Open Access Article
Open Access ArticleIsothermal recombinase polymerase amplification and silver nanoparticle assay: a sustainable approach for ultrasensitive detection of Klebsiella pneumoniae†
Naresh
Patnaik‡
,
Nidhi
Orekonday‡
and
Ruchi Jain
Dey
 *
*
Department of Biological Sciences, BITS Pilani Hyderabad Campus, Telangana State 500078, India. E-mail: ruchij80@hyderabad.bits-pilani.ac.in
First published on 20th September 2024
Abstract
Our study addresses the urgent need for effective detection of Klebsiella pneumoniae, a recognized threat by the World Health Organization (WHO). Current challenges in managing K. pneumoniae infections include the lack of rapid and affordable detection tools, particularly in resource-limited point-of-care (POC) settings. To tackle this, we developed an innovative molecular detection pipeline combining three POC-compatible methods. Firstly, we employed Insta DNA™ card-based sample collection and DNA extraction for simplicity and ease of use. Next, we utilized recombinase polymerase amplification (RPA) targeting the Klebsiella hemolysin gene, khe, specific to the K. pneumoniae species complex (KpSC). Finally, we integrated a silver nanoparticle (AgNP) aggregation assay for visual detection, offering a rapid, sensitive, and specific method capable of detecting as few as ∼3 bacteria of K. pneumoniae within ∼45 minutes. This approach eliminates the need for complex equipment, making it highly suitable for field and resource-limited POC applications. Moreover, our method introduces an environmentally significant detection strategy. The method developed minimizes chemical reagent usage and reduces the carbon footprint associated with sample transportation. Furthermore, our method reduces waste compared to the traditional detection techniques, offering a safer alternative to ethidium bromide or other DNA dyes which are often genotoxic and mutagenic in nature. Silver nanoparticles, being environmentally safer, can also be recycled from the waste, contributing to sustainability in nanoparticle production and disposal. Overall, our technique presents a promising solution for detecting K. pneumoniae in various settings, including environmental, water, and food samples, as well as industrial or hospital effluents. By aligning with global efforts to improve public health and environmental sustainability, our approach holds significant potential for enhancing disease management and reducing environmental impact.
Introduction
The escalation of antimicrobial resistance (AMR) constitutes a significant global health threat, particularly within the notorious ESKAPE group of pathogens. This acronym encompasses six-opportunistic bacterial pathogens with high capabilities of acquiring antibiotic resistance: Enterococcus species (spp.), Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.1 Within the ESKAPE group of pathogens, the World Health Organization (WHO) recognizes K. pneumoniae as a significant pathogen of concern, primarily due to its association with increasing AMR.1–3K. pneumoniae species complex (KpSC) comprises seven phylogroups termed as phylogroups Kp1 to Kp7 (ref. 4 and 5) (Table S1†). KpSC is a major contributor to nosocomial infections, especially in vulnerable or immune-compromised individuals leading to diverse pathologies such as necrotizing pneumonia, urinary tract infections (UTIs), wound or surgical site infections, bloodstream infections, septicaemia, meningitis, and pyogenic liver abscesses.6–9Klebsiella infections are also important in the context of maternal and child health causing complications such as chorioamnionitis, increased likelihood of pre-term labour/birth,10 neonatal infections leading to increased infant mortality.11,12 Hence, preventive measures and early detection of Klebsiella is paramount for timely intervention and effective management of these infections in maternal and child healthcare settings.Early intervention facilitates prompt initiation of appropriate medical measures, reducing the risk of severe complications and preventing transmission. Conventional methods for detecting K. pneumoniae, such as culture-based and biochemical techniques, are time-consuming and provide limited information on abundance and strain diversity within samples, with uncertain sensitivity.4,5,13 The recent introduction of whole metagenomics sequencing in clinical diagnostics has significantly enhanced diagnostic reliability13,14 compared to traditional culture-based methods, offering comprehensive insights into antibiotic resistance profiles, virulence features, and evolutionary analysis of bacterial strains.13,15 Nevertheless, the exorbitant cost, time involvement and requirement of highly skilled individuals to perform and analyse the results currently restricts its routine application in diverse healthcare settings. In contrast, PCR-based assays, particularly real-time quantitative PCR(RT-qPCR),4 have shown 100% sensitivity and specificity, with a limit of detection of ∼15 genomes (Table 1). However, the cost of detection is extremely high due to requirement of quantitative PCR (qPCR) equipment. Subsequent developments introduced isothermal amplification methods, including loop-mediated isothermal amplification (LAMP),19,20 recombinase polymerase amplification (RPA)22–25 and multiple cross displacement amplification (MCDA),21 for rapid K. pneumoniae detection without the requirement of expensive thermal cyclers or qPCR. LAMP alone or in combination with clustered regularly interspaced short palindromic repeats (CRISPR) and MCDA combined with lateral flow strip (LFS) has demonstrated comparable sensitivity (1–20 bacteria) with respect to qPCR. These methods achieve detection within 60–90 minutes under isothermal conditions (∼60–65 °C)19–21 (Table 1). In comparison, RPA provides further advantage of rapid (15–30 min) isothermal amplification at lower temperatures (∼37–42 °C) permitting quicker detection.26 RPA when combined with LFS is capable of detecting as low as 100–1000 bacteria, highlighting an improved sensitivity of detection post RPA-based DNA amplification.22,23 RPA has been applied in recent past for developing non-instrumented point-of-care (POC) detection for several human,27 animal28 and plant pathogens.29 For instance, RPA-based detection was developed for HIV-1 and plant viruses harnessing the body heat.27,29
| Sl. no. | Method of amplification | Method of detection | Sensitivity | Time complexity of detection | References |
|---|---|---|---|---|---|
| 1 | PCR | AGE | 1.2 × 102 bacteria | ∼4–5 hours | 16 |
| 2 | Real-time PCR | Fluorescence | 15 bacteria | ∼2 hours | 4 |
| 3 | Real-time PCR | Fluorescence resonance energy transfer (FRET) | 10 bacteria | ∼2 hours | 17 |
| 4 | PCR | Gold nanoparticles | 16 × 101 bacteria (1 pg gDNA) | ∼2 hours | 18 |
| 5 | LAMP | AGE | 1 bacterium | ∼25 min (LAMP) + 1 hour AGE | 19 |
| 6 | LAMP | CRISPR-based detection | 16 × 101 bacteria (1 pg gDNA) | ∼1 hour | 20 |
| 7 | MCDA | Gold nanoparticle-LFS | 16 bacteria (100 fg gDNA) | ∼30 min (estimated, not reported) | 21 |
| 8 | RPA | LFS | 103 bacteria | ∼15 min | 22 |
| 9 | RPA | LFS | 16 × 103 bacteria (0.1 ng μl−1) | ∼33 min | 23 |
| 10 | Real-time RPA | Fluorescence probes | 100–1000 bacteria | ∼12 min | 24 |
| 11 | RPA | AGE | 102–103 bacteria | ∼1 hour | 25 |
Despite the advancements in a wide variety of methods of isothermal amplification under ambient settings, visualization of the amplified products still remains a major constraint in POC setting. A recent study highlights utility of tailor-made plasmonic aptamer-gold nanoparticle (AuNP) assay for detection of Klebsiella without the need of amplification, however, the sensitivity of this methods is ∼3400 bacteria.18 However, the high cost associated with fluorescent primer probes, aptamers, additional reagents like CRISPR, LFS and AuNP presents accessibility challenges for low-income and resource-poor settings, highlighting the need for cost-effective visual detection tools. Another hurdle in POC settings is safe and simple methods of sample collection, storage, transport and genomic DNA (gDNA) extraction for molecular detection. InstaDNA card have recently been developed to overcome these challenges. This card consists of a special filter paper impregnated with a proprietary formula containing reagents that promote cell lysis and protein denaturation with subsequent release of nucleic acids that are entrapped within the matrix of the card and stabilized at room temperature, allowing long-term storage and ensuring DNA integrity for downstream molecular detection.30–33 Many advantages have been described for InstaDNA card, including low-cost, simple extraction protocols, easy transportation, minimal storage space and no special infrastructure being required.33 However, there are no previous reports for application of InstaDNA cards for K. pneumoniae detection.
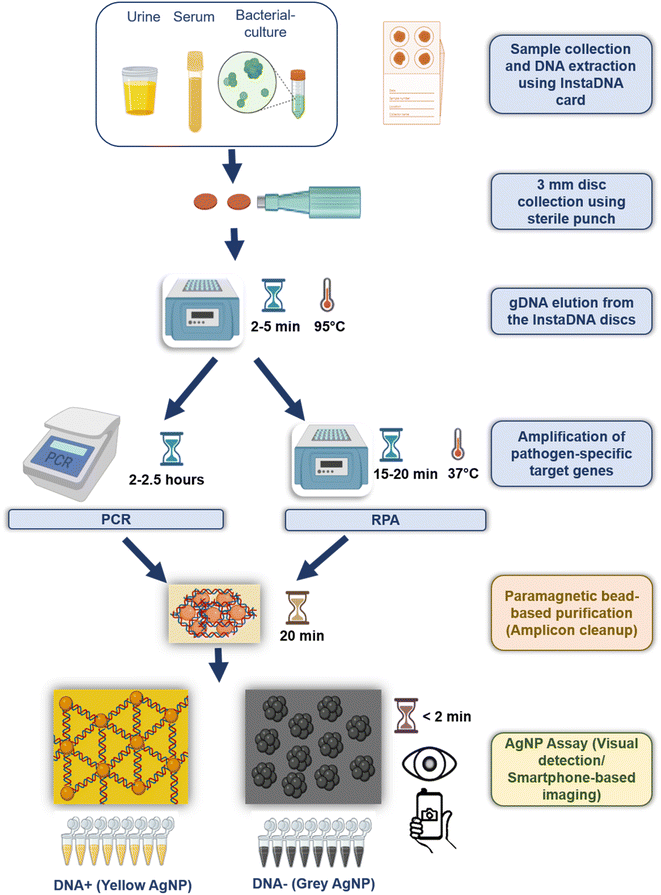
This study offers POC friendly molecular diagnostic pipeline combining three methods, (a) InstaDNA card, for easy sample collection and DNA extraction (b) RPA-based isothermal amplification in addition to PCR (c) application of silver nanoparticle (AgNP) aggregation assay developed in our laboratory34 for rapid, ultrasensitive visual molecular detection of K. pneumoniae. This unique combination is particularly suitable for K. pneumoniae detection in resource-poor settings without the need of costly thermal cycler and equipment like centrifuge or qPCRs and is compatible with clinical samples like serum and urine. The integration of these methods is anticipated to address the clinical needs to mitigate the impact of Klebsiella infections.
Results
Specific molecular detection of K. pneumoniae
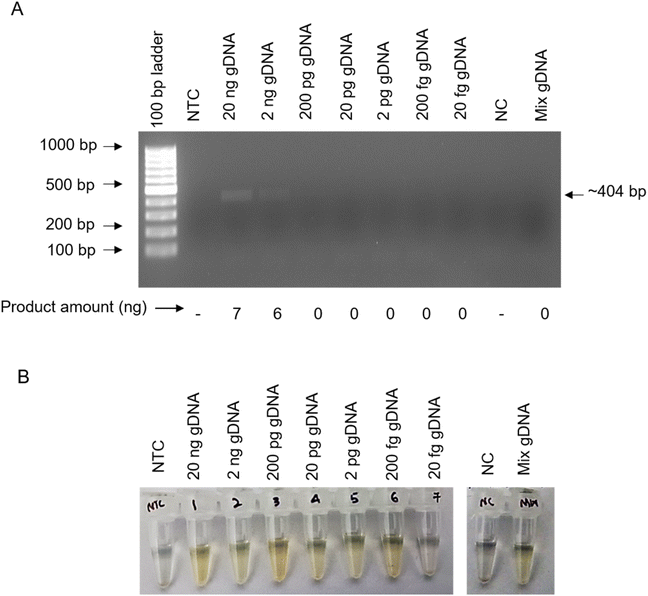
In this study, we designed primers specific to two virulence gene, namely, Klebsiella hemolysin gene (khe)35,36 and uridine diphospho (UDP)-D-galacturonate 4-epimerase gene (uge)37 (Table 2). These two loci are unique to K. pneumoniae (Fig. 1A and B). We then elucidated the sensitivity of molecular detection of the bacteria using PCR followed by agarose gel electrophoresis (AGE) (called PCR-AGE hereafter) with all the primer sets. PCR-AGE performed using the highly specific primer sets against khe and uge genes, showed detection sensitivity of 33 × 101 (Fig. 2A) and 33 × 103 (ESI Fig. S1†) K. pneumoniae bacteria, respectively. Further, using in silico BLAST analysis we show that the primers sets designed against khe and uge can detect all the phylogroups of KpSC (Kp1 to Kp7) (ESI Table S1†). Given the low sensitivity of uge based PCR compared to khe (Fig. 2A and ESI Fig. S1†), hereafter we exclusively utilized khe gene target for comparative assessment of various POC molecular detection strategies for K. pneumoniae.| Si no. | Target gene (amplicon size) | Primer sequence (5′ → 3′) | PCR conditionsb |
|---|---|---|---|
| a All the primers were designed using PrimerQuest tool (Integrated DNA Technologies) in this study. b Annealing temperatures obtained using gradient PCR. | |||
| 1 | khe gene (404 bp) | (F) CGGGATTGAGCGGGTAATAA | 95 °C, 30 s; 64 °C, 30 s; 72 °C, 30 s |
| (R) GATGAAACGACCTGATTGCATTC | |||
| 2 | uge gene (222 bp) | (F) CATATTCTTCTGCGCTTCCATTC | 95 °C, 30 s; 64 °C, 30 s; 72 °C, 30 s |
| (R) AAGCGCGACTTCACCTATATC | |||
Since AGE requires equipment for UV-based visualisation, we next assessed the utility of an equipment-free and naked eye visual detection method, namely, AgNP assay following PCR. In addition, we also compared the sensitivity of detection of this method with respect to PCR-AGE. PCR-AgNP assay, show highly sensitive and specific detection of as low as ∼3 bacteria (Fig. 2B).
InstaDNA card-based sample collection and DNA extraction followed by PCR based amplification and visual detection of K. pneumoniae by AgNP assay
Due to several advantages of InstaDNA card pertaining to their ease of sample collection, storage, logistics, cost and time effectiveness, efficiency of extraction without need of any high-end equipment (Table 3), compatibility with various amplification techniques and AgNP visual detection assay, we found InstaDNA cards to be most suited for POC application.30–33 Hence, we next assessed the utility of InstaDNA card-based DNA isolation for PCR based molecular detection of K. pneumoniae as shown in Fig. 3.| Parameters | Advantages of InstaDNA card |
|---|---|
| Sample collection | Sample collection in the card itself |
| Sample pre-processing | No pre-processing is necessary |
| Sample storage and transportation | InstaDNA cards allow room temperature storage and transportation |
| Equipment necessary for DNA isolation | Heat block for DNA isolation |
| Compatibility with PCR and RPA amplification techniques | Yes. Discs cut from the InstaDNA card containing sample can be directly used for PCR, without the need for DNA elution. DNA elution step is necessary before RPA |
| Compatibility with AgNP assay | Yes |
| Cost | Low cost. Sample collection, storage, transportation is done on the InstaDNA cards at room temperature. No requirement for sample pre-processing and equipment. Recurring cost involves only InstaDNA cards (<1$ per sample) and a simple heat block or water bath |
| Time for DNA extraction | ∼15–30 minutes. ∼2 minutes for DNA elution. Samples collected on the InstaDNA cards are usually air dried, which takes ∼15–30 minutes |
| DNA extraction & amplification efficiency | High |
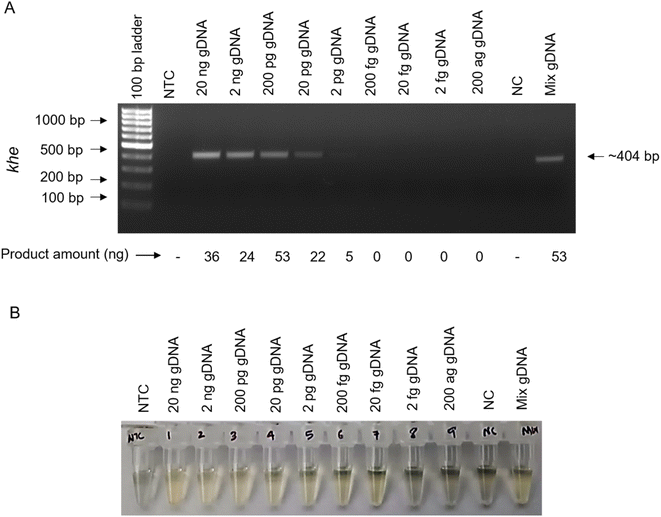
InstaDNA-PCR-AGE combination shows a sensitivity of detection as low as ∼33 × 102 bacterial cells of K. pneumoniae when khe gene is used as the gene targets for PCR amplification (Fig. 4A). The advantage of using PCR, following an InstaDNA card-based DNA extraction is that we do not require any additional elution step, as elution is automatically favoured during the initial denaturation step performed during the PCR cycles. AgNP assay following InstaDNA-PCR using primer targeting khe gene (InstaDNA-PCR-AgNP) assay (Fig. 4B) showed highly sensitive and specific detection as low as ∼3 bacteria similar to that observed in case of PCR-AgNP (Fig. 2B). Altogether, we report highly sensitive and specific detection of K. pneumoniae utilizing AgNP following PCR and InstaDNA-PCR.
POC compatible ultrasensitive molecular detection pipeline for K. pneumoniae using RPA and AgNP assay
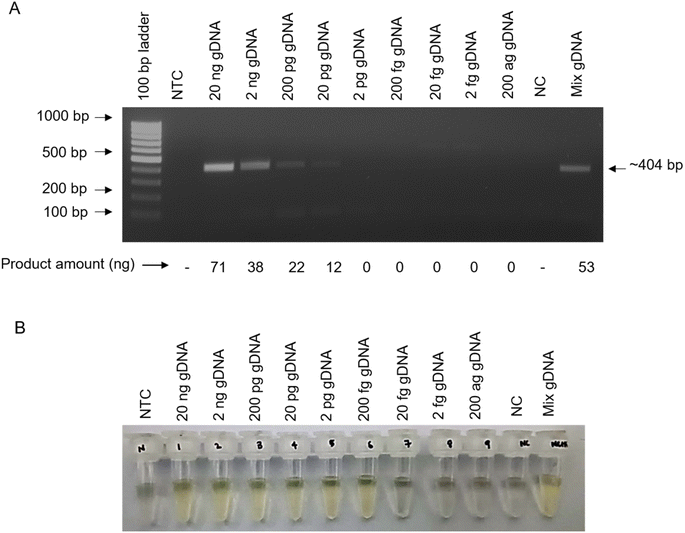
Since PCR requires a high-end thermocycler which is often unavailable in resource poor POC settings, we next assessed the utility of isothermal amplification method using RPA, that performs DNA amplification under ambient temperature (25–42 °C) and can be conducted either on table-top or in an inexpensive heat-block or water-bath (Fig. 3). We performed the RPA amplification of khe gene of K. pneumoniae at 37 °C (Table 2), following standard gDNA isolation. We then compared two methods for visualization of amplified DNA products obtained on RPA, namely, AGE and AgNP assay (called as RPA-AGE and RPA-AgNP assay, respectively, hereafter). The RPA-AGE utilizing khe gene shows a detection sensitivity of ∼33 × 101 bacteria (Fig. 5A).We next assessed the sensitivity of DNA detection using RPA-AgNP assay. AgNP dramatically improves visual detection of DNA obtained on amplification of khe gene with sensitivity as low as ∼3 bacteria when compared to AGE (∼33 × 101 bacteria) (Fig. 5A and B). AgNP assay thus, is suitable for molecular detection post-RPA-based DNA amplification of the target genes and offers a useful alternative to AGE-based detection following RPA-based isothermal amplification techniques.
We then validated the compatibility of the InstaDNA-PCR-AGE & AgNP and InstaDNA-RPA-AGE & AgNP detection workflows in clinical settings. To simulate the protein-rich environment of clinical samples, we spiked serum from a healthy subject with varying amounts of K. pneumoniae gDNA on 3 mm InstaDNA card discs.
These discs were subsequently used for downstream PCR and RPA-based DNA amplification, followed by visualization using AGE (Fig. 6A and 7A) and AgNP (Fig. 6B and 7B), respectively. Additionally, all four assays were also validated using urine samples (ESI Fig. S2 and S3†). The results support the clinical applicability of InstaDNA-PCR-AgNP and InstaDNA-RPA-AgNP.
Discussion
Our study addresses the critical need for rapid, affordable, and effective detection of Klebsiella pneumoniae, a pathogen recognized as a significant threat by the WHO. Current diagnostic challenges include the lack of tools that are both cost-effective and suitable for POC settings, particularly in resource-limited environments. To overcome these challenges, we developed an innovative detection pipeline that integrates three POC-compatible methods: InstaDNA™ card-based sample collection and DNA extraction, RPA targeting the khe gene specific to KpSC, and an AgNP aggregation assay for visual detection.34Our combined approach offers several key advantages. The InstaDNA™ card allows for simple and safe sample collection, storage, and transport, providing instant DNA extraction without the need for sophisticated equipment or additional reagents. The RPA method, targeting the khe gene, provides rapid amplification, while the AgNP assay facilitates visual detection, capable of identifying as few as 3 bacteria within approximately 45 minutes. This system eliminates the need for expensive thermal cycling equipment, centrifuges, and gel documentation facilities, making it highly suitable for field and POC applications. Furthermore, the non-labelled primer sets we developed are compatible with both PCR and RPA, offering flexibility and cost saving. RPA and InstaDNA card further offer multiple benefits enlisted in Tables 1 and 3.4,16–25
Importantly, our method aligns with environmental sustainability goals. The reduced use of hazardous dyes like ethidium bromide, along with the potential for recycling magnetic beads and silver nanoparticles, contributes to a lower environmental impact by eliminating the need for harmful chemicals. This approach not only supports global public health efforts but also promotes sustainable diagnostic practices.
AgNP assays present significant advancements in diagnostic capabilities, especially in resource-limited settings. Citrate-stabilized AgNPs are particularly advantageous due to their enhanced stability against photo-oxidation, which ensures a consistent and reliable signal over time. This stability, combined with their cost-effectiveness compared to gold nanoparticles, makes AgNPs a practical choice for DNA detection assays.34 The unique properties of citrate-stabilized AgNPs, including their colorimetric response, offer a simple and visual means of detection without requiring complex instruments, making the method accessible even in POC settings. Furthermore, when coupled with isothermal amplification techniques like LAMP and RPA, the label-free AgNP aggregation approach could revolutionize POC diagnostics. These techniques could drastically reduce the cost, time, and complexities associated with diagnosis of infectious diseases, potentially improving the management of the disease in under-resourced areas. While visual detection is promising, it is important to note that it may not provide quantitative results, which are crucial for monitoring disease progression and treatment response. The integration of AgNP-based assays with isothermal amplification methods promises to enhance diagnostic capabilities in challenging settings such as hospitals, war zones, sewage treatment plants, food, water and pharmaceutical industries, remote or resource-poor areas, environmental monitoring sites, and even in-field testing during disease outbreaks or bioterrorism threats.
While our in silico analysis identified primers against khe and uge are suitable for detecting all seven phylogroups of K. pneumoniae, our experiments were limited to a laboratory strain (MTCC 109) and have not yet been validated on other phylogroups and in clinical settings. To address this, we conducted experiments using clinical mimic samples to simulate clinical conditions. These experiments demonstrate the effectiveness of the RPA and AgNP assays in serum and urine, further validating the method's potential for real-world clinical applications. Table 4 and ESI Table S3,† highlight the detection sensitivity of K. pneumoniae in clinical mimics. Future research in this direction will make this tool more robust and affordable for widespread applications.
| Si. no | Input amount of DNA | Input amount of bacteria | InstaDNA-PCR-AGE | InstaDNA-PCR-AgNP | InstaDNA-RPA-AGE | InstaDNA-RPA-AgNP |
|---|---|---|---|---|---|---|
| 1 | 20 ng | ∼33 × 105 | + | + | + | + |
| 2 | 2 ng | ∼33 × 104 | + | + | + | + |
| 3 | 200 pg | ∼33 × 103 | + | + | − | + |
| 4 | 20 pg | ∼33 × 102 | + | + | − | + |
| 5 | 2 pg | ∼33 × 101 | − | + | − | + |
| 6 | 200 fg | ∼33 | − | + | − | + |
| 7 | 20 fg | ∼3 | − | − | − | − |
| 8 | 2 fg | ∼3 × 10−1 | − | − | − | − |
| 9 | 200 ag | ∼3 × 10−2 | − | − | − | − |
Conclusion
This study presents an ultrasensitive, rapid, and cost-effective method for specific molecular detection of K. pneumoniae by combining Insta-DNA card-based DNA extraction, RPA isothermal amplification, and AgNP visual detection. We developed unique primers targeting the virulence gene khe that are compatible with both PCR and RPA assays. The method eliminates the need for expensive equipment, making it highly suitable for point-of-care (POC) settings with a sensitivity as low as ∼3 bacteria per sample and a total cost under $5. Experiments using clinical mimic samples demonstrate the effectiveness of the PCR and RPA along with AgNP visual detection assays in serum and urine, further validating the method's potential for real-world clinical applications.Materials and methods
Ethics statement
The study was conducted in accordance with the guidelines and approval of the Institutional Biosafety Committee (BITS/IBSC/2019-1/7 and BITS/IBSC/2019-1/8), and on ethical approval of the Institutional Human Ethics Committee (IHEC), the Institutional Review Board (IRB) of Birla Institute of Technology and Science (BITS), Hyderabad (IHEC approval number: BITS-HYD/IHEC/2022/01) with appropriate biosafety level 2 measures.Reagents and bacterial strains
All reagents used in this study were obtained from Himedia Laboratories Pvt. Ltd, unless specified otherwise. The oligonucleotide primers listed in Table 2 were purchased from Sigma-Aldrich. Paramagnetic beads were sourced from MagGenome, and AgNP (20 nm size, stabilized in citrate buffer, 20 mg ml−1) were also obtained from Sigma-Aldrich. Taq polymerase and related reagents were acquired from Takara Clontech. The Twist Amp Liquid Basic RPA Kit came from TwistDX.Cultures of E. faecalis (ATCC 29212), S. aureus (MTCC 96), K. pneumoniae (MTCC 109), A. baumanii (MTCC 12889), P. aeruginosa (MTCC 424), Enterobacter spp. (MTCC 7104), and Neisseria gonorrhoeae (ATCC 19424) were sourced from the American Type Culture Collection (ATCC, distributed by Himedia Laboratories Pvt. Ltd) or the Microbial Type Culture Collection and Gene Bank (MTCC) at the Institute of Microbial Technology (IMTECH), Chandigarh, India. The WHO BCG Danish 1331 vaccine sub-strain was obtained from the National Institute for Biological Standards and Control (NIBSC), UK.
Sample collection and DNA extraction
Genomic DNA was extracted from pure bacterial cultures using a standard kit-based method, specifically the DNEasy Blood and Tissue Kit (Qiagen) and NucleoSpin Tissue DNA, RNA and protein purification kit (Macherey Nagel), with minor adjustments to the manufacturer's protocol. For cost-effective and rapid gDNA isolation, we employed InstaDNA card method.30–32 The resulting gDNA was quantified using NanoDrop Microvolume Spectrophotometers (Thermo Fisher Scientific) and utilized for amplification reactions with species-specific primers.In the InstaDNA card-based DNA isolation method, a known bacterial culture of colony forming units (CFU), e.g., 33 × 104 (in 2 μL volume), was blotted onto a 3 mm InstaDNA card disc, allowed to dry for 15–30 minutes at room temperature, washed in sterile water by just dipping the disc for 15 seconds, and transferred to the PCR reaction mix for amplification. Alternatively, the disc can be transferred to elution buffer (1× Tris–EDTA) for DNA elution by heating at 95 °C for 2–5 minutes. The eluent can further be used for RPA-based DNA amplification. Similarly, 10-fold serially diluted bacterial samples were blotted onto separate discs and processed accordingly in order to perform sensitivity assays using InstaDNA card followed by PCR and RPA, and various detection methods (gDNA dilutions and their corresponding bacterial number are provided in Table S2†).
In order to mimic the protein-rich environment of clinical samples, waste or leftover serum and urine from healthy subject was collected on InstaDNA card, air dried. We then punched out 3 mm discs using sterile punching machine. Each disc was then spiked with 20 ng gDNA of K. pneumoniae. The disc was then transferred to elution buffer (1× Tris–EDTA) for DNA elution by heating at 95 °C for 5 minutes. The eluent was estimated using NanoDrop Microvolume Spectrophotometers (Thermo Fischer Scientific), serially diluted and used for PCR and RPA-based DNA amplification.
PCR-AGE-based molecular detection
Specific oligonucleotide primers targeting K. pneumoniae were custom-designed and are detailed in Table 2. However, considerations during primer designing for RPA should include a higher GC content (≥50%), longer primer length (20–35 bp), and maintaining the amplicon size between 200-500 bp. Primers specific to KpSC strains were designed against khe gene (∼404 bp) and uge gene (∼222 bp). Primer specificity for all the gene targets was confirmed in silico through NCBI-BLAST analysis (Table S1†). Based on in silico analysis, primers against khe and uge are suitable for molecular detection of all the seven phylogroups of K. pneumoniae, however, current study has utilized the laboratory strain of K. pneumoniae (MTCC 109). PCR amplification using these primer sets was carried out in a Veriti Thermal Cycler from Applied Biosystems, with Taq Polymerase sourced from Takara Clontech, following the manufacturer's instructions with minor adjustments. Each 50 μl PCR reaction was prepared with a 1 μM primer mix in the reaction mixture, and thermal cycling included an initial denaturation step at 95 °C for 10 minutes, followed by 35 cycles of PCR conditions outlined in Table 2. The process concluded with a final extension at 72 °C for 10 minutes. The verification of PCR products was performed through AGE and DNA was visualized by ethidium bromide dye-based staining. The DNA products on the gel were quantified using ImageJ software (Version 1.8.0).In order to validate the in vitro primer specificity for K. pneumoniae, a mix of gDNA samples obtained from diverse organisms which could possibly cause respiratory infections or pneumonia, were utilized as negative controls (NC) in PCR, such as Mycobacterium tuberculosis (M. tb), Mycobacterium bovis (M. bovis), M. bovis BCG, N. gonorrhoeae, Chlamydia trachomatis, Ureaplasma urealyticum, E. faecalis, S. aureus, A. baumanii, P. aeruginosa, Enterobacter spp. and human DNA. NC is prepared by mixing 5 ng gDNA obtained from each of these organisms, except K. pneumoniae. In order to determine interference in amplification due to presence of other pathogens or human host DNA (as anticipated in a clinical setting), we also included a control named, “Mix gDNA”, which contains a mix of NC and genomic DNA of K. pneumoniae. This mix is prepared by mixing 5 ng gDNA of K. pneumoniae to the above-mentioned NC mix. For amplification reactions, 2 μl of NTC and Mix gDNA is used in case of PCR and RPA. For determination of detection sensitivity of PCR-AGE, serial ten-fold dilutions of gDNA templates of K. pneumoniae ranging from 20 nanograms, ng (3.3 × 106 bacteria) to 200 attogram, ag (3.3 × 10−2 bacteria) were employed in PCR reactions (Table S2†).
RPA-AGE-based molecular detection of K. pneumoniae
Isothermal nucleic acid amplification was performed for the pathogens using Twist Amp Liquid Basic RPA Kit (TwistDX, TALQBAS01 kit) as recommended by the manufacturer with minor modifications. Briefly, all the reagents provided in the kit are mixed with defined quantity of DNA, reaction is initiated by addition of magnesium acetate, followed by incubation at 37 °C for 15–20 minutes. The RPA products are immediately cleaned up using XpressPure PCR Cleanup kit (MagGenome, MG20Pcr-50) as recommended by the manufacturer with slight modifications. Briefly, we used 0.8× volume/volume (v/v) of magnetic beads while purifying the RPA products and 1× v/v in case of PCR products. These modifications allowed elimination of smaller size primers and primer dimers from the amplification mix. Altogether, it offers simple, user-friendly, cost and time-efficient method for DNA purification without requiring any high-end electrical equipment. Similar to PCR-AGE assay, we determined the sensitivity or limit of detection of the RPA-AGE using serial ten-fold dilutions of gDNA templates ranging from (20 ng (3.3 × 106 bacteria)) to 200 ag (0.03 bacteria) (Table S2†). DNA was visualized by ethidium bromide dye-based staining of agarose gel.Visual detection of amplified products using citrate-stabilized AgNPs
AgNP-based visual detection assay was performed using PCR amplified products as described by us previously,34 wherein presence of DNA prevents the aggregation of AgNPs upon addition of aggregating agent, sodium chloride (NaCl) causing sliver dispersion to maintain yellow colour. However, in absence of DNA, addition of NaCl leads to AgNP aggregation causing grey coloration of the dispersion.Briefly, 20 μl of RPA or PCR reaction products were made up to a volume of 50 μl in case of PCR and a volume of 100 μl in case of RPA using sterile water. Amplified products were then cleaned-up using MagGenome PCR clean up kit as per the manufacturer's instructions with slight modifications as mentioned earlier. Finally, DNA was eluted in 20 μl of elution buffer (10 mM Tris, 1 mM EDTA, pH 8.0) followed by incubation with 50 μl of the citrate-stabilized AgNP dispersion at room temperature for ∼1 minute. Following this, 5 μl of aggregating reagent (5 M NaCl) was added to the assay tubes. The onset of AgNP aggregation is promptly revealed (in <1 min) by a grey colour dispersion in tubes lacking amplicons (negative controls with DNA from non-target organisms, NC or no-template controls, NTC). Conversely, in tubes with amplified DNA, the addition of NaCl prevents AgNP aggregation, resulting in the retention of a yellow colour. The entire process of purification and visual detection is completed within a time frame of 20–25 minutes. The sensitivity of detection was assessed by executing AgNP aggregation-based assay using amplicons derived from RPA conducted with varying amounts of gDNA isolated from K. pneumoniae.
Abbreviations
| M. tb | Mycobacterium tuberculosis |
| M. bovis | Mycobacterium bovis |
| POC | point-of-care |
| ml | millilitre |
| μl | microlitre |
| ng | nanogram |
| pg | picogram |
| fg | femtogram |
| ag | attogram |
| M | molar |
| mM | millimolar |
| μM | micromolar |
| gDNA | genomic DNA |
| khe | Klebsiella hemolysin gene |
| uge | uridine diphospho (UDP)-D-galacturonate 4-epimerase gene |
| KpSC | K. pneumoniae species complex |
| PCR | polymerase chain reaction |
| DNA | deoxyribonucleic acid |
| qPCR | Quantitative PCR |
| RT-PCR | real time PCR |
| NTC | no-template-control |
| AGE | agarose gel electrophoresis |
| UV | ultraviolet |
| NaAc | sodium acetate |
| PEG | polyethylene glycol |
| AgNP | silver nanoparticles |
| Tris | tris(hydroxymethyl)aminomethane |
| EDTA | ethylene diamine tetra-acetic acid |
| NaCl | sodium chloride |
| LAMP | loop-mediated isothermal amplification |
| RPA | recombinase polymerase amplification |
| MCDA | multiple cross displacement amplification |
| BCG | Bacille Calmette-Guerin |
| WHO | World Health Organisation |
| CFU | colony forming unit |
| UTIs | urinary tract infections |
| LFS | lateral flow strip |
| HIV-1 | human immunodeficiency virus-1 |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| v/v | volume/volume |
Data availability
All data that supports the findings of this study are available from corresponding author upon reasonable request.Author contributions
NP, NO and RJD have conceptualized the study. NP and NO have equally contributed to the work. NP and NO designed methodologies, curated data, performed analysis, designed figures for visualization of results. RJD and NP acquired the funding. NP, NO and RJD have written the original draft of the manuscript. All the authors reviewed and edited the manuscript.Conflicts of interest
Authors declare no conflict of interest.Acknowledgements
RJD is thankful to Birla Institute of Science and Technology (BITS) Pilani, Hyderabad campus, India for their funding support through intramural funding under Research Initiation Grant (RIG) and Centre for Human Disease Research (CHDR). RJD is thankful to Defence Research and Development Organization (DRDO), India for supporting through research grant (LSRB/81/48222/LSRB-367/BTB/2020). RJD is thankful to the overall infrastructure support by Department of Biological Sciences, BITS Pilani Hyderabad. NP is thankful to Indian Council of Medical Research (ICMR), Govt. of India for providing Senior Research Fellowship (RBMH/FW/2019/13). NO is thankful to LSRB, DRDO and BITS Pilani Hyderabad for fellowship. We acknowledge Dr Raghunand R Tirumalai, Centre for Cellular and Molecular Biology (CCMB), Hyderabad, India for providing gDNA for M. tuberculosis H37Rv. Dr Bappaditya Dey, National Institute of Animal Biotechnology (NIAB), Hyderabad, India is acknowledged for M. bovis gDNA. We are grateful to Dr Benu Dhawan, Department of Microbiology, All India Institute of Medical Sciences (AIIMS, New Delhi, India) for giving gDNA from Ureaplasma urealyticum. We thank Dr Karthika Rajeeve, Rajiv Gandhi Centre for Biotechnology, Trivandrum, India for providing Chlamydia trachomatis (L2 serovar) gDNA.References
- V. Ballén, Y. Gabasa, C. Ratia, R. Ortega, M. Tejero and S. Soto, Front. Cell. Infect. Microbiol., 2021, 11, 738223 CrossRef PubMed.
- M. K. Paczosa and J. Mecsas, Microbiol. Mol. Biol. Rev., 2016, 80, 629–661 CrossRef CAS PubMed.
- K. H. P. Riwu, M. H. Effendi, F. A. Rantam, A. R. Khairullah and A. Widodo, Vet. World, 2022, 15, 2172 CAS.
- E. Barbier, C. Rodrigues, G. Depret, V. Passet, L. Gal, P. Piveteau and S. Brisse, Appl. Environ. Microbiol., 2020, 86, e02711–e02719 CAS.
- K. L. Wyres, M. M. Lam and K. E. Holt, Nat. Rev. Microbiol., 2020, 18, 344–359 CrossRef CAS PubMed.
- R. Podschun and U. Ullmann, Clin. Microbiol. Rev., 1998, 11, 589–603 CrossRef CAS PubMed.
- C. Y. Effah, T. Sun, S. Liu and Y. Wu, Ann. Clin. Microbiol. Antimicrob., 2020, 19, 1–9 CrossRef CAS PubMed.
- S. Navon-Venezia, K. Kondratyeva and A. Carattoli, FEMS Microbiol. Rev., 2017, 41, 252–275 CrossRef CAS PubMed.
- C. N. Murphy and S. Clegg, Future Microbiol., 2012, 7, 991–1002 CrossRef CAS PubMed.
- M. P. Bonasoni, A. Palicelli, G. Dalla Dea, G. Comitini, P. Nardini, L. Vizzini, G. Russello, M. Bardaro and E. Carretto, Microorganisms, 2021, 9, 96 CrossRef PubMed.
- J. P. Seliga-Siwecka and M. K. Kornacka, Early Hum. Dev., 2013, 89, 271–275 CrossRef PubMed.
- K. Puri, D. H. Taft, N. Ambalavanan, K. R. Schibler, A. L. Morrow and S. G. Kallapur, PLoS One, 2016, 11, e0162734 CrossRef PubMed.
- K. Lindstedt, D. Buczek, T. Pedersen, E. Hjerde, N. Raffelsberger, Y. Suzuki, S. Brisse, K. Holt, Ø. Samuelsen and A. Sundsfjord, Gut Microbes, 2022, 14, 2118500 CrossRef PubMed.
- J. R. Bowers, D. Lemmer, J. W. Sahl, T. Pearson, E. M. Driebe, B. Wojack, M. A. Saubolle, D. M. Engelthaler and P. Keim, J. Clin. Microbiol., 2016, 54, 2582–2596 CrossRef CAS PubMed.
- K. D. Chavda, L. Chen, D. E. Fouts, G. Sutton, L. Brinkac, S. G. Jenkins, R. A. Bonomo, M. D. Adams and B. N. Kreiswirth, mBio, 2016, 7(6), e02093 CrossRef CAS PubMed.
- Y. Liu, C. Liu, W. Zheng, X. Zhang, J. Yu, Q. Gao, Y. Hou and X. Huang, Int. J. Food Microbiol., 2008, 125, 230–235 CrossRef CAS PubMed.
- P. Kurupati, C. Chow, G. Kumarasinghe and C. L. Poh, J. Clin. Microbiol., 2004, 42, 1337–1340 CrossRef CAS PubMed.
- S. Ahmadi, H. Kamaladini, F. Haddadi and M. R. Sharifmoghadam, J. Fluoresc., 2018, 28, 987–998 CrossRef CAS PubMed.
- R. Nakano, A. Nakano, Y. Ishii, T. Ubagai, T. Kikuchi-Ueda, H. Kikuchi, S. Tansho-Nagakawa, G. Kamoshida, X. Mu and Y. Ono, J. Infect. Chemother., 2015, 21, 202–206 CrossRef CAS PubMed.
- X. Qiu, X. Liu, X. Ma, R. Wang, S. Chen, F. Li and Z. Li, Microbiol. Spectrum, 2022, 10, e01545–e01522 Search PubMed.
- L. Niu, F. Zhao, J. Chen, J. Nong, C. Wang, J. Wang, N. Gao, X. Zhu, L. Wu and S. Hu, PLoS One, 2018, 13, e0204332 CrossRef PubMed.
- P. Hemwaranon, A. Srisrattakarn, A. Lulitanond, P. Tippayawat, R. Tavichakorntrakool, L. Wonglakorn, J. Daduang and A. Chanawong, Antibiotics, 2022, 11, 1499 CrossRef CAS PubMed.
- N. Li, L. Wang, F. Wang, H. Chen, S. Tao, Q. Zhu, L. Liu, W. Liang and F. Ma, Front. Cell. Infect. Microbiol., 2022, 12, 877649 CrossRef CAS PubMed.
- B. Raja, H. J. Goux, A. Marapadaga, S. Rajagopalan, K. Kourentzi and R. C. Willson, J. Appl. Microbiol., 2017, 123, 544–555 CrossRef CAS PubMed.
- S. Lapa, S. Surzhikov, S. Blagodatskikh, V. Shershov and A. Chudinov, Mol. Biol., 2023, 57, 544–549 CrossRef CAS.
- R. K. Daher, G. Stewart, M. Boissinot and M. G. Bergeron, Clin. Chem., 2016, 62, 947–958 CrossRef CAS PubMed.
- Z. A. Crannell, B. Rohrman and R. Richards-Kortum, PLoS One, 2014, 9, e112146 CrossRef PubMed.
- Q. Guo, K. Zhou, C. Chen, Y. Yue, Z. Shang, K. Zhou, Z. Fu, J. Liu, J. Lin and C. Xia, Front. Cell. Infect. Microbiol., 2021, 11, 791997 CrossRef CAS PubMed.
- S. J. Priti, V. Baranwal, R. G. Dietzgen and A. Ghosh, J. Pest Sci., 2021, 94, 219–229 CrossRef PubMed.
- R. Alyethodi, U. Singh, S. Kumar, R. Alex, G. Sengar, T. Raja, R. Deb and B. Prakash, BMC Biotechnol., 2021, 21, 36 CrossRef CAS PubMed.
- C. Ismailova, E. Golkocheva-Markova, T. Tenev and S. Krumova, PROBLEMS of Infectious and Parasitic Diseases, 2019, vol. 47, pp. 16–20 Search PubMed.
- S. Krumova, E. Golkocheva-Markova, S. Angelova, S. Voleva, A. Pavlova, I. Georgieva and P. Genova-Kalou, Am. Sci. Res. J. Eng. Technol. Sci., 2019, 51, 183–191 Search PubMed.
- G. da Cunha Santos, Arch. Pathol. Lab. Med., 2018, 142, 308–312 CrossRef PubMed.
- N. Patnaik and R. J. Dey, ACS Infect. Dis., 2023, 10(2), 426–435 CrossRef PubMed.
- C. Yin-Ching, S. Jer-Horng, L. Ching-Nan and C. Ming-Chung, Microb. Pathog., 2002, 33, 1–6 CrossRef PubMed.
- L. J. Hartman, E. B. Selby, C. A. Whitehouse, S. R. Coyne, J. G. Jaissle, N. A. Twenhafel, R. L. Burke and D. A. Kulesh, J. Mol. Diagn., 2009, 11, 464–471 CrossRef CAS PubMed.
- M. Regué, B. Hita, N. Piqué, L. Izquierdo, S. Merino, S. Fresno, V. J. Benedí and J. M. Tomás, Infect. Immun., 2004, 72, 54–61 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ay00993b |
| ‡ Equal contribution as co-first authors. |
| This journal is © The Royal Society of Chemistry 2024 |