Open Access Article
Open Access ArticleBoosting the catalytic performance of a marine yeast in a SpinChem® reactor for the synthesis of perillyl alcohol†
Silvia
Donzella
ab,
Concetta
Compagno
a,
Francesco
Molinari
a,
Francesca
Paradisi
 *b and
Martina Letizia
Contente
*b and
Martina Letizia
Contente
 *a
*a
aDepartment of Food, Environmental and Nutritional Sciences (DeFENS), University of Milan, via Celoria 2, 20133, Milan, Italy. E-mail: martina.contente@unimi.it
bDepartment of Chemistry, Biochemistry and Pharmaceutical Sciences, University of Bern, Freiestrasse 3, 3012, Bern, Switzerland. E-mail: francesca.paradisi@unibe.ch
First published on 1st November 2023
Abstract
A sustainable approach for the reduction of perillaldehyde to perillyl alcohol (POH) through alginate immobilized yeast cell beads has been here developed. The process was optimized in small-scale batch reactions and then scaled up in a rotating bed reactor (SpinChem®), enhancing productivity while reducing catalyst loading thanks to better mass transfer and catalyst/substrate interaction (i.e., 90% molar conversion, 8 hours). The biocatalyst biomass was also grown on waste material (molasses) and cultivated using seawater to minimize the environmental impact. By harnessing the potential of immobilized yeast cells in a rotating reactor and utilizing eco-friendly resources, this study exemplifies a sustainable biocatalytic approach that can be extended to other natural terpenes. The incorporation of waste materials and seawater into the process showcases the commitment to increase the sustainability of chemical reactions and aligns with the principles of circular economy.
Developing sustainable energy- and cost-efficient bio-based processes to produce chemicals, fuels and materials is increasingly attractive due to increasing environment concern. Conventional chemical syntheses are generally high-yielding processes; however, they are often environmentally unfriendly, thus increasing downstream costs for both product purification and reaction waste disposal. In comparison to chemical catalysis, whole cell biocatalysis offers some unique advantages. Among them selectivity, high catalytic efficiency, ability to perform multi-step reactions using a single strain as a catalyst and mild operational conditions made this technique widely employed for the efficient biosynthesis of value-added fine and bulk chemicals, as well as pharmaceutically active ingredients (APIs).1 Moreover, whole cell biocatalysts do not require time-consuming and material-intensive purification steps for the obtainment of the catalysts which can be easily recovered from the culture medium in a cheap and abundant manner. In addition to this upstream simplification, downstream processing can also be simplified, thus reducing production time and costs.2 Furthermore, the use of whole cells is usually preferred for reactions involving cofactors as they can synthesize and recycle these expensive molecules themselves.3,4
However, cells present intrinsic metabolic pathways, so competitive secondary reactions due to the presence of other enzymes may occur with the risk of reducing final yields as well as forming undesired by-products. Other common drawbacks of whole cell biocatalysis include possible substrate or product inhibition and the presence of the cell membrane acting as a mass transport barrier.5
To enhance the usability of whole cells, immobilization techniques can be employed.6 Among them, encapsulation within a hydrogel is noteworthy. In fact, hydrogel beads provide a protective environment for the catalyst, allowing it to retain its activity and stability during the reaction process. The immobilization also facilitates product isolation and catalyst separation from the reaction mixture, further simplifying downstream processing for industrial scale-up.7 Moreover, the development of the process in the SpinChem® reactor system, which efficiently facilitates liquid percolation through a packed particle bed within the stirring element, presents additional benefits.8
The use of yeast whole cells for the biotransformation of certain monoterpenoids, including carvone, geraniol and limonene, into highly valuable flavoring derivatives has been studied due to their economic potential in the food, pharmaceutical and cosmetic industries.9,10 For instance, perillaldehyde is a terpenoid found in various plants and essential oils, with perilla herb being the most abundant source. It is commonly used as a food additive for flavoring and in perfumery to provide a spicy aroma. However, in 2015, the European Food Safety Authority (EFSA) conducted an evaluation and determined that perillaldehyde demonstrated genotoxic potential in vivo, raising safety concerns as a flavoring substance.11,12 Subsequently, the European Commission announced its intention to remove perillaldehyde from the EU list of flavorings (http://eur-lex.europa.eu/eli/reg/2015/1760/oj).
However, this compound can be used to produce the corresponding natural derivative, perillyl alcohol (POH), which has been investigated in animal models as a possible therapeutic agent in the prevention and treatment of cancer (i.e. cytotoxic activity).13,14 As this natural compound is present only in low quantities in only a few plant oils, an alternative source of POH is becoming necessary.14 Although POH can be produced via classical organic synthesis, for example, through the hydrogenation of α,β-unsaturated aldehydes, in recent years there has been a significant preference towards biotransformation processes carried out using microbial cells as biocatalysts for the production of fine chemicals.15 Accordingly, molecules obtained by such bioprocesses can be labelled as ‘natural’ and GRAS (‘generally regarded as safe’), thus increasing their market value.16
A total of 14 yeast wild-type strains (full list in ESI,† Table S1), belonging to the genera Saccharomyces, Dekkera, Kluyveromyces, Trichosporon, Debaryomyces, Candida, Lipomyces, Rhodosporidium and Rhodotorula, were screened for their ability to modify monoterpenes for the achievement of interesting APIs. In the first screening, (S)-perillaldehyde was added to growing cells at the concentration of 1 g L−1 after 24 hours of cultivation in rich YPD medium (10 g L−1 yeast extract, 20 g L−1 peptone, 20 g L−1 glucose). The cultural broth was then collected after 24 hours and analyzed by TLC and NMR techniques. The desired product POH, resulting from the reduction of carbonyl by dehydrogenases (DHs) associated with the yeast cells, yielding the corresponding alcohol (Scheme 1), was identified in three yeast strains: CEN-PK and the non-conventional yeast strains Bio1 and CCAT2, with Bio1 as the best POH producer (see Table S1 in the ESI†).
Bio1 strain was identified by sequencing of the D1/D2 region (see ESI,† Table S2) as Candida viswanathii (UBOCC-A-208001), belonging to the Saccharomycetes order and isolated for the first time from hydrothermal vent waters in the South-West Pacific.17 Due to its unique characteristics as a halotolerant yeast, capable of living in challenging environments, as well as its potential tolerance to harsh industrial reaction conditions, Bio1 strain was selected for further studies.
To obtain a more stable and easier-to-use biocatalyst, Bio1 cells, grown in YPD medium, were immobilized into Ca-alginate hydrogel beads (see the ESI†). The medium for the biotransformation was composed of physiological solution (NaCl 0.9%) supplemented with glucose (40 g L−1) to enhance the cofactor recycling system.5,18 Working on small volumes (2 mL), different concentrations of immobilized catalyst (Table 1) and substrate (Table 2) were firstly tested. The best result (90% molar conversion) was achieved using 1 g mL−1 of immobilized biocatalyst with 4 mM (S)-perillaldehyde (corresponding to 0.5 g L−1) after 6-hour reaction time (entry 1, Table 2). After 24 hours, the molar conversion remained unchanged. To minimize costs and energy consumption, mild conditions of temperature (23–25 °C) and pH (7.0) were employed.
| Entry | Immobilized catalyst (g mL−1) | POH (mM) | Molar conversion (%) |
|---|---|---|---|
| 1 | 0.1 | 2.8 ± 0.2 | 35 |
| 2 | 0.5 | 3.5 ± 0.3 | 44 |
| 3 | 1 | 4.2 ± 0.2 | 53 |
| Entry | (S)-Perillaldehyde (mM) | POH (mM) | Molar conversion (%) |
|---|---|---|---|
| 1 | 4 | 3.4 ± 0.3 | 90 |
| 2 | 7.5 | 4.2 ± 0.2 | 54 |
| 3 | 14 | 5.3 ± 0.1 | 37 |
Interestingly, immobilization extended the shelf-life of the biocatalyst: after 1 month of storage at 4 °C, the immobilized yeast cells kept more than 80% of the initial activity calculated under optimized conditions (catalyst concentration 1 g mL−1, substrate concentration 4 mM, reaction time 6 hours), further reducing the costs associated with the biocatalyst preparation. In contrast, non-immobilized cells lost more than 50% of their activity just after 48 hours.
In an initial attempt to scale up the process, we performed the reaction under continuous conditions. To achieve this, a packed-bed reactor (4 mL final volume) was prepared with 4 g of alginate beads containing the biocatalyst (Bio1 cells). After 15 minutes of residence time, poor conversions have been observed since (S)-perillaldehyde adhered to the plastic tubing of the instrument, and just a minimal substrate concentration came into contact with the biocatalyst. This phenomenon was observed previously with a similarly hydrophobic substrate.19
As an alternative system, the SpinChem® reactor was used. SpinChem® is a rotating bed reactor (RBR) that exploits centrifugal acceleration to enhance mixing and mass transfer in the system.8 Additionally, this reactor allows for easier scalability, making it suitable for both laboratory-scale and industrial-scale applications. The SpinChem® reactor is ideal to be used with cells immobilized in hydrogel beads as catalysts. Using this system, more than 90% of molar conversion was reached after 8 hours in a working volume of 200 mL (Fig. 1). More interestingly, this result was obtained using 10 times less biocatalyst compared to small scale reactions (0.1 g mL−1 compared to 1 g mL−1). The efficient design of the system allowed for effective interaction between the catalyst and the reaction mixture, ensuring uniform distribution of reactants and enhancing reaction kinetics and overall performance.
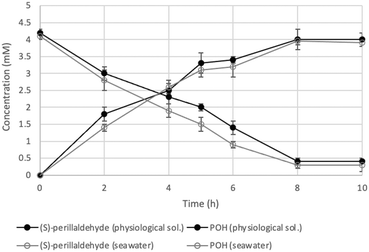 | ||
| Fig. 1 Time course of (S)-perillyl alcohol production in the SpinChem® reactor with immobilized yeast cells. Full symbols: biotransformation with cells grown and used in fresh water; hollow symbols: biotransformation with cells grown and used in seawater. Concentration detected by GC analysis (see the ESI†). | ||
Finally, to adhere to circular economy principles and prioritize sustainable resource management, we evaluated the growth ability of Bio1 strain on a waste-derived medium based on molasses, by-products of sugar beet refining, instead of the expensive YPD synthetic medium. In addition, considering that Bio1 is a marine yeast strain, we chose to employ filtered and sterilized seawater. Seawater contains essential minerals needed for microorganism growth, reduces the risk of contamination due to the salt presence, and allows saving of distilled freshwater, which is a precious source produced via energy-consuming procedures.20 Marine yeasts have proved a relevant source of DHs to be used in seawater.21
Bio1 strain was not very efficient at metabolizing sucrose contained in molasses, therefore an acidic pre-treatment (H2SO4 1.5% (v/v)) was carried out to promote the hydrolysis of sucrose into glucose and fructose. In addition, a small amount of yeast extract (1 g L−1, 10 times less than the synthetic medium YPD) was added to stimulate growth. With this strategy, we were able to produce more than 35 g L−1 (dry weight) of biocatalyst in 70 hours of process in a 2 L bioreactor (see the ESI†). To further enhance the sustainability of the overall process, we also replaced the physiological solution used as biotransformation medium with seawater, obtaining similar results in terms of both molar conversion and product concentration (Fig. 1). Among other challenges, water consumption is an important aspect to consider for the assessment of the sustainability of biocatalytic processes, especially when large scale operations are involved (e.g. fermentation and SpinChem® technologies). Taking into account the current research efforts to develop efficient biocatalytic systems with improved ecological footprints, the here obtained results highlighted the successful use of seawater as both the reaction environment and a convenient ingredient for yeast growth medium.
In summary, this study represents a successful proof of concept to demonstrate the viability and potential applicability of immobilized yeast cells for the modification of natural terpenes through SpinChem® technology.
The results show the high potential of non-conventional extremophilic yeasts as robust biocatalysts for the biotransformation of cheap and largely available monoterpenes to high-value derivatives employable as active pharmaceutical ingredients (APIs). Following the European and US regulation, due to the use of a natural starting material and biocatalytic approaches, the obtained products can be claimed as ‘natural’, thus maximizing their market value. Furthermore, the incorporation of waste materials into the production process emphasizes the commitment to sustainability and aligns with the principles of circular economy. By utilizing molasses and seawater as feedstocks the overall process becomes more environmentally friendly and resource efficient.
Author contributions
Conceptualization M. L. C., F. P.; methodology S. D., M. L. C.; investigation S. D.; resources C. C., F. M., F. P.; data curation, S. D.; first draft writing S. D., manuscript review and editing all authors. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
M. L. C. acknowledges funding from University of Milan (Piano di Sostegno UNIMI Linea 2-2021) (Italy) through the project W-BioFlow “From agro-food waste to valuable bioactives through flow biocatalysis”. S. D. thanks the Swiss-European Mobility Programme (SEMP) for the financial support during the stay abroad and the SNSF (200021_192274, F. P.) for the facilities provided in Bern.Notes and references
- B. Lin and Y. Tao, Microb. Cell Fact., 2017, 16, 106 CrossRef PubMed.
- M. Schrewe, M. K. Julsing, B. Bühler and A. Schmida, Chem. Soc. Rev., 2013, 42, 6346–6377 RSC.
- R. U. Haque, F. Paradisi and T. Allers, Appl. Microbiol. Biotechnol., 2019, 103, 3807–3817 CrossRef CAS PubMed.
- S. Filippucci, G. Tasselli, F. Z. Kenza Labbani, B. Turchetti, M. R. Cramarossa, P. Buzzini and L. Forti, Fermentation, 2020, 6(1), 29 CrossRef CAS.
- T. Johannes, M. R. Simurdiak and H. Zhao, Biocatalysis, in Encyclopedia of chemical processing, 2006, pp. 101–110 Search PubMed.
- A. Pinto, M. L. Contente and L. Tamborini, Curr. Opin. Green Sustainable Chem., 2020, 25, 2452 Search PubMed.
- C. J. C. Rodrigues and C. C. C. R. de Carvalho, Microorganisms, 2022, 10, 966 CrossRef CAS PubMed.
- H. Mallin, J. Muschiol, E. Byström and U. T. Bornscheuer, ChemCatChem, 2013, 5, 3529–3532 CrossRef CAS.
- M. Goretti, C. Ponzoni, E. Caselli, E. Marchegiani, M. R. Cramarossa, B. Turchetti, L. Forti and P. Buzzini, Bioresour. Technol., 2011, 102, 3993–3998 CrossRef CAS PubMed.
- J. B. van Beilen, R. Holtackers, D. Lüscher, U. Bauer, B. Witholt and W. A. Duetz, Appl. Environ. Microbiol., 2005, 71(4), 1737–1744 CrossRef CAS PubMed.
- C. A. Hobbs, S. V. Taylor, C. Beevers, M. Lloyd, R. Bowen, L. Lillford, R. Maronpot and S. Hayashi, Food Chem. Toxicol., 2016, 97, 232–242 CrossRef CAS PubMed.
- F. Erhunmwunsee, C. Pan, K. Yang, Y. Li, M. Liu and J. Tian, Crit. Rev. Food Sci. Nutr., 2022, 62(23), 6328–6340 CrossRef CAS PubMed.
- S. Shojaei, A. Kiumarsi, A. R. Moghadam, J. Alizadeh, H. Marzban and S. Ghavami, Enzymes, 2014, 36, 7–32 CAS.
- T. C. Chen, C. O. Fonseca and A. H. Schönthal, Am. J. Cancer Res., 2015, 5(5), 1580–1593 Search PubMed.
- L. Forti, S. Di Mauro, M. R. Cramarossa, S. Filippucci, B. Turchetti and P. Buzzini, Molecules, 2015, 20(6), 10377–10398 CrossRef CAS PubMed.
- M. L. Contente, L. Tamborini, F. Molinari and F. Paradisi, J. Flow Chem., 2020, 10, 235–240 CrossRef CAS.
- G. Burgaud, N. T. M. Hue, D. Arzur, M. Coton, J. M. Perrier-Cornet, M. Jebbar and G. Barbier, Res. Microbiol., 2015, 166(9), 700–709 CrossRef PubMed.
- M. Goretti, C. Ponzoni, E. Caselli, E. Marchigiani, M. R. Cramarossa, B. Turchetti, P. Buzzini and L. Forti, Enzyme Microb. Technol., 2009, 45, 463–468 CrossRef CAS.
- A. I. Benítez-Mateos, A. Schneider, E. Hegarty, B. Hauer and F. Paradisi, Nat. Commun., 2022, 13, 6269 CrossRef PubMed.
- P. Domínguez de María, ChemCatChem, 2013, 5, 1643–1648 CrossRef.
- I. Serra, B. Guidi, G. Burgaud, M. L. Contente, P. Ferraboschi, A. Pinto, C. Compagno, F. Molinari and D. Romano, ChemCatChem, 2016, 8, 3254–3260 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3re00474k |
| This journal is © The Royal Society of Chemistry 2023 |