Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
5-(Hydroxymethyl)uracil and -cytosine as potential epigenetic marks enhancing or inhibiting transcription with bacterial RNA polymerase†
Martina
Janoušková‡
a,
Zuzana
Vaníková‡
b,
Fabrizia
Nici
b,
Soňa
Boháčová
b,
Dragana
Vítovská
a,
Hana
Šanderová
a,
Michal
Hocek
 *bc and
Libor
Krásný
*bc and
Libor
Krásný
 *a
*a
aDept. of Molecular Genetics of Bacteria, Institute of Microbiology of the Czech Academy of Sciences, CZ-14220 Prague 4, Czech Republic. E-mail: krasny@biomed.cas.cz
bInstitute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences, Flemingovo nam. 2, 16610 Prague 6, Czech Republic. E-mail: hocek@uochb.cas.cz
cDept. of Organic Chemistry, Faculty of Science, Charles University in Prague, Hlavova 8, CZ-12843 Prague 2, Czech Republic
First published on 22nd November 2017
Abstract
DNA templates containing 5-hydroxymethyluracil or 5-hydroxymethylcytosine were used in an in vitro transcription assay with RNA polymerase from Escherichia coli. A strong enhancement of transcription was observed from DNA containing the Pveg promoter whereas a decrease was observed from DNA containing the rrnB P1 promoter, suggesting that they may act as epigenetic marks.
Modifications of DNA by epigenetic bases (i.e. 5-methylcytosine, 5mC) play critical roles in the regulation of gene expression both in eukaryotes and prokaryotes and their dysregulation may lead to diseases.1 Recent advances in detection techniques have resulted in the discovery of the new bases 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxycytosine (5caC).2,3 More recently, also 5-hydroxymethyluracil (5hmU), previously detected in bacteriophages,4,5 dinoflagellates6 and leishmania,7 has been found in eukaryotic genomes8 where its level appears to be cell type-specific.9 The level of 5-hydroxymethyl-2′-deoxyuridine in blood DNA was investigated as a marker for breast cancer.10 It is being hotly debated whether these modifications function as regulators of gene expression or whether they are just intermediates in active demethylation of DNA or products of oxidative damage. While the role of 5hmC in regulation of transcription has been demonstrated,11 the role of 5hmU remains unclear.12 The 5hmU base can be generated by oxidation/hydroxylation of thymine by the Ten-Eleven-Translocation (TET) proteins13 or result from deamination of 5hmC.14 The DNA containing 5hmU was reported to be more flexible and hydrophilic.15 The Schultz lab has evolved16 a bacterium which replaced T with 5hmU in the genome showing that this base in principle is compatible with replication and transcription, as has been also previously reported by Herala and Vilpo from biochemical experiments.17
In order to shed light on the possible regulatory role of 5hmU, we performed an in vitro transcription study on modified DNA templates (containing 5hmU, 5hmC, 5mC or U) with Escherichia coli RNA polymerase (RNAP). The assay was previously developed for the systematic study of the effects of non-natural DNA modifications in the major groove on prokaryotic transcription.18 Here we used four templates with different promoters (Fig. 1) but the same transcribed region. The first two promoters were Pveg and rrnB P1, respectively, both from Bacillus subtilis and both well-recognized by the E. coli RNAP holoenzyme containing the primary sigma factor, σ70.19 The other two promoters were reciprocal chimeras made from the two former promoters to identify regions within the promoters where the modifications affected transcription the most.
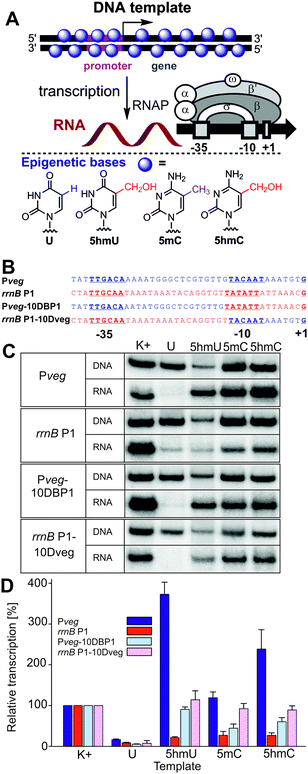 | ||
| Fig. 1 (A) Scheme of the in vitro transcription experiment using a fully base-modified DNA template and structures of the epigenetic bases. (B) Promoters and hybrid sequences used. (C) Gel electrophoresis of the 32P-labelled fully-modified DNA templates and the corresponding RNA transcripts. (D) Quantitation of transcriptions from fully-modified templates containing different modifications. K+ is the natural (non-modified) DNA template. The graph (also in Fig. 2) shows quantitation from at least three independent experiments. K+ was set to be 100%. The error bars are ±SD. | ||
We started by studying transcription on the four fully modified DNA templates, where all T or C bases were replaced with the epigenetic bases (Fig. 1). The templates were prepared by PCR using d5hmUTP20 or commercial d5hmCTP,21 d5mCTP or dUTP (see Fig. S1–S4 in the ESI†). An initial challenge was how to quantify the relative concentrations of the modified DNA templates since the modified bases may influence the extinction coefficient (NanoDrop) or the intercalation or fluorescence of DNA dyes (DNA gel staining). Since the use of 5′-FAM-labeled primers18 and fluorescence quantification of PCR products gave results of limited reproducibility, we prepared radioactively labelled DNA templates (using 32P-labelled primers for PCR) and found that the concentrations measured by radioactivity did not correlate with those measured on the NanoDrop but they correlated reasonably well (±10–15%) with the relative DNA amounts obtained from the fluorescence intensity of the DNA spots on gels stained by GelRed. These two methods were then used for determination of the relative concentrations of the DNA templates.
The first in vitro transcription experiments with the fully modified templates with the Pveg promoter surprisingly showed that the presence of 5hmU or 5hmC enhanced transcription by a factor of 3.5 or 2.3, respectively, whereas the template containing 5mC was transcribed with similar efficiency as the natural DNA (Fig. 1). On the other hand, and in accordance with our previous observations,18 the presence of U in the template significantly decreased transcription. It should be noted that in these experiments, the level of the 5hmU template was usually lower than the levels of the other templates due to technical problems with the 5hmU-DNA template preparation (lower efficiency of the PCR using d5hmUTP). To exclude the possibility that normalization to the relatively low level of the 5hmU DNA could have overrated the apparent stimulation, we performed transcription experiments with U and 5hmU DNA where we used equal amounts of DNA templates based on their prior quantitation from DNA gels. The results of this experiment (Fig. S35, ESI†) were in accordance with the observation in Fig. 1 confirming the significant enhancement of transcription with hmU-modified DNA. When using modified DNA templates containing the rrnB P1 promoter, the presence of any modified base decreased transcription significantly. With model hybrid sequences containing part of Pveg and part of rrnB P1, we observed transcription at about the same or just slightly decreased level compared to natural DNA templates for 5hmU, 5hmC and 5mC, implying that both parts of the promoter might be required for the observed effects (Fig. 1).
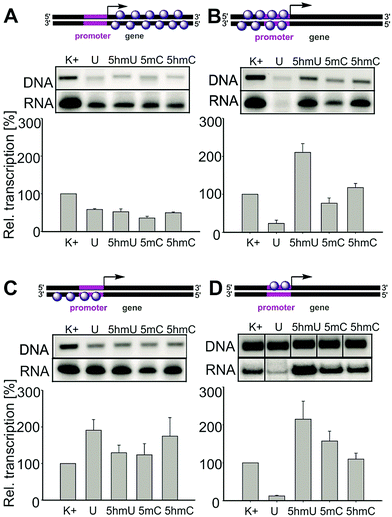
In order to identify which parts of the template are responsible for the unexpected enhancement of transcription, we prepared and systematically tested a series of partially modified templates containing the Pveg promoter (Fig. 2). For the schemes of synthesis of the partially modified templates by a combination of PCR, primer extension and ligations, see the ESI,† Fig. S5–S9. Templates modified in the gene region (with unmodified promoter) showed slightly decreased transcription but the effect was not very strong and appeared to be nonspecific (A). On the other hand, templates modified in both strands of the promoter region displayed similar trends as the fully modified templates (significant enhancement of transcription for 5hmU and moderate enhancement for 5hmC templates, B). Transcription of templates modified in the template strand of the promoter showed an increase for U, a slight enhancement for 5hmU, and an even more pronounced enhancement for 5hmC (C). Transcription of the template modified in the non-template strand of the promoter (D) showed strong inhibition for U as previously predicted,18 a significant enhancement for 5hmU and a less significant enhancement for 5hmC. This clearly demonstrates that in the case of 5hmU it is the non-template strand of the promoter that is critically important for the regulation of transcription by this epigenetic mark. Further detailed structural and functional studies will be needed to understand the mechanism of the transcription enhancement and inhibition.
To conclude, we have shown here that both 5hmC and 5hmU affect transcription by bacterial RNAP depending on the promoter sequence. For the first time, we directly observed strong enhancement of transcription of templates containing 5hmU or 5hmC in an in vitro enzymatic assay. In the case of 5hmU, both the enhancement and inhibition are mediated predominantly by interactions of the promoter nontemplate strand with RNAP. We note that while we used for our studies a well-characterized promoter (Pveg), other promoters may exist in the genome where random modifications of even single bases may have even more pronounced effects on transcription. Taken together, this illustrates the strong potential of 5hmU to alter gene expression. Therefore, we speculate that the presence of 5hmU in regulatory DNA sequences in the bacterial cell may contribute to diversification of the population, enhancing its chances of survival. As 5hmU occurs in bacteriophage DNA4,5 where it was described to facilitate binding of some transcription factors,22 it also may give it an advantage in transcription over the bacterial DNA. Notwithstanding these speculations, artificial modification of DNA templates by 5hmU may in principle serve for chemical regulation of transcription (chemical epigenetics) or even for the development of chemical switches.20,23 Studies along these lines, as well as investigation of the effects of these epigenetic bases on eukaryotic transcription, are under way.
This work was supported by the Czech Academy of Sciences (Praemium Academiae award to M. H.), and by the Czech Science Foundation (17-03419S to M. J., Z. V., S. B., L. K. and M. H.). Part of the project was supported by the C4Sys infrastructure.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) K. Chen, B. S. Zhao and C. He, Cell Chem. Biol., 2016, 23, 74–85 CrossRef CAS PubMed; (b) J. H. Gommers-Ampt and P. Borst, FASEB J., 1995, 9, 1034–1042 CAS; (c) E. A. Raiber, R. Hardisty, P. van Delft and S. Balasubramanian, Nat. Rev. Chem., 2017, 1, 0069 CrossRef.
- X. Lu, B. S. Zhao and C. He, Chem. Rev., 2015, 115, 2225–2239 CrossRef CAS PubMed.
- (a) M. Wagner, J. Steinbacher, T. F. J. Kraus, S. Michalakis, B. Hackner, T. Pfaffeneder, A. Perera, M. Müller, A. Giese, H. A. Kretzschmar and T. Carell, Angew. Chem., Int. Ed., 2015, 54, 12511–12514 CrossRef CAS PubMed; (b) T. Pfaffeneder, B. Hackner, M. Truss, M. Münzel, M. Müller, C. A. Deiml, C. Hagemeier and T. Carell, Angew. Chem., Int. Ed., 2011, 50, 7008–7012 CrossRef CAS PubMed; (c) D. Globisch, M. Münzel, M. Müller, S. Michalakis, M. Wagner, S. Koch, T. Brückl, M. Biel and T. Carell, PLoS One, 2010, 5, e15367 CAS.
- H. Witmer, J. Virol., 1981, 39, 536–547 CAS.
- R. G. Kallen, M. Simon and J. Marmur, J. Mol. Biol., 1962, 5, 248–250 CrossRef CAS PubMed.
- P. M. M. Rae, Science, 1976, 194, 1062–1064 CAS.
- F. Kawasaki, D. Beraldi, R. E. Hardisty, G. R. McInroy, P. van Delft and S. Balasubramanian, Genome Biol., 2017, 18, 23 CrossRef PubMed.
- T. Pfaffeneder, F. Spada, M. Wagner, C. Brandmayr, S. K. Laube, D. Eisen, M. Truss, J. Steinbacher, B. Hackner, O. Kotljarova, D. Schuermann, S. Michalakis, O. Kosmatchev, S. Schiesser, B. Steigenberger, N. Raddaoui, G. Kashiwazaki, U. Müller, C. G. Spruijt, M. Vermeulen, H. Leonhardt, P. Schär, M. Müller and T. Carell, Nat. Chem. Biol., 2014, 10, 574–581 CrossRef CAS PubMed.
- J. Guz, D. Gackowski, M. Foksinski, R. Rozalski and R. Olinski, Biol. Reprod., 2014, 91, 55 Search PubMed.
- Z. Djuric, L. K. Heilbrun, M. S. Simon, D. Smith, D. A. Luongo, P. M. LoRusso and S. Martino, Cancer, 2006, 77, 691–696 CrossRef.
- (a) A. Perera, D. Eisen, M. Wagner, S. K. Laube, A. F. Künzel, S. Koch, J. Steinbacher, E. Schulze, V. Splith, N. Mittermeier, M. Muller, M. Biel, T. Carell and S. Michalakis, Cell Rep., 2015, 11, 283–294 CrossRef CAS PubMed; (b) C. M. Greco, P. Kunderfranco, M. Rubino, V. Larcher, P. Carullo, A. Anselmo, K. Kurz, T. Carell, A. Angius, M. V. G. Latronico, R. Papait and G. Condorelli, Nat. Commun., 2016, 7, 12418 CrossRef CAS PubMed; (c) C. You, D. Li, X. Dai and Y. Wang, Sci. Rep., 2014, 4, 7052 CrossRef CAS PubMed; (d) L. Wang, Y. Zhou, L. Xu, R. Xiao, X. Lu, L. Cheng, J. Chong, H. Li, C. He, X.-D. Fu and D. Wang, Nature, 2015, 523, 621–625 CrossRef CAS PubMed; (e) M. W. Kellinger, C.-X. Song, J. Chong, X.-Y. Lu, C. He and D. Wang, Nat. Struct. Mol. Biol., 2012, 19, 831–833 CrossRef CAS PubMed.
- R. Olinski, M. Starczak and D. Gackowski, Mutat. Res., 2016, 767, 59–66 CAS.
- M. Yu, C.-X. Song and C. He, Methods, 2015, 72, 16–20 CrossRef CAS PubMed.
- S. Carson, J. Wilson, A. Aksimentiev, P. R. Weigele and M. Wanunu, Nucleic Acids Res., 2016, 44, 2085–2092 CrossRef CAS PubMed.
- F. Kawasaki, P. Murat, Z. Li, T. Santner and S. Balasubramanian, Chem. Commun., 2017, 53, 1389–1392 RSC.
- A. P. Mehta, H. Li, S. A. Reed, L. Supekova, T. Javahishvili and P. G. Schultz, J. Am. Chem. Soc., 2016, 138, 7272–7275 CrossRef CAS PubMed.
- A. M. Herala and J. A. Vilpo, Biochemistry, 1981, 28, 8274–8277 CrossRef.
- V. Raindlová, M. Janoušková, M. Slavíčková, P. Perlíková, S. Boháčová, N. Milisavljevič, H. Šanderová, M. Benda, I. Barvík, L. Krásný and M. Hocek, Nucleic Acids Res., 2016, 44, 3000–3012 CrossRef PubMed.
- L. Sojka, T. Kouba, I. Barvík, H. Šanderová, Z. Maderová, J. Jonák and L. Krásný, Nucleic Acids Res., 2011, 39, 4598–4611 CrossRef CAS PubMed.
- Z. Vaníková and M. Hocek, Angew. Chem., Int. Ed., 2014, 53, 6734–6737 CrossRef PubMed.
- B. Steigenberger, S. Schiesser, B. Hackner, C. Brandmayr, S. K. Laube, J. Steinbacher, T. Pfaffeneder and T. Carell, Org. Lett., 2013, 15, 366–369 CrossRef CAS PubMed.
- J. R. Greene, L. M. Morrissey, L. M. Foster and E. P. Geiduschek, J. Biol. Chem., 1986, 27, 12820–12827 Search PubMed.
- P. Kielkowski, H. Macíčková-Cahová, R. Pohl and M. Hocek, Angew. Chem., Int. Ed., 2011, 50, 8727–8730 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Extended results and discussion, experimental part, sequencing of all modified DNA templates. See DOI: 10.1039/c7cc08053k |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |