Open Access Article
Open Access ArticleDetection of specific antibodies against SARS-CoV-2 spike protein via ultra-sensitive bio-functionalized carbonnitride-reduced graphene oxide electrochemical immunosensing platform in real samples†
Mohd. Abubakar
Sadique
 ab,
Shalu
Yadav
ab,
Pushpesh
Ranjan
ab,
Raghuraj Singh
Chouhan
ab,
Shalu
Yadav
ab,
Pushpesh
Ranjan
ab,
Raghuraj Singh
Chouhan
 *c,
Ivan
Jerman
*c,
Ivan
Jerman
 d,
Ashok
Kumar
e,
Saurabh
Saigal
e,
Sagar
Khadanga
e,
Raju
Khan
d,
Ashok
Kumar
e,
Saurabh
Saigal
e,
Sagar
Khadanga
e,
Raju
Khan
 *ab and
Avanish K.
Srivastava
*ab and
Avanish K.
Srivastava
 a
a
aCSIR–Advanced Materials and Processes Research Institute (AMPRI), Hoshangabad Road, Bhopal-462026, India. E-mail: khan.raju@gmail.com
bAcademy of Scientific and Innovative Research (AcSIR), Ghaziabad-201002, India
cJožef Stefan Institute, Jamova Cesta-39, Ljubljana-1000, Slovenia. E-mail: raghuraj.singh@ijs.si
dNational Institute of Chemistry, Hajdrihova 19, Ljubljana-1000, Slovenia
eAll India Institute of Medical Sciences (AIIMS), Bhopal-462020, India
First published on 11th October 2023
Abstract
In this work, we fabricate a facile de novo synthesis route to prepare bio-functionalized carbon nitride-reduced graphene oxide with chitosan (Chi) nanocomposite (C3N4/RGO/Chi). The nitrogen-rich C3N4 has been synthesized through facile and low-cost thermal oxidation and offers a metal-free and highly active surface for synergistic modification with RGO and Chi. Moreover, the synergistically incorporated C3N4/RGO and biopolymer improve the electrocatalytic activity, provide surface functionalities, and enhance electroconductivity for immunosensing applications. Consequently, the bio-functionalized C3N4/RGO nanocomposite provides a suitable substrate for the immobilization of SARS-CoV-2 RBD spike protein. The immunosensor detects SARS-CoV-2 antibodies in a wide detection range from 10 zg mL−1 (10 × 10−21 g mL−1) to 100 ng mL−1 with an ultra-low detection limit (LOD) of 3.31 zg mL−1. The validation of the immunosensor was also carried out on spiked serum samples, which show a linear detection range of 100 ag mL−1 to 100 ng mL−1 and a LOD of 1.73 ag mL−1. Nevertheless, the clinical diagnostic application of the immunosensor is validated through the examination of real serum samples from COVID-19 patients. The results suggest that the immunosensor can be efficiently used as a screening platform to distinguish between positive and negative samples with high accuracy. The findings of the work have the potential to translate the immunosensing strategy into next-generation point-of-care testing (POCT) of various infectious diseases.
Introduction
The global pandemic of coronavirus 2019 (COVID-19) has been shrinking and re-emerging randomly in the last two years with mutations in the variants of the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2).1 The concerning situation had increased the number of cases due to a poor understanding of the SARS-CoV-2, inadequate detection techniques, and weak vaccination efficacy.2–4 Standard diagnostic techniques like reverse transcription polymerase chain reaction (RT-PCR), enzyme-linked immunosorbent assay (ELISA), clustered regularly interspaced short palindromic repeats (CRISPR), etc. have been extensively utilized for primarily testing and screening of COVID-19.5,6 Even after significant modifications in conventional diagnostic techniques as per modern requirements, they still lack the potential to accomplish the growing demands of disease monitoring.7,8 The boom in cases has drawn attention to the hunt for alternative measures to diagnose COVID-19. In the last two years, the advancements in material science and nanotechnology have shifted the diagnostic developments in the regime of biosensors with the use of materials having better sensitivity and stability.9,10 Various biosensors like optical,11,12 lateral flow,13 thermoelectric,14 piezoelectric,15 field-effect transistor,16 electrochemical,17 colorimetric,18 and mass-based19 have been explored for the detection capabilities of the SARS-CoV-2 virus.20 Among these, few have the potential to be translated from lab scale to point-of-care testing (POCT). In particular, electrochemical biosensors have recently been developed with suitable and superior sensing probes and enhanced biosensor performance. In electrochemical biosensors, the main emphasis has been given to the modification of working electrodes with smart and effective nanomaterials.21,22Several two-dimensional (2D) nanomaterials such as graphene-based nanomaterials,23–25 metal nanocomposites,26 transition metal dichalcogenides,27 metal oxides,28 metal sulfides,29 and ionic liquids30–32 have been investigated for their biosensing properties. Particularly, graphitic-carbon nitride (C3N4) and its nanocomposites have been extensively used in vast sensing applications. The C3N4 exhibits superior electron kinetics behavior, high surface area, conjugation structure, catalytic properties, and bond formation due to the existence of hydrogen atoms bonded with nitrogen at the edges determining its usefulness as an alternative in carbon-based biosensing applications.33 For instance, an analog of graphitic carbon nitride copper single-atom (CuSAC6N6) demonstrated strong catalytic oxidation and a 3.6-fold increase under 50 mW cm−2 domestic light, outperforming control systems and temperature stimuli. The fabricated glucose biosensor demonstrated dynamic sensitivity and linear detection range in vitro.34 Alternatively, a carbon nitride film with exceptional resilience and electrochemiluminescence (ECL) efficiency-based biosensor has been developed via double crystallization showing minimal pinholes, and high transparency for DNA detection with high sensitivity.35 In Another study, efficient polymeric carbon nitride films emitting ECL in blue to green were synthesized, with high ECL quantum efficiency (ΦECL) due to surface-trapped electrons and electron–hole recombination kinetics. Simultaneous detection of miRNA-21 and miRNA-141 was carried out using a multiplexed ECL biosensor.36 Furthermore, mercury monitoring has been reported in water using C3N4 nanosheets.37 Recently, a photoelectrochemical genosensor has been reported for the detection of SARS-CoV-2 using C3N4 and Au nanoparticles.38 However, C3N4 is a desirable material for biosensors due to its surface area, biomolecule binding, and chemical stability. However, its low reactivity limited electron transfer, poor conductivity, deficient functionality, and weak film-forming capacity make functionalization with biomolecules challenging and limit the applicability of C3N4 in biosensing platforms. To improve the properties of C3N4, the composition, and morphology have been modified by introducing advanced functional carbon-based nanomaterials.
Reduced graphene oxide (RGO) is a form of graphene that has been chemically reduced, making it more hydrophilic and easier to functionalize with biomolecules. RGO-based nanocomposites have been exhaustively used in immunosensing applications.39 RGO has remarkable properties including an abundance of oxygen functionalities, a large surface-to-volume ratio, and great biocompatibility.40 In addition, RGO imparts some complementary characteristics such as electron-donating groups, functional groups for covalent binding, electro-catalytic properties, conductivity, stability, and film-forming capabilities that are necessary for the fabrication of electrochemical biosensors.41 Therefore, the RGO has been utilized to modify the C3N4 to overcome the drawbacks of C3N4 and enhance the electrochemical biosensing properties.42 However, the C3N4-based nanocomposites lack some attributes like structural disorder, low conductivity, and poor dispersibility that need further modifications to improve the working efficiency of the immunosensing platform.43 When compared to previous investigations on C3N4 biosensors, the use of C3N4 and RGO in the nanocomposite offers various advantages. The addition of RGO improves biocompatibility, improved electrical conductivity. The addition of C3N4 and RGO increases the overall surface area. The improved stability is ascribed to the synergistic impacts of C3N4 and RGO combination properties.
Chitosan (Chi) is a naturally occurring alkaline cationic polymer that possesses notable attributes such as exceptional biocompatibility, absence of toxicity, and biodegradability. Chi is a biopolymer derived from chitin by the process of deacetylation. It possesses a notable presence of amino and hydroxy groups within its molecular structure, which contribute to its exceptional immobilization and adsorption properties.44 Moreover, various studies have exploited the peculiar properties of Chi, such as the abundance of functional groups that support the binding of biomolecules and act as a substrate in electrochemical immunosensing applications.45 The effective surface density for the receptors is governed by the number of amino groups of Chi available for the covalent binding of biomolecules on the substrate.46 The electrocatalytic, adhesive, binding, and film-forming capabilities of Chi have been extensively utilized in the development of electrochemical biosensors. Moreover, the presence of hydroxyl groups enables easy cross-linking and binding capabilities with other nanomaterials and biomolecules providing a suitable immobilization matrix for electrochemical immunosensing applications.47
Particularly, for the detection of SARS-CoV-2 infections, few works have been reported on C3N4-based nanocomposites. Recently, Tabrizi et al. reported a photoelectrochemical (PEC) aptasensor based on Chi/CdS-C3N4 nanocomposite to detect the SARS-CoV-2 spike antigen. They obtained a LOD of 0.12 nM and validated the performance in human saliva samples.48 In a study by Yin and his group, they presented a resonance energy transfer (RET) based on a ratiometric ECL biosensor using C3N4 nanosheets and Ru–SiO2@folic acid for the detection of the SARS-CoV-2 virus (RdRp gene). The biosensor exhibits a LOD of 0.18 fM in a wide detection range of 1 fM to 10 nM.49 The fluorine-doped tin oxide (FTO) coated glass slide was modified with strontium titanate (SrTiO3 or ST), sulfur-doped carbon nitride (C3N4–S or CNS) and palladium NPs entrapped in aluminum hydroxide matrix (PdAlO(OH) or PdNPs) (PdNPs/CNS/ST/FTO) was used as a sensing probe for SARS-CoV-2 spike antigen in the detection range 1 fg mL−1 to 1000 pg mL−1 with an acceptable detection limit.50 In another study, bismuth tungstate/bismuth sulfide composite (Bi2WO6/Bi2S3) was used as a sensing platform and C3N4 decorated with Au NPs and tungsten trioxide sphere composite (C3N4/Au/WO3) was used as a signal amplifier for the detection of SARS-CoV-2 nucleocapsid protein (SARS-CoV-2 NP) having LOD of 3 fg mL−1.51 A comparative table of recent SARS-CoV-2 antibody electrochemical immunosensors is given in ESI† as Table S1.
Herein, after considering all the aforestated particulars, we designed an ultrasensitive immunosensing platform for the detection of antibodies specific to SARS-CoV-2 spike antigen based on bio-functionalized C3N4/RGO nanocomposite for the first time as per the literature. The fabricated immunosensor exhibits various merits which include facile synthesis steps, rapid and accurate analysis, selective, reproducible, and stable immunosensing platform. The obtained results affirm the ultrasensitive detection limit of the immunosensor is 3.31 zg mL−1. The immunosensor has a LOD of 1.73 ag mL−1 in serum samples and can even distinguish between positive and negative real patient samples with 100% accuracy. On that account, this work paves the way for its translation into miniaturized portable devices for POCT of COVID-19 patients and further for other infectious diseases as well.
Experimental details
Materials and solvents
Recombinant SARS-CoV-2 Spike RBD His-tag protein (Cat # 10500-CV-100), and the SARS-CoV- 2 Spike RBD antibody (Cat # MAB10540-100 SARS-CoV-2 antibodies) were purchased from R&D Systems (United States). Graphite powder, dicyandiamide, N-(3-dimethyl aminopropyl)-N′-ethyl carbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide sodium (NHS), and bovine serum albumin (BSA) was purchased from Sigma Aldrich, USA. Chitosan (Chi) (low molecular weight), and Hydrogen peroxide (H2O2; ∼30%) were purchased from Central Drug House (P) Ltd (CDH), India. Sulfuric acid (H2SO4), orthophosphoric acid (H3PO4), and hydrochloric acid (HCl) were purchased from RANKEM, India. Potassium hydroxide (KOH), isopropyl alcohol (IPA), Potassium permanganate (KMnO4), potassium ferricyanide(III) (K3[Fe(CN)6]), potassium ferrocyanide trihydrate (K4[Fe(CN)6].3H2O), sodium dihydrogen phosphate 2-hydrate (NaH2PO4·2H2O), disodium hydrogen phosphate (Na2HPO4), and potassium chloride (KCl), were procured from SRL Chemical, India. Glacial acetic acid and sodium hydroxide (NaOH pellets) were purchased from Merck, India. All solutions were prepared in ultrapure water from a Merck Milli-Q instrument. Milli-Q water (18.2 ΩM cm) was used throughout the experiment.Instrumentations
For optical characterization, crystallinity, roughness, binding energy, vibrational bonding, molecular states, and morphology of the synthesized nanomaterials; UV-visible spectroscopy (Evolution 220 UV-vis spectrophotometer, Thermo Fisher Scientific), X-ray diffraction (Rigaku Miniflex-1; Cu Kα radiation, λ = 1.54 Å), atomic force microscopy (Witec alpha300 A-AFM microscope), X-ray photoelectron spectroscopy (targeted factor analysis, TFA (Physical Electronics)), Fourier-transform infrared spectroscopy (Bruker, Alpha-II), Raman Spectroscopy (IndiRAM- CTR 300, Technos Instruments), Scanning electron microscopy (JEOL, JCM-6000PLUS & JED-2300, Analysis Station) were used respectively. For the electrochemical characterization and detection, electrochemical workstation (Autolab P/G electrochemical analyzer; Metrohm, PGSTAT204) was employed.Synthesis and preparation of C3N4 nanosheets
The C3N4 nanosheet powder was synthesized by thermal oxidation of a dicyandiamide monomer as reported by Singh et al. with minor modifications.52 Briefly, 5 g dicyandiamide was dried and preheated in an oven for 2 h at 600 °C. The product was cooled at ambient temperature, which was then ground into fine powder. The obtained 3 g powder was again heated at a rate of 5 °C min−1 and kept at 550 °C for 2 h in a ceramic boat. A 2.6 g of yellowish color powder was obtained which was collected as C3N4 nanosheets. This approach is simple, green, and has a high yield.Synthesis of C3N4/RGO nanocomposite
The GO was prepared by the Tour's method,53 the synthesis steps in detail are given in supplementary information. The GO was reduced to RGO using ethanol in an alkaline medium (pH adjusted by KOH) by keeping it in an oven at 80 °C for 2 hours. Next, the synthesized C3N4 was dispersed in isopropanol and sonicated to make a solution. Further, C3N4/RGO nanocomposites were prepared by mixing 500 μL RGO (0.15 mg mL−1) suspension with 300 μL C3N4 (0.6 mg mL−1) solution. The mixture was sonicated for 30 min for thorough mixing.54 The obtained C3N4/RGO was further air-dried and kept in a desiccator before further use.Synthesis of bio-functionalized C3N4/RGO nanocomposite
The bio-functionalized C3N4/RGO was prepared through one-step mixing of C3N4/RGO with the Chi as shown in Scheme 1. For this, 1 mg mL−1 of C3N4/RGO in Milli-Q water was sonicated for 10 min for homogeneous mixing. Subsequently, 300 μL of glacial acetic acid was added while stirring, and then 20 mg of Chi was mixed under constant stirring. The above solution was further sonicated for 10 min to obtain a uniform bio-functionalized C3N4/RGO nanocomposite solution.55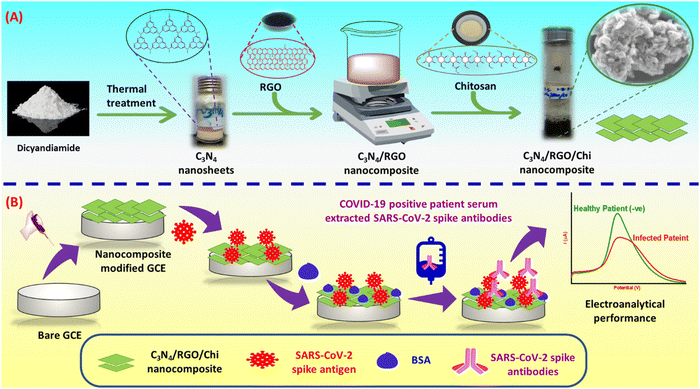 | ||
| Scheme 1 (A) Illustration of the steps involved in the synthesis of C3N4/RGO/Chi nanocomposite and (B) fabrication of the immunosensor for the detection of SARS-CoV-2 spike antibodies. | ||
Electrochemical studies
The electrochemical analysis of the synthesized nanomaterials has been done on a three-electrode electrochemical workstation. Here, the Ag/AgCl reference electrode and Pt wire counter electrode were used along with a nanomaterials-modified glassy carbon electrode (GCE) as the working electrode. The techniques such as cyclic voltammetry (CV), and differential pulse voltammetry (DPV) were carried out in the potential window of −0.3 to +0.8 V, and electrochemical impedance spectroscopy (EIS) in the frequency range of 0.1 Hz to 0.1 MHz. The electrode rinsing buffer was made up of 0.1 M PBS (pH 7) and 0.1 M KCl. The 0.1 M PBS comprising 5 mM ferricyanide/ferrocyanide redox couple and 0.1 M KCl of pH 7.4 is used as PBS for electrochemical characterizations and detection purposes. All the optimization and detection studies were done with triplicate repetition.SARS-CoV-2 sample preparation
The initial performance analysis of the fabricated immunosensors was carried out in PBS (pH 7.4) solution. SARS-CoV-2 spike antibodies detection samples were prepared in PBS (pH 7.4) solution and spiked in diluted serum samples. The synthetic SARS-CoV-2 spike antibodies detection samples (100 ng to 10 zg) were prepared from the 5 μg mL−1 stock solution of SARS-CoV-2 spike antibodies (Cat# MAB10540-100; diluted from 100 μg) in the PBS (pH 7.4) solution by serial dilution from concentrations of 10 zg mL−1 to 100 ng mL−1. In brief, the initial antibody concentration in the stock solution is 100 μg mL−1. To create a 1 mL solution with a final concentration of 10 μg mL−1, add 100 μL to 900 μL of diluent, achieving a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 dilution. Repeat this process to achieve the final solution concentration of 10 zg mL−1. For the spiked serum samples, human serum was diluted in a 1
9 dilution. Repeat this process to achieve the final solution concentration of 10 zg mL−1. For the spiked serum samples, human serum was diluted in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio in PBS, and then different concentrations of SARS-CoV-2 spike antibodies ranging from 100 ag mL−1 to 100 ng mL−1 were made by serial dilution. Further, for the validation of immunosensor performance, the real patient serum samples (N = 20, 10 +ve (P1–P10), 10 −ve (N1–N10)) were received from AIIMS, Bhopal. Approval of the Institutional Human Ethics Committee of AIIMS, Bhopal, and the Institutional Biosafety Committee of CSIR-AMPRI, Bhopal were obtained. Blood samples were collected with written informed consent. The real patient samples were diluted in a 1
10 ratio in PBS, and then different concentrations of SARS-CoV-2 spike antibodies ranging from 100 ag mL−1 to 100 ng mL−1 were made by serial dilution. Further, for the validation of immunosensor performance, the real patient serum samples (N = 20, 10 +ve (P1–P10), 10 −ve (N1–N10)) were received from AIIMS, Bhopal. Approval of the Institutional Human Ethics Committee of AIIMS, Bhopal, and the Institutional Biosafety Committee of CSIR-AMPRI, Bhopal were obtained. Blood samples were collected with written informed consent. The real patient samples were diluted in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio in PBS (pH 7.4) for detection. All the samples were kept at −20 °C in a deep freezer before further use.
10 ratio in PBS (pH 7.4) for detection. All the samples were kept at −20 °C in a deep freezer before further use.
Fabrication of SARS-CoV-2 immunosensor
The synthesized nanocomposite was used for several steps of modification of the GCE for the fabrication of the immunosensor. At first, 4 μL of C3N4/RGO/Chi nanocomposite was drop cast on a 3 mm diameter of the GCE and left for drying at ambient temperature for 24 h. Afterward, 5 μL of N-(3-dimethyl aminopropyl)-N′-ethyl carbodiimide hydrochloride, N-hydroxysulfosuccinimide sodium, (EDC: NHS) (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was drop cast on the nanocomposite as an amide cross-linker for 1.5 h. The Milli-Q water was used to wash the excess cross-linker. Then 5 μL of 5 μg mL−1 SARS-CoV-2 spike antigen (Cat # 10500-CV-100; diluted from 100 μg stock solution) was immobilized and kept at 4 °C overnight for incubation. Further, the unbounded spike antigen was removed using a rinsing buffer. At last, 5 μL of 1% bovine serum albumin (BSA) was used as a blocking agent of the reactive sites of the electrode to avoid unspecific binding, and excess BSA was removed with a rinsing buffer. The working electrodes were electrochemically characterized after each surface modification step. The C3N4/RGO/Chi/SARS-CoV-2 spike antigen/BSA is termed an immunosensor. The fabricated immunosensor is ready for the detection of SARS-CoV-2 spike antibodies and the steps involved in the fabrication of the immunosensor are depicted in Scheme 1.
1) was drop cast on the nanocomposite as an amide cross-linker for 1.5 h. The Milli-Q water was used to wash the excess cross-linker. Then 5 μL of 5 μg mL−1 SARS-CoV-2 spike antigen (Cat # 10500-CV-100; diluted from 100 μg stock solution) was immobilized and kept at 4 °C overnight for incubation. Further, the unbounded spike antigen was removed using a rinsing buffer. At last, 5 μL of 1% bovine serum albumin (BSA) was used as a blocking agent of the reactive sites of the electrode to avoid unspecific binding, and excess BSA was removed with a rinsing buffer. The working electrodes were electrochemically characterized after each surface modification step. The C3N4/RGO/Chi/SARS-CoV-2 spike antigen/BSA is termed an immunosensor. The fabricated immunosensor is ready for the detection of SARS-CoV-2 spike antibodies and the steps involved in the fabrication of the immunosensor are depicted in Scheme 1.
Results and discussion
Optical characterization
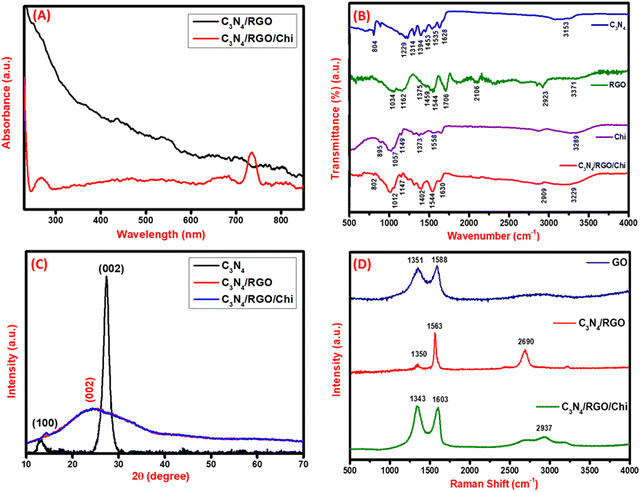
In the UV-vis spectroscopy, the C3N4/RGO composite does not have any significant peak other than that of RGO at about 240 nm. Whereas the C3N4/RGO/Chi nanocomposite has a characteristic peak of C3N4/RGO at about 240 nm and an absorption peak of Chi at around 730 nm. The UV-vis spectra of the nanocomposites are shown in Fig. 1A. FTIR spectroscopy was used to characterize the functional groups, and bond strength of the nanomaterials with major emphasis on the presence of vibrational bonds. As shown in Fig. 1B, the base material C3N4 consists of an N–H bond stretching at a 3152 cm−1 vibrational band. For the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N group, the vibrational stretching bands have peaks appearing at 1229, 1314, 1394, and 1453 cm−1. However, the C–N bond has a stretching band at 1535 and 1628 cm−1. A sharp peak is visible at 804 cm−1 for out-of-plane bending of the tri-s-triazine allotrope of C3N4.52 The FTIR spectra of C3N4 suggest the presence of primary and secondary amines. Further, the FTIR spectra of RGO have significant peaks for the stretching vibrational bands of the –OH, C–H (sp3), C
N group, the vibrational stretching bands have peaks appearing at 1229, 1314, 1394, and 1453 cm−1. However, the C–N bond has a stretching band at 1535 and 1628 cm−1. A sharp peak is visible at 804 cm−1 for out-of-plane bending of the tri-s-triazine allotrope of C3N4.52 The FTIR spectra of C3N4 suggest the presence of primary and secondary amines. Further, the FTIR spectra of RGO have significant peaks for the stretching vibrational bands of the –OH, C–H (sp3), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and C
O, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond at 3371, 2923, 1706, and 1544 cm−1 respectively.56–58 The bending vibrations of C–OH, C–H, C–O–C, and C–O bonds are observed at 1375, 1459, 1162, and 1034 cm−1 respectively suggesting the successful reduction of GO.59 In Chi, the stretching vibrational of –NH, –NH2, C–H, and C–O groups is observed at 3289, 1558, 1373, and 1057 cm−1 respectively.60 The polysaccharide structure of Chi has vibrational bands at 895 and 1149 cm−1.61 Subsequently, the presence of all the characteristic peaks at 802, 1012, 1147, 1402, 1544, 1630, 2909, and 3229 cm−1 implies the coexistence of C3N4, RGO, and Chi in the C3N4/RGO/Chi nanocomposite.
C bond at 3371, 2923, 1706, and 1544 cm−1 respectively.56–58 The bending vibrations of C–OH, C–H, C–O–C, and C–O bonds are observed at 1375, 1459, 1162, and 1034 cm−1 respectively suggesting the successful reduction of GO.59 In Chi, the stretching vibrational of –NH, –NH2, C–H, and C–O groups is observed at 3289, 1558, 1373, and 1057 cm−1 respectively.60 The polysaccharide structure of Chi has vibrational bands at 895 and 1149 cm−1.61 Subsequently, the presence of all the characteristic peaks at 802, 1012, 1147, 1402, 1544, 1630, 2909, and 3229 cm−1 implies the coexistence of C3N4, RGO, and Chi in the C3N4/RGO/Chi nanocomposite.
Crystallinity characterization: X-ray diffraction spectroscopy
For the analysis of the crystallinity of the synthesized nanomaterials, XRD was done. The base nanomaterial C3N4 has a distinctive crystalline peak at 13.08° and 27.37° and RGO has a broad peak at 24.28°. Fig. 1C shows the (100) and (002) plane of C3N4 corresponding to tri-s-triazine units and conjugated aromatic system indicating the formation of the layered structure. In C3N4/RGO the (002) plane of both C3N4 and RGO is seen which suggests stable hybridization of both two-dimensional nanomaterials in the form of C3N4/RGO nanosheets.54 In the case of the C3N4/RGO/Chi nanocomposite, the characteristic peak of C3N4 has diminished due to the coated surface by Chi. The Chi covering in the C3N4/RGO/Chi nanocomposite is well seen in SEM images as well.55Vibrational analysis: Raman spectroscopy
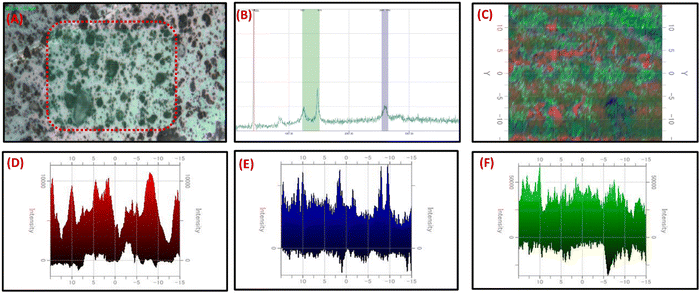
For the analysis of carbon-based nanocomposites, Raman spectroscopy is an effective tool for measuring the D, G, and 2D band peaks in terms of Raman shift. The disorder in sp2 carbon nanomaterials is observed in the Raman analysis. GO exhibited the D and G bands at 1351 cm−1 and 1588 cm−1, respectively. On adding C3N4 nanosheets, the shift in the D and G band peaks at 1350, and 1563 cm−1, and the presence of a 2D band at 2690 cm−1 evidenced a high reduction of GO to RGO and covalent binding of C3N4 with RGO Fig. 1D. The difference in D and G band intensities and the presence of a 2D band at 2690 cm−1 in C3N4/RGO suggests the highly conducting nature of the nanocomposite after the reduction of GO.62 The C-NH bending region lies around 1290–1350 cm−1 and amide-I around 1635 cm−1.63 The paranoid ring containing the methylene group in Chi has C–H stretching vibrations at around 2885–2945 cm−1.64 After the addition of Chi, the peak shifts in D, and G bands at 1343 and 1603 cm−1 respectively, and the presence of broad 2D peaks around 2937 cm−1 suggest the successful formation of Chi-covered C3N4/RGO nanocomposite.After Raman characterization of the synthesized bio-functionalized C3N4/RGO nanocomposite, Raman mapping was carried out to evaluate the nanomaterial distribution on the surface and the arrangement of laminar structures of C3N4 and RGO along with the covering of Chi on C3N4/RGO nanocomposite. A larger region was mapped for the identification of C3N4, RGO, and Chi dispersion in nanomaterials. The confocal Raman maps were created by raster scanning spectra at intervals of 0.1 μm throughout a specific region. The spectra collected can be plotted to identify the peak strength and spatial distribution of nanocomposites. The Raman mapping data of bio-functionalized C3N4/RGO nanocomposite shown in Fig. 2 depicts the Chi-coated layered structure of C3N4 and RGO nanomaterials. As seen from the optical image (Fig. 2(A)), Raman spectra (Fig. 2(B)), and mapping image (Fig. 2(C)), it is clear that the synthesized bio-functionalized C3N4/RGO nanocomposite has uniformly distributed C3N4/RGO sheets which are decorated by Chi on the surface. The 2D Raman mapping image in Fig. 2(D–F) shows the distribution of Si/SiO2, Chi, and C3N4/RGO nanocomposite during Raman mapping.
Morphological studies of the nanocomposites
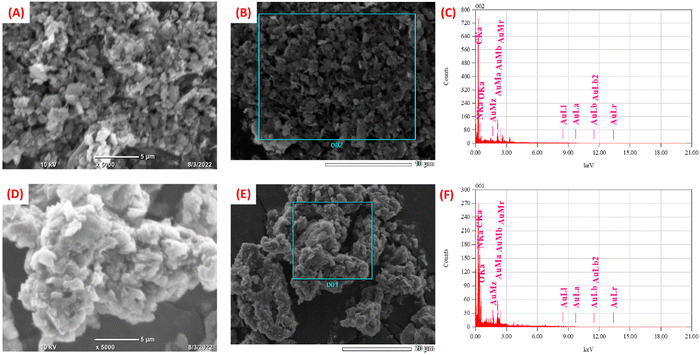
The morphological characterization of synthesized nanomaterials was carried out using SEM. The SEM image of C3N4 nanosheets is shown in Fig. S3 (ESI†). The morphology of C3N4/RGO was observed to be flaky sheets as seen in Fig. 3(A and B). A large number of nanocomposite bundles show the bulky nature of the nanocomposite that acts as a suitable substrate comprising abundant functional groups.65 In the case of bio-functionalized C3N4/RGO nanocomposite the surface morphology of the nanocomposite reveals that Chi has covered C3N4/RGO making it more stable (Fig. 3(D, E)). The presence of Chi on the surface of C3N4/RGO facilitates its film-forming capabilities.66 The EDX plot of C3N4/RGO (Fig. 3(C)) and C3N4/RGO/Chi (Fig. 3(F)) clearly distinguishes the presence of characteristic elements in the nanocomposites.Electrochemical studies
Electrochemical characterizations
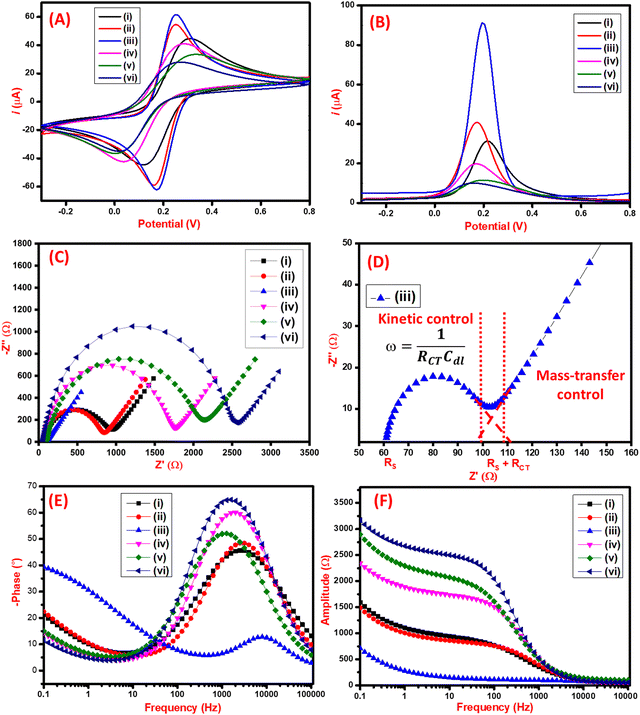
The electrochemical performance of the synthesized nanomaterials was analyzed by CV, DPV, and EIS techniques. As seen in Fig. 4A, the CV characterization of the C3N4 had an anodic peak current of 44 μA. On further modification with RGO, the increase in anodic peak current to 54 μA was observed due to the restoration of π electrons that facilitate electron transfer on the surface of RGO. Furthermore, the film-forming capacity and stability of the nanocomposite were improved by adding Chi which also electrocatalytic effect on the final nanocomposite having the anodic peak current of 61 μA. The C3N4/RGO/Chi nanocomposite has a maximum peak current of 61 μA which was slightly reduced to 41 μA after the immobilization of SARS-CoV-2 spike antigen due to the biolayer formation. Finally, after blocking with BSA, the current was further decreased to 33 μA suggesting the successful fabrication of the immunosensor. The binding of the SARS-CoV-2 spike antibodies was also analyzed which has the least current of 28 μA which suggests proper binding of the antibody–antigen complex. The comparative assessment of the modified electrodes was also carried out by the DPV technique (Fig. 4B) which shows a similar pattern as observed in CV peak current values for the modified electrodes. The change in peak current and peak potential after each modification step was due to the electrochemical characteristics of synthesized nanomaterials and their interaction with immobilized biomolecules that were visible in both CV and DPV curves.The CV analysis was used to determine the effective surface area from the Randles–Sevcik equation. The peak current (Ip) is expressed as:
| Ip = (2.69 × 105) × (n)3/2 × A × (D)1/2 × C × (υ)1/2 | (1) |
For the C3N4/RGO/Chi nanocomposite-modified electrode, the calculated effective surface area was 12 × 10−2 cm2. This proves the presence of a large effective surface area for the fabrication of the immunosensor. However, for the fabricated immunosensor the peak current observed was 33 μA, which gave the effective surface area of the immunosensor to be 6.53 × 10−2 cm2 for targeted binding of SARS-CoV-2 spike antigen-specific antibodies.
EIS studies were conducted to further study the electrochemical interfacial process of the modified electrode surfaces, and the Nyquist plots are displayed in Fig. 4(C). The interpretation of the EIS data may vary upon modification with nanocomposites or biological components. Subsequently, it is important to carefully understand the unique physical characteristics of each system before selecting the appropriate equivalent circuit model to represent the electrical effects of the electrochemical systems and to explain the mechanism of the reaction. The results presented here were fitted using an equivalent circuit model made up of a combination of resistors and capacitors to interpret the EIS results of nanocomposites, also considering the factors governing the electrochemical impedance on the biomolecules-modified surface.
The Nyquist plot in Fig. 4(C) illustrates each step of modification of the electrode surface, wherein the semi-circular segment is related to the charge-transfer resistance and the double-layer capacitance, and the linear segment is related to the diffusion-controlled during the redox processes at the electrode–electrolyte interface. The increase of the semicircle in the high-frequency region is due to the incorporation of biomolecules into the nanocomposite film (C3N4/RGO/Chi). To assess this effect, EIS data were fitted with the equivalent circuit model shown in Fig. S3 (ESI†). This equivalent circuit consists of a series resistance contribution (Rs), which accounts for electrolyte and current collector resistance, and a charge-transfer resistance (RCT), which is associated with the interfacial faradaic process, and Warburg impedance (Rw). The obtained RCT of C3N4 and C3N4/RGO was 825 Ω, and 705 Ω respectively. The lower RCT (46 Ω) of C3N4/RGO/Chi nanocomposite evidence that the C3N4/RGO/Chi nanocomposite sensing probe has superior electrochemical properties. Further on surface alteration with SARS-CoV-2 spike antigen, and blocking agent (BSA), the increase in RCT values (from 1581 Ω to 1862 Ω) is due to the steric hindrance in electron transfer from the addition of biological compounds on the surface which indicates successful fabrication of the immunosensor. The mechanism behind the extremely high electro-conductive nature is depicted in the Nyquist plot (Fig. 4(D)). The semicircular region at low frequencies projects towards the kinetic control mechanism that occurs at the electrode–electrolyte interface. On the other side, the linear line at a higher frequency region provides the diffusion-controlled mass transfer reaction at the electrode–electrolyte junction. The binding of an antibody–antigen complex is also proposed by the increased RCT value of 2352 Ω.67 The respective EIS fitting data is represented in Table 1.
| Electrode | R s (Ω) | R p (kΩ) | R CT (Ω) | CPE (μMho*s^N) | N | Rw (mMho*s1/2) | Convergence fit (χ2) | Number of iterations | Convergence |
|---|---|---|---|---|---|---|---|---|---|
| C3N4 nanosheets | 45.4 | 0.871 | 825.6 | 3.72 | 0.74 | 1.56 | 0.009 | 46 | ✓ |
| C3N4/RGO | 53.4 | 0.759 | 705.6 | 1.55 | 0.82 | 1.58 | 0.158 | 300 | ✓ |
| C3N4/RGO/Chi nanocomposite | 60.5 | 0.107 | 46.5 | 1.39 | 0.92 | 1.98 | 0.024 | 35 | ✓ |
| C3N4/RGO/Chi/SARS-CoV-2 spike antigen | 58.8 | 1.64 | 1581.2 | 0.76 | 0.89 | 1.55 | 0.011 | 33 | ✓ |
| C3N4/RGO/Chi/SARS-CoV-2 spike antigen/BSA | 97.7 | 1.96 | 1862.3 | 1.46 | 0.82 | 1.20 | 0.019 | 39 | ✓ |
| C3N4/RGO/Chi/SARS-CoV-2 spike antigen/BSA/SARS-CoV-2 spike antibodies | 57.3 | 2.41 | 2352.7 | 0.66 | 0.90 | 1.40 | 0.010 | 34 | ✓ |
The bode diagram provides a clear explanation of how the electrochemical system responds to changes in frequency and how this affects the dispersion of experimental data. The most practical method for extrapolating and analyzing impedance data at low frequencies is using the bode plot.68 The correlation between the frequency and the shift in phase values and amplitude is shown in Fig. 4(E and F) respectively. The bode plot also decreased the dispersion of experimental results and clarified the electrochemical behavior of electrodes in low-frequency ranges (less than 1000 Hz). The solution resistance corresponds to a plateau with a phase angle of 0° on the bode plot for the higher frequency area (more than 1000 Hz). The classification of the median frequencies (1000–10 Hz) and the determination of the electrode's capacitive behavior, which specifies the dielectric characteristics of an electrically conductive electrode interface, were done using the rise in phase angles up to 65°. Mass transport processes, electron charge transfer phenomena, and other relaxation events happened at the interface between the electrode and redox solution at frequencies lower than 10 Hz.69
The correlation between CV, DPV, and EIS data suggests that the surface modification of the working electrode has been done. The scan rates studies of the C3N4/RGO/Chi nanocomposite and fabricated immunosensor show good linearity as shown in Fig. S5A and B (ESI†) respectively.
Optimization studies for fabrication of immunosensor
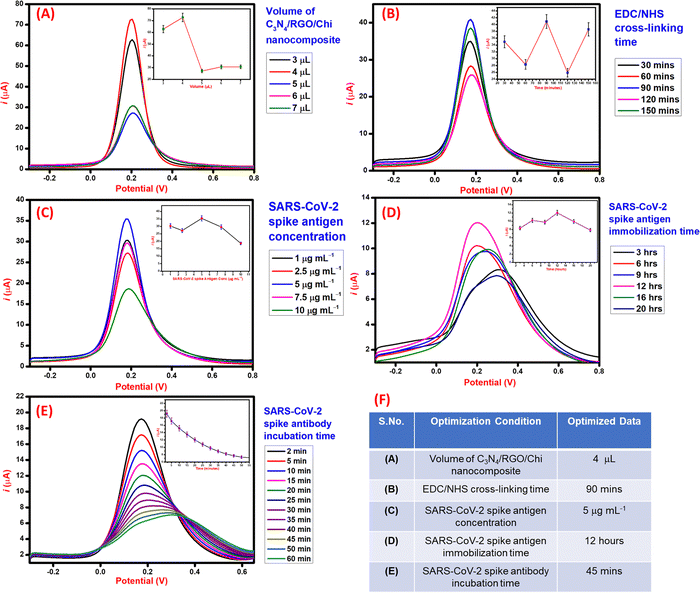
The process parameters involved in the fabrication of the immunosensor were optimized using the DPV technique for higher performance and sensitivity. Initially, the volume study of C3N4/RGO/Chi nanocomposite was done to optimize the volume to be drop cast on the electrode for the maximum electrochemical response. As seen in Fig. 5A, the maximum DPV peak current response was observed for 4 μL, hence 4 μL of C3N4/RGO/Chi nanocomposite was selected for drop cast and further surface modifications. Further, the EDC/NHS cross-linker incubation time was optimized. As seen in Fig. 5B, the cross-linker incubation time was varied from 30–150 minutes and the maximum DPV current response was obtained for 90 minutes. Hence, 90 minutes was selected as the optimal incubation time for the cross-linker for further electrode modification. Subsequently, the varied concentration of SARS-CoV-2 spike antigen from 1 to 10 μg mL−1 was optimized for the modification of the electrode surface. As seen in Fig. 5C, the maximum DPV current response was observed for 5 μg mL−1. Hence the optimal concentration of the SARS-CoV-2 spike antigen was selected to be 5 μg mL−1 for further modification. Moreover, the incubation time of the SARS-CoV-2 spike antigen plays an important role in the effective binding of the spike antigen to the surface. The incubation time study was carried out for 3–20 h. As seen from Fig. 5D, the maximum DPV current response was obtained for 12 h, hence 12 h was selected as the optimum incubation time for the SARS-CoV-2 spike antigen. At last, after the fabrication of the immunosensor, the spike antibodies incubation study was carried out to examine the antigen–antibody interaction. The SARS-CoV-2 spike antibodies were incubated for 2–50 minutes and recorded after a fixed interval. The DPV current response achieved a saturation at 45 mins and negligible signal change was observed at 50 mins. Hence, 45 mins was noted as the incubation time of the SARS-CoV-2 spike antibodies as seen in Fig. 5E.Based on the optimization studies during the fabrication steps of the immunosensor as summarized in tabular form Fig. 5F, the optimal values were selected to evaluate the performance of the immunosensor for the detection of the SARS-CoV-2 spike antibodies.
Detection of SARS-CoV-2 spike antibodies in PBS solution
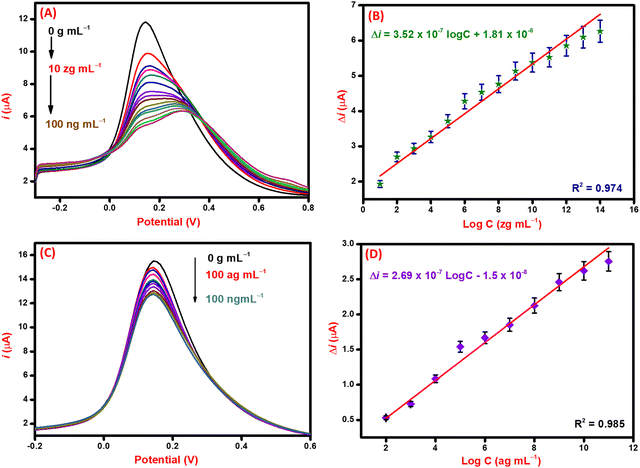
For the detection of the SARS-CoV-2 spike antibodies using the fabricated immunosensor, the DPV technique was employed. During the analysis of SARS-CoV-2 spike antibodies in the wide concentration range of 10 zg mL−1 to 100 ng mL−1, the current response decreases with an increase in the concentration of the SARS-CoV-2 spike antibodies. As shown in Fig. 6A, the immunosensor attained saturation at a concentration of 100 ng mL−1. In addition, Δi values were calculated using the corresponding current values of each concentration and its difference from the response obtained for immunosensor (without analyte). The maximal current of the immunosensor (in the absence of SARS-CoV-2 spike antibodies) was 11 μA. This was used to determine the respective Δi values for each concentration, which were then plotted on the calibration curve (Fig. 6B) in the range of 10 zg mL−1 to 100 ng mL−1. The error bars in Fig. 6B represent the reproducibility of the method which was measured individually. The LOD and LOQ were calculated using the standard equations,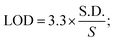 | (2) |
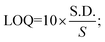 | (3) |
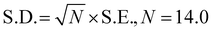 , and S is the slope of the calibration curve (3.52 × 10−7), S.E. = (9.44 × 10−8) standard error of intercept; S.D. = 3.53 × 10−7. The obtained values of LOD and LOQ are 3.31 zg mL−1 and 10.03 zg mL−1 in PBS (pH 7.4) solution respectively.
, and S is the slope of the calibration curve (3.52 × 10−7), S.E. = (9.44 × 10−8) standard error of intercept; S.D. = 3.53 × 10−7. The obtained values of LOD and LOQ are 3.31 zg mL−1 and 10.03 zg mL−1 in PBS (pH 7.4) solution respectively.
Particularly, for the detection of SARS-CoV-2 infections, few works have been reported on C3N4-based nanocomposites. However, this work presents the lowest detection limit among all reported studies. A comparative table of recent SARS-CoV-2 antibody electrochemical immunosensors is given in ESI† as Table S1.
Analytical performance in spiked serum samples
After the successful detection of the SARS-CoV-2 spike antibodies in PBS (pH 7.4) solution. The performance of the immunosensor was validated in spiked serum samples via the DPV technique taking 100 ag mL−1 as the lowest detectable concentration. The current response curve is shown in Fig. 6C, where the current response decreases with an increase in the concentration of the target (SARS-CoV-2 spike antibodies) analyte. This decrease is attributed to the hindrance in electron mobility due to the addition of bulky biomolecules that restrict the transfer of electrons. The change in the current of each concentration of analyte concerning the immunosensor was calculated and plotted against the log of concentration to draw the corresponding calibration curve (Fig. 6D). From the calibration curve, the statistical analysis of the immunosensor was done to evaluate the performance in spiked serum samples. The correlation coefficient (R2) value of the curve suggests significant sensitivity and favorable performance of the immunosensor. The obtained LOD and LOQ values for spiked serum samples are 1.73 ag mL−1 and 5.25 ag mL−1, respectively. The performance of the immunosensor in PBS (pH 7.4) and spiked serum samples validates it for use in the examination of patient samples.Selectivity, reproducibility, and stability studies of the immunosensor
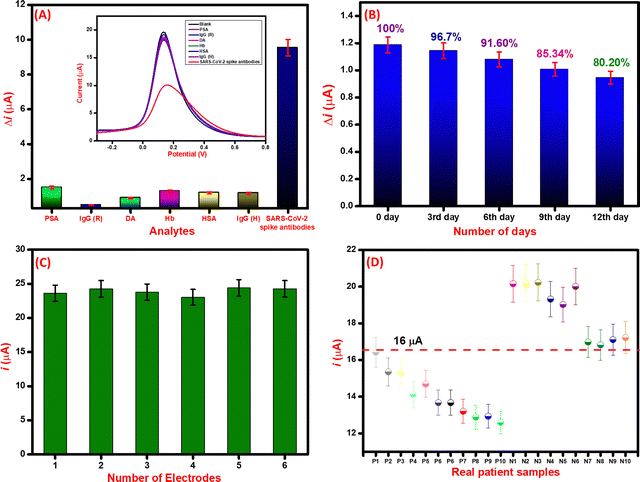
The selectivity of the immunosensor was evaluated by using various biomolecules present in biological fluids as interfering agents. Various proteins such as prostate-specific antigen (PSA) (100 ng mL−1), immunoglobin G (rabbit) (IgG (R)) (100 ng mL−1), immunoglobin G (human) (IgG (H)) (100 ng mL−1), dopamine (DA) (100 ng mL−1), hemoglobin (Hb) (100 ng mL−1), and human serum albumin (HSA) (100 ng mL−1) were used as potential interfering analytes. As seen in Fig. 7A, the immunosensor response was similar to that of the interfering analytes whereas, for the SARS-CoV-2 spike antibodies (10 ng mL−1), the obtained current response is significantly less than the immunosensor. The change in current (Δi) for SARS-CoV-2 spike antibodies and insignificant difference in Δi for interfering analytes suggests that the immunosensor is highly selective towards SARS-CoV-2 spike antibodies.70Further, the stability study of the immunosensor was conducted by keeping the immunosensor at 4 °C and evaluating its detection capability with 100 ng mL−1 SARS-CoV-2 spike antibody. Results in Fig. 7B reveal that the immunosensor performs well within a week time and then gradually the performance decays in the second week. This suggests that the immunosensor is stable for a week for sensitive detection of SARS-CoV-2 spike antibodies. Moreover, the reproducibility was evaluated by constructing six different immunosensors by the same protocol. All the immunosensors were electrochemically characterized by the DPV technique and their respective peak currents were analyzed. As seen in Fig. 7C, the electrode-to-electrode standard variation of DPV peak current was calculated to be less than 5% which suggests that the fabricated immunosensors are reproducible in performance.
Detection of SARS-CoV-2 spike antibodies in real serum samples
After the successful detection of SARC-CoV-2 spike antibodies in PBS (pH 7.4) and spiked serum samples, the performance of the immunosensor was further analyzed in real patient samples. For the initial validation of the immunosensor, 10 positive and 10 negative samples of SARS-CoV-2 infected patients were selected that were previously screened by RT-PCR and in-house ELISA kit. The samples were analyzed through DPV, and the respective current responses were recorded. For the positive samples, the current value lies between 12–16 μA, whereas for negative samples the current values were in the range of 16–20 μA. The details are included in tabular form as Table 2. The threshold was estimated according to the current response of the control (blank) sample.71 The threshold for the screening of positive and negative was set at 16.5 μA. The positive COVID-19 samples had current values of <16.5 μA, whereas the negative had >16.5 μA. The testing data of all positive and negative samples based on threshold values are depicted in Fig. 7D.| S.No. | Sample ID | SARS-CoV-2 antibodies | ELISA result | DPV current value (μA) | Electrochemical Immunosensor | Validated |
|---|---|---|---|---|---|---|
| 1. | P1 | YES | + | 16.42 | + | ✓ |
| 2. | P2 | YES | + | 15.35 | + | ✓ |
| 3. | P3 | YES | + | 15.33 | + | ✓ |
| 4. | P4 | YES | + | 14.12 | + | ✓ |
| 5. | P5 | YES | + | 14.69 | + | ✓ |
| 6. | P6 | YES | + | 13.67 | + | ✓ |
| 7. | P7 | YES | + | 13.67 | + | ✓ |
| 8. | P8 | YES | + | 13.20 | + | ✓ |
| 9. | P9 | YES | + | 12.88 | + | ✓ |
| 10. | P10 | YES | + | 12.93 | + | ✓ |
| 11. | N1 | NO | − | 20.15 | − | ✓ |
| 12. | N2 | NO | − | 20.19 | − | ✓ |
| 13. | N3 | NO | − | 20.23 | − | ✓ |
| 14. | N4 | NO | − | 19.32 | − | ✓ |
| 15. | N5 | NO | − | 19.02 | − | ✓ |
| 16. | N6 | NO | − | 20.01 | − | ✓ |
| 17. | N7 | NO | − | 16.98 | − | ✓ |
| 18. | N8 | NO | − | 16.82 | − | ✓ |
| 19. | N9 | NO | − | 17.10 | − | ✓ |
| 20. | N10 | NO | − | 17.22 | − | ✓ |
Conclusion
In summary, the utilization of bio-functionalized C3N4/RGO nanocomposite has demonstrated promising prospects in the field of immunosensing. The inclusion of C3N4 and RGO inside the nanocomposite offers enhanced electrical conductivity and a significantly expanded surface area, facilitating the effective binding of biomolecules. Additionally, the presence of biopolymer, Chi within the nanocomposite contributes to its biocompatibility and enhances the stability of the biomolecules that are bound to its surface (Aeff = 12 × 10−2 cm−2). Subsequently, bio-functionalized C3N4/RGO nanocomposite showed highly sensitive electrochemical immunosensing capabilities for the detection of SARS-CoV-2 spike antibodies in PBS (sensitivity = 2.9 μA mL zg−1 cm−2), spiked serum samples, and even in real patient samples. The electrochemical analysis reveals that the immunosensor performs very efficiently with an ultra-low detection limit of antibodies against the SARS-CoV-2 spike antigen and adequately differentiates between positive and negative samples. Moreover, the immunosensor also showed high selectivity, reproducibility, and high accuracy. Probable limitations of the proposed work that need to be further studied are the improvement of its stability and detection linearity. Nonetheless, the real sample results being validated by RT-PCR and ELISA techniques suggest the successful development of the immunosensor for the effective detection of antibodies against the SARS-CoV-2 spike antigen in real samples. However, further work is needed for the standardization of the results and translation into portable immunosensors for use in POCT of COVID-19 patients.Author contributions
Mohd. Abubakar Sadique: conceptualization, methodology, writing – original draft preparation, validation, software, writing – reviewing and editing; Shalu Yadav: software, writing – original draft preparation, data curation, writing – reviewing and editing, validation; Pushpesh Ranjan: data curation, validation, writing – reviewing and editing; Raghuraj Singh Chouhan: methodology, data curation, writing – reviewing and editing, software, validation; Ivan Jerman: writing – reviewing and editing, validation; Ashok Kumar: visualization, investigation, validation, writing – reviewing and editing; Saurabh Saigal: validation, writing – reviewing and editing; Sagar Khadanga: writing – reviewing and editing, validation; and Raju Khan: conceptualization, reviewing and editing, validation, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the Director, Council of Scientific and Industrial Research (CSIR)-AMPRI, Bhopal for his encouragement for this work. M.A.S. is obliged to the Science and Engineering Research Board (SERB) for JRF. R.K. would like to acknowledge SERB and CSIR for financial aid for this work under the project IPA/2020/000130 and MLP-0301, respectively. S. Y. and P. R. are thankful to the CSIR for providing the SRF. The authors would express gratitude to CSIR-AMPRI for materials characterization facilities. The Slovenian Research Agency is acknowledged through the funding of projects ARRS N1-0100, J1-1716, P2-0393, BI-US/22-24-162, IsoCont J1-3033, and program P1-0143.References
- A. L. Lorenzen, A. M. dos Santos, L. P. dos Santos, L. da Silva Pinto, F. R. Conceição and F. Wolfart, Electrochim. Acta, 2022, 404, 139757 CrossRef CAS PubMed.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. C. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. W. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS PubMed.
- G. Alhamid, H. Tombuloglu, A. A. Rabaan and E. Al-Suhaimi, Saudi J. Biol. Sci., 2022, 29, 103465 CrossRef CAS PubMed.
- Y. P. Chong, K. W. Choy, C. Doerig and C. X. Lim, Mol. Diagn. Ther., 2023, 27, 303–320 CrossRef PubMed.
- L. J. Carter, L. v Garner, J. W. Smoot, Y. Li, Q. Zhou, C. J. Saveson, J. M. Sasso, A. C. Gregg, D. J. Soares, T. R. Beskid, S. R. Jervey and C. Liu, ACS Cent. Sci., 2020, 6, 591–605 CrossRef CAS PubMed.
- J. Guo, J. Ge and Y. Guo, J. Clin. Lab. Anal., 2022, 36, e24178 CrossRef CAS PubMed.
- M. A. Sadique, S. Yadav, P. Ranjan, S. Verma, S. T. Salammal, M. A. Khan, A. Kaushik and R. Khan, J. Mater. Chem. B, 2021, 9, 4620–4642 RSC.
- A. Parihar, S. Yadav, M. A. Sadique, P. Ranjan, N. Kumar, A. Singhal, V. Khare, R. Khan, S. Natarajan and A. K. Srivastava, Bioeng. Transl. Med., 2023, e10481 CrossRef CAS PubMed.
- D. Namdeo, A. K. Singh, A. Meher, A. K. Yadav and D. Biswas, COVID-19 diagnosis: approaches and challenges, in Advanced Biosensors for Virus Detection, ed. R. Khan, A. Parihar, A. Kaushik and A. Kumar, Academic Press, 2022, pp. 61–77 Search PubMed.
- L. Tessaro, A. Aquino, A. C. de Morais Mirres and C. A. Conte-Junior, Nanotechnology advancements in antiviral coatings to combat viral infection surfaces, in Smart Nanomaterials to Combat the Spread of Viral Infections, ed. R. Khan and M. A. Sadique, Elsevier, 2023, pp. 125–146 Search PubMed.
- S. Yadav, A. Parihar, M. A. Sadique, P. Ranjan, N. Kumar, A. Singhal and R. Khan, ACS Appl. Opt. Mater., 2023, 1(7), 1245–1262 CrossRef CAS.
- G. Simone, ArXiv, 2020, 1–8 Search PubMed.
- C. Wang, X. Yang, B. Gu, H. Liu, Z. Zhou, L. Shi, X. Cheng and S. Wang, Anal. Chem., 2020, 92, 15542–15549 CrossRef CAS PubMed.
- P. E. Guevara-Pantoja, R. Rodriguez-Moncayo, O. G. Chavez-Pineda, D. F. Cedillo-Alcantar, K. B. Reynoso-Hernandez, J. A. Ramirez-Pool, L. A. Nuñez-Muñoz, B. Xoconostle-Cazares and J. L. Garcia-Cordero, Adv. Mater. Technol., 2023, 8, 2200902 CrossRef CAS.
- D. Mandal, M. M. Indaleeb, A. Younan and S. Banerjee, Sens.Biosensing Res., 2022, 37, 100510 CrossRef PubMed.
- P. Fathi-Hafshejani, N. Azam, L. Wang, M. A. Kuroda, M. C. Hamilton, S. Hasim and M. Mahjouri-Samani, ACS Nano, 2021, 15, 11461–11469 CrossRef CAS PubMed.
- B. A. Braz, M. Hospinal-Santiani, G. Martins, J. L. Gogola, M. G. P. Valenga, B. C. B. Beirão, M. F. Bergamini, L. H. Marcolino-Junior, V. Thomaz-Soccol and C. R. Soccol, Talanta, 2023, 257, 124348 CrossRef CAS PubMed.
- B. Della Ventura, M. Cennamo, A. Minopoli, R. Campanile, S. B. Censi, D. Terracciano, G. Portella and R. Velotta, ACS Sens., 2020, 5, 3043–3048 CrossRef PubMed.
- C. Mann, J. S. Hoyle and K. M. Downard, Analyst, 2022, 147, 1181–1190 RSC.
- M. A. Sadique, P. Ranjan, S. Yadav and R. Khan, Advanced high-throughput biosensor-based diagnostic approaches for detection of severe acute respiratory syndrome-coronavirus-2, Computational Approaches for Novel Therapeutic and Diagnostic Designing to Mitigate SARS-CoV2 Infection, Academic Press, 2022, pp. 147–169 Search PubMed.
- R. Kour, S. Arya, S.-J. Young, V. Gupta, P. Bandhoria and A. Khosla, J. Electrochem. Soc., 2020, 167, 37555 CrossRef CAS.
- K. Kerman, M. Saito, E. Tamiya, S. Yamamura and Y. Takamura, TrAC, Trends Anal. Chem., 2008, 27, 585–592 CrossRef CAS.
- S. Yadav, M. A. Sadique, A. Kaushik, P. Ranjan, R. Khan and A. K. Srivastava, J. Mater. Chem. B, 2022, 10, 1146–1175 RSC.
- M. Abubakar Sadique, S. Yadav, P. Ranjan, M. Akram Khan, A. Kumar and R. Khan, Mater. Lett., 2021, 305, 130824 CrossRef CAS PubMed.
- W. Zhang, Y. Qin, W. Wang, F. Liu, F. Meng, F. Chen, N. Zhu, A. Aihaiti and M. Zhang, Electrochim. Acta, 2021, 391, 138970 CrossRef CAS.
- M. A. Sadique, S. Yadav, P. Ranjan, R. Khan, F. Khan, A. Kumar and D. Biswas, ACS Appl. Bio. Mater., 2022, 5, 2421–2430 CrossRef CAS PubMed.
- S. Yadav, M. A. Sadique, P. Ranjan, R. Khan, N. Sathish and A. K. Srivastava, J. Mater. Chem. B, 2022, 10(41), 8478–8489 RSC.
- M. Sadique, S. Yadav, V. Khare, R. Khan, G. K. Tripathi and P. S. Khare, Diagnostics, 2022, 12 Search PubMed.
- M. A. Kachouei, F. Hekmat, H. Wang, G. A. J. Amaratunga, H. E. Unalan and S. Shahrokhian, Electrochim. Acta, 2022, 428, 140952 CrossRef CAS.
- P. Ranjan, M. Abubakar Sadique, S. Yadav and R. Khan, ACS Appl. Mater. Interfaces, 2022, 14, 20802–20812 CrossRef CAS PubMed.
- Y. Wang, J. Ma, X. Ye, W. Wong, C. Li and K. Wu, Electrochim. Acta, 2018, 271, 551–559 CrossRef CAS.
- P. Ranjan, S. Yadav, M. A. Sadique, R. Khan and A. K. Srivastava, ACS Appl. Nano Mater., 2022, 5, 14999–15010 CrossRef CAS.
- A. O. Idris, E. O. Oseghe, T. A. M. Msagati, A. T. Kuvarega, U. Feleni and B. Mamba, Sensors, 2020, 20, 1–28 CrossRef PubMed.
- Q. Hong, H. Yang, Y. Fang, W. Li, C. Zhu, Z. Wang, S. Liang, X. Cao, Z. Zhou, Y. Shen, S. Liu and Y. Zhang, Nat. Commun., 2023, 14, 2780 CrossRef CAS PubMed.
- Y. Hou, Y. Fang, Z. Zhou, Q. Hong, W. Li, H. Yang, K. Wu, Y. Xu, X. Cao, D. Han, S. Liu, Y. Shen and Y. Zhang, Adv. Opt. Mater., 2023, 11, 2202737 CrossRef CAS.
- Y. Fang, Z. Zhou, Y. Hou, C. Wang, X. Cao, S. Liu, Y. Shen and Y. Zhang, Anal. Chem., 2023, 95, 6620–6628 CrossRef CAS PubMed.
- R. Chouhan, J. Gačnik and M. Horvat, in EGU General Assembly Conference Abstracts, AA(Jožef Stefan Institute, Department of Environmental Sciences, Slovenia), AB(Jožef Stefan Institute, Department of Environmental Sciences, Slovenia), AC(Jožef Stefan Institute, Department of Environmental Sciences, Slovenia), 2022, pp. EGU22–8308.
- L. G. da S. Catunda, T. Martimiano do Prado, T. R. de Oliveira, D. J. Almeida dos Santos, N. O. Gomes, D. S. Correa, R. C. Faria and S. A. S. Machado, Electrochim. Acta, 2023, 451, 142271 CrossRef CAS PubMed.
- A. V. Police Patil, Y.-S. Chuang, C. Li and C.-C. Wu, Biosensors, 2023, 13, 125 CrossRef CAS PubMed.
- N. Singh, D. S. Dkhar, P. Chandra and U. P. Azad, Biosensors, 2023, 13, 166 CrossRef CAS PubMed.
- A. Koyappayil, A. K. Yagati and M. H. Lee, Biosensors, 2023, 13, 91 CrossRef CAS PubMed.
- R. S. Chouhan, I. Jerman, D. Heath, S. Bohm, S. Gandhi, V. Sadhu, S. Baker and M. Horvat, Nano Select, 2021, 2, 712–743 CrossRef CAS.
- Q. Cao, B. Kumru, M. Antonietti and B. V. K. J. Schmidt, Mater. Horiz., 2020, 7, 762–786 RSC.
- C. Zhao, Q. Yan, S. Wang, P. Dong and L. Zhang, RSC Adv., 2018, 8, 27516–27524 RSC.
- S. Amirthalingam and J. Rangasamy, in Advances in Polymer Science, ed. R. Jayakumar and M. Prabaharan, Springer International Publishing, Cham, 2021, vol. 287, pp. 233–255 Search PubMed.
- W. Suginta, P. Khunkaewla and A. Schulte, Chem. Rev., 2013, 113, 5458–5479 CrossRef CAS PubMed.
- Y. Jiang and J. Wu, Electrophoresis, 2019, 40, 2084–2097 CrossRef CAS PubMed.
- M. Amouzadeh Tabrizi, L. Nazari and P. Acedo, Sens. Actuators, B, 2021, 345, 130377 CrossRef CAS PubMed.
- T. Yin, Y. Ye, W. Dong and G. Jie, Biosens. Bioelectron., 2022, 215, 114580 CrossRef CAS PubMed.
- C. N. Botelho, S. S. Falcão, R.-E. P. Soares, S. R. Pereira, A. S. de Menezes, L. T. Kubota, F. S. Damos and R. C. S. Luz, Biosens. Bioelectron X, 2022, 11, 100167 CAS.
- C. Karaman, B. B. Yola, O. Karaman, N. Atar, İ. Polat and M. L. Yola, Mikrochim. Acta, 2021, 188, 425 CrossRef CAS PubMed.
- R. S. Chouhan, G. Žitko, V. Fajon, I. Živković, M. Pavlin, S. Berisha, I. Jerman, A. Vesel and M. Horvat, Sensors, 2019, 19, 3432 CrossRef CAS PubMed.
- D. C. Marcano, D. v Kosynkin, J. M. Berlin, A. Sinitskii, Z. Sun, A. S. Slesarev, L. B. Alemany, W. Lu and J. M. Tour, ACS Nano, 2018, 12, 2078 CrossRef CAS PubMed.
- J. Wang, B. Yang, S. Li, B. Yan, H. Xu, K. Zhang, Y. Shi, C. Zhai and Y. Du, J. Colloid Interface Sci., 2017, 506, 329–337 CrossRef CAS PubMed.
- C. Guo, C. Wang, H. Sun, D. Dai and H. Gao, RSC Adv., 2021, 11, 29590–29597 RSC.
- R. Maharsi, A. F. Arif, T. Ogi, H. Widiyandari and F. Iskandar, RSC Adv., 2019, 9, 27896–27903 RSC.
- I. O. Faniyi, O. Fasakin, B. Olofinjana, A. S. Adekunle, T. V. Oluwasusi, M. A. Eleruja and E. O. B. Ajayi, SN Appl. Sci., 2019, 1, 1181 CrossRef CAS.
- N. Sharma, R. Vyas, V. Sharma, H. Rahman, S. K. Sharma and K. Sachdev, Appl. Nanosci., 2020, 10, 517–528 CrossRef CAS.
- M. Ruidíaz-Martínez, M. A. Álvarez, M. V. López-Ramón, G. Cruz-Quesada, J. Rivera-Utrilla and M. Sánchez-Polo, Catalysts, 2020, 10, 520 CrossRef.
- H. Zhang, L. Peng, A. Chen, C. Shang, M. Lei, K. He, S. Luo, J. Shao and Q. Zeng, Carbohydr. Polym., 2019, 214, 276–285 CrossRef CAS PubMed.
- S. Anicuta, L. Dobre, M. Stroescu and I. Jipa, Analele Universitatii din Oradea Fascicula: Ecotoxicologie, Zootehnie si Tehnologii de Industrie Alimentara, 2010, 1234–1240.
- Y. Fu, J. Zhu, C. Hu, X. Wu and X. Wang, Nanoscale, 2014, 6, 12555–12564 RSC.
- S. Zhang, N. T. Hang, Z. Zhang, H. Yue and W. Yang, Nanomaterials, 2017, 7, 1–11 Search PubMed.
- C. C. Ryan, M. Bardosova and M. E. Pemble, J. Mater. Sci., 2017, 52, 8338–8347 CrossRef CAS.
- D. Adekoya, M. Li, M. Hankel, C. Lai, M. S. Balogun, Y. Tong and S. Zhang, Energy Storage Mater., 2020, 25, 495–501 CrossRef.
- M. Azmana, S. Mahmood, A. R. Hilles, A. Rahman, M. A. Bin Arifin and S. Ahmed, Int. J. Biol. Macromol., 2021, 185, 832–848 CrossRef CAS PubMed.
- S. Yadav, M. A. Sadique, P. Ranjan and R. Khan, Anal. Chim. Acta, 2023, 341326 CrossRef CAS PubMed.
- B. P. Charitha and P. Rao, Int. J. Biol. Macromol., 2018, 112, 461–472 CrossRef CAS PubMed.
- D. Debnath, S. H. Kim and K. E. Geckeler, J. Mater. Chem., 2009, 19, 8810–8816 RSC.
- J. Zhao, Z. Fu, H. Li, Y. Xiong, S. Cai, C. Wang, Y. Chen, N. Han and R. Yang, Electrochim. Acta, 2022, 404, 139766 CrossRef CAS PubMed.
- D. Najjar, J. Rainbow, S. Sharma Timilsina, P. Jolly, H. De Puig, M. Yafia, N. Durr, H. Sallum, G. Alter, J. Z. Li, X. G. Yu, D. R. Walt, J. A. Paradiso, P. Estrela, J. J. Collins and D. E. Ingber, Nat. Biomed. Eng., 2022, 6, 968–978 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, synthesis of GO, nanomaterial characterization (AFM, XPS, SEM), scan rate studies. See DOI: https://doi.org/10.1039/d3ma00399j |
| This journal is © The Royal Society of Chemistry 2023 |