Development of a novel and useful methodology for the simultaneous analysis of multiclass contaminants in bovine fat†
Rodrigo
Souza
ab,
Natalia
Gérez
 a,
Natalia
Besil
c,
María Verónica
Cesio
a,
Natalia
Besil
c,
María Verónica
Cesio
 ad,
Horacio
Heinzen
ad,
Horacio
Heinzen
 *ad and
Lucia
Pareja
*ad and
Lucia
Pareja
 *c
*c
aGrupo de Análisis de Compuestos Traza, Departamento de Química Orgánica, Facultad de Química, Universidad de la República, General Flores 2124, 11700, Montevideo, Uruguay. E-mail: heinzen@fq.edu.uy
bGraduate Program in Chemistry, Facultad de Química, Universidad de la República, Montevideo, Uruguay
cGrupo de Análisis de Compuestos Traza, Departamento de Química del Litoral, Cenur Litoral Norte, Universidad de la República, Ruta 3 Km 363, 60000, Paysandú, Uruguay. E-mail: lpareja@fq.edu.uy
dGrupo de Análisis de Compuestos Traza, Cenur Litoral Norte, Universidad de la República, Ruta 3 Km 363, 60000, Paysandú, Uruguay
First published on 12th September 2024
Abstract
The instrumental development of tandem mass spectrometers fosters the actual trend in the trace analysis of organic compounds to the development of methods that allow the analysis of contaminants of the most diverse origin in a single analytical sample. The multiclass methods are aligned with the Who's One Health initiative while accomplishing the concepts of green chemistry. However, there are few reports of wide scope multiclass methods for the analysis of contaminants in this matrix. In this work, a method for the simultaneous determination of 62 compounds in bovine fat, was developed and validated following DG-SANTE/11813/2021 guidance. Liquid nitrogen milled fat was extracted with toluene and acetonitrile. Then a clean-up in a cryogenic bath, followed by dispersive solid phase extraction was performed. Residue determination was done using liquid chromatography for 44 compounds and gas chromatography for 18 compounds, both coupled to tandem mass spectrometry in MRM mode. The method was first developed and validated for two ectoparasiticides, then the scope was expanded for the analysis of 13 veterinary drugs and 49 pesticides. Recovery percentages were in the range of 60–134%, high matrix effect was observed in 50% of the scope of the method. Most compounds presented limits of quantification of 10 μg kg−1 in compliance with international requirements. The method was applied to monitor 49 commercial samples to evaluate its performance. Eighty percent of samples contained ethion and 10% had fluazuron, both within MRLs, highlighting the need for proper withdrawal times.
Introduction
Pest control in agriculture and livestock involves treatments with a wide variety of synthetic compounds such as pesticides and veterinary drugs. Pesticides can be transferred from plants to animals through the food chain, subsequently contaminating by-products used for human consumption (muscle, milk, liver, and others).1 Veterinary drugs employed for animal disease control can also contaminate the final products. To frame the contaminant contents within Good Agricultural and Veterinary Practices, the Codex Alimentarius Commission, multinational organizations such as the European Commission, and regulation bodies of several countries, have established Maximum Residue Levels (MRLs) in different food matrices, including, the bovine ones.2–5Distinct regulations govern foods of animal and plant origin, labeled as “pesticides” and “veterinary drugs,” respectively, despite often being the same molecule (codex, EU). Compounds exclusively used in agriculture, lack established MRLs, defaulting to 10 parts per billion (ppb) for enforcement. This unharmonized regulations, has shaped diverse approaches to food safety. Residue analysis for animal-derived foods focuses on specific compound families (antibiotics, antiparasitics, among others), while plant origin analysis relies on multiresidue methods for fungicides, herbicides, and insecticides.
Given the potential for contamination through feed intake and sanitary management, it is essential to consider pesticide exposure in animals within the overall analytical scope of potentially dangerous contaminants in food of animal origin.
The complexity of analyzing contaminants in animal products, particularly in fat matrices, has led to the development of various analytical methods. Fat is a challenging matrix due to its ability to bioaccumulate lipophilic compounds, posing a risk of exposure through food.1 Particularly, organophosphorus and pyrethroids pesticides, antibiotics such as chloramphenicol, parasiticides like ivermectin, with high Kow tend to accumulate in fat tissues and partitionate between the extracting solvent and the fat itself.6
In addition, fat co-extractives are known to pollute the chromatographic system, poisoning electron capture and mass detectors, lowering the overall sensibility of the method; and forcing exhaustive clean-up steps to decrease the levels of co-extractives in the final extract.7,8
Moreover, there are at least two types of fat samples with different water content and lipid composition, the one composed of 100% triglycerides, which is used to fry purposes, and fat tissues, with variable fat content. The U.S. Food and Drug Administration (FDA) defines fatty foods as those with fat content over 2%,9 but Lehotay et al., considered that matrices with a fat content over 20% should be cataloged as high fatty foods.9
Sample treatment can influence the final concentration expressed for each compound. For fatty tissues, old methodologies preferred to melt the sample before extraction, and report the findings on the basis of the melted fat.10 The assumption made is that all the lipophilic compounds are concentrated in fat and their occurrence in other tissues is negligible. This is not the actual trend in the analysis of veterinary drugs and pesticide residues analysis. During the analysis of high fat matrices, it is recommended to express the concentration levels on a lipid adjusted basis, to obtain more representative results in comparison with unadjusted values.10,11 To compare results, the concentration might be expressed as milligrams or micrograms per kilogram of fat.10
Several methods have been developed to address this issue. Akre and Macneil, 2006 utilized Solid Phase Extraction (SPE) and gas chromatography electron capture detection for analyzing synthetic pyrethroids.12 Sartarelli et al., 2014 adapted the QuEChERS method for analyzing pesticides in bovine fat and meat, involving an extraction with acetonitrile and hexane, followed by a clean-up step with PSA and MgSO4, and GC-MS analysis.13 Also, Hmadamin and Hassan developed a variation of the QuEChERS method for the analysis of six pesticides in bovine fat.14 Unterluggauer et al., introduced the FATchers method, a QuEChERS-based approach suitable for fat and tricky matrices, which is widely used in the European Union Reference Laboratory for Food of Animal Origin.15 This method allows the determination of a wide range of compounds with varying polarity, using a combination of liquid–liquid extraction and dispersive solid phase extraction. Other methods, such as those developed by Castillo et al. (2012) and Hoyos et al. (2013), also focus on specific families of contaminants, typically those that are highly lipophilic and GC-amenable.16,17 However, these methods often concentrate on a limited range of compounds, leaving out other relevant contaminants.
A general thought in the past was that this type of methods should be developed only to monitor highly lipophilic compounds, such as organochlorine, organophosphate or pyrethroids insecticides, all GC amenable compounds, leaving out other families of contaminants.9 Nowadays, new lipophilic compounds that are heat sensitive and which also tend to bioaccumulate in fat tissues,18,19 can only be analysed by HPLC (ivermectins, spinosads, spirotetramat and metabolites) and must be determined as well.9
In line with green chemistry concepts, the present work aims to develop and validate a simple and comprehensive multiclass method for the simultaneous analysis of 62 pesticides and veterinary drugs in fat. The proposed method seeks to address the limitations of previous approaches by including a broader range of contaminants, particularly those that are lipophilic, heat-sensitive, and tend to bioaccumulate in fat tissues. By integrating various extraction and clean-up steps, the method offers a more holistic approach to food safety, providing a more representative analysis of potentially hazardous compounds in animal-derived products. This comprehensive approach allows for the simultaneous determination of multiple contaminants at trace levels after a single sample preparation step, enhancing the overall assessment of food safety in the production chain.
Experimental
Materials
Samples of perirenal fat for method development and validation were obtained from local butcheries, then stored at −20 °C until analysis. Analytical standards in a range of purity of 96–99.9%, were purchased from Dr Ehrenstorfer (Augsburg, Germany). All solvents were of pesticide analysis grade, EtOAc was obtained from J.T Baker (NJ, USA), dichloromethane (CH2Cl2), n-hexane, MeCN and methanol (MeOH) from Merck (Darmstadt, Germany) and toluene was purchased from Pharmco (CT, USA). Water used for HPLC-MS/MS analysis was obtained from a Barnstead EASY pure RoDi from Thermo Scientific (Waltham, USA). Ammonium Formate (HCOONH4) and MgSO4 were acquired from J.T Baker (NJ, USA). Sodium Chloride (NaCl), PSA and 40–60 μm RPC-18 were purchased from Scharlau (Barcelona, Spain). Alumina and Florisil were provided by Carlo Erba (Sabadell, Spain). Individual stock standard solutions of the targeted compounds at 2000 mg L−1 were prepared according to solubility properties in EtOAc, MeCN, and MeOH, and stored at −18 °C. Working solutions of 100 and 10 mg L−1 for the spiking procedure and matrix-matched calibration, were prepared by dilution of the stock standard solutions.Instrumental analysis
Liquid chromatography analysis was conducted with an Agilent 1200 series coupled to a Sciex 4000 Qtrap (Concord, Canada). Quadrupole-linear ion trap (QTrap®) was operated in triple quadrupole MS/MS mode, in positive and negative ESI polarity. Two transitions for the correct identification of each compound were selected. Source voltages were set at 4000 and −4500 V for the positive and negative ionization modes, respectively. Solvent evaporation in the source was assisted by heated nitrogen as drying gas (500 °C per 50 psi). Collision Energies (CE) and Declustering Potentials (DPs) for each investigated compound were optimized, after direct infusion, using Analyst Software v 1.5.1. Scheduled™ MRM mode was selected to increase compound sensitivity, as it is shown in Table ESI 1 and 2.†For the analysis of pesticides and veterinary drugs the positive MS/MS mode was performed using a mobile phase (MP) of 0.1% formic acid and 5 mM of HCOONH4 in water (A) and 0.1% formic acid and 10 mM of HCOONH4 in MeOH (B). The gradient was 0 min, 98% A, 12 min, 0% A (4 min hold time), and 21 min, 98% A. The re-equilibration time was 5 min.
For negative MS/MS mode, the MP was a mixture of 0.1% of formic acid in water (A) and MeCN (B). The chromatographic method held the initial mobile phase composition 0 min, 50% B; 5 min, 100% B (4 min hold time); 10 min, 50% B. The re-equilibration time until the next injection took 5 min. The flow rate used was 0.6 mL min−1 for positive and negative mode, and the injection volume was 5 μL.
GC amenable analytes were determined using a GC-MS/MS with a Shimadzu GC-2010 plus coupled to MS/MS TQ 8050 spectrometer in Electron Impact ionization (EI) mode. The target analytes were separated in an RTX-5MS (5% diphenyl/95% dimethyl polysiloxane) analytical column (30 m × 0.25 mm i.d. × 0.25 m film thickness), from Restek, (Bellfonte, PA, USA). Tandem mass spectrometry detection was performed in the multiple reaction monitoring (MRM) mode using transitions and collision energies previously selected for each compound using Smart Pesticide Database (SPDB) provided by Shimadzu Corporation.20
The injector temperature was set at 240 °C, interface temperature at 300 °C, and Helium at 14 mL s−1 as carrier gas. Oven temperature: 0 min, 80 °C (2 min hold); 7 min, 180 °C (20 °C min−1 rate); 31 min, 300 °C (5 °C min−1 rate, 3 min hold). The injection volume was 1 μL in splitless mode. The retention times for each pesticide were set using the Automatic Adjustment Retention Time function (AART) ™ (Shimadzu). The AART function stablish the retention times of the target components based on Linear Retention Indices (LRI) and the retention times of n-alkanes. The final MRM method parameters are shown in Table ESI 1.†
Sample preparation
Pieces of frozen perirenal fat samples of about 1 × 1 cm were placed in a mixer with liquid nitrogen and the mixture milled to obtain a fine powder. N2 was allowed to evaporate and the comminuted fat sample was transferred to polypropylene flasks and stored at −18 °C. Blank samples for all fortification experiments and matrix-matched calibration curves were analyzed using the same analytical method, confirming that no pesticides or veterinary drugs from the scope, were present above the quantification limits.Method development and validation
Preliminary tests: Three different strategies were evaluated in terms of recovery percentages and relative standard deviations during method development: QuEChERS, Ethyl Acetate method and Matrix Solid Phase Dispersion (MSPD).15,21,22 For every combination, two sample pre-treatments were tested; cutting the sample in small pieces with a knife or cryogenic freezing with liquid N2. All the combinations are summarized in Table 1.| Method | Critical step | Variables |
|---|---|---|
| MSPD | Sample homogenization | With or without N2 |
| Dispersant | Alumina or florisil | |
| Elution | Type of solvent | |
| Volume amount | ||
| Concentration factor | ||
| QuEChERS | Sample homogenization | With or without N2 |
| Sample amount | 2 or 5 g | |
| Extraction | Type of solvent | |
| Purification | Amount and type of sorbent | |
| Ethyl acetate | Sample homogenization | With or without N2 |
| Sample amount | 2 or 5 g | |
| Extraction | Type of solvent | |
| Purification | Amount and type of sorbent |
QuEChERS and ethyl acetate methods
For QuEChERS and ethyl acetate, some variations of the original methods were evaluated. Two sample amounts (2 and 5 g) were tested. As extraction agents EtOAc, MeCN, and MeCN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Toluene (2
Toluene (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) were assessed. Moreover, two different clean-up sorbents, (PSA and RPC-18) were selected, in two amounts (25 and 50 mg mL−1).
1, v/v) were assessed. Moreover, two different clean-up sorbents, (PSA and RPC-18) were selected, in two amounts (25 and 50 mg mL−1).
MSPD
For MSPD the selection of an appropriate dispersive agent and the types of solvents for the elution step are critical. Therefore, to obtain the best extraction conditions, method comparison was performed selecting alumina and florisil as sorbents and two different volumes (10 and 15 mL) of EtOAc and hexane saturated in MeCN as elution solvent.Selected sample preparation methodology
The final method consisted of weighting (2 ± 0.5) g of liquid N2 milled perirenal fat previously homogenized in a 50 mL-polypropylene falcon tube followed by the addition of 5.0 mL of toluene and 5 minutes vortex. Then, 10.0 mL of MeCN were added and shaked in a vortex for 1 minute. Then the tubes were centrifuged at 4400 × g for 4 minutes, placed in a cryogenic bath at −42 °C for 15 minutes and centrifuged again for 2 minutes. Finally, eight mL of the supernatant were transferred to a 15 mL-falcon tube containing 360 mg of PSA, 360 mg of RPC-18 and 750 mg of anhydrous MgSO4, vortexed for 1 minute and centrifuged at 4400 × g for 4 minutes (Fig. 1). For GC-MS/MS analysis, 2 mL aliquot was concentrated under nitrogen stream at 55 °C for 20 minutes and re-dissolved in one mL of EtOAc. For HPLC-MS/MS analysis, 2 mL aliquot was concentrated in the same conditions, re-dissolved in one mL of MeCN, and stored overnight at −20 °C. The extract was filtered with a 0.45 μm PVDF filter and transferred to an amber vial for injection. | ||
| Fig. 1 Scheme of the validated methodology for multiclass analysis of veterinary drugs and pesticides in bovine fat. | ||
Total lipidic content determination was made by gravimetry, as in the method developed by Folch et al.23 This percentage was used in the eqn (1) to express the concentration of each sample in 100% fat.
 | (1) |
Method validation
The evaluation of the selectivity was performed through the injection of blank matrix extractions spiked with a mix of the selected compounds, to check the presence of potential interferences.
Trueness and precision were studied through the determination of recoveries (%Rec) and relative standard deviations (%RSD), at 10, 50 and 100 μg kg−1 by quintuplicate.
Recoveries were calculated based on the comparison between the concentration calculated with the matrix-matched calibration curve and the true concentration for the spiked analytes at each fortification level.
Repeatability and intermediate precision were tested on different days, a %RSD < 20 was considered acceptable.24
The linearity was studied by preparing six-point calibration curves; in pure solvent and in matrix extract, in the range of 5–100 μg kg−1 for HPLC and GC-MS/MS. Coefficient of variations (r2) and Back Calculated Concentration (BCC) residuals of each calibration curve were determined for all the compounds being less than ± 20%.
Matrix effect (ME) was calculated by comparing the slopes of the curves in pure solvent and in the matrix-matched calibration curve. SANTE guidelines suggest that for every compound with a %ME > 20 in absolute value, matrix-matched calibration is mandatory.24 Compounds were classified in three ranges of ME percentages according to Kmellar et al., (2008).25
LOQs were determined as the lowest concentration of each analyte that has been validated with acceptable trueness and precision by applying the complete analytical method for both instruments.24
Results and discussion
As stated above, lipophilic contaminants in animal origin foods, might come either from the feed, which is normally some agricultural waste, or from veterinary drugs used to prevent diseases and pests. It is well known that adipose tissue is a relevant reservoir of lipophilic compounds.26 These contaminants, at relatively high concentrations, can bioaccumulate within the fatty tissues. When consumed by human they can be released by the enzymes of the digestive tract. To perform an efficient and comprehensive evaluation of food safety, the development of multiclass methods to analyze multiple contaminants of diverse origin in a single chromatographic run is of great importance. Also, the use of multiclass methods enhances the productivity of routine laboratories.Scope of the validated method
Being fat a highly lipidic matrix, non and middle-polar compounds with middle to high Kow, tend to accumulate in this tissue.7,9,27 Assuming that compounds with high pKow would tend to bioaccumulate, two criteria were considered for the selection of the scope. First, the Uruguayan National Program of Biologic Residues (PNRB), which establishes the directives for local residue monitoring.28 Second, other compounds used in cattle breeding, as well as those agricultural productive chains whose by-products are used as feed for cattle.18,19 The final list was checked with the international requirements of Codex Alimentarius and EU regulations to comprehensively include important MRLs.2–5 Based on these criteria, we selected 70 compounds (Table 2) either classified as pesticides or veterinary drugs to be analyzed in a single run. Fig. 2 shows the HPLC-MS/MS MRM transitions at 100 μg kg−1 of a fat extract and the Extracted Ion Chromatograms (XIC) at 10, 50 and 100 μg kg−1 of three compounds.| Compound | Instrumental method | 10 μg kg−1 | 50 μg kg−1 | 100 μg kg−1 | LOQ (μg kg−1) | Matrix effect | MRL (μg kg−1) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| % rec | % RSD | % rec | % RSD | % rec | % RSD | |||||
| Abamectin | HPLC-MS/MS | 88 | 6 | 84 | 10 | 80 | 9 | 10 | 109 | 10 |
| Alachlor | HPLC-MS/MS | 81 | 14 | 94 | 2 | 90 | 5 | 10 | 1.3 | 10 |
| Ametryn | HPLC-MS/MS | 101 | 4 | 113 | 4 | 106 | 4 | 10 | −8.2 | 10 |
| Atrazine | HPLC-MS/MS | 97 | 5 | 116 | 3 | 103 | 3 | 10 | −6.3 | 10 |
| Azinphos methyl | GC-MS/MS | 91 | 16 | 104 | 15 | 94 | 18 | 10 | 92 | 10 |
| β-Cyfluthrin | GC-MS/MS | nd | nd | 112 | 7 | 101 | 9 | 50 | 71 | 200 |
| Byfenthrin | GC-MS/MS | 102 | 54 | 116 | 12 | 96 | 14 | 10 | 49 | 3000 |
| Boscalid | HPLC-MS/MS | 143 | 13 | 117 | 5 | 98 | 2 | 10 | 16 | 300 |
| Carbofuran | HPLC-MS/MS | 67 | 14 | 115 | 6 | 110 | 10 | 10 | 15 | 10 |
| Cypermethrin | GC-MS/MS | 77 | 19 | 90 | 15 | 97 | 11 | 10 | 66 | 2000 |
| Clomazone | HPLC-MS/MS | 96 | 4 | 87 | 3 | 84 | 3 | 10 | −1.3 | 10 |
| Chlorantraniliprole | HPLC-MS/MS | 104 | 10 | 115 | 5 | 100 | 5 | 10 | 7.1 | 200 |
| Chlorfenvinphos | GC-MS/MS | 79 | 11 | 114 | 7 | 106 | 9 | 10 | 54 | 10 |
| Chlorpyrifos methyl | GC-MS/MS | 72 | 8 | 105 | 8 | 99 | 12 | 10 | 54 | 10 |
| Coumaphos | HPLC-MS/MS | 108 | 4 | 106 | 6 | 97 | 12 | 10 | 74 | 10 |
| Diazinon | HPLC-MS/MS | 81 | 6 | 112 | 8 | 96 | 10 | 10 | 38 | 700 |
| Difenoconazole | HPLC-MS/MS | 106 | 13 | 133 | 13 | 128 | 19 | 10 | −6.3 | 50 |
| Deltamethrin | GC-MS/MS | 70 | 12 | 107 | 10 | 99 | 12 | 10 | 66 | 500 |
| Doramectin | HPLC-MS/MS | 88 | 6 | 69 | 10 | 73 | 11 | 10 | 104 | 150 |
| Emamectin benzoate | HPLC-MS/MS | 84 | 10 | 80 | 18 | 78 | 9 | 10 | −3.9 | 20 |
| Endosulfan sulfate | GC-MS/MS | 120 | 3 | 98 | 15 | 50 | 35 | 50 | ||
| Eprinomectin | HPLC-MS/MS | 92 | 13 | 89 | 10 | 76 | 13 | 10 | 107 | 250 |
| Ethion | HPLC-MS/MS | 90 | 8 | 101 | 3 | 90 | 7 | 10 | 83 | 10 |
| Fenhexamid | HPLC-MS/MS | 63 | 19 | 108 | 16 | 66 | 19 | 10 | −11 | 50 |
| Fenthion | HPLC-MS/MS | 70 | 4 | 87 | 18 | 88 | 11 | 10 | 59 | 50 |
| Fenvalerate | GC-MS/MS | 70 | 10 | 106 | 10 | 100 | 9 | 10 | 62 | 250 |
| Fipronil | GC-MS/MS | 86 | 4 | 134 | 7 | 111 | 11 | 10 | 56 | 30 |
| Fluazuron | HPLC-MS/MS | 130 | 12 | 103 | 8 | 96 | 25 | 10 | 66 | 7000 |
| Flutriafol | HPLC-MS/MS | 116 | 7 | 127 | 3 | 104 | 1 | 10 | −7.2 | 10 |
| Fluvalinate | GC-MS/MS | 70 | 7 | 120 | 15 | 109 | 12 | 10 | 53 | 300 |
| Haloxyfop methyl | HPLC-MS/MS | 100 | 5 | 119 | 4 | 97 | 10 | 10 | 7.2 | 10 |
| Hexythiazox | HPLC-MS/MS | 96 | 18 | 87 | 7 | 82 | 9 | 10 | 105 | 50 |
| Iprodione | GC-MS/MS | 67 | 19 | 105 | 10 | 104 | 11 | 10 | 69 | 10 |
| Isoprothiolane | HPLC-MS/MS | 100 | 4 | 116 | 4 | 110 | 4 | 10 | 1.8 | 10 |
| Ivermectin | HPLC-MS/MS | 86 | 8 | 73 | 16 | 60 | 8 | 10 | 59 | 100 |
| Kresoxim methyl | HPLC-MS/MS | 96 | 7 | 119 | 2 | 111 | 8 | 10 | −1.8 | 50 |
| Lambda cyhalothrin | GC-MS/MS | 104 | 19 | 124 | 8 | 112 | 14 | 10 | 49 | 3000 |
| Linuron | HPLC-MS/MS | 91 | 4 | 111 | 3 | 108 | 5 | 10 | −12 | 10 |
| Malaoxon | HPLC-MS/MS | 111 | 3 | 115 | 3 | 98 | 2 | 10 | −3.5 | 20 |
| Malathion | HPLC-MS/MS | 108 | 5 | 111 | 6 | 97 | 2 | 10 | 5 | 20 |
| Mebendazole | HPLC-MS/MS | 104 | 4 | 117 | 3 | 106 | 4 | 10 | −2.3 | 60 |
| Metconazole | HPLC-MS/MS | 123 | 11 | 81 | 10 | 80 | 6 | 10 | −1.8 | 20 |
| Methiocarb | HPLC-MS/MS | 89 | 6 | 104 | 4 | 96 | 5 | 10 | −12 | 30 |
| Metolachlor | HPLC-MS/MS | 94 | 5 | 101 | 3 | 98 | 4 | 10 | −4.3 | 10 |
| Mirex | GC-MS/MS | 60 | 17 | 81 | 7 | 68 | 11 | 10 | 34 | 10 |
| Monensin | HPLC-MS/MS | 83 | 15 | 71 | 10 | 74 | 13 | 10 | 101 | 10 |
| Moxidectin | HPLC-MS/MS | 72 | 19 | 60 | 20 | 50 | 21 | 10 | 101 | 500 |
| p,p′-DDD | GC-MS/MS | 64 | 24 | 97 | 14 | 110 | 12 | 10 | 90 | 10 |
| p,p′-DDT | GC-MS/MS | 67 | 12 | 65 | 12 | 61 | 17 | 10 | 39 | 10 |
| p,p′-DDE | GC-MS/MS | 61 | 10 | 79 | 5 | 77 | 8 | 10 | 38 | 10 |
| Parathion methyl | GC-MS/MS | 82 | 5 | 117 | 11 | 106 | 10 | 10 | 74 | 10 |
| Pendimetalin | HPLC-MS/MS | 91 | 11 | 79 | 5 | 76 | 9 | 10 | 109 | 200 |
| Permethrin | GC-MS/MS | 80 | 29 | 114 | 16 | 94 | 13 | 10 | 57 | 500 |
| Pyraclostrobin | HPLC-MS/MS | 106 | 9 | 138 | 5 | 102 | 19 | 10 | −12 | 50 |
| Pirimifos methyl | HPLC-MS/MS | 76 | 17 | 115 | 10 | 105 | 12 | 10 | 41 | 10 |
| Propiconazole | HPLC-MS/MS | 96 | 8 | 92 | 5 | 95 | 8 | 10 | −0.73 | 10 |
| Spinosyd A | HPLC-MS/MS | 63 | 27 | 84 | 14 | 73 | 7 | 10 | −24 | 3000 |
| Spinosyd D | HPLC-MS/MS | 96 | 23 | 87 | 20 | 104 | 16 | 10 | −35 | 3000 |
| Tebuconazole | HPLC-MS/MS | 115 | 12 | 109 | 8 | 114 | 11 | 10 | 0.6 | 100 |
| Tebufenozide | HPLC-MS/MS | 116 | 8 | 128 | 3 | 106 | 4 | 10 | −11 | 50 |
| Tetraconazole | HPLC-MS/MS | 115 | 18 | 117 | 4 | 104 | 3 | 10 | −12 | 200 |
| Triclabendazole | HPLC-MS/MS | 101 | 6 | 97 | 9 | 99 | 3 | 10 | −0.7 | 100 |
| Tryfloxystrobin | HPLC-MS/MS | 110 | 6 | 128 | 4 | 105 | 8 | 10 | −11 | 60 |
| Triflumuron | HPLC-MS/MS | 133 | 32 | 99 | 12 | 71 | 38 | 10 | −11 | 100 |
| Triticonazole | HPLC-MS/MS | 107 | 27 | 100 | 18 | 86 | 10 | 10 | −5 | 10 |
| Vinclozolin | GC-MS/MS | 81 | 9 | 129 | 9 | 99 | 10 | 10 | 49 | 10 |
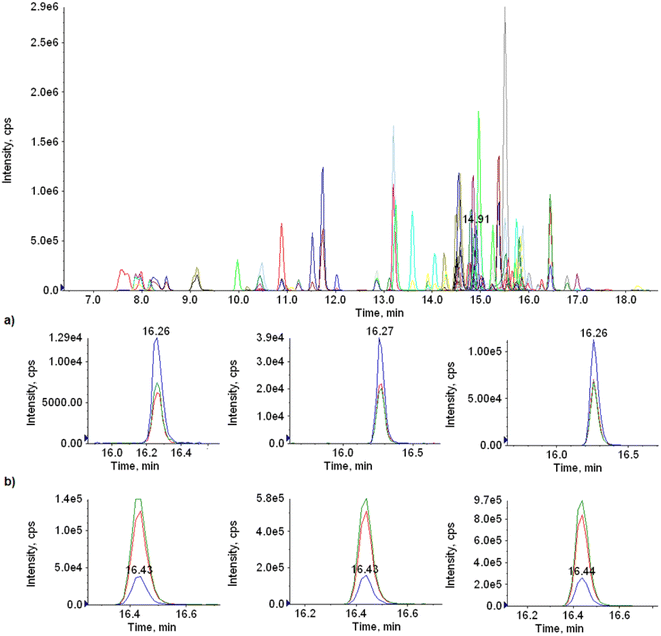 | ||
| Fig. 2 HPLC-MS/MS MRM transitions at 100 μg kg−1 of a fat extract and the Extracted Ion Chromatograms (XIC) of (a) fluazuron and (b) ethion at, from left to right, 10, 50 and 100 μg kg−1. | ||
Sample preparation
The particle size of the sample is critical for the quantitative extraction of the analytes from the fatty matrix.29 The cryo-milling with liquid N2, provided a good homogeneous sample, of small particle size (fine powder), which was impossible to achieve using other types of milling strategies. Without this milling process, poor extraction repeatability was observed.Method development
Initially, the method was developed for the analysis of ethion and cypermethrin, two compounds used either as veterinary drugs or pesticides which are of high relevance in Uruguayan agriculture and livestock production. According to their pKow, both acaricides are good models for test method's suitability. Then, the scope of the method was expanded to include the compounds listed in Table 2. Many trials were assayed to optimize the final method, such as MSPD and variations of MeCN and ethyl acetate-based extraction protocols. All of them failed to yield acceptable results for two reasons. The first one was due to the inefficient extraction of some pesticides from the fatty tissue when a polar solvent such as MeCN or its mixtures with apolar ones were assayed. Additionally high amounts of co-extractives were obtained when a less polar solvent was employed.Based on our previous experience when developing an analytical methodology for beeswax, we decided to make a liquid extraction with MeCN but without melting the fat. The fat was dissolved in toluene and then MeCN was added. A single phase was formed, ensuring that the whole sample was in contact with the extracting solvent. The next step was to eliminate the excess of lipids in the solution. To achieve this goal, the solution was kept for 15 min in a cryocooler at −42 °C. The cooled, solid lipid precipitated and the solution was centrifuged immediately. These steps need to be performed very fast to avoid the melting of the lipids. A refrigerated centrifuge could improve lipid separation. The supernatant was cleaned-up and leaded to the chromatographic-tandem mass spectrometry analysis. It is important to mention that, although good results were obtained, the instruments came dirty very fast. Therefore, a further clean-up was added. After comparing an n-hexane extraction of the MeCN solution and a freeze-out step, the latter proved to be easier and reproducible to perform for routine analysis. So, the solution was kept at −18 °C overnight, filtered and analyzed.
Method validation
After sample pretreatment and method extraction were optimized, five replicates of spiked blanks of fat at different concentration levels (10, 50, and 100 μg kg−1) were analyzed to evaluate trueness (%Rec), repeatability, and intermediate precision (%RSD) of the procedure. According to Document N° SANTE/11813/2021, 62 out of the 70 compounds analyzed presented acceptable recovery percentages (70–120%) (Table 2). Regarding reproducibility all compounds showed %RSD <20. The LOQ was set as 10 μg kg−1 for all compounds except for β-cyfluthrin and endosulfan sulfate which presented a limit of 50 μg kg−1. As the MRLs for these compounds in fat are usually higher than in other matrices, the LOQs fairly comply with the international requirements of the EU and Codex Alimentarius.2–5Almost all compounds showed good linearity between 5–100 μg kg−1 for both instruments in pure solvent and matrix-matched, only β-cyfluthrin and endosulfan sulphate had a linear range between 10–100 μg kg−1. Residuals expressed as BCC were below 20% for all the studied compounds.
Although the exhaustive clean-up of the method, matrix effects (ME) were observed for 35 out of the 62 compounds (Fig. 3). Around 37% of the analytes had strong matrix effect (over 50%), whereas 17% showed medium ME, (between 20 and 50%). No relationship between physicochemical properties and the ME profile was found, being organophosphorus pesticides, avermectins and pyrethroids the group with higher ME. As expected, in GC-MS/MS the predominant effect observed was signal enhancement. This may be since certain co-extractives are acting as analyte protectants, preventing compound degradation in the injector or along the chromatographic run.
In HPLC-MS/MS, ME alternated between signal suppression and enhancement depending on each compound. Based on these results, matrix-matched calibration was chosen for the quantification.
Real samples analysis
The validated method was checked against 49 samples obtained before the ethion restriction in Uruguay, to verify the compliance with the MRLs and generate information about the occurrence of these contaminants in local final products. Ethion was present in 80% of the selected samples, in most of them at concentrations below the LOQ (10 μg kg−1) complying with both national and EU MRLs (10 μg kg−1). However, it is important to state that ethion is a non-authorized compound for Codex Alimentarius, so these results enhance the importance of this method to establish correct withdrawal times. Fluazuron was observed in 10% of the samples, in all cases at concentrations below the MRL (7000 μg kg−1).All results were corrected according to the lipidic contents of the samples as reported by ref. 13. This correction is important to compare the results with classic methodologies which use melting as sample pre-treatment, to eliminate water and thus, expressing the final concentration as 100% fat, with a deviation between 20–30% compared to the proposed method.13
Conclusions
A simple multiclass methodology for the simultaneous analysis of pesticides and veterinary drugs was developed and validated. It consists of the extraction of the selected compounds with (MeCN/toluene), and a clean-up with a fast deep freeze-out, PSA, RPC-18, and MgSO4. Due to the complexity of the matrix, few methods for the analysis of these compounds are reported, so this work represents a real solution to routine and field analysis of multiclass contaminants in bovine fat. Sample treatment with liquid N2 is much milder compared to the classical melting pre-treatments, and produces a fine powder easier to handle, improving the extraction efficiency and the recovery of heat sensible compounds. Also, lipid elimination through a fast freeze-out by a cryogenic bath (15 min), shortens the time of analysis and lowers the cost compared to lipid elimination through n-hexane washes, turning the whole process into a greener methodology. Finally, the presence of several classes of compounds, with different physicochemical properties, shows the flexibility of the developed method to face its scope enlargement to other polar or semi-polar contaminants.Data availability
The data supporting the findings of this study are available within the article and its ESI.†Author contributions
Souza and Gérez were responsible for sample preparation and measurement collection. Souza, Gérez and Besil were responsible for the data processing and its interpretation. Pareja, Heinzen and Cesio were responsible for ideation, funding acquisition, experimental design, data visualisation, writing and editing. Souza prepared the first draft of the manuscript and performed all the revisions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors would like to thank Programa de Desarrollo de las Ciencias Básicas (PEDECIBA). HH, VC, LP and NB thanks Sistema Nacional de Investigadores (SNI)-ANII.References
- M. LeDoux, J. Chromatogr. A, 2011, 1218(8), 1021 CrossRef CAS PubMed.
- European Commission, Regulation (EC) No.299/2008 of the European Parliament and of the Council of 11 March 2008 amending Regulation (EC) No. 396/2005 on maximum residue levels of pesticides in or on food and feed of Plant and animal origin, Off J Eur Commun, L9 Search PubMed.
- European Commission, Regulation (EU) No 37/2010 of the Commission of 22 December 2009 on pharmacologically active substances and their classification as regards the maximum residue limits in foodstuffs of animal origin, L15, 1, pp. , pp. 1–72 Search PubMed.
- Codex Alimentarius Commission, Codex Pesticides Residues in Food Online Database, 2018, https://www.fao.org/fao-who-codexalimentarius/codex-texts/dbs/pestres/en/, Accessed 18.02.2024 Search PubMed.
- Codex Alimentarius Commission, Codex Veterinary Drug Residue in Food, 38th Session, 2015, https://www.codexalimentarius.org/standards/vetdrugs/veterinary-drugs/en/, Accessed 19.02.2019 Search PubMed.
- A. Valdes, A. Beltrán, C. Mellinas, A. Jiménez and M. C. Garrigós, Trends Food Sci. Technol., 2018, 77, 120 CrossRef CAS.
- A. D. Gil, S. M. Huertas and O. Rampoldi, https://xdoc.mx/documents/composicin-de-la-grasa-de-cobertura-en-bovinos-5ef2669c6c8a1, Accessed 10.08.2024.
- M. Hostens, V. Fievez, J. L. M. R. Leroy, J. Van Ranst, B. Vlaeminck and G. Opsomer, J. Dairy Sci., 2012, 95, 3756 CrossRef CAS PubMed.
- S. J. Lehotay and K. Mastovska, J. AOAC Int., 2005, 88, 630 CrossRef CAS PubMed.
- Department of human health and human services centers for disease control and prevention USA (CDC), Fourth National Report on Human Exposure to Environmental Chemicals, 2009 Search PubMed.
- K. Lichtmannegger, R. Fischer, F. X. Steemann, H. Unterluggauer and S. Masselter, Anal. Bioanal. Chem., 2015, 407(13), 3727 CrossRef CAS PubMed.
- C. J. Akre and J. D. MacNeil, J. AOAC Int., 2006, 89(5), 1425 CrossRef CAS PubMed.
- N. C. Saratelli, A. N. de Macedo, J. P. de Sousa, A. R. de Araujo Nogueira and S. H. Govoni Brondi, Anal. Bioanal. Chem., 2015, 407, 3727 CrossRef PubMed.
- A. Y. Hmadamin and K. I. Hassan, Saudi J. Biol. Sci., 2020, 27(3), 887 CrossRef PubMed.
- H. Unterluggauer, FATchers – a novel, simple and fast sample preparation in food of animal origin, 14th European Pesticide Residue Workshop, 2022 Search PubMed.
- M. Castillo, C. González and A. Miralles, Anal. Bioanal. Chem., 2011, 400, 1315 CrossRef CAS PubMed.
- O. Hoyos, E. Duvan, O. Cuartas, A. Yudy, M. Peñuela and A. Gustavo, Food Chem., 2018, 221, 891 CrossRef PubMed.
- University of Hertfordshire, Pesticide properties database, https://sitem.herts.ac.uk/aeru/ppdb/ Search PubMed.
- University of Hertfordshire, Veterinary substances database, https://sitem.herts.ac.uk/aeru/vsdb/index.htm Search PubMed.
- Shimadzu Automatic Adjustment of Retention Time Function (AART), https://www.shimadzu.eu/automatic-adjustment-retention-time-function-aart/. Accessed 10.08.2022 Search PubMed.
- M. Anastassiades, S. J. Lehotay, D. Stajnbaher and F. Schenck, J. AOAC Int., 2003, 86, 412 CrossRef CAS PubMed.
- H. G. J. Mol, A. Rooseboom, R. Van Dam, M. Roding, K. Arondeus and S. Sunarto, Anal. Bioanal. Chem., 2007, 389, 1715 CrossRef CAS PubMed.
- J. Folch, M. Lees and G. H. Sloane-Standley, J. Biol. Chem., 1957, 226, 497 CrossRef CAS PubMed.
- European Commission DG-SANTE, Method validation and quality control procedures for pesticide residue analysis in food and feed, Document No. SANTE/12682/2019, 2021, https://ec.europa.eu/food/plant/pesticides/guidance_documents/docs/qualcontrol_en.pdf/, Accessed 20 September 2023 Search PubMed.
- B. Kmellar, P. Fodor, L. Pareja, C. Ferrer, M. A. Martínez-Uroz, A. Valverde and A. R. Fernández-Alba, J. Chromatogr. A, 2008, 1215, 37 CrossRef CAS PubMed.
- Y.-M. Lee, K.-S. Kim, D. R. Jr Jacobs and D.-H. Lee, Obes. Rev., 2017, 18, 129 CrossRef PubMed.
- A. Pérez, G. R. González, J. González and H. Heinzen, J. AOAC Int., 2010, 93(2), 712 CrossRef.
- Ministerio de Ganadería Agricultura y Pesca, MGAP, Manual del Programa Nacional de Residuos Biológicos, 2009, https://www.mgap.gub.uy/portal/page.aspx?2,dgsg,dgsg-programa-nacional-decontrol-de-residuos-de-medicamentos-veterinarios-y-contaminantes-en-alimentos-de-origen-animal,O,es,0/ Search PubMed.
- S. Niell, V. Cesio, J. Hepperle, D. Doerk, L. Kirsch, D. Kolberg, E. Scherbaum, M. Anastassiades and H. Heinzen, J. Agric. Food Chem., 2014, 62(17), 3675 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ay00516c |
| This journal is © The Royal Society of Chemistry 2024 |

