussed as follows.2.1. AuNP synthesis via chemical methods
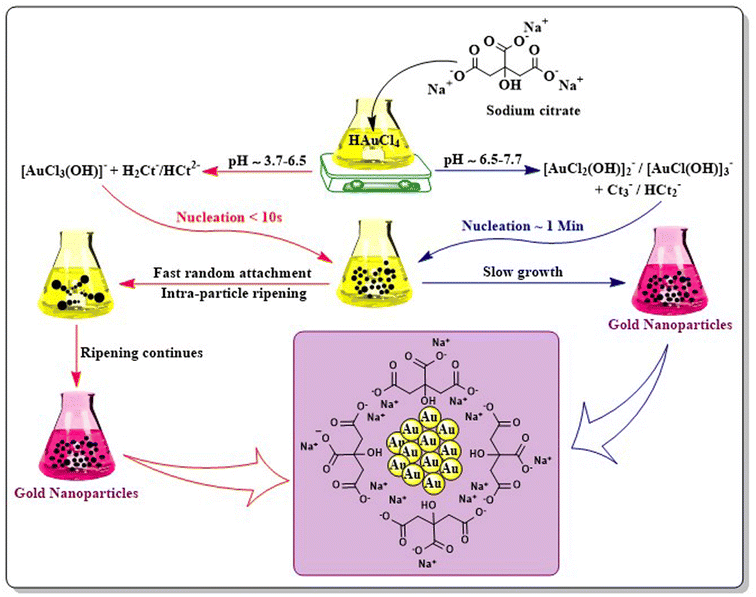
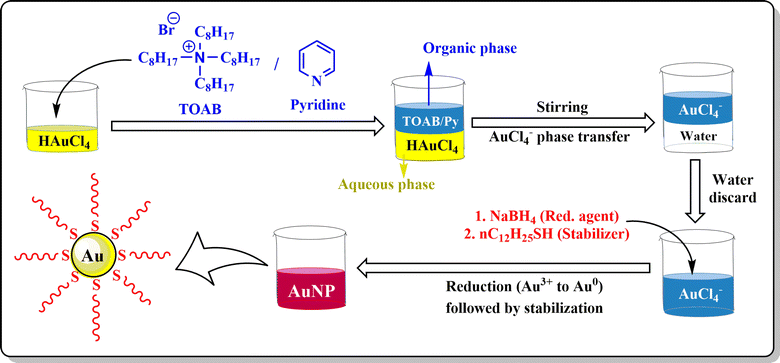
Chemical synthesis of AuNPs involves reduction as well as stabilization with different chemicals using different techniques. The Turkevich method, Fren's method of modified synthesis, and the Brust–Schiffrin method are generally applied for the synthesis of AuNPs.The modification of the Turkevich method by Frens modified synthesis in 1973 successfully produced the narrow size range of AuNPs by tuning the molar ratio of citrate to Au salt.40 Several modifications were applied to the technique with different pH values to control the size and mechanism of AuNPs (Fig. 4).41 The Turkevich method generally forms spherical to quasi-spherical nanoparticles but when the particle dimension exceeds 30 nm, it tends to exert a broader size distribution pattern with considerably low yield.42
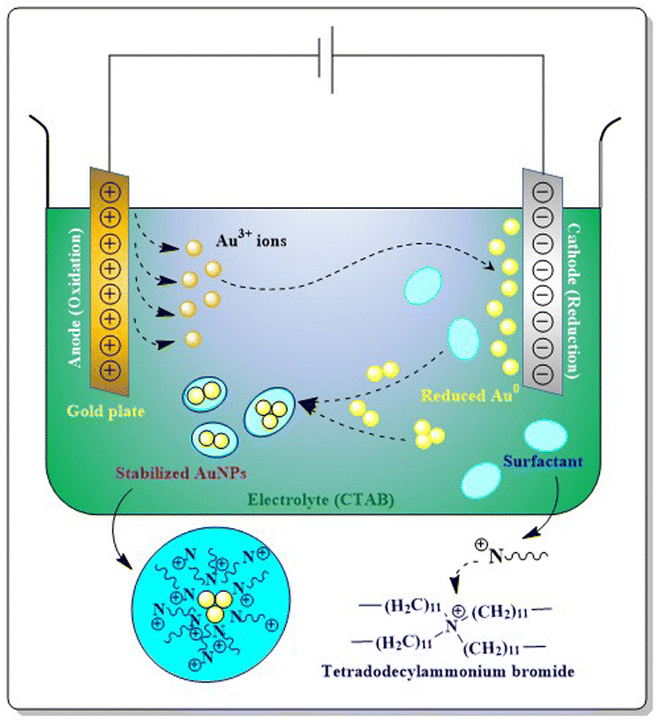
2.2. AuNP synthesis via a physical method
The physical method for AuNP synthesis involves different physical or physicochemical or electrical or photochemical approaches.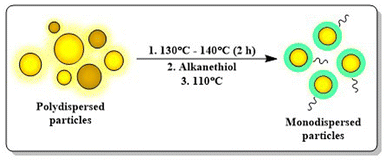 | ||
| Fig. 8 Digestive ripening in AuNP synthesis. This method is well known for making monodispersed particles using alkanethiols at an elevated temperature. | ||
Apart from all the physical methods discussed above, ultrasound mediated synthesis,26 UV-induced photochemical reduction31 or laser ablation techniques29 are also employed for the synthesis of AuNPs. Microwave53 and solvothermal reduction of HAuCl4 (ref. 54) are also considered as physicochemical approaches in synthesising AuNPs.
2.3. AuNP synthesis via a biological method
The chemical and physical methods applied for the synthesis of AuNPs are associated with expensive and toxic substances, which limit their biological applications. Biological methods on the other hand provide eco-friendly green approaches and have recently gained much more importance because of easy availability, biocompatibility, induced bio-applications, and easy and ready synthesis at normal suitable temperatures and pressure. Biological methods for AuNP synthesis can be achieved using different plants and their extracts, micro-organisms, biomolecules etc.55Nowadays, biosynthesis of AuNPs using fungi rather than using bacteria is becoming a more popular approach due to more frequent and less tedious work plans with successful outcomes. Fungi produce more enzymes than bacteria which make them more suitable for the faster conversion of metallic salts to NPs. Using fungi for the microbial synthesis of metallic nanoparticles is more advantageous than that with bacteria due to the presence of mycelia, a thread-like hyphae which causes greater surface for interaction with metallic salts.59 Yeasts and algae can also perform biological synthesis of AuNPs. Several studies have suggested that the hydroxyl and carbonyl groups present in algal biomass are responsible for the reduction as well as stabilization of AuNPs.60
Extracellular microbial or fungal enzymes like nicotinamide adenine dinucleotide (NADH) and nicotinamide adenine dinucleotide phosphate (NADPH)-reliant enzymes viz. nitrate reductase act as electron carriers essential for the conversion of Au3+ to Au0.61 Fungi also produce enzymes like acetyl xylan esterase, cellobiohydrolase D, glucosidase and β-glucosidase, hemicellulose, 3-glucanase, cell wall lytic enzyme β-1 etc. for extracellular mycosynthesis of AuNPs.62Rhodococcus or Thermomonospora sp. (actinomycetes) and Verticillium sp. (fungi) are involved in the intracellular synthesis of AuNPs. Phanerochaete chrysosporium, a fungi producing laccase enzyme, acts as an extracellular reducing agent, whereas ligninase from the same sp. helps in intracellular bioreduction of gold ions.63 Recent developments in biosynthesis of AuNPs using microorganism are summarized in Table 1.
| Genus | Microorganism | Temperature | Synthesis type | Size and shape of AuNPs | Reference |
|---|---|---|---|---|---|
| Bacteria | Enterococcus sp. RMAA | 37 °C | Intracellular | 5–10 nm; spherical | 64 |
| Vibrio alginolyticus | 40 °C | Extracellular | 50–100 nm; irregular shape | 65 | |
| Lysinibacillus odysseyi | 45 °C | Extracellular | 12–16 nm; spherical | 66 | |
| Bacillus marisflavi | r.t. | Extracellular | 14 nm; spherical | 67 | |
| Amycolatopsis sp. | 30 °C | Extracellular | 44 nm; spherical and irregular | 68 | |
| Fungi | Fusarium oxysporum | 30 °C | Extracellular | 22–30 nm; spherical or hexagonal | 69 |
| Agaricus bisporus | 80–100 °C | Extracellular | 53 nm; spherical, oval, drum-like, hexagonal, and triangular shapes | 70 | |
| Fusarium solani | 28 °C | Intracellular | 40–45 nm, spindle shape | 71 | |
| Yeast | Candida parapsilosis | 37 °C | Intracellular | < 30 nm, spherical | 72 |
| Algae | Acanthophora spicifera | 60 °C | Extracellular | < 20 nm, spherical and oval | 73 |
| Gracilaria crassa | r.t. | Extracellular | 28–36 nm; spherical | 60 | |
| Halymenia dilatata | 60 °C | Extracellular | 16 nm, triangular and spherical | 74 | |
| Chondrus crispus, Gelidium corneum and Porphyra linearis | 30 °C | Extracellular | 16.9–44.2 nm, spherical and polyhedral | 75 | |
| Bread mold | Neurospora crassa | 28 °C | Intracellular | 32 nm; spherical | 76 |
| Part of plant used | Plant | Active components | Size of AuNPs | Shape of particles | Reference |
|---|---|---|---|---|---|
| Seed | Mangifera indica | Hydroxyl and carboxyl groups in flavanoids, terpenoids, tannins | 50 nm | Spherical | 85 |
| Garcinia kola | OH (polyphenolic compounds) and NH (proteins) of phytates, tannin, oxalate, cyanate, saponins and anthraquinones | 2–17 nm | Spherical | 86 | |
| Citrus reticulata | Vitamins, citric acid, amino acids, proteins, terpenes, ascorbic acid | 11.14–32.76 nm | Spherical | 87 | |
| Parkia speciosa Hassk | OH, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–H, and C–N functional groups of polyphenol, phytosterol, and flavonoids C, C–H, and C–N functional groups of polyphenol, phytosterol, and flavonoids |
5–20 nm | Spherical | 88 | |
| Leaves | Terminalia arjuna | Arjunetin, leucoanthoc-yanidins, hydrolyzable tannins | 15–30 nm | Spherical | 89 |
| Saturejarechingeri | Geraniol, geranyl acetate, geranial and neral | 15.1 ± 3.7 nm | Spherical | 90 | |
| Cyanthillium cinereum | Phenols, flavonoids, steroids, tannins, saponins and phlobatannins | 14.90 nm | Spherical | 91 | |
| Jasminum auriculatum | Amine, hydroxyl and amide present in flavonoids, polyalcohols and terpenoids | 8–37 nm | Spherical | 92 | |
| Combretum erythrophyllum | Flavonoids, alkaloids, terpenoids, proteins, and water-soluble biomolecules | 13.20 nm | Spherical | 93 | |
| Flower | Clitoriaternatea | Flavonol and anthocyanin | 18–50 nm | Spherical | 94 |
| Jatropha integerrima | Anthocyanin, carbohydrate, coumarin, glycoside, phenol, protein, saponin, and tannin | 28–43 nm | Spherical | 95 | |
| Polianthes tuberosa L. | Amines, phenol, alcohol, ester linkages, and carboxylic acid | 38.76 nm | Spheres, triangles, pentagons, hexagons, and rods | 96 | |
| Stigma | Crocus sativus | NH or OH groups for reduction, COOH for stabilization | 25–35 nm | Spherical, rod and triangle | 97 |
| Fruit | Rosa canina | Polyphenols, flavanols, carboxylic acid, alkenes | 20 nm | Pseudo-spherical | 98 |
| Carica papaya | Flavonoids, tannins, alkaloids, polyphenols, carotenoids, papain and chymopapain | 12 ± 2.31 nm | Spherical | 99 | |
| Piper nigrum | NH or OH groups for reduction, COOH for stabilization | 40–60 nm | Spherical and oval | 100 | |
| Peel | Spondias dulcis | Carboxyl, hydroxyl, and amide groups | 36.75 ± 11.36 nm | Spherical | 101 |
| Ananas comosus and Passiflora edulis | Proteins, minerals, lipids, vitamin, phenolic compounds, flavonoids and carotenoid | 20.71 ± 7.44 nm and 18.68 ± 5.55 nm | Spherical | 102 | |
| Plant | Physalis minima | OH or NH groups found in carbohydrates or proteins | 36 nm | Cubical | 103 |
| Turnera diffusa | Hydroquinone-β-D-glucoside (arbutin) | 24 nm | Spherical | 104 | |
| Stem | Brassica oleracea var. acephala | OH and NH groups of proteins, polysaccharides or polyphenols, sulphoraphane and glucosinolates | 25.08 ± 3.73 nm | Spherical | 105 |
| Euphorbia neriifolia L. | Sugar, tannins, flavonoids, alkaloids, 24-methylene cycloartenol, triterpennoidal saponins | 23–25 nm | Spherical | 106 | |
| Root | Glycyrrhiza glabra | NH, OH and CH groups present in flavonoids, phenolics, glycosides, organic acid, proteins, amino acids and fatty acid | 2.6–16.25 nm | Spherical | 107 |
| Hemidesmus indicus L. | Proteins, lipids and polyphenols | 37 nm | Hexagonal | 108 | |
| Rhizome | Corallocarpus epigaeus | Hydroxylamine and proteins | 30 nm | Spherical | 109 |
| Kaempferia parviflora | Polymethoxy flavanones, hydroxyl and carbonyl groups | 44 ± 3 nm | Spherical | 110 | |
| Bark | Cinnamomum cassia | Terpenoids, carbohydrates, flavones, and proteins | 35 nm | Spherical | 111 |
| Salix alba L. | Hexose (fructose, glucose, mannose and xylose), tannins and mineral substances | 8–25 nm | Spherical | 112 | |
| Nut shells | Juglans regia | Reducing sugars, alkaloid, tannins, phenols and saponins | 10–50 nm | Spherical and triangular | 113 |
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and Au3+ reduces to Au0. Generally, for flavonoids, the preferred chelating site is found to be at the 3rd or 5th positioned –OH or the 4th positional carbonyl group possibly due to the comparatively lower bond dissociation energy than that of the –OH at other positions.116 In the case of reducing sugars, the electron is transferred when oxidation of aldehyde (–CHO) groups takes place to produce corresponding carboxylic acids (–COOH) in the synthesis of nanoparticles.117
O) and Au3+ reduces to Au0. Generally, for flavonoids, the preferred chelating site is found to be at the 3rd or 5th positioned –OH or the 4th positional carbonyl group possibly due to the comparatively lower bond dissociation energy than that of the –OH at other positions.116 In the case of reducing sugars, the electron is transferred when oxidation of aldehyde (–CHO) groups takes place to produce corresponding carboxylic acids (–COOH) in the synthesis of nanoparticles.117
| Phytometabolite | Class | Size of AuNPs | Shape of AuNPs | Activity | Reference |
|---|---|---|---|---|---|
| Epigallocatechin-3-gallate (EGCG) | Catechin | 135 and 39 nm | Sea-urchin (135 nm) and spherical (39 nm) | Uptake increases on HeLa (135 nm AuNPs) and MDA-MB-231 cells (with 39 nm AuNPs) | 119 |
| Kaempferol 3-O-β-D-apiofuranosyl-7-O-α-L-rhamnopyranoside | Flavonol linked with sugar | 37 ± 11 nm | Spherical | Catalytic, antioxidant and anticancer activities | 120 |
| Arbutin | Glycoside | 10.30–17.13 nm | Spherical | Enhanced anti-inflammatory activity and whitening capabilities | 121 |
| Apigenin | Flavonoid (tri-hydroxy flavone) | 21.4 ± 11.6 nm | Not mentioned | Cardioprotective activity against Dox-induced cardiotoxicity. Boost myocardial performance | 122 |
| Chrysin | Dihydroxy flavone | 32–38 nm | Spherical | Anti-cancer, antioxidant and anti-microbial | 123 |
| Quercetin | Flavonoid (flavonol) | 20–50 nm | Spherical | Anticancer activities against A549 and HeLa | 124 |
| Curcumin | Phenolic pigments (diarylheptanoid) | 18 ± 3.3 nm | Spherical | ROS mediated apoptosis in MCF-7 cancer cells | 125 |
| Resveratrol | Polyphenolic phytoalexin (stilbenoid) | 11.9 ± 3.1 nm | Spherical | Caspase mediated apoptosis in pancreatic cancer cells BxPC-3 | 126 |
| Baicalein | Trihydroxyflavone | 20–40 nm | Spherical | Anti-cancer, antioxidant and anti-microbial | 127 |
| Genistein | 7-Hydroxyisoflavone | 10–23 nm | Spherical | Anti-oxidative and anti-proliferative activity in prostate cancer cell lines | 128 |
| Luteolin | Tetrahydroxyflavone | 25–30 nm | Spherical | Anti-cancer activity against TNBC cell line | 26 |
| Naringenin | Flavanone | 12.53 nm | Spherical | Anti-cancer activity against prostate cancer | 129 |
| Anthracene | Alkaloid | 76 nm (DLS) | Not mentioned | 1O2 generation and local X-ray dose enhancement efficacy | 130 |
| Phenylalanine conjugated cholic acid | Protein | 21.4 nm | Spherical | Colorimetric probe for the detection of Hg2+, Pb2+ and Cr6+ ions | 31 |
| Methionine | Protein | 13 nm | Spherical | Tumor targeting efficacy and SPECT-based imaging capacity | 131 |
| Cysteine, tyrosine, tryptophan | Protein | 8–27 nm | Spherical | Increased cellular uptake by HeLa cancer cells | 132 |
| Histidine | Protein | 3 ± 0.3 nm | Spherical | Novel detection platform for barbaloin and temperature sensor | 133 |
| Calix[4]resorcinarene | Cyclic oligomer | 20 nm | Spherical, hexagonal, rods | Selective and sensitive fluorescent probe for copper and leucine | 134 |
| Thymol | Terpenoid | 10 nm | Granular | Reduce toxicity of bacteria resistant last-resort antibiotics | 135 |
| Cinnamic acid | Organic acid | 89 nm | Spherical | Anti-amoebic activity against Naegleria fowleri | 136 |
| Ferulic acid | Hydroxycinnamic acid | 34.2 ± 1.3 nm | Spherical | Inducing ROS mediated apoptosis on skin cancer cells (A431) by increasing MMP | 137 |
| Chlorogenic acid | Hydroxycinnamic acid | 35–40 nm | Flower-shaped | Antioxidant activity | 138 |
| Gallic acid | Phenolic acid | 14.1 ± 2.2 nm | Spherical | Multifunctional theranostic agent with Doxorubicin loading capacity | 139 |
| Rosmarinic acid | Polyphenolic ester | 100 nm | Spherical or quasi-spherical shape | NIR-mediated PTT system for the treatment of breast cancer | 140 |
While synthesizing nano-gold, determining the optimized conditions i.e. optimal concentration, temperature, reaction time or pH should be done very carefully as these factors significantly affect the physical, morphological and even chemical characteristics of the synthesized particles. In our previous study with luteolin engineered AuNPs, we found that even with a small change in optimized concentration (molar ratio), temperature or reaction time, particle size and zeta potential values change drastically.26 The reason behind the controlled NP properties with respect to these factors are well explained by Zuhrotun et al. (2023).116 Kuppusamy et al. (2016) reported that the number of hydrogen ions in the hydroxyl group possessed by a phytochemical could affect the size and shape of synthesized NPs.118
2.4 Stabilization of the synthesised AuNPs
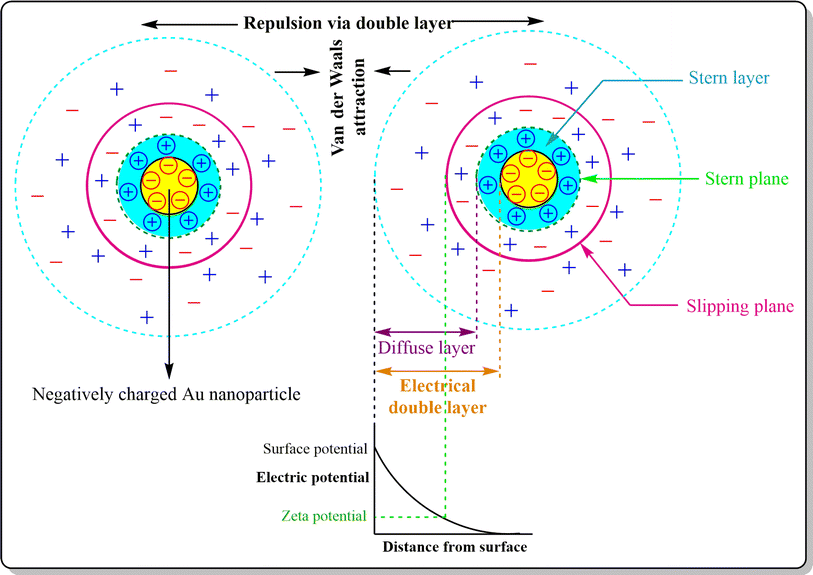
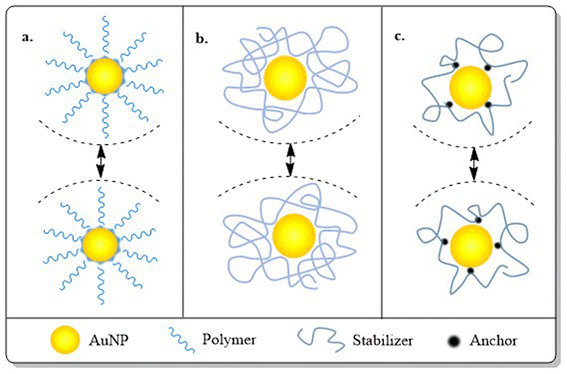
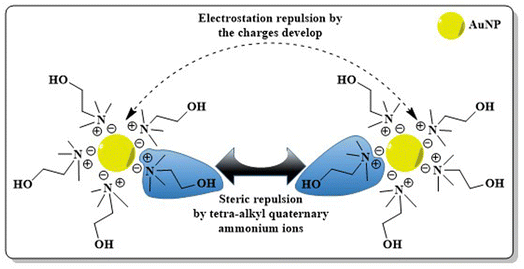
After synthesis, metallic NPs possess high surface energy (up to 2000 mJ m−2) due to their surface area enhancement resulting in thermodynamic instability.141 By their nature, NPs undergo aggregation which reduces the interfacial tension of nanoconjugates via decreased surface area and effective volume fraction, thus compensating for the disordered circumstances of NPs in a colloidal state. Despite the relative high surface area and high energy, van der Waals forces serve as the main attractive force which leads to nanoparticle aggregation, with co-ordination unsaturation and unstable atomic environment as other proven contributors. To perform the stabilization of colloidal NPs, stabilizing agents, capping agents come into play which mainly work via creating repulsive barrier between the particles to overcome the influence of van der Waals forces among the particles.142 The stabilization process of NPs can be achieved by three different stabilization phenomena i.e. electrostatic, steric and electrosteric techniques.3. Engineering AuNPs with diverse moieties for BC management
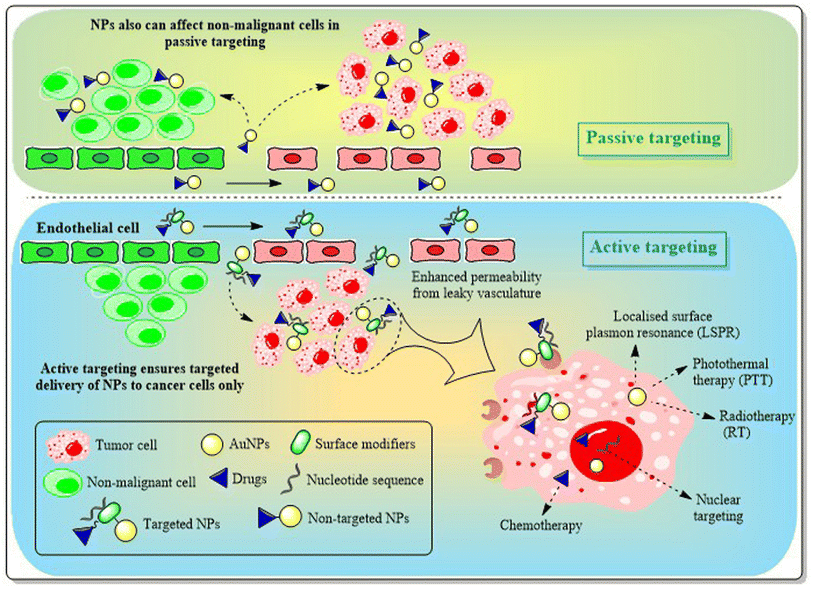
The primary limitation of AuNPs without functionalization lies in their in vivo drug delivery applications. AuNPs with their ease of functionalization offer remarkable clinical opportunities, which provides solution to the problems present in first generation AuNPs.150 Engineering AuNPs with different modifiers like PEG, antibodies, proteins, oligonucleotides, sugars etc. ensures selective or site-specific targeting approaches and improved pharmacokinetics with the desired renal clearance.Targeting AuNPs in cancer cells without hampering healthy cells can be achieved through two different targeting approaches i.e. passive and active targeting. Tumor cells possess larger vasculature gaps (100 nm–2 μm) than normal cells. Being nano-sized, AuNPs can easily penetrate through these gaps and are retained in tumor cells for a much longer time, as tumor cells possess disorganized lymphatic systems, restricting lymphatic clearance and resulting in “enhanced permeability and retention” (EPR) effects. Passive targeting of AuNPs into cancer cells exploring EPR facilitates selective targeting and improved retention assisting in enhanced therapeutic efficacy151,152 (Fig. 12). Though passive targeting ensures preferential accumulation of AuNPs in tumors, active targeting offers an enhanced therapeutic response with maximized chances of protecting healthy tissues compared to the passive approach. Attaching different targeting ligands like antibodies, cell surface markers, oligonucleotide sequences, or proteins with AuNPs helps with targeting and interacting with specific receptors over-expressed on the BC cell surface152 (Fig. 12). Thus, the active targeting technique follows a dual approach, one through EPR and the other through ligand–receptor interactions.
Modification of AuNPs with different ligands requires suitable attachment either by covalent or by non-covalent interaction between the particle and the ligands. Gold has a strong gold–thiol (Au–S) bond. Love et al. (2005) calculated Au–S bond energies between Au–thiol and Au–disulfide, in which they reported Au–S (disulfide) to be stronger (∼60 kcal mol−1) than Au–S (thiol) (∼45 kcal mol−1).153 On the other hand, non-covalent interaction can be classified as two types-electrostatic attachment or by layer-by-layer (LbL) assembly. Electrostatic attachment/adsorption generally involves attachment of antibodies, DNAs or proteins. Proteins being zwitterionic biomolecules, possess charge when a suitable pH system with a similar isoelectric point is applied. The developed charged molecules are absorbed on the charged surface of AuNPs easily.154 Electrostatic adsorption can alter, weaken or even denature the protein structure, which plays a disadvantageous role along with weak interaction. LbL assembly of multiple polymers with alternative charge allows AuNPs to bear tunable surface charges or enables introduction of hydrophobic or lipophilic regions in AuNPs, thus ensuring increased solubility and clearance of drugs with low solubility.155
3.1. Engineering AuNPs with polymers or PEG
The main hurdle AuNPs face in biological applications is their opsonization. The reticuloendothelial system (RES) engulfs the opsonized NPs by phagocytosis which limits their targeting approach. PEG is the polymer most commonly used to functionalize AuNPs to achieve a stealth effect.156 PEG can be attached directly with AuNPs or through a linker i.e. thiols. In a study, it was suggested that thiol-PEGylated AuNPs not only offered cargo (Tamoxifen) delivery but also increased intracellular drug import which led to 2.7 fold more potency of the drug in estrogen receptor positive BC cells.157 Along with stabilization, PEG block polymer can also be used as a reducing agent to synthesize AuNPs. Sarkar et al. (2017) used a non-toxic, non-ionic amphiphilic tri-block copolymer made up of PEG and polypropylene glycol (PPG) (PEG–PPG–PEG) as a reducing and stabilizing agent for the synthesis of Au-nanomicelles. The nanomicelle was further conjugated with ZD6474 (a tyrosine kinase inhibitor) and the nano-conjugate was targeted to tumor cells in MDA-MB-231 xenograft mice models where the authors observed decreased tumor size with promising conclusions.158 In a recent study, Wang et al. (2021) also synthesized Au-nanorods functionalized with copolymer PLGA–PEG. The biodegradable and biocompatible copolymeric system carried water insoluble Paclitaxel to the targeted vicinity.159 Mahalunkar et al. (2015) used another polymer polyvinylpyrrolidone (PVP) to functionalize with curcumin–gold nanoconjugate and further tagged with folic acid (FA). FA–curcumin–PVP–AuNC exhibited promising in vivo anti-cancer activity without any significant toxicity.160 Some of the recent studies of polymer functionalized Au-nanosystems for the treatment of BC are given in Table 4.| Functionalization system | Coating agent or ligand | Conjugated system | Synthesis/conjugation technique | Key features of the functionalized ligand | Main outcome delivered by the conjugated system | Reference |
|---|---|---|---|---|---|---|
| Polymers | PEG | Triptorelin (TRP)-functionalized PEG-AuNPs | Layer by layer assembly | Adhesion with plasma membrane | Specific targeting achieved through TRP-LHRH interaction present in TNBC surface | 197 |
| AuNPs-KT2-PEG conjugates | Covalent technique | Increases colloidal stability of KT2 peptide | Rapid internalization and increased cytotoxicity in MDA-MB-231 cell line | 198 | ||
| PVP | PVP-co-2-dimethylaminoethyl methacrylate modified celastrol-folic acid-AuNPs | PVP capping induced acid–base reaction | High encapsulating efficiency, improves solubility and loading content | Induced cellular uptake, apoptosis with low colony-forming assay unit in 2D and 3D breast cancer models | 199 | |
| PLGA | AuNP encapsulated PLGA modified nanoclusters | Single step nano-precipitation method | Encapsulation of AuNPs into polymer nanoconstructs enhance Specific Absorption Rate (SAR) value | Photothermal ablation in SUM159 BC cells | 200 | |
| Poly(ethylimine) (PEI) | PEI–PEG-AuNPs | Layer by layer assembly | Low mol wt of PEI reduce toxicity, helps in delivering gapmers in cancer cells | Reduce TNBC cell proliferation, reduce Gemcitabin resistance in mutant p53 cancer cells (MDA-MB-231) | 201 | |
| Polylysine | Poly-L-lysine (PLL)-AuNS-miRNA | Layer by layer assembly | Deliver miR-34a to TNBC cells | Suppress proliferation | 202 | |
| Peptides and proteins | Albumin (HSA or BSA) | Gd2O3@BSA-Au NPs | Chemical reduction | Forms metal ion complex with Au and Gd during synthesis and enhanced stability observed | Enhanced tumor suppression when exposed to X-ray radiation (5 Gy) in 4T1 bearing mice | 203 |
| HSA-AuNPs-TMX | Chemical reduction | Template to fabricate AuNPs with better TMX delivery | Dose dependent cytotoxicity in BT-474 and MDA MB-231 BC cell line | 163 | ||
| Glutamine | Glucose-BSA-AuNPs, glutamine-BSA-AuNPs, and folic acid-BSA-AuNPs | Chemical conjugation | Increase biocompatibility, better tumor targeting efficacy | Glutamine-BSA-AuNPs showed better targeting, better gold accumulation and enhanced radiosensitizing property in 4T1 bearing BALB/c mice | 204 | |
| Folate | AuNP-CS-FA-His | Carbodiimide coupling | CS induced stability, histidine enhanced endosomal escape and folic acid for targeting | Less cytotoxicity observed. Conjugation of His and FA to Au-CS produced a significant increase in the luciferase activity in SKBR-3 and MCF-7 cells, respectively | 205 | |
| RGD, TAT, NLS | DOX, RGD and nuclear localization peptides (NLS) tagged AuNPs (DRN AuNPs) | Chemical reduction | Targeting- RDG ensures cell penetration, NLS ensures cargo release inside nucleus | More efficacy observed in HeLa cells than in MCF-7 | 206 | |
| RGD-Gd@Au core–shell tecto dendrimer CSTDs-PS-nanocomplex | Carbodiimide coupling | Targeting specificity to αvβ3 integrin-overexpressing tumors because of RDG | Enhanced CT/MR imaging of 4T1 BC model in vivo and metabolism capability with good biosafety | 207 | ||
| 111In-Match-AuNP-Tat | Carbodiimide coupling | TAT increased nuclear localization | DNA double-strand breaks and caused a dose-dependent reduction in clonogenic survival of telomerase-positive (MDA-MB-435) cells | 208 | ||
| EGF | EGF-luteolin tagged AuNPs | Carbodiimide coupling | Targeting and enhanced cellular uptake | Protein tagging leads to better cytotoxicity with AuNPs delivered into TNBC (MDA-MB-231) cell line in vitro | 26 | |
| Tumor-penetrating peptide (TPP) | Hsp70 (TPP)-PEG4-FeAuNP | Covalent linker coupling | Increased uptake | Induced G2/M cell cycle arrest and ROS mediated apoptosis in MDA-MB-231 and 4T1 cell line | 209 | |
| Antibodies | Trastuzumab | G5-AuNP-Gd-trastuzumab nanoconjugate | Click reaction using G5-PEG-alkyne-DOTA-NHAc and trastuzumab-azide | Targeting | Enhanced MRI signal intensity by 20% and improved CT resolution and contrast by two-fold in vivo | 16 |
| HER-2 | BSA-AuNCs-FA-HER | Carbodiimide coupling | Targeting and radiosensitizing effect | Increased cellular internalization with improved radiation therapy in SK-BR3 BC cells | 210 | |
| Cetuximab | Ctxb-AuNPs | Using coupling reagent (COMU) | Targeting | Enhanced proton irradiation based cytotoxicity in A431 cells but not in MDA-MB-453 cells | 211 | |
| ER-α | Anti-ERα-AuNPs | Carbodiimide coupling | Targeting | 1,2-Bis(4-pyridyl) ethylene (BPE)-ERα-AuNPs Raman nanotags highly accumulated with a strong SERS signal to ensure detection of spheroid BC cells (MCF-7) | 212 | |
| EpCAM or TARP | (Paclitaxel-thiol-AuNPs-thiol)-TARP and EpCAM | Carbodiimide coupling | Cargo loading and targeting | AuNPs-thiol-TARP/EpCAM is a non-toxic drug delivery system. Attachment of TARP antibody to paclitaxel-thiol-AuNPs showed more activity than EpCAM attached system | 174 | |
| Hsp-70 | cmHsp70.1-AuNPs | Maleimide coupling reaction | In vivo tumor targeting | Accumulation in intracellular vesicles observed without any toxicity | 213 | |
| CA 15-3 | CA 15-3/Au based screen printed electrode | Carbodiimide coupling | Selectivity towards CA 15-3 BC antigen | Immunosensor successfully quantified CA 15-3 BC antigen in artificial serum samples with wide linear response | 214 | |
| Affibody | Dox@affi-F/AuNPs (affibody used: ZhcHER2:342) | Covalent conjugation | Targeting for HER2 | Enhanced co-delivery of 5-fluorodeoxyuridine and doxorubicin | 215 | |
| Nucleotides or aptamers | DNA | Dual-miRNA activated DNA walker comprised of carboxyfluorescein (FAM) and cyanine5 (Cy5) attached Mg2+-specific DNAzyme (Dz) hybridized AuNPs | Freeze mediated conjugation bypassing salt-aging technique | FAM fluorescence enhancement, recovers the Cy5 fluorescence | Depending on the expression levels of miR-21 and miR-31, DNA walker can successfully distinguish malignant (noninvasive) MCF-7, metastatic (highly invasive) MDA-MB-231, and nontumorigenic MCF-10A cell lines | 216 |
| RNA | TectoRNAtrimer:AuNP | Thiolated conjugation via salt-aging technique | Targeting against the CopGFP gene | Regulates the GFP gene expression, biocompatible system | 217 | |
| Peptide-AuNR-siRNA | Thiolated conjugation | Targeting and gene silencing effect | Gene silencing, and inhibition of metastasis in TNBC MDA-MB-231 | 218 | ||
| Aptamer | AS1411-chitosan-AuNPs loaded with methotrexate | pH mediated conjugation | Nucleolin targeting | Chitosan gives better biocompatibility, AS1411 ensures targeting, MTX induce cytotoxicity in vitro and in vivo | 219 | |
| Monosaccharides & polysaccharide | Chitosan | Janus chitosan/gold nanoparticles (J-Au-CS NP) | Nonsolvent-aided counterion complexation method | Selective surface fabrication | Enhanced cytotoxicity and intracellular localization achieved. PA imaging-guided synergistic PTT/gene therapy was achieved after CD-PGEA decoration upon J-Au-CS NP | 220 |
| Glucose | 2-Amino-2deoxy glucose (2DG) conjugated Au-loaded apoferritin nanoparticles (Au-HoSAF-2DG NP) | COOH activated EDC/NHS coupling | Cell surface glucose transport protein specific targeting | Selective targeting for MCF-7, selective cell death, enables in vitro X-ray and computed tomography (CT) imaging | 221 | |
| Cellulose | Carboxy methyl cellulose (CMC)-AuNPs | Chemical reduction | Reduction, providing chemical stability | Nanocomposite confers promising antibacterial, antifungal and anticancer activities. Induces apoptosis and necrosis by increasing caspase-8/-9 and decreasing VEGFR-2 activity | 222 | |
| Gum acacia | Gemcitabin (GEM)-GA-AuNPs | Chemical reduction | Reduction, drug loading capacity improvement | Enhanced aqueous solubility and drug release rate of GEM, enhanced cytotoxicity by the nanosystem | 223 | |
| β-Cyclodextrin | AuPEI-β-CD-Pep NPs | Microwave radiation | Decorated the carrier with targeting moieties using “host–guest” inclusion complexation | Enhanced transfection efficacy, targeted therapeutic delivery of nucleic acids to MCF-7 BC cells | 224 | |
| Enzymes | Collagenase | COL-AuNPs | Carbodiimide chemistry | Improved cellular interaction/penetration of Metformin (MET)-AuNPs in mammospheres | Increased apoptosis and reduced CD24−/CD44+ CSCs by COL-AuNPs and MET-AuNPs co-treatment against JIMT-1 BC cell | 225 |
| L-Asparaginase | AuNPs-PEG-L-asparaginase-RGD | Thiol mediated conjugation | Enhanced anti-cancer efficacy | Remarkable antioxidant effects with high tumor targeting efficacy and distribution in MCF-7 cells. Decreased cell proliferation and clonogenicity of MCF-7, apoptosis through intrinsic pathway and cell cycle arrest at the G2/M phase | 226 | |
| Horseradish Peroxidase | HRP-AuNPs | Carbodiimide chemistry | Ensured enzyme prodrug therapy | Co-treatment with HRP-AuNCs and IAA efficiently triggers cell death induced by oxidative stress | 227 |
3.2. Engineering AuNPs with proteins and peptides
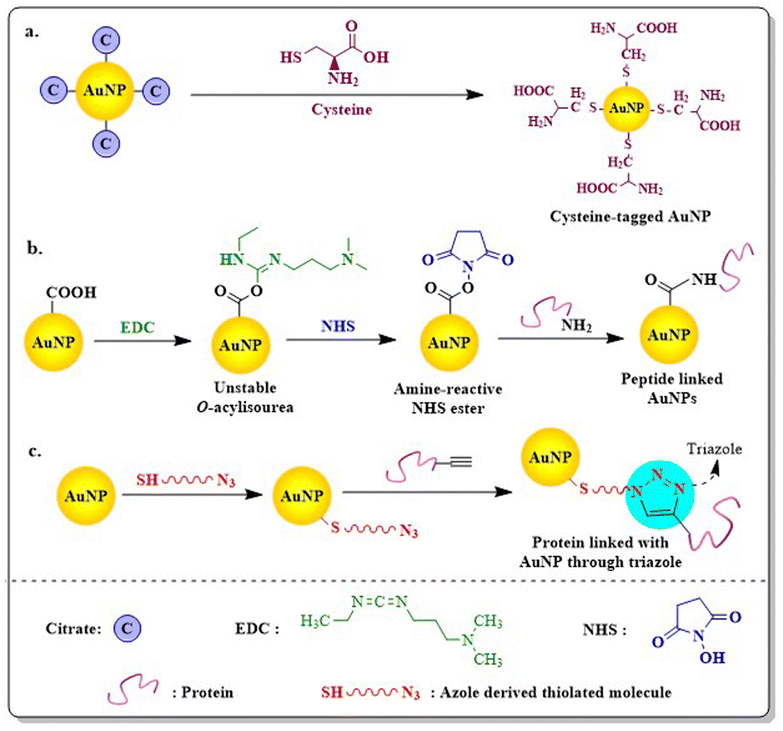
The most effective way to achieve targeted drug delivery using AuNPs is to conjugate different proteins or peptides via amino acid conjugation. Different proteins i.e. essential amino acids (aspartic acid, glutamic acid, phenylalanine, tryptophan, tyrosine, and L-cysteine), plasma proteins (albumins), epidermal growth factor (EGF), protein sequences i.e. RGD, NLS or TAT or even vitamins (folic acid) can be easily conjugated with AuNPs via covalent or non-covalent interactions to favour the intended application (Table 4). The amine group present in an amino acid or peptide bind with negatively charged AuNPs through ionic interaction, whereas the negatively charged carboxylic group helps in stabilizing AuNPs.The direct conjugation or ligand exchange method, amine carboxylate coupling or carbodiimide coupling technique and click chemistry are the three main techniques are used in covalent attachment of peptides to NPs. The ligand exchange method, which is the most commonly used approach to prepare peptide engineered AuNPs, was first explored by Hostetler et al. (1999).161 Due to their strong affinity to Au, sulphur containing amino acids i.e. cysteine, can easily undergo a ligand exchange method to substitute itself with citrate and produce stable cys-AuNPs162 (Fig. 13). Certain basic amino acids like tyrosine or cysteine also reduce AuCl4− to Au0 and form peptide conjugated AuNPs through the chemical reduction method.163 Although direct attachment is an easy technique, multivalent interactions between numerous residues of protein and the NP surface often lead to protein structure deformation, which is a major disadvantage.164 The most important method to functionalize proteins or peptides to carboxylated AuNPs through chemical conjugation technique is using “carbodiimide coupling chemistry”, where EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) reacts with COOH to form an active intermediate i.e. O-acylisourea. The unstable O-acylisourea then gets easily displaced by nucleophilic attack from primary amine groups present in the protein, leading to the formation of an amide bond and thus conjugates proteins with AuNPs. N-hydroxysuccinimide (NHS) or Sulfo-NHS is often used in carbodiimide coupling reaction (EDC/NHS coupling) to generate another amine-reactive ester intermediate which more efficiently conjugates with proteins (Fig. 13).165 Raposo et al. (2017) conjugated BSA (bovine serum albumin) to PEG-AuNPs via the carbodiimide coupling reaction and then AuNPs@PEG@BSA were further functionalized with Zn(II) based complex TS262 ([Zn(1,10-phenanthroline-5,6-dione)2]Cl) and Co(II) based complex TS265 ([CoCl(H2O)(1,10-phenanthroline-5,6-dione)2][BF4]). BSA ensures high loading of cargo in the nanosystems (AuNPs@PEG@BSA@TS262 and AuNPs@PEG@BSA@TS265), which exhibited remarkable cytotoxicity in canine mammary FR37-CMT cell line with IC50 values better than the standard drugs Doxorubicin and Cisplatin.166
Recently, click chemistry has been used by researchers for allowing site specific protein conjugation to NPs through carbon-hetero-atom bond formation giving high and selective yield. Though click chemistry generally suggests the copper-catalyzed azide–alkyne 1,3-dipolar cycloaddition reaction (CuAAC), the presence of copper is detrimental to living cells. Keeping this in mind, Liu et al. (2017) used a strain-promoted azide–alkyne cycloaddition reaction (called copper-free click chemistry or SPAAC) to synthesise folate receptor (FR)-targeted SERS active Au-nanoprobe without observable cytotoxicity. The SERS nanoprobe is created by modifying hollow AuNPs with a monolayer of a Raman-active label known as tris-aza-5,5′-dithiobis(2-nitrobenzoic acid) or N3-DNBA. These labels are comprised of disulfides, enabling them to be covalently attached to the surface of AuNPs and azide groups, facilitating their connection with folate bicyclo[6.1.0]nonynes derivatives (BCN-Folate) through click chemistry. The click coupling reaction facilitated by cyclooctynes activated by ring strain proceeds rapidly under mild conditions and does not necessitate a cytotoxic copper catalyst167 (Fig. 13). AuNPs functionalized with lysine or glycine bind to DNA efficiently and ensure efficient gene delivery without any toxicity. One of the major hurdles associated with protein engineered AuNPs is the pH dependency of the tagged protein which often lead to the aggregation of AuNPs.168
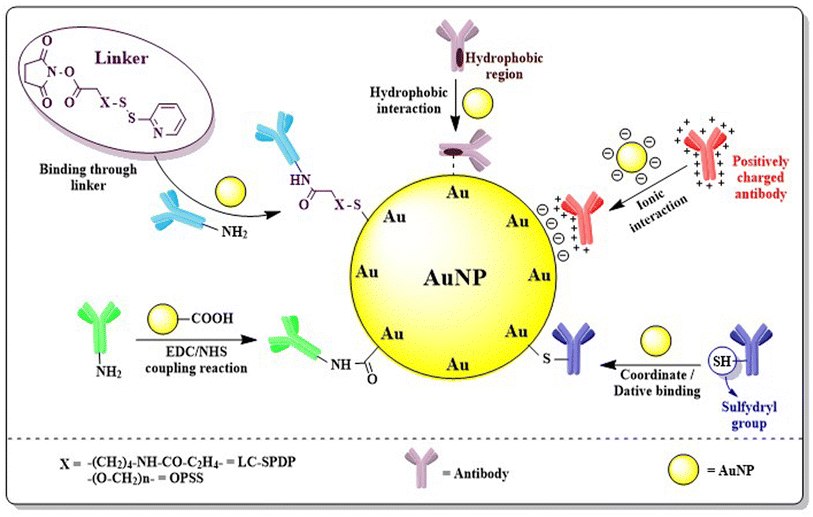
3.3. Engineering AuNPs with antibodies
Antigens present in cancer cells are able to assist in treating cancer via immunotherapy. Specific cancer cells produce their own significant antigens i.e. tumor-associated antigens (TAAs) by cellular gene mutation of aberrantly expressed normal genes. In BC, mutated as well as genes those are over-expressed in breast tissues produce different antigens. The antigens found in BC include human epidermal growth factor receptor 2 (HER2), carcinoembryonic antigen (CEA), mucin-1 (MUC-1), carbohydrate antigens (CA-15, Tn, TF, and STn) which are over-expressed and produced by abnormal or immature glycosylation of different amino acids i.e. serine and threonine, human telomerase reverse transcriptase (hTERT), and p53 – the most common tumor suppressor gene found mutated in cancers.169 AuNPs engineered with specific antibodies bind to definite antigens present in adenocarcinoma cells and induce an antigen–antibody reaction, thus improving biosensing efficacy and site-specificity of nanoparticles.Antibodies can be coupled with AuNPs through covalent, non-covalent or coordinate (dative) interactions (Fig. 14). Generally, the EDC/NHS coupling reaction involved in covalent bonding between antibodies and the COOH/NH2 group activates AuNPs but often leads to aggregation and polymerization. Another problem arises from disordered orientation or wrapping of antibodies on the surface of nanoparticles blocking free antigen binding sites.170 The problem can be solved by incorporating external pH near the isoelectric point of antibody which favors ionic or electrostatic adsorption of antibodies to the surface of nanoparticles. In ionic interaction, although the net charge distribution and the asymmetry of the antibody allows the antigen binding sites to be free for binding, it has certain limitations like a high antibody requirement to conjugate with non-magnetic nanoparticles, instability in different pH conditions and replacement of antibody with other biomolecules by electrostatic interactions.171 Another non-covalent attachment is the hydrophobic interaction often formed when the hydrophobic area of the antibody interacts with the metal surface. Dative binding or coordinate bonding is one type of physical interaction that occurs between the antibody and AuNP surface, when only the free sulfhydryl group present in the antibody shares its outermost electron and both of them use it covalently.170 Antibodies can also be attached to AuNPs through a linker. Liao et al. (2005) and Loo et al. (2005) used pyridyl cross-linkers i.e. long chain succinimidyl 6-[3(2-pyridyldithio)propionamido]hexanoate (LC-SPDP) or orthopyridyl disulfide (OPSS)–PEG–NHS to conjugate antibodies with Au-nanosystems.172,173
Protein EpCAM over-expression in BC was targeted by antibody engineered AuNPs with paclitaxel payload. They exhibited significant diminution of BC cell viability compared to AuNPs designed without antibodies.174 In another comparative study report it was observed that lectin jacalin or anti HER-2 antibody conjugated AuNPs exhibited a similar localisation pattern in the acidic compartments of both human colorectal adenocarcinoma (HT-29) and HER2+ BC cells (SK-BR-3).175 Penon et al. (2017) reported that engineered gold nanoparticles with a porphyrin derivative ligand, a PEG derivative ligand and an anti-erbB2 antibody resulted in a site-directed photodynamic response against SK-BR-3 human breast cancer cells.176 Recent advancement of antibody linked AuNPs in BC treatment is disclosed in Table 4.
3.4. Engineering AuNPs with oligonucleotides
Oligonucleotide (DNA or RNA) or aptamer functionalized AuNPs find use in a wide range of applications, with the most common being as probes in DNA-based biosensing assays, or in therapy for nucleic acid delivery including cell-related applications.177 DNA hybridized AuNPs have become a fascinating area of research because of the attachment of a definite and sequential probe which possesses a programmable assembly process followed by a specific action. Functionalization of AuNPs with DNA is easy because of the tendency of DNA bases to form coordinate bonds with Au.178 For direct conjugation or covalent conjugation, oligonucleotides are first introduced to thiol linkers at either the 5′- or 3′-end of the aptamer to form thiol-modified oligonucleotide. Since both oligonucleotides and AuNPs are negatively charged, this conventional technique needs to follow “salt-aging” and delicate control of the ionic strength to compensate for the charge repulsion between the nanoparticles surface and the DNA strands with a longer incubation period (usually overnight).179 DNA absorption onto the AuNP surface also depends on pH, a freezing environment, presence of anions like arsenates, or small molecules.180 The study by Shin et al. (2012) suggested DNA-functionalized AuNPs were very stable in buffer but they lost stability in PEG and became easily aggregated.181 Dougan et al. (2009) used disulphide-(thioctic acid) modified oligonucleotide probes which suggested significant improvements in the stability of oligo-Au/Ag nanoparticles.182 Apart from the conventional technique, non-thiolated DNA functionalized AuNPs have also been reported.183 Some recent studies with oligo-AuNPs in breast cancer treatment are given in Table 4.PEGylated AuNPs engineered with the mucin1 (MUC1) aptamer loaded with Paclitaxel resulted in synergised photothermal therapy and targeted drug delivery leading enhanced cytotoxicity compared to photothermal therapy or chemotherapy alone.184 Nano-complex designed with AS1411 aptamers, melittin and AuNPs produced significant selective cytotoxicity to MCF-7 BC cells compared to L929 cells ensuring site-directed delivery of melittin.185 AuNP modified with polyA sequences, AS1411 aptamer and antagomir-155 influences targeted delivery and promotes apoptosis by enhancing the expression of Tp53INP1 in MCF-7 cells.186
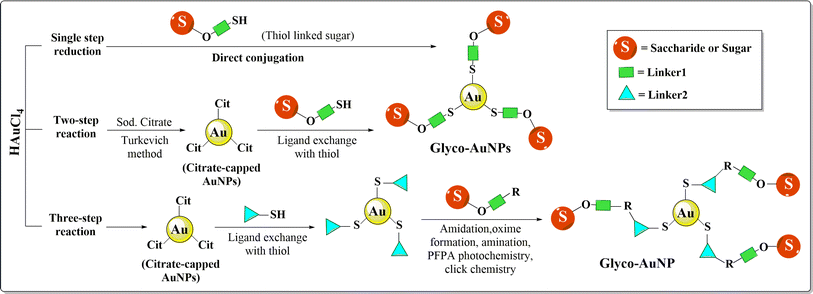
3.5. Engineering AuNPs with saccharides
Apart from proteins, carbohydrates gained much popularity in nano-based treatment as they possess specific but complex structural features, which affect different physiological and pathological circumstances in living systems. Carbohydrate based nanosystems or simply glycol-nanosystems possess the capability to restrict first pass metabolism, counteract p-glycoprotein mediated efflux, improve intestinal lymphatic transport, enhance absorption, facilitate chemical modification, and exhibit mucoadhesive properties make them an excellent solution for drug delivery associated problems in BC treatment. According to the Warburg effect, cancer cells need glucose far more than healthy cells as they reproduce quickly and at a greater rate. Substitution of this energy source with other monosaccharides could retard tumor progression and hence such carbohydrate tagged AuNPs could play an important strategic role in cancer treatment.187 AuNPs attached to the second carbon of glucose propound special features like targeting, the ability to differentiate between cancer and inflammatory cells, enabling contrast ability and triggering clathrin-mediated cellular uptake of NPs inside cancer cells depending on GLUT-1 expression.188Fabrication of different carbohydrate polymers into nanosystems can be achieved via the in situ copolymerization technique, solvent casting method or by the electrospinning technique.189 Glyco-AuNPs can be synthesized through a one, two or three step reaction (Fig. 15). Single step functionalization or direct conjugation simply refers to the reduction of Au3+ to Au0 with sugar itself being a reducing agent as well as a stabilizer. Two steps synthesis consists of synthesis of citrate-capped AuNPs commonly via the Turkevich method and then ligand exchange of citrates with thiol ending sugars. Mannose, galactose and glucosamine functionalized Au-nanosystems were synthesized using this approach; however, a longer synthesis time, lower glycan loading, and difficulty in controlling the size and shape of the nanosystem are the main drawbacks associated with the method.190–192 Three step reactions enable synthesis of AuNPs via traditional methods, attachment of linkers through the ligand exchange method followed by functionalization of sugar moieties using different reactions between ligands and carbohydrates i.e. oxime formation, reductive amination, alkyne–azide cycloaddition, perfluorophenylazide (PFPA) photochemistry, amidation etc..193 Katti et al. (2009) functionalized glucose (monosaccharide); sucrose, maltose, or lactose (disaccharides); raffinose (trisaccharide); and starch (polysaccharide) with AuNPs using the non-toxic, water-soluble reducing agent tris(hydroxymethyl)phosphine-alanine (THPAL).194
AuNPs with glycoamino acids ligands bearing the Thomsen–Friedenreich antigen linked to isolipoic acid had a profound influence on stability enhancement of AuNPs without hindering their biological action.195 Phthalocyanine-AuNPs engineered with lactose were reported to produce singlet oxygen and effect cell death upon irradiation as well as be associated with the galectin-1 receptor on the surface of BC cells.196 Carbohydrates as glycolipids or glycoproteins serve as important signalling moieties of our bodies. Among them monosaccharides and oligosaccharides have binding selectivity to protein receptors overexpressed on cancer cells. So, these variants can be engineered with AuNPs to get a favourable clinical outcome as therapeutic, diagnostic or theranostic agents.
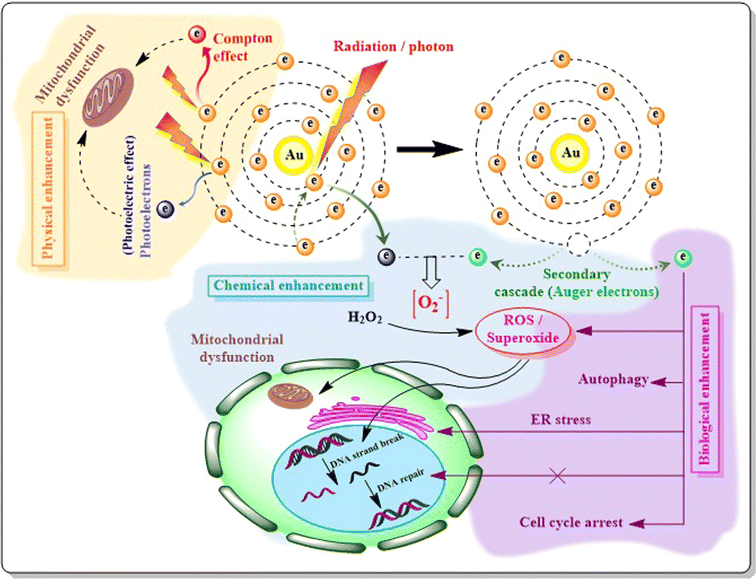
4. Theranostic effects of AuNPs
Conventional therapy for heterogeneous diseases lacks specificity towards particular cell types or tissues, often resulting in suboptimal outcomes. Theranostics, a concept combining “therapy” and “diagnostic” into a single approach, was coined in 1998 by US consultant John Funkhouser, aiming for personalized medicine offering improved prognoses.228 By integrating nanotechnology, specifically nanotheranostics, targeted early disease detection, treatment, and monitoring becomes feasible. AuNPs serve as effective carriers for both imaging and therapeutic agents, enabling precise delivery to target sites while minimizing systemic toxicity. Moreover, AuNPs can be loaded with multiple contrast agents viz. fluorophores, MRI agents, photoacoustic agents, and surface-enhanced Raman spectroscopy (SERS) reporters for multimodal imaging with real-time monitoring of their effects, enhancing disease characterization without overwhelming the patient's immune system.229 Nanoplatforms leverage particle size for enhanced tumor targeting via the EPR effect, exploiting differences in the tumor microenvironment (TME) such as vascular abnormalities, hypoxia, and pH. TME-triggered theranostic gold nanoparticles hold promise for tumor-specific detection and therapy by capitalizing on these unique features.230AuNRs with varying aspect ratios were coated with folic acid (FA)-PEG block copolymer (FAP) and pheophorbide a (Pheo) for tumor targeting and photodynamic therapy (PDT). AuNRs with an aspect ratio of 3.84 and longitudinal plasmon resonance at 873 nm (Pheo-conjugated AuNR100) demonstrated enhanced singlet oxygen generation, photothermal conversion, GSH-mediated Pheo release, tumor targeting, and a synergistic PDT–PTT effect.231 An amphiphilic AuNP was coated with a Raman reporter (BGLA) and made stealthy with PEG, while pH sensitivity was achieved via PMMAVP grafts. DOX was incorporated as cargo, and HER2 antibody enabled specific targeting. The system's cellular binding, uptake, and intracellular disruption were evaluated, highlighting its potential for targeted theranostic applications of AuNPs.232 Nanoparticles face a highly intricate in vivo environment, which differs significantly from in vitro settings. This leads to notable disparities attributed to (i) a limited understanding of how NP properties influence biointeractions and (ii) variations in experimental parameters.233 For instance, Zhang et al. (2020) observed discrepancies where PEG-AuNPs were swiftly cleared from mouse circulation despite in vitro studies showing reduced uptake by RAW 264.7 macrophages.234 Similarly, J. Cancino-Bernardi et al. (2018) found no significant differences in shape or protein coating in vitro, yet AuNP-HSA triggered a myeloperoxidase mediated inflammatory response when orally administered to Wistar rats.235 Citrate-capped AuNPs showed no impact on cytokine secretion in vivo but caused DNA damage in vitro.236 Wang et al. compared the in vitro and in vivo performances of Au-nanohexapods for theranostic applications, revealing discrepancies in tumor accumulation and cytotoxicity.237 Moreover, Dubaj et al. (2022) noted significant variations in the internalized gold concentrations between cell lines and corresponding tissues in vivo, emphasizing the absence of natural barriers in in vitro setting as the primary cause for these inconsistencies.238 The discrepancy of in vitro and in vivo results are well discussed in the reviews of Pavithra Natarajan et al.239 and Khlebtsov et al.233
While various nanoparticles like carbon nanotubes, quantum dots, iron oxide nanoparticles, and silica nanoparticles can be utilized for theranostic applications, AuNPs hold a prominent position due to their distinct features such as strong surface plasmon absorption, stability, biosafety, and ease of modification. Unlike quantum dots, which face concerns about the fragility of disulfide binding during surface modification, stable ligand anchoring of AuNPs typically requires only a monodentate thiol. However, there is debate regarding whether residual carbon nanotubes may cause long-term damage to the host, and the absence of a standardized protocol for preparing high-purity carbon nanotubes at a large scale presents a challenge for clinical translation.240 Despite the advantages of each nano-scale agent, they also present their own set of drawbacks, like toxicity of quantum dots, high-cost of gold nanoparticles, the limited sensitivity of iron oxide nanoparticles as MRI contrast agents, non-biodegradability of carbon nanotubes, and the size and self-destructing properties of silica NPs.241 The study of Alexander Vasil’kov with Au and iron-oxide NPs in conjugation with Methotrexate clearly suggested the better stability of AuNPs over iron-oxide NPs as iron was involved in oxidation at higher temperature.242 Research for improvement of these limitations is ongoing and could be an interesting brainstorming option for future endeavours. Here, in this review, we would like to address the theranostic applications of AuNPs part by part, first its diagnostic applications, then therapeutic strategies including photothermal therapy, radiotherapy and AuNPs as cargo delivery.
4.1. AuNPs in BC diagnosis
The unique optical properties of AuNPs aptly supported by sophisticated instrumentation setting facilitate BC management by exploring them in a dual platform of therapy and diagnostics.243 Reports of AuNPs tailored with different moieties like peptides or nucleic acids, antibodies, photo-enhancers, radiolabeled agents, aptamers, oligonucleotide sensors, antigen detectors and immunosensors have assisted molecular probing or imaging of BC cells with high accuracy. AuNPs have been reported for their integration with BC surface receptors i.e. HER-2 (human epidermal growth factor receptor-2), integrin β, and CD-44 (cluster of differentiation 44), in detection of over-expressed intracellular receptors i.e. ERα (estrogen receptor α) and also in genetic alterations i.e. mutated BRCA1 (breast cancer gene 1), p53 over-expression, tumor mi-RNA etc..244| Nanoparticle variant | Conjugation | Target biomarker | Detection technique | Detection limit (lower) | Additional findings | Reference |
|---|---|---|---|---|---|---|
| GCE modified ErGO-SWCNT/AuNPs | Thiolated anti-HER2 aptamer | HER2 | Cyclic voltammetry (CV), differential pulse voltammetry (DPV) and electrochemical impedance spectroscopy (EIS) | 50 fg mL−1; analytical range (0.1 pg mL−1–1 ng mL−1) | — | 258 |
| Antibody tagged AuNPs (2018) | Anti-HER2 antibody | HER2 | ELISA | 0.01 ng mL−1 | — | 259 |
| BPE-tagged AuNPs | Anti-ERα antibody | ERα | Surface enhanced Raman spectroscopy (SERS) | — | Identification and distinguishing of MCF-7 (Erα/+) and SK-BR-3 (Erα/−) BC cells | 212 |
| Graphene oxide (GO)-AuNPs and Bi2Se3-AuNPs | BRCA1 nonsense mutated ssDNA | BRCA1 | Colorimetric analysis | 1 aM (GO-AuNPs); 1 pM (Bi2Se3-AuNPs) | — | 260 |
| DNA-functionalized AuNPs | cfDNA probe | BRCA1 | Metal-enhanced fluorescence (MEF); colorimetric analysis | 0.34 fM; linear range: 1 fM–100 pM | Activated CRISPR-Cas12a cleaves ssDNA but dsDNA cannot be cleaved | 261 |
| Thiolated DNA conjugated AuNP duplex | Thiolated DNA | BRCA1 | Surface plasmon coupling electrochemiluminescence (SPC-ECL) | 0.83 fM; linear range: 1 fM–1 nM | Au–Au dimers with a gap distance of 2 nm is superior in enhancing ECL signal of GCN QDs | 262 |
| Tetrahedral DNA framework (TDF)-modified AuNPs | cfDNA probe | BRCA1 | CV and amperometry | 1 aM; linear range: 1 aM–1 pM | Biosensor with TDF-26 showed unique and best response with selective stability | 263 |
| AuNPs/Thi/MSNP-NH2 | HPR-anti-CA 15-3 antibody | CA 15-3 | DPV | 0.001 U mL−1; range: 0.002–125 U mL−1 | Quantification of MCF-7 cells; range: 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–50,000 cells per mL 000–50,000 cells per mL |
264 |
| Polypyrrole-luminol-AuNPs | Anti-CA153 antibody | Carbohydrate antigen 153 | ECL | 5.8 × 10−4 U mL−1; linear range: 0.001–700 U mL−1 | Good film-forming property, quantifies CA-153 in serum plasma | 265 |
| AuNPs@Cu7S4@Cu/Mn-AzoPPOP | MUC1 aptamer | MUC1 | CV and CA | 0.72 fg mL−1 (DPV) and 0.82 fg mL−1 (CA); linear concentration range: 1 fg mL−1–10 pg mL−1 | Quantifies MUC1 in serum plasma | 266 |
| Polyethylenimine coated-gold nanoparticles AuNPs | DNA probes | CA 15-3, MUC1 and HER2 antibody | Electrochemical redox analysis | 0.21 U mL−1 (CA 15-3), 0.53 ng mL−1 (MUC1) and 0.50 ng mL−1 (HER2); range: 0.10–100 U mL−1 (CA 15-3) and 0.10–100 ng mL−1 (MUC1 and HER2) | Detect three tumor markers in human serum also | 267 |
| GO-IL-AuNPs | Anti-CD44 antibody | CD-44 | DPV, EIS | 2 fg mL−1 (DPV) and 1.90 fg mL−1 (EIS); linear detection range: 5.0 fg ml−1-50.0 μg mL−1 | — | 268 |
| Diphenylalanine-AuNPs | CD44BP | CD-44 | EIS | 2.17 pg mL−1; linear range: 0.01 ng ml−1–100 ng mL−1 | Limit of detection: 8 cells per mL for CD44-positive BC stem cells | 269 |
| PEGylated AuNPs | Cyclic 4-aminoproline-RGD semipeptides | Integrin αvβ3 | Confocal laser microscopy | — | Potent inhibitors of integrin-mediated melanoma tumor cell and are selectively internalized via receptor-mediated endocytosis | 270 |
| Raman reporter tagged AuNPs | ssDNA probe | miR-200c | SERS | — | The expression level of miR-200c in SK-BR-3 cells was 5 times greater than that in MCF-7 | 271 |
| AuNPs/GQDs/GO | Thiol-modified miRNA probes | miR-21, miR-210, miR-155 | Square wave voltammetry (SWV) | 0.04 fM (miRNA-21), 0.33 fM (miRNA-155), and 0.28 fM (miRNA-210); linear dynamic range: 0.001 to 1000 pM | High selectivity and applicability for the detection of miRNAs in human serum samples | 272 |
| Hydroxyapatite nanorods (HApNRs) decorated with AuNPs | Thiolated AS1411 aptamer and 1-ethyl-3-methylimidazolium alanine | Nucleolin cell surface protein | DPV | 8 ± 2 cells per mL | Highly sensitive and selective detection of surface nucleolin on MCF-7 | 273 |
| Graphene oxide-chitosan-gold nanoparticles | AS1411 aptamer | Nucleolin cell surface protein | CV and EIS | Linear range: 1 × 101–1 × 106 cells per mL | MCF-7 LDQ: 4 cells per mL | 274 |
| MoS2-AuNPs modified carbon paper (CP) | DNA S1 probe and biotinylated DNA S3 probe | p53 | CV and EIS | 68 fM | Linear ranges: 10−15–10−12 M for wild type p53 in MCF-7 and 10−12–10−6 M for mutated p53 in SK-BR-3 | 275 |
4.2. AuNPs as probing agents
The strong interactions of AuNPs with visible light (SPR properties) are often explored as labeling agents for sensing cancer cells. Site-directed AuNPs accumulated in cancer cells exhibit optical scattering properties, which helps in visualization of cancer cells which can be tracked using different sophisticated techniques i.e. TEM, phase contrast optical properties, dark field microscopy, photoacoustic or photothermal imaging.276 Sensitive probing of cancer cells with the help of AuNPs also can be achieved using SERS imaging. External radiation passing through materials results in elastic and inelastic collision. In inelastic collision of photons with the matter, the molecular vibrations cause scattering of photons with a different wavelength which is known as Raman scattering. Nanoparticles like Au and Ag, can enhance the energy of scattering photons or simply amplify the Raman signals due to the spikes present on their surface acting as “SERS hot-spots” and therefore can be used as SERS sensors to detect even extremely low concentrations of biomarkers for disease quantification and identification when appropriately targeted.277 Raman reporters (dye molecules) were usually immobilized or embedded onto plasmonic nanoparticles to produce SERS nanotags which exhibit attractive properties like generating multiple sets of narrow peaks, low spectral overlap, negligible photobleaching, high sensitivity and multiplexing ability or low background noise etc. making the nanotags much more efficient than conventional fluorescence probes.278The major disadvantage of using plasmonic nanoparticles in Raman nanotags arises when adsorption of Raman code onto the reporter causes colloidal instability of the nanoparticles. This problem was solved by using a self-assembled monolayer (SAM) of long chain thiolated polymers but the problem with SAMs is its constant desorption and degradation upon laser excitation.279 Keeping the several stability issues, researchers nowadays are focused on applying different chemical approaches in making synthetic compounds which can be attached to plasmonic AuNPs with strong covalent bonds. Li et al. (2019) developed alkyne and nitrile based background free SERS reporters also acting as anchors for citrate capped AuNPs as well as for different antibodies. Radiation of the 633 nm laser on alkyne/nitrile SERS Au-nanoprobes significantly produced sharp Raman signals in the 2000–2230 cm−1 region against ER, PR and EGFR over-expressed human BC cells, thus acting as biosensors for multiplex detection and imaging.280 Song et al. (2012) developed a suitable plasmonic vesicle using SERS active amphiphilic AuNPs and BGLA Raman reporter, which not only amplify the Raman signal given by SERS active AuNPs, but also induce Doxorubicin loading and its release inside cancer cells. The probing of cancer cells was tracked down using Raman spectroscopy as well as by plasmonic imaging.232
Using SERS spectra, Zhu et al. (2013) in their study, reported the interaction of AuNPs with intracellular components inside cancer cells. Raman spectroscopy combined with TEM study revealed the interaction of AuNPs with the phenylalanine present in MDA-MB-231 with a sharp peak at 1030 cm−1 could be useful in cancer cell SERS mapping.281 Cellular components of MDA-MB-231 TNBC cells were also well observed using Raman spectroscopy due to amplified SERS signal by rhodamine 6G reporter tagged Au-aryl nanostars.13 Rhodamine 6G-AuNP nanotags also exerted their effectiveness in distinguishing different BC cells by altering two-photon scattering intensity. In the MDA-MB-231 cell line, the scattering intensity changed 2.2 times while in the case of the SK-BR-3 cell line, the scattering intensity differed 13 times.282 In a recent study, Gao et al. (2021) successfully attached 785 porous silicon photogenic crystals which acted as a SERS substrate to distinguish the blood serum in healthy people from the serum obtained from BC patients.283 In spite of the various advantages driven by AuNPs as biosensors, its applicability is somehow limited to cancer cells close to the skin surface.284
4.3. Treatment of BC using AuNPs
Due to recent availability of advanced treatment facilities like gene expression profiling and endocrine therapy, the incidences of BC and its associated lymphedema have declined, with enhancement of survival rate. However, maintaining patients' cosmetic appearance and associated challenges still remained unsolved. Chemotherapy related adverse effects, lack of targets, metastasis of tumor cells or managing TNBC are some of the major problems that continuously invite questions about effective treatment strategies in BC. In the nanotechnology guided era, AuNPs can provide answers to those issues as they are biologically inert or non-reactive and can offer suitable drug delivery in vivo compared to relatively toxic (cadmium) Cd or (silver) Ag-NPs.285 Besides, controlled synthesis for maintaining appropriate size and surface chemistry, strong optical and electrical properties due to localized surface plasmon resonance (LSPR), multifunctional domain serving ability pushes AuNPs ahead of other NPs in the road to BC management.Once AuNPs accumulate in tumor cells with the help of EPR effect or by active targeting, they can damage cell membranes or DNA when illuminated by a NIR laser. NIR light can be transmitted through tissue components with minimal absorption. When NIR radiation is applied, AuNPs accumulated in tumor cells generate heat via localized surface plasmon resonance (SPR) due to AuNPs' special optical effect of light absorption and scattering in the NIR region (650–900 nm). The heat generated in cancer cells successfully ablates the tumor cells without or with minimal effects in normal tissues, thus ensuring the limitation exerted by conventional chemotherapy i.e. death of cancer as well as normal cells.286 Targeted therapy in BC and the thermal ablation method achieved by AuNPs ensure increased heat distribution throughout the cancer cells as well as the surrounding perivasculature area. External heating treatments often face inappropriate ablation as the blood flow by the adjacent capillaries cool down the perivasculature area acting as a ‘heat sink’ but when AuNPs accumulate in tumor cells as well as in the surrounding vasculature, not only do they cause heating of the tumor upon NIR irradiation, but they also destroy the surrounding capillaries. However, certain episodes of bleeding, skin burning or inflammation, and pain induction have been claimed as side effects when this application of AuNPs has been explored.24
A study by Zharov et al. (2005) suggested effective laser (λ = 1064 nm) induced selective cell damage to TNBC cell line (MDA-MB-231). Antigens attached to 40 nm AuNPs self-assembled to form nanoclusters on the tumor cell membrane which destroyed cancer cells through laser induced bubble formation.288 In a similar study with Au-nanoshells and SK-BR-3 BC cells, Loo et al. (2005) reported that Anti-HER2 immuno-targeted Au-silica nanoshells could be used effectively for the detection as well as treatment of BC cells when NIR laser beam of 800 nm was applied.289
Mohammed and his group in their research study prepared multi-walled carbon nanotube (MWCNT) conjugated AuNPs and tested for anti-cancer efficacy using laser irradiation on the MCF-7 cell line. They found that the thermal ablation and cytotoxicity effect of AuNP/CNT was boosted compared to simple MWCNT alone, establishing the PPTT efficacy of nanogold.290 In another study by Yang et al. (2015), the MUC1-aptamer conjugated AuNP based graphene-oxide (GO) nanocomposite exhibited specific targeting efficacy and increased bioavailability along with remarkable photo-ablation capacity by controlled heat generation under laser irradiation. In the same study, the group also suggested that fabrication of heat shock protein (HSP-70) inhibitor to aptamer-AuNPs destroyed MCF-7 cells in a synergistic manner.291 Another study involving anti-EGFR engineered AuNPs, irradiation with NIR laser not only significantly increased different autophagy associated protein and gene expression i.e. light chain-3 (LC3), beclin-1 and Atg5, but also inhibited Akt-mTOR pathway pointing out the photothermal efficacy of AuNPs in inducing autophagy related cell death in TNBC (MDA-MB-231) cell in vitro. The group also performed a gene expression study with MDA-MB-231 xenograft mouse models where they found that NIR laser induced AuNP mediated PPTT also increased LC3 and beclin-1 level in vivo.292 Chen et al. (2020) performed a similar study with folic acid (FA)-functionalized gelatin based AuNPs for TNBC management in vitro and in vivo. The study results indicated a complete photothermal ablation of MDA-MB-231 cells with NIR laser irradiation but the damage to the tumor cells depended on the shape of the NPs, irradiation time and the laser intensity applied.293 An interesting study with AuNRs by Ali et al. (2019) in canines and felines suggested that PPTT treatment before tumor removal effectively decreases the risk of metastasis and blood loss during surgery due to altered blood vessels in mammary tumors.294 In a similar study with AuNRs previously, they had successfully ablated mammary tumors in BC xenograft cat and dog models without any signs of toxicity on liver, kidney and blood profiles after NIR laser irradiation suggesting the biocompatibility achieved using AuNR-PPTT treatment.295
Combination of chemotherapy and photothermal therapy is one of the finest treatment strategies involved in BC prevention. In one recent study by Faid et al. (2022), it was suggested that combination of chemotherapy and PTT of 6-marcaptopurine (6 MP) alone or loaded in highly stable chitosan AuNPs could be effective in damaging MCF-7 BC cells in vitro when DPPS laser beam was irradiated.296 Thiol modified mannoside tagged AuNP was synthesized which offered targeted delivery to human TNBC cells due to the over-expressed mannose receptor on MDA-MB-231 and resulted in laser (λ = 808 nm) ablated cell death as well as metastasis reduction by Lin et al. (2020).297 Another example of combination of chemo-photothermal therapy was given by Poursalehi et al. (2019). According to their research, terpolymer modified smaller AuNPs (30 nm) along with Doxorubicin showed a synergistic effect by completely abolishing MCF-7 cells that underwent PTT.298 Similar combination treatment was performed in MCF-7 cells, in which it was observed that chemo-photothermal therapy was shape dependent as Doxorubicin-loaded polymer coated Au-nanostars showed more efficacy in thermal ablation than Au-nanocages.299 The shape dependency of NIR laser induced thermal treatment can be quite conflicting as upon intense laser illumination, AuNRs sometimes get converted to Au-nanoshells which possess different optical properties to AuNRs, thus altering BC management efficacy.300
One of the major disadvantages of PPTT using AuNPs in BC management is that for effective photothermal ablation, the size of NPs should be larger (∼50 nm). Xia et al. (2018) developed a suitable method to solve the above problem associated with smaller particles by inter-conversion of modifying agents to another. They synthesized 2-5-diphenyl-tetrazole modified methacrylic group linked AuNPs of smaller (23 nm) diameter. A shorter wavelength laser (λ = 405 nm) irradiation effectively converted 2,5-diphenyltetrazole to nitrile-imine dipole which covalently cross-linked with methacrylic groups present on the surface of AuNPs which resulted in particle aggregates. The resultant aggregates exhibited enhanced photothermal efficacy against 4T1-murine BC upon 808 nm laser irradiation while the cross-linked exempted particles did not.301 To enhance the efficacy of the smaller particles, different strategies like tuning the charge shared by the surface or utilizing an internal or external stimuli also can be performed.302–304
The experimental studies both in vitro and in vivo investigations suggest the plausible mechanisms by which AuNPs act as radiosensitizers. Though mainly physical enhancement was found to be the primary attribute, researchers also found chemical and biological radiosensitization of AuNPs in different studies. The physical dose enhancement effect of AuNPs as a radiosensitizer mainly involves the ‘Compton and photoelectric effect’ of AuNPs, responsible for absorption co-efficient differences between AuNPs and soft tissues.307 The Compton effect relies on the collision between incident photons and weakly bound outer electrons of Au atom, in which the energy transfers from photons to electrons cause them to be released from the Au atoms. In the photoelectric mechanism, incident photons are absorbed and energize the electrons present in the K, L or M shells of the Au atom to be released from Au. Once the shells are vacant, they are usually filled up by the outermost electrons which results in the lower energy fluorescence and secondary Auger electrons to be released. These lower energy electrons (LEE) play a critical role in radiosensitization.307 An 111In-radiolabeled EGF-coated AuNP with a diameter of 14 nm (111In-EGF/AuNP) was developed by Song et al. (2016) and was tested for anti-cancer activity in TNBC (MDA-MB-231) and MCF-7 cell lines through RT. They suggested that the uptake of AuNPs solely depended on the EGF/Au molar ratio. A higher internalization of 111In was observed for MDA-MB-231 cells than MCF-7. The clonogenic assay suggested dose enhancement by 111In-EGF/AuNPs along with decreased survival fraction to MDA-MB-468 compared to non-labeled AuNPs.308
Chemical radio enhancement of AuNPs deals with catalysis of a radical reaction by surface activation of AuNPs. The low energy electrons with very little energy (<10 eV) generated after radiation energy is applied weakens H-bonding in DNA and makes DNA more vulnerable to radiation damage. The photoelectric effect of AuNPs also induces catalytic cascade where the energized surface electrons of AuNPs transfer to biological O2, creating ROS (reactive oxygen species) or free radicals which in turn damage DNA, RNA, mitochondria and cell membranes of cancer cells.309 According to some studies, small and positively charged AuNPs with a large surface area can lead to chemical enhancement through both DNA damage and radical cascade following superoxide or ROS production.310
While physical dose enhancement by lower (kV) energy is used to treat superficial tumors, biological radio-enhancing capability of AuNPs offers deep penetrating treatment with higher energy irradiation (MV).311 Biological radio enhancing can be induced by ROS production, cell cycle arrest, inhibiting DNA repair or by inducing autophagy and (endoplasmic reticulum) ER stress312 (Fig. 16). A study with dihydrorhodamine (DHR)-123 radiolabeled PEG-AuNPs by Choi et al. (2019) suggested not only a significant increase (7–14 fold in vitro and 3–6 fold in vivo than control) of ROS in MDA-MB-231 cells with a radiation dose of 6 Gy, but also induced fluorescence imaging of TNBC cells in vivo.313 In another study, thioglucose tagged AuNPs of 49 nm in size induced more radiation than smaller particles along with radiative cell cycle arrest in the G2/M phase in MDA-MB-231 cell line in vitro.314 Various recent studies with a radiosensitizing effect of AuNPs in treatment of BC are tabulated in Table 6.
| Nanoparticle system | Particle size | Cell line/tumor models | Radiation energy | Outcomes | Reference |
|---|---|---|---|---|---|
| Citrate, glutathione and aminodextran reduced AuNPs | 8.8–21.9 nm | In vitro: MDA-MB-231 | 6 MV; dose: 4 Gy | Biological radio enhancement observed with accumulation of cyclin B and cyclin D and over-expression of p53. S and G2/M arrest after irradiation | 315 |
| Mercaptosuccinic acid (MSA) coated AuNPs | 14 nm | In vitro: MDA-MB-232; in vivo: NCRNU f sp/sp athymic mice | 160 kV; dose: 15 Gy | Size-dependent RT enhancement in vitro; RT induced immunogenic cell death (ICD), enhanced macrophage infiltration in vivo | 316 |
| Glutamine and folic acid functionalized BSA-AuNPs | 12.2 ± 3.1 nm, 12.5 ± 3.5 nm | In vivo: 4T1 breast tumor-bearing female BALB/c mice | 6 MV; dose: 6 Gy | Physical dose enhancement with increased cytotoxicity | 204 |
| AS1411 aptamer-conjugated AuNPs | 10 ± 1.5 nm | In vitro: MCF-7, MDA-MB-231 | 4 MeV electron beams | Physical dose enhancement (DEF) 1.35 and 1.66 for MCf-7, MDA-MB-231 | 317 |
| Doxorubicin-loaded chitosan capped AuNPs | 18 nm | In vitro: MCF-7 | 6 MV; dose 0.5, 1, 3 Gy | Physical or chemical enhancement observed for combination of chemo-radiotherapy; necrosis occurred due to DNA DSB even at lowest dose (0.5 Gy) | 318 |
| Bleomycin loaded RDG modified AuNPs | 10 nm | In vitro: MDA-MB-231 | 6 MV; dose: 2 Gy | Biological enhancement observed due to combination therapy as p53 binding protein found to cause DSB of DNA. DEF: 1.31 | 319 |
| 177Lu-dendrimer conjugated folate and bombesin modified AuNP | Not mentioned | In vitro: T47D | Dose: 63.16 ± 4.20 Gy | 177Lu–DenAuNP–folate–bombesin is about 4 times more lethal than 177Lu–DenAuNP; 177Lu–DenAuNP is more lethal than 177LuCl3 | 320 |
| DSPE PEGylated Au nanowares (AuNWs) and nanospheres (AuNSs) | AuNW: 3.6 nm (diameter) and 1.3 μm (length); AuNS: 14 nm | In vitro and in vivo: 4T1 and 4T1 xenograft BALB/c mice | 5 Gy | Biological enhancement observed for increased hydroxyl radical and superoxide dismutase levels. AuNS showed better radiosensitized cell viability than AuNW | 321 |
| PEGylated curcumin modified AuNPs | 73.8 ± 6.76 nm | In vitro and in vivo: 4T1 and 4T1 xenograft BALB/c mice | 6 MV; dose: 4 Gy | Enhanced ROS generation via chemical radio-enhancement | 322 |
| Cisplatin loaded RDG AuNPs | 10.04 ± 0.89 nm | In vitro: MDA-MB-231 | 6 MV; dose: 2 Gy | Biological radio-enhancement observed with DNA DSB and induced p53BP levels | 323 |
| MSA coated AuNPs | 4 and 14 nm | In vitro: MCF-7 | 160 kV, 2.5 MV or 10 MV; dose: 4 Gy | Radiation induced inhibition of cell proliferation at 160 kV for 4 nm AuNPs and for 14 nm AuNPs at 2.5 or 10 MV. Biological dose enhancement suggested by DNA DSBs | 324 |
Suitable active drug targeting and delivery using AuNPs depend upon the attachment as well as the release of drugs in suitable sites. Covalent attachment, electrostatic attachment, hydrophilic–lipophilic interactions etc. offer suitable drug conjugation with AuNPs, whereas the release of the drugs can be achieved using SERS, photothermal release, pH sensitive release, light and radiation mediated release, glutathione mediated release etc. Controlled release of drugs from AuNPs can also be achieved by conjugating a time specific release linker between the AuNP surface and drugs.325–327 Polymers such as poly(acrylic acid) (PAA) or poly(ethylene glycol) (PEG) can be used to coat AuNPs. At acidic pH, the protonation or deprotonation of acidic or basic functional groups within these polymers can trigger changes in their conformation or solubility, resulting in the disruption of nanoparticle-drug interactions and subsequent drug release. Alternatively, acid/base labile pH-sensitive linkers can be incorporated into the nanoparticle–drug conjugate, where the linker undergoes cleavage or hydrolysis in response to changes in pH, releasing the drug payload. According to the study by Joshi et al. (2012), chloroquine loaded AuNPs when incubated with MCF-7 cells, the drug release occurred at the lower pH environment and exhibited concentration dependent cytotoxicity after being taken up by lysosome or endosome in BC cells. In a lower pH tumor microenvironment, approximately 26% of the drug was released after 2 h, which increased to 81% after 48 h, indicating a sustained drug release over time. These levels may be the result of the release of non-specifically bound drugs from the surface of the nanoparticles.328 Cisplatin release mediated by an endosomal lower pH environment using dumbbell shaped Au–Fe3O4 magnetic nanoparticles in SK-BR-3 cells was reported by Xu and co-researchers.329 Another example of drug release on SK-BR-3 cells with doxorubicin-loaded biohybrid AuNPs was achieved using SERS. Dox can be released from biohybrid nanoparticle surfaces by intracellular GSH. GSH is the most abundant thiol species in the cell cytoplasm, with a concentration range of 1–10 mM, and has been used as an in situ releasing reagent in living cells, owing to its biochemical reducing capability.330 The thiol group in the GSH has strong affinity to AuNPs and can bind with the AuNPs through covalent coupling. For this reason, amine containing Dox can easily be replaced from AuNP surfaces.
When AuNPs are irradiated with light of a specific wavelength (usually in the near-infrared region), they absorb photons, leading to an increase in temperature at the NP surface. This localized heating triggers the release of the drug payload either by disrupting the surrounding matrix (e.g., polymeric coating) or by inducing a phase transition in thermosensitive drug carriers. Light responsive molecules or photosensitive linkers can be designed to break or cleave in response to specific wavelengths of light, resulting in the release of the drug payload. Higher drug accumulation and increased cytotoxicity was observed when 785 nm NIR laser of 3 mW power was irradiated.331 NIR laser mediated drug release by Au-nanoshells for the treatment of TNBC was also studied by You et al. (2012). NIR laser irradiation triggers local Doxorubicin release at the tumor site because of the photothermal conducting effect of hollow AuNS.332 Another group explained the involvement of dual NIR/pH dependent sustained release of paclitaxel (PTX) from the PTX-loaded PEG-AuNPs@antiMUC1 system in treatment of BC. PTX was physically loaded into the PEG-AuNPs through the non-covalent interactions between the OH group of PTX and the oxygen of PEG. The authors suggested that at higher pH, the repulsive ionic interactions between the carboxyl groups caused prolonged encapsulation of the PTX onto the AuNPs surface, assisting drug ligands dispersion. However, in acidic pH (pH 5.4), because of Coulomb repulsion forces and protonation of OH group of PTX and PEG-AuNPs, electrostatic interaction was mitigated resulting in an increased PTX release.223
Drug loading onto AuNPs can offer a solution for by-passing several barriers i.e. the RES system, blood–brain barrier (BBB) and blood tumor barrier.333 Doxorubicin-loaded TAT (trans-activating transcriptional activator)-modified AuNPs were reported to have BBB penetrating ability which was confirmed after accumulation of the drug in brain for the treatment of brain metastatic TNBC (MDA-MB-231/Br). Our study implemented an acid-labile hydrazine bond between Dox and the surface of the gold nanoparticle, which has been shown to cleave in response to an acidic environment such as that found in endosomal compartments (late stage endosome pH 5.5, lysosomes pH 4.8).334 Development of multi-drug resistance (MDR) tumor cells has become a complex threat for drug development scientists for over a decade. The unique targeting capability of AuNPs ensures suitable drug release in the specific site of action enhancing the chemotherapeutic efficacy even against MDR. Wang et al. (2011) developed a nanosystem (Dox-hyd@AuNP) with Dox, PEG-coated AuNP and an acid-labile-linker. The acidic environment of MCF-7/MDR helps in cleaving the linker followed by release of Dox which ultimately resulted in increased cytotoxicity in MDR BC cells.25
Though several studies have been performed exploring the area of active drug targeting using different nanosystems, AuNP-based drug targeting to BC cells is relatively limited. Jiang et al. (2008) perfectly demonstrated how the size and antibody conjugation greatly affects the uptake and internalization of drug loaded AuNPs in the tumor site. Their group suggested that the smaller particles became dissociated from the cell membrane whereas trastuzumab loaded AuNPs of 40–50 nm diameter reduced the membrane wrapping, thus permitting the AuNPs to enter inside SK-BR-3 cells. They also suggested that the targeting approach is achieved in a larger extent for HER2-conjugated Trastuzumab/AuNPs than non-targeted drug loaded NPs alone with a 2-fold increase in chemotherapeutic efficacy for Trastuzumab, being released by the lowered pH of lysosomal compartments inside the cell.335 In another study by Garcia Calavia et al. (2018), increased ROS production and apoptosis was observed with lactose-phthalocyanine-AuNPs after irradiation. The attached lactose in AuNPs helped in targeting MDA-MB-231 and SK-BR-3 cells and in binding with the galactose binding receptor (galactin-1) present in the surface of BC cells.196 Similar studies exploring AuNPs as targeted therapeutics in BC management are tabulated in Table 7.
| Anticancer drug | AuNP system | Study method | Drug release | Experimental outcomes | Reference |
|---|---|---|---|---|---|
| Doxorubicin (Dox) | DOX-conjugated levan-capped AuNPs | In vitro: MCF-7 | pH responsive biphasic release. Initial burst release disrupting electrostatic and hydrogen bonding interaction between drugs and particles followed by sustained release disrupting levan-particle strong hydrophobic interaction | Dox-levan linked AuNPs showed better cytotoxicity due to specific targeting | 336 |
| EpCAM Apt-peptosome conjugated AuNRs loaded with Dox | In vitro: MCF-7 and 4T1; in vivo: 4T1 xenograft BALB/c mice | pH responsive sustained, controlled release of DOX | EpCAM-Apt-Pep@AuNR-Dox showed higher cellular uptake with increased cytotoxicity against 4T1 | 337 | |
| Docetaxel (DTX) | Magnetic Au/Fe3O4/PVA-10%DTX nanocomposite | In vitro and in vivo: MCF-7 and xenograft mouse model | Release via NIR irradiation onto nanosystem through temperature sensitive PVA swelling after heating with green light | In vitro apoptosis 75% and in vivo apoptosis 70% achieved with minimal side effect | 338 |
| Epirubicin (EPI) | EPI-folic acid-AuNPs complex | In vitro: MCF-7 | pH dependent controlled release | 200% increased cytotoxicity observed for EPI-FA-AuNPs than free drug | 339 |
| Gemcitabine (GEM) | GEM loaded gum acacia (GA) influenced AuNPs | In vitro: MDA-MB-231 | Acidic pH mediated drug release | GEM-GA-AuNPs displayed more cytotoxicity than free GEM | 223 |
| Letrozole (LTZ) | LTZ loaded GA-AuNPs | In vitro: MCF-7 | Acidic pH mediated release without burst effect | Higher and dose dependent cytotoxicity than free LTZ was observed | 340 |
| Linalool (LIN) | LIN-AuNPs-modified with CALNN peptide | In vitro: MCF-7 | Not reported | LIN-AuNPs-CALNN were remarkably more cytotoxic than LIN or AuNPs alone | 341 |
| Methotrexate (MTX) | Gelatin-coated and MTX loaded AuNPs and AuNRs | In vitro: MCF-7 | pH mediated size and shape dependent drug release. Drug released more with small sized AuNR | AuNRs exhibited more drug release efficacy and cytotoxicity than AuNPs | 342 |
| 99mTc-MTX-AuNPs | In vitro: MCF-7 | Acidic environment controlled biphasic drug release; initially from the core shell followed by release from the core | 99mTc-MTX-AuNPs showed three times better anticancer activity (IC50 = 0.098 mL/100 mL) than MTX alone (IC50 = 0.3 mL/100 mL) | 343 | |
| Paclitaxel (PTX) | PTX-loaded PEG-AuNPs@antiMUC1 system | In vitro: MCF-7 and MDA-MB-231 | PTX was physically loaded into the PEG-AuNPs through the non-covalent interactions. Dual NIR/pH-dependent sustained release due to mitigation of electrostatic interaction by coulomb repulsion forces and OH group protonation of the drug and PEG-AuNPs | PTT and targeted drug delivery indicated a synergistic effect | 184 |
| Tamoxifen (TMX) | TMX loaded HAS-AuNPs | In vitro: BT-474 and MDA MB-231 | pH responsive biphasic release due to disruption of the disulfide bonds of albumin and the protonation of drug | Effective eradication of tumor cells achieved with targeted NPs | 163 |
| Curcumin (CUR) | CUR-loaded FA-functionalized Au-PVP NCs | In vitro: MDA-MB-231 and MCF-7; in vivo: 4T1 xenograft mice | pH responsive release via the protonation of drug | Inhibitory effects on cell migration and high antitumor efficacy | 160 |
| Berberine (BB) | BB loaded collagen (Col)-fabricated AuNPs | In vitro: HER-2 positive BC | Not defined | AuNPs-Col-BB significantly inhibited cell migration in HER-2 BC via inducing MMP-9 expression | 344 |
| 5-Fluorouracil (5-FU) | 5FU-complexed cyclodextrin conjugated AuNPs | In vitro: MDA-MB-231 and MCF-7 | Not defined | 5-FU/cyclodextrin/AuNPs showed enhanced EPR and increased proliferation inhibitory effect | 345 |
| Bleomycin (BLM) | BLM-RGD-AuNPs | In vitro: MDA-MB-231 | pH mediated release | More internalization, higher DNA damage and less survival than free BLM | 346 |
| Cisplatin (CPT) | Hyaluronic acid decorated CPT loaded AuNPs | In vitro: MCF-7; in vivo: MCF-7 xenograft mice | pH dependent release associated with polymer coating disruption by chloride ion, and due to electrophilicity of platinum | Enhanced cytotoxicity with encapsulated cisplatin AuNPs was noticed in comparison to free drug in vitro and in vivo | 347 |
5. Clinically developed nano-gold products: the major challenge for clinical application
Global research impetus on cancer management has crossed many decades but a permanent curative solution still eludes healthcare management in spite of cutting-edge technological breakthroughs in both diagnosis and management of the disease. Curative success to date is limited mostly to the 3rd stage. Research on nano-medicine in cancer control is now the sought after approach because of possibilities of site-specific delivery and evasion of drug resistance. Plenty of nano-formulations are on the market for cancer treatment with a crowded pipeline of such candidates currently undergoing preclinical or clinical investigations. Some obvious hurdles exist in the translation of such products to clinical use like complexity in scaling up, stability issues, cost-effectiveness etc. However the Nanotechnology Characterisation Laboratory (NCL) is responsible for analysis of the pre-clinical trial data of various nano-formulations targeted for cancer therapy. The Nanotechnology Characterisation Laboratory (NCL) is a federally funded facility in United States, collectively established by the National Cancer Institute (NCI), the National Institute of Standards and Technology (NIST), the Food and Drug Administration (FDA), and the National Institutes of Health (NIH) in purview of the fast-growing interest in clinical translation of nanotherapeutics/nanodiagnostics/nanotheranostics. NCL supports physicochemical characterization, in vitro testing, and in vivo studies to assess the safety, efficacy, and pharmacokinetics of nano-scale agents targeted for clinical application. By offering standardized protocols, reference materials, and expertise in nanotechnology characterization, the NCL plays a crucial role in safe and effective translation of nanotechnology-based solutions for diverse clinical applications. To date the NCL has performed different standardisation activity both in vitro and in vivo for more than 170–180 such drugs.348,349The exclusive optical, electrical and magnetic properties expressed by AuNPs are widely explored for clinical applications in different cancer variants (Table 8). Aurimune (CYT-6091) was the first developed product in this domain and was applied to solid tumour diagnosed patients (NCT00356980 and NCT00436410). CYT-6091, a recombinant human tumour necrosis factor alpha (TNF-α)-functionalized polymer (PEG) coated nano-gold with a size of 30 nm, is another such agent which modulated immune responses to treat cancer. The phase I clinical trial (NCT00356980) of this agent was among the earliest ones in this segment. The observed result revealed a tolerance dose range of 50–600 μg m−2 as well as the maximum tolerated dose of 600 μg m−2 with no acute side effects.333
| Sl. no. | Name of the gold nanoparticle | No. of participants | Stage | Clinical trial identifier | Application | Reference |
|---|---|---|---|---|---|---|
| 1 | Aurimune CYT-6091 | 60 | Phase I completed (April 2009) | NCT00356980 | Advanced solid tumors | 333 |
| Aurimune CYT-6091 (PEGylated colloidal AuNP-bound rhTNFα) | 108 | Phase I completed (August 2009) | NCT00436410 | Adrenocortical carcinoma breast, colorectal, gastrointestinal, kidney, liver, skin, ovarian, pancreatic cancer | ||
| 2 | Auroshell (AuroLase™) | 11 | Interventional pilot study completed (August 2014) | NCT00848042 | Recurrent tumour and cancers (head & neck) | 355 |
| AuroShell particle infusion | 45 | Phase I (October 2020) | NCT02680535 | Neoplasms of the prostate | 356 | |
| 3 | NU-0129 (spherical nucleic acid-AuNPs) | 8 | Early phase 1 completed (August 19, 2020) | NCT03020017 | Gliosarcoma and Recurrent glioblastoma | 357 |
| 4 | Organically functionalized AuNP and carbon nanotubes | 1000 | Observational completed | NCT01420588 | Patients associated with malignant or benign gastric lesions | 354 |
| 5 | CD24-Au nanocomposite | 60 | Observational completed (February 3, 2021) | NCT04907422 | Carcinoma ex pleomorphic adenoma of salivary glands | 358 |
| 6 | Nano swarna bhasma | 6 | Pilot study completed | DNA_SPN_B001_17 (AYUSH, India) | Breast cancer | 352 |
Auroshell is a silica-gold nanoconjugation product of size ∼150 nm which in under clinical trials using photothermal ablation technique in cancer therapy. Auroshell was applied in 22 patients of prostate cancer and provided excellent control with minimal adverse symptoms like itching and burning sensation in epigastrium.350 Aurolase is the modification of Auroshell therapy which also adds the NIR laser approach for inducing thermal ablation of recurrent head and neck cancers.351 Khoobchandani et al. (2020) in his research investigated a sustainable newer biosafe gold nano-formulation for breast cancer management from preclinical to clinical trials. This product was designed by conjugation of mangiferin with AuNPs with subsequent engineering with siRNA & thiolated PEG and was named “Nano-Swarna-Bhasma”.352 Another spherical nucleic acid-Au-nanoconjugate named NU-0129 (NCT03020017) has completed an early phase I study intended for the treatment of glioblastoma, with minimal toxicity.353 Another example of AuNP under clinical evaluation is organically functionalized AuNPs and single-walled carbon nanotubes (NCT01420588) which is in observation I stage and targeted for patients with malignant or benign gastric lesions. The nano-complex exclusively diagnose gastric cancer and distinguishes it from other gastric diseases.354
Despite continuous efforts, Au-nano based products' clinical status is limited to early phase I or phase I clinical stages. Compared to other nano-formulations viz. liposomes and polymeric nanoparticles, AuNP-based products or formulations or nanodevices failed to enter the late phase clinical stage and they have not been marketed. One of the major limitations contributing to the failure of Au-based nanoformulations include toxicity, which is discussed next at length. Despite using a targeted approach, AuNPs cause significant side-effects to non-malignant tissues due to several factors like size, shape, chemical functionalities, biodistribution alterations etc.355 AuNPs can produce long-term toxicity as they are non-degradable entities. CYT-6091 and NU-0129, phase I cleared AuNP based products, pre-clinically suggested detectable amounts of gold in the liver and tumor cells even after 120 or 174 days of treatment.359 The pilot study with AuroLase™ in primary or metastatic lung cancer was terminated in 2014.355
To develop encouraging and improved results of cancer management using AuNPs, a length of detailed understanding of NP-target interaction and the toxicity parameters are highly required. More pre-clinical studies are needed prior to starting any early stage clinical trials associated with Au-based nanosystems against cancer for successful clinical outcomes.
6. Toxicity issues of AuNPs
The major question in scientific minds while exploring the versatile applications of AuNPs is their toxicity and biosafety especially in the case of clinical applications. Interestingly, gold in bulk form exhibits inertness and biocompatibility most of the time. However, its behavior changes contrastingly when Au is in nano-dimensional form. Scientific reports related to its formulation development to date failed to provide a concrete idea regarding to what extent or at which concentration AuNPs exert toxicity or to what extent the toxicity arises when the effective concentration is applied. Repeated dose administration of AuNPs in animals revealed interesting facts with no prominent signs of toxicity. Firstly, even after increasing the dose, gold concentration in systemic circulation did not increase, although its presence in the different organs increased. Secondly, different studies reported there were no toxicity signs such as life-span, behavioral pattern, body-weight, organ morphology, blood parameters and tissue histo-architecture.360Diverse parameters which influence the toxicity of AuNPs include: (i) dimension, (ii) morphology, (iii) chemical functionalization and coating agents defining their surface chemistry, (iv) type of assay used, (v) incubation conditions related to time of exposure and concentration used and (vi) their interaction with biological materials like proteins, fluids etc.361 However, the complexity enhances when in assessment of toxicity of AuNPs because of the overlapping factors where multi-variants come to play with distinctive roles in contributing the biocompatibility assessments of AuNPs.
6.1. Size dependent AuNPs toxicity
The primary concern related to the toxicity of AuNP is the particle size and shape. Particle size as well as the stabilizing agents used in its synthesis significantly affects agglomeration as well as the sedimentation of AuNPs which in-turn hampers the passage of the AuNPs through the cell membrane. Generally, the smaller particles with a large surface area tend to bio-accumulate more, leading to enhanced toxicity manifestations.362 Dependency of cytotoxicity on the size of AuNPs has yielded somewhat inconclusive results, as shown in the investigations by Pan et al. (2007). Their group synthesized tri-phenylphosphine stabilized AuNPs with particle size ranges from 0.8–15 nm, in which particularly AuNPs with 1.4 nm diameter showed toxicity to different cell lines.363 These peculiarity in toxicity levels by distinct particles most likely occur due to the well-fitting of AuNPs of a particular size with the major DNA groove. Different in vivo studies in rats and mice by De Jong et al. (2008), Abdelhalim et al. (2013) and Sonavane et al. (2008) confirmed that smaller particles tended to accumulate more in the organs like the liver, lung, spleen, kidney and heart as well as blood, indicating signs of toxicity.364–366 Irrespective of increased cellular uptake with smaller AuNPs, matching toxicity with size may not produce definite conclusions. Studies by Connor et al. (2005) and Murphy et al. (2008) suggested that AuNPs 4, 12 or 18 nm in diameter synthesized with different coating agents exhibited no significant toxicity at all in K562 human leukemia cells though the particles were well taken up by the cells as observed in TEM.367,368 AuNPs of size 3, 5, 50, and 100 nm administered to BALB/C mice i.p. at a dose of 8 mg per kg per week did not exhibit a toxic response however similar AuNPs with 8 to 37 nm diameter produced fatigue, loss of appetite, change of fur color and weight loss, subsequently causing mortality of the majority of the population after three weeks.3696.2. Shape dependent AuNPs toxicity
Different geometries of AuNPs play a critical role in their toxicity presentations. Among the different shapes of AuNPs, some researchers suggest that Au-nanostars with their specific shape provide additional advantages i.e. large surface area and high drug load which ultimately reduce their quantity used and thereby reduce toxicity compared to other shapes.361 However, AuNRs possess two different plasmon responses depending on the longer (longitudinal) and shorter (transverse) axis, hence possessing a different physicochemical property which complexes their cytotoxicity influence.370 Wang et al. (2013) developed naked and PEGylated Au-nanocages, nanorods and nanohexapods and compared their toxicities in the MDA-MB-435 BC cell line. According to the study, AuNRs were found to be most toxic whereas, Au-nanohexapods showed the lowest toxicity in spite of being well taken up by the cells.237 However, some studies clearly indicate that cellular uptake takes place more rapidly in spherical AuNPs than AuNRs. In this context, Li et al. (2015) suggested fastest internalization capability of Au-nanospheres followed by AuNRs, Au-nanocubes and Au-nanodisks.371 The researcher also suggested that Au-nanostars can definitely compete with Au-nanospheres in case of internalization as they were observed to be readily taken up by the cell membrane. In an in vivo study with the murine EMT-6 breast cancer model, Black et al. (2014) also confirmed that radioactive 198Au-doped nanospheres were better taken up by cancer cells than 198AuNRs and 198Au-nanocages of a similar size despite their good distribution pattern.372 AN in vivo study using a zebrafish model also confirmed this fact to be related to Au-nanospheres.3736.3. Surface functionalization and surface charge dependent AuNPs toxicity
Surface functionalization is one of the most important factors that governs different properties as well as toxicity of AuNPs. Different ligand functionalization or surface modification facilitates distribution as NP circulation in the bloodstream increases. PEG-coating on the surface of AuNPs not only defends particles against opsonization, but also develops a non-toxic AuNP delivery platform against healthy cells. Mioc et al. (2018) observed no toxicity against HaCaT and IBR-3 non-malignant cells. The author also suggested botulin-modified PEG-coated AuNPs indeed exerted toxicity against human and murine melanoma (A375 and B164A5) cell lines.374 However, Zhang et al. (2011) in their in vivo research study showed that the toxicity of PEG-coated AuNPs is somehow perplexing with respect to particle size. The toxicity exerted by 10 nm and 60 nm PEG-AuNPs was higher than that of the 5 nm and 30 nm particles.375Gum arabic coated cumin derived AuNPs of 13 nm size when tested for a viability assay against human fibroblast cells turned out to be non-toxic also after 24 h at a dose of 150 μM.376 Like PEG, surface modification with chitosan not only exerts excellent biocompatibility but also offers stability in the circulation period as well. The presence of sodium citrate residue in case of citrate capped AuNPs exposed to different cells generally induced toxicity as confirmed by different studies. However, according to Connor et al. (2005), AuNPs with varying sizes of 4–18 nm and varying functionalization i.e. citrate, glucose, biotin or cysteine or CTAB did not induce toxicity against the K562 leukemia cell line despite its higher internalization.367 A similar result with no toxicity was observed with citrate modified AuNPs against human dermal fibroblast-fetal cells even at high concentration of 300 μM but they had an impact on their morphology.377 On the other hand, conjugating phytochemicals with AuNPs has recently attracted the attention of researchers because of their ability to reduce the toxic impact of AuNPs.378
The net surface charge carried by NPs is another important aspect that modulates toxicity. NPs with a positive charge on their surface can bind to the negatively charged DNA, specifically targeting the minor and major grooves of DNA via electrostatic interaction causing DNA to bend around AuNPs which finally results in strand separation of the organized DNA structure.379 Damage to the DNA was also observed when the hydrophobic ligands of NPs attached to DNA due to van der Waals interactions. As electrostatic attraction strongly induces cellular uptake, the NPs with a negative charge on the surface can be internalized within the cellular microenvironment specifically in the vacuoles.380 Goodman et al. (2004) showed that the electrostatic interaction between the negative charges carried by cationic AuNPs was responsible for the lysis of the cell membrane which made them toxic as compared to the anionic AuNPs.381 However, interestingly Schaeublin et al. (2011) suggested that both positively and negatively charged AuNPs were equally responsible for inducing oxidative stress via altering of mitochondrial membrane potential.382
6.4. AuNP interaction with biological proteins
While determining cytotoxicity in vitro, the commonly used culture medium bypasses the use of serum or when even used, the media contains a lower amount of serum. For that reason, the cellular viability does not represent the actual toxicity. In case of in vivo study, the toxicity represents quite different results as serum and different proteins influence aggregation of particles hampering particle concentration. Interaction between biological proteins and AuNPs alter the surface charge of NPs.To decrease the high surface energy of NP, after entering the biological system, they adsorb biomolecules especially plasma proteins to form a protein shell called a “protein corona” (PC), which alters the original interfacial physicochemical properties of the NPs. In the circulatory environment, speedy protein corona formation occurs, influencing interactions with blood cells and potentially modulating their behavior in vivo383 [6–10 of protein corona]. The interaction between AuNPs and proteins is driven by various forces including van der Waals forces, hydrophobic and electrostatic interaction, solvation forces, coordination, steric hindrance, and hydrogen bonding.384 For instance, bovine serum albumin (BSA) interacts with AuNPs, via at least 12 strong Au–S bonds, where ubiquitin forms corona with citrate-coated Au NPs mainly via short-range hydrogen bonds.385 Proteins with high abundance initially adsorb onto nanoparticles, but are eventually replaced by high-affinity proteins, forming NP-protein complexes. The protein corona can be categorized as hard, with tightly bound proteins, or soft, with proteins less tightly bound and dynamically exchanging with those in the surrounding media over time.386
Various factors, including NP size, hydro/lipophilicity, surface properties, and incubation time, influence the composition of the protein corona, subsequently impacting the biological behavior of the nanomaterials. M. Schäffler et al. (2013) used gel electrophoresis and a combination of matrix-assisted laser desorption/ionization and time-of-flight mass spectrometry for quantitative identification of mouse serum proteins adsorbed on 5, 15 and 80 nm phosphine stabilized AuNPs with negative surface charges and reported that smaller nanoparticles exhibit lower protein adsorption due to their higher curvature.387 Reports by Cheng et al. (2015) also confirm that cellular uptake inhibition in phagocytic cells by PC is inversely proportional to the size of AuNPs.388 Surface modification with hydrophilic PEGs can resist complement protein adsorption, prolonging nanoparticle circulation time by evading uptake by the reticuloendothelial system (RES).387 Serum proteins interact with the particle surface and facilitate uptake via clathrin-mediated endocytosis, which is suggested to be linked to hydrophobicity or dispersibility of AuNPs.361 These findings underscore the importance of understanding protein corona dynamics in nanoparticle interactions with biological systems for optimizing their biomedical applications.
The interaction between gold nanoparticles (Au NPs) and proteins induces structural changes in both the nanoparticles and the adsorbed proteins, with the protein corona dynamically evolving over time due to continuous adsorption, desorption, and exchange influenced by various factors including protein concentration, surface chemistry of GNPs, and environmental conditions. Understanding how nanoparticle properties affect protein corona composition is crucial, and analytical techniques such as TEM, DLS, UV-Vis-NR spectra, CD spectra, QCM, and NMR, as well as conventional methods like chromatography and electrophoresis are essential for studying NP–protein interactions and characterizing the NP-protein complex. Advanced techniques like synchrotron radiation X-ray near edge absorption spectroscopy (XANES) provide insights into the bound interface structure of proteins.389
The formation of a protein corona can significantly alter the physical characteristics of AuNPs, including size, charge, and morphology and subsequently their bioactivities. Lai et al. (2017) demonstrated how proteins from human plasma induce negative surface charge to overall corona formed over 20 nm AuNPs. Around 300 proteins were identified on the coronas, while 99 are commonly found on each Ag and Au-nanomaterial.390 A biocorona can render them effectively invisible to the immune system, potentially masking the functionality of functionalized AuNPs. Although many reports convey that PC enhances the nanoparticle cellular localization. Carnovale et al. (2019) reported that serum-pre-incubation of AuNPs assisted in minimization of cellular uptake and subsequent toxicity. The same study reported that tyrosine capped AuNPs exhibited different cellular viabilities in diverse media conditions like serum-free (52.2%), serum-pre-incubated (79.9%) and serum-supplemented (106.1%).391 Moreover, the composition of the biocorona can change over time due to its dynamic nature, making it challenging to predict the behavior of NPs after exposure to a particular host environment.392 For example, interactions between fibrinogens and poly(acrylic acid)-coated AuNPs can induce protein unfolding, exposing its cryptic peptide which specifically interact with the Mac-1 receptor, leading to inflammation and cytokine release mediated by the NF-κB pathway.393
The attachment of opsonins like immunoglobulins and complements, can lead to rapid immune recognition and removal of the corona from the blood by macrophages, eventually leading to loss of its therapeutic effects.384 Studies suggested rapid AuNP agglomeration and associated toxicities due to an increased hydrodynamic size of formed biocorona complexes.394 The dynamic interaction of different proteins with the NPs can cover or eliminate the targeting moieties on NP surfaces, and effectively mitigate the recognition of the targeting ligands or functional groups by different targets in vivo.395,396 Also, changes in the protein secondary structure can reveal hydrophobic regions and new epitopes, affecting its native function, including protein fibrillation and enzymatic activity. These alterations are closely linked to potential biological toxicity.397 Zhifang Ma and his research group reported that the cytotoxicity exposed by AuNP–PC complex depends upon the degradation of PC degradation process. They suggested that HSA corona degrades faster than γ-globulin (HGG), and serum fibrinogen (HSF) corona and AuNP–HSA PC complex exhibit highest cytotoxicity by elevating ROS level, slowing down ATP production and lowering MMP.398 The research group of Yang et al. (2019) showed how conformational transition of HSA from partial β-sheet to α-helix upon AuNP–HSA PC formation led to increased thermal stability of the corona and increased cytotoxic nature of NP–PC complex via inducing apoptosis.399
However, tuning protein corona to possess beneficial applications like increasing residence time and reduced accumulation in the liver and spleen, ensuring drug delivery targeting also holds considerable appeal. It is widely recognized that AuNPs with an albumin-enriched surface exhibit a strong affinity for tumors because of the transcytosis mediated by albumin, suggesting significant promise for cancer-targeted therapy along with long–circulation properties, better biocompatibility and rapid clearance bypassing efficacy.400 The detailed impact of protein corona is well reviewed by Liu et al.384 and Wang et al.389
6.5. Variation in cell line and assay technique
Differences in toxicity with respect to use of different cell lines can be well observed in the study of Patra et al. (2007). Citrate capped AuNPs with 13 nm diameter exhibited remarkable anti-proliferative activity in human lung carcinoma cell line A549 while the toxicity vanished when the same dose was applied against the HepG2 human hepatocarcinoma cell line.401 One of our previous studies with luteolin tagged AuNPs goes in line with the difference in toxicity with respect to variable cell lines as the non-targeted nanosystem produced toxicity against TNBC cells while remained biocompatible with fibroblast cells (NIH-3T3).26While determining cytotoxicity, choosing an appropriate assay method is one of the major tasks to be carried out. There are different assays i.e. 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT) assay, lactate dehydrogenase (LDH) leakage test, calcein AM assay, colony forming (CFE) assay, 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium (WST-1) assay, tryptan blue assay etc. which are used routinely for cytotoxicity assessments. The conflicting result of toxicity of AuNPs also arises with varying assay methods used in experimental study. An example of the toxicity dependency on the assay method was reported by Coradeghini et al. (2013). The group showed that 5 nm AuNPs exerted toxicity after 72 h of treatment at a dose above 50 μM against Balb/3T3 mouse fibroblast cell line when determined through CFE assay while no toxicity was observed applying same sized AuNP, same dose and same cell line in case of cytotoxicity evaluation using tryptan blue assay. Even if the dose was escalated to a higher dose (300 μM) no toxic influence was observed.402
Besides the above discussed parameters, nanoparticle dose and concentration as well as administration route greatly affects the toxicity in a definite cell line.403 While evaluating toxicity relationship, it is better to consider not only LD50, but also ED50. NP concentration in the cells can be measured using two different but related concentration parameters i.e. numerical concentration and mass concentration which are expressed as below: 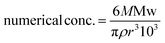 and
and 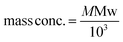 [where M = gold molar conc., Mw = Molar mass in g mol−1, r = radius of NP, ρ = density in g mol−1].404 Also, the clearance of the nanoparticles after completion of therapeutic action is also a parameter which can raise a question in defining toxicity states. According to Huang et al. (2008) the size may contribute to the clearance as they observed the persistence of >10 nm AuNPs in mice up to 6 months although no significant toxicity was observed.405
[where M = gold molar conc., Mw = Molar mass in g mol−1, r = radius of NP, ρ = density in g mol−1].404 Also, the clearance of the nanoparticles after completion of therapeutic action is also a parameter which can raise a question in defining toxicity states. According to Huang et al. (2008) the size may contribute to the clearance as they observed the persistence of >10 nm AuNPs in mice up to 6 months although no significant toxicity was observed.405
Thus, we can well understand that determining toxicity of AuNPs is a complex task to perform as lots of factors govern the toxic impact of the particle. And it is also highly required to design a suitable pathway or formula by which the toxicity of NPs can be addressed accurately. In Table 9, we generalize some of the studies with different toxicities of AuNPs as well as their governing attributes.
| Parameter | System and functionalization | Size | Cell line and animal used | Assay method (for in vitro) | Results obtained | Reference |
|---|---|---|---|---|---|---|
| Particle size | AuNPs without any surface modifiers | 1–3 nm, 3–5 nm, 5–7 nm, 7–10 nm | In vitro: mouse melanoma (B16F10) and non-malignant human keratinocyte (HaCaT) | MTT, crystal violet assay, trypan blue exclusion assay | Smaller particles (1–3 nm) produce more toxicity | 406 |
| Mono and tri-sulfonated triphenylphosphine functionalized AuNPs | 0.8–15 nm | HeLa, SK-Mel-28, L929, J774A1 | MTT assay | NPs with 1.4 nm size showed most toxicity; however, 15 nm AuNPs caused no toxicity | 363 | |
| Bare AuNPs | 20 and 100 nm | In vitro: human retina microvascular endothelial cells (HRMECs), CTX-TNA2, human retinoblastoma cells, SNUOT-Rb1; in vivo: C57BL/6 mice; dose given iv | MTT assay for in vitro; TUNEL assay for in vivo | No toxicity observed in in vitro analysis. However, 20 nm AuNPs penetrates trough BRB (blood retinal barrier) in vivo | 407 | |
| Citrate capped AuNPs | 13 ± 1.1 and 45 ± 3.2 nm | In vitro: human dermal fibroblasts (CF-31) | Apoptosis and colony formation assay | Larger particles were taken up in cell by clathrin mediated endocytosis and the smaller ones by phagocytosis | 408 | |
| Citrate stabilized AuNPs | 10, 30, 60 nm | In vitro: HepG2 and HT-29; in vivo model: Wister rats | LDH assay, Comet assay | Smaller particles damaged DNA via induced ROS and oxidative stress; larger particles got accumulated in liver and spleen though no inflammatory cytokine levels altered in rats | 236 | |
| Particle shape | Gold-silica nanoshells | Core 55 nm, shell 10 nm | In vitro: SK-BR-3 cells; in vivo: canine TVT cells inoculated female non-obese diabetic CB17-Prkd c SCID/J mice models | Fluorescent viability stain assay | No toxicity observed | 409 |
| Surface functionalization | Tea phytochemicals-and gum arabic-stabilized AuNPs | 15–45 nm | In vitro: PC-3, MCF-7 | MTT assay | Nontoxic | 410 |
| AuNPs coated with PEG and betulin | 10 nm, 50 nm | In vitro: HaCaT, 1BR3 cells | MTT assay | PEG coating diminished cytotoxicity in both the non-malignant cell lines | 374 | |
| AuNPs coated with citrate, starch, and gum arabic | 20 ± 1 nm | In vitro: PC-3, MCF-7, CHO22 cells | MTT, neutral red cell, and LDH assay | Gum arabic-AuNPs most biocompatible and citrate-AuNPs showed toxicity w.r.t. dose and time of exposure | 411 | |
| AuNRs coated with CTAB, phosphatidylcholine (PC) | 65 ± 5 nm × 11 ± 1 nm | In vitro: HeLa | MTT | Toxicity decreases remarkably for PC-AuNRs than CTAB-AuNRs | 412 | |
| Cumin-gum arabic-AuNPs | 13 nm | In vitro: human fibroblast cells | MTT assay | No toxicity observed | 376 | |
| Administered dose | Citrate AuNPs | 12.5 nm | In vivo: male C57/BL6 mice; ip given | N/A | Dose-dependent tissue accumulation without any toxicity | 360 |
| Route of administration | Citrate AuNPs | 13.5 nm | In vivo: male ICR mice | N/A | Oral and ip route causes more toxicity | 403 |
| Shape and surface chemistry | Spherical, rod-shaped, prismatic, and cubic AuNPs, citrate, tryptophan, tyrosine and CTAB coated NPs | Varying size | In vitro: human prostate cancer cells (PC3) | MTT assay | CTAB-stabilized spherical and prismatic AuNPs produced more toxicity where rod and cube shapes found to be bio-compatible. Uptake increased observed with amino acid coated NPs | 391 |
| Size and surface chemistry | BPEI-lipoic acid-PEG coated AuNPs forming biocorona with HSA | 40 and 80 nm | In vitro: HUVEC | AlamarBlue assay | 40 nm BPEI-AuNPs showed most uptake and toxicity. Biocorona formation led to non-toxic nature. HAS tagging increased uptake | 413 |
Development of surface defects during nanomaterial development like constitutional arrangement, disarray, grain margins, and diverse surface topography would modulate its interactions with cellular environment thus influencing toxicity signs.361 The impact of such deformities in formation and subsequently their effect on toxicity in case of AuNPs could be an area of interest for future investigations.
Conclusions
Gold mediated nanotechnological interventions have revolutionized cancer management by facilitating transport of drug cargo along with particle engineering mediated pin-point selectivity and magnified uptake in cancerous cells. The role of AuNPs has undergone a paradigm shift in the last decade making them the go-to candidate not only for therapy and diagnosis but also as a multifunctional device with theranostic attributes. Breast cancer management often involve single chemotherapeutics or their combination and, in both cases, AuNPs serve as an efficient carrier. The unique properties of gold at the nanoscale, the influence of particle engineering and ease of metamorphosis of AuNPs into potent multifunctional structures, as well as their inertness, biocompatibility, chemical and thermal stability, and resistance to oxidation or corrosion support their superiority over other nanoparticles and have propelled their journey from laboratories to the market. Moreover, the entry of AuNPs into newer avenues like PTT and PDT therapy or their use as contrast agents in X-ray, CT and multimodal imaging has further prompted clinicians to explore nanodimensional gold in cancer management. As stated in the review, AuNPs in clinical practice has now become a reality. In addition to this fact, a huge pipeline of nano-scale gold products are currently undergoing Phase I, Phase II or Phase III trials and may soon reach the market. This review focuses on unravelling the various synthesis routes and tailoring of gold nanoparticles for their targeted use in breast cancer management.Immense scientific variations in synthesis procedures of AuNPs have occurred from the route initiated by Frens or Turkevich, which have significantly favoured their pharmacodynamics but the toxicity aspects of nano-scale gold are a major concern in their translation to clinics. Even toxicity assessments of engineered or multifunctional AuNPs pose a complex challenge to nano-toxicologists. However, biocompatibility issues, scale-up obstacles and the development of proper regulatory framework to cater these nano-dimensional products still require brainstorming ideas from all connected scientific professionals to strengthen AuNPs' clinical presence.
Gold nanoparticles enable the combination of chemotherapy, photothermal therapy, and radiotherapy, enhancing efficacy while minimizing side effects. Gold nanoparticles' unique properties circumvent drug resistance mechanisms, prolonging therapy effectiveness. AuNP-based immunotherapies leverage the immune system, improving patient outcomes and long-term survival rates. Selectively heating and destroying cancer cells with laser-activated AuNPs allows precise tumor removal, reducing damage to healthy tissue and improving patient recovery. Future research may focus on optimizing targeting strategies and investigating novel ligands for improved specificity. Additionally, exploring synergies between gold nanoparticles and conventional cancer therapies could enhance efficacy while minimizing side effects. Investigating AuNPs' potential to overcome drug resistance mechanisms may lead to more effective combination therapies. Developing multifunctional AuNPs capable of imaging cancer cells while delivering therapeutic payloads is a promising avenue. Elucidating the mechanisms underlying the immunomodulatory effects of AuNPs and integrating AuNPs with emerging technologies for cancer screening are areas of interest. Last but not least, optimizing the physicochemical properties of AuNPs to enhance biocompatibility and safety, along with long-term studies assessing systemic effects, are crucial for clinical translation.
Conflicts of interest
The authors declare no relevant financial or non-financial conflict of interests.Acknowledgements
This work was financially supported by the “Indian Council of Medical Research, Government of India” (Grant Number: AMR/Adhoc/301/2022-ECD-II) to Prof. Sudhir Kumar Paidesetty; the “Royal Society of Chemistry, UK”, (Grant No. R21-1248177714) to Prof. Suvadra Das & Prof. Partha Roy and SOADU-PhD fellowship to Mr Suvadeep Mal (Registration No. 2081606007) in Pharmacy. Continuous support from Dean Research and Dean SPS, SOA (Deemed to be University), Chancellor UEM Kolkata and Dean GITAM (Deemed to be University) throughout the entire work tenure are heartily acknowledged.References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- G. N. Sharma, R. Dave, J. Sanadya, P. Sharma and K. Sharma, J. Adv. Pharm. Technol. Res., 2010, 1, 109 Search PubMed.
- M. Mahapatra, P. Mohapatra, K. Pakeeraiah, R. K. Bandaru, I. Ahmad, S. Mal, R. Dandela, S. K. Sahoo, H. Patel and S. K. Paidesetty, Int. J. Biol. Macromol., 2023, 249, 126084 CrossRef CAS PubMed.
- K. W. Powers, M. Palazuelos, B. M. Moudgil and S. M. Roberts, Nanotoxicology, 2007, 1, 42–51 CrossRef CAS.
- M. E. Davis, Z. Chen and D. M. Shin, Nat. Rev. Drug Discovery, 2008, 7, 771–782 CrossRef CAS PubMed.
- K. Pakeeraiah, S. Mal, M. Mahapatra, S. K. Mekap, P. K. Sahu and S. K. Paidesetty, Int. J. Biol. Macromol., 2023, 128402 Search PubMed.
- B. Mekuye and B. Abera, Nano Sel., 2023, 486–501 CrossRef CAS.
- G. D. Nash, Calif. Hist., 1998, 77, 276–292 CrossRef.
- M. Zaimy, N. Saffarzadeh, A. Mohammadi, H. Pourghadamyari, P. Izadi, A. Sarli, L. Moghaddam, S. Paschepari, H. Azizi and S. Torkamandi, Cancer Gene Ther., 2017, 24, 233–243 CrossRef CAS PubMed.
- M. Yafout, A. Ousaid, Y. Khayati and I. S. El Otmani, Sci. Afr., 2021, 11, e00685 CAS.
- Á. I. López-Lorente, Anal. Chim. Acta, 2021, 1168, 338474 CrossRef PubMed.
- W. Yang, H. Liang, S. Ma, D. Wang and J. Huang, Sustainable Mater. Technol., 2019, 22, e00109 CrossRef CAS.
- M. K. Hameed, J. B. Parambath, M. T. Gul, A. A. Khan, Y. Park, C. Han and A. A. Mohamed, Appl. Surf. Sci., 2022, 583, 152504 CrossRef CAS.
- M. Fan, Y. Han, S. Gao, H. Yan, L. Cao, Z. Li, X.-J. Liang and J. Zhang, Theranostics, 2020, 10, 4944 CrossRef CAS PubMed.
- L. Rotimi, M. O. Ojemaye, O. O. Okoh, A. Sadimenko and A. I. Okoh, Green Chem. Lett. Rev., 2019, 12, 61–68 CrossRef CAS.
- J. S. Chen, J. Chen, S. Bhattacharjee, Z. Cao, H. Wang, S. D. Swanson, H. Zong, J. R. Baker and S. H. Wang, J. Nanobiotechnol., 2020, 18, 1–9 CrossRef CAS PubMed.
- K. Shrivas, N. Nirmalkar, S. S. Thakur, R. Kurrey, D. Sinha and R. Shankar, RSC Adv., 2018, 8, 24328–24337 RSC.
- W. N. Rahman, N. Bishara, T. Ackerly, C. F. He, P. Jackson, C. Wong, R. Davidson and M. Geso, in Nanomedicine in Cancer, Jenny Stanford Publishing, 2017, pp. 737–752 Search PubMed.
- S. Chakraborty, S. Mal, A. Halder, S. Das, K. K. Sen, A. Mukherjee and P. Roy, Part. Sci. Technol., 2024, 42, 145–163 CrossRef CAS.
- N. Sarfraz and I. Khan, Chem.–Asian J., 2021, 16, 720–742 CrossRef CAS PubMed.
- V. Ramalingam, Adv. Colloid Interface Sci., 2019, 271, 101989 CrossRef CAS PubMed.
- S. Murthy, P. Effiong and C. C. Fei, in Metal Oxide Powder Technologies, Elsevier, 2020, pp. 233–251 Search PubMed.
- N. R. S. Sibuyi, K. L. Moabelo, A. O. Fadaka, S. Meyer, M. O. Onani, A. M. Madiehe and M. Meyer, Nanoscale Res. Lett., 2021, 16, 1–27 CrossRef PubMed.
- J. Lee, D. K. Chatterjee, M. H. Lee and S. Krishnan, Cancer Lett., 2014, 347, 46–53 CrossRef CAS PubMed.
- F. Wang, Y.-C. Wang, S. Dou, M.-H. Xiong, T.-M. Sun and J. Wang, ACS Nano, 2011, 5, 3679–3692 CrossRef CAS PubMed.
- S. Mal, T. Saha, A. Halder, S. K. Paidesetty, S. Das, W. T. Wui, U. Chatterji and P. Roy, J. Drug Delivery Sci. Technol., 2023, 80, 104148 CrossRef CAS.
- V. Patsula, L. Kosinová, M. Lovrić, L. Ferhatovic Hamzić, M. Rabyk, R. Konefal, A. Paruzel, M. Šlouf, V. Herynek and S. k. Gajović, ACS Appl. Mater. Interfaces, 2016, 8, 7238–7247 CrossRef CAS PubMed.
- S. Wirunchit, P. Gansa and W. Koetniyom, Mater. Today: Proc., 2021, 47, 3554–3559 CAS.
- N. Z. A. Naharuddin, A. R. Sadrolhosseini, M. H. A. Bakar, N. Tamchek and M. A. Mahdi, Opt. Mater. Express, 2020, 10, 323–331 CrossRef CAS.
- M. Pišlová, K. Kolářová, B. Vokatá, A. Brož, P. Ulbrich, L. Bačáková, Z. Kolská and V. Švorčík, Mater. Sci. Eng. C, 2020, 115, 111087 CrossRef PubMed.
- M. Annadhasan, J. Kasthuri and N. Rajendiran, ChemistrySelect, 2019, 4, 6557–6567 CrossRef CAS.
- M. Gautam, J. O. Kim and C. S. Yong, J. Pharm. Invest., 2021, 51, 361–375 CrossRef CAS PubMed.
- S. Manivannan and R. Ramaraj, Chem. Eng. J., 2012, 210, 195–202 CrossRef CAS.
- Y. He, E. Du, X. Zhou, J. Zhou, Y. He, Y. Ye, J. Wang, B. Tang and X. Wang, Spectrochim. Acta, Part A, 2020, 230, 118031 CrossRef CAS PubMed.
- R. G. Palgrave and I. P. Parkin, J. Am. Chem. Soc., 2006, 128, 1587–1597 CrossRef CAS PubMed.
- K. X. Lee, K. Shameli, Y. P. Yew, S.-Y. Teow, H. Jahangirian, R. Rafiee-Moghaddam and T. J. Webster, Int. J. Nanomed., 2020, 275–300 CrossRef CAS PubMed.
- A. Rónavári, N. Igaz, D. I. Adamecz, B. Szerencsés, C. Molnar, Z. Kónya, I. Pfeiffer and M. Kiricsi, Molecules, 2021, 26, 844 CrossRef PubMed.
- J. Turkevich, P. C. Stevenson and J. Hillier, Discuss. Faraday Soc., 1951, 11, 55–75 RSC.
- W. Patungwasa and J. H. Hodak, Mater. Chem. Phys., 2008, 108, 45–54 CrossRef CAS.
- G. Frens, Nature Phys. Sci., 1973, 241, 20–22 CrossRef CAS.
- K. Zabetakis, W. E. Ghann, S. Kumar and M.-C. Daniel, Gold Bull., 2012, 45, 203–211 CrossRef CAS.
- J. Dong, P. Carpinone, G. Pyrgiotakis, P. Demokritou and B. Moudgil, KONA Powder Part. J., 2020, 37, 2020011 Search PubMed.
- L. M. Liz-Marzán, Chem. Commun., 2013, 49, 16–18 RSC.
- C. A. Waters, A. J. Mills, K. A. Johnson and D. J. Schiffrin, Chem. Commun., 2003, 540–541 RSC.
- M. Brust, M. Walker, D. Bethell, D. J. Schiffrin and R. Whyman, J. Chem. Soc. Chem. Commun., 1994, 801–802 RSC.
- M. T. Reetz and W. Helbig, J. Am. Chem. Soc., 1994, 116, 7401–7402 CrossRef CAS.
- C.-J. Huang, P.-H. Chiu, Y.-H. Wang, K.-L. Chen, J.-J. Linn and C.-F. Yang, J. Electrochem. Soc., 2006, 153, D193 CrossRef CAS.
- Z.-C. Xu, C.-M. Shen, C.-W. Xiao, T.-Z. Yang, H.-R. Zhang, J.-Q. Li, H.-L. Li and H.-J. Gao, Nanotechnology, 2007, 18, 115608 CrossRef.
- N. Li, P. Zhao and D. Astruc, Angew. Chem., Int. Ed., 2014, 53, 1756–1789 CrossRef CAS PubMed.
- C. Kohout, C. Santi and L. Polito, Int. J. Mol. Sci., 2018, 19, 3385 CrossRef PubMed.
- B. Prasad, S. I. Stoeva, C. M. Sorensen and K. J. Klabunde, Chem. Mater., 2003, 15, 935–942 CrossRef CAS.
- S. J. Amina and B. Guo, Int. J. Nanomed., 2020, 9823–9857 CrossRef CAS PubMed.
- M. S. Punnoose, D. Bijimol and B. Mathew, Environ. Nanotechnol., Monit. Manage., 2021, 16, 100525 Search PubMed.
- J. Choi, S. Park, Z. Stojanović, H.-S. Han, J. Lee, H. K. Seok, D. Uskoković and K. H. Lee, Langmuir, 2013, 29, 15698–15703 CrossRef CAS PubMed.
- M. J. Firdhouse and P. Lalitha, Inorg. Chem. Commun., 2022, 109800 CrossRef.
- U. Shedbalkar, R. Singh, S. Wadhwani, S. Gaidhani and B. Chopade, Adv. Colloid Interface Sci., 2014, 209, 40–48 CrossRef CAS PubMed.
- A. Ahmad, S. Senapati, M. I. Khan, R. Kumar, R. Ramani, V. Srinivas and M. Sastry, Nanotechnology, 2003, 14, 824 CrossRef CAS.
- S. Mukherjee and C. R. Patra, Future Sci. OA, 2017, 3, FSO203 CrossRef PubMed.
- D. Lahiri, M. Nag, H. I. Sheikh, T. Sarkar, H. A. Edinur, S. Pati and R. R. Ray, Front. Microbiol., 2021, 12, 636588 CrossRef PubMed.
- C. Kamaraj, S. Karthi, A. D. Reegan, G. Balasubramani, G. Ramkumar, K. Kalaivani, A. A. Zahir, P. Deepak, S. Senthil-Nathan and M. M. Rahman, Environ. Res., 2022, 213, 113711 CrossRef CAS PubMed.
- S. Pandey, G. Oza, A. Gupta, R. Shah and M. Sharon, Eur. J. Exp. Biol., 2012, 2, 475–483 CAS.
- M. Ovais, A. T. Khalil, M. Ayaz, I. Ahmad, S. K. Nethi and S. Mukherjee, Int. J. Mol. Sci., 2018, 19, 4100 CrossRef PubMed.
- R. Sanghi, P. Verma and S. Puri, Adv. Chem. Eng. Sci., 2011, 1, 154 CrossRef CAS.
- J. T. Nandhini, D. Ezhilarasan and S. Rajeshkumar, Environ. Toxicol., 2021, 36, 24–32 CrossRef CAS PubMed.
- R. Shunmugam, S. R. Balusamy, V. Kumar, S. Menon, T. Lakshmi and H. Perumalsamy, J. King Saud Univ., Sci., 2021, 33, 101260 CrossRef.
- T. Cherian, D. Maity, R. T. Rajendra Kumar, G. Balasubramani, C. Ragavendran, S. Yalla, R. Mohanraju and W. J. Peijnenburg, Nanomaterials, 2022, 12, 2940 CrossRef CAS PubMed.
- N. Y. Nadaf and S. S. Kanase, Arabian J. Chem., 2019, 12, 4806–4814 CrossRef CAS.
- F. Kabiri, S. S. Aghaei, A. A. Pourbabaee, M. Soleimani and T. Komeili Movahhed, Prep. Biochem. Biotechnol., 2023, 53, 265–278 CrossRef CAS PubMed.
- N. Naimi-Shamel, P. Pourali and S. Dolatabadi, J. Mycol. Med., 2019, 29, 7–13 CrossRef CAS PubMed.
- M. A. Dheyab, M. N. Owaid, M. A. Rabeea, A. A. Aziz and M. S. Jameel, Environ. Nanotechnol., Monit. Manage., 2020, 14, 100312 Search PubMed.
- P. Clarance, B. Luvankar, J. Sales, A. Khusro, P. Agastian, J.-C. Tack, M. M. Al Khulaifi, H. A. Al-Shwaiman, A. M. Elgorban and A. Syed, Saudi J. Biol. Sci., 2020, 27, 706–712 CrossRef CAS PubMed.
- S. Krishnan, P. N. Patel, K. K. Balasubramanian and A. Chadha, New J. Chem., 2021, 45, 1915–1923 RSC.
- B. Babu, S. Palanisamy, M. Vinosha, R. Anjali, P. Kumar, B. Pandi, M. Tabarsa, S. You and N. M. Prabhu, Bioprocess Biosyst. Eng., 2020, 43, 2231–2242 CrossRef CAS PubMed.
- M. Vinosha, S. Palanisamy, R. Muthukrishnan, S. Selvam, E. Kannapiran, S. You and N. M. Prabhu, Process Biochem., 2019, 85, 219–229 CrossRef CAS.
- N. González-Ballesteros, L. Diego-González, M. Lastra-Valdor, M. Grimaldi, A. Cavazza, F. Bigi, M. C. Rodríguez-Argüelles and R. Simón-Vázquez, Mar. Drugs, 2022, 20, 182 CrossRef PubMed.
- E. Castro-Longoria, A. R. Vilchis-Nestor and M. Avalos-Borja, Colloids Surf., B, 2011, 83, 42–48 CrossRef CAS PubMed.
- K. K. Bharadwaj, B. Rabha, S. Pati, T. Sarkar, B. K. Choudhury, A. Barman, D. Bhattacharjya, A. Srivastava, D. Baishya and H. A. Edinur, Molecules, 2021, 26, 6389 CrossRef CAS PubMed.
- D. A. Lomelí-Rosales, A. Zamudio-Ojeda, O. K. Reyes-Maldonado, M. E. López-Reyes, G. C. Basulto-Padilla, E. J. Lopez-Naranjo, V. M. Zuñiga-Mayo and G. Velázquez-Juárez, Molecules, 2022, 27, 1692 CrossRef PubMed.
- M. Uzma, N. Sunayana, V. B. Raghavendra, C. S. Madhu, R. Shanmuganathan and K. Brindhadevi, Process Biochem., 2020, 92, 269–276 CrossRef CAS.
- S. Akhtar, S. Asiri, F. A. Khan, S. Gunday, A. Iqbal, N. Alrushaid, O. Labib, G. Deen and F. Henari, Arabian J. Chem., 2022, 15, 103594 CrossRef CAS.
- J. Chen, J. Ding, D. Li, Y. Wang, Y. Wu, X. Yang, A. Chinnathambi, S. H. Salmen and S. A. Alharbi, Arabian J. Chem., 2022, 15, 103479 CrossRef CAS.
- M. S. Majoumouo, J. R. Sharma, N. R. Sibuyi, M. B. Tincho, F. F. Boyom and M. Meyer, Molecules, 2020, 25, 4469 CrossRef CAS PubMed.
- S. Ahmed and S. Ikram, J. Photochem. Photobiol., B, 2016, 161, 141–153 CrossRef CAS PubMed.
- J. Santhoshkumar, S. Rajeshkumar and S. V. Kumar, Biochem. Biophys. Rep., 2017, 11, 46–57 CAS.
- S. Vimalraj, T. Ashokkumar and S. Saravanan, Biomed. Pharmacother., 2018, 105, 440–448 CrossRef CAS PubMed.
- S. O. Anadozie, O. B. Adewale, N. R. Sibuyi, A. O. Fadaka, C. C. Isitua, H. Davids and S. Roux, Process Biochem., 2023, 128, 49–57 CrossRef CAS.
- P. Ahati, T. Xu, L. Chen and H. Fang, Inorg. Chem. Commun., 2022, 143, 109791 CrossRef CAS.
- I. S. Saputra, A. H. Saputro, D. O. B. Apriandanu, Y. N. Permana and Y. Yulizar, Chem. Pap., 2022, 76, 4733–4742 CrossRef CAS.
- A. A. Dudhane, S. R. Waghmode, L. B. Dama, V. P. Mhaindarkar, A. Sonawane and S. Katariya, Int. J. Nanosci. Nanotechnol., 2019, 15, 75–82 CAS.
- S. S. Asl, F. Tafvizi and H. Noorbazargan, Environ. Sci. Pollut. Res., 2023, 30, 20168–20184 CrossRef CAS PubMed.
- M. S. Punnoose and B. Mathew, Res. Chem. Intermed., 2022, 48, 1025–1044 CrossRef CAS.
- S. Balasubramanian, S. M. J. Kala and T. L. Pushparaj, J. Drug Delivery Sci. Technol., 2020, 57, 101620 CrossRef CAS.
- O. T. Fanoro, S. Parani, R. Maluleke, T. C. Lebepe, J. R. Varghese, V. Mavumengwana and O. S. Oluwafemi, Antibiotics, 2021, 10, 893 CrossRef CAS PubMed.
- I. Fatimah, H. Hidayat, B. H. Nugroho and S. Husein, S. Afr. J. Chem. Eng., 2020, 34, 97–106 Search PubMed.
- G. Suriyakala, S. Sathiyaraj, R. Babujanarthanam, K. M. Alarjani, D. S. Hussein, R. A. Rasheed and K. Kanimozhi, J. King Saud Univ., Sci., 2022, 34, 101830 CrossRef.
- M. A. Alghuthaymi, C. Rajkuberan, T. Santhiya, O. Krejcar, K. Kuča, R. Periakaruppan and S. Prabukumar, Plants, 2021, 10, 2370 CrossRef CAS PubMed.
- F. A. Alhumaydhi, I. Khan, A. Rauf, M. N. Qureshi, A. S. Aljohani, S. A. Khan, A. A. Khalil, M. A. El-Esawi and N. Muhammad, Green Process. Synth., 2021, 10, 230–245 CrossRef CAS.
- N. Alikhani, M. Hekmati, B. Karmakar and H. Veisi, Inorg. Chem. Commun., 2022, 139, 109351 CrossRef CAS.
- S. O. Anadozie, O. B. Adewale, A. O. Fadaka, O. B. Afolabi and S. Roux, Biocatal. Agric. Biotechnol., 2022, 42, 102348 CrossRef CAS.
- S. Bawazeer, I. Khan, A. Rauf, A. S. Aljohani, F. A. Alhumaydhi, A. A. Khalil, M. N. Qureshi, L. Ahmad and S. A. Khan, Green Process. Synth., 2022, 11, 11–28 CrossRef CAS.
- C. Pechyen, K. Ponsanti, B. Tangnorawich and N. Ngernyuang, Toxicol Rep, 2022, 9, 1092–1098 CrossRef CAS PubMed.
- C. Pechyen, K. Ponsanti, B. Tangnorawich and N. Ngernyuang, J. Mater. Res. Technol., 2021, 14, 2982–2991 CrossRef CAS.
- V. Sekar, M. M. Al-Ansari, J. Narenkumar, L. Al-Humaid, P. Arunkumar and A. Santhanam, J. King Saud Univ., Sci., 2022, 34, 102197 CrossRef.
- M. Reyes-Becerril, F. Ruvalcaba, V. Sanchez, M. G. López, J. Silva-Jara, L. Hernandez-Adame and C. Angulo, Aquacult. Res., 2021, 52, 3391–3402 CrossRef CAS.
- N. González-Ballesteros, J. Vidal-González and M. C. Rodríguez-Argüelles, J. Nanostruct. Chem., 2021, 1–10 Search PubMed.
- T. ul Haq and R. Ullah, Int. J. Nanosci., 2022, 21, 2250008 CrossRef CAS.
- N. S. Al-Radadi, Arabian J. Chem., 2021, 14, 102956 CrossRef CAS.
- J. Shilpha, V. Meyappan and N. Sakthivel, Process Biochem., 2022, 122, 224–237 CrossRef CAS.
- S. Kandasamy, S. Chinnappan, S. Thangaswamy and S. Balakrishnan, Mater. Res. Express, 2020, 6, 1250c1251 Search PubMed.
- B. A. Varghese, R. V. R. Nair, S. Jude, K. Varma, A. Amalraj and S. Kuttappan, J. Taiwan Inst. Chem. Eng., 2021, 126, 166–172 CrossRef CAS.
- O. S. ElMitwalli, O. A. Barakat, R. M. Daoud, S. Akhtar and F. Z. Henari, J. Nanopart. Res., 2020, 22, 1–9 CrossRef.
- R. C. Sandulovici, M. Carmen-Marinela, A. Grigoroiu, C. A. Moldovan, M. Savin, V. Ordeanu, S. N. Voicu, D. Cord, G. M. Costache and M. L. Galatanu, Pharmaceuticals, 2022, 16, 48 CrossRef PubMed.
- M. Salandari Rabori, M. Noroozi Karimabad and M. R. Hajizadeh, World Cancer Res. J., 2021, 8, e2037 Search PubMed.
- S. Mal and D. Pal, Bioactive Natural Products for Pharmaceutical Applications, 2021, pp. 715–757 Search PubMed.
- A. Halder, S. Das, D. Ojha, D. Chattopadhyay and A. Mukherjee, Mater. Sci. Eng. C, 2018, 89, 413–421 CrossRef CAS PubMed.
- A. Zuhrotun, D. J. Oktaviani and A. N. Hasanah, Molecules, 2023, 28, 3240 CrossRef CAS PubMed.
- S. Durmazel, A. Uzer, B. Erbil, B. Sayın and R. Apak, ACS Omega, 2019, 4, 7596–7604 CrossRef CAS PubMed.
- P. Kuppusamy, M. M. Yusoff, G. P. Maniam and N. Govindan, Saudi Pharm. J., 2016, 24, 473–484 CrossRef PubMed.
- N. Gan, C. Wakayama, S. Inubushi, T. Kunihisa, S. Mizumoto, M. Baba, H. Tanino and T. Ooya, ACS Appl. Bio Mater., 2021, 5, 355–365 CrossRef PubMed.
- M. H. Oueslati, L. B. Tahar and A. H. Harrath, Arabian J. Chem., 2020, 13, 3112–3122 CrossRef CAS.
- J. J. Park, S. J. Hwang, Y. S. Kang, J. Jung, S. Park, J. E. Hong, Y. Park and H.-J. Lee, Arch. Pharmacal Res., 2019, 42, 977–989 CrossRef CAS PubMed.
- Z. Sharifiaghdam, S. M. Amini, F. Dalouchi, A. B. Behrooz and Y. Azizi, Heliyon, 2023, 9(3), e14024 CrossRef CAS PubMed.
- A. J. Jasim, G. M. Sulaiman, H. Ay, S. A. Mohammed, H. A. Mohammed, M. S. Jabir and R. A. Khan, Nanotechnol. Rev., 2022, 11, 2726–2741 CrossRef CAS.
- A. Parthiban, V. Sachithanandam, S. Sarangapany, R. Misra, P. Muthukrishnan, T. C. Jeyakumar, R. Purvaja and R. Ramesh, J. Mol. Struct., 2023, 1272, 134167 CrossRef CAS.
- S. Kondath, R. Rajaram and R. Anantanarayanan, Inorg. Nano-Met. Chem., 2020, 51, 601–613 CrossRef.
- B. S. Inbaraj, L.-H. Hua and B.-H. Chen, Pharmaceutics, 2021, 13, 1871 CrossRef CAS PubMed.
- L. Chen, Y. Huo, Y. X. Han, J. F. Li, H. Ali, I. Batjikh, J. Hurh, J. Y. Pu and D. C. Yang, Appl. Phys. A, 2020, 126, 1–12 CrossRef.
- V. V. Vodnik, M. Mojić, U. Stamenović, M. Otoničar, V. Ajdžanović, D. Maksimović-Ivanić, S. Mijatović, M. M. Marković, T. Barudžija and B. Filipović, Mater. Sci. Eng. C, 2021, 124, 112078 CrossRef CAS PubMed.
- K. Pearce, V. C. Thipe, R. R. Henkel and K. V. Katti, J. Drug Delivery Sci. Technol., 2023, 80, 104100 CrossRef CAS.
- J. d. S. Oliveira and E. J. Guidelli, Mater. Sci. Eng. C, 2021, 126, 112122 CrossRef CAS PubMed.
- A. Gupta, R. Mathur, S. Singh, N. Bag, U. A. Khan, F. J. Ahmad, G. A. Gabr, P. Kesharwani and G. K. Jain, J. Pharm. Sci., 2021, 110, 888–897 CrossRef CAS PubMed.
- M. K. Hameed, I. M. Ahmady, C. Han and A. A. Mohamed, Amino Acids, 2020, 52, 941–953 CrossRef CAS PubMed.
- S. Zhang, M. Bai, J. Qian and Y. Guo, Microchem. J., 2021, 169, 106564 CrossRef CAS.
- B. A. Makwana, D. J. Vyas, K. D. Bhatt and V. K. Jain, Sens. Actuators, B, 2017, 240, 278–287 CrossRef CAS.
- Z. Huang, X. Zhang, Z. Yao, Y. Han, J. Ye, Y. Zhang, L. Chen, M. Shen and T. Zhou, Msphere, 2023, e00549 Search PubMed.
- K. Rajendran, A. Anwar, N. A. Khan, M. R. Shah and R. Siddiqui, ACS Chem. Neurosci., 2019, 10, 2692–2696 CrossRef CAS PubMed.
- I. Rajendran, T. Ponrasu, R. Rajaram and L. Suguna, J. Drug Delivery Sci. Technol., 2021, 63, 102478 CrossRef CAS.
- R. Chen, F. Chen, M. Sun, R. Zhang, S. Wu and C. Meng, Inorg. Nano-Met. Chem., 2022, 52, 1345–1351 CrossRef CAS.
- W. I. El-Ghareb, M. M. Swidan, I. T. Ibrahim, A. Abd El-Bary, M. I. Tadros and T. M. Sakr, Int. J. Pharm., 2020, 586, 119514 CrossRef CAS PubMed.
- T. Ferreira-Gonçalves, M. M. Gaspar, J. M. Coelho, V. Marques, A. S. Viana, L. Ascensão, L. Carvalho, C. M. Rodrigues, H. A. Ferreira and D. Ferreira, Biomolecules, 2022, 12, 71 CrossRef PubMed.
- W. Caseri, Macromol. Rapid Commun., 2000, 21, 705–722 CrossRef CAS.
- H. Kang, J. T. Buchman, R. S. Rodriguez, H. L. Ring, J. He, K. C. Bantz and C. L. Haynes, Chem. Rev., 2018, 119, 664–699 CrossRef PubMed.
- A. Kraynov and T. E. Müller, Applications of Ionic Liquids in Science and Technology, 2011, vol. 9, pp. 235–260 Search PubMed.
- S. Levine and G. Dube, Trans. Faraday Soc., 1939, 35, 1125–1140 RSC.
- A. M. Kalsin, M. Fialkowski, M. Paszewski, S. K. Smoukov, K. J. Bishop and B. A. Grzybowski, Science, 2006, 312, 420–424 CrossRef CAS PubMed.
- R. Javed, M. Zia, S. Naz, S. O. Aisida, N. u. Ain and Q. Ao, J. Nanobiotechnol., 2020, 18, 1–15 CrossRef PubMed.
- P. Dash and R. W. Scott, Chem. Commun., 2009, 812–814 RSC.
- K.-S. Kim, S. Choi, J.-H. Cha, S.-H. Yeon and H. Lee, J. Mater. Chem., 2006, 16, 1315–1317 RSC.
- A. Madu, P. Njoku, G. Iwuoha and U. Agbasi, Int. J. Phys. Sci., 2011, 6, 635–640 CAS.
- L. Vigderman and E. R. Zubarev, Adv. Drug Delivery Rev., 2013, 65, 663–676 CrossRef CAS PubMed.
- C. Caro, A. Avasthi, J. M. Paez-Muñoz, M. P. Leal and M. L. García-Martín, Biomater. Sci., 2021, 9, 7984–7995 RSC.
- Z. R. Goddard, M. J. Marín, D. A. Russell and M. Searcey, Chem. Soc. Rev., 2020, 49, 8774–8789 RSC.
- J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Chem. Rev., 2005, 105, 1103–1170 CrossRef CAS PubMed.
- H. Li and L. Rothberg, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14036–14039 CrossRef CAS PubMed.
- A. S. Angelatos, K. Katagiri and F. Caruso, Soft Matter, 2006, 2, 18–23 RSC.
- N. Hadjesfandiari and A. Parambath, in Engineering of Biomaterials for Drug Delivery Systems, Elsevier, 2018, pp. 345–361 Search PubMed.
- E. C. Dreaden, S. C. Mwakwari, Q. H. Sodji, A. K. Oyelere and M. A. El-Sayed, Bioconjugate Chem., 2009, 20, 2247–2253 CrossRef CAS PubMed.
- S. Sarkar, S. Konar, P. N. Prasad, S. Rajput, B. P. Kumar, R. R. Rao, A. Pathak, P. B. Fisher and M. Mandal, Langmuir, 2017, 33, 7649–7659 CrossRef CAS PubMed.
- Y. Wang, M. Pasternak, K. Sathiyamoorthy and M. C. Kolios, Biomed. Opt. Express, 2021, 12, 2171–2185 CrossRef PubMed.
- S. Mahalunkar, A. S. Yadav, M. Gorain, V. Pawar, R. Braathen, S. Weiss, B. Bogen, S. W. Gosavi and G. C. Kundu, Int. J. Nanomed., 2019, 8285–8302 CrossRef CAS PubMed.
- M. J. Hostetler, A. C. Templeton and R. W. Murray, Langmuir, 1999, 15, 3782–3789 CrossRef CAS.
- R. Lévy, N. T. Thanh, R. C. Doty, I. Hussain, R. J. Nichols, D. J. Schiffrin, M. Brust and D. G. Fernig, J. Am. Chem. Soc., 2004, 126, 10076–10084 CrossRef PubMed.
- O. Akbal Vural, Int. J. Polym. Mater. Polym. Biomater., 2022, 71, 1437–1448 CrossRef CAS.
- M.-E. Aubin-Tam, H. Zhou and K. Hamad-Schifferli, Soft Matter, 2008, 4, 554–559 RSC.
- D. Bartczak and A. G. Kanaras, Langmuir, 2011, 27, 10119–10123 CrossRef CAS PubMed.
- L. Raposo, C. Roma-Rodrigues, J. Jesus, L. Martins, A. Pombeiro, P. Baptista and A. Fernandes, Vet. Comp. Oncol., 2017, 15, 1537–1542 CrossRef CAS PubMed.
- R. Liu, J. Zhao, G. Han, T. Zhao, R. Zhang, B. Liu, Z. Liu, C. Zhang, L. Yang and Z. Zhang, ACS Appl. Mater. Interfaces, 2017, 9, 38222–38229 CrossRef CAS PubMed.
- S. Si and T. K. Mandal, Langmuir, 2007, 23, 190–195 CrossRef CAS PubMed.
- C. Criscitiello, Breast Care, 2012, 7, 262–266 CrossRef PubMed.
- M. K. Yu, J. Park and S. Jon, Theranostics, 2012, 2, 3 CrossRef CAS PubMed.
- S. Puertas, P. Batalla, M. Moros, E. Polo, P. Del Pino, J. M. Guisan, V. Grazu and J. M. de la Fuente, ACS Nano, 2011, 5, 4521–4528 CrossRef CAS PubMed.
- H. Liao and J. H. Hafner, Chem. Mater., 2005, 17, 4636–4641 CrossRef CAS.
- C. Loo, L. Hirsch, M.-H. Lee, E. Chang, J. West, N. Halas and R. Drezek, Opt. Lett., 2005, 30, 1012–1014 CrossRef CAS PubMed.
- Z. Alhalili, J. Shapter, D. Figueroa and B. Sanderson, Drug Deliv. Lett., 2018, 8, 217–225 CrossRef CAS.
- G. Obaid, I. Chambrier, M. J. Cook and D. A. Russell, Photochem. Photobiol. Sci., 2015, 14, 737–747 CrossRef CAS PubMed.
- O. Penon, M. J. Marín, D. A. Russell and L. Pérez-García, J. Colloid Interface Sci., 2017, 496, 100–110 CrossRef CAS PubMed.
- F. Nicolson, A. Ali, M. F. Kircher and S. Pal, Adv. Sci., 2020, 7, 2001669 CrossRef CAS PubMed.
- B. Liu, P. Wu, Z. Huang, L. Ma and J. Liu, J. Am. Chem. Soc., 2018, 140, 4499–4502 CrossRef CAS PubMed.
- W. Zhao, L. Lin and I.-M. Hsing, Bioconjugate Chem., 2009, 20, 1218–1222 CrossRef CAS PubMed.
- R. Wu, L.-P. Jiang, J.-J. Zhu and J. Liu, Langmuir, 2019, 35, 13461–13468 CrossRef CAS PubMed.
- J. Shin, X. Zhang and J. Liu, J. Phys. Chem. B, 2012, 116, 13396–13402 CrossRef CAS PubMed.
- J. A. Dougan, C. Karlsson, W. E. Smith and D. Graham, Nucleic Acids Res., 2007, 35, 3668–3675 CrossRef CAS PubMed.
- X. Zhang, B. Liu, N. Dave, M. R. Servos and J. Liu, Langmuir, 2012, 28, 17053–17060 CrossRef CAS PubMed.
- J. Kadkhoda, A. Aghanejad, B. Safari, J. Barar, S. H. Rasta and S. Davaran, J. Drug Delivery Sci. Technol., 2022, 67, 102954 CrossRef CAS.
- P. Bayat, K. Abnous, S. Balarastaghi, S. M. Taghdisi, M. Saeedi, R. Yazdian-Robati and M. Mahmoudi, Nanomed. J., 2022, 9, 164–169 CAS.
- A. Kardani, H. Yaghoobi, A. Alibakhshi and M. Khatami, J. Cell. Physiol., 2020, 235, 6887–6895 CrossRef CAS PubMed.
- M. V. Liberti and J. W. Locasale, Trends Biochem. Sci., 2016, 41, 211–218 CrossRef CAS PubMed.
- C. Tzror-Azankot, T. Dreifuss, T.-S. Ben-Gal, A. Jacob, T. Sadan, M. Motiei and R. Popovtzer, Nanoscale Imaging, Sensing, and Actuation for Biomedical Applications XVI, March 2019 Search PubMed.
- F. D. Moghaddam, G. Heidari, E. N. Zare, E. Djatoubai, A. C. Paiva-Santos, F. R. Bertani and A. Wu, Carbohydr. Polym., 2022, 120510 Search PubMed.
- R. V. Murthy, P. M. Chaudhary, R. Kikkeri, H. V. Thulasiram, A. Paul and S. Sangabathuni, Chem. Commun., 2015, 51, 15669–15672 RSC.
- S. Combemale, J.-N. Assam-Evoung, S. Houaidji, R. Bibi and V. Barragan-Montero, Molecules, 2014, 19, 1120–1149 CrossRef PubMed.
- T. Fyrner, T. Ederth, D. Aili, B. Liedberg and P. Konradsson, Colloids Surf., B, 2013, 105, 187–193 CrossRef CAS PubMed.
- F. Compostella, O. Pitirollo, A. Silvestri and L. Polito, Beilstein J. Org. Chem., 2017, 13, 1008–1021 CrossRef PubMed.
- K. K. Katti, V. Kattumuri, S. Bhaskaran, K. V. Katti and R. Kannan, Int. J. Green Nanotechnol., 2009, 1, B53–B59 Search PubMed.
- U. K. Mondal and J. J. Barchi Jr, Front. Chem., 2022, 10, 1002146 CrossRef CAS PubMed.
- P. G. Calavia, I. Chambrier, M. J. Cook, A. H. Haines, R. A. Field and D. A. Russell, J. Colloid Interface Sci., 2018, 512, 249–259 CrossRef PubMed.
- V. O. Uzonwanne, A. Navabi, J. D. Obayemi, J. Hu, A. A. Salifu, S. Ghahremani, N. Ndahiro, N. Rahbar and W. Soboyejo, Biomater. Adv., 2022, 136, 212801 CrossRef CAS PubMed.
- P. Maraming, J. Daduang and J. C. Y. Kah, RSC Adv., 2022, 12, 319–325 RSC.
- S. Law, A. W. Leung and C. Xu, Artif. Cells, Nanomed., Biotechnol., 2020, 48, 542–559 CrossRef CAS PubMed.
- C. Iodice, A. Cervadoro, A. Palange, J. Key, S. Aryal, M. R. Ramirez, C. Mattu, G. Ciardelli, B. E. O'Neill and P. Decuzzi, Opt Laser. Eng., 2016, 76, 74–81 CrossRef.
- E. García-Garrido, M. Cordani and Á. Somoza, Pharmaceutics, 2021, 13, 2067 CrossRef PubMed.
- R. Goyal, C. H. Kapadia, J. R. Melamed, R. S. Riley and E. S. Day, Cell. Mol. Bioeng., 2018, 11, 383–396 CrossRef CAS PubMed.
- H. Nosrati, M. Salehiabar, J. Charmi, K. Yaray, M. Ghaffarlou, E. Balcioglu and Y. N. Ertas, ACS Appl. Bio Mater., 2023, 6, 784–792 CrossRef CAS PubMed.
- A. Kefayat, F. Ghahremani, H. Motaghi and M. A. Mehrgardi, Eur. J. Pharm. Sci., 2019, 130, 225–233 CrossRef CAS PubMed.
- C. Joseph, A. Daniels, S. Singh and M. Singh, Pharmaceutics, 2021, 14, 53 CrossRef PubMed.
- H. Q. Yin, G. Shao, F. Gan and G. Ye, Nanoscale Res. Lett., 2020, 15, 1–15 CrossRef PubMed.
- R. Liu, H. Guo, Z. Ouyang, Y. Fan, X. Cao, J. Xia, X. Shi and R. Guo, ACS Appl. Bio Mater., 2021, 4, 1803–1812 CrossRef CAS PubMed.
- B. M. Bavelaar, L. Song, M. R. Jackson, S. Able, O. Tietz, I. Skaripa-Koukelli, P. A. Waghorn, M. R. Gill, R. C. Carlisle and M. Tarsounas, Mol. Pharm., 2021, 18, 3820–3831 CrossRef CAS PubMed.
- Z. Wu, S. Stangl, A. Hernandez-Schnelzer, F. Wang, M. Hasanzadeh Kafshgari, A. Bashiri Dezfouli and G. Multhoff, Cancers, 2023, 15, 1167 CrossRef CAS PubMed.
- R. K. Samani, M. B. Tavakoli, F. Maghsoudinia, H. Motaghi, S. H. Hejazi and M. A. Mehrgardi, Eur. J. Pharm. Sci., 2020, 153, 105487 CrossRef CAS PubMed.
- S. Li, S. Bouchy, S. Penninckx, R. Marega, O. Fichera, B. Gallez, O. Feron, P. Martinive, A.-C. Heuskin and C. Michiels, Nanomedicine, 2019, 14, 317–333 CrossRef CAS PubMed.
- A. Kapara, K. A. Findlay Paterson, V. G. Brunton, D. Graham, M. Zagnoni and K. Faulds, Anal. Chem., 2021, 93, 5862–5871 CrossRef CAS PubMed.
- M. A. Kimm, M. Shevtsov, C. Werner, W. Sievert, W. Zhiyuan, O. Schoppe, B. H. Menze, E. J. Rummeny, R. Proksa and O. Bystrova, Cancers, 2020, 12, 1331 CrossRef CAS PubMed.
- T. S. Rebelo, J. A. Ribeiro, M. G. F. Sales and C. M. Pereira, Sens. Bio-Sens. Res., 2021, 33, 100445 CrossRef.
- C. Zhang, F. Zhang, M. Han, X. Wang, J. Du, H. Zhang and W. Li, Sci. Rep., 2020, 10, 22015 CrossRef CAS PubMed.
- Y. Wu, Y. He, M. Han, D. Zhao, B. Liu, K. Yuan, H. Sun, H.-M. Meng and Z. Li, CCS Chem., 2022, 1–13 Search PubMed.
- A. Graczyk, R. Pawlowska and A. Chworos, Bioconjugate Chem., 2021, 32, 1667–1674 CrossRef CAS PubMed.
- K. Chakraborty, A. Biswas, S. Mishra, A. M. Mallick, A. Tripathi, S. Jan and R. Sinha Roy, ACS Appl. Bio Mater., 2023, 6, 458–472 CrossRef CAS PubMed.
- M. Shahidi, O. Abazari, P. Dayati, A. Bakhshi, A. Rasti, F. Haghiralsadat, S. M. Naghib and D. Tofighi, Nanotechnol. Rev., 2022, 11, 2875–2890 CrossRef CAS.
- X. Dai, X. Zhao, Y. Liu, B. Chen, X. Ding, N. Zhao and F. J. Xu, Small, 2021, 17, 2006004 CrossRef CAS PubMed.
- T. N. Aslan, E. Aşık, N. T. Güray and M. Volkan, JBIC, J. Biol. Inorg. Chem., 2020, 25, 1139–1152 CrossRef CAS PubMed.
- A. S. Doghish, A. H. Hashem, A. M. Shehabeldine, A.-A. M. Sallam, G. S. El-Sayyad and S. S. Salem, J. Drug Delivery Sci. Technol., 2022, 77, 103874 CrossRef CAS.
- L. Devi, R. Gupta, S. K. Jain, S. Singh and P. Kesharwani, J. Drug Delivery Sci. Technol., 2020, 56, 101565 CrossRef CAS.
- B. F. Craciun, L. Clima, D.-I. Bostiog, M. Silion, M. Calin, D. Peptanariu and M. Pinteala, Biomater. Adv., 2023, 144, 213201 CrossRef CAS PubMed.
- E. D. Abdolahinia, S. Nadri, R. Rahbarghazi, J. Barar, A. Aghanejad and Y. Omidi, Life Sci., 2019, 231, 116545 CrossRef CAS PubMed.
- A. G. Al-Dulimi, A. Z. Al-Saffar, G. M. Sulaiman, K. A. Khalil, K. S. Khashan, H. S. Al-Shmgani and E. M. Ahmed, J. Mater. Res. Technol., 2020, 9, 15394–15411 CrossRef CAS.
- G. Vivo-Llorca, Á. Morellá-Aucejo, A. García-Fernández, P. Díez, A. Llopis-Lorente, M. Orzáez and R. Martínez-Máñez, Int. J. Nanomed., 2022, 409–422 CrossRef PubMed.
- S. S. Kelkar and T. M. Reineke, Bioconjugate Chem., 2011, 22, 1879–1903 CrossRef CAS PubMed.
- R. Vinhas, M. Cordeiro, F. F. Carlos, S. Mendo, A. R. Fernandes, S. Figueiredo and P. V. Baptista, Nanobiosensors Dis. Diagnosis, 2015, 11–23 Search PubMed.
- J. Peng, Q. Yang, K. Shi, Y. Xiao, X. Wei and Z. Qian, Adv. Drug Delivery Rev., 2019, 143, 37–67 CrossRef CAS PubMed.
- J. Choi and S. Y. Kim, J. Ind. Eng. Chem., 2020, 85, 66–74 CrossRef CAS.
- J. Song, J. Zhou and H. Duan, J. Am. Chem. Soc., 2012, 134, 13458–13469 CrossRef CAS PubMed.
- N. Khlebtsov and L. Dykman, Chem. Soc. Rev., 2011, 40, 1647–1671 RSC.
- Y. Zhang, A. T. Liu, Y. R. Cornejo, D. Van Haute and J. M. Berlin, PLoS One, 2020, 15, e0234916 CrossRef CAS PubMed.
- J. Cancino-Bernardi, V. Marangoni, J. Besson, M. Cancino, M. Natali and V. Zucolotto, Chemosphere, 2018, 213, 41–52 CrossRef CAS PubMed.
- C. Lopez-Chaves, J. Soto-Alvaredo, M. Montes-Bayon, J. Bettmer, J. Llopis and C. Sanchez-Gonzalez, Nanomed. Nanotechnol. Biol. Med., 2018, 14, 1–12 CrossRef CAS PubMed.
- Y. Wang, K. C. Black, H. Luehmann, W. Li, Y. Zhang, X. Cai, D. Wan, S.-Y. Liu, M. Li and P. Kim, ACS Nano, 2013, 7, 2068–2077 CrossRef CAS PubMed.
- T. Dubaj, K. Kozics, M. Sramkova, A. Manova, N. G. Bastús, O. H. Moriones, Y. Kohl, M. Dusinska, E. Rundén-Pran and V. F. Puntes, Nanomaterials, 2022, 12, 511 CrossRef CAS PubMed.
- P. Natarajan and J. M. Tomich, Arch. Biochem. Biophys., 2020, 694, 108592 CrossRef CAS PubMed.
- M. Witkowska, E. Florek and R. Mrówczyński, Int. J. Mol. Sci., 2022, 23, 15343 CrossRef CAS PubMed.
- J. Xie, S. Lee and X. Chen, Adv. Drug Delivery Rev., 2010, 62, 1064–1079 CrossRef CAS PubMed.
- A. Vasil’kov, A. Voronova, T. Batsalova, D. Moten, A. Naumkin, E. Shtykova, V. Volkov, I. Teneva and B. Dzhambazov, Materials, 2023, 16, 3238 CrossRef PubMed.
- J. Guo, K. Rahme, Y. He, L.-L. Li, J. D. Holmes and C. M. O'Driscoll, Int. J. Nanomed., 2017, 6131–6152 CrossRef CAS PubMed.
- S. S. Khandker, M. S. Shakil and M. S. Hossen, Curr. Drug Metab., 2020, 21, 579–598 CrossRef CAS PubMed.
- S. Gori, S. Rimondini, V. De Angelis, M. Colozza, G. Bisagni, G. Moretti, A. Sidoni, C. Basurto, C. Aristei and P. Anastasi, Oncologist, 2007, 12, 766–773 CrossRef PubMed.
- X. Li, C. Yang, H. Wan, G. Zhang, J. Feng, L. Zhang, X. Chen, D. Zhong, L. Lou and W. Tao, Eur. J. Pharm. Sci., 2017, 110, 51–61 CrossRef CAS PubMed.
- Y. Wu, Y. Feng and X. Li, J. Colloid Interface Sci., 2022, 611, 287–293 CrossRef CAS PubMed.
- P. Yaşar, G. Ayaz, S. D. User, G. Güpür and M. Muyan, Reprod. Med. Biol., 2017, 16, 4–20 CrossRef PubMed.
- R. Ahirwar and P. Nahar, Anal. Bioanal. Chem., 2016, 408, 327–332 CrossRef CAS PubMed.
- Z. Baretta, S. Mocellin, E. Goldin, O. I. Olopade and D. Huo, Medicine, 2016, 95, e4975 CrossRef CAS PubMed.
- J.-H. Oh and J.-S. Lee, Anal. Chem., 2011, 83, 7364–7370 CrossRef CAS PubMed.
- R. Singh and Y.-Y. Mo, Cancer Biol. Ther., 2013, 14, 201–212 CrossRef CAS PubMed.
- F. Hakimian, H. Ghourchian, A. S. Hashemi, M. R. Arastoo and M. Behnam Rad, Sci. Rep., 2018, 8, 2943 CrossRef PubMed.
- M. J. Duffy, C. Duggan, R. Keane, A. D. Hill, E. McDermott, J. Crown and N. O'Higgins, Clin. Chem., 2004, 50, 559–563 CrossRef CAS PubMed.
- J. Peng, Y. Lai, Y. Chen, J. Xu, L. Sun and J. Weng, Small, 2017, 13, 1603589 CrossRef PubMed.
- E. Wolfson, S. Solomon, E. Schmukler, Y. Goldshmit and R. Pinkas-Kramarski, Cell Death Dis., 2018, 9, 47 CrossRef PubMed.
- M. Keshtkar, D. Shahbazi-Gahrouei, S. M. Khoshfetrat, M. A. Mehrgardi and M. Aghaei, J. Med. Signals Sens., 2016, 6, 243 CrossRef PubMed.
- P. F. Rostamabadi and E. Heydari-Bafrooei, Microchim. Acta, 2019, 186, 1–9 CrossRef PubMed.
- S. Sharma, J. Zapatero-Rodríguez, R. Saxena, R. O'Kennedy and S. Srivastava, Biosens. Bioelectron., 2018, 106, 78–85 CrossRef CAS PubMed.
- Y. Bai, H. Li, J. Xu, Y. Huang, X. Zhang, J. Weng, Z. Li and L. Sun, Biosens. Bioelectron., 2020, 166, 112424 CrossRef CAS PubMed.
- J.-H. Choi, J. Lim, M. Shin, S.-H. Paek and J.-W. Choi, Nano Lett., 2020, 21, 693–699 CrossRef PubMed.
- Q. Zhang, Y. Tian, Z. Liang, Z. Wang, S. Xu and Q. Ma, Anal. Chem., 2021, 93, 3308–3314 CrossRef CAS PubMed.
- C. Wang, W. Wang, Y. Xu, X. Zhao, S. Li, Q. Qian and X. Mi, Nanomaterials, 2022, 12, 666 CrossRef CAS PubMed.
- M. Hasanzadeh, E. Solhi, M. Jafari, A. Mokhtarzadeh, J. Soleymani, A. Jouyban and S. Mahboob, Int. J. Biol. Macromol., 2018, 120, 2493–2508 CrossRef CAS PubMed.
- Y. Bao, K. Han, Z. Ding, Y. Li, T. Li, M. Guan and G. Li, Spectrochim. Acta, Part A, 2021, 253, 119562 CrossRef CAS PubMed.
- H. Zhao, T. Liu, L. Cui, Y. Li, F. Yang and X. Zhang, Sens. Actuators, B, 2021, 345, 130332 CrossRef CAS.
- K. Kuntamung, J. Jakmunee and K. Ounnunkad, J. Mater. Chem. B, 2021, 9, 6576–6585 RSC.
- P. Ranjan, M. Abubakar Sadique, S. Yadav and R. Khan, ACS Appl. Mater. Interfaces, 2022, 14, 20802–20812 CrossRef CAS PubMed.
- J. Zhao, Y. Tang, Y. Cao, T. Chen, X. Chen, X. Mao, Y. Yin and G. Chen, Electrochim. Acta, 2018, 283, 1072–1078 CrossRef CAS.
- V. Maggi, F. Bianchini, E. Portioli, S. Peppicelli, M. Lulli, D. Bani, R. Del Sole, F. Zanardi, A. Sartori and R. Fiammengo, Chem. –Eur. J., 2018, 24, 12093–12100 CrossRef CAS PubMed.
- W.-J. Lee, K.-J. Kim, M. K. Hossain, H.-Y. Cho and J.-W. Choi, Biochip J., 2022, 16, 33–40 CrossRef CAS.
- C. Pothipor, J. Jakmunee, S. Bamrungsap and K. Ounnunkad, Analyst, 2021, 146, 4000–4009 RSC.
- L. Farzin, M. Shamsipur, L. Samandari and S. Sheibani, Microchim. Acta, 2018, 185, 1–9 CrossRef PubMed.
- F. Shafiei, R. S. Saberi and M. A. Mehrgardi, Bioelectrochemistry, 2021, 140, 107807 CrossRef CAS PubMed.
- J. Zhao, D. Huo, X. Geng, J. Bao, J. Hou, Z. Shui, H. Yang, Y. Qi, Y. Hu and M. Yang, Sens. Actuators, B, 2021, 332, 129480 CrossRef CAS.
- R. A. Sperling, P. R. Gil, F. Zhang, M. Zanella and W. J. Parak, Chem. Soc. Rev., 2008, 37, 1896–1908 RSC.
- K. Kneipp, H. Kneipp and J. Kneipp, Acc. Chem. Res., 2006, 39, 443–450 CrossRef CAS PubMed.
- Y. Luo, Y. Xiao, D. Onidas, L. Iannazzo, M. Ethève-Quelquejeu, A. Lamouri, N. Félidj, S. Mahouche-Chergui, T. Brulé and N. Gagey-Eilstein, Chem. Commun., 2020, 56, 6822–6825 RSC.
- C. Vericat, M. Vela, G. Benitez, P. Carro and R. Salvarezza, Chem. Soc. Rev., 2010, 39, 1805–1834 RSC.
- M. Li, J. Wu, M. Ma, Z. Feng, Z. Mi, P. Rong and D. Liu, Nanotheranostics, 2019, 3, 113 CrossRef PubMed.
- J. Zhu, J. Zhou, J. Guo, W. Cai, B. Liu, Z. Wang and Z. Sun, Chem. Cent. J., 2013, 7, 1–5 CrossRef PubMed.
- W. Lu, S. R. Arumugam, D. Senapati, A. K. Singh, T. Arbneshi, S. A. Khan, H. Yu and P. C. Ray, ACS Nano, 2010, 4, 1739–1749 CrossRef CAS PubMed.
- N. Gao, Q. Wang, J. Tang, S. Yao, H. Li, X. Yue, J. Fu, F. Zhong, T. Wang and J. Wang, Anal. Bioanal. Chem., 2021, 413, 4775–4784 CrossRef CAS PubMed.
- W. Cai, T. Gao, H. Hong and J. Sun, Nanotechnol., Sci. Appl., 2008, 17–32 CrossRef CAS PubMed.
- N. Lewinski, V. Colvin and R. Drezek, Small, 2008, 4, 26–49 CrossRef CAS PubMed.
- X. Huang, P. K. Jain, I. H. El-Sayed and M. A. El-Sayed, Laser Med. Sci., 2008, 23, 217–228 CrossRef PubMed.
- S. Link and M. A. El-Sayed, Int. Rev. Phys. Chem, 2000, 19, 409–453 Search PubMed.
- V. P. Zharov, E. N. Galitovskaya, C. Johnson and T. Kelly, Lasers Surg. Med., 2005, 37, 219–226 CrossRef PubMed.
- C. Loo, A. Lowery, N. Halas, J. West and R. Drezek, Nano Lett., 2005, 5, 709–711 CrossRef CAS PubMed.
- S. N. Mohammed, A. M. Mohammed and K. F. Al-Rawi, Steroids, 2022, 186, 109091 CrossRef PubMed.
- L. Yang, Y.-T. Tseng, G. Suo, L. Chen, J. Yu, W.-J. Chiu, C.-C. Huang and C.-H. Lin, ACS Appl. Mater. Interfaces, 2015, 7, 5097–5106 CrossRef CAS PubMed.
- M. Zhang, H. S. Kim, T. Jin and W. K. Moon, J. Photochem. Photobiol., B, 2017, 170, 58–64 CrossRef CAS PubMed.
- H. Chen, X. Wang, L. Sutrisno, T. Zeng, N. Kawazoe, Y. Yang and G. Chen, Front. Bioeng. Biotechnol., 2020, 8, 589905 CrossRef PubMed.
- M. R. Ali, H. A. Farghali, Y. Wu, I. El-Sayed, A. H. Osman, S. A. Selim and M. A. El-Sayed, Cancers, 2019, 11, 851 CrossRef PubMed.
- A. S. Abdoon, E. A. Al-Ashkar, O. M. Kandil, A. M. Shaban, H. M. Khaled, M. A. El Sayed, M. M. El Shaer, A. H. Shaalan, W. H. Eisa and A. A. G. Eldin, Nanomed. Nanotechnol. Biol. Med., 2016, 12, 2291–2297 CrossRef CAS PubMed.
- A. H. Faid, S. A. Shouman, N. A. Thabet, Y. A. Badr and M. A. Sliem, J. Pharmaceut. Innovat., 2023, 18, 144–148 CrossRef.
- H.-C. Lin, K.-F. Hsu, C.-L. Lai, T.-C. Wu, H.-F. Chen and C.-H. Lai, Molecules, 2020, 25, 1853 CrossRef CAS PubMed.
- Z. Poursalehi, R. Salehi, N. Samadi, S. H. Rasta, B. Mansoori and H. Majdi, Photodiagnosis Photodyn. Ther., 2019, 28, 25–37 CrossRef CAS PubMed.
- A. Pakravan, M. Azizi, F. Rahimi, F. Bani, F. Mahmoudzadeh, R. Salehi and M. Mahkam, Cancer Nanotechnol., 2021, 12, 1–26 CrossRef PubMed.
- L. C. Kennedy, L. R. Bickford, N. A. Lewinski, A. J. Coughlin, Y. Hu, E. S. Day, J. L. West and R. A. Drezek, Small, 2011, 7, 169–183 CrossRef CAS PubMed.
- H. Xia, Y. Gao, L. Yin, X. Cheng, A. Wang, M. Zhao, J. Ding and H. Shi, ChemBioChem, 2019, 20, 667–671 CrossRef CAS PubMed.
- P. P. Pillai, B. Kowalczyk, K. Kandere-Grzybowska, M. Borkowska and B. A. Grzybowski, Angew. Chem., Int. Ed., 2016, 55, 8610–8614 CrossRef CAS PubMed.
- S. Kang, S. H. Bhang, S. Hwang, J.-K. Yoon, J. Song, H.-K. Jang, S. Kim and B.-S. Kim, ACS Nano, 2015, 9, 9678–9690 CrossRef CAS PubMed.
- T. Bian, L. Shang, H. Yu, M. T. Perez, L. Z. Wu, C. H. Tung, Z. Nie, Z. Tang and T. Zhang, Adv. Mater., 2014, 26, 5613–5618 CrossRef CAS PubMed.
- S.-R. Kim and E.-H. Kim, Int. J. Radiat. Biol., 2018, 94, 8–16 CrossRef CAS PubMed.
- Y. Dou, Y. Guo, X. Li, X. Li, S. Wang, L. Wang, G. Lv, X. Zhang, H. Wang and X. Gong, ACS Nano, 2016, 10, 2536–2548 CrossRef CAS PubMed.
- N. M. Dimitriou, G. Tsekenis, E. C. Balanikas, A. Pavlopoulou, M. Mitsiogianni, T. Mantso, G. Pashos, A. G. Boudouvis, I. N. Lykakis and G. Tsigaridas, Pharmacol. Ther., 2017, 178, 1–17 CrossRef CAS PubMed.
- L. Song, N. Falzone and K. A. Vallis, Int. J. Radiat. Biol., 2016, 92, 716–723 CrossRef CAS PubMed.
- X. Yao, C. Huang, X. Chen, Y. Zheng and L. Sanche, J. Biomed. Nanotechnol., 2015, 11, 478–485 CrossRef CAS PubMed.
- M. Misawa and J. Takahashi, Nanomed. Nanotechnol. Biol. Med., 2011, 7, 604–614 CrossRef CAS PubMed.
- Y. Lin, S. J. McMahon, M. Scarpelli, H. Paganetti and J. Schuemann, Phys. Med. Biol., 2014, 59, 7675 CrossRef PubMed.
- S. Rosa, C. Connolly, G. Schettino, K. T. Butterworth and K. M. Prise, Cancer Nanotechnol., 2017, 8, 1–25 CrossRef PubMed.
- J. Choi, K. O. Jung, E. E. Graves and G. Pratx, Nanotechnology, 2018, 29, 504001 CrossRef PubMed.
- C. Wang, Y. Jiang, X. Li and L. Hu, Breast Cancer, 2015, 22, 413–420 CrossRef PubMed.
- N. Hanžić, A. Horvat, J. Bibić, K. Unfried, T. Jurkin, G. Dražić, I. Marijanović, N. Slade and M. Gotić, Mater. Sci. Eng. C, 2018, 91, 486–495 CrossRef PubMed.
- B. Janic, S. L. Brown, R. Neff, F. Liu, G. Mao, Y. Chen, L. Jackson, I. J. Chetty, B. Movsas and N. Wen, Cancer Biol. Ther., 2021, 22, 124–135 CrossRef CAS PubMed.
- S. S. Mehrnia, B. Hashemi, S. J. Mowla, M. Nikkhah and A. Arbabi, Radiat. Oncol., 2021, 16, 1–12 CrossRef PubMed.
- M. M. Fathy, F. S. Mohamed, N. Elbialy and W. M. Elshemey, Phys. Med., 2018, 48, 76–83 CrossRef PubMed.
- C. Yang, K. Bromma, D. Ciano-Oliveira, G. Zafarana, M. van Prooijen and D. B. Chithrani, Cancer Nanotechnol., 2018, 9, 1–14 CrossRef.
- H. Mendoza-Nava, G. Ferro-Flores, F. d. M. Ramírez, B. Ocampo-García, C. Santos-Cuevas, L. Aranda-Lara, E. Azorín-Vega, E. Morales-Avila and K. Isaac-Olivé, J. Nanomater., 2016, 2016, 1039258 Search PubMed.
- L. Bai, F. Jiang, R. Wang, C. Lee, H. Wang, W. Zhang, W. Jiang, D. Li, B. Ji and Z. Li, J. Nanobiotechnol., 2020, 18, 1–10 CrossRef PubMed.
- H. Nosrati, F. Seidi, A. Hosseinmirzaei, N. Mousazadeh, A. Mohammadi, M. Ghaffarlou, H. Danafar, J. Conde and A. Sharafi, Adv. Healthcare Mater., 2022, 11, 2102321 CrossRef CAS PubMed.
- C. Yang, K. Bromma, W. Sung, J. Schuemann and D. Chithrani, Cancers, 2018, 10, 150 CrossRef PubMed.
- B. Janic, F. Liu, K. Bobbitt, S. Brown, I. Chetty, G. Mao, B. Movsas and N. Wen, J. Nanomed. Nanotechnol., 2018, 9, 1–13 Search PubMed.
- X. Yang, X. Liu, Z. Liu, F. Pu, J. Ren and X. Qu, Adv. Mater., 2012, 24, 2890–2895 CrossRef CAS PubMed.
- P. Shi, K. Qu, J. Wang, M. Li, J. Ren and X. Qu, Chem. Commun., 2012, 48, 7640–7642 RSC.
- R. Hong, G. Han, J. M. Fernández, B.-j. Kim, N. S. Forbes and V. M. Rotello, J. Am. Chem. Soc., 2006, 128, 1078–1079 CrossRef CAS PubMed.
- P. Joshi, S. Chakraborti, J. E. Ramirez-Vick, Z. Ansari, V. Shanker, P. Chakrabarti and S. P. Singh, Colloids Surf., B, 2012, 95, 195–200 CrossRef CAS PubMed.
- C. Xu, B. Wang and S. Sun, J. Am. Chem. Soc., 2009, 131, 4216–4217 CrossRef CAS PubMed.
- K. Ock, W. I. Jeon, E. O. Ganbold, M. Kim, J. Park, J. H. Seo, K. Cho, S.-W. Joo and S. Y. Lee, Anal. Chem., 2012, 84, 2172–2178 CrossRef CAS PubMed.
- M. K. Hossain, H.-Y. Cho, K.-J. Kim and J.-W. Choi, Biosens. Bioelectron., 2015, 71, 300–305 CrossRef CAS PubMed.
- J. You, R. Zhang, G. Zhang, M. Zhong, Y. Liu, C. S. Van Pelt, D. Liang, W. Wei, A. K. Sood and C. Li, J. Contr. Release, 2012, 158, 319–328 CrossRef CAS PubMed.
- S. K. Libutti, G. F. Paciotti, A. A. Byrnes, H. R. Alexander Jr, W. E. Gannon, M. Walker, G. D. Seidel, N. Yuldasheva and L. Tamarkin, Clin. Cancer Res., 2010, 16, 6139–6149 CrossRef CAS PubMed.
- R. A. Morshed, M. E. Muroski, Q. Dai, M. L. Wegscheid, B. Auffinger, D. Yu, Y. Han, L. Zhang, M. Wu and Y. Cheng, Mol. Pharm., 2016, 13, 1843–1854 CrossRef CAS PubMed.
- W. Jiang, B. Y. Kim, J. T. Rutka and W. C. Chan, Nat. Nanotechnol., 2008, 3, 145–150 CrossRef CAS PubMed.
- O. Akturk, Int. J. Biol. Macromol., 2022, 196, 72–85 CrossRef CAS PubMed.
- M. Hasannia, K. Abnous, S. M. Taghdisi, S. Nekooei, M. Ramezani and M. Alibolandi, J. Nanobiotechnol., 2022, 20, 1–27 CrossRef PubMed.
- R. Taheri-Ledari, W. Zhang, M. Radmanesh, S. S. Mirmohammadi, A. Maleki, N. Cathcart and V. Kitaev, Small, 2020, 16, 2002733 CrossRef CAS PubMed.
- C. S. Kumar, A. Mahesh, M. G. Antoniraj, S. Vaidevi and K. Ruckmani, RSC Adv., 2016, 6, 26874–26882 RSC.
- H. M. Aldawsari, S. Singh, N. A. Alhakamy, R. B. Bakhaidar, A. A. Halwani and S. M. Badr-Eldin, Pharmaceutics, 2021, 13, 1554 CrossRef CAS PubMed.
- M. S. Jabir, A. A. Taha, U. I. Sahib, Z. J. Taqi, A. M. Al-Shammari and A. S. Salman, Mater. Sci. Eng. C, 2019, 94, 949–964 CrossRef CAS PubMed.
- B. Khodashenas, M. Ardjmand, A. Rad and M. Esfahani, Mater. Today Chem., 2021, 20, 100474 CrossRef CAS.
- D. El-Safoury, A. B. Ibrahim, D. El-Setouhy, O. Khowessah, M. Motaleb and T. M. Sakr, J. Pharm. Sci., 2021, 110, 2955–2965 CrossRef CAS PubMed.
- C.-F. Chiu, R.-H. Fu, S.-h. Hsu, Y.-H. Yu, S.-F. Yang, T. C.-Y. Tsao, K.-B. Chang, C.-A. Yeh, C.-M. Tang and S.-C. Huang, Cancers, 2021, 13, 5317 CrossRef CAS PubMed.
- J. R. Lakkakula, R. W. M. Krause, D. Divakaran, S. Barage and R. Srivastava, J. Mol. Liq., 2021, 341, 117262 CrossRef CAS.
- C. Yang, J. Uertz and D. B. Chithrani, Nanomaterials, 2016, 6, 48 CrossRef PubMed.
- O. Gotov, G. Battogtokh, D. Shin and Y. T. Ko, J. Ind. Eng. Chem., 2018, 65, 236–243 CrossRef CAS.
- J. E. Bonevich, Nanotechnology Characterization Laboratory, National Cancer Institute, Frederick, MD 21702, (301) 846-6939, 2010 Search PubMed.
- P. Kumari, B. Ghosh and S. Biswas, J. Drug Target., 2016, 24, 179–191 CrossRef CAS PubMed.
- J. M. Stern, V. V. Kibanov Solomonov, E. Sazykina, J. A. Schwartz, S. C. Gad and G. P. Goodrich, Int. J. Toxicol., 2016, 35, 38–46 CrossRef CAS PubMed.
- J. A. Schwartz, A. M. Shetty, R. E. Price, R. J. Stafford, J. C. Wang, R. K. Uthamanthil, K. Pham, R. J. McNichols, C. L. Coleman and J. D. Payne, Cancer Res., 2009, 69, 1659–1667 CrossRef CAS PubMed.
- M. Khoobchandani, K. K. Katti, A. R. Karikachery, V. C. Thipe, D. Srisrimal, D. K. Dhurvas Mohandoss, R. D. Darshakumar, C. M. Joshi and K. V. Katti, Int. J. Nanomed., 2020, 181–197 CrossRef CAS PubMed.
- P. Kumthekar, C. H. Ko, T. Paunesku, K. Dixit, A. M. Sonabend, O. Bloch, M. Tate, M. Schwartz, L. Zuckerman and R. Lezon, Sci. Transl. Med., 2021, 13, eabb3945 CrossRef CAS PubMed.
- Z. Xu, Y. Broza, R. Ionsecu, U. Tisch, L. Ding, H. Liu, Q. Song, Y. Pan, F. Xiong and K. Gu, Br. J. Cancer, 2013, 108, 941–950 CrossRef CAS PubMed.
- Y. Yang, X. Zheng, L. Chen, X. Gong, H. Yang, X. Duan and Y. Zhu, Int. J. Nanomed., 2022, 2041–2067 CrossRef PubMed.
- J. S. Jue, S. Coons, G. Hautvast, S. F. Thompson, J. Geraats, L. Richstone, M. J. Schwartz and A. R. Rastinehad, J. Endourol., 2022, 36, 369–372 CrossRef PubMed.
- P. Kumthekar, A. Rademaker, C. Ko, K. Dixit, M. A. Schwartz, A. M. Sonabend, L. Sharp, R. V. Lukas, R. Stupp and C. Horbinski, J. Clin. Oncol., 2019, 37, 3012 Search PubMed.
- A. F. Farag and N. F. Hassabou, Nanomed. Nanotechnol. Biol. Med., 2022, 46, 102598 CrossRef CAS PubMed.
- L. Yao, D. Bojic and M. Liu, J. Pharmaceut. Biomed. Anal., 2023, 13, 960–967 CrossRef PubMed.
- C. Lasagna-Reeves, D. Gonzalez-Romero, M. Barria, I. Olmedo, A. Clos, V. S. Ramanujam, A. Urayama, L. Vergara, M. J. Kogan and C. Soto, Biochem. Biophys. Res. Commun., 2010, 393, 649–655 CrossRef CAS PubMed.
- I. Fratoddi, I. Venditti, C. Cametti and M. V. Russo, Nano Res., 2015, 8, 1771–1799 CrossRef CAS.
- M. C. Senut, Y. Zhang, F. Liu, A. Sen, D. M. Ruden and G. Mao, Small, 2016, 12, 631–646 CrossRef CAS PubMed.
- Y. Pan, S. Neuss, A. Leifert, M. Fischler, F. Wen, U. Simon, G. Schmid, W. Brandau and W. Jahnen-Dechent, Small, 2007, 3, 1941–1949 CrossRef CAS PubMed.
- W. H. De Jong, W. I. Hagens, P. Krystek, M. C. Burger, A. J. Sips and R. E. Geertsma, Biomaterials, 2008, 29, 1912–1919 CrossRef CAS PubMed.
- M. A. K. Abdelhalim and S. A. A. Moussa, Saudi J. Biol. Sci., 2013, 20, 177–181 CrossRef CAS PubMed.
- G. Sonavane, K. Tomoda and K. Makino, Colloids Surf., B, 2008, 66, 274–280 CrossRef CAS PubMed.
- E. E. Connor, J. Mwamuka, A. Gole, C. J. Murphy and M. D. Wyatt, Small, 2005, 1, 325–327 CrossRef CAS PubMed.
- C. J. Murphy, A. M. Gole, J. W. Stone, P. N. Sisco, A. M. Alkilany, E. C. Goldsmith and S. C. Baxter, Acc. Chem. Res., 2008, 41, 1721–1730 CrossRef CAS PubMed.
- Y.-S. Chen, Y.-C. Hung, I. Liau and G. S. Huang, Nanoscale Res. Lett., 2009, 4, 858–864 CrossRef CAS PubMed.
- J. Pérez-Juste, I. Pastoriza-Santos, L. M. Liz-Marzán and P. Mulvaney, Coord. Chem. Rev., 2005, 249, 1870–1901 CrossRef.
- Y. Li, M. Kröger and W. K. Liu, Nanoscale, 2015, 7, 16631–16646 RSC.
- K. C. Black, Y. Wang, H. P. Luehmann, X. Cai, W. Xing, B. Pang, Y. Zhao, C. S. Cutler, L. V. Wang and Y. Liu, ACS Nano, 2014, 8, 4385–4394 CrossRef CAS PubMed.
- Z. Wang, D. Xie, H. Liu, Z. Bao and Y. Wang, RSC Adv., 2016, 6, 33009–33013 RSC.
- M. Mioc, I. Z. Pavel, R. Ghiulai, D. E. Coricovac, C. Farcaş, C.-V. Mihali, C. Oprean, V. Serafim, R. A. Popovici and C. A. Dehelean, Front. Pharmacol, 2018, 9, 429 CrossRef PubMed.
- X.-D. Zhang, D. Wu, X. Shen, P.-X. Liu, N. Yang, B. Zhao, H. Zhang, Y.-M. Sun, L.-A. Zhang and F.-Y. Fan, Int. J. Nanomed., 2011, 2071–2081 CrossRef CAS PubMed.
- K. Katti, N. Chanda, R. Shukla, A. Zambre, T. Suibramanian, R. R. Kulkarni, R. Kannan and K. V. Katti, Int. J. Green Nanotechnol., 2009, 1, B39–B52 Search PubMed.
- Y. Qu and X. Lü, Biomed. Mater., 2009, 4, 025007 CrossRef PubMed.
- P. Roy, S. Sur, S. Das and W. T. Wui, Breast Cancer, 2022, 29, 761–777 CrossRef PubMed.
- J. G. Railsback, A. Singh, R. C. Pearce, T. E. McKnight, R. Collazo, Z. Sitar, Y. G. Yingling and A. V. Melechko, Adv. Mater., 2012, 24, 4261–4265 CrossRef CAS PubMed.
- A. S. Poulos, D. Constantin, P. Davidson, M. Impéror-Clerc, B. Pansu and S. Rouzière, Europhys. Lett., 2012, 100, 18002 CrossRef.
- C. M. Goodman, C. D. McCusker, T. Yilmaz and V. M. Rotello, Bioconjugate Chem., 2004, 15, 897–900 CrossRef CAS PubMed.
- N. M. Schaeublin, L. K. Braydich-Stolle, A. M. Schrand, J. M. Miller, J. Hutchison, J. J. Schlager and S. M. Hussain, Nanoscale, 2011, 3, 410–420 RSC.
- C. Ge, J. Tian, Y. Zhao, C. Chen, R. Zhou and Z. Chai, Arch. Toxicol., 2015, 89, 519–539 CrossRef CAS PubMed.
- J. Liu and Q. Peng, Acta Biomater., 2017, 55, 13–27 CrossRef CAS PubMed.
- L. Wang, J. Li, J. Pan, X. Jiang, Y. Ji, Y. Li, Y. Qu, Y. Zhao, X. Wu and C. Chen, J. Am. Chem. Soc., 2013, 135, 17359–17368 CrossRef CAS PubMed.
- S. Milani, F. Baldelli Bombelli, A. S. Pitek, K. A. Dawson and J. Radler, ACS Nano, 2012, 6, 2532–2541 CrossRef CAS PubMed.
- M. Schäffler, M. Semmler-Behnke, H. Sarioglu, S. Takenaka, A. Wenk, C. Schleh, S. M. Hauck, B. D. Johnston and W. G. Kreyling, Nanotechnology, 2013, 24, 265103 CrossRef PubMed.
- X. Cheng, X. Tian, A. Wu, J. Li, J. Tian, Y. Chong, Z. Chai, Y. Zhao, C. Chen and C. Ge, ACS Appl. Mater. Interfaces, 2015, 7, 20568–20575 CrossRef CAS PubMed.
- P. Wang, X. Wang, L. Wang, X. Hou, W. Liu and C. Chen, Sci. Technol. Adv. Mater., 2015, 16, 034610 CrossRef PubMed.
- W. Lai, Q. Wang, L. Li, Z. Hu, J. Chen and Q. Fang, Colloids Surf., B, 2017, 152, 317–325 CrossRef CAS PubMed.
- C. Carnovale, G. Bryant, R. Shukla and V. Bansal, ACS Omega, 2019, 4, 242–256 CrossRef CAS.
- K. E. Woods, Y. R. Perera, M. B. Davidson, C. A. Wilks, D. K. Yadav and N. C. Fitzkee, J. Phys. Chem. C, 2016, 120, 27944–27953 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed.
- K. Saha, M. Rahimi, M. Yazdani, S. T. Kim, D. F. Moyano, S. Hou, R. Das, R. Mout, F. Rezaee and M. Mahmoudi, ACS Nano, 2016, 10, 4421–4430 CrossRef CAS PubMed.
- A. Salvati, A. S. Pitek, M. P. Monopoli, K. Prapainop, F. B. Bombelli, D. R. Hristov, P. M. Kelly, C. Åberg, E. Mahon and K. A. Dawson, Nat. Nanotechnol., 2013, 8, 137–143 CrossRef CAS PubMed.
- V. Mirshafiee, M. Mahmoudi, K. Lou, J. Cheng and M. L. Kraft, Chem. Commun., 2013, 49, 2557–2559 RSC.
- A. A. Shemetov, I. Nabiev and A. Sukhanova, ACS Nano, 2012, 6, 4585–4602 CrossRef CAS PubMed.
- Z. Ma, J. Bai and X. Jiang, ACS Appl. Mater. Interfaces, 2015, 7, 17614–17622 CrossRef CAS PubMed.
- H. Yang, M. Wang, Y. Zhang, F. Li, S. Yu, L. Zhu, Y. Guo, L. Yang and S. Yang, RSC Adv., 2019, 9, 4435–4444 RSC.
- A. O. Elzoghby, W. M. Samy and N. A. Elgindy, J. Contr. Release, 2012, 157, 168–182 CrossRef CAS PubMed.
- H. K. Patra, S. Banerjee, U. Chaudhuri, P. Lahiri and A. K. Dasgupta, Nanomed. Nanotechnol. Biol. Med., 2007, 3, 111–119 CrossRef CAS PubMed.
- R. Coradeghini, S. Gioria, C. P. García, P. Nativo, F. Franchini, D. Gilliland, J. Ponti and F. Rossi, Toxicol. Lett., 2013, 217, 205–216 Search PubMed.
- X.-D. Zhang, H.-Y. Wu, D. Wu, Y.-Y. Wang, J.-H. Chang, Z.-B. Zhai, A.-M. Meng, P.-X. Liu, L.-A. Zhang and F.-Y. Fan, Int. J. Nanomed., 2010, 771–781 CrossRef PubMed.
- I. Fratoddi, I. Venditti, C. Cametti and M. V. Russo, Toxicol. Res., 2015, 4, 796–800 CrossRef CAS.
- X.-L. Huang, B. Zhang, L. Ren, S.-F. Ye, L.-P. Sun, Q.-Q. Zhang, M.-C. Tan and G.-M. Chow, J. Mater. Sci.: Mater. Med., 2008, 19, 2581–2588 CrossRef CAS PubMed.
- P.-H. Lu, H.-J. Li, H.-H. Chang, N.-L. Wu and C.-F. Hung, J. Nanopart. Res., 2017, 19, 1–12 CrossRef CAS.
- J. H. Kim, J. H. Kim, K.-W. Kim, M. H. Kim and Y. S. Yu, Nanotechnology, 2009, 20, 505101 CrossRef PubMed.
- T. Mironava, M. Hadjiargyrou, M. Simon, V. Jurukovski and M. H. Rafailovich, Nanotoxicology, 2010, 4, 120–137 CrossRef CAS PubMed.
- L. R. Hirsch, R. J. Stafford, J. Bankson, S. R. Sershen, B. Rivera, R. Price, J. D. Hazle, N. J. Halas and J. L. West, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 13549–13554 CrossRef CAS PubMed.
- S. K. Nune, N. Chanda, R. Shukla, K. Katti, R. R. Kulkarni, S. Thilakavathy, S. Mekapothula, R. Kannan and K. V. Katti, J. Mater. Chem., 2009, 19, 2912–2920 RSC.
- S. Vijayakumar and S. Ganesan, J. Nanomater., 2012, 2012, 14 Search PubMed.
- H. Takahashi, Y. Niidome, T. Niidome, K. Kaneko, H. Kawasaki and S. Yamada, Langmuir, 2006, 22, 2–5 CrossRef CAS PubMed.
- P. Chandran, J. E. Riviere and N. A. Monteiro-Riviere, Nanotoxicology, 2017, 11, 507–519 Search PubMed.
| This journal is © The Royal Society of Chemistry 2024 |