Open Access Article
Open Access ArticleA dynamic DNA nanosponge for triggered amplification of gene-photodynamic modulation†
Dan
Luo‡
,
Xue
Lin‡
,
Yun
Zhao
,
Jialing
Hu
,
Fengye
Mo
,
Gege
Song
,
Zhiqiao
Zou
,
Fuan
Wang
 and
Xiaoqing
Liu
and
Xiaoqing
Liu
 *
*
College of Chemistry and Molecular Sciences, Wuhan University, 430072 Wuhan, P. R. China. E-mail: xiaoqingliu@whu.edu.cn
First published on 28th March 2022
Abstract
Nucleic acid therapeutics has reached clinical utility through modulating gene expression. As a potential oligonucleotide drug, DNAzyme has RNA-cleaving activity for gene silencing, but faces challenges due to the lack of a safe and effective delivery vehicle and low in vivo catalytic activity. Here we describe DNAzyme-mediated gene regulation using dynamic DNA nanomaterials with intrinsic biocompatibility, stability, tumor-targeted delivery and uptake, and self-enhanced efficacy. We assemble programmable DNA nanosponges to package and deliver diverse nucleic acid drugs and therapeutic agents such as aptamer, DNAzyme and its cofactor precursor, and photosensitizer in one pot through the rolling circle amplification reaction, formulating a controllable nanomedicine using encoded instructions. Upon environmental stimuli, DNAzyme activity increases and RNA cleavage accelerates by a supplementary catalytic cofactor. In addition, this approach induces elevated O2 and 1O2 generation as auxiliary treatment, achieving simultaneously self-enhanced gene-photodynamic cancer therapy. These findings may advance the clinical trial of oligonucleotide drugs as tools for gene modulation.
Introduction
Nucleic acids can be used to modulate gene expression for treating or preventing diseases.1–4 In very recent years, oligonucleotide therapeutics has received numerous regulatory approvals.5–7 As an ideal oligonucleotide drug, the RNA-cleaving DNA enzyme (DNAzyme) is a cofactor-dependent DNA that can site-specifically cleave disease genes, and features low-cost synthesis, flexible design, and high specificity.8–10 Compared with other gene regulation approaches, the cleavage activity of DNAzyme does not require accessory cellular proteins, showing a simple, controllable, and stable catalytic cleavage.11–13 However, the efficient and safe oligonucleotide delivery and the in vivo cleavage activity of DNAzyme remain two major challenges for its applications in clinical trials.Specifically, due to the low biological stability and non-specific distribution of oligonucleotides under physiological conditions, the inefficient delivery of DNAzyme is an obstacle to clinical applicability.14,15 To tackle this challenge, various delivery vehicles16–18 have been explored, mainly viral vectors, liposomes,19 inorganic nanoparticles,20,21 and cationic polymers.22 These delivery approaches protect the nucleic acids from degradation and enhance cellular uptake, yet often suffer from potential cytotoxicity or off-target effects.23,24 Even some vectors show improved delivery ability; however, due to the insufficient and inefficient cofactor supply in live cells and tissues, the in vivo catalytic efficacy of DNAzyme is restricted.25,26 This results from the fact that the enzymatic activity of DNAzyme is strongly dependent on the level and type of enzyme cofactor, but the DNAzyme delivery systems mostly rely on the intracellular Mg2+ ion (0.2–2 mM) as the cofactor, failing to reach an ideal level (5 mM) for high catalytic efficiency.27 Besides, the Mg2+ ion is not usually the most efficient metal ion cofactor that can promote the highest DNAzyme cleavage activity.28,29 It has been reported that a low level of Mn2+ ions (0.2 mM) is sufficient to trigger a complete substrate cleavage, while 15 mM Mg2+ ions are needed under the same situation.30 To overcome this limitation, we and others30,31 recently demonstrated a cofactor self-sufficient method via DNA adsorption onto pre-synthesized particles, yet common to the above gene delivery procedures are the limited cargo loading amount and the requirement of multiple-step and labor-intensive synthesis. As such, the existing DNAzyme carriers and their therapeutics can be inefficient and nonspecific.
Advanced DNA materials show promise to overcome these limitations. Beyond being a genetic material for life, DNA is a building block to create new materials, in particular, assembling or organizing nanomaterials with inherent biocompatibility, complexity and function in a programmable way.32–34 Particularly, DNA materials can be encoded with versatile functional oligonucleotide sequences, integrated with unique nanoparticles or molecules of interest,35–37 and endowed with responsiveness to chemical, biological, and physical cues.38–41 These aspects make them emerging delivery vehicles for oligonucleotide drugs and therapeutics. We envisage that a new generation of dynamic DNA materials are ready to effectively package and deliver DNAzyme with perfect compatibility, convenience, and precision in a controllable manner; however, there is a lack of exploration. We hypothesize that assembled DNA nanostructures comprising DNAzyme and its cofactor precursor could enhance cleavage activity, and the corresponding therapeutics may be controlled in a programmable manner using encoded instructions in response to signal molecules or stimuli.
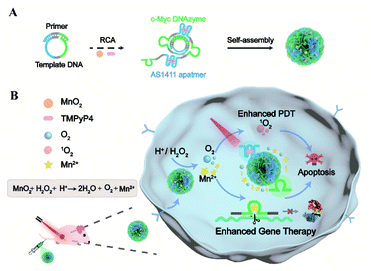
Here we present a nucleic acid delivery approach with biomimetic DNA nanoassemblies in a controllable and self-enhanced manner for gene-photodynamic regulation (Scheme 1). This is achieved with DNA drug-encoded and therapeutic-incorporated dynamic nanosponges via one-pot synthesis using the rolling circle amplification (RCA) reaction, incorporating aptamer, DNAzyme and its cofactor precursor. Upon DNA-mediated crystallization, the synthetic material is encoded with copious different functional oligonucleotides and intercalated photosensitizers or encapsulated particles. Such a densely packed composition ensures large loading capacity and high biological stability. The multivalent nanosponges with AS1411 aptamers allow specific recognition, delivery, and internalization of the Mn2+-dependent DNAzyme. After tumor targeting by the aptamer, the entrapped MnO2 reacts with acidic H2O2 to generate Mn2+ and oxygen in the tumor microenvironment. This self-supplied sufficient and effective DNAzyme cofactors, which is vital for effective silencing of target mRNA in tumor cells. In addition, the stimulus-triggered, self-supplied O2 enhances photosensitizer-mediated ROS generation for auxiliary photodynamic therapy (PDT). This dynamic DNA material achieves self-enhanced gene silencing and self-amplified photodynamic therapy with high tumor specificity, providing a safe and effective delivery approach for DNAzyme based gene regulation and cancer therapy.
Results and discussion
One-pot synthesis and characterization of dynamic DNA nanosponges by biomimetic crystallization
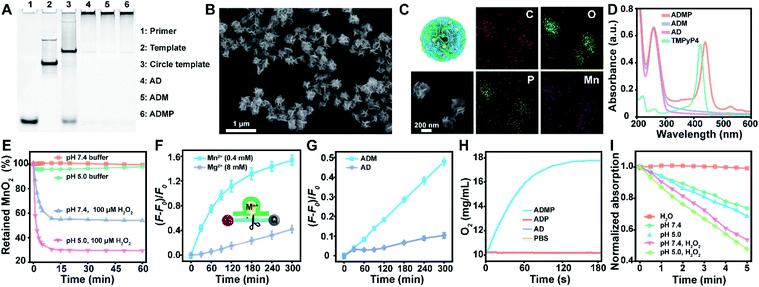
With a designed template DNA comprising sequences complementary to the AS1411 aptamer and Mn2+-dependent DNAzyme, the RCA reaction proceeded in the presence of the DNAzyme cofactor precursor (MnO2) and porphyrin photosensitizer (TMPyP4) (Fig. S1†) via one-pot synthesis (Scheme 1A and Table S1†). The encoded AS1411 aptamer not only acts as a targeting agent to specifically recognize the nucleolin of cancer cells for precise drug delivery, but also offers the binding site to intercalate the photosensitizer. The encoded DNAzyme is a catalytic oligonucleotide that can specifically recognize and then cleave its substrate sequence in the presence of DNAzyme cofactors. Here c-Myc mRNA, one of the proto-oncogenes identified in many cancers,42 is used as an example of target gene. A porous DNA nanoassembly can be formed via anisotropic nucleic acid-driven crystallization,43,44 and the DNAzyme cofactor precursor, MnO2 nanocluster, is spontaneously incorporated into the growing polymer DNA strands and inorganic crystals produced during the RCA process. This enables high loading efficiency and enhanced stability of the respective functional components and drugs, and effective embedding of the enzyme cofactor precursor. Besides, such an approach involves pretty simple and moderate reaction conditions compared to the labor-intensive or postsynthetic modifications related to the preparation of multifunctional DNA nanostructures.45After confirming the successful generation of the circular template and high molecular weight RCA product by native polyacrylamide gel electrophoresis (PAGE) (Fig. 1A), the morphology and size of the particles were investigated by scanning electron microscopy (SEM) and dynamic light scattering (DLS). The SEM image indicated that the DNA nanoassembly incorporating the aptamer, DNAzyme, MnO2, and photosensitizer (ADMP) was spherical in shape with sponge-like porosities and uniformly sized with a 350 nm diameter (Fig. 1B), which was consistent with the DLS results (Table S2†). Then, we calculated the average replicated copies for each template DNA (Table S3†). The high replication efficiency of the RCA method verifies that the DNA nanosponge delivery vehicles ensure functional nucleic acid cargo delivery. Such a monodisperse and densely packed DNA nanostructure may be the result of the porous structure of the assembly and multiple interactions such as van der Waals forces and inner-sphere coordination between the MnO2 nanocluster and nucleic acid.46–48
MnO2 provides Mn2+ and O2 upon tumor microenvironment stimuli, which plays an indispensable role in the catalytic reaction of DNAzyme and therapy enhancement. During the RCA process, the successful incorporation of MnO2 into the polymer DNA was verified using energy dispersive X-ray spectroscopy (EDX) spectra (Fig. S2†) and the corresponding element mapping (Fig. 1C), indicating the presence of homogeneously scattered carbon (C), oxygen (O), phosphorus (P), and manganese (Mn). Besides, X-ray photoelectron spectra (XPS) illustrated that the split energy of Mn 2p3/2 and Mn 2p1/2 was 11.72 eV (Fig. S3A†), which also indicated that the oxidation state of Mn was +4 in ADMP.49 Simultaneously, the mass ratio between the DNA nanosponge and MnO2 was calculated to be 1 mg : 47.3 μg by inductively coupled plasma mass spectrometry (ICP-MS) analysis. These results validated that the one-pot synthesized nanocarriers could co-deliver DNAzyme and its cofactor precursor for self-sufficient gene regulation. Besides MnO2 encapsulation, the successful loading of photosensitizer TMPyP4 in the RCA nanoassemblies was confirmed by UV-vis absorption spectra (Fig. 1D). Compared with the control formulations including DNA nanoassemblies incorporated only with the aptamer and DNAzyme (AD), and DNA nanoassemblies incorporated only with the aptamer, DNAzyme, and MnO2 (ADM), ADMP showed a classic absorption peak similar to TMPyP4. A high encapsulation efficiency of 91.4% and a drug loading capability of 11.4% (wt/wt) were obtained when 0.1 mM TMPyP4 was used (Table S4†).
As a potential drug vehicle, remaining stable under complex physiological conditions is crucial. The stability of the ADMP was evaluated by incubating them in PBS, DNase I (1 U mL−1), and DMEM (10% serum) for 24 h (Table S5 and Fig. S4†). As revealed by DLS and PAGE, the nanoassemblies exhibited high stability, which is essential for drug delivery and therapy.
Enhanced catalytic activity of DNAzyme through the self-supplied Mn2+ cofactor in the presence of signal molecules
To prove the acid/H2O2 responsiveness of the nanosponges, the degradation behavior of the encapsulated MnO2 in DNA nanoassemblies was explored at different pH and H2O2. The degradation of MnO2 could be reflected by the disappearance of its typical absorbance peak at 350 nm (Fig. 1E). Time-dependent absorbance showed that MnO2 was rather stable under physiological conditions (pH = 7.4); in contrast, MnO2 decomposed rapidly in acidic buffer (pH = 5.0) with H2O2. These data were consistent with the elemental (Mn) analysis by XPS (Fig. S3B†) and ICP-MS (Fig. S5†).Such reactivity upon acidic H2O2 stimuli releases Mn2+ ions, which are effective cofactors of DNAzyme for triggering substrate RNA cleavage. The cleavage activity of the DNA nanoassemblies composed of DNAzyme was then investigated by PAGE and fluorescence analysis. We first added exogenous metal ions either Mn2+ or Mg2+ ions at different concentrations to AD, a DNA nanoassembly incorporated with the aptamer and DNAzyme, in the presence of substrate RNA (Fig. S6, S7† and 1F). Denaturing PAGE analysis indicated that 0.4 mM of Mn2+ ions were sufficient to trigger effective substrate cleavage (cleavage efficiency, 85.61%), while 8 mM of Mg2+ ions only induced a cleavage efficiency of 55.23% (Fig. S6 and S7†). Similarly, with the extension of cleavage time, the fluorescence intensity of substrate RNA labeled with both a fluorophore and quencher was significantly increased when 0.4 mM of Mn2+ ions were used as the cofactor, while 8 mM of Mg2+ resulted in limited fluorescence change (Fig. 1F). These results suggest that Mn2+ ions could greatly enhance hydrolysis activity, and promote a dramatically higher DNAzyme activity than Mg2+ ions.
To verify the self-enhanced cleavage activity, we designed and took ADM, a DNA nanoassembly incorporated with the aptamer, DNAzyme and MnO2 nanocluster, as an example (Fig. S8† and 1G). Both denaturing PAGE and fluorescence results demonstrated efficient substrate cleavage by ADM in an acidic H2O2-rich medium, and stronger cleavage was obtained with an elevated amount of ADM (Fig. S8†). Comparison of time-dependent fluorescence changes of ADM and AD indicated that the DNA nanoassembly without MnO2 (AD) had a negligible cleavage capacity, while ADM had a drastic catalytic activity (Fig. 1G). This resulted from the fact that stimuli of acidic H2O2 on ADM induce sufficient Mn2+ generation, and these self-supplied Mn2+ ions act as enzyme cofactors to increase enzymatic activity. All the data indicate that the one-pot synthesized DNA nanosponges have promising potential in self-promoted RNA-cleaving activity.
Elevated 1O2 generation via the acidic H2O2-triggered O2 production
As a crucial component, MnO2 can not only be reduced into Mn2+ as an efficient DNAzyme cofactor for gene regulation, but also generate oxygen for alleviating hypoxia and further enhance PDT that requires the interaction of light, a photosensitizer, and molecular oxygen. Correspondingly, O2 generation under acidic H2O2 conditions was investigated using an oxygen electrode (Fig. 1H). As expected, ADMP showed a significant increase in oxygenation. In contrast, AD and the DNA nanoassembly incorporated with the aptamer, DNAzyme, and photosensitizer (ADP) only led to background signals. This clearly evidenced that due to the incorporated MnO2, ADMP could catalyze H2O2 into O2.To verify the nanoassembly-mediated photocytotoxic reactions, we evaluated singlet oxygen (1O2) production by the activated photosensitizer using 2,2,6,6-tetramethylpiperidine (TEMP) and 1,3-diphenylisobenzofuran (DPBF) (Fig. S9, S10† and 1I). Upon laser irradiation, the typical 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 triplet signal characteristic was measured by electron spin resonance (ESR) spectroscopy, which indicated that the reactive oxygen species produced by TMPyP4, ADP and ADMP were 1O2.50 The DPBF could be oxidized by 1O2 which caused a decrease of its characteristic absorbance peak at 410 nm. The maximum absorption of DPBF was decreased to 73.72% within 5 min upon exposing ADMP to light irradiation under neutral conditions (pH = 7.4). Comparatively, the decrease of DPBF absorption accelerated significantly after the addition of H2O2, particularly in acidic media (48.01%), indicating an elevated generation of 1O2, which was ascribed to the generation of oxygen under acidic H2O2 conditions. Besides, compared with free TMPyP4, ADMP showed improved 1O2 generation efficiency (Fig. S11†). Thus, these results demonstrated that ADMP could be utilized as a multifunctional and simultaneously self-enhanced therapeutic nanoplatform for gene silencing and auxiliary PDT.
1 triplet signal characteristic was measured by electron spin resonance (ESR) spectroscopy, which indicated that the reactive oxygen species produced by TMPyP4, ADP and ADMP were 1O2.50 The DPBF could be oxidized by 1O2 which caused a decrease of its characteristic absorbance peak at 410 nm. The maximum absorption of DPBF was decreased to 73.72% within 5 min upon exposing ADMP to light irradiation under neutral conditions (pH = 7.4). Comparatively, the decrease of DPBF absorption accelerated significantly after the addition of H2O2, particularly in acidic media (48.01%), indicating an elevated generation of 1O2, which was ascribed to the generation of oxygen under acidic H2O2 conditions. Besides, compared with free TMPyP4, ADMP showed improved 1O2 generation efficiency (Fig. S11†). Thus, these results demonstrated that ADMP could be utilized as a multifunctional and simultaneously self-enhanced therapeutic nanoplatform for gene silencing and auxiliary PDT.
Targeted and intensified intracellular gene silencing and ROS generation with self-sufficient therapeutic DNA nanosponges in response to environmental cues
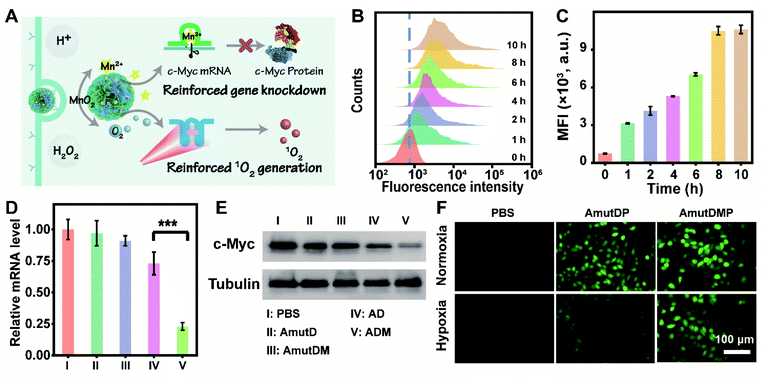
The performance of the dynamic DNA nanomaterials encoded with diverse functionalities and responsiveness was then investigated in live cells (Fig. 2A). Targeted delivery to specific tumor cells is a prerequisite for cancer therapy, and we first tested the cellular uptake capacity of ADMP by flow cytometry and confocal laser scanning microscopy (CLSM). As the encoded AS1411 aptamer in ADMP can specifically recognize the nucleolin of cancer cells, we used 4T1 cells (mouse breast cancer cell line, nucleolin overexpressed) and MCF-10A (human normal breast cell line, low nucleolin expression) as the target and control cells, respectively. Upon incubating 4T1 cells with Cy5-labeled ADMP, the fluorescence intensity of the cells gradually increased and reached a saturation point at 8 h (Fig. 2B and C). Moreover, confocal imaging analysis demonstrated that the fluorescence signal was stronger in cancer cells than in normal cells (Fig. S12†). These results confirmed that the DNA nanoassemblies encoded with the AS1411 aptamer can specifically recognize and target cancer cells, and then achieve selective cellular uptake. To further examine the enhanced uptake mediated by the specific binding between AS1411 and nucleolin, the 4T1 cells were preincubated with an excess amount of free AS1411 for 1 h to block the nucleolin acceptor and then treated with Cy5-labeled ADMP under the same conditions. A noticeably lower fluorescence signal was observed in 4T1 cells than that in an unblocked group under CLSM (Fig. S13†), confirming the reliable cancer cell-specific uptake. Moreover, the intracellular distribution of ADMP was evaluated by incubating 4T1 cells with Cy5-labeled ADMP. After incubation for 6 h, a Pearson's correlation coefficient of 0.78 was calculated from the colocalization of ADMP and lysosomes, which indicated that Cy5-labeled ADMP could be localized in lysosomes after internalization by 4T1 cells (Fig. S14†). As time extended, ADMP could eventually escape from the lysosomes.Next, we explored the DNAzyme-mediated, cofactor-self-supplied mRNA cleavage and O2-evolving 1O2 generation upon light illumination in 4T1 cells. Here c-Myc mRNA, a proto-oncogene, was used as an example of target gene to cleave. Down-regulation of c-Myc mRNA could inhibit the progression of a wide range of cancers and has been identified in many different types of cancers such as breast cancer.42 As demonstrated before, the encapsulated MnO2 in ADM could be decomposed into Mn2+ ions in the tumor microenvironment, which acted as DNAzyme cofactors to improve substrate cleavage efficiency. Correspondingly, we designed control nanoassembly AD (no MnO2 encapsulated) to verify the role of MnO2 in ADM for gene silencing. Meanwhile, to examine the RNA-cleaving activity of DNAzyme, we used mutant DNAzyme (mutD) and designed control nanoassemblies AmutD and AmutDM, which had no DNAzyme activity and were unable to cleave the mRNA substrate.
After incubating with different materials, the relative expression of c-Myc mRNA in 4T1 cells was investigated using quantitative real-time polymerase chain reaction (qRT-PCR) assay (Fig. 2D). The mRNA level of c-Myc was successfully inhibited by 77% after the treatment with ADM. As crucial controls, the catalytically inactive AmutD and AmutDM showed negligible influence on the expression of c-Myc mRNA in 4T1 cells, while cofactor deficient AD reduced the mRNA level of c-Myc mRNA by 27%, showing a weaker mRNA-cleavage ability than ADM. Besides, the expression of protein c-Myc was monitored using western blot (Fig. 2E), and the result was consistent with that of the qRT-PCR assay. These data demonstrated strongly that the crystalized DNAzyme/MnO2 nanosponges could efficiently cleave their target mRNA, as this approach offers a prospective gene regulation that self-supplied effective enzyme cofactors and copious therapeutic DNAzymes.
To explore the feasibility of auxiliary PDT, we further designed a DNA nanoassembly incorporated with the aptamer, mutant DNAzyme and photosensitizer (AmutDP), and a DNA nanoassembly incorporated with the aptamer, mutant DNAzyme, MnO2, and photosensitizer (AmutDMP) and evaluated the intracellular ROS generation under illumination (Fig. 2F). A ROS probe, 2′,7′-dichlorofluorescein diacetate (DCFH-DA), which could be oxidized by 1O2 and generate a fluorescence indicator DCF, was applied to monitor 1O2 generation in 4T1 cells. It should be noted that under normoxic conditions (21% O2) the oxygen level is enough for 1O2 production. Therefore, upon 660 nm laser irradiation, both AmutDP- and AmutDMP-treated 4T1 cells displayed bright green fluorescence under normoxia. However, under hypoxic conditions (1% O2), the cells incubated with AmutDMP showed an evidently stronger fluorescence than those treated with AmutDP, suggesting that the encapsulated MnO2 in DNA nanosponges self-supplied oxygen in situ, and further induced elevated 1O2 generation. Thus, the auxiliary photocytotoxic reaction of the DNA nanosponges could be utilized for enhancing the therapeutic effect.
Simultaneously enhanced and synergistic gene-photodynamic therapy in vitro
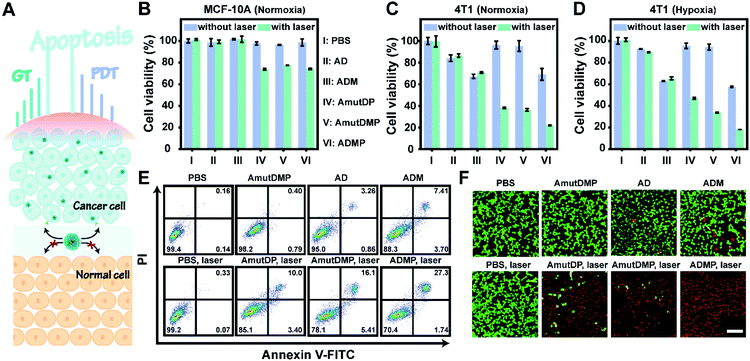
The enhanced gene silencing and intensified ROS generation in cancer cells using the dynamic materials provided an opportunity for tumor-specific treatment with high efficacy; subsequently, we evaluated the in vitro synergistic gene-photodynamic therapy (Fig. 3A). First, cell viability was determined with cell counting kit-8 (CCK-8) assays. In the dark, treating 4T1 cells with different concentrations of AmutDMP caused negligible phototoxicity (Fig. S15†). Besides, when using nucleolin-negative MCF-10A cells as control cells, lower cytotoxicity was obtained after incubating with different DNA materials with or without light exposure (Fig. 3B). These results indicated satisfactory biocompatibility of the nanoparticles.Then the therapeutic performance of the DNA nanoassemblies was evaluated in vitro by comparing the viability of 4T1 cells incubated with different materials with or without irradiation including PBS (I), AD (II), ADM (III), AmutDP (IV), AmutDMP (V), and ADMP (VI) (Fig. 3C and D). Consistent with the results of qRT-PCR and western blot, the cytotoxicity of the AD-treated group was obviously weaker than that of ADM. This is because the MnO2-encapsulated DNAzyme nanoassemblies could release cofactor Mn2+ ions under the tumor microenvironment (acid/H2O2 conditions), to enhance gene therapy (GT) effectively. Moreover, when exposed to laser irradiation, the photocytotoxicity of AmutDP and AmutDMP was similar under normoxia; however, the cytotoxicity of AmutDP was obviously lower than that of the latter under hypoxia, demonstrating that the O2 generated from MnO2 could relieve tumor hypoxia and enhance PDT efficacy. In addition, among all the materials ADMP with activated gene regulation and the auxiliary photodynamic effect caused the highest cell death after 660 nm photoirradiation. Subsequently, the cytotoxicity of ADMP was further evaluated at various concentrations both in the dark and with light exposure (Fig. S16†). A dose-dependent cytotoxicity was observed in the dark via gene regulation, while the viability of 4T1 cells was further decreased once exposed to laser irradiation, suggesting a synergistic gene-photodynamic therapy. Secondly, the in vitro therapeutic performance of ADMP was investigated by flow cytometry assay and live/dead cell staining (Fig. 3E and F). Comparisons of the extent of apoptosis in 4T1 cells using different materials and conditions suggested most remarkable cytotoxicity of ADMP treatment. Also, the determined live/dead ratio of tumor cells with different treatments proved a maximum effect by cofactor self-sufficient gene regulation and O2-supplied photodynamic therapy.
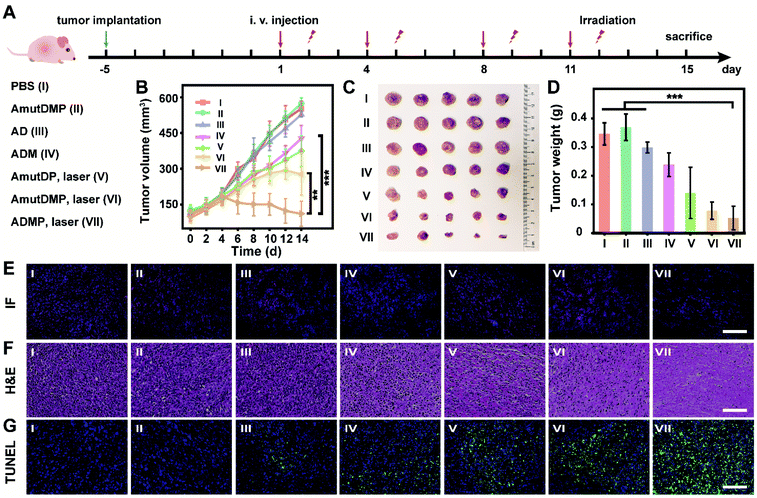
Effective and safe cancer therapy in 4T1 tumor-bearing mice using the self-sufficient DNA nanoassemblies
In 4T1 tumor-bearing BALB/c mice, the in vivo antitumor efficacy of DNA nanosponges was explored using seven mice groups (n = 5) with different treatments (Fig. 4A). As shown in Fig. 4B, neither PBS (group I) nor AmutDMP (group II) could inhibit tumor growth. However, the mice treated with AD (group III) and ADM (group IV) had a relatively slow tumor growth rate, demonstrating the gene regulation effect. Moreover, the treatments with AmutDP plus laser (group V) and AmutDMP plus laser (group VI) effectively inhibited tumor growth, ascribed to the photodynamic therapy. Furthermore, the superior tumor restraining of ADM (group IV) over AD (group III) well-illustrated the MnO2-enhanced gene silencing. Meanwhile, when exposed to laser irradiation, an enhanced PDT performance was observed with AmutDMP (group VI) than with AmutDP (group V) under hypoxic conditions, revealing that the oxygen generated from these nanosponges could promote the generation of 1O2. Among all the groups, the mice treated with ADMP under irradiation (group VI) had the maximum anti-tumor inhibition effect, which was ascribed to the cooperative effort between the DNAzyme cofactor self-sufficient gene regulation and O2-supplement photodynamic therapy. All the data were in accord with the results of size and weight changes of tumors after treatment (Fig. 4C and D).Pathological studies further confirmed the therapeutic outcome (Fig. 4E–G). Immunofluorescence (IF) images displayed a weaker red emission of tumor slices treated with ADM (group IV) than those treated with AD (group III) (Fig. 4E), indicating that the MnO2-encapsulated DNAzyme nanoassemblies could inhibit the expression of c-Myc protein via the self-supplied gene regulation. Subsequently, the pathological morphology of tumor tissue was investigated by hematoxylin-eosin (H&E) staining (Fig. 4F). There was a more severe destruction and more disintegrated nuclei in the tumor cells treated with photoirradiated ADMP, but only partial fibrosis was obtained for the ADM or photoirradiated AmutDMP group. These observations indicated that the combinational therapy achieved the best tumor regression efficacy. Similarly, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining demonstrated that our therapy (group VII) induced the most drastic level of tumor cell apoptosis (Fig. 4G). Finally, we confirmed the tumor-targeting ability by fluorescence imaging using different materials (Fig. S17†), which was ascribed to the aptamer-mediated targeting and the enhanced permeability and retention (EPR) effects. Furthermore, we evaluated the biosafety of the therapy by long-term body weight monitoring, hemolysis test, H&E staining of major organs, and biochemical index analysis, and observed no systemic toxicity (Fig. S18–S21†). These observations indicated that the therapeutic nanoassemblies possess excellent biocompatibility and may be promising for use in clinical applications.
Conclusions
Overall, RNA-cleaving DNAzyme holds great promise for gene regulation, but its clinical utility is hindered because of the inefficient delivery and limited enzymatic activity in vivo. To this end, we have assembled a dynamic DNA nanomaterial as a safe and effective delivery vehicle that can improve catalytic cleavage efficacy and targeting specificity in response to environmental cues. These DNA nanoassemblies can be programmed to package and deliver diverse cargos via one-pot synthesis, leading to densely packed Mn2+-dependent DNAzyme and its cofactor precursor for better stability and DNAzyme activity. In addition, the vehicle is encoded with an aptamer and porphyrin photosensitizer as auxiliary molecular therapeutics for specific and effective cancer cell uptake and apoptosis. Upon tumor microenvironment stimuli, the programmable DNA materials not only self-supply the enzyme cofactor but also generate O2in situ, which leads to targeted and intensified mRNA cleavage and photosensitizer-mediated ROS generation. These data demonstrate a simultaneously self-enhanced gene silencing and self-enhanced adjuvant PDT both in vitro and in vivo. Our study provides a safe and targeted delivery of therapeutics for cancer treatment with programmable and dynamic DNA materials. This delivery approach may improve the utility and translation of DNAzyme to regulate disease-related genes. Furthermore, the delivery platform can be programmed to encode different oligonucleotide drugs and cargos of interest for disease prevention and treatment.Ethical statement
All animal procedures were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Use Committee of the Animal Experiment Center/Animal Biosafety Level-III Laboratory of Wuhan University (license number: WP20210450).Data availability
All data supporting this study are included in ESI.†Author contributions
Conceptualization, X. Liu and D. Luo; investigation, D. Luo, X. Lin and J. Hu; methodology, D. Luo, X. Lin and Y. Zhao; resources, F. Mo, G. Song, Z. Zou and F. Wang; writing – original draft, D. Luo and X. Lin; visualization, D. Luo and Y. Zhao; writing review & editing, X. Liu, D. Luo and X. Lin; project administration, X. Liu; funding acquisition, X. Liu; supervision, X. Liu.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (21874103 and 22074112).Notes and references
- J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt, P. R. Cullis and R. van der Meel, Nat. Nanotechnol., 2021, 16, 630–643 CrossRef CAS PubMed.
- C. E. Dunbar, K. A. High, J. Keith Joung, D. B. Kohn, K. Ozawa and M. Sadelain, Science, 2018, 359, eaan4672 CrossRef.
- K. A. High and M. G. Roncarolo, N. Engl. J. Med., 2019, 381, 455–464 CrossRef CAS.
- G. Rancati, J. Moffat, A. Typas and N. Pavelka, Nat. Rev. Genet., 2018, 19, 34–49 CrossRef CAS.
- T. C. Roberts, R. Langer and M. J. A. Wood, Nat. Rev. Drug Discovery, 2020, 19, 673–694 CrossRef CAS.
- J. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS.
- H. Ledford, Nature, 2018, 560, 291–292 CrossRef CAS PubMed.
- S. K. Silverman, Trends Biochem. Sci., 2016, 41, 595–609 CrossRef CAS PubMed.
- A. A. Fokina, D. A. Stetsenko and J.-C. François, Expert Opin. Biol. Ther., 2015, 15, 689–711 CrossRef CAS PubMed.
- Q. Wang, K. Tan, H. Wang, J. Shang, Y. Wan, X. Liu, X. Weng and F. Wang, J. Am. Chem. Soc., 2021, 143, 6895–6904 CrossRef CAS PubMed.
- H. K. Karnati, R. S. Yalagala, R. Undi, S. R. Pasupuleti and R. K. Gutti, Tumor Biol., 2014, 35, 9505–9521 CrossRef CAS PubMed.
- Y. Wang, K. Nguyen, R. C. Spitale and J. C. Chaput, Nat. Chem., 2021, 13, 319–326 CrossRef CAS PubMed.
- M. Liu, D. Chang and Y. Li, Acc. Chem. Res., 2017, 50, 2273–2283 CrossRef CAS PubMed.
- J. van Haasteren, J. Li, O. J. Scheideler, N. Murthy and D. V. Schaffer, Nat. Biotechnol., 2020, 38, 845–855 CrossRef CAS PubMed.
- K. Nguyen, Y. Wang, W. E. England, J. C. Chaput and R. C. Spitale, J. Am. Chem. Soc., 2021, 143, 4519–4523 CrossRef CAS PubMed.
- I. Lostalé-Seijo and J. Montenegro, Nat. Rev. Chem., 2018, 2, 258–277 CrossRef.
- H. Yin, K. J. Kauffman and D. G. Anderson, Nat. Rev. Drug Discovery, 2017, 16, 387–399 CrossRef CAS PubMed.
- M. T. Manzari, Y. Shamay, H. Kiguchi, N. Rosen, M. Scaltriti and D. A. Heller, Nat. Rev. Mater., 2021, 6, 351–370 CrossRef CAS PubMed.
- J. Buck, P. Grossen, P. R. Cullis, J. Huwyler and D. Witzigmann, ACS Nano, 2019, 13, 3754–3782 CrossRef CAS.
- H. Wang, Y. Chen, H. Wang, X. Liu, X. Zhou and F. Wang, Angew. Chem., Int. Ed., 2019, 58, 7380–7384 CrossRef CAS PubMed.
- X. Gong, H. Wang, R. Li, K. Tan, J. Wei, J. Wang, C. Hong, J. Shang, X. Liu, J. Liu and F. Wang, Nat. Commun., 2021, 12, 3953 CrossRef CAS PubMed.
- W. Shen, R. Wang, Q. Fan, Y. Li and Y. Cheng, Gene Ther., 2020, 27, 383–391 CrossRef CAS PubMed.
- D. C. Luther, R. Huang, T. Jeon, X. Zhang, Y. W. Lee, H. Nagaraj and V. M. Rotello, Adv. Drug Delivery Rev., 2020, 156, 188–213 CrossRef CAS PubMed.
- J. Liu, T. Liu, P. Du, L. Zhang and J. Lei, Angew. Chem., Int. Ed., 2019, 58, 7808–7812 CrossRef CAS PubMed.
- J. Li, C. Zheng, S. Cansiz, C. Wu, J. Xu, C. Cui, Y. Liu, W. Hou, Y. Wang, L. Zhang, I. T. Teng, H. H. Yang and W. Tan, J. Am. Chem. Soc., 2015, 137, 1412–1415 CrossRef CAS.
- Y. Jin, Z. Li, H. Liu, S. Chen, F. Wang, L. Wang, N. Li, K. Ge, X. Yang, X.-J. Liang and J. Zhang, NPG Asia Mater., 2017, 9, e365 CrossRef CAS.
- M. Cieslak, J. Szymanski, R. W. Adamiak and C. S. Cierniewski, J. Biol. Chem., 2003, 278, 47987–47996 CrossRef CAS PubMed.
- S. W. Santoro and G. F. Joyce, Biochemistry, 1998, 37, 13330–13342 CrossRef CAS.
- W. Zhou, R. Saran and J. Liu, Chem. Rev., 2017, 117, 8272–8325 CrossRef CAS PubMed.
- H. Fan, Z. Zhao, G. Yan, X. Zhang, C. Yang, H. Meng, Z. Chen, H. Liu and W. Tan, Angew. Chem., Int. Ed., 2015, 54, 4801–4805 CrossRef CAS PubMed.
- X. Gong, R. Li, J. Wang, J. Wei, K. Ma, X. Liu and F. Wang, Angew. Chem., Int. Ed., 2020, 59, 21648 CrossRef CAS PubMed.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2019, 119, 6459–6506 CrossRef CAS PubMed.
- B. R. Madhanagopal, S. Zhang, E. Demirel, H. Wady and A. R. Chandrasekaran, Trends Biochem. Sci., 2018, 43, 997–1013 CrossRef CAS.
- A. R. Chandrasekaran, Nat. Rev. Chem., 2021, 5, 225–239 CrossRef CAS.
- Y. Gu, M. E. Distler, H. F. Cheng, C. Huang and C. A. Mirkin, J. Am. Chem. Soc., 2021, 143, 17200–17208 CrossRef CAS PubMed.
- S. Liu, Q. Jiang, X. Zhao, R. Zhao, Y. Wang, Y. Wang, J. Liu, Y. Shang, S. Zhao, T. Wu, Y. Zhang, G. Nie and B. Ding, Nat. Mater., 2021, 20, 421–430 CrossRef CAS PubMed.
- Z. Wang, L. Song, Q. Liu, R. Tian, Y. Shang, F. Liu, S. Liu, S. Zhao, Z. Han, J. Sun, Q. Jiang and B. Ding, Angew. Chem., Int. Ed., 2021, 60, 2594 CrossRef CAS PubMed.
- S. Jiang, Z. Ge, S. Mou, H. Yan and C. Fan, Chem, 2021, 7, 1156–1179 CAS.
- J. Gačanin, C. V. Synatschke and T. Weil, Adv. Funct. Mater., 2019, 30, 1906253 CrossRef.
- M. Pan, Q. Jiang, J. Sun, Z. Xu, Y. Zhou, L. Zhang and X. Liu, Angew. Chem., Int. Ed., 2020, 59, 1897–1905 CrossRef CAS PubMed.
- X. Quyang, M. De Stefano, A. Krissanaprasit, A. L. B. Kodal, C. B. Rosen, T. Liu, S. W. Helmig, C. Fan and K. V. Gothelf, Angew. Chem., Int. Ed., 2017, 56, 14423–14427 CrossRef PubMed.
- S. K. Madden, A. D. de Araujo, M. Gerhardt, D. P. Fairlie and J. M. Mason, Mol. Cancer, 2021, 20, 3 CrossRef PubMed.
- E. Kim, L. Zwi-Dantsis, N. Reznikov, C. S. Hansel, S. Agarwal and M. M. Stevens, Adv. Mater., 2017, 29, 1701086 CrossRef PubMed.
- E. Kim, S. Agarwal, N. Kim, F. S. Hage, V. Leonardo, A. Gelmi and M. M. Stevens, ACS Nano, 2019, 13, 2888–2900 CrossRef CAS PubMed.
- W. Sun, W. Ji, J. M. Hall, Q. Hu, C. Wang, C. L. Beisel and Z. Gu, Angew. Chem., Int. Ed., 2015, 54, 12029–12033 CrossRef CAS PubMed.
- Q. Chen, L. Zhang, F. Jiang, B. Wang, T. Lv, Z. Zeng, W. Wu and S. Sun, Sens. Actuators, B, 2017, 244, 1138–1144 CrossRef CAS.
- L. Wang, Z. Huang, Y. Liu, J. Wu and J. Liu, Langmuir, 2018, 34, 3094–3101 CrossRef CAS.
- Y. Zhou, L. Zou, G. Li, T. Shi, S. Yu, F. Wang and X. Liu, Anal. Chem., 2021, 93, 13960–13966 CrossRef CAS PubMed.
- C. Zhang, W.-H. Chen, L.-H. Liu, W.-X. Qiu, W.-Y. Yu and X.-Z. Zhang, Adv. Funct. Mater., 2017, 27, 1700626 CrossRef.
- Y. Jin, H. Wang, X. Li, H. Zhu, D. Sun, X. Sun, H. Liu, Z. Zhang, L. Cao, C. Gao, H. Wang, X.-J. Liang, J. Zhang and X. Yang, ACS Appl. Mater. Interfaces, 2020, 12, 26832–26841 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details; additional characterization and in vitro and in vivo analysis. See DOI: 10.1039/d2sc00459c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |