Open Access Article
Open Access Article2′-O-Methyl modified guide RNA promotes the single nucleotide polymorphism (SNP) discrimination ability of CRISPR–Cas12a systems†
Yuqing
Ke
a,
Behafarid
Ghalandari
a,
Shiyi
Huang
a,
Sijie
Li
a,
Chengjie
Huang
a,
Xiao
Zhi
*a,
Daxiang
Cui
 b and
Xianting
Ding
b and
Xianting
Ding
 *a
*a
aState Key Laboratory of Oncogenes and Related Genes, Institute for Personalized Medicine, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai 200030, China. E-mail: dingxianting@sjtu.edu.cn
bShanghai Engineering Centre for Intelligent Diagnosis and Treatment Instrument, School of Electronic Information and Electrical Engineering, Shanghai Jiao Tong University, Shanghai 200240, China
First published on 1st February 2022
Abstract
The CRISPR–Cas12a system has been widely applied to genome editing and molecular diagnostics. However, off-target cleavages and false-positive results remain as major concerns in Cas12a practical applications. Herein, we propose a strategy by utilizing the 2′-O-methyl (2′-OMe) modified guide RNA (gRNA) to promote the Cas12a's specificity. Gibbs free energy analysis demonstrates that the 2′-OMe modifications at the 3′-end of gRNA effectively suppress the Cas12a's overall non-specific affinity while maintaining high on-target affinity. For general application illustrations, HBV genotyping and SARS-CoV-2 D614G mutant biosensing platforms are developed to validate the enhanced Cas12a's specificity. Our results indicate that the 2′-OMe modified gRNAs could discriminate single-base mutations with at least two-fold enhanced specificity compared to unmodified gRNAs. Furthermore, we investigate the enhancing mechanisms of the 2′-OMe modified Cas12a systems by molecular docking simulations and the results suggest that the 2′-OMe modifications at the 3′-end of gRNA reduce the Cas12a's binding activity to off-target DNA. This work offers a versatile and universal gRNA design strategy for highly specific Cas12a system development.
Introduction
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins are unique for bacteria and archaea, constituting an adaptive immune system against foreign mobile genetic elements.1,2 CRISPR–Cas systems are classified into Class 1 (use multiple Cas proteins) and Class 2 systems (use a single multidomain Cas protein), which are subdivided into six types (types I to VI) based on the complexity and feature proteins.3 As a member of Class 2 systems, the type II-A CRISPR–Cas9 has been most extensively studied and developed as a genome editing and therapeutic tool.4 Cas9 possesses two nuclease sites – His–Asn–His (HNH) and RuvC-like domains to achieve the precise double-stranded DNA (dsDNA) cleavage at a specific site mediated by a dual-CRISPR RNA (crRNA) and trans-activating crRNA (transcrRNA) guide.5,6 The dual – transcrRNA:crRNA guide of Cas9 can also be engineered as a single RNA chimera fused by a short loop.7 Another recently characterized Class 2 CRISPR–Cas system, the type V-A Cas12a (also known as Cpf1), is a single RNA (41–44 nt) guided endonuclease that can also recognize, unwind and induce a double-strand break (DSB) in the target DNA with high specificity.8 Distinct from Cas9, Cas12a possesses a single nuclease pocket in the RuvC domain and has been shown to exhibit indiscriminate single-stranded DNase activity after binding with target DNA.9,10 Besides, Cas12a shows a thymine (T)-rich (typically 5′-TTTV-3′, V = A, C, or G) protospacer adjacent motif (PAM) preference, while Cas9 recognizes guanine (G)-rich sequences.11–13 Based on the precise target DNA cleavage and indiscriminate ssDNA degradation activities, CRISPR–Cas12a systems are widely exploited for molecular diagnostics14–17 and gene editing.12,18 However, a recent research study using nuclease digestion and deep sequencing (NucleaSeq) to compare Streptococcus pyogenes (Sp)Cas9 and Acidaminococcus species (As)Cas12a cleavage activities revealed that Cas12a has similar cleavage specificities to Cas9 and tolerates similar mismatches in vitro,19,20 though Cas12a is reported to show less off-target effects in vivo.21,22 Cas12a proteins are more sensitive to mismatches at PAM-proximal positions 1–8 within the protospacer (target recognition sequence) but tolerable to PAM-distal mismatches, especially for single nucleotide polymorphisms (SNPs).19,23 Therefore, it's necessary to suppress the off-target cleavages of Cas12a in case of false positives or safety issues.To increase the efficacy and specificity of CRISPR systems, developing high-fidelity Cas proteins24,25 and optimizing gRNA design26–28 are two main strategies. The molecular structures and activation mechanisms of Cas12a have been clarified, allowing the development of high-performance proteins.23,29,30 Recently, an enhanced Acidaminococcus sp. Cas12a (enAsCas12a) variant has been engineered to expand the targeting range and increase the genome editing activity of Cas12a, and a high-fidelity version of enAsCas12a has also been developed to improve the specificity.24,31 Considering the safety issues and easy modifications, (g)RNAs are more accessible to be engineered than proteins. Chemical modifications32–34 and hairpin structures35 are applied to promote the stability of gRNAs and improve the editing efficacy of Cas proteins. For example, chemical modifications (such as 2′-O-methyl, bridge nucleic acids, locked nucleic acids and 2′-O-methyl-3′-phosphonoacetate, etc.) incorporated at specific sites of Cas9-gRNAs can dramatically reduce off-target cleavages while maintaining high on-target performance and enhancing the Cas9 genome editing efficiency in human cells.33,36,37 Besides, Cas12a-gRNAs with chemically modified terminal nucleotides are found to be exonuclease resistant while retaining gene-editing activity.38
Studies show that chemical modifications of RNA are effective to probe the RNA–protein relationships in ribonucleoprotein (RNP) enzymes39 and increase the stability of RNA against degradation by RNase.40 Moreover, the 2′-position modifications of the sugar ring in RNA are beneficial to enhance the stability and binding activity of RNA.41 2′-O-Methyl (2′-OMe, also known as Nm) modifications play an important role in regulating the gene expression at the post-transcriptional level in vivo42–44 and can increase the stability of secondary structures of paired RNA.45,46 Due to the commercial availability and low cost, 2′-OMe modifications have emerged as an attractive strategy to modulate RNA ensembles.45 However, little is known about the effects of 2′-OMe modifications for Cas12a-gRNA to improve the Cas12a's SNP discrimination ability in vitro, and the enhancing mechanisms of utilizing modified gRNAs for Cas12a systems remain to be elucidated yet. Herein, we propose a 2′-O-methyl modified gRNA (mgRNA) strategy to suppress the binding affinity between Lachnospiraceae bacterium ND2006 (Lba)Cas12a and off-target DNA, thus improving the overall on-target specificity of the CRISPR–Cas12a systems (mg-CRISPR). The enhanced system shows great potential for establishing highly specific molecular diagnostic tools such as pathogen genotyping platforms.
Results and discussion
2′-OMe RNA modifications at the 3′-end of gRNA suppressed Cas12a's affinity to off-target DNA while maintaining high affinity to on-target DNA
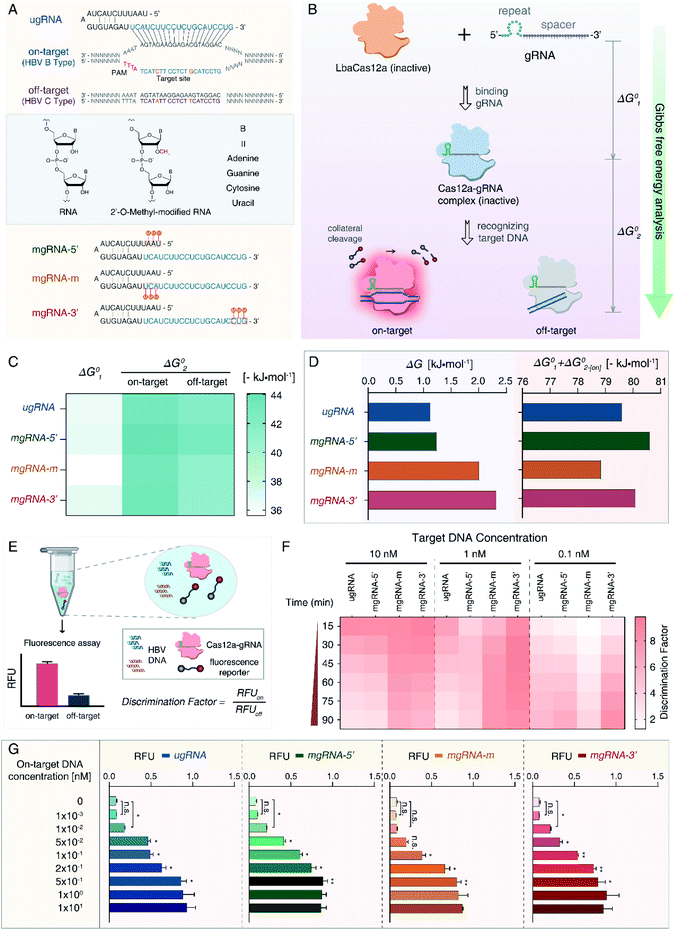
For proof-of-concept, we first selected HBV genotypes B and C as our targets. HBV remains a worldwide health concern and HBV genotyping is significant for clinical therapy, whereas the B-type and C-type are major distributions in Oceania and Asia.47 As shown in Fig. 1A, a 20 nt sequence of the B-type (on-target) containing two single nucleotide polymorphisms (SNPs) differing from the C-type (off-target) was determined as the target site. We designed an unmodified gRNA (ugRNA) and three various mgRNAs with 2′-OMe modifications at different positions (5′-end for mgRNA-5′, the middle region for mgRNA-m, and 3′-end for mgRNA-3′, respectively).To evaluate the effects of 2′-OMe modifications on the activity of Cas12a, we performed the Gibbs free energy analysis (Fig. 1B). Fluorescence quenching methods have been widely exploited to study protein dynamics and interactions.48,49 Cas12a proteins have natural fluorophores (tryptophan, tyrosine and phenylalanine residues), allowing the measurement of conformational changes through intrinsic fluorescence spectroscopy. The Gibbs free energy alteration during Cas12a to Cas12a/gRNA complex transition is termed “ΔG01” (Fig. 1C, S1 and S2†). For target DNA recognition, the Gibbs free energy alteration from Cas12a/gRNA to Cas12a/gRNA/on-target DNA is denoted as “ΔG02-[on]” (Fig. S3 and S4†), while that of off-target DNA is “ΔG02-[off]” (Fig. S5 and S6†).
Then, to evaluate the Cas12a's overall affinity to off-target DNA, we denoted the difference between ΔG02-[on] and ΔG02-[off] as “ΔG” (ΔG = ΔG02-[off] − ΔG02-[on]). Besides, the sum of ΔG01 and ΔG02-[on] was referred to as “SG” (SG = ΔG01 + ΔG02-[on]), representing the Cas12a's overall on-target affinity. According to the results shown in Fig. 1D and Table S1,† the ΔG value of mgRNA-3′ (2.32 kJ mol−1) was higher than those of ugRNA (1.13 kJ mol−1), mgRNA-5′ (1.24 kJ mol−1) and mgRNA-m (2.01 kJ mol−1), indicating that the 3′-end modifications of mgRNA-3′ suppressed the Cas12a's overall affinity to off-target DNA. As shown in Fig. 1D, the SG values of mgRNA-5′ (−80.62 kJ mol−1) and mgRNA-3′ (−80.09 kJ mol−1) were similar to that of ugRNA (−79.61 kJ mol−1). These observations demonstrated that mgRNA-5′ and mgRNA-3′ maintained high-level on-target affinity as ugRNA. According to the standard Gibbs free energy analysis, 2′-OMe modifications at the 3′-end of mgRNA-3′ suppressed the Cas12a's overall off-target affinity without sacrificing the on-target affinity.
We hypothesized that the Cas12a's affinity to on-target and off-target DNA may be associated with the sensitivity and specificity of the Cas12a system, so we established an HBV genotyping platform (Fig. 1E). For signal readout, we used a single-stranded DNA linked quenched fluorophore (5′-HEX-A(12)-BHQ1-3′) as the reporter. The reactions were processed at 37 °C for 1.5 h with fluorescence intensity measured every 2 minutes. To evaluate the specificity of the Cas12a system, we calculated the discrimination factor (DF) using eqn (1):
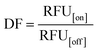 | (1) |
As shown in Fig. 1F and Table 1, for target DNA at high concentrations (10 nM and 1 nM), the DF values of mgRNA-m (6.54 ± 0.31 and 8.11 ± 0.18, respectively) and mgRNA-3′ (5.77 ± 0.64 and 8.64 ± 0.81, respectively) were higher than those of ugRNA (2.93 ± 0.54 and 2.87 ± 0.18, respectively) and mgRNA-5′ (3.2 ± 0.5 and 3.89 ± 0.77, respectively). For target DNA at a lower concentration (0.1 nM), the DF value of mgRNA-3′ (8.52 ± 0.4) was higher than that of ugRNA (5.62 ± 0.61), mgRNA-5′ (6.53 ± 0.11) and mgRNA-m (3.89 ± 0.28). These observations demonstrated that 2′-OMe modifications at the 3′-end of gRNA increased the Cas12a's discrimination efficacy by at least two-fold compared to unmodified gRNA. Notably, for high target concentrations (10 nM and 1 nM), the DF values of ugRNA were reducing along with the nuclease assay time, which was caused by the increasing off-target signal and rapidly saturated on-target signal. For a lower target concentration (0.1 nM), the DF values of all gRNAs increased with the assay time due to the weak off-target signal and slowly increasing on-target signal (Fig. S7†). mgRNA-m showed high DF values for high concentration targets but decreased DF values for a low concentration target, which may be caused by the low on-target sensitivity of mgRNA-m. In summary, mgRNA-3′ possessed the highest final DF values at 90 minutes compared to ugRNA and other mgRNAs. The specificity result of mgRNA-3′ was consistent with our Gibbs free energy analysis that mgRNA-3′ suppressed the Cas12a's overall affinity to off-target DNA compared to other gRNAs.
| Groups | DF values for different target DNA concentrations | |||
|---|---|---|---|---|
| 10 nM | 1 nM | 0.1 nM | ||
| a The 2′-OMe modifications of mgRNA-5′ are located at the 5′-end. b The 2′-OMe modifications of mgRNA-m are located at the middle region. c The 2′-OMe modifications of mgRNA-3′ are located at the 3′-end. | ||||
| Target (Fig. 1) | ugRNA | 2.93 ± 0.54 | 2.87 ± 0.18 | 5.62 ± 0.61 |
| mgRNA-5′a | 3.2 ± 0.5 | 3.89 ± 0.77 | 6.53 ± 0.11 | |
| mgRNA-mb | 6.54 ± 0.31 | 8.11 ± 0.18 | 3.89 ± 0.28 | |
| mgRNA-3′c | 5.77 ± 0.64 | 8.64 ± 0.81 | 8.52 ± 0.4 | |
| Target-11th | ugRNA-11th | 1.13 ± 0.03 | 1.41 ± 0.05 | 2.57 ± 0.11 |
| mgRNA-11th-5′a | 1.64 ± 0.09 | 2.07 ± 0.09 | 2.85 ± 0.08 | |
| mgRNA-11th-mb | 1.48 ± 0.02 | 2.22 ± 0.26 | 3.66 ± 0.16 | |
| mgRNA-11th-3′c | 2.46 ± 0.09 | 4.09 ± 0.18 | 4.85 ± 0.66 | |
| Target-19th | ugRNA-19th | 3.48 ± 0.31 | 4.75 ± 0.14 | 5.35 ± 0.16 |
| mgRNA-19th-5′a | 4.1 ± 0.42 | 4.65 ± 0.27 | 5.19 ± 0.4 | |
| mgRNA-19th-mb | 4.52 ± 0.16 | 8.49 ± 0.08 | 6.75 ± 0.21 | |
| mgRNA-19th-3′c | 6.33 ± 0.3 | 6.53 ± 0.89 | 4.47 ± 0.56 | |
Moreover, we investigated the sensitivity of the Cas12a system. Various concentrations of on-target DNA (0 to 10 nM) were used as input DNA. According to the sensitivity results shown in Fig. 1G, mgRNA-5′ and mgRNA-3′ possessed a low LOD of 10 pM like ugRNA, while mgRNA-m showed higher LOD (100 pM) than ugRNA, which could explain the decreased DF values of mgRNA-m at a lower target DNA concentration (Fig. 1F). These observations were consistent with our Gibbs free energy analysis that mgRNA-3′ maintained the Cas12a's high-level on-target affinity as ugRNA, while mgRNA-m decreased the overall on-target affinity compared to ugRNA. Our results illustrated that the ΔG values were positively correlated with the Cas12a's overall specificity while the SG values were positively correlated with the Cas12a's sensitivity.
2′-OMe modifications at the 3′-end of gRNA increased the single nucleotide polymorphism (SNP) discrimination efficacy of the Cas12a system
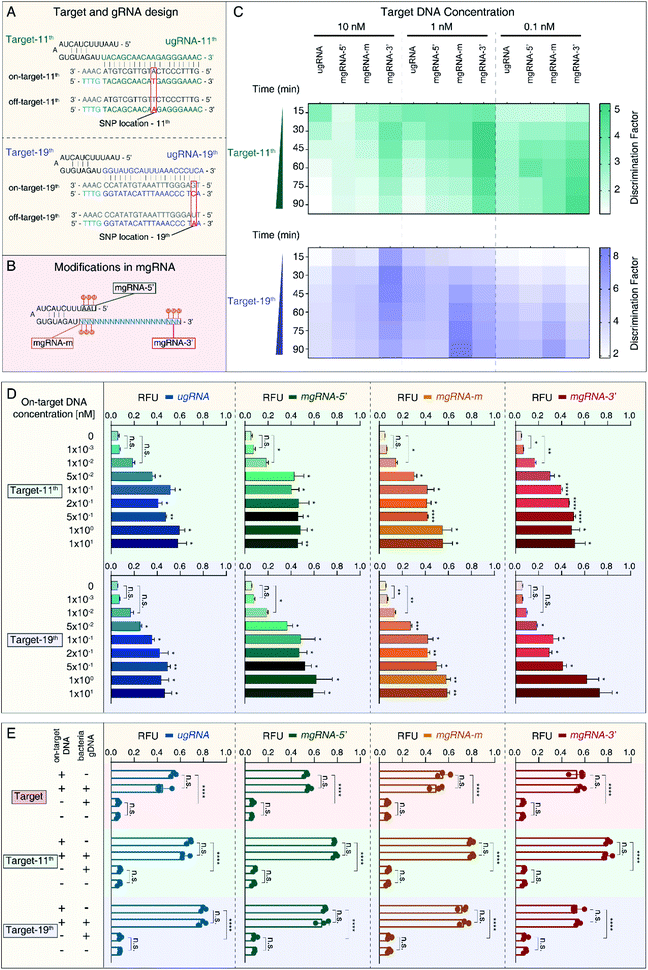
To verify the feasibility of 2′-OMe modification at the 3′-end of gRNA to improve the SNP discrimination ability of the Cas12a system, we selected another two target sequences in HBV B-type (on-target) and C-type (off-target) genomes containing only one SNP. As shown in Fig. 2A, target-11th contains just one SNP at position 11, while target-19th has single SNP at position 19. Accordingly, we designed another two unmodified gRNAs (ugRNA-11th for target-11th and ugRNA-19th for target-19th) and related modified gRNAs (mgRNA-11th for target-11th and mgRNA-19th for target-19th). Similar to the targets described in Fig. 1A, we modified gRNAs at different positions: the 5′-end for mgRNAs-5′, middle region for mgRNAs-m, and 3′-end for mgRNAs-3′, respectively. Three adjacent RNAs were modified for each mgRNA (Fig. 2B).At first, we investigated the specificity of the Cas12a system based on unmodified and modified gRNAs. The specificity results are provided in Fig. 2C and Table 1. For target-11th DNA at 10 nM, mgRNA-11th-3′ (DF value of 2.46 ± 0.09) increased the discriminating ability of Cas12a by two fold compared to ugRNA-11th (1.13 ± 0.03). For target-11th DNA at 1 nM, mgRNA-11th-3′ (4.09 ± 0.18) improved the Cas12a's specificity by about three-fold compared to ugRNA-11th (1.41 ± 0.05). For target-11th DNA at 0.1 nM, mgRNA-11th-3′ (4.85 ± 0.66) showed a two-fold higher specificity than ugRNA-11th (2.57 ± 0.11). These results illustrated that the 2′-OMe modifications at the 3′-end of gRNA enhanced the Cas12a's specificity even for single SNP discrimination. Besides, for target-11th DNA at various concentrations (10 nM, 1 nM and 0.1 nM), mgRNA-11th-5′ (1.64 ± 0.09, 2.07 ± 0.09 and 2.85 ± 0.08, respectively) and mgRNA-11th-m (1.48 ± 0.02, 2.22 ± 0.26 and 3.66 ± 0.16, respectively) showed a slightly improved specificity of about 1.5 fold compared to ugRNA-11th. These observations demonstrated that 2′-OMe modifications at the 5′-end and middle region of gRNA caused minor effects on the Cas12a's activity for target discrimination with only one SNP at the middle position of the spacer (position 11 in a total of 20 nucleotides).
Moreover, we evaluated the Cas12a's activity for target-19th using ugRNA-19th and mgRNAs-19th. According to the results shown in Fig. 2C (bottom) and Table 1, mgRNA-19th-3′ (6.33 ± 0.3) showed about two-fold specificity compared to ugRNA-19th (3.48 ± 0.31) for target DNA at 10 nM. Besides, for target DNA at 1 nM, mgRNA-19th-m (8.49 ± 0.08) increased the Cas12a's specificity by about two-fold compared to ugRNA-19th (4.75 ± 0.14). And for target DNA at 0.1 nM, mgRNA-19th-m (6.75 ± 0.21) showed higher DF values than ugRNA-19th, mgRNA-19th-5′ and mgRNA-19th-3′ (5.35 ± 0.16, 5.19 ± 0.4 and 4.47 ± 0.56, respectively). These results indicated that 2′-OMe modifications at the middle region could promote the Cas12a's specificity for targets containing one SNP proximal to the 3′-end of gRNA.
Next, we tested the sensitivity of the Cas12a system for target-11th and target-19th. As shown in Fig. 2D, mgRNA-11th-3′ showed a lower LOD of 1 pM than ugRNA-11th, mgRNA-11th-5′ and mgRNA-11th-m (50 pM, 10 pM and 10 pM, respectively). For target-19th, mgRNA-19th-3′ showed a similar LOD of 50 pM to ugRNA-19th. These results demonstrated that 2′-OMe modifications at the 3′-end of mgRNA wouldn't sacrifice the sensitivity of the Cas12a system even for various target sites containing different SNP locations (at position 11 or 19).
Furthermore, to explore the performance of the Cas12a system based on ugRNAs and mgRNAs for detection in complex samples, we mixed HBV on-target DNA with bacterial gDNA and tested the final fluorescence intensity. The concentrations of HBV target DNA were 1 nM, while the amounts of bacterial gDNA were 80 ng for each test. As shown in Fig. 2E, all mgRNAs and ugRNAs for the three target sites possessed high stability and specificity even in complex samples.
2′-OMe modifications of gRNA increased the specificity for the SARS-CoV-2 D614G mutant detection
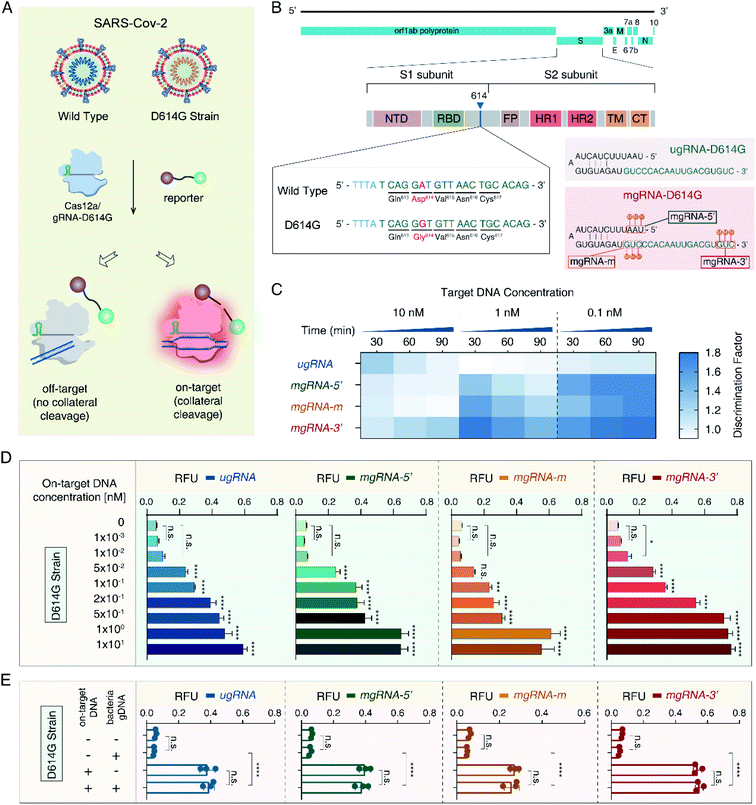
Then, to validate the versatility of the 2′-OMe modifications to improve the Cas12a's specificity for different applications, we developed a SARS-CoV-2 D614G mutant detection platform (Fig. 3A). The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a worldwide pandemic and the mutant strain with one amino acid change at position 614 (D614G) has been reported to be dominant.50,51 As shown in Fig. 3B, we selected the 20 nt sequence containing the D614G mutant from the S1 subunit of the SARS-CoV-2 genome as the target site. Accordingly, we designed one unmodified gRNA complementary to the SARS-CoV-2 D614G mutant (ugRNA-D614G). Three modified gRNAs (mgRNAs) with 2′-OMe modifications at various positions were also designed (5′-end modifications for mgRNA-D614G-5′, the middle region for mgRNA-D614G-m and 5′-end for mgRNA-D614G-3′, respectively). At first, we tested the specificity of the Cas12a based SARS-CoV-2 biosensing platform. The results are provided in Fig. 3C. For target DNA at 1 nM and 0.1 nM, the final DF values of mgRNA-D614G-3′ (1.43 ± 0.08 and 1.8 ± 0.1 for 1 nM and 0.1 nM, respectively) were about two-fold higher than those of ugRNA-D614G (0.94 ± 0.04 and 1.04 ± 0.08, respectively), illustrating that 2′-OMe modifications improved the Cas12a's specificity for D614G mutant detection even at low target concentrations. Moreover, to investigate the sensitivity of the Cas12a based D614G mutant detection system, we used D614G mutant DNA (concentrations varying from 0 to 10 nM) as input DNA. As shown in Fig. 3D, the LOD of mgRNA-D614G-3′ (10 pM) was lower than that of ugRNA-D614G, mgRNA-D614G-5′ and mgRNA-D614G-m (50 pM), indicating that 2′-OMe modifications at the 3′-end of gRNA enhanced the Cas12a's sensitivity for D614G mutant detection. Furthermore, to explore the performance of the Cas12a system in a complex sample, we mixed the SARS-CoV-2 D614G mutant DNA (200 pM per reaction) with HBV B-type DNA (600 pM per reaction) and analysed the fluorescence intensity readout. According to the results shown in Fig. 3E, all unmodified and modified gRNAs-D614G possessed high specificity even in the complex sample, demonstrating the stability of the Cas12a based D614G mutant biosensing platform.Docking simulations of Cas12a interacting with gRNA and off-target DNA
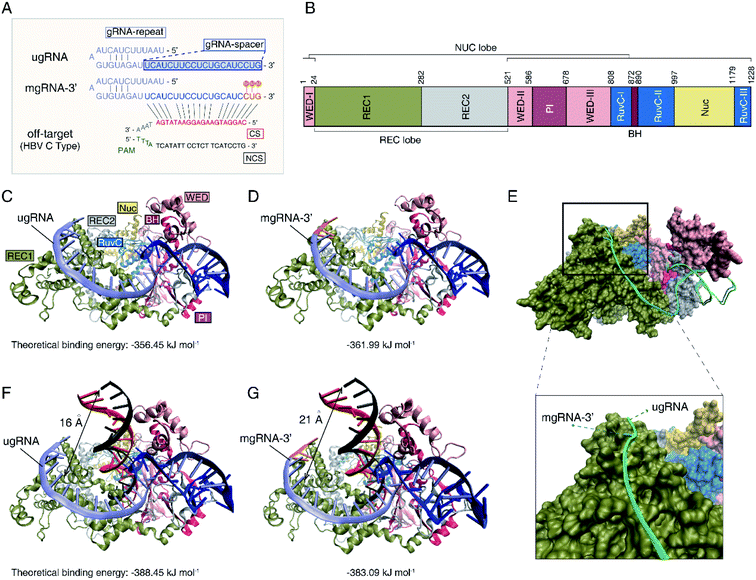
To investigate the mechanisms of the increased Cas12a's specificity based on the 2′-OMe modified gRNA, molecular docking simulations were further performed to illustrate the interactions between Cas12a, gRNA and off-target DNA. Herein, we selected unmodified gRNA (ugRNA), mgRNA-3′ (with 2′-OMe modifications at the 3′-end) and the target site as shown in Fig. 1 for demonstrations (Fig. 4A). Protein Data Bank (PDB) ID-5XUS was selected for Cas12a. The structure of the Cas12a protein is provided in Fig. 4B. The docking result of Cas12a binding with ugRNA is shown in Fig. 4C, while that of mgRNA-3′ is provided in Fig. 4D. In the binary system of Cas12a interacting with gRNAs, the docking analysis showed that gRNAs are located in the groove of the REC lobe (consisting of REC1 and REC2 domains) and the NUC lobe (WED and PI domains) of the Cas12a protein (Fig. 4C and D). The 3′ flanks of ugRNA and mgRNA-3′ are oriented to the REC1 domain surface, close to the channel between the REC2, RuvC1 and Nuc domains. The 5′ flanks of gRNAs are oriented to the groove of the REC2, WED and PI domains, close to the BH domain. Previous studies11,23 suggest that gRNA distribution on the Cas12a's surface is significant for the subsequent catalysis processes, since the REC, RuvC, WED, PI and BH domains act as a clamp to trap gRNA. The theoretical binding energies of Cas12a interacting with ugRNA and mgRNA-3′ are −356.45 kJ mol−1 and −361.99 kJ mol−1, respectively, demonstrating that mgRNA-3′ possesses higher affinity to the Cas12a protein than ugRNA. To better elucidate the difference between ugRNA and mgRNA-3′ interacting with Cas12a, we performed the docking calculations of ugRNA and mgRNA-3′ in a single figure (Fig. 4E). The green strand refers to ugRNA while the blue strand represents mgRNA-3′. As shown in Fig. 4E (bottom), the 3′ flank of mgRNA-3′ is oriented towards penetration Cas12a protein, while the 3′ flank of ugRNA gets more freedom on the REC1 surface. In other words, there is a greater tendency for mgRNA-3′ to form the Cas12a/mgRNA-3′ complex compared to ugRNA due to the high stability of the Cas12a/mgRNA-3′ structure. We believe the difference between the Cas12a/ugRNA and Cas12a/mgRNA-3′ structures are caused by the 2′-OMe modifications at the 3′-end of mgRNA-3′.Moreover, we performed the docking calculations between the Cas12a/gRNA complex and off-target DNA in the ternary system. As shown in Fig. 4F and G, the gRNAs recognize the 5′ flank of target DNA through the 3′ flanks of gRNAs. The calculated binding energies of Cas12a/ugRNA/off-target DNA and Cas12a/mgRNA-3′/off-target DNA complexes are −388.45 kJ mol−1 and −383.09 kJ mol−1, respectively, suggesting that the Cas12a/mgRNA-3′ complex suppressed the affinity to off-target DNA compared to Cas12a/ugRNA. Besides, the relative distance between the off-target DNA and the surface of REC1 in the Cas12a/mgRNA-3′ complex (21 angstroms) increases by 5 angstroms compared to that of the Cas12a/ugRNA complex (16 angstroms). In other words, the activity between Cas12a/mgRNA-3′/off-target DNA is weaker than that of Cas12a/ugRNA/off-target DNA due to the structural differences between ugRNA and mgRNA-3′.
Conclusions
In this study, we proposed an enhanced CRISPR–Cas12a system by utilizing 2′-O-methyl modified gRNA, termed mg-CRISPR, to promote the specificity of the Cas12a protein. Three main improvements were achieved. Firstly, modified gRNAs with 2′-OMe modifications at various positions of the gRNAs were developed to increase the specificity of the Cas12a system. The enhanced Cas12a system suppressed the off-target binding activity while maintaining high on-target affinity compared to unmodified gRNAs. Secondly, HBV genotyping and SARS-CoV-2 D614G mutant biosensing platforms were established to validate the high specificity and versatility of the Cas12a system. 2′-OMe modified gRNAs increased the Cas12a's specificity by up to three fold with high sensitivity (10 pM) compared to unmodified gRNAs. Besides, the enhanced Cas12a system was capable of single SNP discrimination. Current technologies for SNP analysis mainly include enzymatic methods,52 fluorescence-based assays,53 PCR-based detections54,55 and electrochemical methods.56 Existing SNP detection methods mostly suffer from complex procedures, expensive instruments or low specificity. Moreover, CRISPR-based methods for SNP analysis are capable of sensitive, specific, programmable and rapid detection.16 By combining the ordinary CRISPR system with the 2′-OMe modified gRNA studied in this research, the discrimination factors have been increased by three fold for single SNP detection. Thirdly, docking simulations were performed to investigate the mechanisms of 2′-OMe modified gRNA suppressing the non-specific activity of the Cas12a protein. Our results demonstrate that 2′-OMe modifications of gRNA are versatile strategies to improve the Cas12a's specificity for diverse applications.Experimental section
Reagents and instruments
PCR primers and the fluorophore quencher-labelled ssDNA reporter were ordered from Sangon Biotechnology Co., Ltd (Shanghai, China). Primers and ssDNA fluorescence reporters used in this study are listed in Table S2.† PCR reactions were performed following instructions of OneTaq Quick-Load 2X Master Mix with Standard Buffer from New England Biolabs (Ipswich MA, United States). All unmodified and 2′-OMe-modified gRNAs were synthesized by Shanghai DNA Bioscience Co., Ltd (https://dnabio.cn/, Shanghai, China). The sequences of gRNAs are provided in Table S3.† HBV plasmids containing the whole genome of genotype B or C were provided by Prof. Yumei Wen (Fudan University, Shanghai, China). SARS-CoV-2 plasmids containing the wild-type and D614G mutant spike gene (catalog numbers: GS-200519_A001 and GS-200519_A012, respectively) were ordered from GENEWIZ Biotechnology Co., Ltd (Suzhou, China). The detailed information of HBV and SARS-CoV-2 plasmids used in this study are provided in Table S4.† The PCR products were purified using a MinElute PCR purification kit from Qiagen (Hilden, Germany). The LbaCas12a protein and NEBuffer 2.1 were purchased from New England Biolabs (Ipswich MA, United States). RNase-free water was ordered from Thermo Fisher (Waltham, United States).PCR experiments were carried out on a LightCycler 96® (Roche). The real-time fluorescence intensity of Cas12a collateral cleavage assay was measured by the fluorescence channel of a LightCycler 96® (Roche, Switzerland). Electrophoresis was conducted using a Tanon EPS-100 electrophoresis system. Fluorescence of the Cas12a protein was obtained by using an Agilent Technologies Cary Eclipse fluorescence spectrometer.
Target DNA preparation and purification
Firstly, HBV and SARS-CoV-2 plasmids were amplified by PCR using a OneTaq Quick-Load 2× Master Mix. PCR primers for HBV and SARS-CoV-2 targets were designed by using NCBI Primer – Blast. Secondly, the amplified products were evaluated by agarose gel electrophoresis and then purified by using a MinElute PCR purification kit (Qiagen, German). Finally, the concentration of purified target DNA was evaluated by a Nanodrop (ALLSHENG, China).gRNA design
The SNP sites for HBV B-type DNA and HBV C-type DNA were searched using the NCBI Nucleotide – Blast (BLAST: Basic local alignment search tool (nih.gov)), then the sequences containing both PAM (5′-TTTN) and SNP sites were determined to be the candidate protospacers for HBV gRNA. Finally, three HBV target sequences were selected in this study. For the HBV target used in Fig. 1, B-type (on-target) DNA contains two SNPs differing from C-type (off-target) DNA. HBV target-11th contains one SNP at position 11, and HBV target-19th has one SNP at position 19. HBV gRNAs are designed according to the selected target sequences, the gRNA spacers are complementary with the HBV B-type DNA. For SARS-CoV-2 gRNA design, the target sequence containing both the PAM (5′-TTTN) and the D614G mutant site is selected as the protospacer. For both HBV and SARS-CoV-2 gRNAs, chemically modified gRNAs were designed by inserting three adjacent 2′-OMe modifications at the 5′-end, middle region and 3′-end of gRNAs. The detailed information of all gRNAs is provided in Table S3.†Fluorescence spectroscopy measurement
Firstly, the LbaCas12a protein (2.5 μM) was incubated with gRNA (concentrations varying from 0 to 1.25 μM) in a cuvette. Then, the intrinsic fluorescence spectra of the Cas12a/gRNA complex were measured using a fluorescence spectrometer (Cary Eclipse, Agilent, Australia). Afterwards, the Cas12a/gRNA complex was incubated with HBV DNA (0–200 nM) and emission spectra of the complex were recorded. The wavelength of excitation was set at 285 nm and the scan range of emission wavelength was 295–500 nm. All measurements were conducted at 25 °C and 37 °C.Binding energetics and thermodynamic parameter calculations
Fluorescence quenching methods were used to analyse the binding energetics of Cas12a-gRNA and Cas12a-gRNA-target DNA complexes. The quenched fluorescence is described using the Stern–Volmer equation: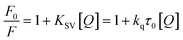 | (2) |
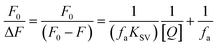 | (3) |
According to the quenching mechanism, the double logarithmic equation was used to obtain thermodynamic parameters for the interactions of the Cas13a protein with crRNAs and HBV RNA:
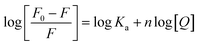 | (4) |
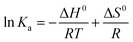 | (5) |
ΔG0 = ΔH0 − TΔS0 = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka Ka | (6) |
Cas12a collateral cleavage assay
The LbaCas12a based fluorescence assay was conducted with a 50 nM LbaCas12a protein, 22.5 nM gRNA, a 100 nM fluorophore quencher-labelled ssDNA reporter and various amounts of target DNA in 1 × NEB2.1 buffer (containing 5 mM NaCl, 1 mM Tris–HCl, 1 mM MgCl2, and 10 μg mL−1 BSA, pH 7.9). The total volume was 20 μL for a single reaction. Assays were proceeded for 1.5 h at 37 °C. Meanwhile, the fluorescence intensity of each assay was collected every minute using the fluorescence channel of a qPCR instrument (Roche, LightCycler® 96, Switzerland).Molecular docking study of the Cas12a/gRNA and Cas12a/gRNA-target DNA complex
The HDOCK webserver (http://hdock.phys.hust.edu.cn/) was used to perform the docking calculations.59,60 The 3D structure of the LbaCas12a protein was acquired from Protein Data Bank ID 5XUS. The RNAComposer server (http://rnacomposer.cs.put.poznan.pl/) was utilized to generate the 3D structures of gRNAs.61 The 3D structure of HBV target DNA was obtained using the HyperChem v8.0.6 program. The gRNAs and target DNA geometries were optimized using the CHARMM36 force field implemented in the NAMD 2.10 package.62,63 The gRNAs and target DNA structures were immersed within 60 Å and 80 Å cubic boxes of TIP3P water molecules and neutralized by 25 and 35 Na+ ions, respectively. Classical molecular dynamics simulations were performed considering calculation parameters based on preparation instructions.64 Next, docking calculations were conducted to search the complex formation between the Cas12a protein and guide RNA (binary system), followed for the Cas12a/gRNA complex and target DNA (ternary system). The Visual Molecular Dynamics (VMD) package was used to investigate the docking results.65 The optimal position of each molecule was analyzed according to the energy score and the condition of interaction was selected based on the minimum energy HDOCK score. We obtained the binding energy using the PM7 semiempirical method implemented in the MOPAC 2016 package.66,67Statistical analysis
Statistical analysis was performed by one-way ANOVA-multiple comparisons using GraphPad Prism 8.0. All assays were repeated three times. The results were presented as mean ± S.D.Author contributions
Conceptualization: Y. K., X. D., X. Z. D. C.; methodology: Y. K., S. L.; investigation: Y. K., B. G., S. H.; visualization: Y. K., B. G., C. H.; supervision: X. D., X. Z.; writing—original draft: Y. K.; writing—review & editing: Y. K., X. D., X. Z., B. G.Conflicts of interest
There are no conflicts to declare.Data availability
All data associated with the research are available from the Royal Society of Chemistry online or from the author.Acknowledgements
We thank the Nature Scientific Foundation of China (No. 81871448 and 81971736), National Key Research and Development Program of China (No. 2017FYA0205301), Nature Scientific Foundation of Shanghai (No. 21ZR1435000, 22N31900400), Interdisciplinary Program of Shanghai Jiao Tong University (No. ZH2018QNA51, YG2021QN58, and Agri-X20200101), Shanghai Clinical Medical Research Center Project (No. 19MC1910800), Shanghai Municipal Health Commission Project (No. 2019CXJQ03), Center of Advanced Electronics Materials and Devices (AEMD), and Shanghai Jiao Tong University for supporting this work.Notes and references
- F. Hille, H. Richter, S. P. Wong, M. Bratovic, S. Ressel and E. Charpentier, Cell, 2017, 172, 1239–1259 CrossRef PubMed.
- M. Kirchner and S. Schneider, Angew. Chem., Int. Ed., 2015, 54, 13508–13514 CrossRef CAS PubMed.
- S. Shmakov, A. Smargon, D. Scott, D. Cox, N. Pyzocha, W. Yan, O. O. Abudayyeh, J. S. Gootenberg, K. S. Makarova, Y. I. Wolf, K. Severinov, F. Zhang and E. V. Koonin, Nat. Rev. Microbiol., 2017, 15, 169–182 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS PubMed.
- S. H. Sternberg, B. LaFrance, M. Kaplan and J. A. Doudna, Nature, 2015, 527, 110–113 CrossRef CAS PubMed.
- H. Nishimasu, L. Cong, W. X. Yan, F. A. Ran, B. Zetsche, Y. Li, A. Kurabayashi, R. Ishitani, F. Zhang and O. Nureki, Cell, 2015, 162, 1113–1126 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed.
- S. Y. Li, Q. X. Cheng, J. K. Liu, X. Q. Nie, G. P. Zhao and J. Wang, Cell Res., 2018, 28, 491–493 CrossRef CAS PubMed.
- B. Paul and G. Montoya, Biomed. J., 2020, 43, 8–17 CrossRef PubMed.
- T. Yamano, B. Zetsche, R. Ishitani, F. Zhang, H. Nishimasu and O. Nureki, Mol. Cell, 2017, 67, 633–645 CrossRef CAS PubMed.
- R. Yao, D. Liu, X. Jia, Y. Zheng, W. Liu and Y. Xiao, Synthetic and Systems Biotechnology, 2018, 3, 135–149 CrossRef PubMed.
- D. C. Swarts and M. Jinek, Wiley Interdiscip. Rev.: RNA, 2018, 9, e1481 Search PubMed.
- Y. Xiong, J. Zhang, Z. Yang, Q. Mou, Y. Ma, Y. Xiong and Y. Lu, J. Am. Chem. Soc., 2020, 142, 207–213 CrossRef CAS PubMed.
- A. Suea-Ngam, P. D. Howes and A. J. deMello, Chem. Sci., 2021, 12, 12733–12743 RSC.
- Y. Chen, Y. Mei and X. Jiang, Chem. Sci., 2021, 12, 4455–4462 RSC.
- Y. Li, H. Mansour, C. J. F. Watson, Y. Tang, A. J. MacNeil and F. Li, Chem. Sci., 2021, 12, 2133–2137 RSC.
- Y. Tang and Y. Fu, Cell Biosci., 2018, 8, 59 CrossRef CAS PubMed.
- S. K. Jones Jr, J. A. Hawkins, N. V. Johnson, C. Jung, K. Hu, J. R. Rybarski, J. S. Chen, J. A. Doudna, W. H. Press and I. J. Finkelstein, Nat. Biotechnol., 2021, 39, 84–93 CrossRef CAS PubMed.
- P. D. Hsu, D. A. Scott, J. A. Weinstein, F. A. Ran, S. Konermann, V. Agarwala, Y. Li, E. J. Fine, X. Wu, O. Shalem, T. J. Cradick, L. A. Marraffini, G. Bao and F. Zhang, Nat. Biotechnol., 2013, 31, 827–832 CrossRef CAS PubMed.
- D. Kim, J. Kim, J. K. Hur, K. W. Been, S. H. Yoon and J. S. Kim, Nat. Biotechnol., 2016, 34, 863–868 CrossRef CAS PubMed.
- B. P. Kleinstiver, S. Q. Tsai, M. S. Prew, N. T. Nguyen, M. M. Welch, J. M. Lopez, Z. R. McCaw, M. J. Aryee and J. K. Joung, Nat. Biotechnol., 2016, 34, 869–874 CrossRef CAS PubMed.
- D. C. Swarts, J. van der Oost and M. Jinek, Mol. Cell, 2017, 66, 221–233 CrossRef CAS PubMed.
- B. P. Kleinstiver, A. A. Sousa, R. T. Walton, Y. E. Tak, J. Y. Hsu, K. Clement, M. M. Welch, J. E. Horng, J. Malagon-Lopez, I. Scarfo, M. V. Maus, L. Pinello, M. J. Aryee and J. K. Joung, Nat. Biotechnol., 2019, 37, 276–282 CrossRef CAS PubMed.
- B. P. Kleinstiver, V. Pattanayak, M. S. Prew, S. Q. Tsai, N. T. Nguyen, Z. Zheng and J. K. Joung, Nature, 2016, 529, 490–495 CrossRef CAS PubMed.
- D. D. Kocak, E. A. Josephs, V. Bhandarkar, S. S. Adkar, J. B. Kwon and C. A. Gersbach, Nat. Biotechnol., 2019, 37, 657–666 CrossRef CAS PubMed.
- H. Kim, W. J. Lee, Y. Oh, S. H. Kang, J. K. Hur, H. Lee, W. Song, K. S. Lim, Y. H. Park, B. S. Song, Y. B. Jin, B. H. Jun, C. Jung, D. S. Lee, S. U. Kim and S. H. Lee, Nucleic Acids Res., 2020, 48, 8601–8616 CrossRef CAS PubMed.
- L. Taemaitree, A. Shivalingam, A. H. El-Sagheer and T. Brown, Nat. Commun., 2019, 10, 1610 CrossRef PubMed.
- D. Dong, K. Ren, X. Qiu, J. Zheng, M. Guo, X. Guan, H. Liu, N. Li, B. Zhang, D. Yang, C. Ma, S. Wang, D. Wu, Y. Ma, S. Fan, J. Wang, N. Gao and Z. Huang, Nature, 2016, 532, 522–526 CrossRef CAS PubMed.
- A. Saha, P. R. Arantes, R. V. Hsu, Y. B. Narkhede, M. Jinek and G. Palermo, J. Chem. Inf. Model., 2020, 60, 6427–6437 CrossRef CAS PubMed.
- L. Gao, D. B. T. Cox, W. X. Yan, J. C. Manteiga, M. W. Schneider, T. Yamano, H. Nishimasu, O. Nureki, N. Crosetto and F. Zhang, Nat. Biotechnol., 2017, 35, 789–792 CrossRef CAS PubMed.
- D. O'Reilly, Z. J. Kartje, E. A. Ageely, E. Malek-Adamian, M. Habibian, A. Schofield, C. L. Barkau, K. J. Rohilla, L. B. DeRossett, A. T. Weigle, M. J. Damha and K. T. Gagnon, Nucleic Acids Res., 2019, 47, 546–558 Search PubMed.
- D. E. Ryan, D. Taussig, I. Steinfeld, S. M. Phadnis, B. D. Lunstad, M. Singh, X. Vuong, K. D. Okochi, R. McCaffrey, M. Olesiak, S. Roy, C. W. Yung, B. Curry, J. R. Sampson, L. Bruhn and D. J. Dellinger, Nucleic Acids Res., 2018, 46, 792–803 CrossRef CAS PubMed.
- B. Li, W. Zhao, X. Luo, X. Zhang, C. Li, C. Zeng and Y. Dong, Nat. Biomed. Eng., 2017, 1, 0066 CrossRef CAS PubMed.
- Y. Ke, S. Y. Huang, B. Ghalandari, S. Li, A. R. Warden, J. Dang, L. Kang, Y. Zhang, Y. Wang, Y. Sun, J. Wang, D. Cui, X. Zhi and X. Ding, Adv. Sci., 2021, 8, 2003611 CrossRef CAS PubMed.
- A. Hendel, R. O. Bak, J. T. Clark, A. B. Kennedy, D. E. Ryan, S. Roy, I. Steinfeld, B. D. Lunstad, R. J. Kaiser, A. B. Wilkens, R. Bacchetta, A. Tsalenko, D. Dellinger, L. Bruhn and M. H. Porteus, Nat. Biotechnol., 2015, 33, 985–989 CrossRef CAS PubMed.
- C. R. Cromwell, K. Sung, J. Park, A. R. Krysler, J. Jovel, S. K. Kim and B. P. Hubbard, Nat. Commun., 2018, 9, 1448 CrossRef PubMed.
- M. A. McMahon, T. P. Prakash, D. W. Cleveland, C. F. Bennett and M. Rahdar, Mol. Ther., 2018, 26, 1228–1240 CrossRef CAS PubMed.
- S. P. Ryder and S. A. Strobel, METHODS, 1999, 18, 38–50 CrossRef CAS PubMed.
- G. F. Deleavey and M. J. Damha, Chem. Biol., 2012, 19, 937–954 CrossRef CAS PubMed.
- X. Shen and D. R. Corey, Nucleic Acids Res., 2018, 46, 1584–1600 CrossRef CAS PubMed.
- L. Ayadi, A. Galvanin, F. Pichot, V. Marchand and Y. Motorin, Biochim. Biophys. Acta, Gene Regul. Mech., 2019, 1862, 253–269 CrossRef CAS PubMed.
- B. A. Elliott, H. T. Ho, S. V. Ranganathan, S. Vangaveti, O. Ilkayeva, H. Abou Assi, A. K. Choi, P. F. Agris and C. L. Holley, Nat. Commun., 2019, 10, 3401 CrossRef PubMed.
- D. G. Dimitrova, L. Teysset and C. Carre, Genes, 2019, 10, 117 CrossRef CAS PubMed.
- H. Abou Assi, A. K. Rangadurai, H. Shi, B. Liu, M. C. Clay, K. Erharter, C. Kreutz, C. L. Holley and H. M. Al-Hashimi, Nucleic Acids Res., 2020, 48, 12365–12379 CrossRef PubMed.
- S. Kumar, K. Mapa and S. Maiti, Biochemistry, 2014, 53, 1607–1615 CrossRef CAS PubMed.
- C. L. Lin and J. H. Kao, J. Gastroenterol. Hepatol., 2011, 26(suppl. 1), 123–130 CrossRef PubMed.
- B. Ghalandari, K. Asadollahi, A. Shakerizadeh, A. Komeili, G. H. Riazi, S. K. Kamrava and N. Attaran, J. Photochem. Photobiol., B, 2019, 192, 131–140 CrossRef CAS PubMed.
- N. M. Fekri, A. Divsalar, A. Seyedarabi and A. A. Saboury, J. Mol. Liq., 2018, 249, 265–271 CrossRef.
- Y. Zhang, H. Xi and M. Juhas, Trends Genet., 2021, 37, 299–302 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C. Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- N. C. Nelson, P. W. Hammond, E. Matsuda, A. A. Goud and M. M. Becker, Nucleic Acids Res., 1996, 24, 4998–5003 CrossRef CAS PubMed.
- M. Varona and J. L. Anderson, Anal. Chem., 2019, 91, 6991–6995 CrossRef CAS PubMed.
- H. O. Hashim and M. B. Al-Shuhaib, J. Appl. Biotechnol. Rep., 2019, 6, 137–144 CrossRef CAS.
- K. Matsuda, Adv. Clin. Chem., 2017, 80, 45–72 CAS.
- C. Gu, X. Kong, X. Liu, P. Gai and F. Li, Anal. Chem., 2019, 91, 8697–8704 CrossRef CAS PubMed.
- R. L. Joseph, Principles of fluorescence spectroscopy, Springer, Boston, MA, 2006, pp. 331–351 Search PubMed.
- A. Dadras, G. H. Riazi, A. Afrasiabi, A. Naghshineh, B. Ghalandari and F. Mokhtari, In vitro study on the alterations of brain tubulin structure and assembly affected by magnetite nanoparticles, J. Biol. Inorg Chem., 2013, 18(3), 357–369 CrossRef CAS PubMed.
- Y. Yan, H. Tao, J. He and S. Y. Huang, Nat. Protoc., 2020, 15, 1829–1852 CrossRef CAS PubMed.
- Y. Yan, D. Zhang, P. Zhou, B. Li and S. Y. Huang, Nucleic Acids Res., 2017, 45, 365–373 CrossRef PubMed.
- M. Popenda, M. Szachniuk, M. Antczak, K. J. Purzycka, P. Lukasiak, N. Bartol, J. Blazewicz and R. W. Adamiak, Nucleic Acids Res., 2012, 40, e112 CrossRef CAS PubMed.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes, I. Vorobyov and A. D. Mackerell Jr, J. Comput. Chem., 2010, 31, 671–690 CAS.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kale and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- T. Saito and Y. Takano, Chemphyschem, 2021, 22, 561–568 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- J. J. Stewart, J. Mol. Model., 2013, 19, 1–32 CrossRef CAS PubMed.
- J. J. Stewart, MOPAC2016, http://openmopac.net.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc06832f |
| This journal is © The Royal Society of Chemistry 2022 |