Recent developments and applications of ambient mass spectrometry imaging in pharmaceutical research: an overview
Bharath Sampath
Kumar
 *
*
Independent researcher, 21, B2, 27th Street, Lakshmi Flats, Nanganallur, Chennai 600061, India. E-mail: bskumar80@gmail.com
First published on 23rd November 2023
Abstract
The application of ambient mass spectrometry imaging “MSI” is expanding in the areas of fundamental research on drug delivery and multiple phases of the process of identifying and developing drugs. Precise monitoring of a drug's pharmacological workflows, such as intake, distribution, metabolism, and discharge, is made easier by MSI's ability to determine the concentrations of the initiating drug and its metabolites across dosed samples without losing spatial data. Lipids, glycans, and proteins are just a few of the many phenotypes that MSI may be used to concurrently examine. Each of these substances has a particular distribution pattern and biological function throughout the body. MSI offers the perfect analytical tool for examining a drug's pharmacological features, especially in vitro and in vivo effectiveness, security, probable toxic effects, and putative molecular pathways, because of its high responsiveness in chemical and physical environments. The utilization of MSI in the field of pharmacy has further extended from the traditional tissue examination to the early stages of drug discovery and development, including examining the structure–function connection, high-throughput capabilities in vitro examination, and ex vivo research on individual cells or tumor spheroids. Additionally, an enormous array of endogenous substances that may function as tissue diagnostics can be scanned simultaneously, giving the specimen a highly thorough characterization. Ambient MSI techniques are soft enough to allow for easy examination of the native sample to gather data on exterior chemical compositions. This paper provides a scientific and methodological overview of ambient MSI utilization in research on pharmaceuticals.
Introduction
It is essential to have the ability to track the distribution and evolution of therapeutic compounds in preclinical investigations using in vivo treatment studies or more fundamental in vitro models when developing new medication compounds and compositions for the pharmaceutical sector.1 By dissecting the tissue, extracting the substance, and then analyzing the extract chemically using the liquid chromatography-mass spectrometric technique, the existence and location of a substance can be evaluated on a coarser level.1 However, it can also be identified through an imaging research study, which visually reveals substantially more data concerning compound localization. Microscopes offer excellent spatial clarity for all imaging methods, particularly fluorescence microscopy, which, by using luminescent markers, may also offer certain drug selectivity.1 However, a variety of problems appear when selectivity is attained via tagging. The binding of a fluorophore to a molecular structure fundamentally modifies its chemical and biological characteristics, potentially leading to an entirely distinct response in an organism compared to what a non-labeled molecular structure would have. Additionally, only a small number of substances can be observed concurrently.1When visualizing by autoradiography, for instance, radioactive tags are utilized instead of fluorescent tags to address a few of these challenges. However, chemical production and working with radiolabeled molecules are expensive and resource intensive.1 Additionally because many medications are ultimately transformed in the body before they are expelled, the radioactive label, rather than the actual molecule that was marked, is what is observed in autoradiography. This is a significant restriction when medications are created as therapeutics to enhance uptake in the body.1 Consequently, autoradiography does not reveal the location of the organism where the drug is transformed into the alleged active drug ingredient or which of the two substances ultimately accesses the drug target.1
For specimens, especially those involving tissue samples, MSI can immediately provide the molecular fingerprint and map the distribution pattern of every ionized molecule. In addition to locating the abundance of drugs and metabolites, MSI, arguably among the most promising and evolving approaches to researching spatial multi-omics, also sheds light on the phenotypic alterations underpinning illness development and medication administration.2–4 This study offers an analytical and methodological review of the use of ambient MSI in pharmacological research. Table 1 presents a list of the nature of the MSI application, drug type, sample, ambient ionization technique, MS analyzer, and supplementary analytical techniques.
| Authors | Application | Drug | Sample | Q | Ionization | Analyzer | ST | SR |
|---|---|---|---|---|---|---|---|---|
| a Q – quantification; NA – not applicable; ST – supplementary techniques; Q-TOF – quadrupole time of flight; FT-ICR – Fourier transform ion cyclotron resonance; TOF – time of flight and SR – spatial resolution. | ||||||||
| Guo et al. (2021) | Drug distribution | Fluoropezil (DC20) | Rat brain coronal tissue | Yes | DESI-MSI | IMS | LC-ESI-MS | NR |
| Danhorn et al. (2022) | Drug distribution | Terfenadine, losartan, diphenhydramine and dextromethorphan | Rat kidney and liver tissues | Yes | DESI-MSI | Q-orbitrap | MALDI-MSI | 100 μm |
| Yamamoto et al. (2021) | Drug distribution & spatial localization | Ciclesonide | Rat lung tissue | Yes | DESI-MSI | TOF | LC-MS, H&E staining | 50 μm |
| Akakpo et al. (2022) | Spatial localization, hepatotoxicity & drug distribution | 4-Methyl pyrazole (aka 4MP, fomepizole) | Mice liver tissue | Yes | DESI-MSI | Q-TOF | LC-MS | 50 μm |
| Liu et al. (2022) | Pharmacometabolomics, drug metabolism & distribution | Olanzapine (OLZ) | Brain tissue | No | AFADESI-MSI | Q-orbitrap | H&E staining | 200 μm |
| Jin et al. (2023) | Drug targets for central nervous system | Sedative-hypnotic drug candidate YZG-331 | Rat sagittal brain tissue | Yes | AFADESI-MSI | Q-orbitrap | LC-MS, H&E staining, nissl staining | NR |
| Li et al. (2023) | Metabolism & hepatotoxicity | Amiodarone (AMI) | HepG2 spheroids | No | AFADESI-MSI | Q-orbitrap | H&E staining & IHC | 40 μm |
| Luo et al. (2019) | Spatial metabolomics | Sedative-hypnotic drug candidates YZG-331 and YZG-330 | Rat whole body tissue sections | No | AFADESI-MSI | Q-orbitrap | NA | NR |
| Brown et al. (2020) | Drug metabolism | Diclofenac | Liver and kidney tissues | Yes | Nano-DESI-MSI | Q-orbitrap | LC-MS | 10 μm |
| Matsumoto et al. (2020) | Brain distributions | Geissoschizine methyl ether (GM) | Brain sagittal tissue section | Yes | DESI-MSI | FT-ICR & Q-TOF | MALDI-MSI, LC-MS | 200 μm |
| Quartier et al. (2021) | Drug distribution, drug penetration pathways | Econazole nitrate (ECZ),D-α-tocopheryl succinate polyethylene glycol 1000 (TPGS) | Porcine skin section | No | DESI-MSI | TOF | NA | ∼80 μm |
| Vallianatou et al. (2018) | Drug interactivity | Propranolol, loperamide | Mouse brain tissue | Yes | DESI-MSI | Q-orbitrap | MALDI, H&E staining | 50 μm |
| Colclough et al. (2021) | Blood brain barrier, metastatic brain disease | Osimertinib | Rat brain tissue | Yes | DESI-MSI | Q-orbitrap | LC-MS, H&E staining | NR |
| Zhang et al. (2020) | Targeting efficiency | Paclitaxel derivatives | Xenografted tumor | Yes | AFADESI-MSI | Q-orbitrap | H&E staining | 40 μm |
| Kokesch-Himmelreich et al. (2022) | Targeting efficiency | Clofazimine, moxifloxacin, bedaquiline, pyrazinamide, rifampicin | Necrotic granulomas | No | AP-MALDI | Q-orbitrap | H&E staining | NR |
| Morosi et al. (2022) | Drug distribution | Pioglitazone | Neoplastic and normal liver tissues | Yes | AP-MALDI | Q-orbitrap | HPLC-UV | 25 μm |
| Pang et al. (2021) | Metabolic network mapping | Scopolamine | Rat brain tissue | No | AFADESI-MSI | Q-orbitrap | H&E staining | ∼71 μm |
| Holm et al. (2022) | Drug metabolism | Cyclosporine | Whole body mouse kidney and intestine | Yes | DESI-MSI | Q-orbitrap | MALDI-MSI | ∼50 μm |
| Gao et al. (2022) | Drug distribution | Uncaria alkaloids | Brain tissue | Yes | DESI-MSI | Q-TOF | H&E staining | NR |
| Lin et al. (2020) | Disease biomarker | NA | Lung tissue & luminal B breast tumor tissue | — | Nano-DESI-MSI | TOF | MALDI-MSI & IHC | NR |
| Islam et al. (2022) | Drug metabolism | Imipramine, chloroquine | Sagittal sections of mice kidneys and brains | Yes | AP-MALDI-MSI | Q-TOF, FT-ICR | DESI-MSI, MALDI-MSI, H&E staining, LC-MS | 50 μm |
| Tan et al. (2023) | Chemical composition | Radix aconiti | R-TBC, H-TBC, and F-TBC | Yes | DESI-MSI | Q-TOF | HPTLC | 150 μm |
Mass spectrometry imaging (MSI)
MSI is a group of imaging technologies that don't necessitate molecule labeling. It has been shown that MSI is an effective method for in situ investigation of the spatial organization of molecular species.1 In a study in which the mass spectrometric [MS] assessment is carried out entirely from the sample as opposed to from isolates of the specimen as performed using traditional chromatographic methodologies, it makes use of the responsiveness and sensitivity of MS, which are superior to the capabilities of spectroscopic methods.1The spatial variation of biological components can be examined in situ using MSI, which has proven to be a highly effective method for doing so. In contrast to other more well-known biochemical imaging methodologies, MSI offers label-free screening without necessitating any prior knowledge of the potential target species. The sample preparation can be tailored for various categories of the relevant molecules of concern, even though MSI evaluations are untargeted.5 An ex vivo molecular visualization technique, MSI helps to better understand the biochemical processes taking place in the tissue by highlighting important components. These approaches do not entail anatomical visualization of the specimen, in contrast to conventional imaging modalities that do. A common characteristic among several clinical disorders is molecular downregulation.6–9 The MSI studies of the biomarkers linked to such disorders provide novel management and therapeutic prospects. When the morphological indicators in traditional histopathology are ambiguous, the evaluation of tissue chemicals typically provides more tangible information on the underlying condition.10 The MS-targeted methodology differs from conventional molecular visualization using chemical discoloration, immunostaining, fluorescence, and radiolabeling in that it does not require prior knowledge of the target species to be used.10 Researchers can more closely investigate the chemical makeup of tissues with the aid of this MSI property in order to appreciate the operation of many biochemical mechanisms that are crucial to biological systems and identify any anomalies that might be related to them.10
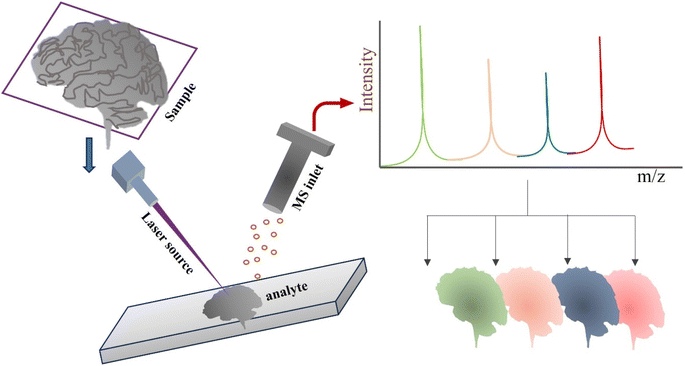
Another aspect to bear in mind is that MSI differs considerably from traditional MS-based techniques in that it examines how chemicals are dispersed on the specimen in space rather than employing isolated tissues.11 MSI has created new potential in the field of clinical MS by foreseeing the switch from the time-consuming LC-MS technology to an efficient and spatially defined in situ procedure. In fact, it has been demonstrated that MSI is quite helpful for assessing the spread of biomarkers in tissue sections to determine the boundaries of malignancy.12–15 Following screening the specimen's surface in 2D for imaging, mass spectrometric (MS) data are recorded pixel-by-pixel (Fig. 1). As a result, an immense quantity of MS data is gathered throughout the surface of the specimen, and any detected chemical entities' ion signals can be combined with their spatial information to produce their ion pictures.16 Due to the ability to spatially describe each ion signal, MSI can generate a substantial number of molecular pictures in a single scan. This allows for the continuous visualization of the spread of multiple endogenous and foreign species within the tissue samples, which can aid in the discovery of biomarkers, the detection of diseases, the development of innovative therapeutics, and pharmaceutical research.17
To assess the efficiency and suitability of MSI for the investigation of various molecular species in tissues, two factors are often prioritized: (i) spatial resolution and (ii) m/z span. The desorption process, which comprises the size of the probe, the layout of the mass detector, and foremost, the sample preparation, has an impact on both of these variables 4.18 The several ionization processes employed in MSI, according to McDonnell and Heeren (2007), are individually defined by complementary benefits and drawbacks that mainly concern the degree of spatial resolution, mass accuracy, chemical sensitivity, specificity, and m/z span.18 When measuring gaseous ionic species in an MS, sample ionization is necessary to move the analyte from the condensed to the gas phase.19
Without specialized sample pretreatment procedures, ambient ionization approaches permit the instantaneous evaluation of intricate materials under ambient (atmospheric) conditions.20–25 Sample processing happens in ambient ionization MS while the data is being analyzed. Analysis can be significantly accelerated and any potential impacts of sample preparation on the localization of molecules in the specimen are eliminated.20–23,26–28 On the basis of the sample processing steps required, Venter et al. (2014) divided ambient ionization approaches into three major categories: In liquid extraction techniques, analyte molecules are taken out of the specimen and retrieved into a solvent before being ionized; in desorption approaches, free ions can be produced straight from substrates; and in desorption techniques, larger fragments are produced that are then taken in through an electrospray cloud and ionized.29 Liquid extraction methods provide a number of important benefits, such as being simple to use, allowing materials to be analyzed in their natural settings, accelerating analysis, and allowing the extraction solvent makeup to be adjusted to the challenge in question. Solvent makeup, for instance, may be tuned for the effective extraction of several analyte categories from the substance, for estimation, or for online derivation via dynamic detection.28–30 Additionally, the capacity to conduct quantitative MSI creates exceptional prospects for analyzing variations in the concentration of chemicals in intricate biological analytes through the addition of meticulously chosen internal standards of established concentrations to the extraction solvent. Liquid extraction MSI methods are particularly appropriate for a variety of purposes because of their unique characteristics.
Although liquid extraction methods were the primary focus of nearly all the empirical investigations covered by this study, laser ablation methods were used in fewer studies. Using laser ablation approaches, analytes can be removed from the specimen's surface by ablation and desorption using IR or UV lasers.31 This study offers a scientific and methodological review of the use of ambient MSI in pharmacological research.
Ambient mass spectrometry (AMS)
The application of mass spectrometry (MS) in analytical and bioanalytical research has grown significantly. This enormous success and breadth are largely due to MS's unrivaled capacities for detecting, recording, and characterizing molecules and atoms of various kinds, formulations, and dimensions.32 High sensitivity, selectivity, and speed together have long been considered key benefits of MS. In recent years, MS has also greatly expanded in terms of the kinds of molecules and compounds that it can handle, enabling it to contend with almost all types of biological molecules, inorganic and organic salts, organometallic complexes, supramolecular objects, and biological entities like viruses and bacteria, in addition to relatively small and thermally inert organic molecules.33–39However, to handle the wide range of atoms, molecules, matrices, and mixtures, MS must promote effective ionization to produce diagnostic ions, preferably for every single component, which can be passed on to the high vacuum setting of MS in which they are identified and measured.40 The transfer of the analyte compounds from their “natural” ambient setting, where target molecules are typically found frequently in condensed forms along with matrices and in complicated mixtures, into the clean but relatively “inhospitable” extreme vacuum MS environment presented significant challenges.40 In MS analysis of mixtures, pre-separation processes were frequently necessary as well, and thermally unsettling and less volatile materials could not be handled.40
Analytical chemistry has undergone an upheaval with the advent of ambient ionization mass spectrometry (AMS), which makes it possible to analyze samples swiftly and directly in their natural form. The analytical sector has been instrumental in advancing AMS since its debut, spurred on by the plethora of benefits it offers in addition to conventional MS.41 The absence of chromatography permits rapid sample assessment; the versatility of ambient ionization sources allows them to be perfect for remote analysis when combined with portable devices; and time-consuming extraction and concentration procedures can be reduced or even removed. As a result, a growing variety of scientific disciplines are becoming aware of the potential uses of AMS, particularly in the fields of illness diagnosis, forensics, and environmental.42
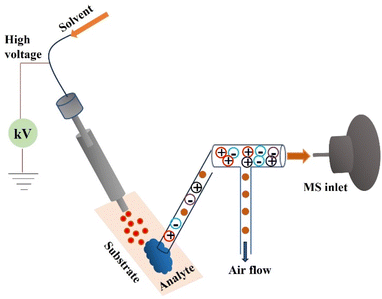
AMS methods can be divided into three main categories. (a) Techniques for solid–liquid extraction include the extraction or desorption of molecules from a sample's surface. (b) Ionization is often accomplished via an electrospray ionization (ESI) method.43 (c) By employing plasma and mechanisms like those in [APCI] air pressure chemical ionization, plasma desorption techniques produce ions in the plasma.44 Lastly, laser ablation approaches remove analytes from the specimen's surface by ablation and desorption using infrared or ultraviolet lasers.45 Methods of solid–liquid extraction encompass a number of well-known AMS techniques. These methods (DESI, AFADESI, nano-DESI, etc.) commonly employ electrospray ionization (ESI).
Ambient liquid extraction techniques and applications
The three main kinds of ambient liquid extraction methods are as follows: (1) spray-based approaches, whereby analyte particles are removed from the specimen via aerosol bombardment; (2) direct liquid extraction procedures, which also include liquid capture of analytical substances that have been desorbed; and (3) substrate spray approaches, in which electrospray ionization is carried out directly from the specimen.21–23,26–29,46Spray based techniques
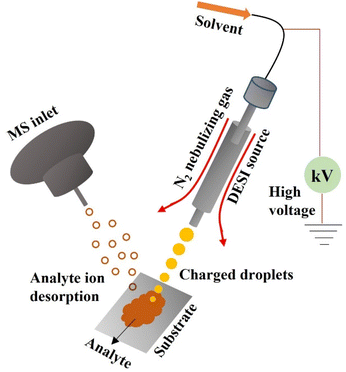
In general, a tissue segment having a layer thickness ranging between 10 and 15 microns serves as the substrate for the DESI-MSI. The resulting tissue segment was exposed to a stream of rapidly moving charged microdroplets to electrospray a solvent at elevated voltages while employing nitrogen as the nebulizing gas.51,52 The solvent droplet dampens the tissue surface as a result, dissolving biological components that exist in the tissue (mainly metabolites and lipids). The ionization by electrospray procedure and the desolvation method both produce ionic species in the gas phase by the same mechanism, which involves evaporation of the solvent and Coulomb fission of the subsequent little droplets, which include the analyte species.51 The biological imaging technique bombards the surface of the tissue with a spray of charged microdroplets while collecting MS spectra across the sample and scanning it in the x and y dimensions. The biochemical makeup of the tissue can be mapped using the two-dimensional image made from the pixel-to-pixel MS spectra.53
Major determinants of pharmacological effectiveness and adverse effects are the levels and spatial distribution of a medication and its byproducts in target tissues. A sensible comparison between the pharmacology of medications in tissues and features of medication formulations is made possible by MSI, which permits viewing of the spatial distribution and amounts of drugs in a tissue segment without radio-labeling.54 The capacity of MSI to assess spatially restricted drugs and their derivatives simultaneously is a significant benefit.54 This has been demonstrated to be especially crucial in drug toxicology research, as the drug and its derivatives may localize to various organs and have various toxicological implications.55
When inhaled corticosteroid clesonide [CIC] is transformed into an active metabolite [des-CIC] desisobutyryl-ciclesonide, which is then reversibly transformed into fatty acid esters inside the alveolar cells of the lung.56 Des-CIC is formed reversibly by fatty acid esters such as des-CIC-oleate, which preferentially aggregate in the lung and extend the pharmacological effects of CIC in vivo.57 Utilizing a hydrofluoroalkane metered-dose inhaler, CIC is administered to the lung in the form of locally functioning medication for the medical management of asthma. Particles with a diameter of less than 3 m are more inclined to settle deep within the lungs, because the airways are smaller.58 Although particles smaller than 3 μm have a probability of roughly eighty percent of reaching the lower airways and a fifty percent probability of settling in the alveoli, prior investigations have found that particles between 1.5 and 6 μm in diameter have a considerably broader distribution inside the lung.59
According to Labiris and Dolovich (2003), particles with “MMAD” [mass median aerodynamic diameter] less than 3 m are ideal for effective medication administration to the lung.60 According to Van Der Wiel et al. (2013), CIC particles with a size of less than 2 m are more inclined to enter the narrow airways of individuals and exert their pharmacological impact there.61 Research studies haven't documented the use of MSI to identify CIC in tissues; this is probably because CIC is a neutral molecule that is challenging to determine with adequate sensitivity to assess spatial localization. Following a single exposure of a CIC aerosol, Yamamoto et al. (2021) used DESI-MSI to examine the 2D spatial localization of CIC and its byproducts in rat lungs with a spatial resolution of 50 μm.54Fig. 3 illustrates how the concurrent presence of CIC, des-CIC, and des-CIC-oleate on the airway epithelium as well as the abundance of des-CIC and des-CIC-oleate in distant lung regions were discovered using MSI imaging.54 It's improbable that CIC and its derivatives were significantly delocalized following CMC injection to the lungs as well as during inflation before MSI analysis. The airway epithelium also saw an autonomous localization of a portion of CIC.54 These findings imply that the spatial dispersion of CIC and its derivatives in the lungs is influenced by the aerosols' targeted delivery to the pulmonary alveoli and surrounding areas as well as the propensity for CIC particles to lodge on the airway epithelium.54
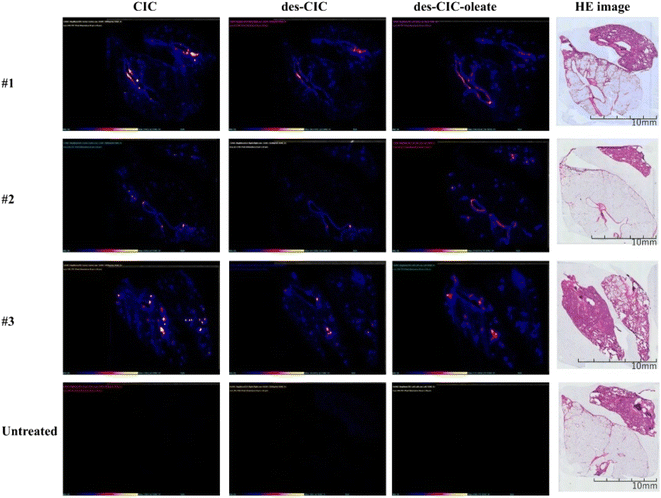 | ||
| Fig. 3 DESI-MS/MS and stained images of CIC, des-CIC, and des-CIC-oleate in lung tissues of rats after 1 h of inhalation of 1 μm CIC aerosols and untreated lung tissue—#: identification number of CIC exposed rats. Reproduced with permission [ref. 54] @ ELSEVIER. | ||
Drug-associated liver damage “DILI” is a big problem for the development of medicines globally and a substantial public health concern.62 In the industrialized nations of the globe, an excessive dosage of acetaminophen (APAP) causes DILI.63 The medicine is benign at therapeutic concentrations considering glucuronidation and sulfation procedures in the liver efficiently breakdown APAP to generate the nonoxidative byproducts APAP-glucuronide [APAP-GLUC] and APAP-sulfate [APAP-SULF]. Oxidative metabolites APAP-glutathione (APAP-GSH), APAP-cysteine (APAP-CYS), and APAP-N-acetylcysteine (APAP-NAC) are quickly picked up by hepatic glutathione reserves to stop liver damage. In the meantime, a small percentage of APAP can create N-acetyl-p-benzoquinone imine.64
In research investigating the chemical breakdown of acetaminophen, LC-MS has frequently been utilized to detect drug metabolic signatures in biofluids, including plasma, urine, or entire tissue homogenates.65–67 While this investigation provides some insight into the frequency of medicines or their derivatives, it fails to offer information on specific variations in drug metabolism across tissues or about localized amounts of drug-derived byproducts that might be hazardous to regions inside certain organs. In the past, conventional methods to reveal the localization of pharmaceuticals have utilized radiolabels or antibodies.68,69 Nevertheless, these approaches frequently fail to detect additional metabolites, that might accumulate in the tissue, leading to harmful adverse effects.70–73 Akakpo et al. (2022) revealed the spatial mapping of APAP breakdown inside the liver post APAP overload in the C57BL/6J mouse model and demonstrated modifications caused by 4-methylpyrazole (4MP) treatment (Fig. 4).64 This was done utilizing DESI-MSI technique. Mouse liver slices were studied after various APAP dosages to determine the spatial concentration and proliferation of oxidative and nonoxidative byproducts.64 Signal intensity variations associated with metabolite concentrations could be clearly seen in tissues, and 4MP therapy drastically altered this spatial distribution. The work offered a unique label- and matrix-free framework with outstanding sensitivity to evaluate the regional fluctuations in APAP metabolite concentration post 4MP therapy with a spatial resolution of 50 micron. This state-of-the-art imaging technique may also be used to study the impact of innovative therapeutic approaches on molecular targets and advance the development of medications to avoid the occurrence of APAP hepatotoxicity.64
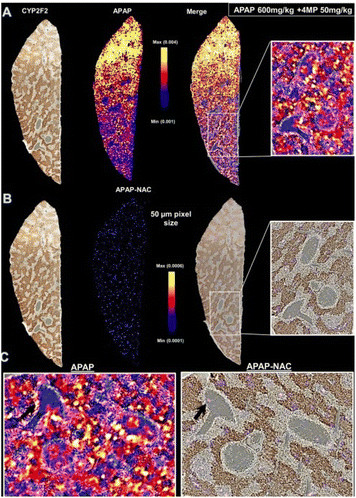 | ||
| Fig. 4 Effect of 4MP on biodistribution of APAP and APAP-NAC captured with DESI-MSI analysis of liver tissue sections at 50 μm pixel resolution after severe (600 mg kg−1) APAP overdose. Fasted male C57BL/6J mice received 600 mg kg−1 APAP followed by sacrifice 30 min later. A representative photomicrograph of the cryo-section used for DESI-MSI and then immunostained for CYP2F2 is shown on left. DESI ion images of APAP (A) and APAP-NAC (B) were then overlaid onto the same liver immunostained for CYP2F2 to generate merged images enlarged in panels on the right and expanded with rotation below (C). Reproduced with permission [ref. 64]@ACS. | ||
Currently, the preferred sample kind for metabolic MSI studies is fresh frozen tissues. Recent investigations, nevertheless, have made use of tissues that are fixed, including formalin fixed paraffin embedded [FFPE] specimen.74,75 Recent investigations have demonstrated the applicability of FFPE tissues for proteomics,76 tissue categorization,77 and N-glycosylation78 research. Micromolecular species, particularly lipids,79 is removed from tissues as part of the FFPE specimen preparation process. The substance's concentrations will eventually shift as a result, and the measurable distributions may be affected. There is frequently little consideration given to the repeatability of preparation and the assessment of the overall impact on the tissue metabolome. In addition to the known deterioration of proteins, lengthy RNA and DNA strands,80,81 and lipids in FFPE tissues,82,83 residual activity of enzymes84–87 (acetyltransferase, glutamic acid decarboxylase, and phospholipases etc.) in unprepared tissue sections during the slow kinetics of fixation may also have a direct effect on the rapidly evolving tissue metabolome.
Dannhorn et al. (2022) investigated the impact of formalin-fixation and FFPE on the observable metabolome, particularly xenobiotics with DESI-MSI and MALDI-MSI.88 Rat kidney and liver tissues were separated during the study and subsequently snap-frozen or preserved in formalin until analysis, depending on the amount of terfenadine, diphenhydramine, dextromethorphan, and losartan.88 To assess the impact of subsequent treatment of the fixed tissues, tissues were also treated in an FFPE-multi-tissue block.88 The discharge of polar constituents, such as salts, amino acids, organic compounds, and carnitine species, and the oxidation of endogenous lipids and the production of products from reactions between lipids and the fixing agent's components, were all found to be considerably affected by formalin fixation (Fig. 5).88 According to the time-related development of fixation, the slow fixation dynamics under ambient circumstances led to enhanced lipid breakdown in the tissue's interior. The deformation of the revealed biodistributions was caused by a partial disappearance of structural lipids during paraffin fixing.88
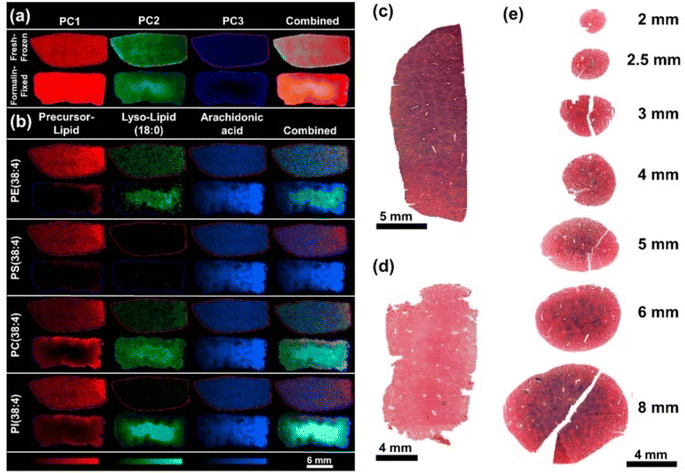 | ||
Fig. 5 Comparison of representative fresh-frozen and formalin-fixed rat liver specimens. (a) Pixel-wise PCA of the tissue sections analyzed by DESI-MSI in negative ion mode. PC1 (red) contributed 16.65% PC2 (green) 9.38% and PC3 (blue) 5.52% of the total observed variance. (b) The regional differences found within the PC1–3 were found to correlate with the degradation of phospholipids in the tissue core and occurrence of the hydrolysis products lyso-lipids (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) and FA(20 0) and FA(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). (c) LDH stain of a frozen liver section compared to (d) LDH stain of a tissue section after fixation of the tissue for 24 h. (e) LDH stained tissue sections of rat liver punches in various sizes after 1 h of fixation. Reproduced with permission [ref. 88]@MDPI. 4). (c) LDH stain of a frozen liver section compared to (d) LDH stain of a tissue section after fixation of the tissue for 24 h. (e) LDH stained tissue sections of rat liver punches in various sizes after 1 h of fixation. Reproduced with permission [ref. 88]@MDPI. | ||
A common aconitum Tibetan medicine is Radix Aconiti, sometimes referred to as Tie-bang-chui or “TBC.” It must be transformed and utilized as a medicine because of its severe adverse effects and amazing performance.89 Highland barley wine [HBW] and fructus chebulae soup [FCS] are two TBC procedures for processing that exclude heating. Aconitum alkaloid constituents were successfully identified using a variety of analytical techniques, including thin layer chromatography [TLC], high-performance liquid chromatography [HPLC], liquid chromatography paired with MS, and others.89 Although separation with HPLC remains a popular technique, it comes with significant drawbacks, including a lengthy analytical period, difficult mobile phase preparation, and substantial solvent usage.90,91 The alkaloid makeup of TBC that hasn't been heated can therefore be visualized using an approach that takes less time and uses fewer chemicals.
Tan et al. (2023) recently conducted an investigation to further comprehend the variations in chemical makeup among raw TBC and non-heat processed versions.89 The chemical makeup of TBC generated by HBW [H-TBC] and FCS [F-TBC] was examined through the investigation using HPLC coupled with DESI-MSI.89 52 chemical compounds were found in unprocessed and processed items, and F-TBC and H-TBC had slightly different chemical profiles from raw TBC. Due to the substantial concentration of caustic tannins in FCS, the degradation procedure of H-TBC also proved distinct from that of F-TBC.89 It was discovered that following treatment by FCS and HBW, all six alkaloids—deoxyaconitine, hypaconitine, acetylaconitine, aconitine, mesaconitine, and benzoylaconine—had their contents drop except for aconitine (Fig. 6). In a nutshell the non-heating treatment technique for TBC differed substantially compared to the heating treatment approach in terms of the chemical alteration pathway.89 The major chemical alteration method of the processing of F-TBC was essentially obvious through this investigation. In comparison to the heating treatment technique, the non-heating process's chemical alteration process proved to be more multifaceted and varied.89 The main paragraph text follows directly on here.
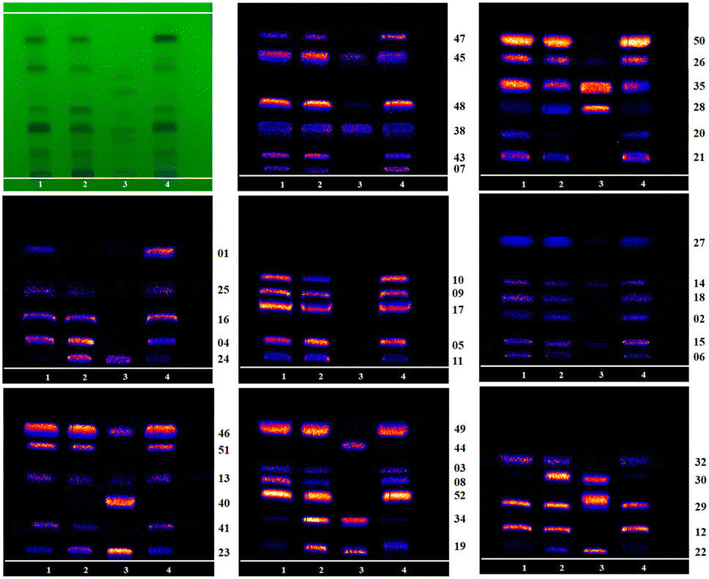 | ||
| Fig. 6 Mass spectrometry imaging comparison of 46 other identifiable alkaloid components on HPTLC (1: R-TBC, 2: F-TBC, 3: D-TBC, 4: H-TBC). Reproduced with permission [ref. 89]@Frontiers. | ||
Antineoplastic medications have rapidly progressed beyond general cytotoxic drugs to specific, mechanistic oriented treatments, and novel immunomodulators, which render them one of the most frequently utilized aggressive cancer treatments across the techniques at hand.96 These medications' effectiveness is severely constrained, nonetheless, by large off-target hazards,97 inferior cancer permeability,98 and often occurring developing resistance.99 To address such drawbacks, scientists have created several targeted approaches that may improve cancer discrimination ability to accomplish the goal of eliminating cancers while preventing major adverse effects. In any case, the spread of medications in the intended and non-intended zones forms the fundamental premise of drug effectiveness and possible adverse effects, irrespective of the kind of anticancer therapy used. In a nutshell, medications can achieve their beneficial curative properties if they remain near a cancer over an adequate length of duration and at the right concentrations; by comparison, an unintended buildup of a potential medication or its byproducts in normal tissues will result in the related toxic effects.100,101 It will be easier to understand the processes underlying the effectiveness of drugs and resistance with the aid of complete and comprehensive studies regarding the intratumor dispersion of chemotherapy drugs with diverse properties, which will encourage the creation of clinically practical compositions that inhibit the spread of tumors.102 Consequently, early drug discovery and study is crucial for determining the uptake of antitumor medications in the native organism. This will also help enhance our comprehension of drug breakdown and kinetics.102
Utilizing a label-free AFADESI-MSI to measure the spread of paclitaxel [PTX] as well as the associated prodrug [PTX-R] in whole-body animal segments, Zhang et al. (2020) assessed the tumor-targeting effectiveness and intra-tumoral variability of a novel anticancer drug choice.102 Using mice with epidermal A549 xenograft cancers, tumor targeting effectiveness was compared between 3 therapy categories: PTX-injection, PTX-liposome, and PTX-R.102 According to these findings (Fig. 8), both the PTX-liposome cohort and the PTX-injection cohort had high levels of PTX in numerous organs throughout the injected body. In particular, the corresponding targeting performance of the PTX-R cohort rose by about 50-fold, significantly reducing widespread harmful effects.102 Both the prodrug and processed PTX accumulated primarily in cancerous tissue in the PTX-R cohort, which distinguished itself from the PTX-liposome cohort. Additionally, PTX-R demonstrated a large aggregation in the necrotic region and an indistinguishable intratumor region.102 Additionally, it was demonstrated that the AFADESI-MSI technology is a highly sensitive and comprehensive method that renders it possible to simultaneously visualize different kinds of drug derivatives both at the whole-body scale and in microregions.102 The creation and improvement of drugs in the initial phases of pharmaceutical R & D might be greatly aided by this technique.102
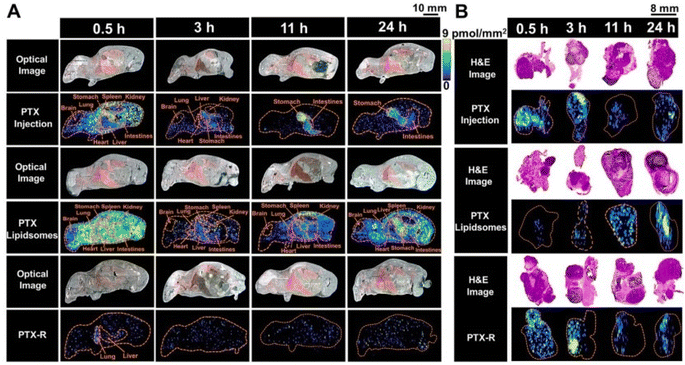 | ||
| Fig. 8 The spatial-temporal and quantitative distribution of PTX in whole-body animals (A) and the corresponding flank tumors (B) at different time points in the three treatment groups visualized by AFADESI-MSI. Reproduced with permission [ref. 102]@IVYSPRING. | ||
The CNS [central nervous system] possesses arguably the most intricate design and operation of any organ, controlling all facets of actions, including speech, thought, consciousness, action, and emotions.103 Drug effectiveness and distribution vary depending on the functional variation in enzymes used for metabolism, carriers, and blood circulation in the brain microregions.104 The distribution of many endogenous active metabolites in the various microregions of the brain is also uneven.105 Because many CNS medications only have an impact upon accessing the brain, it can be particularly difficult to create CNS medications and explain when and how the biological processes might take place for a specific medical treatment.106 While operational and structural diagnostic imaging (MRI, fMRI, CT, and PET) serves an increasingly important role in clinical investigation as well as the creation of CNS drugs, these methods merely offer visuals of the physical makeup of brain tissue instead of molecular analysis,107 have inadequate spatial detail, and can track a tiny amount of different substances.108 The LC-MS analytical technique's preliminary processing of tissue samples is intrusive and unsafe for the microregions of the brain. Consequently, without knowledge of the molecule's spatial distribution across the brain, the outcomes that ensue merely represent the mean metabolic level in the microregion sampled.109,110
To better comprehend the molecular processes of drugs performing on the microregions of the CNS, Liu et al. (2022) used AFADESI-MSI and a temporo-spatial pharmacometabolomic technique to assess the impact of olanzapine (OLZ) on the tissue segments of the brain (Fig. 9).106 They also showed how efficient these methods were at describing the microregional pharmacodynamics and pharmacokinetics of OLZ.106 AFADESI-MSI centered temporo-spatial pharmacometabolomic method possesses the benefit of precisely capturing in situ and microregional molecular phenomena and identifying the fluctuation in levels of endogenous and exogenous compounds within the various microregions of the CNS.106 The drug's pharmacokinetics of the novel medication and its derivative, 2-hydroxymethyl OLZ, in several regions of the rat brain were clearly demonstrated by this technique.106 Furthermore, the visualization of metabolic routes demonstrated the microregional pharmaceutical dynamics of OLZ.106 It was discovered that certain neurotransmitters showed different metabolic routes relevant to pharmaceutical treatment, notably GABA, adenosine, and histamine. Additionally, the beneficial and adverse consequences of OLZ usage in clinical settings may be related to the control of aspartate, glutamate, and the glycerophospholipid metabolic route, which is important knowledge for comprehending the molecular basis of its function.106
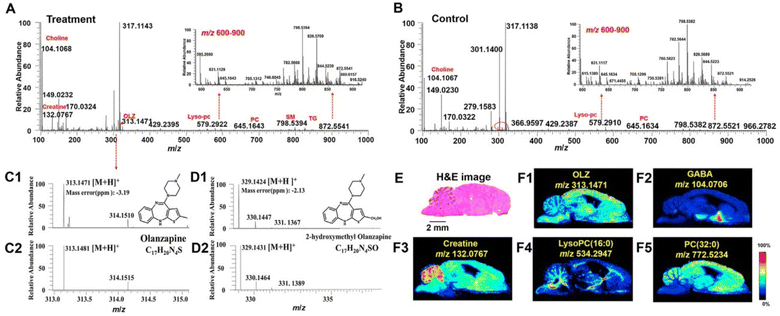 | ||
Fig. 9 Imaging results for exogenous drug and endogenous metabolites obtained using AFADESI-MSI from brain tissue section. The representative mass spectra acquired using AFADESI-MSI from treatment (A) and control groups (B) brain tissue section in positive ion mode. C1 and D1 are experimental mass spectra of olanzapine (OLZ) and its metabolite, 2-hydroxymethyl OLZ, respectively. C2 and D2 are the calculated isotopic peaks of OLZ and 2-hydroxymethyl OLZ, respectively. (E) represents the hematoxylin-eosin (H&E) staining of sections of the brain. Scale bar Z 2 mm. (F1)–(F5) show ion images of OLZ, g-aminobutyric acid (GABA), creatine, lysoPC (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PC (32 0), PC (32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), respectively. Reproduced with permission [ref. 106] @ ELSEVIER. 0), respectively. Reproduced with permission [ref. 106] @ ELSEVIER. | ||
The assessment of possible liver toxicity is a crucial phase in early medication discovery. For choosing among the most viable new drugs for usage in clinical investigations that exhibit superior effectiveness while presenting minimal adverse effects, in vitro research techniques are essential.111–113 Because these platforms allow for the rapid identification of numerous drugs, hepatocyte single layers in different configurations are now frequently utilized to look for drug-induced liver damage or examine the underpinning molecular mechanisms.114 Nevertheless, increasing evidence suggests that two-dimensional single layer cultures give up the biological signals and cell-to-cell interactions vital for sustaining the physiological characteristics and functions that have been specific to the liver in cells, making them poor predictors for substances that cause injury to the liver by drugs.112,115 The viability of cells testing, which depends on activities mediated via cellular enzymes that regulate metabolism and inadvertently reflects cellular metabolic processes, remains by far the most frequently utilized toxicity evaluation approach in 3D spheroid models. The difficulty of quantifying several mechanistic factors that encompass a broad spectrum of prospective cytopathological alterations, nevertheless restricted development.116 Once liver injury develops, specific metabolic routes and activities can be determined thanks to metabolomics ability to provide enormous quantities of information that depict the intricate controls governing internal metabolism.117–119 MSI-based spatially determined metabolomics is being utilized to spatially sort out the abundances of endogenous and exogenous molecules in 3D cell spheroids120–122 as well as preserve the location-specific data for a variety of both known and unidentified molecules in diverse biological specimens with no labeling.123–125 There is still a critical need for a broad-coverage MSI technique for segmenting the metabolic processes and identifying different cell states.
The antiarrhythmic medication amiodarone [AMI] is frequently prescribed and has been linked to liver fibrosis, cirrhosis, and steatohepatitis.126 According to several research studies, AMI metabolism has been linked to drug-induced liver toxicity,127,128 although the exact molecular processes are still poorly understood. AFADESI-MSI and 3D HepG2 spheroids were combined in a recent study by Li et al. (2023) to evaluate the liver injury and metabolism of AMI.129 The spatial and temporal patterns of extrinsic drug breakdown throughout the course of therapy for AMI were studied using AMI as a model drug to identify possible drug metabolic routes. In addition, fluctuations in metabolism during AMI therapy were examined to spot developing modifications linked to medication action.129 To define the effects of hepatotoxic substances, metabolic indicators connected to the viability of cells were assessed and built into biomarker categories.129 Upon AMI therapy at various intervals, 15 metabolites of AMI participating in metabolic processes were discovered, and the metabolic routes of AMI have been proposed based on their spatiotemporal behavior properties (Fig. 10).129 Then, using metabolomic assessment, it was possible to determine the temporal and spatial variations in the metabolic changes brought on by drug intake within spheroids.129 Arachidonic acid and glycerophospholipid breakdown were the two main malfunctioning routes of metabolism, and they provided substantial proof for the underlying cause of AMI liver injury.129 Additionally, a collection of eight fatty acids was chosen as an indicator since they offered an improved prediction of cell survival and may describe the hepatic damage of AMI.129 An efficient technique for assessing the in vitro liver toxicity of medicines is provided by the pairing of AFADESI-MSI with HepG2 spheroids, which may concurrently gather spatiotemporal data concerning medications, drug derivatives, and intrinsic metabolites post AMI therapy.129
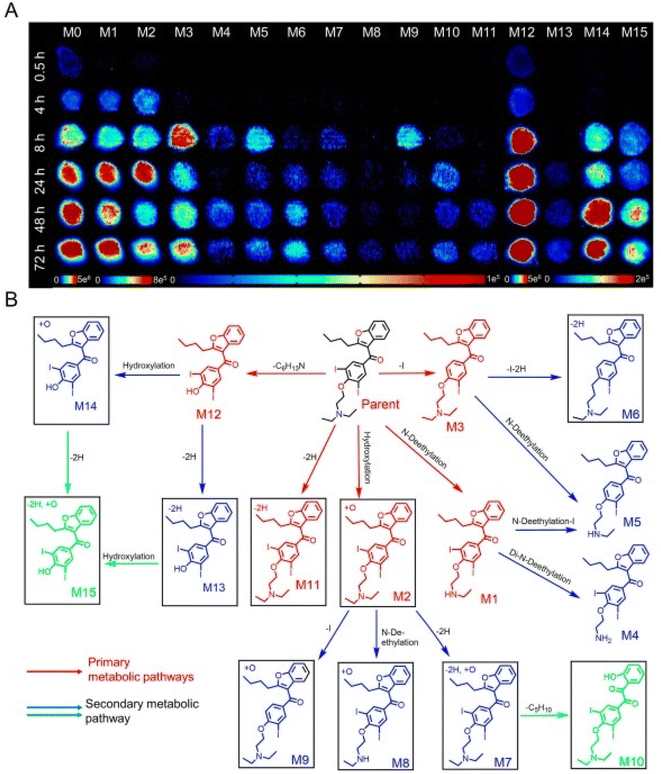 | ||
| Fig. 10 Time-dependent penetration and metabolism of amiodarone (AMI) in HepG2 spheroids analyzed using airflow-assisted desorption electrospray ionization-mass spectrometry imaging (AFADESI-MSI). (A) AFADESI-MS images of AMI and metabolite distribution at different treatment times. HepG2 spheroids were treated with 50 mM AMI for 0.5, 4, 8, 24, 48, and 72 h. (B) Postulated metabolic pathways of AMI in the HepG2 spheroids. Reproduced with permission [ref. 129]@ELSEVIER. | ||
Direct liquid extraction
Medications and xenobiotics frequently undergo the stage II metabolic process of glucuronidation, which makes them more soluble for excretion. An anti-steroidal and non-inflammatory medicine called diclofenac is widely employed to alleviate swelling and discomfort.134–137 4′-hydroxydiclofenac, 5-hydroxydiclofenac, and diclofenac acyl glucuronide [DCG] are the three main diclofenac products in both people and rodents.132–136 A group of cytochrome P450 enzymes breakdown diclofenac to generate both hydroxydiclofenac derivatives. A stage II uridine diphosphate glucuronosyl transferase enzyme converts diclofenac into diclofenac acyl glucuronide throughout the process of metabolism.138–140 In making drugs more water soluble, these biological processes are required to hasten the elimination of drugs via bile and urine. Diclofenac glucuronide is primarily eliminated from the body through the kidneys in human beings as opposed to laboratory animals, where it is primarily eliminated via the bile.141,142 Both the diclofenac acyl glucuronide and the quinone-imine precursors of the hydroxy derivatives have been connected to liver toxicity and intestinal damage in humans as well as animals.135–140,143 Elevated local concentrations of the metabolite, the higher alkaline pH of bile, or the protein's high level of expression all promote covalent connections involving acyl glucuronides and proteins.144 It is suggested that the sensitivity of diclofenac's derivatives causes adverse reactions and unfavorable medication responses.145
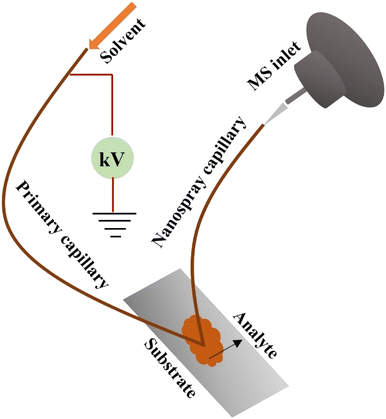
To investigate the distribution of diclofenac and its derivatives in mice kidney and liver tissues, Sanchez et al. (2022) utilized nano DESI-MSI.146 In the absence of sample pretreatment, label-free visualization with outstanding spatial accuracy and responsiveness is possible using nano-DESI MSI.146 Ion visuals for diclofenac and its main derivatives were created using nano-DESI-MSI. The drug and its metabolites had relatively weak signals in MSI data that was collected over a wide mass to charge range.146 With selective ion monitoring “SIM”, with mass to charge intervals concentrated near the low-abundance ions of concern, signal measurements were improved by not less than one order of magnitude.146 Findings (Fig. 12) show that hydroxy diclofenac is restricted to the cortex of the kidney and DCG to the inner medulla.146 The determined concentrations among the two derivatives roughly correspond to the locations of the enzymes that convert diclofenac into each of its corresponding derivatives as previously described.146 The presence of DCG in the medulla suggests that the harmful metabolite is leaving the tissue. Alternatively, the liver tissue showed consistent levels of diclofenac, hydroxy diclofenac, and the DCG derivative. The amounts of metabolite to diclofenac proportions determined from nano-DESI and LC-MS studies were confirmed to be in accord by semiquantitative evaluation.146
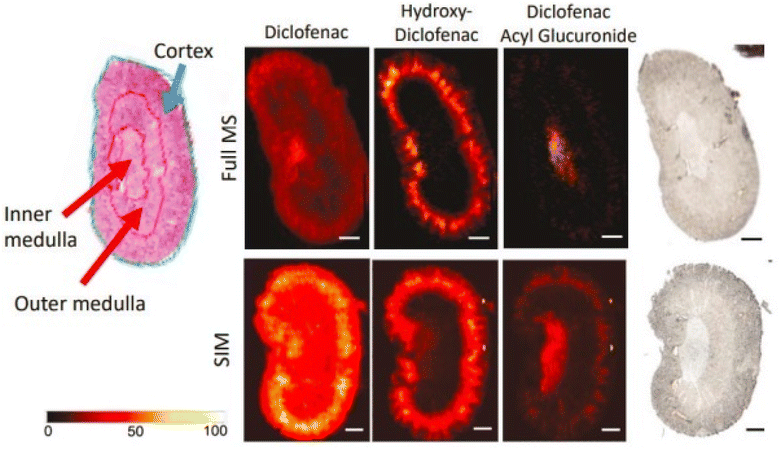 | ||
| Fig. 12 H&E, optical, and nano-DESI ion images of diclofenac dosed mouse kidney tissue. Relevant regions of H&E stained kidney section are annotated for comparison with ion images. Ion images of diclofenac, acyl glucuronide, and hydroxydiclofenac are shown for both broadband (full MS) and SIM imaging modes on kidney. Ion images are self-normalized to the internal standard, diclofenac-d4. Scale bar represents 1 mm. Intensity scale ranges from 0% intensity (black) to 100% (white). Reproduced with permission [ref. 146]@ChemRxiv. | ||
Determining the spread of experimental anti-TB agents inside the lesions as well as their capacity to get past necrotic granulomas is therefore necessary for a thorough experimental assessment of these medications.153 To examine the absorption and uptake patterns of anti-TB medications (clofazimine [CFZ], pyrazinamide [PZA], and rifampicin [RIF]) within mice lung tissue, Kokesch-Himmelreich et al. (2022) designed an AP-MALDI-MSI process.153 The research employed an unusual animal model that closely resembled the pathophysiology of human TB. By employing a procedure that was streamlined to work with superior spatial accuracy MSI, MTb was rendered inactive in the lung regions of these animals by irradiation.153 Given a spatial accuracy of 30-micron, distinct distributions of PZA and RIF have been seen within the tumors of Mtb-infected mice, adding additional confirmation to support the micro histological characteristics of mice tumors.153 Perhaps for the inaugural time, CFZ was visualized in the present investigation using an animal model. The research demonstrates the appropriateness of the animal model for experimental anti-TB medication development by confirming the dispersion characteristics of CFZ and PZA in the tumors of individuals with TB.153 According to the data, CFZ can penetrate the granuloma and build up within the cellular sections of macrophages. The results obtained suggest that experimental animal research's ability to predict is increased by the imaging system described in the study.153
Another widely recognized medication called imipramine is prescribed for treating anxiety and depressive symptoms. It inhibits D2, antimuscarinic, and adrenergic receptors and reduces serotonin and norepinephrine reuptake.154 Another prominent medication, chloroquine, is prescribed for treating viral illnesses, primarily malaria.155 It prevents some viruses from completing their pH-sensitive multiplication processes. While either of these little medications is often utilized in contemporary human medical care, they each have several adverse reactions.156,157 With an improved comprehension of the mechanism of action and hazardous qualities, understanding the location of these given medicines and their derivatives in living tissues is of utmost importance. But there is currently little proof of their location in many organs.158 Islam et al. (2022) developed AP-MALDI-MSI, also referred to as iMScopeTM QT, in a recent study to quickly map imipramine, chloroquine, and its derivatives in mice that are wild-type. Findings from the AP-MALDI-MSI methodology were contrasted with those from the traditional DESI-MSI and MALDI-MSI methods.158
It was discovered that whereas chloroquine and its derivatives were substantially collected in the medulla of the kidneys and pelvis of mice, imipramine and its derivatives were greatly deposited in the cortex of the kidney of mice.158 Various sections of mice's brains exhibited a larger buildup of imipramine (Fig. 13). However, the lateral ventricle, fourth ventricle, and fornix of the mice's brains displayed a considerable buildup of chloroquine and its derivatives. Evaluating these medications' therapeutically significant characteristics, effectiveness, and possible adverse reactions can be aided by the results of the investigation.158
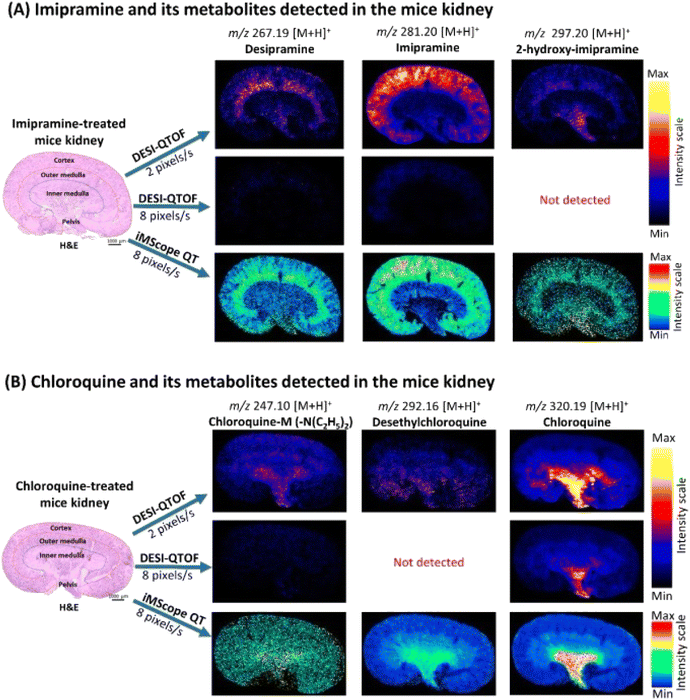 | ||
| Fig. 13 Localization of imipramine, chloroquine, and their metabolites in the kidney of treated mice applying DESI-QTOF and iMScope QT at different data acquisition speeds. (A) Distribution of imipramine and its metabolites in the treated mice kidney. (B) Distribution of chloroquine and its metabolites in the treated mice kidney. Here, spatial resolution was 50 μm × 50 μm (X, Y), and CHCA was used as the matrix for iMScope QT samples s: second. Reproduced with permission [ref. 158]@MDPI. | ||
Advantages and limitations of ionization sources
Table 2 contrasts the advantages and shortcomings of MALDI, DESI, and AFADESI ionization techniques. A fresh perspective on the altered relationships between various biological processes, small molecules, and enzymes in tumor tissues may be obtainable through metabolomics, or the research of small molecules. Spatial metabolomics offers the possibility of discovering molecules' localizations in addition to their relative abundances. In this manner, changes in tiny molecules can be linked directly to anatomical characteristics. Consequently, it might lead to the establishment of individualized treatment, quicker diagnostic techniques, and increased knowledge of complex disorders.159,160 Because spatial metabolomics maintains the spatial information of metabolites, it necessitates a concurrent, in-depth analysis of several small molecules. MSI is a label-free, highly efficient technology that can identify significantly more molecules simultaneously than conventional staining techniques.161 One of the extensively used ionization techniques associated with MSI is MALDI because it provides an excellent balance between sample preparation, spatial resolution, and sensitivity.162 In a recent research investigation, atmospheric pressure (AP)-MALDI was employed to achieve a spatial resolution of 1.4 μm.163 However, the ion yield often decreases when the dimension of the laser's focal area shrinks to attain a superior spatial resolution.162,164 Consequently, to carry out spatial metabolomics investigations, researchers still need to strike a balance between obtaining sufficient signal intensities and having improved spatial resolutions. One of MALDI-MSI's additional limitations is a drop in resolution brought on by delocation, which is the result of molecules diffusing via and far from the tissue.165 Additionally, MALSI-MSI has been shown in some investigations to have difficulty recognizing low molecular weight molecules. This is because the chemical characteristics of the matrix ions resemble several lower weight metabolite ions and may be incompatible with metabolite visualizations.166,167| Technique | MALDI-MSI | DESI-MSI | AFADESI-MSI |
|---|---|---|---|
| a FFPE – formalin-fixed, paraffin-embedded. | |||
| Environment | Vacuum | Ambient | Ambient |
| Maximum spatial resolution | ∼1.4 micron | ∼50–200 micron | ∼100 micron |
| Sample type | Frozen or FFPE | Frozen or FFPE | Frozen or FFPE |
| Pros | Higher spatial resolution | High throughput | Atmospheric conditions |
| Ideal for small specimens | Atmospheric conditions | Minimal sample preparation | |
| Reliable results | Minimal sample preparation | Varying field of coverage | |
| Swift analysis | Better sensitivity and resolution over DESI-MSI | ||
| Cons | Tedious sample preparation process | Low spatial resolution and sensitivity | Low reproducibility |
| Non-ambient conditions | |||
DESI-MSI is the inaugural ambient MSI technique, as opposed to MALDI-MSI, which utilizes a vacuum-based MSI approach. Real-time diagnosis may be possible thanks to ambient MSI's simpler operational requirements and capacity to evaluate larger samples quickly.168–171 Additionally, DESI does not demand a matrix like MALDI does. As a result, it doesn't experience the MALDI technique's typical significant risk of spatial assignment inaccuracy brought on by sample movement.172 DESI additionally renders the samples accessible for observation and additional operations throughout the analysis due to its minimal pretreatment requirements.173 Research investigations implementing the approach, however, have faced a significant hurdle in increasing the sensitivity of DESI. In addition, due to variables such as solvent makeup, capillary dimensions, and gas flow rate, DESI-MSI also has a lesser spatial resolution compared to MALDI-MSI (30–50 μm).174 Recent investigations have utilized nano-DESI-MSI to overcome these restrictions to attain enhanced spatial resolutions (10 μm), highlighting the possible future developments of DESI-MSI.175,176
AFADESI, an approach derived from DESI that substantially increases and widens the imaging area, was recently studied by Abliz et al. (2022)92 AFADESI uses an electrospray plume to ionize the analyte, utilizing the same technique as DESI. Finally, for MSI, airflow is used to transport ions across extensive distances.172 AFADESI-MSI can achieve an extremely broad coverage of the metabolites being studied, along with acquiring the benefits of DESI-MSI. In an untargeted study, it can simultaneously identify thousands of molecules.177 Whole-body sections can also be imaged using this technology.178 Due to the ability to simultaneously study and compare so many more metabolites, this coverage considerably expands the range of potential utility of spatial metabolomics. Furthermore, when the gap that exists between the ion transport tubing and the MSI opening is tuned, AFADESI can attain picomolar sensitivity.179 Future studies can produce several uniform tissues to test the appropriate settings and solvent compositions that can be utilized repeatedly to increase sensitivity and reproducibility.
While improved MSI methodologies are continually being developed, mass and spatial resolutions continue to present a problem in the advancement of spatial metabolomics. The area remains unexplored but has great promise for understanding drug metabolism and advancing diagnostic and therapeutic approaches.
Conclusions and perspectives
A significant and expanding field of biological research is MSI. The relative abundances of a wide range of chemicals in different specimens, like tissue slices, can be readily observed when carried out in ambient settings. These procedures are benign and ambient, making it simple to analyze the native specimen for data on surface chemical compositions. These benefits guarantee that in the future, interest in biochemical MSI via ambient settings will expand significantly.MSI can be improved as an improved multimodal molecular visualization technology in the areas described below.
Isobaric ion identification
The in situ detection technique used by MSI makes it challenging to identify isobaric ions. It is impossible to distinguish between ions having the same chemical formula. Due to its function in gas-phase chromatographic separation, the development of [IMS] ion mobility spectrometry offers a workable solution to this problem. Ion characterization can be improved with chemometrics based on numerous complementing factors, such as isotope affluence, and highlighted fragments.180–182 Online wet experiments, in addition to the post-acquisition dry method, also offer supplementary chemical characteristics to aid in differentiating isobaric ions, including hydrogen-deuterium exchange throughout the course of ionization in the liquid state or the post-ionization procedure in the gaseous phase.183,184Sensitivity
To obtain molecular fingerprints, particularly from a sub-micron-sized sample, without suffering a significant sensitivity loss, an improved delicate in situ benign ionization technique must be developed. The production of novel nanomaterials for mounting analytes, carefully constructed auxiliary ionization, complex ion centering lens systems, and sample-friendly preparatory processing ought to be continually pursued. This constitutes a crucial area of investigation that could further the work on MSI research. In addition to technological improvement, the area of appropriate research into pharmaceuticals will also grow.Spatial resolution
This can be regarded as another crucial MSI statistic to assess the accuracy with which an ion map can be captured to obtain greater precision in spatial data. Since a decrease in sampling size typically entails a very small molecule concentration and the related signal output provides a uniform ionization performance, this is typically a contradictory issue when combined with the sensitivity. Currently, a multi-modular blending of images can enhance spatial resolution. When co-registering pixels derived from a crude MS scan alongside other cell-resolved digital representations obtained from H&E, IF, and MRI, it is possible to anticipate the molecular data within a narrower region.185–188 In contrast to being noticed, such a high-resolution image gets strategically predicted. An improved and economically viable MSI approach for sub-micron-resolved metabolite visualization won't be a barrier for spatial omics until benign ionization achieves a major developmental milestone in resolution.Data acquisition
A simpler and more consistent process is anticipated to be established by an automated robotic device for the automatic handling of tissue specimens, expedited MSI data collection, and quality assurance. It is astonishing to see that efforts are being undertaken to advance this field of commercial design.189 The current standard method for collecting MSI data uses a microprobe-oriented spot-by-spot or one line at a scan mode, that may require several hours to an entire day for completion for bigger samples or whenever a superior image resolution is needed. The MSI's operating efficiency will unquestionably increase with a quick, miniature snapshot-mode collection of data.190 This necessitates a micro-arrayed ion detection approach with a more complex MS equipment setup.Artificial intelligence and omics database
To gain insight into the chemical characteristic design and direct exact spatial detection, machine learning had already been introduced.191 In order to facilitate the merging of spatial data from several omics, especially spatial transcriptomics and genomics, additional advanced data science approaches and computational techniques are anticipated to be introduced. To facilitate unrestricted exchange and investigation among MSI researchers, an open-access MSI information system should be developed.192 Additionally, an open-access spatial omics library with biochemical phenotypic atlases from essential organs and tumor tissues is anticipated. Naturally, this requires cooperation and agreement among the broader MSI and omics societies.Diversity of specimens and functional imaging
There are fewer, more frequently utilized biological samples in pathology studies that can be made compliant with MSI. This is especially true for imaging investigations using ambient ionization that are aimed at drug metabolites. There are several biological associations underlying the peaks in the mass spectral signature obtained by an MSI experiment, which is not just a list of isolated, insignificant peaks. These biologically meaningful links may shed light on the relationship between a drug's traits and its spatially defined molecular composition and function. By assessing and arranging the collected information, it is anticipated that MSI may also evolve into a functional imaging tool to visualize biological events, such as growth, spread, autophagy, iron metabolism, and enzyme catalysis.193–197Considering all the benefits listed in the studies above (Table 1), MSI should only be used when standard modalities, especially liquid chromatographic (LC) methods combined with MS, are unable to provide the required analytical data. LC procedures coupled with MS are often more accurate and quantitative than MSI. Therefore, LC-MS is probably a superior option unless precise spatial details are required, such as when confirming or quantifying the presence of a medication or metabolite in a specimen. This suggests that there is little reason to expect that a chemical will be identifiable by MSI, which lacks the capability for pre-concentration and isolation as in traditional bioanalysis, if the substance cannot be found by LC-MS after thorough specimen extraction and workup.
Author contributions
Each author contributed to the article's design and conceptualization. All authors read the final draft before giving their approval.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the professors who provided helpful criticism and feedback.References
- J. R. Granborg, A. M. Handler and C. Janfelt, Mass spectrometry imaging in drug distribution and drug metabolism studies–Principles, applications and perspectives, TrAC, Trends Anal. Chem., 2022, 146, 116482 CrossRef CAS.
- A. Nilsson, R. J. Goodwin, M. Shariatgorji, T. Vallianatou, P. J. Webborn and P. E. Andrén, Mass spectrometry imaging in drug development, Anal. Chem., 2015, 87(3), 1437–1455 CrossRef CAS PubMed.
- R. J. Goodwin, Z. Takats and J. Bunch, A critical and concise review of mass spectrometry applied to imaging in drug discovery, SLAS Discovery, 2020, 25(9), 963–976 CrossRef CAS PubMed.
- X. Song, C. Li and Y. Meng, Mass spectrometry imaging advances and application in pharmaceutical research, Acta Mater. Med., 2022, 1, 507–533 Search PubMed.
- W. Michno, P. M. Wehrli, K. Blennow, H. Zetterberg and J. Hanrieder, Molecular imaging mass spectrometry for probing protein dynamics in neurodegenerative disease pathology, J. Neurochem., 2019, 151(4), 488–506 CrossRef CAS PubMed.
- M. P. Mattson and S. L. Chan, Dysregulation of cellular calcium homeostasis in Alzheimer's disease: bad genes and bad habits, J. Mol. Neurosci., 2001, 17, 205–224 CrossRef CAS PubMed.
- K. A. Fujita, M. Ostaszewski, Y. Matsuoka, S. Ghosh, E. Glaab, C. Trefois, I. Crespo, T. M. Perumal, W. Jurkowski, P. M. Antony and N. Diederich, Integrating pathways of Parkinson's disease in a molecular interaction map, Mol. Neurobiol., 2014, 49, 88–102 CrossRef CAS PubMed.
- T. D. Prickett and Y. Samuels, Molecular pathways: dysregulated glutamatergic signaling pathways in cancer, Clin. Cancer Res., 2012, 18(16), 4240–4246 CrossRef CAS PubMed.
- M. Telias, Molecular mechanisms of synaptic dysregulation in fragile X syndrome and autism spectrum disorders, Front. Mol. Neurosci., 2019, 12, 51 CrossRef CAS PubMed.
- A. Ajith, Y. Sthanikam and S. Banerjee, Chemical analysis of the human brain by imaging mass spectrometry, Analyst, 2021, 146(18), 5451–5473 RSC.
- X. W. Zhang, Q. H. Li and J. J. Dou, Mass spectrometry-based metabolomics in health and medical science: A systematic review, RSC Adv., 2020, 10(6), 3092–3104 RSC.
- S. Banerjee and S. K. Manna, Assessment of metabolic signature for cancer diagnosis using desorption electrospray ionization mass spectrometric imaging, Cancer Metabolism: Methods and Protocols, 2019, pp. 275–297 Search PubMed.
- S. Banerjee, Ambient ionization mass spectrometry imaging for disease diagnosis: Excitements and challenges, J. Biosci., 2018, 43(4), 731–738 CrossRef CAS PubMed.
- L. Hänel, M. Kwiatkowski, L. Heikaus and H. Schlüter, Mass spectrometry-based intraoperative tumor diagnostics, Future Sci. OA, 2019, 5(3), FSO373 CrossRef PubMed.
- M. Woolman, L. Katz, G. Gopinath, T. Kiyota, C. M. Kuzan-Fischer, I. Ferry, M. Zaidi, K. Peters, A. Aman, T. McKee and F. Fu, Mass spectrometry imaging reveals a gradient of cancer-like metabolic states in the vicinity of cancer not seen in morphometric margins from microscopy, Anal. Chem., 2021, 93(10), 4408–4416 CrossRef CAS PubMed.
- D. S. Cornett, M. L. Reyzer, P. Chaurand and R. M. Caprioli, MALDI imaging mass spectrometry: molecular snapshots of biochemical systems, Nat. Methods, 2007, 4(10), 828–833 CrossRef CAS PubMed.
- K. Schwamborn and R. M. Caprioli, Molecular imaging by mass spectrometry—looking beyond classical histology, Nat. Rev. Cancer, 2010, 10(9), 639–646 CrossRef CAS PubMed.
- L. A. McDonnell and R. M. Heeren, Imaging mass spectrometry, Mass Spectrom. Rev., 2007, 26(4), 606–643 CrossRef CAS PubMed.
- W. Michno, P. M. Wehrli, K. Blennow, H. Zetterberg and J. Hanrieder, Molecular imaging mass spectrometry for probing protein dynamics in neurodegenerative disease pathology, J. Neurochem., 2019, 151(4), 488–506 CrossRef CAS PubMed.
- C. Wu, A. L. Dill, L. S. Eberlin, R. G. Cooks and D. R. Ifa, Mass spectrometry imaging under ambient conditions, Mass Spectrom. Rev., 2013, 32(3), 218–243 CrossRef CAS PubMed.
- R. G. Cooks, Z. Ouyang, Z. Takats and J. M. Wiseman, Ambient mass spectrometry, Science, 2006, 311(5767), 1566–1570 CrossRef CAS PubMed.
- D. J. Weston, Ambient ionization mass spectrometry: current understanding of mechanistic theory; analytical performance and application areas, Analyst, 2010, 135(4), 661–668 RSC.
- (a) G. A. Harris, A. S. Galhena and F. M. Fernandez, Ambient sampling/ionization mass spectrometry: applications and current trends, Anal. Chem., 2011, 83(12), 4508–4538 CrossRef CAS PubMed; (b) M. E. Monge, G. A. Harris, P. Dwivedi and F. M. Fernandez, Mass spectrometry: recent advances in direct open air surface sampling/ionization, Chem. Rev., 2013, 113(4), 2269–2308 CrossRef CAS PubMed.
- C. Wu, A. L. Dill, L. S. Eberlin, R. G. Cooks and D. R. Ifa, Mass spectrometry imaging under ambient conditions, Mass Spectrom. Rev., 2013, 32(3), 218–243 CrossRef CAS PubMed.
- L. Davis, Basic Methods in Molecular Biology, Elsevier, 2012 Search PubMed.
- G. J. Van Berkel, S. P. Pasilis and O. Ovchinnikova, Established and emerging atmospheric pressure surface sampling/ionization techniques for mass spectrometry, J. Mass Spectrom., 2008, 43(9), 1161–1180 CrossRef CAS PubMed.
- D. R. Ifa, C. Wu, Z. Ouyang and R. G. Cooks, Desorption electrospray ionization and other ambient ionization methods: current progress and preview, Analyst, 2010, 135(4), 669–681 RSC.
- A. K. Badu-Tawiah, L. S. Eberlin, Z. Ouyang and R. G. Cooks, Chemical aspects of the extractive methods of ambient ionization mass spectrometry, Annu. Rev. Phys. Chem., 2013, 64, 481–505 CrossRef CAS PubMed.
- A. R. Venter, K. A. Douglass, J. T. Shelley, G. Hasman Jr and E. Honarvar, Mechanisms of real-time, proximal sample processing during ambient ionization mass spectrometry, Anal. Chem., 2014, 86(1), 233–249 CrossRef CAS PubMed.
- R. D. Espy, M. Wleklinski, X. Yan and R. G. Cooks, Beyond the flask: reactions on the fly in ambient mass spectrometry, TrAC, Trends Anal. Chem., 2014, 57, 135–146 CrossRef CAS.
- C. C. Hsu and P. C. Dorrestein, Visualizing life with ambient mass spectrometry, Curr. Opin. Biotechnol., 2015, 31, 24–34 CrossRef CAS PubMed.
- E. De Hoffmann and V. Stroobant, Mass Spectrometry: Principles and Applications, John Wiley & Sons, 2007 Search PubMed.
- H. Oberacher, On the use of different mass spectrometric techniques for characterization of sequence variability in genomic DNA, Anal. Bioanal. Chem., 2008, 391, 135–149 CrossRef CAS PubMed.
- I. A. Kaltashov, M. Zhang, S. J. Eyles and R. R. Abzalimov, Investigation of structure, dynamics and function of metalloproteins with electrospray ionization mass spectrometry, Anal. Bioanal. Chem., 2006, 386, 472–481 CrossRef CAS PubMed.
- M. W. Nielen, MALDI time-of-flight mass spectrometry of synthetic polymers, Mass Spectrom. Rev., 1999, 18(5), 309–344 CrossRef CAS.
- J. B. Fenn, M. Mann, C. K. Meng, S. F. Wong and C. M. Whitehouse, Electrospray ionization for mass spectrometry of large biomolecules, Science, 1989, 246(4926), 64–71 CrossRef CAS PubMed.
- F. Hillenkamp, M. Karas, R. C. Beavis and B. T. Chait, Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers, Anal. Chem., 1991, 63(24), 1193A–203A CrossRef CAS PubMed.
- Electrospray and MALDI Mass Spectrometry: Fundamentals, Instrumentation, Practicalities, and Biological Applications, ed. R. B. Cole, 2011 Search PubMed.
- D. Kuck, Reactive Intermediates: MS Investigations in Solution, ed. L. S. Santos, 2010 Search PubMed.
- R. M. Alberici, R. C. Simas, G. B. Sanvido, W. Romão, P. M. Lalli, M. Benassi, I. B. Cunha and M. N. Eberlin, Ambient mass spectrometry: bringing MS into the “real world”, Anal. Bioanal. Chem., 2010, 398, 265–294 CrossRef CAS PubMed.
- D. J. Weston, Ambient ionization mass spectrometry: current understanding of mechanistic theory; analytical performance and application areas, Analyst, 2010, 135(4), 661–668 RSC.
- S. Rankin-Turner, J. C. Reynolds, M. A. Turner and L. M. Heaney, Applications of ambient ionization mass spectrometry in 2021: An annual review, Anal. Sci. Adv., 2022, 3(3–4), 67–89 CrossRef CAS.
- N. Li, H. Nie, L. Jiang, G. Ruan, F. Du and H. Liu, Recent advances of ambient ionization mass spectrometry imaging in clinical research, J. Sep. Sci., 2020, 43(15), 3146–3163 CrossRef CAS PubMed.
- A. Albert, J. T. Shelley and C. Engelhard, Plasma-based ambient desorption/ionization mass spectrometry: state-of-the-art in qualitative and quantitative analysis, Anal. Bioanal. Chem., 2014, 406, 6111–6127 CrossRef CAS PubMed.
- P. Nemes and A. Vertes, Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry, Anal. Chem., 2007, 79(21), 8098–8106 CrossRef CAS PubMed.
- T. S. Hamid, D. Lostun, E. C. Cabral, R. Garrett, D. K. Bohme and D. R. Ifa, Comparisons of ambient spray ionization imaging methods, Int. J. Mass Spectrom., 2015, 377, 736–743 CrossRef CAS.
- Z. Takats, J. M. Wiseman and R. G. Cooks, Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology, J. Mass Spectrom., 2005, 40(10), 1261–1275 CrossRef CAS PubMed.
- D. R. Ifa, C. Wu, Z. Ouyang and R. G. Cooks, Desorption electrospray ionization and other ambient ionization methods: current progress and preview, Analyst, 2010, 135(4), 669–681 RSC.
- J. M. Wiseman, D. R. Ifa, Q. Song and R. G. Cooks, Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry, Angew. Chem., Int. Ed., 2006, 45, 7188–7192 CrossRef CAS PubMed.
- S. Santagata, L. S. Eberlin, I. Norton, D. Calligaris, D. R. Feldman, J. L. Ide, X. Liu, J. S. Wiley, M. L. Vestal, S. H. Ramkissoon and D. A. Orringer, Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 11121–11126 CrossRef CAS PubMed.
- S. Banerjee and S. Mazumdar, Electrospray ionization mass spectrometry: a technique to access the information beyond the molecular weight of the analyte, Int. J. Anal. Chem., 2012, 2012, 282574 Search PubMed.
- J. B. Fenn, M. Mann, C. K. Meng, S. F. Wong and C. M. Whitehouse, Electrospray ionization for mass spectrometry of large biomolecules, Science, 1989, 246, 64–71 CrossRef CAS PubMed.
- L. S. Eberlin, C. R. Ferreira, A. L. Dill, D. R. Ifa and R. G. Cooks, Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2011, 1811, 946–960 CrossRef CAS PubMed.
- E. Yamamoto, Y. Taquahashi, M. Kuwagata, H. Saito, K. Matsushita, T. Toyoda, F. Sato, S. Kitajima, K. Ogawa, K. I. Izutsu and Y. Saito, Visualizing the spatial localization of ciclesonide and its metabolites in rat lungs after inhalation of 1-μm aerosol of ciclesonide by desorption electrospray ionization-time of flight mass spectrometry imaging, Int. J. Pharm., 2021, 595, 120241 CrossRef CAS PubMed.
- R. J. Goodwin, A. Nilsson, C. L. Mackay, J. G. Swales, M. K. Johansson, M. Billger, P. E. Andrén and S. L. Iverson, Exemplifying the screening power of mass spectrometry imaging over label-based technologies for simultaneous monitoring of drug and metabolite distributions in tissue sections, J. Biomol. Screening, 2016, 21(2), 187–193 CrossRef CAS PubMed.
- N. A. Reynolds and L. J. Scott, Ciclesonide, Drugs, 2004, 64, 511–519 CrossRef CAS PubMed.
- R. Nave, H. Watz, H. Hoffmann, H. Boss and H. Magnussen, Deposition and metabolism of inhaled ciclesonide in the human lung, Eur. Respir. J., 2010, 36(5), 1113–1119 CrossRef CAS PubMed.
- T. C. Carvalho, J. I. Peters and R. O. Williams III, Influence of particle size on regional lung deposition–what evidence is there?, Int. J. Pharm., 2011, 406(1–2), 1–10 CrossRef CAS PubMed.
- M. Biddiscombe, H. Kalsi, S. Meah, A. Allen and O. Usmani, The fate of inhaled drug: corticosteroid particle size effects in asthma.
- N. R. Labiris and M. B. Dolovich, Pulmonary drug delivery. Part I: physiological factors affecting therapeutic effectiveness of aerosolized medications, Br. J. Clin. Pharmacol., 2003, 56(6), 588–599 CrossRef CAS PubMed.
- E. Van Der Wiel, N. H. Ten Hacken, D. S. Postma and M. Van Den Berge, Small-airways dysfunction associates with respiratory symptoms and clinical features of asthma: a systematic review, J. Allergy Clin. Immunol., 2013, 131(3), 646–657 CrossRef PubMed.
- P. Iruzubieta, M. T. Arias-Loste, L. Barbier-Torres, M. L. Martinez-Chantar and J. Crespo, The need for biomarkers in diagnosis and prognosis of drug-induced liver disease: does metabolomics have any role?, BioMed Res. Int., 2015, 2015, 386186 Search PubMed.
- E. S. Björnsson, Clinical management of patients with drug-induced liver injury (DILI), United Eur. Gastroenterol. J., 2021, 9(7), 781–786 CrossRef PubMed.
- J. Y. Akakpo, M. W. Jaeschke, Y. Etemadi, A. Artigues, S. Toerber, H. Olivos, B. Shrestha, A. Midey, H. Jaeschke and A. Ramachandran, Desorption Electrospray Ionization Mass Spectrometry Imaging Allows Spatial Localization of Changes in Acetaminophen Metabolism in the Liver after Intervention with 4-Methylpyrazole, J. Am. Soc. Mass Spectrom., 2022, 33(11), 2094–2107 CrossRef CAS PubMed.
- M. R. McGill and H. Jaeschke, Metabolism and disposition of acetaminophen: recent advances in relation to hepatotoxicity and diagnosis, J. Pharm. Res., 2013, 30, 2174–2187 CrossRef CAS PubMed.
- J. Y. Akakpo, A. Ramachandran, L. Duan, M. A. Schaich, M. W. Jaeschke, B. D. Freudenthal, W. X. Ding, B. H. Rumack and H. Jaeschke, Delayed treatment with 4-methylpyrazole protects against acetaminophen hepatotoxicity in mice by inhibition of c-Jun N-terminal kinase, Toxicol. Sci., 2019, 170(1), 57–68 CrossRef CAS.
- J. Y. Akakpo, A. Ramachandran, S. E. Kandel, H. M. Ni, S. C. Kumer, B. H. Rumack and H. Jaeschke, 4-Methylpyrazole protects against acetaminophen hepatotoxicity in mice and in primary human hepatocytes, Hum. Exp. Toxicol., 2018, 37(12), 1310–1322 CrossRef CAS PubMed.
- A. McEwen and C. Henson, Quantitative whole-body autoradiography: past, present and future, Bioanalysis, 2015, 7(5), 557–568 CrossRef CAS PubMed.
- E. G. Solon, Use of radioactive compounds and autoradiography to determine drug tissue distribution, Chem. Res. Toxicol., 2012, 25(3), 543–555 Search PubMed.
- B. Prideaux and M. Stoeckli, Mass spectrometry imaging for drug distribution studies, J. Proteomics, 2012, 75(16), 4999–5013 CrossRef CAS PubMed.
- O. Karlsson and J. Hanrieder, Imaging mass spectrometry in drug development and toxicology, Arch. Toxicol., 2017, 91, 2283–2294 CrossRef CAS PubMed.
- Y. N. Ho, L. J. Shu and Y. L. Yang, Imaging mass spectrometry for metabolites: technical progress, multimodal imaging, and biological interactions, Wiley Interdiscip. Rev.: Syst. Biol. Med., 2017, 9(5), e1387 Search PubMed.
- B. Balluff and L. A. McDonnell, Mass spectrometry imaging of metabolites, Metabolomics, 2018, 345–357 CAS.
- A. L. Bruinen, C. van Oevelen, G. B. Eijkel, M. Van Heerden, F. Cuyckens and R. M. Heeren, Mass spectrometry imaging of drug related crystal-like structures in formalin-fixed frozen and paraffin-embedded rabbit kidney tissue sections, J. Am. Soc. Mass Spectrom., 2015, 27(1), 117–123 CrossRef PubMed.
- A. Buck, A. Ly, B. Balluff, N. Sun, K. Gorzolka, A. Feuchtinger, K. P. Janssen, P. J. Kuppen, C. J. van de Velde, G. Weirich and F. Erlmeier, High-resolution MALDI-FT-ICR MS imaging for the analysis of metabolites from formalin-fixed, paraffin-embedded clinical tissue samples, J. Pathol., 2015, 237(1), 123–132 CrossRef CAS PubMed.
- D. Bonnel, R. Longuespee, J. Franck, M. Roudbaraki, P. Gosset, R. Day, M. Salzet and I. Fournier, Multivariate analyses for biomarkers hunting and validation through on-tissue bottom-up or in-source decay in MALDI-MSI: application to prostate cancer, Anal. Bioanal. Chem., 2011, 401, 149–165 CrossRef CAS PubMed.
- M. C. Djidja, E. Claude, M. F. Snel, S. Francese, P. Scriven, V. Carolan and M. R. Clench, Novel molecular tumour classification using MALDI–mass spectrometry imaging of tissue micro-array, Anal. Bioanal. Chem., 2010, 397, 587–601 CrossRef CAS PubMed.
- A. V. Everest-Dass, M. T. Briggs, G. Kaur, M. K. Oehler, P. Hoffmann and N. H. Packer, N-glycan MALDI imaging mass spectrometry on formalin-fixed paraffin-embedded tissue enables the delineation of ovarian cancer tissues, Mol. Cell. Proteomics, 2016, 15(9), 3003–3016 CrossRef CAS PubMed.
- S. Cacciatore, G. Zadra, C. Bango, K. L. Penney, S. Tyekucheva, O. Yanes and M. Loda, Metabolic profiling in formalin-fixed and paraffin-embedded prostate cancer tissues, Mol. Cancer Res., 2017, 15(4), 439–447 CrossRef CAS PubMed.
- V. Vincek, M. Nassiri, M. Nadji and A. R. Morales, A tissue fixative that protects macromolecules (DNA, RNA, and protein) and histomorphology in clinical samples, Lab. Invest., 2003, 83(10), 1427–1435 CrossRef CAS PubMed.
- C. Williams, F. Pontén, C. Moberg, P. Söderkvist, M. Uhlén, J. Pontén, G. Sitbon and J. Lundeberg, A high frequency of sequence alterations is due to formalin fixation of archival specimens, Am. J. Pathol., 1999, 155(5), 1467–1471 CrossRef CAS PubMed.
- M. Gaudin, M. Panchal, S. Ayciriex, E. Werner, A. Brunelle, D. Touboul, C. Boursier-Neyret, N. Auzeil, B. Walther, C. Duyckaerts and O. Laprévote, Ultra performance liquid chromatography–mass spectrometry studies of formalin-induced alterations of human brain lipidome, J. Mass Spectrom., 2014, 49(10), 1035–1042 CrossRef CAS PubMed.
- D. N. Vos, A. P. Bowman, R. M. Heeren, B. Balluff and S. R. Ellis, Class-specific depletion of lipid ion signals in tissues upon formalin fixation, Int. J. Mass Spectrom., 2019, 446, 116212 CrossRef CAS.
- G. Liguri, P. Nassi, N. Taddei, C. Nediani and G. Ramponi, Post-mortem modifications of the specific activity of some brain enzymes, Neurosci. Lett., 1988, 85(2), 244–248 CrossRef CAS PubMed.
- S. Fahn and L. J. Cote, Stability of Enzymes in Post-Mortem Rat Brain 1, J. Neurochem., 1976, 26(5), 1039–1042 CrossRef CAS PubMed.
- T. Ritchie, S. A. Scully, J. de Vellis and E. P. Noble, Stability of neuronal and glial marker enzymes in post-mortem rat brain, Neurochem. Res., 1986, 11, 383–392 CrossRef CAS PubMed.
- W. F. Gattaz, A. Maras, N. J. Cairns, R. Levy and H. Förstl, Decreased phospholipase A2 activity in Alzheimer brains, Biol. Psychiatry, 1995, 37, 13–17 CrossRef CAS PubMed.
- A. Dannhorn, J. G. Swales, G. Hamm, N. Strittmatter, H. Kudo, G. Maglennon, R. J. Goodwin and Z. Takats, Evaluation of formalin-fixed and FFPE tissues for spatially resolved metabolomics and drug distribution studies, Pharmaceuticals, 2022, 15(11), 1307 CrossRef CAS PubMed.
- X. Tan, Q. He, Z. Pei, Y. Liu, Z. Feng, C. Li, C. Tang and Y. Zhang, Rapid visual characterization of alkaloid changes in traditional processing of Tibetan medicine Aconitum pendulum by high-performance thin-layer chromatography coupled with desorption electrospray ionization mass spectrometry imaging, Front. Pharmacol, 2023, 14, 1104473 CrossRef CAS PubMed.
- D. Csupor, M. Strömberg, L. Bohlin, K. Woelkart, E. M. Wenzig, J. Hohmann and R. Bauer, The effect of the extraction and processing methods on the alkaloid composition of roots of aconitum carmichaeli, Planta Med., 2007, 73(09), P_391 Search PubMed.
- M. R. Zhi, X. R. Gu, S. Han, K. Y. Liu, Z. Q. Liu, Y. N. Tang, X. T. Han, F. Li, Z. G. Yang, P. Tan and H. Y. Zhao, Chemical variation in Aconti Kusnezoffii Radix before and after processing based on UPLC-Orbitrap-MS, China J. Chin. Mater. Med., 2020, 45(5), 1082–1089 Search PubMed.
- Z. Luo, J. He, Y. Chen, J. He, T. Gong, F. Tang, X. Wang, R. Zhang, L. Huang, L. Zhang and H. Lv, Air flow-assisted ionization imaging mass spectrometry method for easy whole-body molecular imaging under ambient conditions, Anal. Chem., 2013, 85(5), 2977–2982 CrossRef CAS PubMed.
- K. Qi, L. Wu, C. Liu and Y. Pan, Recent advances of ambient mass spectrometry imaging and its applications in lipid and metabolite analysis, Metabolites, 2021, 11(11), 780 CrossRef CAS PubMed.
- C. Sun, T. Li, X. Song, L. Huang, Q. Zang, J. Xu, N. Bi, G. Jiao, Y. Hao, Y. Chen and R. Zhang, Spatially resolved metabolomics to discover tumor-associated metabolic alterations, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(1), 52–57 CrossRef CAS PubMed.
- C. Sun, T. Li, X. Song, L. Huang, Q. Zang, J. Xu, N. Bi, G. Jiao, Y. Hao, Y. Chen and R. Zhang, Spatially resolved metabolomics to discover tumor-associated metabolic alterations, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(1), 52–57 CrossRef CAS PubMed.
- M. Vanneman and G. Dranoff, Combining immunotherapy and targeted therapies in cancer treatment, Nat. Rev. Cancer, 2012, 12, 237–251 CrossRef CAS PubMed.
- Q. Liang, L. Kong, Y. Du, X. Zhu and J. Tian, Antitumorigenic and antiangiogenic efficacy of apatinib in liver cancer evaluated by multimodality molecular imaging, Exp. Mol. Med., 2019, 51, 76 Search PubMed.
- E. C. Randall, K. B. Emdal, J. K. Laramy, M. Kim, A. Roos and D. Calligaris, et al., Integrated mapping of pharmacokinetics and pharmacodynamics in a patient-derived xenograft model of glioblastoma, Nat. Commun., 2018, 9, 4904 CrossRef PubMed.
- D. Chen, X. Lin, C. Zhang, Z. Liu, Z. Chen and Z. Li, et al., Dual PI3K/mTOR inhibitor BEZ235 as a promising therapeutic strategy against paclitaxel-resistant gastric cancer via targeting PI3K/Akt/mTOR pathway, Cell Death Dis., 2018, 9, 123 CrossRef PubMed.
- A. Nilsson, R. J. A. Goodwin, M. Shariatgorji, T. Vallianatou, P. J. H. Webborn and P. E. Andrén, Mass Spectrometry Imaging in Drug Development, Anal. Chem., 2015, 87, 1437–1455 CrossRef CAS PubMed.
- C. Sun, T. Li, X. Song, L. Huang, Q. Zang and J. Xu, et al., Spatially resolved metabolomics to discover tumor-associated metabolic alterations, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 52–57 CrossRef CAS PubMed.
- J. Zhang, Q. Du, X. Song, S. Gao, X. Pang, Y. Li, R. Zhang, Z. Abliz and J. He, Evaluation of the tumor-targeting efficiency and intratumor heterogeneity of anticancer drugs using quantitative mass spectrometry imaging, Theranostics, 2020, 10(6), 2621 CrossRef CAS PubMed.
- H. J. Park and K. Friston, Structural and functional brain networks: from connections to cognition, Science, 2013, 342(6158), 1238411 CrossRef PubMed.
- R. Zhao and G. M. Pollack, Regional differences in capillary density, perfusion rate, and P-glycoprotein activity: a quantitative analysis of regional drug exposure in the brain, Biochem. Pharmacol., 2009, 78(8), 1052–1059 CrossRef CAS PubMed.
- X. Pang, S. Gao, M. Ga, J. Zhang, Z. Luo, Y. Chen, R. Zhang, J. He and Z. Abliz, Mapping metabolic networks in the brain by ambient mass spectrometry imaging and metabolomics, Anal. Chem., 2021, 93(17), 6746–6754 CrossRef CAS PubMed.
- D. Liu, J. Huang, S. Gao, H. Jin and J. He, A temporo-spatial pharmacometabolomics method to characterize pharmacokinetics and pharmacodynamics in the brain microregions by using ambient mass spectrometry imaging, Acta Pharm. Sin. B, 2022, 12(8), 3341–3353 CrossRef CAS PubMed.
- B. C. Brahimaj, R. B. Kochanski, J. J. Pearce, M. Guryildirim, C. S. Gerard, M. Kocak, S. Sani and R. W. Byrne, Structural and functional imaging in glioma management, Neurosurgery, 2021, 88(2), 211–221 CrossRef PubMed.
- M. Ganesana, S. T. Lee, Y. Wang and B. J. Venton, Analytical techniques in neuroscience: recent advances in imaging, separation, and electrochemical methods, Anal. Chem., 2017, 89(1), 314–341 CrossRef CAS PubMed.
- G. Theodoridis, H. G. Gika and I. D. Wilson, LC–MS-based methodology for global metabolite profiling in metabonomics/metabolomics, TrAC, Trends Anal. Chem., 2008, 27, 251e60 CrossRef.
- H. G. Gika, G. A. Theodoridis, R. S. Plumb and I. D. Wilson, Current practice of liquid chromatography–mass spectrometry in metabolomics and metabonomics, J. Pharm. Biomed. Anal., 2014, 87, 12e25 CrossRef PubMed.
- A. S. Serras, J. S. Rodrigues, M. Cipriano, A. V. Rodrigues, N. G. Oliveira and J. P. Miranda, A critical perspective on 3D liver models for drug metabolism and toxicology studies, Front. Cell Dev. Biol., 2021, 9, 626805 CrossRef PubMed.
- V. M. Lauschke, D. F. Hendriks, C. C. Bell, T. B. Andersson and M. Ingelman-Sundberg, Novel 3D culture systems for studies of human liver function and assessments of the hepatotoxicity of drugs and drug candidates, Chem. Res. Toxicol., 2016, 29(12), 1936–1955 Search PubMed.
- D. Zhang, G. Luo, X. Ding and C. Lu, Preclinical experimental models of drug metabolism and disposition in drug discovery and development, Acta Pharm. Sin. B, 2012, 2(6), 549–561 Search PubMed.
- M. J. Gómez-Lechón, L. Tolosa, I. Conde and M. T. Donato, Competency of different cell models to predict human hepatotoxic drugs, Expert Opin. Drug Metab. Toxicol., 2014, 10(11), 1553–1568 Search PubMed.
- R. Nudischer, K. Renggli, A. Hierlemann, A. B. Roth and C. Bertinetti-Lapatki, Characterization of a long-term mouse primary liver 3D tissue model recapitulating innate-immune responses and drug-induced liver toxicity, PLoS One, 2020, 15(7), e0235745 CrossRef CAS PubMed.
- M. J. Gomez-Lechon, A. Lahoz, L. Gombau, J. V. Castell and M. T. Donato, In vitro evaluation of potential hepatotoxicity induced by drugs, Curr. Pharm. Des., 2010, 16(17), 1963–1977 CrossRef CAS PubMed.
- L. Pan, P. Han, S. Ma, R. Peng, C. Wang, W. Kong, L. Cong, J. Fu, Z. Zhang, H. Yu and Y. Wang, Abnormal metabolism of gut microbiota reveals the possible molecular mechanism of nephropathy induced by hyperuricemia, Acta Pharm. Sin. B, 2020, 10(2), 249–261 CrossRef CAS PubMed.
- A. M. Araújo, M. Carvalho, F. Carvalho, M. D. Bastos and P. Guedes de Pinho, Metabolomic approaches in the discovery of potential urinary biomarkers of drug-induced liver injury (DILI), Crit. Rev. Toxicol., 2017, 47(8), 638–654 CrossRef PubMed.
- L. Goracci, A. Valeri, S. Sciabola, M. D. Aleo, W. Moritz, J. Lichtenberg and G. Cruciani, A novel lipidomics-based approach to evaluating the risk of clinical hepatotoxicity potential of drugs in 3D human microtissues, Chem. Res. Toxicol., 2019, 33(1), 258–270 Search PubMed.
- X. Liu and A. B. Hummon, Mass spectrometry imaging of therapeutics from animal models to three-dimensional cell cultures, Anal. Chem., 2015, 87(19), 9508–9519 CrossRef CAS PubMed.
- L. E. Flint, G. Hamm, J. D. Ready, S. Ling, C. J. Duckett, N. A. Cross, L. M. Cole, D. P. Smith, R. J. Goodwin and M. R. Clench, Characterization of an aggregated three-dimensional cell culture model by multimodal mass spectrometry imaging, Anal. Chem., 2020, 92(18), 12538–12547 CrossRef CAS PubMed.
- P. Xie, H. Zhang, P. Wu, Y. Chen and Z. Cai, Three-dimensional mass spectrometry imaging reveals distributions of lipids and the drug metabolite associated with the enhanced growth of colon cancer cell spheroids treated with triclosan, Anal. Chem., 2022, 94(40), 13667–13675 CrossRef CAS PubMed.
- T. Greer, R. Sturm and L. Li, Mass spectrometry imaging for drugs and metabolites, J. Proteomics, 2011, 74(12), 2617–2631 CrossRef CAS PubMed.
- A. Nilsson, R. J. Goodwin, M. Shariatgorji, T. Vallianatou, P. J. Webborn and P. E. Andrén, Mass spectrometry imaging in drug development, Anal. Chem., 2015, 87(3), 1437–1455 CrossRef CAS PubMed.
- Z. Wang, W. Fu, M. Huo, B. He, Y. Liu, L. Tian, W. Li, Z. Zhou, B. Wang, J. Xia and Y. Chen, Spatial-resolved metabolomics reveals tissue-specific metabolic reprogramming in diabetic nephropathy by using mass spectrometry imaging, Acta Pharm. Sin. B, 2021, 11(11), 3665–3677 CrossRef CAS PubMed.
- M. Babatin, S. S. Lee and P. T. Pollak, Amiodarone hepatotoxicity, Curr. Vasc. Pharmacol., 2008, 6(3), 228–236 CrossRef CAS PubMed.
- S. Endo, Y. Toyoda, T. Fukami, M. Nakajima and T. Yokoi, Stimulation of human monocytic THP-1 cells by metabolic activation of hepatotoxic drugs, Drug Metab. Pharmacokinet., 2012, 27(6), 621–630 CrossRef CAS PubMed.
- K. M. Waldhauser, M. Török, H. R. Ha, U. Thomet, D. Konrad, K. Brecht, F. Follath and S. Krähenbühl, Hepatocellular toxicity and pharmacological effect of amiodarone and amiodarone derivatives, J. Pharmacol. Exp. Ther., 2006, 319(3), 1413–1423 CrossRef CAS PubMed.
- L. Li, Q. Zang, X. Li, Y. Zhu, S. Wen, J. He, R. Zhang and Z. Abliz, Spatiotemporal pharmacometabolomics based on ambient mass spectrometry imaging to evaluate the metabolism and hepatotoxicity of amiodarone in HepG2 spheroids, J. Pharm. Anal., 2023, 13(5), 483–493 CrossRef PubMed.
- P. J. Roach, J. Laskin and A. Laskin, Nanospray desorption electrospray ionization: an ambient method for liquid-extraction surface sampling in mass spectrometry, Analyst, 2010, 135(9), 2233–2236 RSC.
- I. Lanekoff, B. S. Heath, A. Liyu, M. Thomas, J. P. Carson and J. Laskin, Automated platform for high-resolution tissue imaging using nanospray desorption electrospray ionization mass spectrometry, Anal. Chem., 2012, 84(19), 8351–8356 CrossRef CAS PubMed.
- J. Laskin, B. S. Heath, P. J. Roach, L. Cazares and O. J. Semmes, Tissue imaging using nanospray desorption electrospray ionization mass spectrometry, Anal. Chem., 2012, 84(1), 141–148 CrossRef CAS PubMed.
- P. J. Roach, J. Laskin and A. Laskin, Molecular characterization of organic aerosols using nanospray-desorption/electrospray ionization-mass spectrometry, Anal. Chem., 2010, 82(19), 7979–7986 CrossRef CAS PubMed.
- H. Stierlin, J. W. Faigle, A. Sallmann, W. Kung, W. J. Richter, H. P. Kriemler, K. O. Alt and T. Winkler, Biotransformation of diclofenac sodium (Voltaren®) in animals and in man: I. Isolation and identification of principal metabolites, Xenobiotica, 1979, 9(10), 601–610 CrossRef CAS PubMed.
- J. R. Kenny, J. L. Maggs, X. Meng, D. Sinnott, S. E. Clarke, B. K. Park and A. V. Stachulski, Syntheses and characterization of the acyl glucuronide and hydroxy metabolites of diclofenac, J. Med. Chem., 2004, 47(11), 2816–2825 CrossRef CAS PubMed.
- A. K. Daly, G. P. Aithal, J. B. Leathart, R. A. Swainsbury, T. S. Dang and C. P. Day, Genetic susceptibility to diclofenac-induced hepatotoxicity: contribution of UGT2B7, CYP2C8, and ABCC2 genotypes, Gastroenterology, 2007, 132(1), 272–281 CrossRef CAS PubMed.
- R. W. Sparidans, J. S. Lagas, A. H. Schinkel, J. H. Schellens and J. H. Beijnen, Liquid chromatography–tandem mass spectrometric assay for diclofenac and three primary metabolites in mouse plasma, J. Chromatogr. B, 2008, 872(1–2), 77–82 CrossRef CAS PubMed.
- S. Shen, M. R. Marchick, M. R. Davis, G. A. Doss and L. R. Pohl, Metabolic activation of diclofenac by human cytochrome P450 3A4: role of 5-hydroxydiclofenac, Chem. Res. Toxicol., 1999, 12(2), 214–222 Search PubMed.
- M. Whirl-Carrillo, E. M. McDonagh, J. M. Hebert, L. Gong, K. Sangkuhl, C. F. Thorn, R. B. Altman and T. E. Klein, Pharmacogenomics knowledge for personalized medicine, Clin. Pharmacol. Ther., 2012, 92(4), 414–417 CrossRef CAS PubMed.
- R. Bort, K. Macé, A. Boobis, M. J. Gómez-Lechón, A. Pfeifer and J. Castell, Hepatic metabolism of diclofenac: role of human CYP in the minor oxidative pathways, Biochem. Pharmacol., 1999, 58(5), 787–796 CrossRef CAS PubMed.
- U. A. Boelsterli, Diclofenac-induced liver injury: a paradigm of idiosyncratic drug toxicity, Toxicol. Appl. Pharmacol., 2003, 192(3), 307–322 CrossRef CAS PubMed.
- S. L. Regan, J. L. Maggs, T. G. Hammond, C. Lambert, D. P. Williams and B. K. Park, Acyl glucuronides: the good, the bad and the ugly, Biopharm. Drug Dispos., 2010, 31(7), 367–395 CrossRef CAS PubMed.
- A. D. Vaz, W. W. Wang, A. J. Bessire, R. Sharma and A. E. Hagen, A rapid and specific derivatization procedure to identify acyl-glucuronides by mass spectrometry, Rapid Commun. Mass Spectrom., 2010, 24(14), 2109–2121 CrossRef CAS PubMed.
- M. A. Alam, F. I. Al-Jenoobi and A. M. Al-Mohizea, High-Throughput Ultra-Performance LC–MS-MS Method for Analysis of Diclofenac Sodium in Rabbit Plasma, J. Chromatogr. Sci., 2015, 53(1), 47–53 CAS.
- Y. Zhang, Y. H. Han, S. P. Putluru, M. K. Matta, P. Kole, S. Mandlekar, M. T. Furlong, T. Liu, R. A. Iyer, P. Marathe and Z. Yang, Diclofenac and its acyl glucuronide: determination of in vivo exposure in human subjects and characterization as human drug transporter substrates in vitro, Drug Metab. Dispos., 2016, 44(3), 320–328 CrossRef CAS PubMed.
- D. M. Sanchez, H. M. Brown, R. Yin, B. Chen, M. Vavrek, M. T. Cancilla, W. Zhong, B. Shyong, N. R. Zhang, F. Li and J. Laskin, Mass spectrometry imaging of diclofenac and its metabolites in tissues using nanospray desorption electrospray ionization, Anal. Chim. Acta, 2022, 1233, 340490 CrossRef PubMed.
- P. Nemes and A. Vertes, Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry, Anal. Chem., 2007, 79(21), 8098–8106 CrossRef CAS PubMed.
- V. V. Laiko, M. A. Baldwin and A. L. Burlingame, Atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry, Anal. Chem., 2000, 72(4), 652–657 CrossRef CAS PubMed.
- S. Rankin-Turner and L. M. Heaney, Applications of ambient ionization mass spectrometry in 2020: An annual review, Anal. Sci. Adv., 2021, 2(3–4), 193–212 CrossRef.
- World Health Organization, Global tuberculosis report 2020: executive summary, in, Global Tuberculosis Report 2020: Executive Summary 2020 Search PubMed.
- C. R. Horsburgh Jr, C. E. Barry III and C. Lange, Treatment of tuberculosis, N. Engl. J. Med., 2015, 373(22), 2149–2160 CrossRef PubMed.
- V. Dartois, The path of anti-tuberculosis drugs: from blood to lesions to mycobacterial cells, Nat. Rev. Microbiol., 2014, 12(3), 159–167 CrossRef CAS PubMed.
- J. Kokesch-Himmelreich, A. Treu, A. M. Race, K. Walter, C. Hölscher and A. Römpp, Do anti-tuberculosis drugs reach their target?— high-resolution matrix-assisted laser desorption/ionization mass spectrometry imaging provides information on drug penetration into necrotic granulomas, Anal. Chem., 2022, 94(14), 5483–5492 CrossRef CAS PubMed.
- R. Fayez and V. Gupta, Imipramine, StatPearls Publishing, Treasure Island, FL, USA, 2021 Search PubMed.
- A. Cortegiani, G. Ingoglia, M. Ippolito, A. Giarratano and S. Einav, A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19, J. Crit. Care, 2020, 57, 279–283 CrossRef CAS PubMed.
- C. Doyno, D. M. Sobieraj and W. L. Baker, Toxicity of chloroquine and hydroxychloroquine following therapeutic use or overdose, Clin. Toxicol., 2021, 59(1), 12–23 CrossRef CAS PubMed.
- E. Obuchowicz, A. Bielecka-Wajdman, M. Zieliński, G. Machnik, M. Gołyszny and T. Ludyga, Imipramine and venlafaxine differentially affect primary glial cultures of prenatally stressed rats, Front. Pharmacol, 2020, 10, 1687 CrossRef PubMed.
- A. Islam, T. Sakamoto, Q. Zhai, M. M. Rahman, M. A. Mamun, Y. Takahashi, T. Kahyo and M. Setou, Application of AP-MALDI imaging mass microscope for the rapid mapping of imipramine, chloroquine, and their metabolites in the kidney and brain of wild-type mice, Pharmaceuticals, 2022, 15(11), 1314 CrossRef CAS PubMed.
- A. Buck, M. Aichler, K. Huber and A. Walch, In situ metabolomics in cancer by mass spectrometry imaging, Adv. Cancer Res., 2017, 134, 117–132 CrossRef CAS PubMed.
- K. Schwamborn and R. M. Caprioli, Molecular imaging by mass spectrometry—looking beyond classical histology, Nat. Rev. Cancer, 2010, 10(9), 639–646 CrossRef CAS PubMed.
- T. Li, J. He, X. Mao, Y. Bi, Z. Luo, C. Guo, F. Tang, X. Xu, X. Wang, M. Wang and J. Chen, In situ biomarker discovery and label-free molecular histopathological diagnosis of lung cancer by ambient mass spectrometry imaging, Sci. Rep., 2015, 5(1), 14089 CrossRef CAS PubMed.
- M. Tuck, F. Grélard, L. Blanc and N. Desbenoit, MALDI-MSI towards multimodal imaging: challenges and perspectives, Front. Chem., 2022, 10, 904688 CrossRef CAS PubMed.
- M. Kompauer, S. Heiles and B. Spengler, Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution, Nat. Methods, 2017, 14(1), 90–96 CrossRef CAS PubMed.
- K. Ščupáková, B. Balluff, C. Tressler, T. Adelaja, R. M. Heeren, K. Glunde and G. Ertaylan, Cellular resolution in clinical MALDI mass spectrometry imaging: the latest advancements and current challenges, Clin. Chem. Lab. Med., 2020, 58(6), 914–929 CrossRef PubMed.
- M. J. He, W. Pu, X. Wang, W. Zhang, D. Tang and Y. Dai, Comparing DESI-MSI and MALDI-MSI mediated spatial metabolomics and their applications in cancer studies, Front. Oncol., 2022, 12, 891018 CrossRef CAS PubMed.
- E. K. Neumann, L. G. Migas, J. L. Allen, R. M. Caprioli, R. Van de Plas and J. M. Spraggins, Spatial metabolomics of the human kidney using MALDI trapped ion mobility imaging mass spectrometry, Anal. Chem., 2020, 92(19), 13084–13091 CrossRef CAS PubMed.
- O. Klein, A. Haeckel, U. Reimer, G. Nebrich and E. Schellenberger, Multiplex enzyme activity imaging by MALDI-IMS of substrate library conversions, Sci. Rep., 2020, 10(1), 15522 CrossRef CAS PubMed.
- S. Banerjee, Ambient ionization mass spectrometry imaging for disease diagnosis: Excitements and challenges, J. Biosci., 2018, 43(4), 731–738 CrossRef CAS PubMed.
- R. L. Griffiths, K. I. Kocurek and H. J. Cooper, Ambient surface mass spectrometry–ion mobility spectrometry of intact proteins, Curr. Opin. Chem. Biol., 2018, 42, 67–75 CrossRef CAS PubMed.
- C. Keller, J. Maeda, D. Jayaraman, S. Chakraborty, M. R. Sussman, J. M. Harris, J. M. Ané and L. Li, Comparison of vacuum MALDI and AP-MALDI platforms for the mass spectrometry imaging of metabolites involved in salt stress in Medicago truncatula, Front. Plant Sci., 2018, 9, 1238 CrossRef PubMed.
- B. Rocha, C. Ruiz-Romero and F. J. Blanco, Mass spectrometry imaging: a novel technology in rheumatology, Nat. Rev. Rheumatol., 2017, 13(1), 52–63 CrossRef CAS PubMed.
- Z. Luo, J. He, Y. Chen, J. He, T. Gong, F. Tang, X. Wang, R. Zhang, L. Huang, L. Zhang and H. Lv, Air flow-assisted ionization imaging mass spectrometry method for easy whole-body molecular imaging under ambient conditions, Anal. Chem., 2013, 85(5), 2977–2982 CrossRef CAS PubMed.
- Z. Takats, J. M. Wiseman and R. G. Cooks, Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology, J. Mass Spectrom., 2005, 40(10), 1261–1275 CrossRef CAS PubMed.
- K. Qi, L. Wu, C. Liu and Y. Pan, Recent advances of ambient mass spectrometry imaging and its applications in lipid and metabolite analysis, Metabolites, 2021, 11(11), 780 CrossRef CAS PubMed.
- R. Yin, J. Kyle, K. Burnum-Johnson, K. J. Bloodsworth, L. Sussel, C. Ansong and J. Laskin, High spatial resolution imaging of mouse pancreatic islets using nanospray desorption electrospray ionization mass spectrometry, Anal. Chem., 2018, 90(11), 6548–6555 CrossRef CAS PubMed.
- D. Unsihuay, R. Yin, D. M. Sanchez, M. Yang, Y. Li, X. Sun, S. K. Dey and J. Laskin, High-resolution imaging and identification of biomolecules using Nano-DESI coupled to ion mobility spectrometry, Anal. Chim. Acta, 2021, 1186, 339085 CrossRef CAS PubMed.
- C. Sun, T. Li, X. Song, L. Huang, Q. Zang, J. Xu, N. Bi, G. Jiao, Y. Hao, Y. Chen and R. Zhang, Spatially resolved metabolomics to discover tumor-associated metabolic alterations, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(1), 52–57 CrossRef CAS PubMed.
- J. He, C. Sun, T. Li, Z. Luo, L. Huang, X. Song, X. Li and Z. Abliz, A sensitive and wide coverage ambient mass spectrometry imaging method for functional metabolites based molecular histology, Advanced Science, 2018, 5(11), 1800250 CrossRef PubMed.
- F. Tang, Y. Chen, J. M. He, Z. G. Luo, Z. Abliz and X. H. Wang, Design and performance of air flow-assisted ionization imaging mass spectrometry system, Chin. Chem. Lett., 2014, 25(5), 687–692 CrossRef CAS.
- A. Palmer, P. Phapale, I. Chernyavsky, R. Lavigne, D. Fay, A. Tarasov, V. Kovalev, J. Fuchser, S. Nikolenko, C. Pineau and M. Becker, FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry, Nat. Methods, 2017, 14(1), 57–60 CrossRef CAS PubMed.
- S. Tortorella, P. Tiberi, A. P. Bowman, B. S. Claes, K. Ščupáková, R. M. Heeren, S. R. Ellis and G. Cruciani, LipostarMSI: comprehensive, vendor-neutral software for visualization, data analysis, and automated molecular identification in mass spectrometry imaging, J. Am. Soc. Mass Spectrom., 2019, 31(1), 155–163 CrossRef PubMed.
- L. Sementé, G. Baquer, M. García-Altares, X. Correig-Blanchar and R. P. rMSIannotation, A peak annotation tool for mass spectrometry imaging based on the analysis of isotopic intensity ratios, Anal. Chim. Acta, 2021, 1171, 338669 CrossRef PubMed.
- F. A. van Geenen, F. W. Claassen, M. C. Franssen, H. Zuilhof and M. W. Nielen, Laser ablation electrospray ionization hydrogen/deuterium exchange ambient mass spectrometry imaging, J. Am. Soc. Mass Spectrom., 2020, 31(2), 249–256 CrossRef CAS PubMed.
- X. Song, J. Li, M. Mofidfar and R. N. Zare, Distinguishing between isobaric ions using microdroplet hydrogen–deuterium exchange mass spectrometry, Metabolites, 2021, 11(11), 728 CrossRef CAS PubMed.
- X. Tian, B. Xie, Z. Zou, Y. Jiao, L. E. Lin, C. L. Chen, C. C. Hsu, J. Peng and Z. Yang, Multimodal imaging of amyloid plaques: fusion of the single-probe mass spectrometry image and fluorescence microscopy image, Anal. Chem., 2019, 91(20), 12882–12889 CrossRef CAS PubMed.
- Z. Yuan, Q. Zhou, L. Cai, L. Pan, W. Sun, S. Qumu, S. Yu, J. Feng, H. Zhao, Y. Zheng and M. Shi, SEAM is a spatial single nuclear metabolomics method for dissecting tissue microenvironment, Nat. Methods, 2021, 18(10), 1223–1232 CrossRef CAS PubMed.
- R. Van de Plas, J. Yang, J. Spraggins and R. M. Caprioli, Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping, Nat. Methods, 2015, 12(4), 366–372 CrossRef CAS PubMed.
- X. Yan, X. Zhao, Z. Zhou, A. McKay, A. Brunet and R. N. Zare, Cell-type-specific metabolic profiling achieved by combining desorption electrospray ionization mass spectrometry imaging and immunofluorescence staining, Anal. Chem., 2020, 92(19), 13281–13289 CrossRef CAS PubMed.
- I. Rosas-Román, C. Ovando-Vázquez, A. Moreno-Pedraza, H. Guillén-Alonso, R. Winkler, Open LabBot and RmsiGUI, Community development kit for sampling automation and ambient imaging, Microchem. J., 2020, 152, 104343 CrossRef.
- A. Körber, J. D. Keelor, B. S. Claes, R. M. Heeren and I. G. Anthony, Fast Mass Microscopy: Mass Spectrometry Imaging of a Gigapixel Image in 34 Minutes, Anal. Chem., 2022, 94(42), 14652–14658 CrossRef PubMed.
- L. Guo, X. Liu, C. Zhao, Z. Hu, X. Xu, K. K. Cheng, P. Zhou, Y. Xiao, M. Shah, J. Xu and J. Dong, iSegMSI: an interactive strategy to improve spatial segmentation of mass spectrometry imaging data, Anal. Chem., 2022, 94(42), 14522–14529 CrossRef CAS PubMed.
- O. Rübel, A. Greiner, S. Cholia, K. Louie, E. W. Bethel, T. R. Northen and B. P. Bowen, OpenMSI: a high-performance web-based platform for mass spectrometry imaging, Anal. Chem., 2013, 85(21), 10354–10361 CrossRef PubMed.
- C. Sun, T. Li, X. Song, L. Huang, Q. Zang, J. Xu, N. Bi, G. Jiao, Y. Hao, Y. Chen and R. Zhang, Spatially resolved metabolomics to discover tumor-associated metabolic alterations, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(1), 52–57 CrossRef CAS PubMed.
- Z. Wang, R. Yang, Y. Zhang, X. Hui, L. Yan, R. Zhang, X. Li and Z. Abliz, Ratiometric Mass Spectrometry Imaging for Stain-Free Delineation of Ischemic Tissue and Spatial Profiling of Ischemia-Related Molecular Signatures, Front. Chem., 2021, 9, 807868 CrossRef CAS PubMed.
- B. R. Hamilton, D. L. Marshall, N. R. Casewell, R. A. Harrison, S. J. Blanksby and E. A. Undheim, Mapping enzyme activity on tissue by functional mass spectrometry imaging, Angew. Chem., 2020, 132(10), 3883–3886 CrossRef.
- E. Takeo, E. Fukusaki and S. Shimma, Mass spectrometric enzyme histochemistry method developed for visualizing in situ cholinesterase activity in Mus musculus and Drosophila melanogaster, Anal. Chem., 2020, 92(18), 12379–12386 CrossRef CAS PubMed.
- B. S. Kumar, Desorption Electrospray Ionization Mass Spectrometry Imaging (DESI-MSI) in Disease Diagnosis: An Overview, Anal. Methods, 2023, 15, 3768–3784 RSC.
| This journal is © The Royal Society of Chemistry 2024 |