Open Access Article
Open Access ArticleThe biomolecule corona of lipid nanoparticles contains circulating cell-free DNA†
Lois
Gardner
 ab,
Jessica
Warrington
ab,
Jessica
Warrington
 a,
Jane
Rogan
c,
Dominic G.
Rothwell
a,
Jane
Rogan
c,
Dominic G.
Rothwell
 b,
Ged
Brady
b,
Caroline
Dive
b,
Kostas
Kostarelos
b,
Ged
Brady
b,
Caroline
Dive
b,
Kostas
Kostarelos
 *ade and
Marilena
Hadjidemetriou
*ade and
Marilena
Hadjidemetriou
 *a
*a
aNanomedicine Lab, Faculty of Biology, Medicine and Health, The University of Manchester, AV Hill Building, Manchester, UK. E-mail: marilena.hadjidemetriou@manchester.ac.uk; kostas.kostarelos@manchester.ac.uk
bCancer Research UK Manchester Institute Cancer Biomarker Centre, The University of Manchester, Manchester, UK
cManchester Cancer Research Centre Biobank, The Christie NHS Foundation Trust, CRUK Manchester Institute, Manchester, UK
dManchester Cancer Research Centre, The University of Manchester, 555 Wilmslow Road, Manchester M20 4GJ, UK
eCatalan Institute of Nanoscience and Nanotechnology (ICN2), UAB Campus, Bellaterra, Barcelona, Spain
First published on 27th August 2020
Abstract
The spontaneous adsorption of biomolecules onto the surface of nanoparticles (NPs) in complex physiological biofluids has been widely investigated over the last decade. Characterisation of the protein composition of the ‘biomolecule corona’ has dominated research efforts, whereas other classes of biomolecules, such as nucleic acids, have received no interest. Scarce, speculative statements exist in the literature about the presence of nucleic acids in the biomolecule corona, with no previous studies attempting to describe the contribution of genomic content to the blood-derived NP corona. Herein, we provide the first experimental evidence of the interaction of circulating cell-free DNA (cfDNA) with lipid-based NPs upon their incubation with human plasma samples, obtained from healthy volunteers and ovarian carcinoma patients. Our results also demonstrate an increased amount of detectable cfDNA in patients with cancer. Proteomic analysis of the same biomolecule coronas revealed the presence of histone proteins, suggesting an indirect, nucleosome-mediated NP–cfDNA interaction. The finding of cfDNA as part of the NP corona, offers a previously unreported new scope regarding the chemical composition of the ‘biomolecule corona’ and opens up new possibilities for the potential exploitation of the biomolecule corona for the enrichment and analysis of blood-circulating nucleic acids.
New conceptsThe aim of this study was to interrogate experimentally the fundamental question whether circulating cell-free DNA (cfDNA) exists in the biomolecule corona that spontaneously forms around nanoparticles upon incubation with complex biological fluids. Despite recent advances in the characterisation of the protein content of the biomolecule corona, to date there has been no experimental evidence of the spontaneous interaction between cfDNA and plasma-incubated nanoparticles. To test this hypothesis and analyse such genomic content in the nanoparticle biomolecule corona, we incubated clinically-used liposomes with human plasma samples, retrieved their coronas and subsequently quantified the total corona cfDNA content using two different real-time quantitative PCR (qPCR) assays. Our data revealed the presence of cfDNA in the liposomal biomolecule corona upon their incubation with human plasma samples obtained from healthy donors and ovarian carcinoma patients. More interestingly, an increased amount of corona cfDNA was detected in cancer patients. Finally, proteomic analysis of the same biomolecule coronas revealed the presence of histone proteins, suggesting an indirect, nucleosome-mediated NP–cfDNA interaction. The revelation of cfDNA as a component of the nanoparticle corona, opens up new possibilities for the potential exploitation of the biomolecule corona for the enrichment and analysis of blood-circulating nucleic acids and the genomic information they carry. |
Introduction
Over the last decade, biomedical applications of nanoparticles (NPs) have been challenged due to the spontaneous adsorption of biomolecules onto their surface upon incubation with complex biofluids, known as the ‘protein’ or ‘biomolecule corona’.1,2 The bio-nanotechnology field has since invested considerable resources investigating the corona composition in an attempt to prevent NP–protein interactions and consequently limit opsonisation-mediated clearance from blood and ‘masking’ of surface ligands.3–6 Protein corona formation is now a widely accepted phenomenon and has been documented for a wide range of NPs, including lipid-, metal-, polymer- and carbon-based nanomaterials, with their composition and surface chemistry altering the specific classes of proteins adsorbed.7More recently, attempts have been made to purposefully manipulate the NP corona to modify the function of NPs4,8 and this approach has since been exploited as an application in its own right.9–12 For example, altering the surface properties of NPs not only alters the corona composition but has also been shown to control the loading and release of drugs and interaction with target molecules.13,14 Manipulating the corona by engineering the surface chemistry of NPs can therefore enhance their biological efficacy.4,6 In an alternative proposition, our laboratory has illustrated the potential exploitation of protein corona as a proteomic biomarker discovery platform that enables a higher-definition, in-depth analysis of the blood proteome and the enrichment of low abundant disease-specific molecules.9–12
Despite the comprehensive characterisation of the blood-derived NP protein corona in mice and humans (both ex vivo and in vivo),10,13,15–19 little attention has been placed on other types of biomolecules that may constitute the biomolecule corona. Growing evidence suggests that lipids and metabolites also interact with the surface of NPs.20–29 For example, a comprehensive metabolomic analysis at the nano-bio interface between mineralo-organic NPs and human biofluids (serum, plasma, saliva, and urine) revealed a range of metabolites including amino acids, sugars, amides, fatty acids and glycerophospholipids.24 More recently, the formation of a ‘metabolite’ corona has been shown using different NPs including, copper oxide, titanium dioxide, zinc oxide, zirconium dioxide, carbon25 and amino-functionalised polystyrene NPs.26 The lipid and metabolite composition of the liposomal corona has also been characterised in human plasma, revealing a complex lipid and metabolite profile.27 Notably, a recent study investigating the metabolite ‘small molecule’ corona formed around a range of nanomaterials revealed the presence of charged metabolites including amino acids and nucleosides.22 These studies highlight the diverse and complex composition of the biomolecule corona.
Scarce speculative statements exist in the literature referencing the presence of nucleic acids in the biomolecule corona.30–32 However, the genomic content of the blood-derived NP corona has never been investigated and there is no experimental evidence to support that the NP corona contains cell-free DNA (cfDNA). This could be partially due to the experimental challenges in identifying minute levels of blood-circulating nucleic acids on the NPs surfaces. The very few studies that have tried to investigate any role that nucleic acids may play in the biomolecule corona, focused on miRNAs using rat urine samples,33 or attempted to understand the interaction of NPs with added synthetic DNA fragments in solution.31,34 NPs have also been extensively used as biosensors or as nano-carriers to deliver nucleic acids, with positively charged lipid-, polymeric-, gold- and carbon-based (among many other) NPs used to deliver electrostatically-complexed nucleic acids for therapeutic applications.34–42 In an alternative approach, Liang et al., intravenously administered cationic polymeric NPs to complex cfDNA in vivo to alleviate the symptoms of inflammation in rheumatoid arthritis (RA).53 Despite these approaches of electrostatic complexation of nucleic acids with cationic NPs, the spontaneous association of blood-derived cfDNA molecules with NPs has largely been ignored, presumably because it was thought that other blood components competitively self-assemble into layers around NPs. It is also worth mentioning that the majority of NP-based colorimetric cfDNA biosensors require the purification of cfDNA from plasma samples prior to the incubation with NPs.43,44
The aim of the present study was to offer experimental evidence on the fundamental question as to whether cfDNA exists in the biomolecule corona formed around NPs in human plasma. In order to investigate this, we incubated clinically-used types of liposomes with plasma samples, retrieved the corona-coated liposomes, and subsequently quantified the total corona cfDNA content using two different real-time quantitative PCR (qPCR) assays. Our data confirmed the presence of cfDNA molecules in the biomolecule corona. In addition, analysis of the liposome corona formed in plasma samples obtained from ovarian carcinoma patients revealed higher total cfDNA content compared to healthy controls, suggesting a disease-specific biomolecule corona. Finally, correlation with the proteomic analysis of the same liposomal coronas revealed nucleosome-derived histone proteins, which suggested an indirect, protein-mediated cfDNA interaction with NPs.
Results
Plasma incubation and biomolecule corona formation
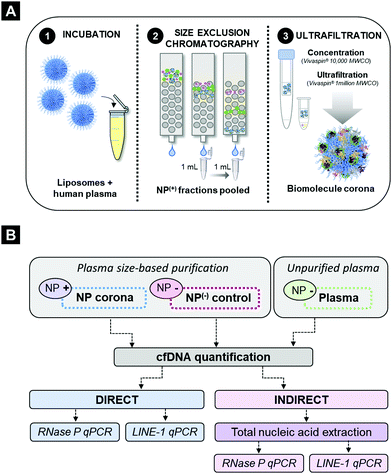
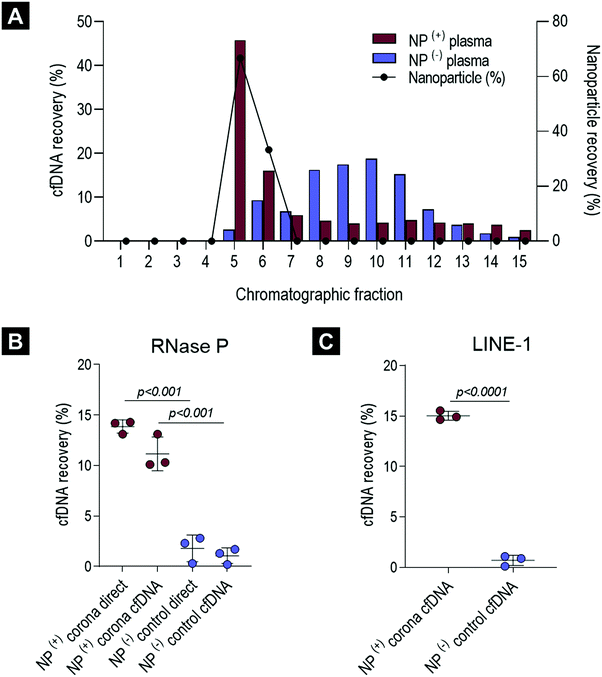
To evaluate the cfDNA content of the biomolecule corona, human plasma samples obtained from healthy volunteers were incubated (37 °C, 10 minutes, 250 rpm) with PEGylated liposomes (HSPC:Chol:DSPE-PEG2000), a formulation which constitutes the basis of the anti-cancer agent Doxil® (Fig. S1A and B, ESI†). Liposomes were employed in this study due to their extensive protein corona characterisation, their use in nucleic acid-based biotechnology applications and more recently due to their promise as a proteomic enrichment tool.10,38,39,45 Our previous ex vivo and in vivo investigations on protein corona revealed that even though PEGylation reduces protein adsorption, it does not completely eliminate binding. PEGylated liposomes were found to reproducibly interact with low molecular weight (MW) plasma proteins of low abundance.9–12,15In order to assess the potential interaction of cfDNA with PEGylated liposomal surfaces, plasma-incubated liposomes were purified by size exclusion chromatography (SEC), shown in Fig. 1A, as described previously.15 Plasma control samples (without prior incubation with liposomes) were subjected to the exact same purification process. SEC column-eluted cfDNA was extracted from chromatographic fractions 1–15, using a QIAamp® circulating nucleic acid extraction kit (QIAGEN) and subsequently quantified using robust and highly sensitive LINE-1 real-time qPCR assay (Fig. 2A). Stewart assay was also performed in order to quantify the amount of liposomes eluted.
As illustrated in Fig. 2A and in agreement with our previous studies,15 corona-coated liposomes were eluted in chromatographic fractions 5 and 6, while no detectable lipid content was found in the fractionated plasma control. Distribution of cfDNA across chromatographic fractions 1–15 revealed significant differences between plasma-incubated liposomes and the matched plasma control. In the case of the plasma-incubated liposome sample the majority of cfDNA (45.8%) was eluted in chromatographic fraction 5, which also contained the largest population of liposome NPs (66.7%), while liposome-free fractions 7–15 contained relatively small quantities of cfDNA (<6%). In contrast, a normal distribution of cfDNA was evident in the fractionated plasma control, with the highest amount of cfDNA detected in fraction 10 (18.8%). Notably, in the absence of NPs, only 2.6% of the cfDNA content was detected in fraction 5. The striking difference in cfDNA distribution between corona-coated liposomes and the fractionated plasma control suggests that a significant proportion of cfDNA eluted in fraction 5 could be associated with the eluted liposomes.
Quantitative detection of cfDNA in the liposome corona
To further purify corona-coated liposomes from any remaining protein complexes and/or unbound cfDNA, chromatographic fractions 5 and 6 were pooled, concentrated and subsequently washed three times using a membrane ultrafiltration column (Vivaspin®, 1 million MWCO). This protocol has been previously developed by us and also used successfully by others to purify corona-coated NPs from unbound plasma proteins.9,10,13To determine the total cfDNA content of the liposomal corona two different real-time qPCR assays were utilised, as outlined in Fig. 1B. A real-time qPCR approach was chosen as the concentration of cfDNA in blood commonly falls below the lower limit of detection for absorbance and fluorescence-based DNA quantification methods. Initially, a standardised TaqMan® RNase P detection real-time qPCR assay (Applied Biosystems®) was used to quantify the cfDNA content of the biomolecule corona in healthy plasma samples. As illustrated in Fig. 2B, the concentration of cfDNA measured in the corona samples was significantly higher in comparison to plasma control samples that underwent the full purification process (adjusted p-value < 0.0001). A small amount of cfDNA was identified in purified plasma controls, suggesting a co-elution of a small population of cfDNA molecules complexed with large proteins or within extracellular vesicles (Fig. 2B). These data suggested that most of the cfDNA quantified in corona samples is associated (directly or indirectly) with the surface of liposomes and was not passively co-eluted in a size-dependent manner.
In order to investigate whether the presence of proteins and/or other molecules in the biomolecule corona affects the direct quantification of cfDNA, we compared the amount of cfDNA with and without prior extraction (QIAGEN's QIAamp® circulating nucleic acid extraction kit). Comparable amounts of cfDNA were detected using the TaqMan® RNase P assay both in corona-coated liposome samples and in cfDNA subsequently purified from the same corona samples (Fig. 2B). These data indicated that the real-time qPCR assay was not significantly inhibited by other molecules present in the corona, allowing direct cfDNA measurements in the presence of lipid-based NPs and complex biofluid contaminants. To further investigate qPCR inhibition in NP-corona samples, a 2-fold dilution was performed prior to real-time qPCR quantification (Fig. S2A and B, ESI†). The cfDNA quantity of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 diluted corona sample was approximately half that of the original measurement (48%), providing further evidence to support the lack of RNase P qPCR inhibition in these direct real-time PCR measurements. The concentration of cfDNA in the NP-corona samples and plasma controls (with no NPs) was confirmed with a robust and sensitive LINE-1 qPCR assay (Fig. 2C). Both assays produced similar values, with RNase P and LINE-1 quantification methods consistently detecting significantly more cfDNA in corona samples when compared to plasma controls, as shown in Fig. 2C.
2 diluted corona sample was approximately half that of the original measurement (48%), providing further evidence to support the lack of RNase P qPCR inhibition in these direct real-time PCR measurements. The concentration of cfDNA in the NP-corona samples and plasma controls (with no NPs) was confirmed with a robust and sensitive LINE-1 qPCR assay (Fig. 2C). Both assays produced similar values, with RNase P and LINE-1 quantification methods consistently detecting significantly more cfDNA in corona samples when compared to plasma controls, as shown in Fig. 2C.
In terms of reproducibility, the percentage of cfDNA recovered with liposomal NPs was consistent across healthy plasma and liposome batches (Fig. S3A, ESI†). In addition, plasma linearity experiments revealed a significant reduction in total cfDNA content when plasma input volume was lowered, while the plasma![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NP ratio was maintained (adjusted p-values < 0.01 for both 410 μL & 205 μL of plasma when compared to 810 μL) (Fig. S3B, ESI†). In contrast to the linear relationship observed between plasma volume and cfDNA concentration, altering the concentration of liposome NPs did not significantly affect the amount cfDNA recovered (Fig. S2C, ESI†). Combined, these data suggested that at the NP concentrations investigated, liposomes interacted reproducibly with a sub-population of plasma cfDNA molecules and that a NP
NP ratio was maintained (adjusted p-values < 0.01 for both 410 μL & 205 μL of plasma when compared to 810 μL) (Fig. S3B, ESI†). In contrast to the linear relationship observed between plasma volume and cfDNA concentration, altering the concentration of liposome NPs did not significantly affect the amount cfDNA recovered (Fig. S2C, ESI†). Combined, these data suggested that at the NP concentrations investigated, liposomes interacted reproducibly with a sub-population of plasma cfDNA molecules and that a NP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) plasma [μL
plasma [μL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μL] ratio of 0.2 was found optimal to recover this fraction of cfDNA.
μL] ratio of 0.2 was found optimal to recover this fraction of cfDNA.
Detection of cfDNA in ovarian carcinoma liposomal corona samples
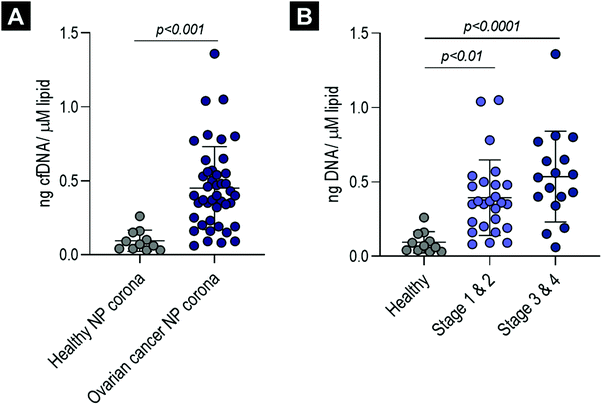
To establish whether cfDNA could also be detected on the surface of liposomes incubated ex vivo with plasma obtained from cancer patients, corona-coated liposomes were prepared upon incubation and purification from plasma samples obtained from 43 patients with ovarian cancer (18 patients with FIGO stage I, 8 with stage II, 12 with stage III and 5 with stage IV) (Table S1, ESI†). Patients with ovarian cancer classified across all stages of the disease were included in the study to determine whether cfDNA could be detected in NP corona samples both at early stages and as the disease progressed. These samples were quantified directly using a robust high sensitivity LINE-1 qPCR assay and compared to corona samples from 11 healthy aged matched females (Fig. 3). When normalised to post-purification liposome concentration, cfDNA was significantly higher in ovarian cancer samples (all stages, early stage (I and II) and late-stage (III and IV)) compared to healthy controls (p values = <0.001, <0.01 and <0.0001, respectively) (Fig. 3). In addition, average cfDNA content increased from early (FIGO stage I and II) to late stage (FIGO stage III and IV), although this was not statistically significant (Fig. 3B). These data are consistent with previous studies that have proposed quantification cfDNA as a diagnostic and prognostic biomarker for ovarian cancer, with increased cfDNA levels detected with disease progression.46,47To determine whether direct cfDNA quantification in ovarian cancer corona samples would be inaccurate and skewed, with real-time qPCR inhibition increasing disproportionately with cancer stage, we compared cfDNA concentration in purified and unpurified samples for eight late-stage (stage III n = 6, stage IV n = 2) high-grade serous ovarian cancer samples (details provided in Fig. S2E, ESI†). Similar cfDNA concentrations were measured for both unpurified ovarian cancer corona samples and their respective purified cfDNA samples (Fig. S2C, ESI†). This suggests that real-time qPCR was not significantly inhibited in these biomolecule corona qPCR reactions and that no significant cfDNA loss occurred during cfDNA extraction using QIAGEN's QIAamp® circulating nucleic acid extraction kit. We were also able to measure the cfDNA content directly in ovarian cancer plasma samples (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40), which again showed no significant difference from the respective purified plasma cfDNA samples (Fig. S2D, ESI†).
40), which again showed no significant difference from the respective purified plasma cfDNA samples (Fig. S2D, ESI†).
Mass spectrometry (LC-MS/MS) proteomic analysis was then performed on the 43 samples from ovarian cancer patients and the 11 samples from healthy controls to investigate whether proteins known to associate with cfDNA could be detected in the biomolecule corona (Fig. S4, ESI†). Histone proteins, H2A, H2B and H4, which are found within the core nucleosome complex, were detected in the biomolecule corona and were identified at significantly higher levels in ovarian cancer samples relative to healthy controls (Fig. S4A, ESI†). Two additional nucleosome-interacting proteins were identified in these samples, namely histone-lysine N-methyltransferase 2D and histone PARylation factor 1 (Fig. S4B, ESI†)48 Combined, these data confirmed the presence of cfDNA in the biomolecule corona of liposomes and suggested an indirect interaction which is potentially mediated via the nucleosome complex.
Discussion
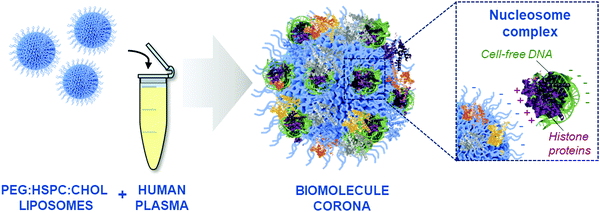
Understanding the composition of the biomolecule corona, spontaneously adsorbed onto the surface of nanoparticles (NPs), remains challenging due to the diverse physiochemical characteristics of NPs, as well as the complex multi-molecular composition of biological fluids.1 To date, the focus has been to elucidate the blood protein corona composition of various NPs with different surface chemistry and physiochemical properties.49 However, limited attention has been placed in profiling other biomolecules present in the NP corona. Recent studies have highlighted the interaction of NPs with lipids and metabolites,21–23 yet the contribution of blood-circulating cell-free DNA (cfDNA) to the formation of the NP corona remains almost completely unexplored.Fragmented cell-free DNA can be actively secreted into the bloodstream and is also passively released into circulation during cell death (i.e. apoptosis, necrosis).50 In healthy individuals, cfDNA levels are usually extremely low, with elevated concentrations of cfDNA commonly triggered by pathological disease states, such a tumorigenesis, inflammation, ischemia, trauma and sepsis.50 Cell-free DNA is widely protected from nuclease digestion by its complexation with a core of histone proteins, known as nucleosomes.51 Genomic analysis of the nucleic acid content in blood is of growing interest for diagnostic and disease monitoring applications, particularly in the context of liquid biopsies for cancer.52
Despite recent advances in analysing the blood-circulating genome, very little attention has been placed on the utilisation of the spontaneous interaction of NPs with nucleic acids upon incubation with biological fluids. Recently, Qian and colleagues investigated the interaction of negatively-charged, carboxylated magnetic NPs with miRNAs in rat urine samples and suggested an RNA-induced silencing complex (RISC)-mediated adsorption of miRNA.33 In a different study, Yun et al. developed a planar-substrate based protocol in order to investigate the interaction of metal–phenolic network (MPN)-based nanomaterials with multiple biomolecules including synthetic single stranded 40 bp DNA fragments.31 It is important to note however, that these DNA molecules do not reflect the size and structure of endogenous plasma cfDNA, nor the molecular complexity of blood.31
Despite the above-mentioned preliminary indications of the interaction of nucleic acids with the surface of NPs, no studies have demonstrated the presence of cfDNA in the biomolecule corona formed when NPs come into contact with human plasma. The present study aimed to answer this fundamental question and establish whether cfDNA participates in the formation of the biomolecule corona in a non-specific, untargeted manner. In order to investigate this, we used a real-time qPCR-based approach to quantify cfDNA in the biomolecule corona of lipid-based NPs upon incubation with human plasma. The NP corona samples underwent a two-step purification process prior to cfDNA quantification (with and without an additional cfDNA purification procedure). Our data provide the first experimental evidence of the presence of cfDNA in the NP corona samples and show that the majority of cfDNA detected is associated with the surface of liposomes and is not passively co-eluted during purification (Fig. 2A–C). Direct quantification of cfDNA was possible within complex lipid-based biomolecule corona samples without prior cfDNA extraction using the QIAamp circulating nucleic acid extraction kit (QIAGEN). In addition, cfDNA was successfully purified from lipid NPs using a standard cfDNA extraction kit, highlighting the compatibility of lipid-based NPs with downstream purification and quantification methods. Consistent cfDNA recovery across NP batches (Fig. S3A, ESI†) suggested its reproducible and stable interaction with the liposomal surface as part of the biomolecule corona.
The PEGylated liposomes used in this study have a negative surface charge (Fig. S1A, ESI†), therefore it was considered unlikely that DNA molecules would be bound directly onto the liposome surface via electrostatic interactions. Considering that cfDNA is protected within nucleosome complexes in the blood,51 we hypothesised that cfDNA may not be directly bound onto the liposome surface, but through the adsorption of DNA–protein complexes (illustrated in Fig. 4). This indirect mechanism of adsorption was further supported by the identification of positively charged nucleosome core proteins, including histone proteins H2A, H2B and H4, in the biomolecule corona by LC-MS/MS analysis (Fig. S4, ESI†). Our group has previously detected histone proteins in human and mouse liposomal corona, both ex vivo and in vivo.9,11,12 Moreover, human histone proteins (H2B and H4) have also been found to interact with colloidal gold NPs upon incubation with human plasma.54 Finally, De Paoli and colleagues demonstrated that calf thymus histone H1 can bind onto carboxylated-multiwalled carbon nanotubes.55
Our data demonstrated that the corona-containing cfDNA levels were significantly higher in the biomolecule coronas formed upon incubation with plasma samples obtained from ovarian cancer patients (both early- and late-stages) in comparison to healthy controls (Fig. 3). It has been widely reported that the total amount of cfDNA increases with disease progression in many different cancer types, such as colorectal, glioblastoma, colorectal and breast cancer.47,56–59 It is important to clarify that circulating DNA originating from the tumour (ctDNA) commonly accounts for only a small proportion of the total cfDNA, with the majority of DNA molecules released from non-malignant cells.51,60 Moreover, cfDNA detected in individuals with cancer is commonly of heamatopoietic origin and can be attributed to increased white blood cell turnover and chemotherapeutic- and/or radiation-induced cell death.51,56 Overall our data here demonstrated that the total amount of corona cfDNA increases in the presence of ovarian cancer, which renders the need for further studies to estimate the fraction of ctDNA by analysing tumour-specific mutations. This approach could offer significant advantages over current cfDNA purification methods, which lack the sensitivity required to detect ctDNA in small volumes of human plasma in patients with low tumour burden, especially pertinent to the challenge of early cancer detection.
Previous observations have shown that the composition of the biomolecule corona is directly affected by the presence of a disease.9 Comprehensive comparison of ‘healthy’ and ‘diseased’ protein coronas has been proposed for the discovery of diagnostic biomarkers.9–12 For example, we have previously shown that protein corona quantitatively and qualitatively changed in the presence of tumorigenesis, with higher total amount of protein found to interact with intravenously injected liposomes recovered from melanoma and lung adenocarcinoma tumour-bearing mice in comparison to healthy controls.9 Further analysis revealed that histone H2A was significantly upregulated in the in vivo lung adenocarcinoma corona samples.9 Therefore, the increased amount of nucleosome-related proteins identified in ovarian cancer patients in this study is likely to extend to other cancer types and NP classes, as a general reflection of increased histone levels commonly seen in cancer.61–64
In recent years, other cell-free nucleic acids, such as miRNAs, have received growing interest as disease biomarkers65 and although extensive characterisation of the NP corona total nucleic acid content was beyond the scope of this study, it remains an important avenue of future research. Blood-circulating RNA molecules are commonly complexed with proteins, including those forming the RNA-induced silencing complex (RISC).66 The potential protein-mediated interaction of RNA with plasma-incubated NPs remains to be explored, but if confirmed would add another layer of complexity to the NP biomolecule corona. In addition, epigenetic analysis of ctDNA, such as differential methylation profiles can also provide cancer-specific signatures.67 Intriguingly, methylated tumour DNA isolated from the plasma of cancer patients has been shown to display a strong affinity to gold nanoparticles and forms the basis of a ctDNA detection tool.68,69 The molecular complexes of cell-free nucleic acids contained within the biomolecule corona need to be fully elucidated in order to establish the scope for a sensitive blood-based biomarker enrichment tool.
The molecular information contained within the NP corona is far richer than originally described and has been shown to contain a diverse array of biomolecules including proteins, lipids, metabolites and now cfDNA. This complex coating on the surface of NPs has the potential to be able to enhance nano-drug delivery and NP uptake, but perhaps most significantly, offers the potential to provide greater sensitivity for liquid biopsies. Further studies are required to determine whether tumour-derived nucleic acids (ctDNA, miRNAs, RNAs) are detectable in the biomolecule corona and whether NPs can enrich the tumour-specific content obtained from human blood. Additional studies are also necessary to determine how the physiochemical properties of different nanomaterials affect their interaction with nucleic acids. Understanding the complexity of the biomolecular shell coating the surface NPs is fundamental in developing new and effective biomedical applications. Only now, is the diagnostic and therapeutic potential of the NP corona finally being realised, entering the nanomedicine field into an exciting new era.
Conclusion
This study has shown that cell-free DNA is present in the biomolecule corona that forms around lipid-based NPs, upon incubation with human plasma. The cfDNA content of the biomolecule corona could be directly quantified in the presence other biomolecules (e.g. proteins) using conventional real-time qPCR assays. Furthermore, proteomic analysis of the biomolecule corona by LC-MS/MS revealed the presence of nucleosome complex proteins, suggesting an indirect protein-mediated interaction of cfDNA with NPs. Notably, the amount of cfDNA was found to be significantly higher in the coronas formed in early- and late-stage cancer patient plasma samples compared to healthy controls, indicating a disease-specific biomolecule corona formation. This work is thought to pave the way for future studies, to further understand the mechanism, type and multi-molecular complex formation and adsorption of blood-circulating nucleic acids onto the surface of NPs. Equally, our study highlights the potential exploitation of the biomolecule corona as a novel blood-analysis nanoscale tool.Experimental
Plasma samples
Healthy human female pooled K2EDTA plasma samples were purchased from BioIVT (West Sussex, UK) (Lot #HMN2528). All ovarian cancer K2EDTA plasma samples were collected by the MCRC Biobank (details provided in Table S1 and Fig. S2E, ESI†). Individual age- and sex-matched K2EDTA plasma controls (female, 45–85 years old) were purchased from BioIVT (West Sussex, UK) (Table S1, ESI†). All plasma samples were stored at −80 °C.Liposome preparation
HSPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Chol
Chol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DSPE-PEG2000 (56.3
DSPE-PEG2000 (56.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38.2
38.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5.5) liposomes (Doxil® formulation) liposomes were prepared using the thin lipid film method followed by extrusion as described previously.15 All liposome batches were diluted to 12.5 mM, with the same batch of liposomes used for group comparisons. The physiochemical characteristics of the liposome batches are shown in Fig. S1 (ESI†).
5.5) liposomes (Doxil® formulation) liposomes were prepared using the thin lipid film method followed by extrusion as described previously.15 All liposome batches were diluted to 12.5 mM, with the same batch of liposomes used for group comparisons. The physiochemical characteristics of the liposome batches are shown in Fig. S1 (ESI†).
Dynamic light scattering (DLS) for size and zeta-potential measurements
Liposome size and surface charge were measured as described previously.15 Liposomes were diluted in distilled water and measured in size or capillary cuvettes using the Zetasizer Nano ZS (Malvern, Instruments, UK).Biomolecule corona formation (liposome plasma incubation and purification)
Liposome and plasma incubations and purifications were performed as described previously.15 In brief, 820 μL human plasma and 180 μL PEGylated liposomes were incubated for 10 min at 37 °C, shaking at 250 rpm. Unbound proteins and other unknown biomolecules were removed by size exclusion chromatography (SEC) (Sepharose CL-4B columns (Sigma-Aldrich)) followed by membrane ultrafiltration (Vivaspin® columns (Sartorious, Fisher Scientific)). Samples were concentrated to 100 μL for characterisation or downstream processing. For characterisation of individual chromatographic fractions, samples were concentrated to 100 μL using 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecular weight cut off (MWCO) Vivaspin® membrane ultrafiltration columns (Sartorious, Fisher Scientific). Plasma controls were subjected to the same purification process for comparison.
000 molecular weight cut off (MWCO) Vivaspin® membrane ultrafiltration columns (Sartorious, Fisher Scientific). Plasma controls were subjected to the same purification process for comparison.
Circulating cell-free nucleic acid extraction
Cell-free nucleic acids were purified from ex vivo plasma samples, liposomal corona samples and plasma control samples using a QIAamp® Circulating Nucleic Acid Extraction kit and QIAvac 24 Plus vacuum manifold according to manufacturer's instructions (QIAGEN, Hilden, Germany). After an initial sample lysis step, cell-free nucleic acids were bound onto a silica-based purification column (QIAGEN mini column). Multiple washing steps were performed prior to elution of cell-free nucleic acids in buffer AVE (QIAGEN). All samples were eluted in a final volume of 50 μL.Cell-free DNA quantification
Cell-free DNA was measured using two real-time quantitative PCR (qPCR) assays. The single-copy RNase P probe real-time assay was performed using TaqMan® RNase P Detection Reagents kit (Life Technologies) and SensiFAST Probe Hi-ROX master mix (Bioline, Meridian Bioscience). All real-time qPCR reactions included 7.5 μL of 2× SensiFAST probe mastermix, 0.75 μL 20× RNase P primer/probe mix, 1.75 μL nuclease-free water (Ambion, Texas, USA) and 5 μL of sample. Cycling conditions included (95 °C, 5 min) × 1, (95 °C, 10 s; 60 °C, 50 s) × 40 and were performed on a LightCycler® 96 (Roche, Basel, Switzerland).The multi-locus LINE-1 real-time qPCR assay was performed using primers described previously70 purchased from Integrated DNA Technologies (desalted, 25 nmol scale) using a robust Terra qPCR Direct SYBR Premix master mix (Takara Bio, USA). All real-time PCR reactions included 7.5 μL of 2× Terra qPCR Direct SYBR Premix master mix, 0.75 μL of each 10 μM (forward and reverse primers), 5.75 μL nuclease-free water (Ambion, Texas, USA) and 1 μL of sample. Cycling conditions included (98 °C, 2 min) × 1, (98 °C, 10 s; 60 °C, 15 s; 68 °C, 30 s) × 35 and were performed on a LightCycler® 96 (Roche, Basel, Switzerland).
Sample input was either corona-coated liposomes, purified cfDNA or plasma samples diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40. Plasma samples were only quantified using the LINE-1 real-time PCR assay in combination with the robust Terra qPCR Direct SYBR Premix master mix.
40. Plasma samples were only quantified using the LINE-1 real-time PCR assay in combination with the robust Terra qPCR Direct SYBR Premix master mix.
Mass spectrometry
In-gel digestion of corona proteins was performed prior to LC-MS/MS analysis, as described previously.15 Digested proteins were analysed by LC-MS/MS using an UltiMate 3000 Rapid Separation LC (RSLC, Dionex Corporation, Sunnyvale, CA) plus Q Exactive Hybrid Quadrupole-Orbitrap (Thermo Fisher Scientific, Waltham, MA, USA) mass spectrometer system. Data were analysed using Mascot (Matrix Science UK) in combination with the SwissProt_2016_04 database (taxonomy human). Progenisis QI software (version 4.3.2, Proteome Software Inc.) was used for relative protein quantification based on spectral counting and statistical analyses (one-way analyses of variance (ANOVA)).Statistical analysis
Statistical comparisons of these data were performed using GraphPad Prism v.8.2.0. For comparisons of three groups or more, one-way ANOVA tests were performed followed by the Tukey's multiple comparison test (adjusted p values < 0.05 were considered significant). For comparisons of two groups unpaired Student t-tests were performed (FDR-adjusted p values < 0.05 were considered significant). All data averages were presented as mean ± standard deviation (SD).Ethical approvals
This project has research ethics approval under the Manchester Cancer Research Centre (MCRC) Biobank Research Tissue Bank Ethics (NHS NW Research Ethics Committee 18/NW/0092). All participants provided written informed consent to participate in this study.Author contributions
L. G., M. H., K. K., G. B. designed and performed all experiments and took responsibility for planning and writing the manuscript. J. W. performed mass spectrometry sample preparation and respective data analysis. J. R. provided access and information from the Christie NHS Hospital Biobank and supported the study. D. R. supervised and provided intellectual input to the genomics experimental work. G. B., C. D., K. K. and M. H. initiated, overall supervised, provided intellectual input and contributed to the writing of the manuscript. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the NIHR Manchester Biomedical Research Centre (BRC). Research samples were obtained from the Manchester Cancer Research Centre (MCRC) Biobank, UK. The role of the MCRC Biobank is to distribute samples and therefore, cannot endorse studies performed or the interpretation of results. The authors also would like to thank the Faculty of Life Sciences Mass Spectrometry facility at the University of Manchester for their assistance. Finally, the authors would like to thank the ovarian cancer patients and healthy volunteers for their blood donations, which supported this research.References
- M. Hadjidemetriou and K. Kostarelos, Nat. Nanotechnol., 2017, 12, 288–290 CrossRef CAS PubMed.
- D. Docter, D. Westmeier, M. Markiewicz, S. Stolte, S. K. Knauer and R. H. Stauber, Chem. Soc. Rev., 2015, 44, 6094–6121 RSC.
- Q. Dai, N. Bertleff-Zieschang, J. A. Braunger, M. Björnmalm, C. Cortez-Jugo and F. Caruso, Adv. Healthcare Mater., 2018, 7, 1700575 CrossRef PubMed.
- D. Chen, S. Ganesh, W. Wang and M. Amiji, Nanoscale, 2019, 11, 8760–8775 RSC.
- M. Debayle, E. Balloul, F. Dembele, X. Xu, M. Hanafi, F. Ribot, C. Monzel, M. Coppey, A. Fragola, M. Dahan, T. Pons and N. Lequeux, Biomaterials, 2019, 219, 119357 CrossRef CAS PubMed.
- J. Müller, K. N. Bauer, D. Prozeller, J. Simon, V. Mailänder, F. R. Wurm, S. Winzen and K. Landfester, Biomaterials, 2017, 115, 1–8 CrossRef PubMed.
- D. Docter, S. Strieth, D. Westmeier, O. Hayden, M. Gao, S. K. Knauer and R. H. Stauber, Nanomedicine, 2015, 10, 503–519 CrossRef CAS PubMed.
- A. Cifuentes-Rius, H. De Puig, J. C. Y. Kah, S. Borros and K. Hamad-Schifferli, ACS Nano, 2013, 7, 10066–10074 CrossRef CAS PubMed.
- M. Hadjidemetriou, Z. Al-ahmady, M. Buggio, J. Swift and K. Kostarelos, Biomaterials, 2019, 188, 118–129 CrossRef CAS PubMed.
- M. Hadjidemetriou, S. McAdam, G. Garner, C. Thackeray, D. Knight, D. Smith, Z. Al-Ahmady, M. Mazza, J. Rogan, A. Clamp and K. Kostarelos, Adv. Mater., 2019, 31, 1–9 Search PubMed.
- L. Papafilippou, A. Claxton, P. Dark, K. Kostarelos and M. Hadjidemetriou, Nanoscale, 2020, 12, 10240–10253 RSC.
- M. Hadjidemetriou, L. Papafilippou, R. D. Unwin, J. Rogan, A. Clamp and K. Kostarelos, Nano Today, 2020, 34, 100901 CrossRef CAS.
- Z. S. Al-Ahmady, M. Hadjidemetriou, J. Gubbins and K. Kostarelos, J. Controlled Release, 2018, 276, 157–167 CrossRef CAS PubMed.
- M. Dalmina, F. Pittella, J. A. Sierra, G. R. R. Souza, A. H. Silva, A. A. Pasa and T. B. Creczynski-Pasa, Mater. Sci. Eng., C, 2019, 99, 1182–1190 CrossRef CAS PubMed.
- M. Hadjidemetriou, Z. Al-Ahmady, M. Mazza, R. F. Collins, K. Dawson and K. Kostarelos, ACS Nano, 2015, 9, 8142–8156 CrossRef CAS PubMed.
- M. Hadjidemetriou, Z. Al-Ahmady and K. Kostarelos, Nanoscale, 2016, 8, 6948–6957 RSC.
- R. García-Álvarez, M. Hadjidemetriou, A. Sánchez-Iglesias, L. M. Liz-Marzán and K. Kostarelos, Nanoscale, 2018, 10, 1256 RSC.
- D. Docter, U. Distler, W. Storck, J. Kuharev, D. Wünsch, A. Hahlbrock, S. K. Knauer, S. Tenzer and R. H. Stauber, Nat. Protoc., 2014, 9, 2030–2044 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos, H. Schild and R. H. Stauber, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS PubMed.
- A. A. Kapralov, W. H. Feng, A. A. Amoscato, N. Yanamala, K. Balasubramanian, D. E. Winnica, E. R. Kisin, G. P. Kotchey, P. Gou, L. J. Sparvero, P. Ray, R. K. Mallampalli, J. Klein-Seetharaman, B. Fadeel, A. Star, A. A. Shvedova and V. E. Kagan, ACS Nano, 2012, 6, 4147–4156 CrossRef CAS PubMed.
- S. S. Raesch, S. Tenzer, W. Storck, A. Rurainski, D. Selzer, C. A. Ruge, J. Perez-Gil, U. F. Schaefer and C. M. Lehr, ACS Nano, 2015, 9, 11872–11885 CrossRef CAS PubMed.
- A. J. Chetwynd, W. Zhang, J. A. Thorn, I. Lynch and R. Ramautar, Small, 2020, 16, 2000295 CrossRef CAS PubMed.
- A. J. Chetwynd and I. Lynch, Environ. Sci.: Nano, 2020, 7, 1041–1060 RSC.
- J. Martel, C. Y. Wu, C. Y. Hung, T. Y. Wong, A. J. Cheng, M. L. Cheng, M. S. Shiao and J. D. Young, Nanoscale, 2016, 8, 5537–5545 RSC.
- M. Pink, N. Verma, C. Kersch and S. Schmitz-Spanke, Environ. Sci.: Nano, 2018, 5, 1420 RSC.
- K. Grintzalis, T. N. Lawson, F. Nasser, I. Lynch and M. R. Viant, Nanotoxicology, 2019, 13, 783–794 CrossRef CAS PubMed.
- G. La Barbera, A. L. Capriotti, G. Caracciolo, C. Cavaliere, A. Cerrato, C. M. Montone, S. Piovesana, D. Pozzi, E. Quagliarini and A. Laganà, Talanta, 2020, 209, 120487 CrossRef CAS PubMed.
- E. Hellstrand, I. Lynch, A. Andersson, T. Drakenberg, B. Dahlbäck, K. A. Dawson, S. Linse and T. Cedervall, FEBS J., 2009, 276, 3372–3381 CrossRef CAS PubMed.
- J. Y. Lee, H. Wang, G. Pyrgiotakis, G. M. DeLoid, Z. Zhang, J. Beltran-Huarac, P. Demokritou and W. Zhong, Anal. Bioanal. Chem., 2018, 410, 6155–6164 CrossRef CAS PubMed.
- Y. Randika Perera, R. A. Hill and N. C. Fitzkee, Isr. J. Chem., 2019, 59, 962–979 CrossRef CAS.
- G. Yun, J. J. Richardson, M. Capelli, Y. Hu, Q. A. Besford, A. C. G. Weiss, H. Lee, I. S. Choi, B. C. Gibson, P. Reineck and F. Caruso, Adv. Funct. Mater., 2020, 30, 1905805 CrossRef CAS.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- S. Xu, S. Hossaini Nasr, D. Chen, X. Zhang, L. Sun, X. Huang and C. Qian, ACS Biomater. Sci. Eng., 2018, 4, 654–662 CrossRef CAS PubMed.
- R. M. Pallares, S. L. Kong, T. Hui Ru, N. T. K. Thanh, Y. Lu and X. Su, Chem. Commun., 2015, 51, 14524–14527 RSC.
- H. Lee, A. K. R. Lytton-Jean, Y. Chen, K. T. Love, A. I. Park, E. D. Karagiannis, A. Sehgal, W. Querbes, C. S. Zurenko, M. Jayaraman, C. G. Peng, K. Charisse, A. Borodovsky, M. Manoharan, J. S. Donahoe, J. Truelove, M. Nahrendorf, R. Langer and D. G. Anderson, Nat. Nanotechnol., 2012, 7, 389–393 CrossRef CAS PubMed.
- F. E. Alemdaroglu, N. C. Alemdaroglu, P. Langguth and A. Herrmann, Adv. Mater., 2008, 20, 899–902 CrossRef CAS.
- M. E. Davis, J. E. Zuckerman, C. H. J. Choi, D. Seligson, A. Tolcher, C. A. Alabi, Y. Yen, J. D. Heidel and A. Ribas, Nature, 2010, 464, 1067–1070 CrossRef CAS PubMed.
- M. Rasoulianboroujeni, G. Kupgan, F. Moghadam, M. Tahriri, A. Boughdachi, P. Khoshkenar, J. J. Ambrose, N. Kiaie, D. Vashaee, J. D. Ramsey and L. Tayebi, Mater. Sci. Eng., C, 2017, 75, 191–197 CrossRef CAS PubMed.
- M. Sioud and D. R. Sørensen, Biochem. Biophys. Res. Commun., 2003, 312, 1220–1225 CrossRef CAS PubMed.
- S. Guo, Y. Huang, Q. Jiang, Y. Sun, L. Deng, Z. Liang, Q. Du, J. Xing, Y. Zhao, P. C. Wang, A. Dong and X. J. Liang, ACS Nano, 2010, 4, 5505–5511 CrossRef CAS PubMed.
- P. V. Jena, M. M. Safaee, D. A. Heller and D. Roxbury, ACS Appl. Mater. Interfaces, 2017, 9, 21397–21405 CrossRef CAS PubMed.
- J. D. Harvey, H. A. Baker, M. V. Ortiz, A. Kentsis and D. A. Heller, ACS Sens., 2019, 4, 1236–1244 CrossRef CAS PubMed.
- S. Khanna, P. Padhan, S. Das, K. Jaiswal, A. Tripathy, S. Smita, S. Tripathy, S. Raghav and B. Gupta, ChemistrySelect, 2018, 3, 11541–11551 CrossRef CAS.
- R. Cao, B. Li, Y. Zhang and Z. Zhang, Chem. Commun., 2011, 47, 12301–12303 RSC.
- K. Hamad-Schifferli, Nanomedicine, 2015, 10, 1663–1674 CrossRef CAS PubMed.
- J. H. No, K. Kim, K. H. Park and Y.-B. Kim, Anticancer Res., 2012, 32, 3467–3471 Search PubMed.
- A. A. Kamat, M. Baldwin, D. Urbauer, D. Dang, L. Y. Han, A. Godwin, B. Y. Karlan, J. L. Simpson, D. M. Gershenson, R. L. Coleman, F. Z. Bischoff and A. K. Sood, Cancer, 2010, 116, 1918–1925 CrossRef CAS PubMed.
- J. Kowal, G. Arras, M. Colombo, M. Jouve, J. P. Morath, B. Primdal-Bengtson, F. Dingli, D. Loew, M. Tkach and C. Théry, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E968–E977 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- H. Schwarzenbach, D. S. B. Hoon and K. Pantel, Nat. Rev. Cancer, 2011, 11, 426–437 CrossRef CAS PubMed.
- M. W. Snyder, M. Kircher, A. J. Hill, R. M. Daza and J. Shendure, Cell, 2016, 164, 57–68 CrossRef CAS PubMed.
- G. D. Sorenson, D. M. Pribish, F. H. Valone, V. A. Memoli, D. J. Bzik and S. L. Yao, Cancer Epidemiol., Biomarkers Prev., 1994, 3, 67–71 CAS.
- H. Liang, B. Peng, C. Dong, L. Liu, J. Mao, S. Wei, X. Wang, H. Xu, J. Shen, H. Q. Mao, X. Gao, K. W. Leong and Y. Chen, Nat. Commun., 2018, 9, 1–14 CrossRef PubMed.
- M. A. Dobrovolskaia, B. W. Neun, S. Man, X. Ye, M. Hansen, A. K. Patri, R. M. Crist and S. E. McNeil, Nanomedicine, 2014, 10, 1453–1463 CrossRef CAS PubMed.
- S. H. De Paoli, L. L. Diduch, T. Z. Tegegn, M. Orecna, M. B. Strader, E. Karnaukhova, J. E. Bonevich, K. Holada and J. Simak, Biomaterials, 2014, 35, 6182–6194 CrossRef CAS PubMed.
- S. Valpione, G. Gremel, P. Mundra, P. Middlehurst, E. Galvani, M. R. Girotti, R. J. Lee, G. Garner, N. Dhomen, P. C. Lorigan and R. Marais, Eur. J. Cancer, 2018, 88, 1–9 CrossRef CAS PubMed.
- S. J. Bagley, S. Ali Nabavizadeh, J. J. Mays, J. E. Till, J. B. Ware, S. Levy, W. Sarchiapone, J. Hussain, T. Prior, S. Guiry, T. Christensen, S. S. Yee, M. P. Nasrallah, J. J. D. Morrissette, Z. A. Binder, D. M. O’Rourke, A. J. Cucchiara, S. Brem, A. S. Desai and E. L. Carpenter, Clin. Cancer Res., 2020, 26, 397–407 CrossRef PubMed.
- T. B. Hao, W. Shi, X. J. Shen, J. Qi, X. H. Wu, Y. Wu, Y. Y. Tang and S. Q. Ju, Br. J. Cancer, 2014, 111, 1482–1489 CrossRef CAS PubMed.
- D. Fernandez-Garcia, A. Hills, K. Page, R. K. Hastings, B. Toghill, K. S. Goddard, C. Ion, O. Ogle, A. R. Boydell, K. Gleason, M. Rutherford, A. Lim, D. S. Guttery, R. C. Coombes and J. A. Shaw, Breast Cancer Res., 2019, 21, 149 CrossRef CAS PubMed.
- K. Mizuno, S. Akamatsu, T. Sumiyoshi, J. H. Wong, M. Fujita, K. Maejima, K. Nakano, A. Ono, H. Aikata, M. Ueno, S. Hayami, H. Yamaue, K. Chayama, T. Inoue, O. Ogawa, H. Nakagawa and A. Fujimoto, Sci. Rep., 2019, 9, 1–11 CrossRef CAS PubMed.
- K. Kuroi, C. Tanaka and M. Toi, Int. J. Oncol., 2001, 19, 143–148 CAS.
- S. Holdenrieder, D. Nagel, A. Schalhorn, V. Heinemann, R. Wilkowski, J. Von Pawel, H. Raith, K. Feldmann, A. E. Kremer, S. Müller, S. Geiger, G. F. Hamann, D. Seidel and P. Stieber, Annals of the New York Academy of Sciences, New York Academy of Sciences, 2008, vol. 1137, pp. 180–189 Search PubMed.
- S. Kumar, R. Guleria, V. Singh, A. C. Bharti, A. Mohan and B. C. Das, Clin. Lung Cancer, 2010, 11, 36–44 CrossRef CAS PubMed.
- S. Holdenrieder, P. Stieber, H. Bodenmüller, M. Busch, G. Fertig, H. Fürst, A. Schalhorn, N. Schmeller, M. Untch and D. Seidel, Int. J. Cancer, 2001, 95, 114–120 CrossRef CAS PubMed.
- D. de Miguel Pérez, A. Rodriguez Martínez, A. Ortigosa Palomo, M. Delgado Ureña, J. L. Garcia Puche, A. Robles Remacho, J. Exposito Hernandez, J. A. Lorente Acosta, F. G. Ortega Sánchez and M. J. Serrano, Sci. Rep., 2020, 10, 1–13 CrossRef PubMed.
- J. D. Arroyo, J. R. Chevillet, E. M. Kroh, I. K. Ruf, C. C. Pritchard, D. F. Gibson, P. S. Mitchell, C. F. Bennett, E. L. Pogosova-Agadjanyan, D. L. Stirewalt, J. F. Tait and M. Tewari, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5003–5008 CrossRef CAS PubMed.
- M. C. Liu, G. R. Oxnard, E. A. Klein, C. Swanton, M. V. Seiden, M. C. Liu, G. R. Oxnard, E. A. Klein, D. Smith, D. Richards, T. J. Yeatman, A. L. Cohn, R. Lapham, J. Clement, A. S. Parker, M. K. Tummala, K. McIntyre, M. A. Sekeres, A. H. Bryce, R. Siegel, X. Wang, D. P. Cosgrove, N. R. Abu-Rustum, J. Trent, D. D. Thiel, C. Becerra, M. Agrawal, L. E. Garbo, J. K. Giguere, R. M. Michels, R. P. Harris, S. L. Richey, T. A. McCarthy, D. M. Waterhouse, F. J. Couch, S. T. Wilks, A. K. Krie, R. Balaraman, A. Restrepo, M. W. Meshad, K. Rieger-Christ, T. Sullivan, C. M. Lee, D. R. Greenwald, W. Oh, C. K. Tsao, N. Fleshner, H. F. Kennecke, M. F. Khalil, D. R. Spigel, A. P. Manhas, B. K. Ulrich, P. A. Kovoor, C. Stokoe, J. G. Courtright, H. A. Yimer, T. G. Larson, C. Swanton, M. V. Seiden, S. R. Cummings, F. Absalan, G. Alexander, B. Allen, H. Amini, A. M. Aravanis, S. Bagaria, L. Bazargan, J. F. Beausang, J. Berman, C. Betts, A. Blocker, J. Bredno, R. Calef, G. Cann, J. Carter, C. Chang, H. Chawla, X. Chen, T. C. Chien, D. Civello, K. Davydov, V. Demas, M. Desai, Z. Dong, S. Fayzullina, A. P. Fields, D. Filippova, P. Freese, E. T. Fung, S. Gnerre, S. Gross, M. Halks-Miller, M. P. Hall, A. R. Hartman, C. Hou, E. Hubbell, N. Hunkapiller, K. Jagadeesh, A. Jamshidi, R. Jiang, B. Jung, T. H. Kim, R. D. Klausner, K. N. Kurtzman, M. Lee, W. Lin, J. Lipson, H. Liu, Q. Liu, M. Lopatin, T. Maddala, M. C. Maher, C. Melton, A. Mich, S. Nautiyal, J. Newman, J. Newman, V. Nicula, C. Nicolaou, O. Nikolic, W. Pan, S. Patel, S. A. Prins, R. Rava, N. Ronaghi, O. Sakarya, R. V. Satya, J. Schellenberger, E. Scott, A. J. Sehnert, R. Shaknovich, A. Shanmugam, K. C. Shashidhar, L. Shen, A. Shenoy, S. Shojaee, P. Singh, K. K. Steffen, S. Tang, J. M. Toung, A. Valouev, O. Venn, R. T. Williams, T. Wu, H. H. Xu, C. Yakym, X. Yang, J. Yecies, A. S. Yip, J. Youngren, J. Yue, J. Zhang, L. Zhang, L. Zhang, N. Zhang, C. Curtis and D. A. Berry, Ann. Oncol., 2020, 31, 745–759 CrossRef.
- A. A. I. Sina, L. G. Carrascosa, Z. Liang, Y. S. Grewal, A. Wardiana, M. J. A. Shiddiky, R. A. Gardiner, H. Samaratunga, M. K. Gandhi, R. J. Scott, D. Korbie and M. Trau, Nat. Commun., 2018, 9, 1–13 CrossRef CAS PubMed.
- A. H. Nguyen and S. J. Sim, Biosens. Bioelectron., 2015, 67, 443–449 CrossRef CAS PubMed.
- C. Rago, D. L. Huso, F. Diehl, B. Karim, G. Liu, N. Papadopoulos, Y. Samuels, V. E. Velculescu, B. Vogelstein, K. W. Kinzler and L. A. Diaz, Cancer Res., 2007, 67, 9364–9370 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nh00333f |
| This journal is © The Royal Society of Chemistry 2020 |