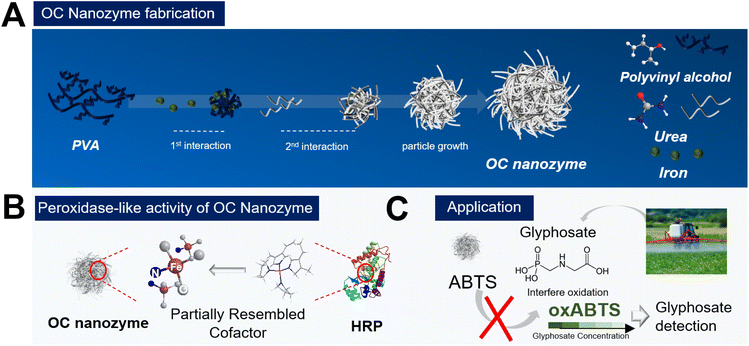
Organic compound-based nanozymes for agricultural herbicide detection†
Dong Hoon
Lee
and
Mohammed
Kamruzzaman
 *
*
Department of Agricultural and Biological Engineering, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA. E-mail: mkamruz1@illinois.edu
First published on 28th July 2023
Abstract
Nanozymes are increasingly being used for agricultural applications, but their adoption is limited as they are generally considered toxic, have low cost-effectiveness, and pose complexity of fabrication. In this study, an organic compound-based, peroxidase-like nanozyme (OC nanozyme) was developed for use in the agricultural environment. This nanozyme was synthesized through a self-assembled one-pot particle synthesis process, interacting with urea and the metal ion to form a homogenous nanoparticle containing partially mimicked cofactors (Fe–N) of the natural enzyme. The OC nanozyme exhibited decent kinetic properties (H2O2/Km:0.056 mM and Vmax:2.19 μM s−1) and pH stability. The OC nanozyme was successfully used to detect glyphosate via integrated colorimetric assay, with a good limit of detection (LOD) of at least 0.001 ng mL−1. The authors envision that this agricultural-friendly OC nanozyme holds great potential for a wide range of agricultural applications.
Introduction
Nanozyme1–5 has recently become a popular research subject in various disciplines, including biomedical, chemical engineering, and even agricultural engineering. One of the most popular application of nanozymes is in biosensors; researchers have been able to integrate this nanozyme into the colorimetric or electrochemical readout platform to detect target molecules.1,4 The catalytic activity of nanozymes facilitates the customization of cascade reaction chains for on-demand detection of various target molecules encompassing biological molecules or related small molecules. In recent years, researchers have also explored the use of nanozymes in agriculture and food science6,7 to detect certain molecules based on their conventional nanozyme-substrate-dependent reactions platform. These are effective, however, the materials that researchers have contrived are not ideal materials, and thus, they may not be directly utilized in agricultural and environmental applications. A majority of nanozymes utilized for agricultural applications are conventional nanozymes, metal-based nanoparticles, engineered carbon structure, MOF (metal–organic framework), and single atom nanozyme (SAN),1,5,8 however, they are generally too toxic and expensive for utilization in agriculture. To strengthen the biological and eco-friendly aspect, researchers try to manipulate the surface of the nanozymes to elevate their biocompatibility, but this is a transitory process and has yet to solve the problem entirely. Also, there are some examples of using MOF or SAN for agricultural biomolecule detection, but the application for further real-world application as MOF or conventional SAN is challenging2,8 as nanozymes require heavy fabrication steps and have low-cost effectiveness. To overcome this drawback, a novel nanozyme that is stable and eco-friendly, and cost-effective for agricultural applications, is desired. As most of the nanozymes deal with dominant inorganic materials, the development of the organic nanoscale structure has been conceptualized which follows the catalytic activity of the natural enzyme but is structurally based on organic compounds. Urea is a selected organic compound with amino and carboxyl groups, which can also function as chelating agents in metallic ions. Urea interacts with the iron ion and promotes the Fe–N bond, therefore, partially mimicking the cofactor of the natural peroxidase, to have a peroxidase-like activity. The particle stabilizer polyvinyl alcohol (PVA) participates in the particle generation process to form stable, spherical nanoparticles which yield a novel organic compound-based nanozyme. In addition, there is a massive demand to detect pesticides or herbicides ‘on demand’. Therefore, researchers have contrived nanozyme-based sensors to detect pesticides/herbicides via colorimetric assays. The detection of agricultural biomolecules using nanozymes is essential and it lessens the process compared to the conventional analytical methods and creates the potential for user-focused on-demand monitoring applications. Among many agricultural biomolecules, glyphosate is selected as the target molecule for detection using OC nanozyme-based colorimetric sensors.Glyphosate is a widely used herbicide in the agricultural industry; detecting its presence in agricultural products can help prevent human damage and protect crop health.6,7,9–11
In this study, the first example of the organic compound-based peroxidase-mimicking nanozyme (OC nanozyme) has been presented. The OC nanozyme has peroxidase-like activity with a decent kinetic profile and a considerably high eco-friendly nature as all its constituent materials are organic. The OC nanozyme has decent stability and cost-effectiveness compared to natural enzymes and conventional nanozymes. It has been utilized for agricultural applications to detect glyphosate using a colorimetric assay. The OC nanozyme successfully detected glyphosate with a decent LOD, highlighting its potential for further agricultural applications.
Materials and methods
Materials
Polyvinyl alcohol (MW 9k–10k) from Sigma-Aldrich (341584), urea from Sigma-Aldrich (U5378), iron(II) sulfate heptahydrate from Sigma-Aldrich (F8633), sodium acetate from Sigma Aldrich (236500), acetic acid solution from Sigma-Aldrich (45474), ABTS (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt) from Sigma-Aldrich (A1888), hydrogen peroxide from Sigma-Aldrich, and glyphosate from Sigma-Aldrich (45521) were used as chemicals in the experiment.Morphological and chemical analysis
The analysis of the operation was conducted at various facilities at the University of Illinois at Urbana-Champaign (UIUC), including the Materials Research Lab (MRL), Beckman Institute, and the High throughput screening facility. STEM (Scanning Transmission Electron Microscope) images were characterized by a Tecnai G2 F20 S-Twin from Thermo Fisher Scientific, which was operated at a maximum voltage of 200 kV. SEM (Scanning Electron Microscopy) images were acquired with a Hitachi S-4800 SEM that was operated with a maximum accelerating voltage of 100 kV. XPS (X-ray photoelectron spectroscopy) measurements were conducted on a Kratos Axis ULTRA with Al Kα X-ray radiation as the X-ray source for excitation. UV-Vis-NIR spectra were acquired by Varian Cary 5G and Agilent Cary 5000. NTA (nanoparticle tracking analysis) data was obtained from Nanosight NS300 from Malvern Panalytic, with sCMOS camera with blue488 laser. DLS/zeta potential data were obtained from Malvern Zetasizer, and Malvern Panalytic. FT-IR analysis profiles were obtained from Nicolet is50 FTIR spectrometer, Thermo Fisher Scientific.Fabrication of organic compound-based nanozyme
The preparation of nanozymes involved several steps. Iron sulfate powder was dissolved in deionized water to achieve a final concentration of 1 mg mL−1 at pH 4.8. PVA was dissolved in deionized water to a final concentration of up to 1 mg mL−1. Urea was prepared as a 1 mg mL−1 solution. A glass vial (10 mL) was placed on a stirring plate and 2 mL of PVA solution was added to the vial. Then, 3 mL of iron sulfate solution was added dropwise to the vial under stirring conditions (180–200 rpm) for 10 minutes. Lastly, 4 mL of urea solution was added to the vial dropwise while stirring fast (280–300 rpm) and the sample was incubated while stirring for up to 30 minutes. This vial was placed in a 4 °C refrigerator for up to 2 hours. The solution was then transferred to a 1.5 mL plastic tube and centrifuged at 25 °C for 5 minutes at 5000 rpm. The supernatant was removed and washed using deionized water several times. Finally, the nanozyme at the bottom was resuspended with deionized water or buffer for further assay.Optimization and characterization of experimental condition
The experimental conditions were determined based on optimization and the previous reference (ESI Ref. 3†). The different pH buffers/temperature buffers were prepared for the colorimetric assay to compare each performance directly (buffer = 0.1 M sodium acetate). The performance was evaluated using absorbance spectroscopy, where the highest absorbance at 417 nm represented a 100% endpoint, while the control buffer showed a 0% endpoint. The entire experiment was conducted at a temperature of 25 °C and a pH of 4; these conditions yielded the maximum absorbance endpoint.Peroxidase-like activity and kinetic studies
Peroxidase activities were determined by ABTS-based colorimetric assays. Fifteen microliters of OC nanozyme (2.01e + 14 M) and 15 μL of hydrogen peroxide (final 100 μM) and final 6 mM ABTS were added into a 96-well plate containing 105 μL of sodium acetate–acetic acid buffer [70 mM final (pH 4.0)]. Other groups, which displaced hydrogen peroxide to deionized water and displaced them from nanozyme to buffer, were prepared in parallel. The catalytic oxidation of ABTS was studied by measuring the absorption changes of the oxidized form of ABTS at λmax = 417 nm. Absorbance spectra were scanned between λ = 400 nm to 500 nm, repeating the measurements at least thrice for each group. The steady-state kinetic profiles were determined by measuring the absorbance signal through the kinetic mode using a Varian Cary 5G and an Agilent Cary 5000. The 700 μL of NaAc buffer, 100 μL of the OC nanozyme, 100 μL of the substrate (e.g. H2O2), and 100 μL of ABTS (varies according to the concentration) were placed in the cuvette to measure the consistent absorbance signal after 1 minute. Kinetic parameters were calculated by the Michaelis–Menten equation. The Michaelis constant (Km) and maximum initial velocity (Vmax) were decreased from the Lineweaver–Burk plots and Michaelis–Menten equations, described as follows: ν = Vmax × [S]/(Km + [S]), where ν is the initial velocity and [S] is the concentration of the substrate. The catalytic constant, known as kcat, was calculated using the equation kcat = Vmax/[E], where [E] is the molar concentration of Fe present in the OC nanozyme. It was calculated by the profile obtained from NTA analysis.Colorimetric detection of hydrogen peroxide
Fifteen microliters of the OC nanozyme (around 2.01e + 14 M) were added into a 96-well plate containing 105 μL of sodium acetate–acetic acid buffer [100 mM (pH 4.0)]. Then, 15 μL of varying concentrations of H2O2 (10 nM to 1000 μM) and 60 mM ABTS were added into the solution (oxABTS λmax = 417 nm). The catalytic oxidation of substrates was studied by measuring the changes in the absorption of the colorimetric assays undertaken with TMB in parallel (if TMB = oxTMB (λmax = 652 nm)). The target time point was set at 5 minutes after all the chemicals were added to the 96-microwell plate.Sample preparation for the stability test
The OC nanozyme (around 2.01e + 14 M) was dissolved in various pH buffers (2 < pH < 8.5) prepared using 0.1 M sodium acetate. After incubation for 5 hours, the nanozyme was collected using a centrifuge (5000 rpm) and resuspended in a buffer of pH 4 (NaAc, 0.1 M). The ABTS-based colorimetric assay was performed following the previously established procedure (endpoint); then, we calculated the relative stability based on the absorbance peak number.Glyphosate detection via colorimetric assay
Glyphosate was dissolved in deionized water and subjected to serial dilution to prepare various concentrations of glyphosate. The column for the colorimetric assay was prepared according to a previously established procedure using 0.1 M pH 4 sodium acetate buffers and 1 mM hydrogen peroxide. A 20 μL solution of glyphosate was added to the column and the solution was mixed with a pipette. For measurements, a 6 mM/final ABTS solution was added to the column. Glyphosate concentrations were performed using 96-well plate-based UV-Vis-NIR spectroscopy (Biotek 5 microplate reader). The endpoint analysis was conducted within 1 minute after adding all the chemicals to the 96-well plate. The absorbance spectra scanning was performed after 5 minutes. For the LOD measurement, a 10 times lower concentration of the peroxidase was used to inhibit the fast oxidation of the ABTS; its absorbance was measured in the scanning mode at 5 minutes after adding all the chemicals in the 96-well plates.Results and discussion
Morphological analyses of the OC nanozyme
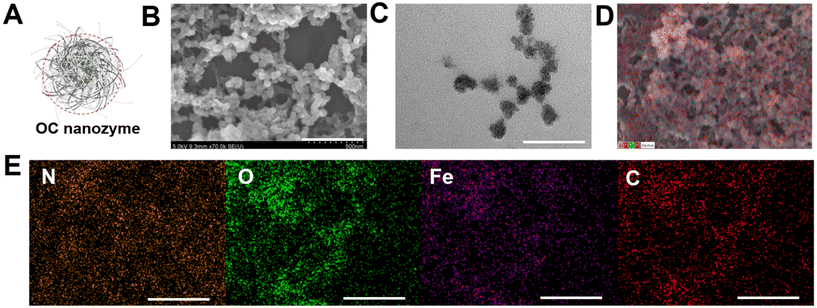
As illustrated in Scheme 1, the OC nanozyme was made in two major steps: the interaction between the functional group of the polymer and the iron ion. The second interaction with urea was essential. The amino group from the urea formed a coordinate covalent bond with iron, which resulted in the consistent growth of its framework, and the final particles formed a spherical nanostructure with a solid unit (Fig. S1†). Particle growth happened due to the chelation of iron; thus, this one-pot synthesis method did not require conventional nanoparticle fabrication methods, including heat increment. The morphological aspect was validated through STEM, SEM, and EDS (energy-dispersive X-ray spectroscopy) analysis (Fig. 1). The TEM and SEM images indicated that the OC nanozyme was a spherical nanoparticle-like structure, and the approximate diameter of the particle was ∼200 nm (Fig. 1B, C and D). The presence of PVA, iron, and urea within the OC nanozyme was intentionally designed, and the homogeneous distribution of these components on the nanozyme was confirmed through EDS analysis. Urea was the only source for a nitrogen group, and it was found that nitrogen and iron signals were enriched in the EDS image (Fig. 1E). The EDS image exhibited a homogenous iron signal (Fig. 1E) and the atomic ratio of iron was quantified by EDS analysis; this was determined to be 9.4% (Fig. S2†). Carbon and nitrogen were obtained from PVA and urea, respectively. The atomic ratio of carbon was determined to be 35.7%, while the atomic ratio of nitrogen was found to be 11.9%. This indicated the homogeneity of the PVA and urea in the OC nanozyme indirectly (Fig. S2†). To understand the size and surface information of the OC nanozyme, DLS (dynamic light scattering) and the zeta potential measurement were conducted. DLS profiles indicated that the OC nanozyme possessed a consistent size distribution with a mean size of around 160 nm with a decent polydispersity index (around 0.17), which implied its considerable homogeneity although it is an OC-based nanoparticle (Fig. S3A†).As the designated interaction of the urea was in the last steps, there is a higher possibility that the urea may be located on the surface of the whole structure; the zeta potential value was 4.85 mV (Fig. S3B†), which was positively charged due to the amino group from the urea. Before further analysis, another characterization was conducted related to the absorbance scanning with the OC nanozyme with two other organic compounds, PVA and urea, to ascertain whether they exhibited intrinsic absorption at specific wavelengths. From this profile, there was no significant absorption in the Vis-NIR range for all elements (Fig. S4†). The nanoparticle tracking analysis was conducted to examine the particle information of the OC nanozyme, particularly, the concentration of the nanozyme, for further assays. The estimated concentration of the nanozyme was obtained and calculated to a molar concentration of ∼2.01e + 14 M.
As it has been argued that the OC nanozyme contains a partially mimicked cofactor of the natural enzyme, further characterization was conducted to find the inner bond within the OC nanozyme. The heme-cofactor model structure was selected as the OC nanozyme aimed to resemble the natural peroxidase's cofactor, which has iron ions with its supporting ligands, including nitrogen. XPS analysis was chosen to understand the chemical bonds of each element. The XPS survey revealed the presence of several elements enriched in the OC nanozyme. Four elements with orbitals, C1s, N1s, O1s, and Fe2p, were selected (Fig. 2). Their XPS quantification-derived atomic ratios were 60.03, 4.52, 34.63, and 0.83, respectively (Fig. S5†). The C1s, 281.25 eV binding energy indicated a C–O bond, and that of 281.85 eV indicated a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, which could be derived from PVA and urea,21,28 respectively. The 289.7 eV result indicated a COOH bond from urea,14 and the 290.4 eV indicated a pi–pi bond of carbon.16 (Fig. 2A) For N1s, 406.3 eV indicated an N–O bond, which could be the three N–O bonds in urea;20 405.9 eV represented oxidized N, resulting in the interaction between Fe ion22,29 (Fig. 2B). The 396.3 eV implied the Fe–N bond,19,26 formed by urea and the iron ion (Fig. 2B). In O1s, 527.1 indicated O2− ion bonding, possibly with the Fe atoms, or the generation of a new iron oxide ion as the result of the Fe–O bond.23,27 The 529.25 eV was associated with the lattice oxygen (O1),15 and the 534.6 eV was related to the hydroxyl surface, in the case of the hydroxyl group on the PVA (Fig. 2C).12,22 In Fe2p, oxidized Fe3+ was found at 725.2 eV, which was a result of the interaction with other atoms.11 The Fe2+ ion in the Fe–N3 coordination structure was found on 709.4 eV,18,25 thus Fe–N peaks13 were located on both N1s and the Fe2p graph (Fig. 2D). Through the XPS analysis, it was concluded that the OC nanozyme contained a partially mimicked, cofactor-like structure. This also implied that the iron was not simply entrapped in the framework but rather provided the certain bond that generated the ‘active site’.
O, which could be derived from PVA and urea,21,28 respectively. The 289.7 eV result indicated a COOH bond from urea,14 and the 290.4 eV indicated a pi–pi bond of carbon.16 (Fig. 2A) For N1s, 406.3 eV indicated an N–O bond, which could be the three N–O bonds in urea;20 405.9 eV represented oxidized N, resulting in the interaction between Fe ion22,29 (Fig. 2B). The 396.3 eV implied the Fe–N bond,19,26 formed by urea and the iron ion (Fig. 2B). In O1s, 527.1 indicated O2− ion bonding, possibly with the Fe atoms, or the generation of a new iron oxide ion as the result of the Fe–O bond.23,27 The 529.25 eV was associated with the lattice oxygen (O1),15 and the 534.6 eV was related to the hydroxyl surface, in the case of the hydroxyl group on the PVA (Fig. 2C).12,22 In Fe2p, oxidized Fe3+ was found at 725.2 eV, which was a result of the interaction with other atoms.11 The Fe2+ ion in the Fe–N3 coordination structure was found on 709.4 eV,18,25 thus Fe–N peaks13 were located on both N1s and the Fe2p graph (Fig. 2D). Through the XPS analysis, it was concluded that the OC nanozyme contained a partially mimicked, cofactor-like structure. This also implied that the iron was not simply entrapped in the framework but rather provided the certain bond that generated the ‘active site’.
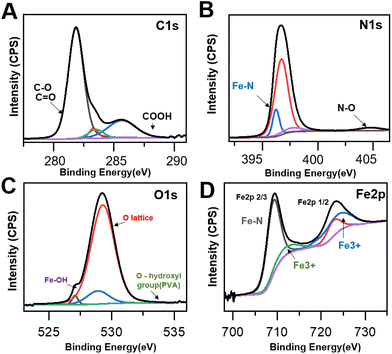 | ||
| Fig. 2 XPS spectra of the OC nanozyme. (A) C1s (277 eV–293 eV), (B) N1s (394 eV–407 eV), (C) O1s (523 eV–537 eV), (D) Fe2p(698 eV–735 eV). | ||
The enzymatic catalytic activity of the OC nanozyme
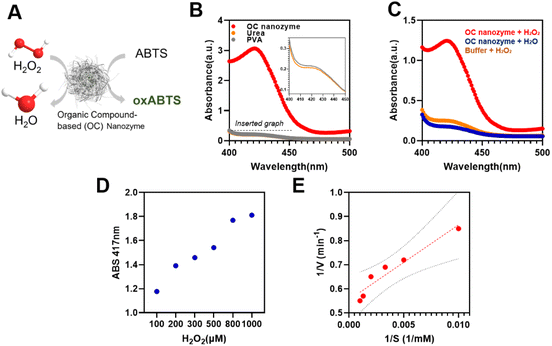
To designate this organic particle as a nanozyme, a colorimetric assay was conducted to confirm its enzyme-like catalytic activity (Fig. 3). As the core metallic ion selected in this OC nanozyme was iron, having peroxidase activity was a promising output of the OC nanozyme. The coloring substrate, ABTS, was used to conduct the two-step colorimetric reaction (Fig. 3A). The TMB also worked on the OC nanozyme as a substrate, but ABTS has more affinity as compared to the TMB because it has less measurement noise, and thus, it better identifies the OC nanozyme's peroxidase-like activity (Fig. S6†). It was confirmed that the iron located in the OC nanozyme was the only resource to exhibit catalytic activity due to the Fenton-like reaction; the PVA and urea did not exhibit catalytic activity themselves (Fig. 3B). The OC nanozyme was tested to verify its peroxidase-like activity, i.e., whether it broke down the H2O2 and led to ABTS oxidation (Fig. 3C). Both absorbance spectra and the absorbance peak were obtained in parallel, and the LOD range was below 100 nM (Fig. S7†). The steady-state kinetic assay was validated utilizing the two substrates: H2O2 and ABTS. To minimize the measurement noise attributed to ABTS for the kinetic assay, a diluted H2O2 concentration was selected for the steady-state kinetic assay. The values were calculated through the conventional Michaelis–Menten equation, obtaining the substrate affinity. The Km of OC nanozyme on H2O2 was 0.056 mM, and the Km of OC nanozyme on ABTS was 1.88 mM (Fig. 3D, E, and ESI Table 1†). To calculate the Kcat, the particle concentration obtained from NTA analysis was used. Thus, the calculated Kcat of the H2O2 on OC nanozyme was 1.6 × 105. In comparison to the references cited in Table S1,† the OC nanozyme exhibited decent substrate affinity and an excellent turnover rate. Additionally, the pH stability was tested to determine whether the nanozyme loses its activity due to incubation under a variety of pH conditions. The pH stability of the OC nanozyme was evaluated by incubating it for 5 hours in various buffers, and the catalytic activity of the nanozyme remained relatively consistent across a wide range of pH, i.e., pH 2–8.5. This indicates that the OC nanozyme exhibits more stability under acidic-to-neutral pH conditions (Fig. S8†).Detection of glyphosate using OC nanozyme integrated with a colorimetric assay
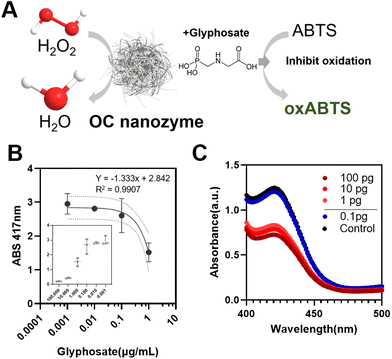
To prove its potential for agricultural applications, a glyphosate–OC nanozyme-H2O2-ABTS assay was conducted (Fig. 4). As it was confirmed that the OC nanozyme had a peroxidase-like activity with a decent kinetic profile, it was applied to agricultural biomolecule detection with the colorimetric assay platform. For certain herbicides, for instance, it was found that glyphosate presents radical scavenging activity indirectly that may interfere with the oxidation of ABTS. In other words, glyphosate compounds reduce the maximum absorbance peak when located on the OC nanozyme/H2O2/coloring substrate system as they interfere with the direct oxidation of coloring substrate derived from the radical generated by hydrogen peroxide (Fig. 4A). Glyphosate detection was conducted with ABTS colorimetric assay with the OC nanozyme–ABTS–H2O2 system (glyphosate) as follows. The oxidation of the ABTS molecule was measured by varying the concentrations of glyphosate range between 1 ng mL−1 to 100 μg mL−1 (Fig. 4B). A linearity between the concentration of glyphosate and the absorbance peak at 417 nm was identified.This was applicable to glyphosate concentrations in the range of 1 ng mL−1 to 1 μg mL−1 with an R2 value was 0.9907. The LOD was obtained from an additional experiment, lowering the H2O2 concentration 10 times to reduce the rapid oxidation of the ABTS.
A differentiable absorbance peak was observed at the 1 pg mL−1 border, leading us to conclude that the LOD for the system was at least 1 pg mL−1 (0.001 ng mL−1) for glyphosate, which implies a decent sensing performance compared to the conventional nanozyme-based sensor for glyphosate detection.9,10,17,24,30
Conclusions
Self-assembled organic compound-based nanostructures have been synthesized that rely on the chelation of iron with functional groups from urea and polyvinyl alcohol, promoting the one-pot synthesis of the nanozyme. Since there were abundant and heterogenous iron-based ligand elements, it was found that the OC-nanozyme exhibits a decent kinetic profile, which corresponds to the high density of the ‘active site’. Due to OC nanozyme's decent enzyme-like catalytic performance, it successfully detects glyphosate with a decent LOD. Since the OC nanozyme was prepared from an organic compound that dominantly forms its framework, the authors expect it to be eco-friendly and suitable for direct agricultural applications.Author contributions
Dong Hoon Lee: conceptualization, formal Analysis, investigation, methodology, writing – Original draft, writing-review & editing. Mohammed Kamruzzaman: conceptualization, supervision, writing – original draft, writing – review & editing.Abbreviation
| DLS | Dynamic light scattering |
| XPS | X-ray photoelectron spectroscopy |
| STEM | Scanning transmission electron microscopy |
| SEM | Scanning electron microscopy |
| EDS | Energy-dispersive X-ray spectroscopy |
| ABTS | (2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) |
| TMB | (3,3′,5,5′-Tetramethylbenzidine) |
| PVA | Poly-(vinyl alcohol) |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We appreciate the experimental and research support provided by the Material Research Laboratory, High-Throughput Screening facility (utilization of a BioTek Cytation 5 Multi-mode Imaging Reader, funded by the office of the Director, National Institutes of Health, award # S10 OD025289), and the Beckman Institute at the University of Illinois at Urbana-Champaign. We appreciate the ABE Department and the Department Head for providing the necessary space for the ‘Illinois Nanozyme Engineering Lab (INEL)’.References
- C. Peng, R. Pang, J. Li and E. Wang, Adv. Mater., 2023, 2211724 CrossRef PubMed.
- H. Wei, L. Gao, K. Fan, J. Liu, J. He, X. Qu, S. Dong, E. Wang and X. Yan, Nano Today, 2021, 40, 101269 CrossRef CAS.
- R. Zhang, X. Yan and K. Fan, Acc. Mater. Res., 2021, 2, 534–547 CrossRef CAS.
- W. Gao, J. He, L. Chen, X. Meng, Y. Ma, L. Cheng, K. Tu, X. Gao, C. Liu, M. Zhang, K. Fan, D. W. Pang and X. Yan, Nat. Commun., 2023, 14, 160 CrossRef CAS PubMed.
- G. Tang, J. He, J. Liu, X. Yan and K. Fan, Exploration, 2021, 1, 75–89 CrossRef PubMed.
- Y. Tang, W. Gou, X. Lv, X. Zhou, J. Hao, C. Sun, T. Sun, L. Hu and Z. Yan, Food Chem., 2023, 408, 135259 CrossRef CAS PubMed.
- J. Kuang, J. Ju, Y. Lu, Y. Chen, C. Liu, D. Kong, W. Shen, H. W. Shi, L. Li, J. Ye and S. Tang, Food Chem., 2023, 416, 135856 CrossRef CAS PubMed.
- D. Jiang, D. Ni, Z. T. Rosenkrans, P. Huang, X. Yan and W. Cai, Chem. Soc. Rev., 2019, 48, 3683–3704 RSC.
- X. Luo, G. Huang, C. Bai, C. Wang, Y. Yu, Y. Tan, C. Tang, J. Kong, J. Huang and Z. Li, J. Hazard. Mater., 2023, 443, 130277 CrossRef CAS PubMed.
- H. Li, S. Zhao, Z. Wang and F. Li, Small, 2023, 19, 2206465 CrossRef CAS PubMed.
- Z. Wang, H. Jin, T. Meng, K. Liao, W. Meng, J. Yang, D. He, Y. Xiong and S. Mu, Adv. Funct. Mater., 2018, 28, 1802596 CrossRef.
- W. Liu, J. Zhou, D. Liu, S. Liu and X. Liu, Mater. Lett., 2022, 307, 131076 CrossRef CAS.
- X. Yin, H. T. Chung, U. Martinez, L. Lin, K. Artyushkova and P. Zelenay, J. Electrochem. Soc., 2019, 166, F3240–F3245 CrossRef CAS.
- J. Bai, W. Ge, P. Zhou, P. Xu, L. Wang, J. Zhang, X. Jiang, X. Li, Q. Zhou and Y. deng, J. Colloid Interface Sci., 2022, 616, 433–439 CrossRef CAS PubMed.
- A. Farajollahi, A. P Marjani, N. N. Pesyan and H. Alamgholiloo, Appl. Surf. Sci., 2023, 622, 156903 CrossRef CAS.
- Y. Peng, H. Wang, Q. Li, L. Wang, W. Zhang, L. Zhang, S. Guo, Y. Liu, S. Liu and Q. Ma, Mater. Des., 2022, 215, 110522 CrossRef CAS.
- X. Luo, G. Huang, C. Bai, C. Wang, Y. Yu, Y. Tan, C. Tang, J. Kong, J. Huang and Z. Li, J. Hazard. Mater., 2023, 443, 130277 CrossRef CAS PubMed.
- B. Mecheri, V. C. A. Ficca, M. A. C. D’Oliveira, A. D’Epifanio, E. Placidi, F. Arciprete and S. Licoccia, Appl. Catal., B, 2018, 237, 699–707 CrossRef CAS.
- Z. Zhang, D. Li, J. Wang and J. Jiang, Appl. Catal., B, 2023, 323, 122164 CrossRef CAS.
- Z. Kamal, M. Z. Ghobadi, S. M. Mohseni and H. Ghourchian, Biosens. Bioelectron., 2021, 188, 113334 CrossRef CAS PubMed.
- H. Sepehrmansourie, H. Alamgholiloo, N. N. Pesyan and M. A. Zolfigol, Appl. Catal., B, 2023, 321, 122082 CrossRef CAS.
- Agarwal and B. R. Sankapal, Chem. Eng. J., 2021, 422, 130131 CrossRef CAS.
- Y. Wang, S. Zhao, Y. Zhu, R. Qiu, T. Gengenbach, Y. Liu, L. Zu, H. Mao, H. Wang, J. Tang, D. Zhao and C. Selomulya, iScience, 2020, 23, 100761 CrossRef CAS PubMed.
- J. Chang, L. Yu, T. Hou, R. Hu and F. Li, Anal. Chem., 2023, 95, 4479–4485 CrossRef CAS PubMed.
- W. Su, N. Yan, F. Liu, Z. Liu, G. Zhu, S. Wang, X. Liu and W. Wang, J. Phys. Chem. C, 2022, 126, 4826–4835 CrossRef CAS.
- S. Zhong, X. Yang, L. Chen, N. Tsumori, N. Taguchi and Q. Xu, ACS Appl. Mater. Interfaces, 2021, 13, 46749–46755 CrossRef CAS PubMed.
- D. Di Girolamo, F. Matteocci, F. U. Kosasih, G. Chistiakova, W. Zuo, G. Divitini, L. Korte, C. Ducati, A. Di Carlo, D. Dini and A. Abate, Adv. Energy Mater., 2016, 6, 1501489 CrossRef PubMed.
- K. Guo, F. Shaik, J. Yang, X. Ren and B. Jiang, J. Electrochem. Soc., 2021, 168, 062512 CrossRef CAS.
- X. W. Song, S. Zhang, H. Zhong, Y. Gao, L. A. Estudillo-Wong, N. Alonso-Vante, X. Shu and Y. Feng, Inorg. Chem. Front., 2021, 8, 109–121 RSC.
- J. Chang, L. Yu, T. Hou, R. Hu and F. Li, Anal. Chem., 2023, 95, 4479–4485 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr02025h |
| This journal is © The Royal Society of Chemistry 2023 |