Interaction of imidazolium-based ionic liquids with supported phospholipid bilayers as model biomembranes†
Massimiliano
Galluzzi
 *ab,
Lorenzo
Marfori
b,
Stefania
Asperti
b,
Alessandro
De Vita
*ab,
Lorenzo
Marfori
b,
Stefania
Asperti
b,
Alessandro
De Vita
 b,
Matteo
Giannangeli
c,
Alessandro
Caselli
b,
Matteo
Giannangeli
c,
Alessandro
Caselli
 c,
Paolo
Milani
c,
Paolo
Milani
 b and
Alessandro
Podestà
b and
Alessandro
Podestà
 *b
*b
aMaterials Interfaces Center, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, Guangdong, China. E-mail: galluzzi@siat.ac.cn
bC.I.Ma.I.Na and Dipartimento di Fisica “Aldo Pontremoli”, Università degli Studi di Milano, via Celoria 16, 20133-Milano, Italy. E-mail: alessandro.podesta@mi.infn.it
cDipartimento di Chimica and CNR-SCITEC, Università degli Studi di Milano, via Golgi 19, 20133-Milano, Italy
First published on 12th October 2022
Abstract
The cytotoxicity of ionic liquids (ILs) has been receiving attention in the context of the biological and environmental impact of their vast field of applications. It has been ascertained that the cell membrane is the main target of ILs when they interact with microorganisms, cells and bacteria; nevertheless, studies at the micro- and nano-scale aiming at better understanding of the fundamental mechanisms of toxicity of ILs are lacking. In this work, we used atomic force microscopy (AFM) to investigate the impact of room-temperature ILs on the mechanical, morphological and electrostatic properties of solid-supported DOPC phospholipid bilayers, taken as models of biomembranes. In particular, we have characterized the concentration-dependent and time-dependent evolution of the morphological, structural and mechanical properties of DOPC lipid membranes in the presence of imidazolium-based ILs with different alkyl chain lengths and hydrophilic/hydrophobic characteristics. The majority of ILs investigated were found to possess the ability of restructuring the lipid bilayer, through the formation of new IL/lipid complexes, showing distinctive morphological features (increase of area and roughness). The nanomechanical analysis of the lipid membrane exposed to ILs revealed a progressive, concentration-dependent perturbation of the structural ordering and rigidity of the membrane, evidenced by a decrease in the breakthrough force, Young's modulus and area stretching modulus. AFM detected a modification of the electrostatic double-layer at the membrane surface, in terms of a reduction of the original negative surface charge density, suggesting a progressive stratification of cations on the exposed leaflet of the lipid membrane. Our findings may be helpful in designing novel ILs with tailored interaction with biological membranes.
Introduction
Room-temperature ionic liquids (briefly ILs) are used as effective and tunable new solvents in several applications.1–3 ILs represent an interesting alternative to the standard aqueous electrolytes in applications aimed at the conversion and storage of energy,4 such as electrochemical supercapacitors,5–8 Graetzel solar cells,9–11 and Li-ion batteries,12–14 as well as good lubricants in micro-electro-mechanical devices15–18 and synthesis media.1,19 Due to the intrinsic controlled flammability, miscibility and negligible vapor pressure of ILS, a ‘green’ label is often assigned to them.20–22 In fact, ILs are not intrinsically green, although in principle they can be designed to be environmentally more sustainable.23 The increasing spread of IL technologies and their applications is raising several concerns upon their accidental leakage into the environment (soil or aqueous ecosystem),24,25 as well as contact with biological organisms and human beings.26,27 It is matter of concern, for example, that the ionic liquid cation [C8MIM]+ has been recently detected in urban landfill sites as an exobiotic and major pollutant, causing toxicity in a liver model cell line.28 In this framework, the investigation of the toxic character of ILs represents a challenge due to their different nature and variety of contacts with different environments and biological organisms. In-depth and rigorous investigations are very important to determine the mechanism of interaction of ILs with living organisms in order to control and design environmentally safe ILs or establish a safety protocol for existing ionic liquids.Investigations focused on biological interaction of ionic liquids have highlighted a moderate to severe effect of several ILs on living organisms.29–31 Moreover, an eco-toxicity potential risk has been forecast due to accumulation in the environment because IL structures are very stable and are not easily biodegradable.27,32 In particular, the general toxic action of ILs on living organisms seems to be connected to the functional modification (or even poration/destruction in severe case) of the plasma membrane due to the nonspecific lipophilic interaction between the lipid membrane of the cells and the alkyl chain of the cations.
Toxicity indicators (in terms of critical concentrations, such as the half maximal effective concentration EC50) were found to be in linear correlation with lipophilicity parameters (i.e. water/octanol partition coefficient), highlighting the primary interaction of ILs on lipid-rich membranes.33,34 In a previous study from our group,35 the limits of detection (LODs) of electrochemical techniques for imidazolium ILs, (along with octanol/water partitioning coefficients) were quantitatively correlated with toxicity data, highlighting the role of the biomembrane in the toxicity mechanisms. The general observation of toxic/pharmacological effects are explained based on the assumption that IL-based compounds adsorb onto and/or intercalate into biomembranes,36,37 resulting in the structural perturbation of the biomembrane, its poration, and even disruption in severe cases.35 A variety of experimental and simulated conditions were investigated,27,32,38,39 confirming a direct link between the IL cation lipophilicity and toxicity, in particular related to the adsorption/intercalation of cations into biomembranes resulting in membrane perturbation/disruption and, eventually, cellular uptake.32–34,36,40 Cations with long hydrocarbon chains showed an increased lipophilic and therefore toxic character in comparison with shorter chain analogues.41 Bhattacharya et al.42 reported that the in-plane elasticity of supported monolayers decreases upon addition of the IL suggesting a relaxation of the rigid structure of lipids in the monolayer. Benedetto et al.43 performed molecular dynamics simulations suggesting the microscopic mechanism of the IL–biomembrane interaction, involving the ingress of cations into a phospholipid bilayer. The uptake of ILs is enhanced by the increasing temperature and fluidity of the lipid phase.44
Studies on the toxic character of ILs pave the way for their exploitation in the biological field attracting noteworthy interest in pharmaceutics.39 The state of art research for pharmaceutical applications is investigating promising paths in using ILs: (1) to enhance the performance of active pharmaceutical ingredients (APIs), providing novel drug-delivery options and serving as a solution for polymorphic molecule solvation;45,46 (2) as APIs themselves, modifying cations or anion chemical structures, in order to exploit their biological related properties.
Due to the importance of the interaction of ILs with the cell membrane in the determination of IL cytotoxicity, reconstituted biomembranes (such as solid-supported phospholipid bilayers) exposed to ILs can be used as model interfaces, to be studied by means of different surface techniques, for example atomic force microscopy (AFM),36,47,48 neutron scattering,49 X-ray,36,42,50 quartz crystal balance,51 nanoplasmonics,52 electrochemical impedance measurements,35 and by molecular dynamic simulations.43,53–55 Given the observed structural modifications induced by ILs on biomembranes, AFM is a promising technique to study the cytotoxicity mechanisms of ILs, since it allows a combined topographic and mechanical investigation of lipid membranes with nanoscale spatial resolution.56–60 Indeed, AFM has recently been used to investigate the morphological and mechanical modifications caused by the interaction of model biomembranes with ILs;48 aiming at targeting a more complex biological interface, we have recently performed a similar study on living cells exposed to ILs.61
In this study, we used AFM to carry out a systematic study of DOPC phospholipid bilayers interacting with imidazolium-based ILs at different concentrations. Combining topographic imaging with the mapping of several interfacial properties, such as Young's modulus of elasticity, adhesion force, and surface charge density, we were able to characterize the concentration dependent morphological, structural and mechanical evolution of the lipid membrane in the presence of ILs with different alkyl chain lengths and hydrophilic/hydrophobic characteristics.
Experimental section
Ionic liquids and imaging solutions
The ionic liquids used in experiments, with their specific nomenclature and abbreviation, are 1-butyl-3-methyl imidazolium chloride ([C4MIM][Cl]), 1-butyl-3-methyl imidazolium tetrafluoroborate ([C4MIM][BF4]), 1-butyl-3-methyl imidazolium bis(trifluoro methyl sulfonyl)imide ([C4MIM][NTf2]), 1-methyl-3-octyl imidazolium chloride ([C8MIM][Cl]), and 1-dodecyl-3-methyl imidazolium bis(trifluoro methyl sulfonyl)imide ([C12MIM][NTf2]). All the ionic liquids were purchased from Sigma Aldrich, each with a purity grade greater than 98.0%. To be stored in a clean way and to reduce water contamination, all the used ILs have been maintained in a desiccator under mild vacuum conditions (around 10−2 mbar).Aqueous electrolytic solutions containing ILs at different concentrations were prepared prior to AFM experiments on phospholipid bilayers. For different ILs, Table 1 reports some of the physicochemical properties relevant in this work, in particular density, melting point, the critical micelle concentration (CMC), the main character (hydrophobic/hydrophilic) and the octanol/water partition coefficient (KOW) as quantification of the hydrophilic character. In principle, the concentration of ILs was chosen based on their hydrophobicity as well as on their CMC. Imaging buffer was prepared with a solution of pH 7.4 phosphate buffered saline (PBS) or HEPES-buffered saline (HBS). Considering hydrophobic ionic liquids, starting solutions at a high concentration were prepared in methanol, while final operative concentrations were prepared maintaining a methanol content below 5%.
| Ionic liquid | Density (g cm−3) | Melting point (°C) | CMC (mM) | Character | K OW |
|---|---|---|---|---|---|
| a Extrapolated from CnMIM series data curve. | |||||
| [C4MIM][Cl] | 1086 | 70 | >100062 | Hydrophilic | 0.00463 |
| [C4MIM][BF4] | 121 | −75 | >100064 | Hydrophilic | 0.00363 |
| [C4MIM][NTf2] | 143 | −4 | 1264 | Hydrophobic | 0.3763 |
| [C8MIM][Cl] | 101 | 12 | 20062 | Hydrophilic | 0.2563 |
| [C12MIM][NTf2] | 125 | 21.9 | 0.0665 | Hydrophobic | 150a |
Phospholipid bilayer deposition
The phospholipid molecules (dioleoyl phosphatidylcholine (DOPC) from Avanti polar lipids) were initially dissolved in a chloroform solution at a concentration of 10 mg mL−1. The dried lipids (0.1 mg) are rehydrated using 1 mL of high ionic strength buffer solution 450 mM KCl, 25 mM HEPES (4-(2-hydroxy ethyl)-1-piperazine ethane sulfonic acid). A high ionic strength is necessary to shield effectively the electrostatic repulsive interaction between the negatively charged mica surface and phospholipid heads.66–68 After rehydration, and just before sample preparation for the AFM investigation, lipids are vortexed and sonicated for 10 minutes. The effect of sonication is to break the bigger multi-layered vesicles, favoring the formation of smaller unilamellar vesicles (better results were obtained using an ultrasonic tip rather than an ultrasonic bath). After sonication, the solution containing the lipids is deposited on a freshly cleaved mica surface in the amount of 40 μL with 15 minutes incubation time. The deposition protocol proceeds with the elimination of deposition buffer (HEPES + KCl), obtained with a gentle flux of nitrogen. Finally, the sample was rehydrated to perform AFM measurement in a fluid environment using the imaging buffer with dissolved ILs. The overall procedure was performed several times in order to ensure repeatability.AFM imaging
AFM imaging experiments were performed in PeakForce Tapping mode in a liquid environment using both Multimode and Bioscope Catalyst (Bruker) AFMs mounting ScanAsyst-Fluid probes (Bruker, nominal elastic constant k = 0.7 N m−1). For each morphological image, scan parameters were optimized in order to minimize the interaction force between probe and sample (F < 0.1 nN), still maintaining the highest quality possible. Topographic maps were typically acquired with a sampling resolution of 512 × 256 points, ramping frequency of 1 kHz and scan rate of 1 Hz, leading to a total acquisition time for each map of approximately 4 min. All AFM raw data were analyzed using custom Matlab routines. AFM images were flattened by subtracting up to 3rd order polynomials line-by-line, upon suitable masking of protruding features and defects, using the exposed substrate as reference, whenever possible, or alternatively the uniform flat bilayer surface.During data analysis, every image can be converted into a quantitative histogram helping to perform quantification of the thickness (h), bilayer coverage (A) and rms roughness (Rq). The relative increase in the surface area of the lipid bilayer upon interaction with the IL was calculated from the height histograms as (A − A0)/A0, where A and A0 are the peak values with and without IL.
As an example, we show in Fig. SI1 (ESI†) the AFM morphology of typical DOPC structures before interaction with the IL, with highlighted height profiles. The vertical profile section (red line, Fig. SI1b, ESI†) shows amorphous aggregates in the upper part of the image. The height and curvature of the aggregates demonstrate they have droplet-like shapes. Double bilayers (such as in the horizontal profile, Fig. SI1c, ESI†) have similar morphology and profile sections of single bilayers, typical of flat two-dimensional structures; in particular, these structures are rather smooth and flat, with heights (10–11 nm) twice the one measured on single bilayers (5–6 nm).
AFM nanomechanical and force spectroscopy measurements
AFM nanomechanical and force spectroscopy experiments were performed using the same Bioscope Catalyst or Multimode Nanoscope (Bruker) instruments equipped with soft silicon nitride MLCT-E probes (Bruker, nominal elastic constant k = 0.1 N m−1, nominal radius R = 20 nm) or other probes with similar or lower stiffness, allowing to cover the 10–104 pN force interaction range. The spring constant of each cantilever was calibrated by means of the thermal noise method.69,70Force curves
Nanomechanical and force spectroscopy analyses are based on the acquisition of raw force curves (FCs), involving the acquisition of the raw cantilever deflection signal from the photodetector (in volts) as a function of the z-piezo approaching (retracting) distance (in nanometers), while the AFM tip is ramped vertically on the sample.71 The tip reverses the motion when a setpoint deflection is reached, typically corresponding to a maximum force large enough to penetrate the lipid bilayer. Force vs. distance curves in physical units of nN and nm, respectively, like those shown in Fig. 1, are obtained by rescaling the raw FCs as explained in detail elsewhere.71,72 Briefly, the tip-sample distance (or indentation) is obtained by adding the cantilever deflection (in nm) to the z-piezo displacement; the force exerted by the tip (in nN) is retrieved by multiplying the raw photodetector deflection signal by the deflection sensitivity S (in nm V−1) and by the cantilever spring constant k (in N m−1). The deflection sensitivity S is measured as the inverse of the slope of the contact part of the FC after the probe breaks through the bilayer, reaching the substrate; the spring constant k is calibrated using the thermal noise method based on the equipartition theorem.70,73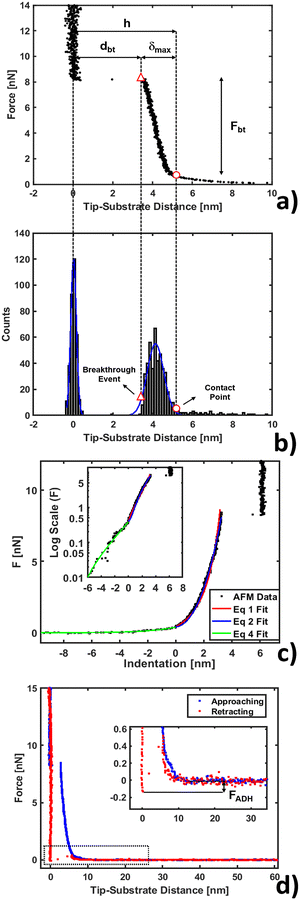 | ||
| Fig. 1 AFM force curve analysis for indentation on a phospholipid bilayer. (a) Example of force vs. tip-substrate distance curve performed on the DOPC bilayer supported on a mica surface, showing the contact point (○) and breakthrough event (△). (b) Corresponding histogram on the distance axis showing the Gaussian fit to individuate the substrate and layer. (c) Example of fit using eqn (1), (2) and (4) (red, blue and green lines) with a log scale inset to magnify the non-contact region. (d) Approaching and retracting curves overlapped with a magnification showing the adhesion well and closure of the phospholipid bilayer. | ||
Force mapping
By collecting several FCs distributed regularly across the sample surface (the dataset is also known as Force Volume, FV) it is possible to perform a spatially resolved force spectroscopic or nanomechanical analysis. From each FC, the local height of the sample, relative to a common point along the z axis, is calculated;72,74 it is therefore possible to reconstruct a topographic map of the sample under investigation, which is in one-to-one correspondence with the maps of other physical observables extracted from the FCs. Each FV, recorded in macroscopically separated locations, typically spans an area of 10 μm2 and consists of 64 × 64 or 10 × 10 FCs, for high-resolution and fast mapping purposes, respectively. Two distinct nanomechanical experiments are then performed. First, we recorded fast maps on freshly prepared bilayers, each time changing the IL concentration. This approach provides no information on the evolution dynamics of the system but allows precise control of both the interaction time and the IL concentration. Secondly, to study the evolution of the IL interaction compared with the control, high-resolution datasets were recorded: the scan area was fixed on a region of interest and datasets were acquired while increasing the IL concentration. Under these conditions, the effective interaction time is longer than the nominal one, because further increments of ILs are relative to a system that has already been exposed to lower concentrations; however, the interaction dynamics can be captured, thus complementing results from high-resolution scans and allowing for a more complete description of the IL–bilayer interaction.Force curve analysis on phospholipid bilayers
A typical FC acquired on a phospholipid bilayer is shown in Fig. 1a. Considering a vertical approach on the phospholipid bilayer, the FC presents specific and recognizable features.75–77 Upon contacting the surface at the contact point, the tip exerts a compressive stress on it, which first induces an elastic deformation of the bilayer, and then the rupture and penetration of the bilayer when a force threshold is reached (breakthrough event, corresponding to a breakthrough force Fbt). The nonlinear force vs. distance dependence in the compressive part of the FC accounts for the elastic resistance of the bilayer (δmax is the maximum compression, or indentation). Upon contact with the underlying hard substrate, the FC becomes vertical (tip-sample distance does not change). The distance between the breakthrough event and substrate is defined as the breakthrough distance dbt. Since we deal with a large number of FCs, we developed a statistical automatic procedure to identify the relevant points and quantities in the FCs (in particular contact point and breakthrough event), which is based on the study of the histogram of distances in the FC (Fig. 1b). The histogram typically contains distinct peaks corresponding to the distances at which the FC is steeper, which are the loading region on the bilayer from the contact point up to the breakthrough, and the contact region on the substrate. The onset of the peak corresponding to the loading of the bilayer can be identified with the contact point. We fit a Gaussian curve to the histogram to characterize the center of the distribution and its width w; the contact point is identified as the point that is found at a distance 2.5 × w before the center. We tested the validity of this empirical procedure of subsets of FCs and fixed it for all FCs; moreover, the same procedure is used for both the control system (bilayer without IL) and the system under study (bilayer with IL). In Table SI1 (ESI†) we present the error propagation on mechanical parameters when a ±0.5 nm uncertainty is artificially introduced in the contact point determined following the described procedure. We found out that the thickness maps reconstructed from the contact points are comparable within the error to the topographic maps acquired in standard imaging modes, including PeakForce Tapping.Mechanical analysis
After contact point determination, the distance axis of the FC is shifted (like in Fig. 1c) so that the zero of the distance axis corresponds to the upper surface of the lipid bilayer. With this representation, the positive distance axis is defined as the indentation δ. Here the force-indentation relationships can be employed to analyze the elastic properties of the lipid bilayers. First, we obtain the Young's modulus by fitting the Hertz model for a parabolic indenter to the force vs. indentation curves, with the finite-thickness correction valid for not bonded thin elastic layers supported on a solid substrate (eqn (1)):59,72,78,79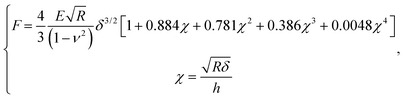 | (1) |
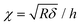 represents the ratio of the contact radius to the bilayer thickness and is the driving parameter of the finite thickness correction. For the calculation of χ, as well as for the fit with eqn (2), we have used the value of thickness h measured as described above. The Hertz equation is reasonably accurate also for spherical indenters as long as the indentation is small compared to the radius. Given the truly nanoscale structuring of the lipid bilayer, the relatively large deformations and the limits of validity of the continuous mechanics models we employ, we must consider the obtained elastic parameters as effective ones.
represents the ratio of the contact radius to the bilayer thickness and is the driving parameter of the finite thickness correction. For the calculation of χ, as well as for the fit with eqn (2), we have used the value of thickness h measured as described above. The Hertz equation is reasonably accurate also for spherical indenters as long as the indentation is small compared to the radius. Given the truly nanoscale structuring of the lipid bilayer, the relatively large deformations and the limits of validity of the continuous mechanics models we employ, we must consider the obtained elastic parameters as effective ones.
Additionally, we also fitted to the data the theoretical model described by Connel et al. for the indentation of a lipid bilayer, which should intrinsically take into account the finite thickness of the lipid structure and its supported nature:60
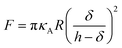 | (2) |
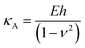 | (3) |
We show in Fig. 1c the results of fitting the loading part of a force vs. indentation curve using eqn (1) and (2) (red and blue lines respectively). In particular, the fit follows the experimental data until indentation reaches about half of the lipid bilayer thickness, at which point the elastic energy of the deformed bilayer overcomes the energetic cost of forming a hole and the tip penetrates the full bilayer in a breakthrough event. The results of the fit from Fig. 1c are shown in Table SI1 (ESI†), specifically the first line with precise determination of the contact point. We notice that the compressibility modulus from eqn (2) and from eqn (3) are very similar (198 ± 12 and 190 ± 9 mN m−1 respectively), therefore we decided to base our analysis on the results of the Hertzian fit (much simpler to implement) and to employ the Young's modulus as the main descriptor of elasticity in lipid bilayers.
It is worth noticing that a thin structured water layer is likely to be present between the hard substrate and the supported phospholipid bilayer. Because of the influence on the results of the mechanical analysis, we included an estimated thickness of the water layer (≈0.5 nm) in the effective thickness of the whole bilayer.80 The model employed with our data was previously confirmed with finite element simulations.78
Electrostatic analysis and adhesion force
We analyzed the double-layer (DL) electrostatic interaction force due to the presence of a net surface charge density (typically negative) on both tip and lipid bilayer surfaces.81 In our measurements, we noticed that before the mechanical contact with the lipid surface, the DL force is small (FDL < 0.5 nN), but not negligible.82 This force can be appreciated only at very small tip-sample separation, because the high ionic strength of the imaging buffer determines a screening Debye length smaller than 1 nm. The DL interaction should be taken into account in the mechanical analysis, because it could affect the determination of the tip-bilayer contact point; moreover, characterization of the net surface charge density of the lipid bilayer upon interaction with the IL can provide important information on the toxicity mechanisms, since it may be related to IL-related charge stratification at the lipid-water interface, and determine osmotic stresses due to charge gradients.61,83 In our case, we used the linearized version of the DL force:81,84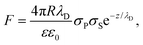 | (4) |
Finally, we show a complete force curve cycle in Fig. 1d, visualizing approaching and retracting branches. In the retracting portion of the force curve, the probe moves away from the sample, and the hole formed in the phospholipid bilayer closes behind the retracting tip. This can be inferred by the fact that after detachment the tip equilibrates under a positive force of 0.4 nN at a distance of 5–6 nm from the hard substrate, matching the bilayer thickness, overlapping the approaching curve (blue), which describes, in this range of distances, the electrostatic repulsion between the tip and bilayer. In addition, the hole filling is witnessed by the fact that we repeatedly observe the same penetration pattern when ramping several times on the same location. In the retracting FC, the adhesion force FADH is defined as the force required by the probe to detach from the substrate and reach the non-interacting ‘zero’ force. A typical adhesion well is visible in the zoom of the inset of Fig. 1d. Additional details about adhesion analysis can be found in Butt et al.71
Statistics
For a single force mapping dataset, the distribution of the measured values of each quantity (such as h, Fbt, E etc.) is represented as a histogram, with the corresponding Gaussian fit. The center and the width σ of the Gaussian curve represent the mean value and the standard deviation, respectively, of the single measurement. The uncertainty associated with a single measurement is calculated by summing in quadrature the standard deviation of the mean σ/sqrt(N), with N being the number of FCs in the map, and an estimated overall uncertainty in the probe calibration procedure (≈10%). Eventually, the best estimator of the true value for each condition (represented by a group of force maps) was obtained as the mean of the single measurement values, and the corresponding error was calculated as the standard deviation of the mean, adding in quadrature the calibration uncertainty. In the case of further operations, such as normalization with respect to the control, error propagation was applied to estimate the final error.Results and discussion
Morphological variations of lipid bilayers interacting with ILs
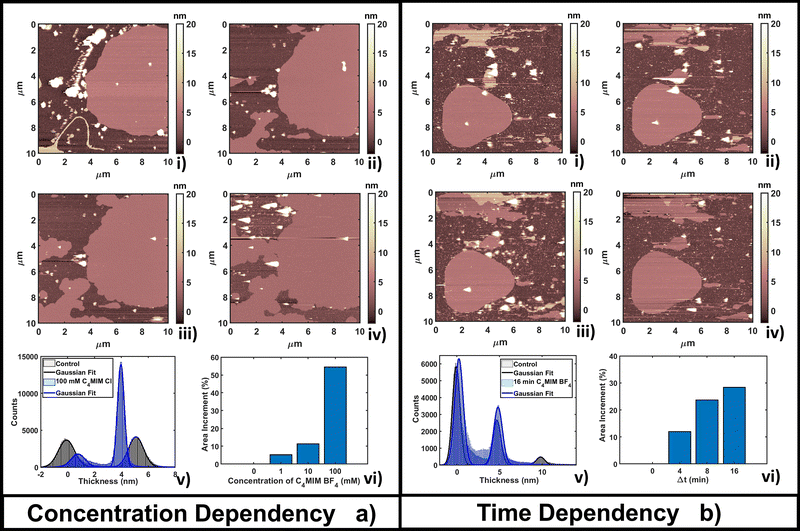
The time-dependent behavior of [C4MIM][BF4] at a fixed concentration of 10 mM is shown in Fig. 2b; frames were captured every 4–8 minutes as shown in Fig. 2b(i–iv). From the statistical analysis in Fig. 2b(vi) we observed an increase of coverage for the single DOPC bilayer up to +28% after 16 minutes (see also Fig. 2b(iv)), while the thickness of the lipid structures is conserved (Fig. 2b(v)). We observed a restructuring of the less ordered DOPC material (amorphous and partially formed double bilayers), increasing the extent of single bilayer coverage. A similar increase of phospholipid bilayer lateral extension was observed by Schaffran et al.87 showing the reorganization of liposomes in solution after adding ILs: short-tailed ILs promoted the reorganization of unilamellar liposomes in the extended bilayer sheets. The authors explained this fact as the short-tailed ILs act as “edge-actants”, stabilizing and bridging open membrane structures in water. The system in our study is supported by a solid surface; we argue that ILs with short lateral tails promote the two-dimensional growth of IL–lipid hybrid bilayers from the edges of the pristine DOPC bilayers. Such hybrid IL–lipid nanostructures were also observed by Jing et al.,36 who studied the influence of ILs at different concentrations on α-PC (L-α-phosphatidylcholine), reporting swelling and formation of α-PC/ILs complexes for concentrations lower then CMC, and bilayer disruption for concentrations higher than CMC. Although carried out under different experimental conditions, a recent AFM study by Rotella et al.48 confirmed how a ‘short tail’ IL such as [C4MIM][Cl] enhanced the recovery of uniform lipid bilayers by filling the holes and irregularities of the original layers.
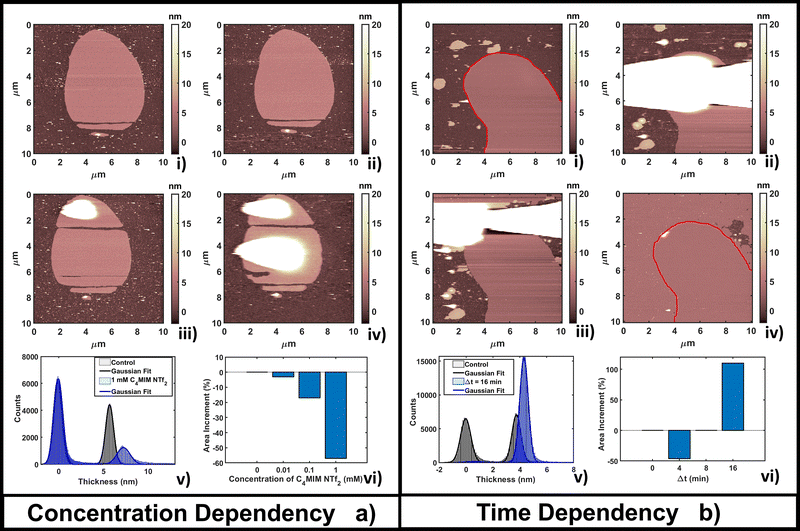
Fig. 3b shows the time evolution of DOPC at a fixed concentration (1 mM) of [C4MIM][NTf2]. It is interesting to observe that a longer interaction time causes a transition from a segregated liquid (Fig. 3b(ii)) to an almost complete bilayer covering the substrate (Fig. 3b(iv)). A red line is used as a guide to the eye to define the profile of the original DOPC bilayer. Generally, we noticed the formation of new layered structures similar to the lipid bilayer but characterized by increased roughness and local irregularities. Because of the good reliability of the flattening procedure on the uniform bilayer surface (Fig. 3b(iv)), the roughness analysis is also reliable, and shows an increase, from Rq = 0.30 nm on the pristine bilayer, to Rq = 0.38 nm on the newly formed layer. Since holes and drops were excluded from the analysis, the roughening process is probably related to the formation of DOPC/ILs hybrid complexes, where ionic liquids act as a bridging material between amorphous and regular bilayer structures. Computational analysis was able to observe segregation because of its short time scale;55 here, by AFM, we could capture the system evolution (lateral growth and roughening of the IL-exposed bilayer) across a much longer time scale. We cannot exclude that these processes are facilitated by the mechanical action of the scanning tip, which may help by redistributing and restructuring loosely bound IL–lipid material.
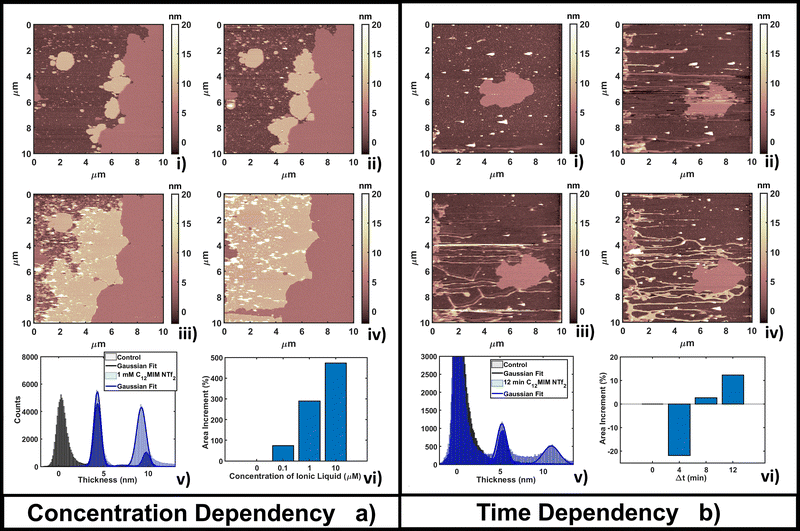
This behavior can be explained by considering that the high lipophilic character of [C12MIM][NTf2] favors the formation of extended (and thicker) composite layers, while the hydrophobic nature favors the segregation of the unreacted fraction (the smallest, brightest spots in Fig. 4a(ii–iv)). Formation of multi-layer structures for ILs with longer alkyl chains in the cation was confirmed in other studies.87 The formation of [C12MIM][NTf2]/DOPC complexes was observed by electrochemical methods and simulations.35,88 A DOPC monolayer on mercury electrodes underwent irreversible structural reorganization upon interaction with [C12MIM][NTf2], indicating a strong integration of ILs in the regular lipid monolayer structure. The electrochemical fingerprint of IL/DOPC complexes was a convolution of single part signatures (pure DOPC and pure [C12MIM][NTf2]).
The time dependent investigation of the IL-DOPC bilayer interaction was performed at the lowest fixed concentration (0.1 μM) of [C12MIM][NTf2]. Fig. 4b shows that the area and the shape of the lipid bilayer patch do not change dramatically across a time span of 16 min. Area analysis (Fig. 4b(vi)) shows that there is an initial area reduction, due to re-equilibration, during the first 8 minutes (Fig. 4b(i–ii)), followed by a gradual recovering of the initial value. The appearance (in Fig. 4b(ii)) and the evolution of filamentous and locally planar structures (such as those in the left border of the scanned area) suggest that loosely bound IL–lipid matter is displaced by the scanning tip and redistributed across the scan area, where it reassembles; the dendritic structures indeed grow preferentially along the fast tip scanning direction, suggesting an active role of the scanning tip. Height histogram analysis (Fig. 4b(v)) shows that the thickness of the formed structures, either elongated or more extended, is compatible with both a single and a double lipid bilayer (a second peak appears at 10.7 nm, about twice the thickness of the single bilayer). Similar dendritic structures were observed by confocal laser scanner microscopy by Jing et al.;36 in particular, the coexistence of fibers/tubes with multi-layer aggregates upon interaction with IL (depending on the concentration and lipophilicity of cations).
The time lapse analysis supports the hypothesis that the IL is able to interact strongly with the phospholipids, forming a composite material that maintains most of the pristine lipid bilayer properties including the typical bilayer structure. At higher concentrations or after a longer interaction time, more extended and less defected layers like those observed in Fig. 3b can form, also depending on the hydrophobicity of the IL.
Mechanical analysis of DOPC bilayers interacting with ILs
We analyzed the nanomechanical properties of hydrophilic ILs only, since the observed segregation phenomena suggest that hydrophobic ILs may not be uniformly interacting with the lipid bilayer, therefore making the measurements unreliable. Moreover, we observed already that an AFM tip displacing an IL droplet can be easily and irreversibly covered by an IL layer that would negatively affect all measurements.89We used [C4MIM][Cl] and [C8MIM][Cl] as the most common water-soluble ILs with short and long lateral tails to explain the mechanical modifications of phospholipid bilayers upon interaction with ILs. The single measurements with their associated errors are presented in Table SI2 (ESI†) (3–5 experiments for [C4MIM][Cl] and 1 for [C8MIM][Cl]), where we also highlighted the percentage variation with respect to the control experiment. Examples of approaching force curves collected on a DOPC bilayer before and after interaction with 1 mM and 100 mM of [C4MIM][Cl] are shown in Fig. 5. The short-scale steps observed before the breakthrough events are artifacts due to either the digitization of the deflection signal or to vibrational noise from the floor, or both, and do not affect the results. Fig. 5 shows that the IL weakens the mechanical stability of the DOPC bilayer, i.e., it reduces the breakthrough force, as well as its rigidity: as a matter of fact, the Young's modulus is proportional to the slope of the loading portion of the force curve, and the FCs are less steep after interaction with the IL.
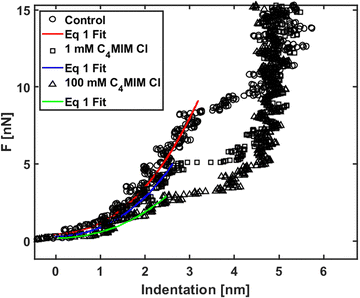 | ||
| Fig. 5 Force curves on phospholipid bilayers interacting with [C4MIM][Cl] at different concentrations after equilibration time Teq = 20 min. Typical examples of force vs. indentation (after contact) with fitting to eqn (1). | ||
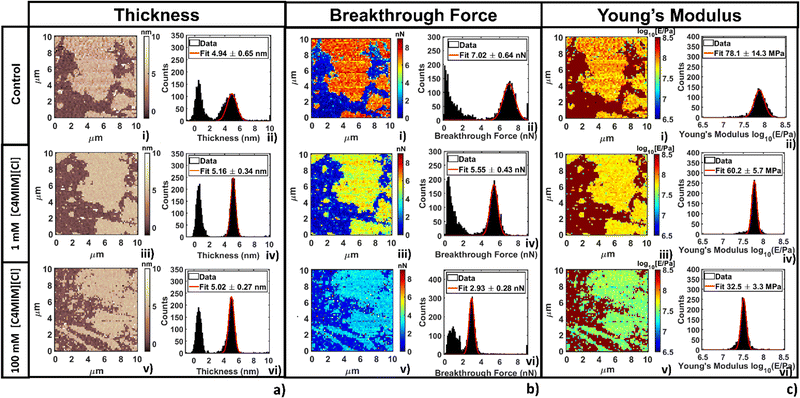
The thickness of phospholipid bilayers can be measured from topographic maps, as explained in the Methods section. No significant modification of the height (thickness) of the bilayers was detected, as shown in Fig. 6a. In particular, it is interesting that the same thickness was measured by either direct topographic imaging (5.5 nm, shown in previous Fig. 2–4) or by nanomechanical imaging (4.9–5.9 nm, Fig. 6a). Both analyses confirmed that modification of the thickness measured by AFM are within the experimental error.
In Fig. 6b we show breakthrough force maps and related force histograms for DOPC bilayers on mica before and after interaction for 20 min with 1 mM and 100 mM [C4MIM][Cl], respectively. Depending on the probe, the average values of breakthrough force can typically vary between 2 and 14 nN. The mean breakthrough force, as extracted through a gaussian fit of the histogram, presents a first −15% variation at 1 mM, and a remarkable −62% variation at 100 mM, with respect to the control system (Fig. 6b(i)).
The maps of Young's modulus and the corresponding histograms for DOPC bilayers on mica before and after interaction for 20 min with 1 mM and 100 mM [C4MIM][Cl], respectively, are shown in Fig. 6c. Depending on the probe used and the different experimental conditions, the Young's modulus of control DOPC is between 20–100 MPa (the Young's modulus of the mica substrate was arbitrarily set at 109 Pa in all maps). Similarly to the breakthrough force, the Young's modulus decreases −28% and −49% as the concentration of [C4MIM][Cl] increases from 1 mM to 100 mM, respectively (see Fig. 6c(iv–vi)).
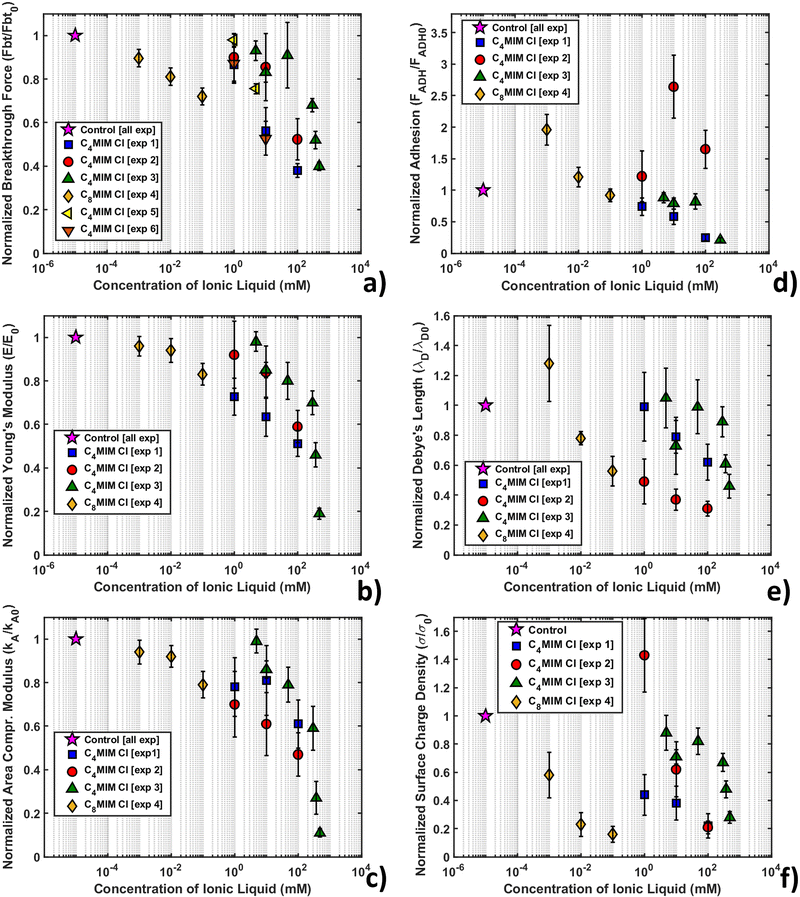
Fig. 7 shows the variation of the measured normalized quantities with the concentration of IL, as observed in the course of several different replicated experiments, for the same nominal conditions. The adopted normalization with respect to the measurements performed with the same AFM probe in the absence of IL should remove the dependence on the tip radius, deflection sensitivity and other experimental parameters, which directly influence the measured forces and distances. Given that the same common trends are observed through all the experiments, a certain quantitative variability is observed in the reported concentration-dependent curves for the same ILs, recorded during different experiments performed in the same nominal conditions.
Fig. SI2 (ESI†) shows the normalized thickness, maximum indentation, and breakthrough distance versus IL concentration. As the IL concentration increases, we observe an increase of the maximum indentation (δmax), while the thickness (h) and breakthrough distance (dbt) are similar to the control values within the experimental error for all concentrations of IL.
All experiments consistently show a remarkable decrease of breakthrough force (Fig. 7a), Young's modulus (Fig. 7b) and area compressibility modulus (Fig. 7c) at progressively increasing concentrations of the IL. The increase of the maximum indentation shown in Fig. SI2 (ESI†) is compatible with the softening and structural weakening of the lipid bilayer. The interaction of ILs with the lipid membrane is concentration dependent and inherent to the lipophilic nature of the cation (through the length of the alkyl chain), as demonstrated by the different concentration ranges of [C4MIM][Cl] (1–100 mM) and [C8MIM][Cl] (1–100 μM). Our results suggest that the hydrocarbon tail of the IL strongly interact with the hydrophobic tails of the phospholipids inside the bilayer, which causes a relaxation of the bilayer structure that makes the layer softer (i.e. reduces the Young's and the area compressibility moduli) and more vulnerable to the tip indentation (i.e. it reduces the breakthrough force).
Although the decreasing trend of the mechanical quantities is obeyed in all experiments, the rate of decrease is not always the same. As mentioned, normalization with respect to the control without IL should remove any residual dependence on the tip radius and/or on the accuracy of the calibration of both the cantilever spring constant and deflection sensitivity, therefore other factors should explain the observed differences. One such factor could be the different timing of the IL–bilayer interaction in the different experiments; for example, in experiments 1, 4 and 6 (see the legend in Fig. 7), the combined topographic and mechanical analysis was performed on the same lipid bilayer domains, which have been sequentially exposed to increasing concentrations of ILs, while large datasets (4096 curves) were acquired (with an acquisition time in the order of 20 min) (as shown for example in Fig. 6). This experimental strategy aimed at studying the kinetics of IL–bilayer interaction on the same location. Clearly, the effective interaction times at a given IL concentration under these experimental conditions were increasingly longer, which may explain why the observed mechanical effects are stronger and faster. Conversely, in experiments 2, 3 and 5, different bilayers have been studied at different IL concentrations, while keeping the interaction time fixed (20 min); smaller datasets (225 curves) were acquired (acquisition time approximately 4 min) in different locations, before and after the interaction with the IL. The effective interaction time under these conditions were both shorter and more precisely controlled, which explains the observed weaker and slower effects.
Nevertheless, we should point out that a supported phospholipid bilayer system is very sensitive to temperature, ionic strength and contamination (for example lipid and IL residues on the AFM tip), especially during the mechanical investigation. Rotella et al.48 performed indentation experiments on supported POPC bilayers interacting with 0.5 M [C4MIM][Cl] in deionized milliQ water (instead of a physiological buffer like in our experiments), measuring a slight increase of thickness (0.5 nm) and slight increase of all mechanical parameters. To explain this behavior, Dusa et al.90 using QCM proposed a “carpet” mechanism where ILs first adsorb onto the phospholipid layer and after a certain amount of time, when a time/concentration threshold value is reached, begin intruding into the lipid bilayer. Different working solutions could cause a modification of this threshold value. It is possible that Rotella et al. characterized the bilayer during the initial carpet regime, which could explain the observed trend, which is opposed to the one reported in this work and in others.42,53,91 To support this hypothesis, we report that our research group recently investigated the interaction of a DOPC bilayer with a common lipid dye (Nile red).78 These hydrophobic molecules are designed to target the lipid layer avoiding structural damage. AFM indentation indeed showed the presence of a stabilizing overlayer of dye on top of the exposed leaflet causing an increase of thickness, breakthrough force and Young's modulus, similarly to Rotella et al.48 In a recent work, Kumari et al.91 showed that after 5 hours interaction with DOPC, [C4MIM][Cl] can overcome the interaction energy threshold, showing a dramatic decrease of stability and mechanical properties as a consequence of cation tails inserting in the bilayer.
The lowered mechanical stability of the lipid membrane found by AFM nanomechanical measurements is in agreement with previous electrochemical results, where inclusions of IL tails inside the lipid layer contributed to destabilize the lipid structure.35 A confirmation of the modification of the mechanical properties of the lipid bilayer after interaction with dispersed IL was provided by simulation works performed by Bingham et al.55 and Yoo et al.53,54 Bingham et al.55 simulated a POPC bilayer in pure water. After the addition of [C4MIM][Cl] (500 mM) to the solution, the area compressibility modulus decreased by −15%, while in the [C4MIM][PF6] case, the effect was quantitatively more important, with the variation reaching −45%. Yoo et al.53,54 simulated a system similar to our experiments using POPC in buffer solution (NaCl) with [C4MIM][Cl]. Variation predicted by simulation showed a −40% decrease of area compressibility modulus, in line with the trend of our results. Using pressure-area isotherms, Bhattacharya et al.42 confirmed the penetration of IL molecules in a lipid monolayer and an IL concentration-dependent decrease in the in-plane rigidity, following the trend of our AFM results. Kundu et al.92 analyzed the effect of ILs on liposomes using dissipative quartz crystal microbalance (QCM-D). Voigt-based viscoelastic modeling and the temperature-dependence of frequency shifts underlined a shear viscosity profile displaying a clear shift downwards of the main phase transition upon the addition of ILs.51
Adhesive and electrostatic behavior of DOPC bilayers interacting with ILs
The maps and histograms of adhesion force relative to the control for a DOPC bilayer interacting with 1 mM and 100 mM [C4MIM][Cl] are shown in Fig. SI3a (ESI†), while the mean adhesion force versus IL concentration curve is shown in Fig. 7d. The adhesion force decreases with increasing IL concentration. To interpret this result, we observe that while, after the breakthrough event, the tip is retracted from the underlying mica substrate, the bilayer closes around it, interacting with the tip walls. We can argue that, as the IL weakens and disrupts the lipid bilayer integrity, the AFM tip encounters less resistance upon extraction. A similar effect was recently visualized studying the interaction with Nile red hydrophobic staining agent on the DOPC bilayer, with and without laser illumination.78 Using Nile red, the adhesion force increases in agreement with the presence of a dye overlayer on top of the DOPC molecules, improving their stability (breakthrough force and Young's modulus). On the other hand, when focusing the laser light on the bilayer, the DOPC structure weakens, leading to a decrease of mechanical parameters and adhesion force.The double layer electrostatic force (eqn (4)) at the water–lipid bilayer interface was studied as a function of the IL concentration. In PBS or HBS at pH 7.4, the Si3N4 AFM tip is (weakly) negatively charged, since the isoelectric point of silicon nitride is typically between 6 and 7.5 pH units; in order to capture the order of magnitude of the surface charge density of the lipid bilayer, we assumed a surface charge density σP = −9 × 10−3 C m−2.85 Nevertheless, the normalization of the surface charge density with respect to the control sample without IL removes the dependence on both the tip radius and the charge density.
As shown in Fig. 1c, a repulsive interaction between the AFM tip and the lipid bilayer is evident in the absence of IL, which suggests a negatively charged DOPC bilayer surface, in agreement with measurements performed under similar conditions.81 We also observed that the measured Debye length (λD) is between 0.5 and 2 nm for all measurements, as expected for high ionic strength electrolytes. The maps and quantification of Debye length are presented in Fig. SI3b (ESI†), showing a slight decrease (although not significant) while increasing the IL concentration.
As shown in Fig. SI3c (ESI†), the surface charge of the DOPC bilayer decreases in absolute value when the IL concentration increases. Considering that cations first interacting with the negatively charged lipid bilayer surface and also through their hydrocarbon tails, our results suggest that the crowding of cations on the bilayer surface is likely, leading to a progressive neutralization of the pristine negative surface charge of the lipid bilayer outer leaflet.
The overall behavior using different probes on [C4MIM][Cl] and [C8MIM][Cl] was averaged and reported in Table SI3 (ESI†) (with absolute values), while the dependence of the normalized surface charge density on the IL concentration is shown in Fig. 7f.
Our results show that besides modifying the mechanical stability of the lipid bilayers, ILs also modify their surface charge density, an effect that can influence the ionic transport in membranes of living cells and/or produce interfacial crowding of ionic species. For example, Ryu et al.93 showed that gramicidin channels in phospholipid membranes are influenced by ILs, depending on their concentration and lipophilicity. In particular, the decreased channel conductance upon interaction with ILs was explained by increased cationic positive charge accumulation, an effect that we likely observed in this work. The increase of cations on the surface of phospholipids was also observed as an increase of surface potential.94 Crowding of cations on phospholipid membranes could induce hyperosmotic shock in a short interaction time. This effect was visualized on metastatic cancer cells after 20 min,61 where a high concentration (100 mM) of [C4MIM][Cl] caused a decrease in cell volume and increase in cell stiffness as a consequence of water expulsion, which increased the viscosity of the cytoplasm. On top of this, a cross-talk interaction between mechanical properties and electrostatic effect on the charged phospholipid membrane was recently observed.95 The evolution of surface charge at the membrane level is thus important to understand the effect of ILs on the natural complex membranes of living cells.
Conclusions
We have studied the interaction of imidazolium-based ILs with model biological membranes consisting of DOPC phospholipid bilayers supported on mica surfaces by means of atomic force microscopy, carrying out combined morphological, mechanical and short-range electrostatic characterization of the IL/lipid bilayer interface. Most of the ILs interacting with DOPC bilayers showed a visible and measurable effect on their morphological and structural properties. Our findings can be summarized as follows:- The strength of the IL-DOPC bilayer interaction is related to the length of the alkyl chain of the IL cation. The experimental evidence supports the hypothesis that the specific affinity of the IL alkyl chains for the hydrophobic tails of the phospholipids leads to (a) crowding of the IL cations at the exposed lipid bilayer leaflet and (b) penetration (tail-first) of the cation into the bilayer structure.
- Upon interaction with imidazolium-based ILs, the pristine DOPC bilayer structure is mechanically and structurally weakened, as witnessed by a reduction in the breakthrough force, as well as of the Young's and area compressibility moduli; at higher concentration, the membrane locally reorganizes into a composite phospholipid-IL material and, eventually, is disrupted.
The noticeable decrease in the breakthrough force and Young's modulus indicates a lowering in mechanical stability but the bilayers are more compliant. This finding is important in the IL toxicity scenario, since it suggests that the primary interaction channel of ILs with living cells is localized at the outer cell membrane, where, depending on their concentration and alkyl chain length, the ILs induce structural reorganization and softening, and likely a permeabilization of the membrane. On the other hand, AFM analysis was able to capture modification of the phospholipid bilayer surface charge density coherent with positive cationic crowding on the exposed leaflet.
The results obtained in this work provide information about the activity of widely used and investigated imidazolium based ILs at the biomembrane level, relating their structure to different degrees of interaction. Tuning the interaction of ILs with the biomembrane, by modifying the chemical structure of ions, would allow the design of novel ILs that satisfy strict environmental sustainability and low cytotoxicity constraints, but also provide novel membrane targeting molecules for biomedical applications.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
M. G. acknowledges the financial support from the National Natural Science Foundation of China (No. 21950410518 and 32071318), the Shenzhen Basic Research Project (No. JCYJ20190807163007525) and the Youth Innovation Promotion Association Chinese Academy of Sciences (2022363). A. P., A. C. and M. Giann. acknowledge the financial support from the EU ERC Advanced grant no MAGIC (No. 695078). We thank Jacopo Nicolò Toffanetti and Davide Piotti for support in the preparation and AFM imaging of lipid bilayers.References
- J. S. Wilkes, P. Wasserscheid and T. Welton, Ionic Liquids in Synthesis, Wiley-VCH Verlag GmbH & Co. KGaA, 2008, pp. 1–6 Search PubMed.
- J. D. Holbrey and K. R. Seddon, Clean Products and Processes, 1999, 1, 223–236 Search PubMed.
- J. P. Hallett and T. Welton, Chem. Rev., 2011, 111, 3508–3576 CrossRef CAS PubMed.
- M. Galiński, A. Lewandowski and I. Stępniak, Electrochim. Acta, 2006, 51, 5567–5580 CrossRef.
- C. Zhong, Y. Deng, W. Hu, J. Qiao, L. Zhang and J. Zhang, Chem. Soc. Rev., 2015, 44, 7484–7539 RSC.
- L. G. Bettini, P. Piseri, F. De Giorgio, C. Arbizzani, P. Milani and F. Soavi, Electrochim. Acta, 2015, 170, 57–62 CrossRef CAS.
- S. Shahzad, A. Shah, E. Kowsari, F. J. Iftikhar, A. Nawab, B. Piro, M. S. Akhter, U. A. Rana and Y. Zou, Glob Chall, 2019, 3, 1800023 CrossRef PubMed.
- S. Pan, M. Yao, J. Zhang, B. Li, C. Xing, X. Song, P. Su and H. Zhang, Front Chem., 2020, 8, 261 CrossRef CAS PubMed.
- S. M. Zakeeruddin and M. Gratzel, Adv. Funct. Mater., 2009, 19, 2187–2202 CrossRef CAS.
- D. K. Pandey, H. L. Kagdada, A. Materny and D. K. Singh, J. Mol. Liq., 2021, 322, 114538 CrossRef CAS.
- C. P. Lee and K. C. Ho, Eur. Polym. J., 2018, 108, 420–428 CrossRef CAS.
- M. A. Navarra, MRS Bull., 2013, 38, 548–553 CrossRef CAS.
- A. Lewandowski and A. Świderska-Mocek, J. Power Sources, 2009, 194, 601–609 CrossRef CAS.
- H. Z. Niu, L. Wang, P. Guan, N. Zhang, C. R. Yan, M. L. Ding, X. L. Guo, T. T. Huang and X. L. Hu, J. Energy Storage., 2021, 40, 102659 CrossRef.
- H. Xiao, Tribol. Trans., 2017, 60, 20–30 CrossRef CAS.
- M.-D. Bermúdez, A.-E. Jiménez, J. Sanes and F.-J. Carrión, Molecules, 2009, 14, 2888–2908 CrossRef PubMed.
- M. Antunes, M. T. Donato, V. Paz, F. Caetano, L. Santos, R. Colaco, L. C. Branco and B. Saramago, Tribol. Lett., 2020, 68, 70 CrossRef CAS.
- J. Song, ACS Omega, 2021, 6, 29345–29349 CrossRef CAS PubMed.
- M. M. Seitkalieva, D. E. Samoylenko, K. A. Lotsman, K. S. Rodygin and V. P. Ananikov, Coord. Chem. Rev., 2021, 445, 213982 CrossRef CAS.
- M. J. Earle and K. R. Seddon, Pure Appl. Chem., 2000, 72, 1391–1398 CrossRef CAS.
- Z. Yang and W. B. Pan, Enzyme Microb. Technol., 2005, 37, 19–28 CrossRef CAS.
- H. Zhao, S. Q. Xia and P. S. Ma, J. Chem. Technol. Biotechnol., 2005, 80, 1089–1096 CrossRef CAS.
- R. D. Rogers and K. R. Seddon, Science, 2003, 302, 792–793 CrossRef PubMed.
- M. Deetlefs and K. R. Seddon, Green Chem., 2010, 12, 17–30 RSC.
- M. C. Bubalo, K. Radosevic, I. R. Redovnikovic, J. Halambek and V. G. Srcek, Ecotoxicol. Environ. Saf., 2014, 99, 1–12 CrossRef PubMed.
- K. S. Egorova and V. P. Ananikov, ChemSusChem, 2014, 7, 336–360 CrossRef CAS PubMed.
- A. Jordan and N. Gathergood, Chem. Soc. Rev., 2015, 44, 8200–8237 RSC.
- P. M. Probert, A. C. Leitch, M. P. Dunn, S. K. Meyer, J. M. Palmer, T. M. Abdelghany, A. F. Lakey, M. P. Cooke, H. Talbot, C. Wills, W. McFarlane, L. I. Blake, A. K. Rosenmai, A. Oskarsson, R. Figueiredo, C. Wilson, G. E. Kass, D. E. Jones, P. G. Blain and M. C. Wright, J. Hepatol., 2018, 69, 1123–1135 CrossRef CAS PubMed.
- S. Stolte, M. Matzke, J. Arning, A. Böschen, W.-R. Pitner, U. Welz-Biermann, B. Jastorff and J. Ranke, Green Chem., 2007, 9, 1170 RSC.
- J. Ranke, S. Stolte, R. Stormann, J. Arning and B. Jastorff, Chem. Rev., 2007, 107, 2183–2206 CrossRef CAS PubMed.
- J. Ranke, K. Molter, F. Stock, U. Bottin-Weber, J. Poczobutt, J. Hoffmann, B. Ondruschka, J. Filser and B. Jastorff, Ecotoxicol. Environ. Saf., 2004, 58, 396–404 CrossRef CAS PubMed.
- S. Stolte, M. Matzke and J. Arning, Ionic Liquids Completely UnCOILed, John Wiley & Sons, Inc., 2015, pp. 189–208 Search PubMed.
- A. Benedetto, Biophys. Rev., 2017, 9, 309–320 CrossRef CAS PubMed.
- J. Ranke, A. Muller, U. Bottin-Weber, F. Stock, S. Stolte, J. Arning, R. Stormann and B. Jastorff, Ecotoxicol. Environ. Saf., 2007, 67, 430–438 CrossRef CAS PubMed.
- M. Galluzzi, S. Zhang, S. Mohamadi, A. Vakurov, A. Podesta and A. Nelson, Langmuir, 2013, 29, 6573–6581 CrossRef CAS PubMed.
- B. Jing, N. Lan, J. Qiu and Y. Zhu, J. Phys. Chem. B, 2016, 120, 2781–2789 CrossRef CAS PubMed.
- J. Ranke, A. Müller, U. Bottin-Weber, F. Stock, S. Stolte, J. Arning, R. Störmann and B. Jastorff, Ecotoxicol. Environ. Saf., 2007, 67, 430–438 CrossRef CAS PubMed.
- M. Petkovic, K. R. Seddon, L. P. Rebelo and C. Silva Pereira, Chem. Soc. Rev., 2011, 40, 1383–1403 RSC.
- K. S. Egorova, E. G. Gordeev and V. P. Ananikov, Chem. Rev., 2017, 117, 7132–7189 CrossRef CAS PubMed.
- M. Alvarez-Guerra and A. Irabien, Green Chem., 2011, 13, 1507 RSC.
- S. Jeong, S. H. Ha, S.-H. Han, M.-C. Lim, S. M. Kim, Y.-R. Kim, Y.-M. Koo, J.-S. So and T.-J. Jeon, Soft Matter, 2012, 8, 5501–5506 RSC.
- G. Bhattacharya, R. P. Giri, H. Saxena, V. V. Agrawal, A. Gupta, M. K. Mukhopadhyay and S. K. Ghosh, Langmuir, 2017, 33, 1295–1304 CrossRef CAS PubMed.
- A. Benedetto, R. J. Bingham and P. Ballone, J. Chem. Phys., 2015, 142, 124706 CrossRef PubMed.
- A. Benedetto and E. G. Kelley, J. Phys. Chem. B, 2022, 126, 3309–3318 CrossRef CAS PubMed.
- S. Jain, N. Patel and S. Lin, Drug Dev. Ind. Pharm., 2015, 41, 875–887 CrossRef CAS PubMed.
- A. Benedetto and P. Ballone, ACS Sustainable Chem. Eng., 2016, 4, 392–412 CrossRef CAS.
- K. O. Evans, J. Phys. Chem. B, 2008, 112, 8558–8562 CrossRef CAS PubMed.
- C. Rotella, P. Kumari, B. J. Rodriguez, S. P. Jarvis and A. Benedetto, Biophys. Rev., 2018, 10, 751–756 CrossRef CAS PubMed.
- A. Benedetto, F. Heinrich, M. A. Gonzalez, G. Fragneto, E. Watkins and P. Ballone, J. Phys. Chem. B, 2014, 118, 12192–12206 CrossRef CAS PubMed.
- I. Kontro, K. Svedstrom, F. Dusa, P. Ahvenainen, S. K. Ruokonen, J. Witos and S. K. Wiedmer, Chem. Phys. Lipids, 2016, 201, 59–66 CrossRef CAS PubMed.
- P. Losada-Perez, M. Khorshid and F. U. Renner, PLoS One, 2016, 11, e0163518 CrossRef PubMed.
- J. Witos, G. Russo, S.-K. Ruokonen and S. K. Wiedmer, Langmuir, 2017, 33, 1066–1076 CrossRef CAS PubMed.
- B. Yoo, B. Jing, S. E. Jones, G. A. Lamberti, Y. Zhu, J. K. Shah and E. J. Maginn, Sci. Rep., 2016, 6, 19889 CrossRef CAS PubMed.
- B. Yoo, Y. Zhu and E. J. Maginn, Langmuir, 2016, 32, 5403–5411 CrossRef CAS PubMed.
- R. J. Bingham and P. Ballone, J. Phys. Chem. B, 2012, 116, 11205–11216 CrossRef CAS PubMed.
- A. Alessandrini and P. Facci, Micron, 2012, 43, 1212–1223 CrossRef CAS PubMed.
- S. Garcia-Manyes, L. Redondo-Morata, G. Oncins and F. Sanz, J. Am. Chem. Soc., 2010, 132, 12874–12886 CrossRef CAS PubMed.
- L. Picas, F. Rico and S. Scheuring, Biophys. J., 2012, 102, L01–03 CrossRef CAS PubMed.
- F. W. S. Stetter, S.-H. Hyun, S. Brander, J. M. Urban, D. H. Thompson and T. Hugel, Polymer, 2016, 102, 326–332 CrossRef CAS.
- C. Das, K. H. Sheikh, P. D. Olmsted and S. D. Connell, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2010, 82, 041920 CrossRef PubMed.
- M. Galluzzi, C. Schulte, P. Milani and A. Podesta, Langmuir, 2018, 34, 12452–12462 CrossRef CAS PubMed.
- M. Blesic, M. H. Marques, N. V. Plechkova, K. R. Seddon, L. S. P. N. Rebelo and A. N. Lopes, Green Chem., 2007, 9, 481 RSC.
- L. Ropel, L. S. Belveze, S. N. V. K. Aki, M. A. Stadtherr and J. F. Brennecke, Green Chem., 2005, 7, 83–90 RSC.
- W. Zhang, Q. Hu, S. Jiang, L. Wang, J. Chai and J. Mei, Environ. Sci. Pollut. Res., 2022, 29, 46325–46340 CrossRef CAS PubMed.
- I. Rodriguez-Palmeiro, O. Rodriguez, A. Soto and C. Held, Phys Chem Chem Phys., 2015, 17, 1800–1810 RSC.
- R. P. Richter and A. R. Brisson, Biophys. J., 2005, 88, 3422–3433 CrossRef CAS PubMed.
- I. Reviakine and A. Brisson, Langmuir, 2000, 16, 1806–1815 CrossRef CAS.
- M. Indrieri, M. Suardi, A. Podesta, E. Ranucci, P. Ferruti and P. Milani, Langmuir, 2008, 24, 7830–7841 CrossRef CAS PubMed.
- H. J. Butt and M. Jaschke, Nanotechnology, 1995, 6, 1–7 CrossRef.
- M. Chighizola, L. Puricelli, L. Bellon and A. Podestà, J. Mol. Recognit., 2021, 34, e2879 CrossRef CAS PubMed.
- H.-J. Butt, B. Cappella and M. Kappl, Surf. Sci. Rep., 2005, 59, 1–152 CrossRef CAS.
- L. Puricelli, M. Galluzzi, C. Schulte, A. Podesta and P. Milani, Rev. Sci. Instrum., 2015, 86, 033705 CrossRef PubMed.
- M. Jaschke and H. J. Butt, Rev. Sci. Instrum., 1995, 66, 1258–1259 CrossRef CAS.
- C. Rotsch and M. Radmacher, Biophys. J., 2000, 78, 520–535 CrossRef CAS PubMed.
- A. Alessandrini, H. M. Seeger, T. Caramaschi and P. Facci, Biophys. J., 2012, 103, 38–47 CrossRef CAS PubMed.
- N. M. Fonseka, F. T. Arce, H. S. Christie, C. A. Aspinwall and S. S. Saavedra, Langmuir, 2022, 38, 100–111 CrossRef CAS PubMed.
- L. M. Lima, M. I. Giannotti, L. Redondo-Morata, M. L. Vale, E. F. Marques and F. Sanz, Langmuir, 2013, 29, 9352–9361 CrossRef CAS PubMed.
- M. Galluzzi, B. Zhang, H. Zhang, L. Wang, Y. Lin, X.-F. Yu, Z. Chu and J. Li, Front. Mol. Biosci., 2021, 8, 669361 CrossRef CAS PubMed.
- E. K. Dimitriadis, F. Horkay, J. Maresca, B. Kachar and R. S. Chadwick, Biophys. J., 2002, 82, 2798–2810 CrossRef CAS PubMed.
- J. Pan, S. Tristram-Nagle, N. Kucerka and J. F. Nagle, Biophys. J., 2008, 94, 117–124 CrossRef CAS PubMed.
- Y. Yang, K. M. Mayer and J. H. Hafner, Biophys. J., 2007, 92, 1966–1974 CrossRef CAS PubMed.
- W. F. Heinz and J. H. Hoh, Biophys. J., 1999, 76, 528–538 CrossRef CAS PubMed.
- S. Pal, A. Sar and B. Dam, Ecotoxicol. Environ. Saf., 2019, 184, 109634 CrossRef CAS PubMed.
- H. J. Butt, Biophys. J., 1991, 60, 777–785 CrossRef CAS PubMed.
- X. H. Yin and J. Drelich, Langmuir, 2008, 24, 8013–8020 CrossRef CAS PubMed.
- K. O. Evans, Int. J. Mol. Sci., 2008, 9, 498–511 CrossRef CAS PubMed.
- T. Schaffran, J. Li, G. Karlsson, K. Edwards, M. Winterhalter and D. Gabel, Chem. Phys. Lipids, 2010, 163, 64–73 CrossRef CAS PubMed.
- H. Lee and T. J. Jeon, Phys Chem Chem Phys., 2015, 17, 5725–5733 RSC.
- M. Galluzzi, S. Bovio, P. Milani and A. Podesta, J. Phys. Chem. C, 2018, 122, 7934–7944 CrossRef CAS.
- F. Duša, S.-K. Ruokonen, J. Petrovaj, T. Viitala and S. K. Wiedmer, Colloids Surf., B, 2015, 136, 496–505 CrossRef PubMed.
- P. Kumari, V. V. S. Pillai, B. J. Rodriguez, M. Prencipe and A. Benedetto, J. Phys. Chem. Lett., 2020, 7327–7333 CrossRef CAS PubMed.
- N. Kundu, S. Roy, D. Mukherjee, T. K. Maiti and N. Sarkar, J. Phys. Chem. B, 2017, 121, 8162–8170 CrossRef CAS PubMed.
- H. Ryu, H. Lee, S. Iwata, S. Choi, M. Ki Kim, Y.-R. Kim, S. Maruta, S. Min Kim and T.-J. Jeon, Sci. Rep-Uk, 2015, 5, 11935 CrossRef PubMed.
- S. Mitra, D. Ray, G. Bhattacharya, R. Gupta, D. Sen, V. K. Aswal and S. K. Ghosh, Eur. Biophys. J., 2019, 48, 119–129 CrossRef CAS PubMed.
- G. Bhattacharya, R. P. Giri, A. Dubey, S. Mitra, R. Priyadarshini, A. Gupta, M. K. Mukhopadhyay and S. K. Ghosh, Chem. Phys. Lipids, 2018, 215, 1–10 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cp02866b |
| This journal is © the Owner Societies 2022 |