Biogenic carbon dots: a novel mechanistic approach to combat multidrug-resistant critical pathogens on the global priority list†
Ajith Manayil
Parambil
 ,
Abhinav
Prasad
,
Abhinav
Prasad
 ,
Anuj Kumar
Tomar
,
Anuj Kumar
Tomar
 ,
Ilora
Ghosh
,
Ilora
Ghosh
 and
Paulraj
Rajamani
and
Paulraj
Rajamani
 *
*
School of Environmental Sciences, Jawaharlal Nehru University (JNU), New Delhi 110067, India. E-mail: paulrajr@yahoo.com
First published on 11th December 2023
Abstract
This study delves into investigating alternative methodologies for anti-microbial therapy by focusing on the mechanistic assessment of carbon dots (CDs) synthesized from F. benghalensis L. extracts. These biogenic CDs have shown remarkable broad-spectrum anti-bacterial activity even against multi-drug resistant (MDR) bacterial strains, prompting a deeper examination of their potential as novel anti-microbial agents. The study highlights the significant detrimental impact of CDs on bacterial cells through oxidative damage, which disrupts the delicate balance of ROS control within the cells. Notably, even at low doses, the anti-bacterial activity of CDs against MDR strains of P. aeruginosa and E. cloacae is highly effective, demonstrating their promise as potent antimicrobial agents. The research sheds light on the capacity of CDs to generate ROS, leading to membrane lipid peroxidation, loss of membrane potential, and rupture of bacterial cell membranes, resulting in cytoplasmic leakage. SEM and TEM analysis revealed time-dependent cell surface, morphological, and ultrastructural changes such as elongation of the cells, irregular surface protrusion, cell wall and cell membrane disintegration, internalization, and aggregations of CDs. These mechanisms offer a comprehensive explanation of how CDs exert their anti-bacterial effects. We also determined the status of plasma membrane integrity and evaluated live (viable) and dead cells upon CD exposure by flow cytometry. Furthermore, comet assay, biochemical assays, and SDS PAGE identify DNA damage, carbohydrate and protein leakage, and distinct differences in protein expression, adding another layer of understanding to the mechanisms behind CDs' anti-bacterial activity. These findings pave the way for future research on managing ROS levels and developing CDs with enhanced anti-bacterial properties, presenting a breakthrough in anti-microbial therapy.
1. Introduction
One of the biggest threats to world health is bacterial infection. The “dark ages” of bacterial illnesses ended with the development of antibiotics.1,2 The main ways in which antibiotics work are by preventing the creation of bacterial cell walls, rupturing cell membranes, obstructing the production of bacterial proteins, and preventing the transcription and replication of bacterial nucleic acids.3,4 However, repeated use of antibiotics can result in biofilms around bacteria, which impedes the interaction of antibiotic molecules with bacteria.5 Many chemical drugs, however, have unique biological toxicity and can destroy normal tissue cells, especially in children. Furthermore, the emergence of multidrug-resistant (MDR) microorganisms presents a significant issue. Antimicrobial-resistant illnesses kill many people yearly and are expected to have severe repercussions if not managed appropriately.6,7 Critical pathogens on the WHO Global Priority List (2017), including Pseudomonas aeruginosa and Enterobacter cloacae, pose substantial public health threats due to their antibiotic resistance and potential for widespread outbreaks.8Finding innovative, effective, anti-bacterial medications that work differently to traditional small-molecule antibiotics is thus critical to avert or at least postpone the looming catastrophe caused by bacterial resistance.9 Developing anti-bacterial nanoparticles created a novel anti-bacterial mechanism and significantly increased the anti-bacterial impact.10,11 Nanomaterials have been discovered to be less likely to produce bacterial resistance than traditional antibiotics due to their higher membrane permeability, which might augment anti-bacterial activity as efflux pump inhibitors.12,13 The anti-bacterial mechanisms of nanoparticles may be divided into three categories-piercing bacteria's drug-resistant biofilm to perform bactericidal tasks, generating oxidative damage to bacteria based on their nanoenzyme activity, and anti-microbial agent loading to improve antibiotic bioavailability and efficiency.14,15
In recent years, there has been a surge of interest in the use of various nanomaterials, such as anti-microbial peptides (AMPs), noble metal nanoparticles (such as Au NPs and Ag NPs), semiconductor nanoparticles, polymer nanostructures, and carbon-based nanomaterials (CNMs), for the management and eradication of bacterial infections, both in laboratory settings (in vitro) and in vivo.16 AMPs are short-cationic and amphiphilic peptides renowned for their broad-spectrum anti-bacterial action, mainly through breaking microbial membranes, making bacteria resistant.17 However, due to limitations such as in vivo protease hydrolysis, vigorous hemolytic activity against red blood cells, and high production costs, AMPs' therapeutic applicability is restricted. One of the difficulties in using nanomaterials, particularly noble metal complexes, is anticipating their pharmacokinetics and assuring their safety. The Food and Drug Administration (FDA) has approved less than sixty nanomedicines for intravenous delivery, underlining the need for increased understanding and management of noble metal structures in biomedical applications.16
To investigate alternate methodologies, this study focuses on the mechanistic assessment of the anti-pathogenic activity of carbon dots (CDs) synthesized from F. benghalensis L. extracts. The biogenic CDs have demonstrated a broad-spectrum anti-bacterial activity, even against multi-drug resistant (MDR) bacterial strains. So, we further focused on their efficacy and mechanistically examined their potential as a novel anti-microbial therapy alternative by delving into the underlying processes. Structural analyses reveal time-dependent morphological alterations and CD-induced membrane disruptions. Evaluating plasma membrane integrity, live–dead cell assessment, and molecular assays elucidated the mechanisms underpinning CDs' anti-bacterial efficacy. These findings indicate a breakthrough in anti-microbial therapy, illuminating avenues for ROS modulation and enhanced CD-based anti-microbial formulations.
2. Experimental
2.1. Synthesis and characterization of CDs
This study utilized CDs synthesized from Ficus benghalensis and characterized them as mentioned in our previously reported studies.18,19 In short, the CDs were synthesized by microwave-assisted thermal carbonization using the aqueous extract of 5 g F. benghalensis L. After homogenization and centrifugation, supernatants are used for synthesis. In a 2 min 350 °C microwave reaction, 5 ml of supernatant was processed. The resulting CD, after cooling, and 5 ml of Milli Q water were filtered through a 0.1 m syringe filter. The purified CDs underwent various physiochemical analytical characterizations such as UV-Visible absorption spectra, photoluminescence emission spectra, high-resolution transmission electron microscopy (HRTEM) imaging, dynamic light scattering (DLS) analysis, zeta potential analysis and Fourier transformation infrared spectroscopy (FTIR).2.2. Maintenance and treatment of bacterial culture
Bacterial sensitive strains – Staphylococcus epidermidis (ATCC 14990) and Escherichia coli DH5α and MDR strains – Pseudomonas aeruginosa (MTCC 1035), and Enterobacter cloacae (MTCC 509) were maintained and sub-cultured in nutrient broth as well as nutrient agar media. In every experiment, strain-specific MICs of CD were added to cultures of bacteria in the exponential phase of growth with an OD600nm 0.6 for varying treatment durations, ranging from 2 to 40 hours (depending on the requirements of the assays), as specified in the corresponding experimental sections (sections 2.3–2.14). For flow cytometry experiments, cells were only treated for a shorter duration (till 8 h) to avoid an excess number of dead cells and debris in the treated samples that can make the samples unsuitable for analysis.2.3. Well diffusion assay
S. epidermidis, a Gram-positive strain, E. coli DH5α, Gram-negative strains and clinical isolates of MDR strains of E. cloacae and P. aeruginosa were used to evaluate the anti-microbial activity of CD. Culture with OD600nm = 0.6 was spread over the agar plates, and 6 mm wells were made using a cork borer. These wells were loaded with 30 μl of 25 μg ml−1 aqueous solution of CDs, and plates were incubated for 20 hours at 37 °C. Simultaneously, 6-in-one ready antibiotic combination module (Himedia Hexa G-minus 3) containing six commercial antibiotic discs of Amoxicillin (AMC), Gentamycin (GEN), Tetracycline (TE), Ofloxacin (OF), Cotrimoxazole (CXM), and Cefuroxime (COT) were applied on MHA plates of bacterial strains to compare the anti-bacterial activity of CDs with the various antibiotics. After incubation, the zone of inhibition was measured, and data were analyzed following standard Clinical and Laboratory Standard Institute (CLSI) guidelines.2.4. Assay for the determination of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)
The MIC and MBC were determined using the standard CLSI-A8 broth serial-dilution method.20 Bacterial cultures of E. cloacae and P. aeruginosa were treated with CDs at concentrations of 100, 50, 25, 12.5, 5, 2.5, and 1.25 μg CDs per ml media. Muller Hinton Broth (MHB) containing the bacterial culture was used as the positive control, while the negative control consisted of only uninoculated broth. The samples were incubated for 20 hours at 37 °C. The visual difference in the turbidity was noted before and after incubation. The MIC is the lowest concentration of CDs that inhibited the visible in vitro growth of the bacteria, preventing the appearance of turbidity. For the MBC, 50 μl cultures from the microtiter well, which corresponded to the MBC value, were spread on an MHA plate. This step aimed to confirm the absence of further bacterial growth.2.5. Growth curve study
The investigation involved analyzing the growth curve of multidrug-resistant strains of E. cloacae and P. aeruginosa to discern deviations from the typical growth curve pattern following inhibition due to exposure to carbon dots (CD), in comparison to a control group, employing a series of experimental steps.21 Inoculums from a 0.5 McFarland-adjusted overnight culture were prepared to begin the study. These inoculums were then introduced into 100 ml of nutrient broth medium. For the E. cloacae strain, a dose of 25 μg ml−1 (MIC) of CDs was administered, while for the P. aeruginosa strain, a dose of 2.5 μg ml−1 (MIC) of CDs was used. The medium containing the inoculums and CDs was incubated up to 12 h, and absorbance readings were taken at regular intervals until the late stationary phase of the growth curve was reached. Absorbance was measured at a wavelength of 600 nm.2.6. Microtiter dish biofilm formation assay
E. cloacae and P. aeruginosa biofilm inhibition assay was performed on pre-sterilized 96 well flat bottom polystyrene microtiter plates in duplicate as described by O'Toole with slight modifications.22 10 μl of cell suspension with O.D600nm = 0.6 was inoculated in a 190 μl medium in each well. Then, the microtiter plate was incubated for 16 h at 37 °C to enable biofilm formation. Then, these bacterial biofilms were treated with their respective MBC of CD and its ten-fold serial dilution concentrations for 20 h. After incubation, unattached cells and media were removed by turning the plate over and shaking the liquid. Then, the plates were submerged in water twice to reduce background staining. Afterward, 125 μl of 0.2% crystal violet solution (prepared in methanol) was added to each well and incubated for 10–15 min at room temperature (RT). The plates were rinsed with water and dried by keeping them upside down. To quantify the biofilm, 125 μl of 33% acetic acid was added to each well to solubilize the crystal violet. After incubating for 10–15 min at RT, the solubilized solution was transferred to respective wells of a new microtiter dish, and the absorbance was measured at 590 nm using 30% acetic acid as the blank in the microplate plate reader. Lastly, plates were photographed for qualitative assays.2.7. Scanning and transmission electron microscopic (SEM and TEM) analysis
SEM and TEM were employed to image bacterial cells, both untreated and carbon dot (CD)-treated, at various time intervals, following established protocols.23 Briefly, one milliliter of an overnight culture of E. cloacae and P. aeruginosa adjusted to the density of 0.5 McFarland standard were inoculated in 100 ml nutrient broth medium and treated with 25 μg ml−1 and 2.5 μg ml−1 of CDs, respectively, for 4, 8 and 12 h. After completion of treatment, bacterial suspensions were centrifuged at 5000 rpm at 4 °C for 30 minutes, and the supernatant was removed. Then, the cell pellet was washed once with phosphate-buffered saline (PBS) by gentle pipetting and centrifugation at 5000 rpm for 10 min at 4 °C. After that, PBS was removed, and the pellets were suspended in appropriate volumes (five times the pellet size) of fixative (2.5% glutaraldehyde) and kept overnight at 4 °C. The next day, glutaraldehyde was removed by centrifugation, and samples were washed twice with PBS. After that, cells were incubated with 1% osmium tetroxide (prepared in PBS) for 2 h, followed by washing with PBS. Then, the samples were dehydrated using increasing concentrations of ethanol, dried, coated on gold, and visualized under SEM (Zeiss EVO40) and TEM (JEOL 2100F).2.8. Cell viability (live–dead) assay by flow cytometry
The percentage of live or dead cells after bacteria exposure to CDs was evaluated by flow cytometry using staining with propidium iodide (PI) dyes.24 Overnight cultures (OD600nm = 0.1) were treated with or without (control sample) MIC50 of CDs for 2, 4, and 8 h, washed with PBS, and incubated with PBS containing 15 μM PI for 5 min. After that, the samples were washed twice with cold PBS, and PI fluorescence from 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells of duplicate samples was captured using a BD FACSLyric™ flow cytometer with a 488 nm laser line for excitation of dyes, and emission was detected in the red channel. Data were analyzed by BD FACSuite v1.5 and FCS Express™ v7 (De Novo Software, CA, USA) software. Appropriate gates were applied on side scatter (SSC) versus PI density plots to determine the percentage of live (PI negative) and dead (PI positive) cells in each sample.
000 cells of duplicate samples was captured using a BD FACSLyric™ flow cytometer with a 488 nm laser line for excitation of dyes, and emission was detected in the red channel. Data were analyzed by BD FACSuite v1.5 and FCS Express™ v7 (De Novo Software, CA, USA) software. Appropriate gates were applied on side scatter (SSC) versus PI density plots to determine the percentage of live (PI negative) and dead (PI positive) cells in each sample.
2.9. Reactive oxygen species (ROS) detection
The ROS generation of E. cloacae and P. aeruginosa following CD administration was assessed using a peroxynitrite indicator, 2′-7′-dichlorodihydrofluorescein diacetate (DCFH-DA) (Sigma-Aldrich, UK), which can detect a wide variety of ROS including nitric oxide and hydrogen peroxide.25 The adjusted bacterial cultures (0.5 McFarland exponential phase bacteria culture) were treated with the MIC of CDs. At 2, 4, 6, and 8 h, the treated bacterial cultures were withdrawn and centrifuged, and the pallets were washed thrice with PBS. The untreated bacterial culture served as a control. After that, 10 μM of DCFH-DA was administered to the PBS-suspended pellets under dark conditions and incubated for 1 h. Then, the cultures were washed, and the fluorescence emission of DCFH-DA was measured at 525 nm with an excitation wavelength of 485 nm.2.10. Lipid peroxidation analysis
Lipid peroxidation was analyzed using thiobarbituric acid as a reactive substance.23 0.6 OD culture of E. cloacae and P. aeruginosa was used to inoculate the media in a 100 ml flask, and a group of these bacteria was treated up to 40 h with MIC −25 μg ml−1 and 2.5 μg ml−1 doses of CDs, respectively, and incubated. The control group was the bacterial solution without treatment. After a regular interval, 2 ml of the sample was withdrawn in a screw cap vial from the flask and kept in a shaker incubator at 37 °C. Sequentially, 100 μl of 20% sodium dodecyl-sulfate (SDS), 300 μl of 2% butyl-hydroxytoluene, 40 μl of 200 mM of (ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid) (EGTA) and 2 ml of 1% thiobarbituric acid (TBA) in 10 ml trichloroacetic acid were added, vigorously mixed and heated at 90 °C for 20 min. After cooling to RT, 2 ml of butanol was added and mixed, and OD was recorded at 532 nm of the supernatant.2.11. Evaluation of changes in bacterial membrane potential
A relative change in bacterial membrane potential was evaluated by flow cytometry using the fluorescent dye DiOC2(3) (3,3′-diethyloxacarbocyanine iodide) following a standard protocol.26 Logarithmically grown cells from overnight culture with an OD600nm of 0.1 were treated with or without CDs (MIC50) for 2, 4, and 8 h. Then, the cells were washed with PBS and incubated with 30 μM of fluorochrome DiOC2(3) for 30 min. The samples were again washed twice with cold PBS, and the fluorescence of the dye from 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells was acquired using a flow cytometer (BD FACSLyric™ equipped with BD FACSuite v1.5 software) and analyzed using FCS Express™ v7 software. Dye was excited using a 488 nm laser line, and emission was detected in green (FITC) and red (PerCP-Cy 5.5) channels. The dye's red/green fluorescence ratios for the treatment and control groups were calculated using the gated populations' median fluorescence (red and green) intensities.
000 cells was acquired using a flow cytometer (BD FACSLyric™ equipped with BD FACSuite v1.5 software) and analyzed using FCS Express™ v7 software. Dye was excited using a 488 nm laser line, and emission was detected in green (FITC) and red (PerCP-Cy 5.5) channels. The dye's red/green fluorescence ratios for the treatment and control groups were calculated using the gated populations' median fluorescence (red and green) intensities.
2.12. DNA damage analysis by comet assay (single cell gel electrophoresis assay)
After the treatment of E. cloacae and P. aeruginosa with the MBCs of CD for 4, 8, and 12 h, bacterial cells were harvested, and the comet assay was performed using a standard protocol,27 with slight modification. In a nutshell, the bacterial cell suspensions were mixed with the pre-warmed agarose solution at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v) and then rapidly transferred the mixture onto a pre-coated microscope slide with a thin layer of agarose. A coverslip was placed on top to create a uniform thickness of the agarose gel, allowing it to solidify at 4 °C for approximately 10–15 min. Then, to lyse the cells and remove the cellular protein, the slides were submerged in ice-cold lysis buffer containing Triton X-100, EDTA, and NaCl and incubated at 4 °C for 1 hour to allow DNA to unwind. Then, the slides immersed in the electrophoresis buffer were subjected to electrophoresis at a low voltage of 25 V for 30 min to allow the DNA fragments to migrate, forming comet-like structures. Then, the slides were removed, and DNA was neutralized by washing them with a Tris–HCl neutralization buffer for 15 min. Then, DNA in the comet structures was stained using ethidium bromide (EtBr) for 15–30 min, and EtBr fluorescence was detected using a fluorescence microscope at 10× magnification using a red emission filter. DNA damage in the treatment and the control groups were compared by analyzing the images using Comet Score software (version IV) that evaluated the parameters used for indicating the DNA damage. Statistical tests were also performed to determine the significance of differences between treatment groups and the control.
10 (v/v) and then rapidly transferred the mixture onto a pre-coated microscope slide with a thin layer of agarose. A coverslip was placed on top to create a uniform thickness of the agarose gel, allowing it to solidify at 4 °C for approximately 10–15 min. Then, to lyse the cells and remove the cellular protein, the slides were submerged in ice-cold lysis buffer containing Triton X-100, EDTA, and NaCl and incubated at 4 °C for 1 hour to allow DNA to unwind. Then, the slides immersed in the electrophoresis buffer were subjected to electrophoresis at a low voltage of 25 V for 30 min to allow the DNA fragments to migrate, forming comet-like structures. Then, the slides were removed, and DNA was neutralized by washing them with a Tris–HCl neutralization buffer for 15 min. Then, DNA in the comet structures was stained using ethidium bromide (EtBr) for 15–30 min, and EtBr fluorescence was detected using a fluorescence microscope at 10× magnification using a red emission filter. DNA damage in the treatment and the control groups were compared by analyzing the images using Comet Score software (version IV) that evaluated the parameters used for indicating the DNA damage. Statistical tests were also performed to determine the significance of differences between treatment groups and the control.
2.13. Carbohydrate leakage analysis
The anthrone method was used to study carbohydrate leakage.28 Culture of E. cloacae (MTCC 509) and P. aeruginosa with 0.6 OD was used to inoculate the media in a 100 ml flask, and the experiment group of the two bacteria was treated up to 40 h with MIC −25 μg ml−1 and 2.5 μg ml−1 dose of CDs, respectively, and incubated at 37 °C. The control group was the bacterial solution without treatment. After a regular period, 2 ml of sample were withdrawn from the flask, kept in a shaker incubator, and centrifuged at 6000 rpm for 10 min. 1 ml of supernatant was taken in a screw cap vial, and an equal volume of distilled water was added. Then, 5 ml of anthrone reagent was added subsequently. After vigorously mixing, vials were incubated in a water bath at 95 °C for 10 min. After cooling to room temperature, OD was recorded at 620 nm. The concentration of carbohydrates was measured from a calibration curve of glucose.2.14. Bacterial cell lysis and protein estimation
Bacteria were lysed using a combination of lysis buffer, lysozyme, and glass beads.29 Briefly, control and 4 h-treated bacterial suspensions were centrifuged at 5000 rpm at 4 °C for 30 min, and the supernatant was discarded. Then, cell pellets were washed twice with PBS by gentle pipetting and centrifugation at 5000 rpm for 10 min at 4 °C. After that, PBS was removed, and the pellets were suspended in appropriate volumes (five times pellet size) of lysis buffer. Additionally, a small quantity of glass beads (0.5 mm) was added, and the samples were vortexed multiple times. Then, the lysate was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C, and the supernatant was collected and processed for protein estimation.
000 rpm for 10 min at 4 °C, and the supernatant was collected and processed for protein estimation.
Protein content in the cell lysate was estimated by Bradford assay following the standard protocol.30 Briefly, 5 μl of bacterial lysis buffer (for blank) or cell lysate (Test Sample) was added to 200 μl of Bradford reagent (BR) in a 96-well plate and incubated at RT for 10 min. Then, the absorbance was measured at 595 nm in a microplate reader (iMarkTM, Bio-Rad). Other than the test samples, the assay included an increasing concentration of bovine serum albumin (1–5 μl of 1 mg ml−1 stock) added to 200 μl of BR (reaction volume made to 205 μl with double distilled water (DDW) to plot a standard curve for quantifying the protein content in test samples.
2.15. Protein expression pattern study
Sodium dodecyl-sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) examines the protein expression pattern. The sample for SDS-PAGE was prepared by mixing the lysate containing 20 μg of protein from each group with an appropriate volume of 5× Laemmli buffer (0.25 M Tris–HCl, pH 6.8 + 10% (w/v) SDS + 40% (v/v) glycerol + 20% (v/v) β-mercaptoethanol + 0.05% (w/v) bromophenol blue) and boiling the mixture for 5 min. Two types of gel, namely 4% stacking gel, pH 6.8 [13.3% (v/v) of 30% (w/v) acrylamide + 0.125 M Tris–(Cl) + 0.1% (w/v) SDS + 35% (v/v) DDW + 1% (v/v) of 10% (w/v) APS + 0.1% (v/v)TEMED] and 12. 5% resolving gel, pH 8.8 [41.7% (v/v) of 30% (w/v) acrylamide + 0.375 M Tris–(Cl) + 0.1% (w/v) SDS + 7.3% (v/v) DDW + 1%(v/v) of 10% (w/v) APS + 0.1% (v/v)TEMED] were prepared to stack and resolve the proteins, respectively. Proteins in the samples and marker were loaded onto the stacking gel and run using a running buffer of pH 8.3 (0.025 M Tris–(Cl) + 0.1% SDS + 0.192 M glycine) at 80 V. Then, the gel was stained overnight with a staining solution [0.1% (w/v) Coomassie brilliant blue R-250 + 4.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.5 mixture of methanol
4.5 mixture of methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetic acid (glacial)
acetic acid (glacial)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDW]. The next day, the gel was destained by destaining solution [2
DDW]. The next day, the gel was destained by destaining solution [2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 mixture of methanol
2 mixture of methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetic acid (glacial)
acetic acid (glacial)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDW)], and the image of the gel was captured and analyzed.
DDW)], and the image of the gel was captured and analyzed.
2.16. Statistical analysis
Except for SEM and TEM, all the experiments were performed thrice. Statistical significance of the changes in data was analyzed by one-way ANOVA and Tukey's test. A change with p-value <0.05, between groups, was considered significant.3. Results and discussion
3.1. Synthesis and characterization
Previously, our laboratory reported a detailed surface and structural characterization of CDs synthesized from F. benghalensis and F. religiosa L. extracts.19,31 In summary, the CDs have a spherical shape with a particle size of around 2.4 nm and a zeta potential of −20 mV. High-resolution transmission electron microscopy (HRTEM) imaging discovered the absence of a clear lattice structure, demonstrating the particles' amorphous nature. The UV-visible spectra of the CDs revealed an absorbance peak at 317 nm, which may be ascribed to the CDs, and another peak at 280 nm, which can be related to the * transition of aromatic amino acids contained in the leaf extract's protein content. Moreover, the CDs' photoluminescence excitation (PLE) spectrum, excited at 317 nm, displayed an emission peak at 455 nm in the green region of the absorption spectrum, indicating a blue-green luminescence characteristic of the CDs. The surface chemistry of CDs was characterized through Fourier-transform infrared (FTIR) analysis.19 The FTIR spectra of CDs revealed distinctive features, including a prominent broad peak at 3360 cm−1 corresponding to the O–H stretch of alcohol. Additionally, a minor peak at 2835 cm−1 indicated the C–H stretch of aldehyde, while another minor peak at 1876 cm−1 signified the C–H bending of aromatic compounds. Notably, three medium-sized and strong peaks observed at 1627 cm−1, 1422 cm−1, and 1093 cm−1 were attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch of alkenes, O–H bending of alcohol, and C–O stretch of alcohol, respectively. Furthermore, a minor peak at 824 cm−1 indicated the C
C stretch of alkenes, O–H bending of alcohol, and C–O stretch of alcohol, respectively. Furthermore, a minor peak at 824 cm−1 indicated the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending of the alkene in the CD structure.
C bending of the alkene in the CD structure.
CDs exhibited robust thermal stability over a six-month testing period, showcasing resistance to both high and low temperatures.19 According to studies, solid CDs remain stable up to 400 °C, with minimal weight loss at temperatures below 100 °C.32 Additionally, CDs synthesized through the green hydrothermal process displayed notable pH stability, making them well-suited for diverse environmental conditions.11 The application of CDs as highly sensitive fluorescent detectors in ionic solutions demonstrated their versatility in detecting various ions, such as Fe ions, in aquatic solutions.18 These findings underscore the CDs' potential for diverse applications and contribute to our understanding of their stability and functionality in different environmental contexts.11
3.2. Anti-microbial activity of CDs
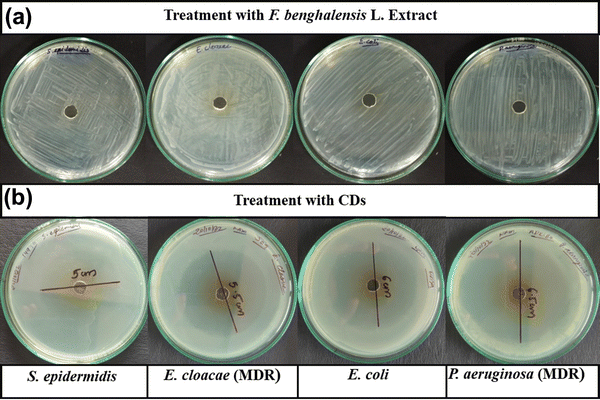 | ||
| Fig. 1 Determination of the inhibition zone of (a) F. benghalensis aqueous extract and (b) CDs for S. epidermidis, E. cloacae, E. coli DH5α and P. aeruginosa. | ||
| Bacteria | Zone of inhibition (mm) | ||||||
|---|---|---|---|---|---|---|---|
| CDs | AMC | GEN | TE | OF | CXM | COT | |
| S. epidermidis (ATCC 14990) | 50 | 17 | 20 | 22 | 30 | 0 | 19 |
| E. cloacae (MTCC 509) – MDR | 55 | 0 | 6 | 12 | 14 | 0 | 3 |
| E. coli DH5α | 60 | 18 | 18 | 21 | 33 | 4 | 21 |
| P. aeruginosa (MTCC 1035) – MDR | 65 | 0 | 5 | 9 | 15 | 0 | 0 |
| Bacteria | MIC (μg ml−1) | MBC (μg ml−1) |
|---|---|---|
| S. epidermidis (ATCC 14990) | 25 | 50 |
| E. cloacae (MTCC 509) – MDR | 25 | 50 |
| P. aeruginosa (MTCC 1035) – MDR | 2.5 | 5 |
| E. coli DH5α | 1.25 | 2.5 |
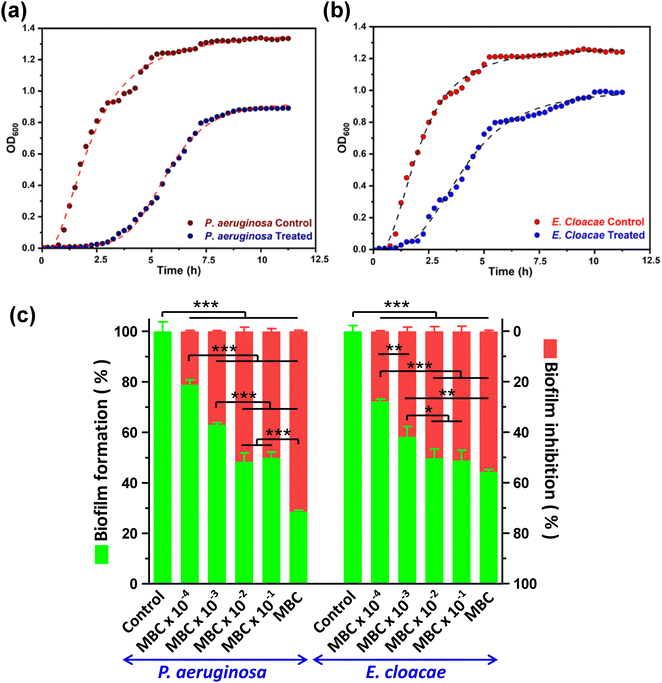
3.3. Analysis of growth curve dynamics and biofilm inhibition
Growth curve analysis showed that bacterial growth has significantly deviated from the characteristic normal pattern after CD exposure was inhibited relative to the control group. The growth curve of the control group shows an obvious time frame for lag, log, and stationary phases of the growth curve. However, the group treated with CDs showed an extension of the lag phase and shortening of the log phase of growth due to the entrapment of CDs inside the cell (Fig. 2a and b). The negative charge and small size of CDs provide ample surface area to volume ratio, facilitating more significant interaction with bacterial cells, consequently reducing the growth of bacteria. P. aeruginosa taking more time to adapt in the presence of CDs than E. cloacae.CDs have demonstrated promising potential as a biofilm inhibitor through their ability to disrupt the extracellular polymeric substance matrix and interfere with bacterial adhesion, ultimately hindering biofilm formation by pathogenic bacteria. For both MDR strains, biofilm formation was significantly inhibited in the order of an increase in CD concentration. The highest biofilm inhibition was observed at MBC, which resulted in 71.46 ± 0.45 and 55.61 ± 0.46% of biofilm inhibition compared to the control, in P. aeruginosa and E. cloacae, respectively. At lesser concentrations, both bacteria displayed a comparable decrease in biofilm formation as evident from a 50.11 ± 1.12 to 21.14 ± 0.41% and 51.16 ± 2.09 to 27.76 ± 0.26% of biofilm inhibition in P. aeruginosa and E. cloacae, respectively, in the CD concentration range of MBC × 10−1 to MBC × 10−4 (Fig. 2c). The 96 well flat bottom polystyrene micro-titer plate showed a decrease in crystal violet colour intensity, indicating biofilm inhibition, in the wells containing CD treatment is shown in the ESI,† Fig. S2. The result indicated that biofilm inhibition caused by CDs at the MBC was more selective toward P. aeruginosa than E. cloacae.
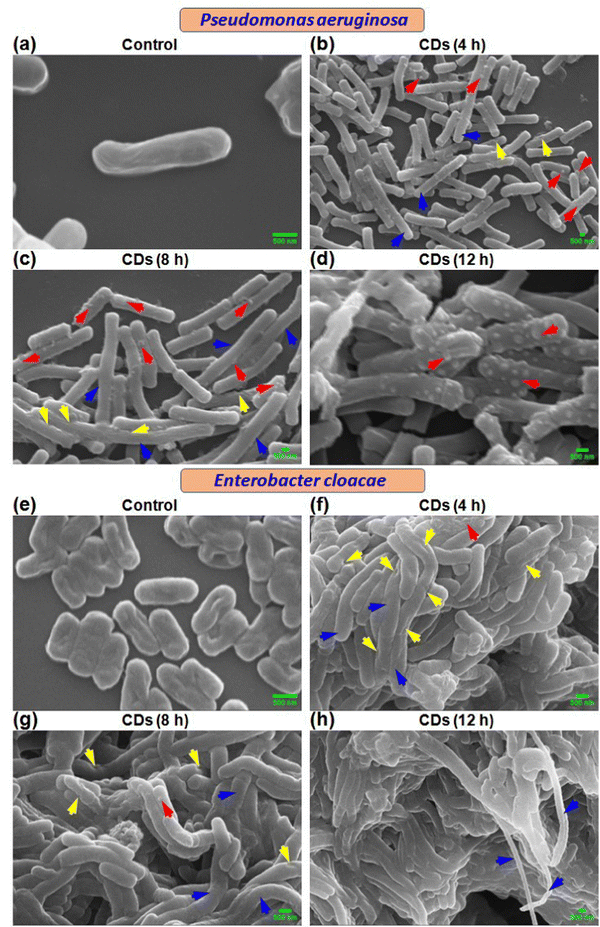
3.4. Investigation of cell surface morphology alteration
Scanning electron microscopic (SEM) analysis revealed time-dependent cell surface and other morphological changes in P. aeruginosa and E. cloacae due to CD exposure. Control cells showed normal size and morphology with smooth surfaces (Fig. 3a and e). Treatment with CDs for 4 h exhibited the presence of elongated cells (blue arrow) with rough surfaces containing small, rounded, or irregular protrusions (red arrow) and dents (yellow arrow) (Fig. 3b and f). Similar but more pronounced alterations were observed at 8 h (Fig. 3c and g) and 12 h (Fig. 3d and h). Time-dependent progressively more damaged cells are observed, indicating the enhanced trend of bacterial necrosis.3.5. Investigation of ultrastructural changes in bacteria
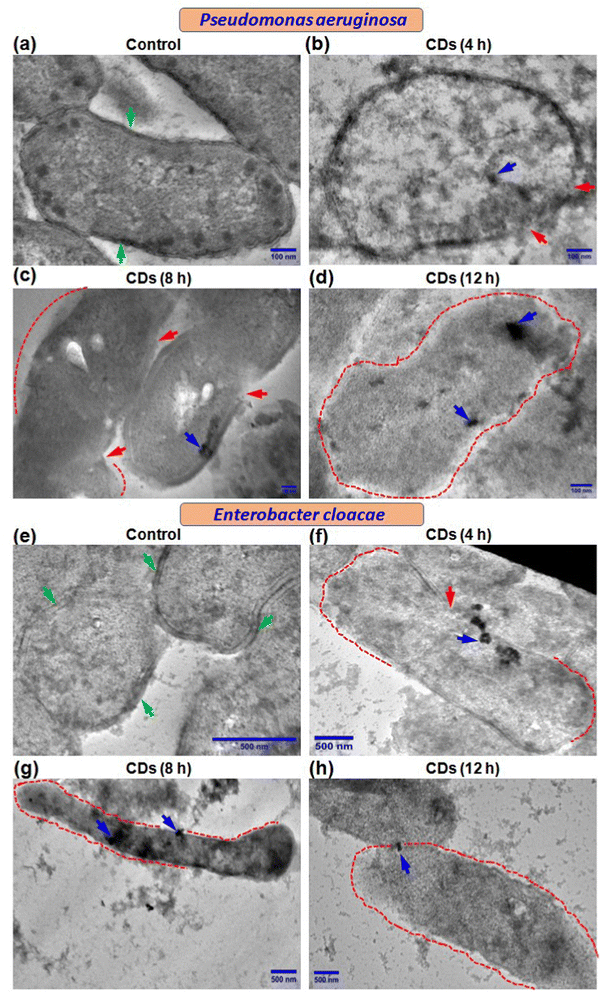
We further performed transmission electron microscopy (TEM) to determine the fate of CDs in terms of their internalization and effect on ultrastructural changes in the two bacteria being studied. Control cells showed a normal and characteristic ultrastructure with intact cell walls, cell membranes (green arrow), and electron-dense cytoplasm (Fig. 4a and e). Both P. aeruginosa and E. cloacae displayed regions with ruptured cell walls and cell membranes (red arrow) when exposed to CDs for 4 h (Fig. 4b and f). Treatment with CDs for 8 h resulted in more degradation of the cell wall and cell membrane (regions marked by the red dot) (Fig. 4c and g), which eventually vanished almost completely at 12 h in both bacteria (Fig. 4d and h). Elongation in cell size was also observed in CD-treated E. cloacae (Fig. 4f–h). These ultrastructural changes can be correlated with the time-dependent internalization of CDs. Though complete intake of CDs, visible as a cluster of CDs (blue arrow), was observed in P. aeruginosa at all three-time points (Fig. 4b–d), maximum accumulation occurred at 12 h (Fig. 4d). However, E. cloacae with 4 h exposure to the CDs exhibited the presence of a cluster of CDs on the cell surface or just inside the cytoplasm (Fig. 4f). In contrast, treatment for 8 h showed the highest CD internalization (Fig. 4g). It can be speculated that excellent hydrophilicity of the biogenic CDs due to phytochemicals present in the F. benghalensis could facilitate entry of these CDs through the channel protein porin present in the outer membrane of these Gram-negative bacteria, followed by binding to and degradation of the peptidoglycan layer, resulting in the anti-microbial effects. The absence of CDs inside the cytoplasm and their limited presence near the ruptured cell wall and cell membrane in the 12 h exposure group corroborated the inability of the bacteria to retain the internalized CDs due to the almost complete removal of the cell wall and cell membrane (Fig. 4h).3.6. Assessment of bacterial cell viability and membrane disruption by live/dead cell quantification
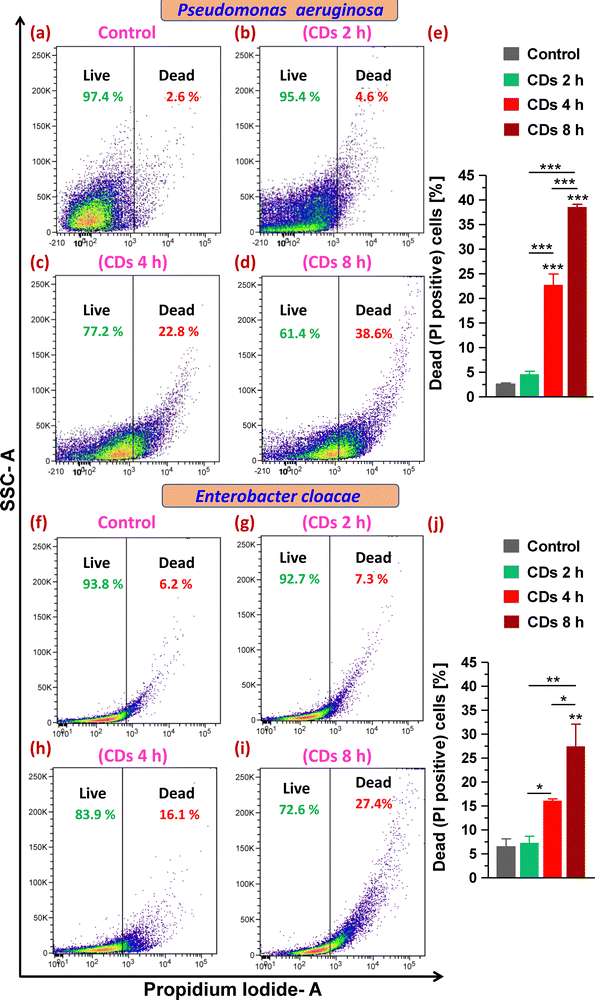
Next, staining with the cell impermeant and red fluorescent fluorophore PI that only stains cells with disrupted and compromised membranes (dead cells) led to determining the status of plasma membrane integrity and evaluating live (viable) and dead cells upon CD exposure. On analysis of gated populations in side scatter (SSC) versus PI scatter plots for P. aeruginosa, it was observed that CDs caused a significant increase in dead cell percentage to 23% and 39% after 4 and 8 h exposure, respectively, compared to 3% in the control group (Fig. 5a and c–e). Treatment with CDs for 2 h was found to be ineffective in decreasing the cell viability (Fig. 5b and e). E. cloacae were observed to be relatively less sensitive to death caused by CDs and showed a 10% and 21% increase in dead cells (from 6% in the control group) when treated for 4 and 8 h, respectively (Fig. 5f and h–j).3.7. Assessment of ROS generation and lipid peroxidation
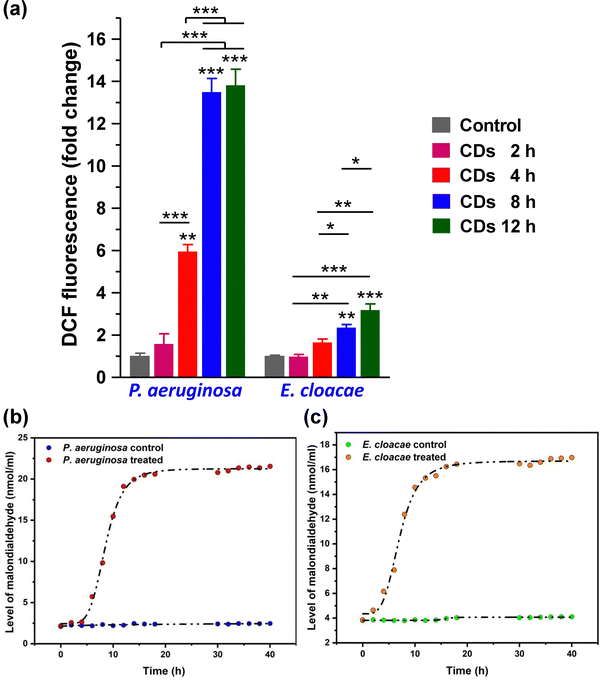
Usually, ROS in bacterial cells is regulated by their oxidoreductase system, maintaining a dynamic balance at a relatively normal level. This balance is disturbed under the conditions of oxidative stress when the antioxidant defense system fails to effectively neutralize excess ROS. Significantly increased DCF fluorescence intensity compared to the control set shows a high level of DCF in the CD-treated bacterial cells, which directly correlated to the increased level of ROS. Both strains showed a time-dependent increase in ROS level after CD treatment (Fig. 6a). However, P. aeruginosa showed a significantly higher ROS level than E. cloacae.A common toxic effect caused by excess ROS due to CD exposure is oxidative damage. Lipid peroxidation is one of the prevalent indications of oxidative damage caused by increased ROS by reacting with lipids in bacteria's cell membrane, leading to membrane damage. Even at low concentrations, the CDs showed good anti-bacterial activity against E. cloacae and P. aeruginosa. As shown in Fig. 6b and c, the malondialdehyde level produced in P. aeruginosa is slightly higher (4.4 nmol ml−1) than E. cloacae at 15 h, corroborating the ROS analysis. P. aeruginosa is more sensitive and has more lipid peroxidation, indicating increased ROS levels in the cell, even much earlier. Their anti-bacterial mechanism originated from the fact that CDs were able to produce ROS, which acts on the surface of bacteria, disrupting the cell membrane and causing cytoplasmic leakage. This is majorly due to the production of ROS, which induces oxidative stress in bacteria, disrupting the integrity of cell membranes and leading to cell death.34 Researchers can regulate the level of ROS in bacteria and introduce environment-responsive mechanisms, which may improve the performance of CDs and bring smart anti-bacterial functions.
3.8. Analysis of bacterial membrane potential alteration
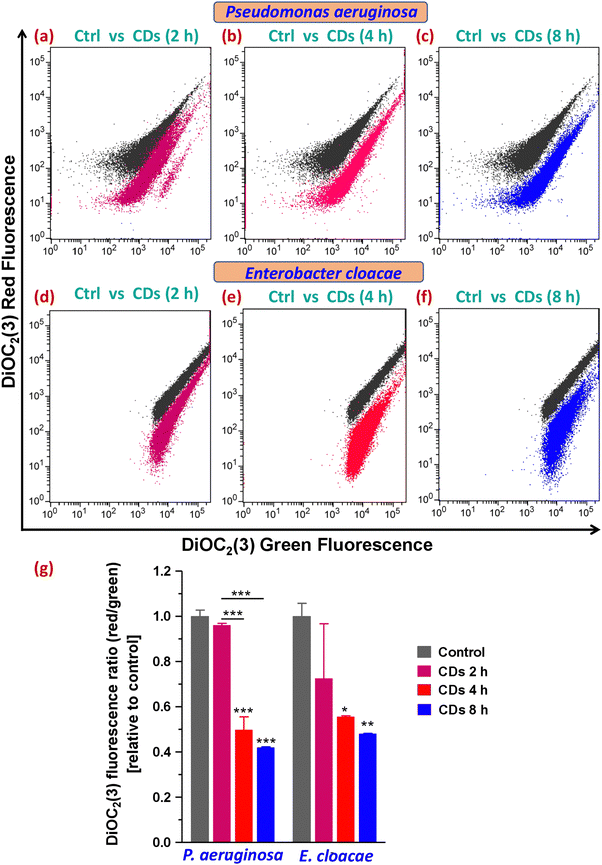
Together with membrane lipid peroxidation, ROS can also depolarize the bacterial membrane by causing a drop in the membrane potential. This happens because of the enhancement in the permeability of the oxidized membrane to ions due to lipid peroxidation, resulting in the altered ion concentration across the membrane. In bacterial cells, DiOC2(3), a probe used to detect the level of bacterial membrane potential, fluoresces green, but as the dye molecules self-associate at higher cytosolic concentrations brought on by larger membrane potentials, the fluorescence shifts toward red emission. Therefore, DiOC2(3) red/green ratios of samples are compared to determine the relative change in bacterial membrane potentials. Treatment with CDs for 4 and 8 h exhibited in a 2.00 ± 0.22 and 2.37 ± 0.02-fold decrease in DiOC2(3) red/green ratio, respectively, relative to the control in P. aeruginosa, indicating a depolarized membrane (Fig. 7b, c and g), whereas E. cloacae led to a 1.79 ± 0.01 and 2.07 ± 0.01-fold decrease, respectively, at these time point (Fig. 7e–g). Both strains were resistant to membrane potential loss after 2 h of CD exposure (Fig. 7a, d and g). Bacteria in the stationary phase of their growth were reported previously to have a significantly low membrane potential (similar to the MPs of MP inhibitor CCCP-treated and heat-killed samples) compared to those in the exponential phase.35 Therefore, our results on membrane potential alterations by CDs in the two MDR bacteria corroborated our growth curve data that showed a delayed plus shortened exponential (log) phase and an early plus prolonged stationary phase.3.9 Investigation of DNA damage
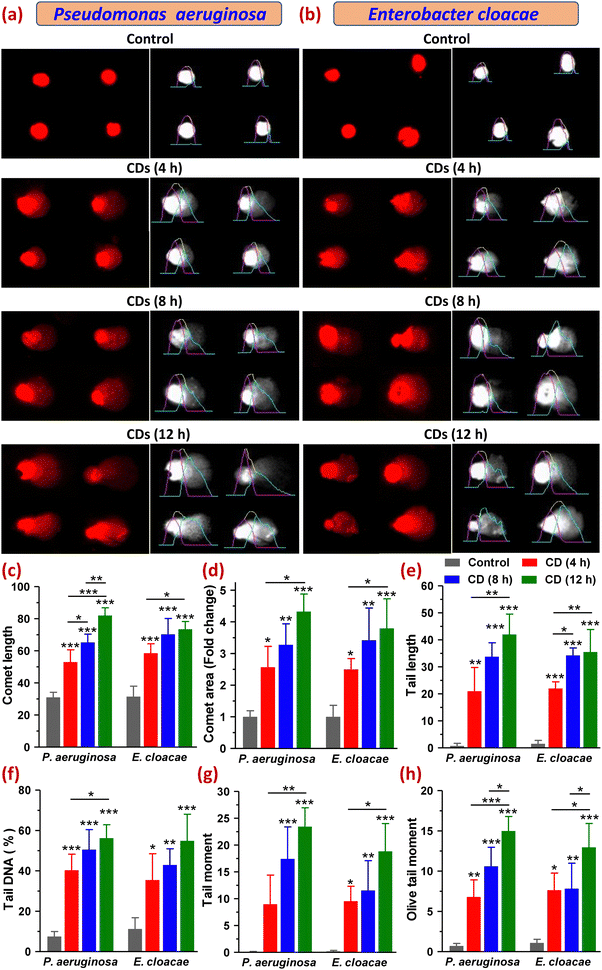
Furthermore, increased intracellular ROS levels may also cause oxidation of DNA, thereby causing oxidative damage leading to breakage in its strands. So, to evaluate the genotoxicity of CDs in P. aeruginosa and E. cloacae, we carried out a comet assay that detects DNA damage based on the electrophoretic movement of DNA from a single cell. A comet with a sharp, undamaged head and a short tail shows only minor DNA damage, whereas one with a diffused head and a protracted tail suggests more significant DNA damage. The extent of DNA damage is also related to the amount of DNA in the tail.36,37 For both bacteria, the control (untreated) group showed almost no comet tails (Fig. 8a, b and e). Among the treated groups exposed to CDs for 4, 8, and 12 h, all six parameters, namely comet length (CL), comet area (CA), tail length (TL), tail DNA percentage (TD%), tail moment (TM) and olive tail moments (OTM), which are indicative of the DNA damage, were found to be varied in increasing order of time points. Both bacteria showed comets with a round head with or without a tail in the 4 h treatment group. However, with the increase in CD exposure time (8 and 12 h), the majority of the comets of both bacterial cells showed a typical comet-like head but a diffused tail with a hedgehog appearance, where the tail resembled a series of spikes radiating outward by multiple sharp projections that meant severe DNA fragmentation. Some of the comets had a distinct appearance as diffused cloud-like heads, and a long, fine, faint tail resembled the apoptotic comet (Fig. 8a and b). The cloudiness of the head resulted from the dispersion of fragmented DNA within the nucleoid region. Some cells in the 8 h treatment group of E. cloacae appeared as a double-headed comet with a wide tail distribution. Comets with radiated tails and oval-shaped heads were also observed within the treatment groups. The highest amount of DNA damage was detected at 12 h of CD treatment in P. aeruginosa, which demonstrated an increase in CL to 82.00 ± 4.83 from 31.00 ± 3.16 (control), CA by 4.33 ± 0.55-fold, TL to 42.00 ± 7.53 from 0.75 ± 0.96, TD% to 56.24 ± 6.64 from 7.49 ± 2.49, TM to 23.44 ± 3.51 from 0.073 ± 0.093 and OTM to 15.00 ± 1.79 from 0.71 ± 0.31. A similar, but relatively smaller, increase in these parameters was observed in E. cloacae, where CL, CA, TL, TD%, TM and OTM were found to be 70.25 ± 9.91, 3.42 ± 1.02-fold, 35.50 ± 8.35, 54.91 ± 13.15, 18.84 ± 5.16, and 12.97 ± 2.95, respectively, from their control values of 31.50 ± 6.45, 1 ± 0.6-fold, 1.50 ± 1.29, 11.20 ± 5.59, 0.19 ± 0.17 and 1.08 ± 0.45, respectively (Fig. 8c–h). DNA migration length in a comet assay is correlated with the fragment size and level of strand breaks in DNA (Gao et al., 2018). All analyzed parameters suggested a high degree of DNA fragmentation, indicating strand breaks of the cell nuclei in both MDR bacteria under this study (Fig. 8a–h). This extensive DNA damage leading to cell death can also be viewed as the harmful oxidative effect of lipid peroxidation-derived reactive aldehydes, as reported in E. coli (Pérez et al., 2008). Thus, the comet assay results established the significant potential of CDs to cause the most effective DNA damage in P. aeruginosa after their 12 h of exposure, which also corroborated our earlier findings that the synthesized CDs possessed relatively superior anti-bacterial and antibiofilm activity in P. aeruginosa than in E. cloacae.3.10. Determination of macromolecular leakage and changes in protein expression
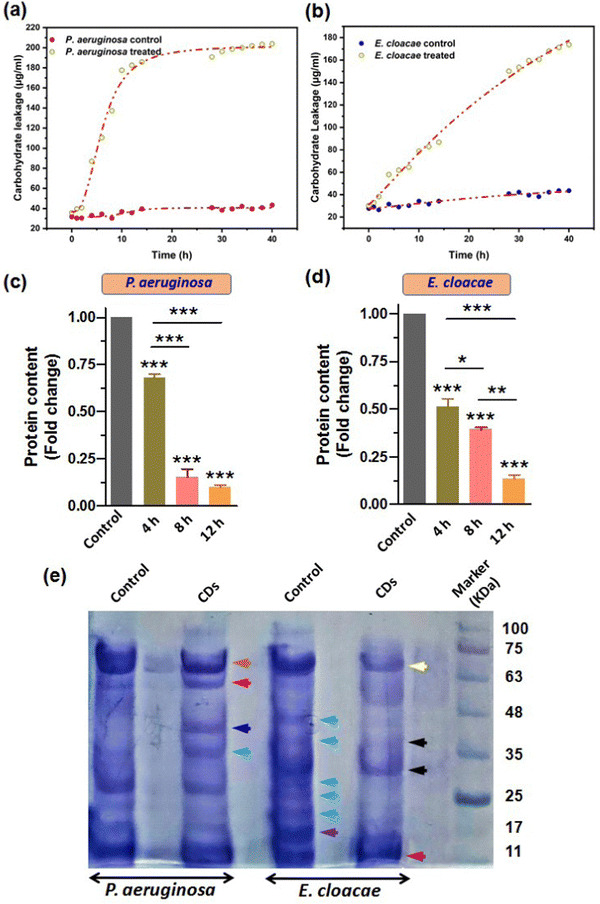
Our results from the previous sections confirmed that the breach in membrane integrity and loss of cell viability of the bacterial cells upon CD exposure was due to the CD-induced oxidative stress, which caused membrane lipid peroxidation and a fall in membrane potential. Also, the increased ROS level may damage other important biomolecules such as carbohydrates and proteins (Phaniendra et al., 2015). This can further aggravate the compromised membrane integrity and facilitate cytosolic leakage of macromolecules. Carbohydrate and their different conjugations with lipids and proteins play vital roles in cells as inside transporting and signaling systems as well as secretary bodies. Therefore, leakage of these biomolecules eventually leads to cell death. So, we further estimated the carbohydrate and protein leakage from the bacteria by measuring the quantity of intracellular carbohydrates and protein. Furthermore, we also reported the CD-induced variation in the overall protein expression patterns.Treatment with CDs showed high carbohydrate leakage in both MDR strains P. aeruginosa and E. cloacae relative to the control. The bacterium P. aeruginosa displayed a relatively high discharge of carbohydrates (the standard curve for determining the amount of carbohydrate is provided in the ESI,† Fig. S3a) than E. cloacae, as evident from Fig. 9a and b, which show a 2.6, 4.5, and 5.13-fold increase (relative to control) in carbohydrate leakage in P. aeruginosa compared with the 1.8, 2.1, and 2.6-fold increase in E. cloacae after their exposure to CDs for 4, 8, and 12 h, respectively. This suggests that P. aeruginosa is more susceptible to CDs. However, E. cloacae also had almost three times more leakage than the control group in the maximum exposure time with particles.
In addition to carbohydrates and lipids, proteins play many vital functions, especially in conferring pathogenicity to bacteria. Therefore, determining an anti-microbial agent's effect on the microbes' total protein content becomes important. Our results of Bradford assay for relative quantification of total protein content based on a standard BSA graph (Fig. S3b, ESI†) showed that 4, 8, and 12 h exposure of P. aeruginosa to CDs caused a 1.5, 6.6, and 10-fold decrease in total protein amount, relative to the control (Fig. 9c). A similar decreasing trend of protein content due to CD treatment for 4, 8, and 12 h (2, 2.6, and 8-fold decrease, respectively) was observed for E. cloacae (Fig. 9d). However, the fold decrease in 4 and 8 h treatment groups were not as large as in the 12 h group.
SDS-PAGE further examined the changes in protein expression pattern in P. aeruginosa and E. cloacae after treatment with CDs for 4 h (Fig. 9e). Compared to untreated cells, a decreased and increased expression of approximately 70 kDa (brown arrow) and 60 kDa (red arrow) proteins were found, respectively, in P. aeruginosa. Interestingly, only this treatment group observed two new protein bands with approximate sizes of 40 kDa (blue arrow) and 35 kDa (green arrow). The control group of E. cloacae showed expression of proteins with approximate molecular weights of 45, 40, 28, 25, 21 kDa (sky blue arrow), and 17 kDa (purple arrow) that were not expressed in the treated group of this bacteria. CD-treated E. cloacae exhibited a decreased expression of proteins of approximately 65 kDa (yellow arrow), 38 and 28 kDa (black arrow), and 11 kDa (dark red arrow).
3.11. Mechanism of anti-pathogenic activity
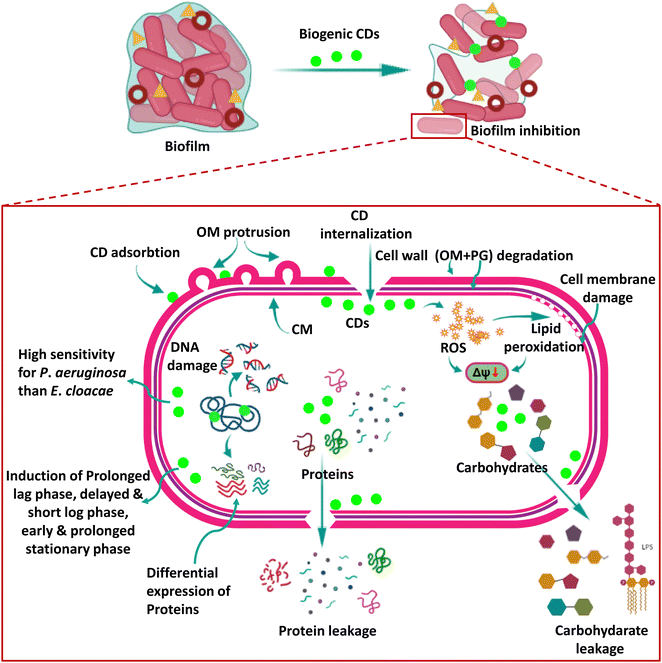
Recently, CDs have been shown to significantly impact bacterial systems by inducing oxidative damage. The oxidoreductase system in bacterial cells carefully regulates ROS levels, which is critical in maintaining dynamic equilibrium within a typical range. This delicate equilibrium is required for cellular activities to function appropriately. However, excessive ROS build-up ensues when this balance is broken, resulting in oxidative stress.In a notable breakthrough, by exploring the anti-bacterial activity of the biogenic CDs of the present study, we not only provide concrete evidence in favour of ROS-mediated killing of bacteria by CDs, as recently studied by other researchers,38 but the present study also delves deeper into the anti-bacterial mechanism of biogenic CDs in two deadly bacterial strains which are critical pathogens on the WHO Global Priority List (2017), E. cloacae and P. aeruginosa, which were not explored earlier. Our CDs exhibited significant anti-bacterial activity, even at low concentrations, against these bacteria. Of particular interest was the heightened sensitivity of P. aeruginosa toward CDs and the concurrent increase in lipid peroxidation levels observed within these cells. The elevated lipid peroxidation levels indicate an augmented presence of ROS, even during the early stages of exposure. This finding implies that P. aeruginosa is more prone to ROS-induced damage, underscoring the potential of CDs as effective anti-microbial agents against this pathogen. Fig. 10 shows the schematic representation of the mechanism of anti-pathogenic action of CDs studied in the present research.
The anti-bacterial mechanism of CDs can be attributed to their intrinsic ability to get adsorbed on the bacterial cell wall and then internalize into the cells to generate intracellular ROS upon interaction with bacterial components. These ROS species then act upon the bacterial cell surface, initiating a cascade of events that disrupt the integrity of the cell membrane. We also observed lipid peroxidation, a typical side effect of oxidative stress caused by the interaction of ROS with lipids found in bacterial cell walls. ROS generated by CDs also induced membrane depolarization. Both of these processes eventually resulted in membrane damage, jeopardizing the structural integrity of the cells.
Consequently, the cell membrane disruption leads to the leakage of vital cytoplasmic contents such as carbohydrates and proteins, triggering cellular dysfunction and ultimately resulting in cell death. The CDs also led to DNA damage, though we could not identify whether it occurred as a ROS-dependent or independent event. Thus, these CDs acted by affecting the functioning of all major macromolecules such as carbohydrates, lipids, nucleic acids (DNA), and proteins, which increases their effectiveness, and they inhibited the two biofilm-forming bacteria of growing concerns at concentrations as low as 2.5–25 μg ml−1. The production of ROS by CDs serves as a pivotal factor in inducing oxidative stress in bacteria, primarily targeting and compromising the integrity of the cell membranes. The ability to regulate the levels of ROS within bacterial cells shows tremendous promise in modulating the efficacy of CDs as anti-microbial agents. Researchers can explore innovative strategies to fine-tune ROS production and mitigate potential cytotoxic effects. By manipulating ROS levels, CDs' anti-microbial activity can be optimized, thereby enhancing their performance and augmenting their potential therapeutic applications. Table 3 shows the reported studies on the anti-microbial activity of CDs.
| CD/CD based anti-microbials | Source and synthesis method | Targeted bacteria | Efficiency (mg mL−1) | Mechanism of action | Ref. |
|---|---|---|---|---|---|
| PCQDs | Papaya L.; hydrothermal | Broad-spectrum | 250 | Photothermal | 38 |
| CDs | Vitamin C; microwave-assisted | Broad-spectrum | S. aureus-100 | Denaturation of DNA | 39 |
| B. subtilis-50 | |||||
| E. coli-75 | |||||
| LCDs | Levofloxacin; hydrothermal | Broad-spectrum | MR S. aureus-250 | ROS, Electrostatic interaction, Retention of anti-bacterial active structures | 40 |
| P. aeruginosa-250 | |||||
| S. epidermidis-500 | |||||
| E. faecalis-500 | |||||
| E. coli-125 | |||||
| CQDGent | Gentamicin sulfate; pyrolysis | Broad-spectrum | S. aureus-1.59 | ROS, Electrostatic interaction, retention of anti-bacterial active structures | 41 |
| E. coli-203.4 | |||||
| CDs-Kan | Kanamycin sulfate; hydrothermal | Broad spectrum | 8 | Retention of anti-bacterial active structures | 42 |
| ACDs | Artemisia argyi L.; combustion | E. coli | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Altering the enzymatic secondary structure | 43 |
| CQDSpds | Spermidine; pyrolysis | Broad spectrum | 2–4 | Electrostatic interaction | 44 |
| CDs | Citric acid + curcumin; hydrothermal broad | Broad spectrum | 375 | Electrostatic interaction | 45 |
| PC-CQDs | L-Glutathione + PEI + citric acid; solvothermal | Broad spectrum | S. aureus-15 | Electrostatic interaction, ROS | 46 |
| E. coli-480 | |||||
| N,P-CDs | Glucose + PEI; hydrothermal | Broad spectrum | S. aureus-7.5 | Electrostatic interaction | 45 |
| E. coli-500 | |||||
| C60-GQD | C60; oxidation | S. aureus | 200 | Gaussian-curvature match | 47 |
| BrCND | HBr, acros organics; combustion | Broad spectrum | — | ROS | 48 |
| CQDs | Citric acid + ethylenediamine; hydrothermal | S. aureus | — | ROS | 49 |
| CDArg | Arginine; pyrolysis | Broad spectrum | S. aureus-62.5 | Electrostatic interaction | 50 |
| E. coli-62.5 | |||||
| Fe-CDs colony counting method | NaFeEDTA; pyrolysis | Broad spectrum | — | ROS | 50 |
| CS/nHA/CD | Citric acid + glycine; microwave-assisted | Broad spectrum | — | Photothermal effect | 51 |
| SCD | Spermine; pyrolysis | E. coli | — | Programmed death | 52 |
| qCQDs | Glucose + dimethyl diallyl ammonium chloride; solvothermal | Broad spectrum | S. epidermidis-12 | Influence on protein activity | 53 |
| S. aureus-25 | |||||
| MR S. aureus-25 | |||||
| E. faecalis-25 | |||||
| E. coli-50 | |||||
| P. aeruginosa-50 | |||||
| CDs | Si-QAC + glycerol solvothermal | S. aureus | 2500 | Electrostatic and hydrophobic interaction | 54 |
| N-CQDs | Polyvinylpyrrolidone; hydrothermal | E. coli | 32 | Electrostatic interaction | 55 |
| N-CQDs | Bis-quaternary ammonium salt; hydrothermal | Broad spectrum | AR E. coli-8 | ROS, electrostatic interaction, Altering bacterial metabolism | 56 |
| E. coli-8 | |||||
| CDLys | Lysine; pyrolysis | Broad spectrum | S. aureus-16 | Electrostatic interaction | 50 |
| E. coli-31.25 | |||||
| FA-CD | Folic acid; solvothermal | S. aureus | 16 | ROS | 57 |
| CDs | F. benghalensis L. extracts | Broad spectrum | S. epidermidis (ATCC 14990)-25 | Cell wall/membrane protrusion and disruption, ROS, Lipid peroxidation, Membrane potential loss, DNA damage | Present work |
| E. cloacae (MTCC 509) MDR-25 | |||||
| P. aeruginosa MDR-2.5 | |||||
| E. coli DH5α-1.25 |
Furthermore, developing environment-responsive mechanisms presents an intriguing avenue to further enhance CDs' anti-microbial function. Environmental factors, such as pH, temperature, and the presence of specific molecules, can be leveraged to trigger controlled ROS release from CDs. This responsive behaviour would allow for precise ROS delivery, ensuring the targeted anti-microbial action while minimizing potential off-target effects. Such innovative anti-microbial functionalities can potentially revolutionize the field of anti-bacterial agents and significantly improve treatment strategies against bacterial infections. It has also been noticed that CDs could alter the protein expression pattern in both E. cloacae (MTCC 509) and P. aeruginosa. We have found up-regulation and down-regulation of some proteins. Interestingly, only this treatment group observed two new protein bands with approximate sizes of 40 kDa (blue arrow) and 35 kDa (green arrow). This suggests that genomic damage/manipulation is also taking place.
The use of CDs in antimicrobial therapy is a promising frontier in biomedical research. CDs showed remarkable antimicrobial properties and proved their efficacy against various pathogens. Their unique physical–chemical properties, such as size, surface load and functional group, contribute to their antimicrobial activity. CDs have been studied for their potential in combating antibiotic-resistant strains and offer alternative or complementary approaches to traditional antimicrobials. Furthermore, CD flexibility allows them to adapt to antimicrobial activity, making them a versatile candidate for targeted therapy. CD integration into antibiotic research represents a step forward and innovative way of addressing the challenges of emerging infectious diseases and antibiotic resistance.
4. Conclusion
CDs have a significant detrimental effect by causing oxidative damage, which disrupts the delicate balance of ROS control in bacterial cells. Despite low doses, the anti-bacterial activity of CDs against E. cloacae and P. aeruginosa demonstrates their potential as effective anti-microbial agents against MDR strains. The capacity of CDs to generate ROS, which then act on the bacterial cell surface, depolarizes and breaks cell membranes, and promotes DNA damage and cytoplasmic leakage of macromolecules, explains how they work. This study has shed light on the potential of CDs as a multifaceted tool for combating biofilm-related infections, thereby offering a new avenue for addressing antibiotic resistance challenges in healthcare settings. Researchers can improve the performance of CDs and unlock their full potential as smart anti-microbial agents by manipulating ROS levels within bacterial cells and introducing environment-responsive mechanisms. We have additionally identified differences in protein expression. These findings lay the groundwork for future research into the management of ROS and developing CDs with increased anti-bacterial properties, suggesting a breakthrough.Conflicts of interest
There are no conflicts of interest to disclose.Acknowledgements
A M P is thankful to the Department of Science and Technology (DST), Ministry of Science and Technology, India for INSPIRE JRF AND SRF fellowship (IF180104).References
- F. Baquero, Int. Microbiol., 2021, 24, 499–506 CrossRef CAS.
- A. Engström, in Understanding the Creeping Crisis, ed. A. Boin, M. Ekengren and M. Rhinard, Springer International Publishing, Cham, 2021, pp. 19–36 Search PubMed.
- M. C. Bagley, J. W. Dale, E. A. Merritt and X. Xiong, Chem. Rev., 2005, 105, 685–714 CrossRef CAS.
- C. Willyard, Nature, 2017, 543, 15 CrossRef CAS PubMed.
- J. M. V. Makabenta, A. Nabawy, C.-H. Li, S. Schmidt-Malan, R. Patel and V. M. Rotello, Nat. Rev. Microbiol., 2021, 19, 23–36 CrossRef CAS.
- K. R. Dalton, C. Rock, K. C. Carroll and M. F. Davis, Antimicrob. Resist. Infect. Control, 2020, 9, 78 CrossRef.
- I. N. Okeke, R. Laxminarayan, Z. A. Bhutta, A. G. Duse, P. Jenkins, T. F. O’Brien, A. Pablos-Mendez and K. P. Klugman, Lancet Infect. Dis., 2005, 5, 481–493 CrossRef CAS PubMed.
- WHO, https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed, (accessed September 14, 2023).
- P. D. Tamma, S. E. Cosgrove and L. L. Maragakis, Clin. Microbiol. Rev., 2012, 25, 450–470 CrossRef CAS.
- A. F. Halbus, T. S. Horozov and V. N. Paunov, ACS Appl. Mater. Interfaces, 2019, 11, 12232–12243 CrossRef CAS.
- M. P. Ajith, S. Pardhiya and P. Rajamani, Small, 2022, 18, 2105579 CrossRef.
- A. Gupta, S. Mumtaz, C.-H. Li, I. Hussain and V. M. Rotello, Chem. Soc. Rev., 2019, 48, 415–427 RSC.
- L. D. Blackman, T. D. Sutherland, P. J. D. Barro, H. Thissen and K. E. S. Locock, Mater. Horiz., 2022, 9, 2076–2096 RSC.
- A. Karnwal, G. Kumar, G. Pant, K. Hossain, A. Ahmad and M. B. Alshammari, ACS Omega, 2023, 8, 13492–13508 CrossRef CAS PubMed.
- A. Sikder, A. K. Pearce, C. M. S. Kumar and R. K. O’Reilly, Mater. Horiz., 2023, 10, 171–178 RSC.
- S. Huang, Y. Song, J.-R. Zhang, X. Chen and J.-J. Zhu, Small, 2023, 17, 2207385 CrossRef.
- A. Cherkasov, K. Hilpert, H. Jenssen, C. D. Fjell, M. Waldbrook, S. C. Mullaly, R. Volkmer and R. E. W. Hancock, ACS Chem. Biol., 2009, 4, 65–74 CrossRef CAS PubMed.
- M. P. Ajith, S. Pardhiya, A. K. Prabhakar and P. Rajamani, Chemosphere, 2022, 303, 135090 CrossRef CAS.
- M. P. Ajith, E. Priyadarshini and P. Rajamani, Bioresour. Technol., 2020, 314, 123786 CrossRef CAS PubMed.
- I. Wiegand, K. Hilpert and R. E. W. Hancock, Nat. Protoc., 2008, 3, 163–175 CrossRef CAS.
- J. S. Sidhu, S. Mayank, T. Pandiyan, N. Kaur and N. Singh, ChemistrySelect, 2017, 2, 9277–9283 CrossRef CAS.
- G. A. O’Toole, J. Visualized Exp., 2011, 2437 Search PubMed.
- P. Bhattacharya, A. Dey and S. Neogi, J. Mater. Chem. B, 2021, 9, 5329–5339 RSC.
- T. Sawada, M. Katayama, S. Takatani and Y. Ohiro, Sci. Rep., 2021, 11, 2873 CrossRef CAS.
- K. S. Ong, Y. L. Cheow and S. M. Lee, J. Adv. Res., 2017, 8, 393–398 CrossRef CAS.
- D. Novo, N. G. Perlmutter, R. H. Hunt and H. M. Shapiro, Cytometry, 1999, 35, 55–63 CrossRef CAS.
- D. Solanky and S. E. Haydel, J. Microbiol. Methods, 2012, 91, 257–261 CrossRef CAS.
- G. Toennies and J. J. Kolb, Anal. Biochem., 1964, 8, 54–69 CrossRef CAS.
- M. Shehadul Islam, A. Aryasomayajula and P. R. Selvaganapathy, Micromachines, 2017, 8, 83 CrossRef.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- A. Manayil Parambil, M. Nabeel Mattath, P. Rajamani, P. V. Pham, G. Kumar and V. K. Ponnusamy, Chem. Eng. J., 2023, 463, 142354 CrossRef CAS.
- S. Dua, P. Kumar, B. Pani, A. Kaur, M. Khanna and G. Bhatt, RSC Adv., 2023, 13, 13845–13861 RSC.
- D. Zhao, X. Li, M. Xu, Y. Jiao, H. Liu, X. Xiao and H. Zhao, Int. J. Biol. Macromol., 2023, 231, 123303 CrossRef CAS PubMed.
- M. Yu, P. Li, R. Huang, C. Xu, S. Zhang, Y. Wang, X. Gong and X. Xing, J. Mater. Chem. B, 2023, 11, 734–754 RSC.
- F. David, A. Berger, R. Hänsch, M. Rohde and E. Franco-Lara, Microb. Cell Fact., 2011, 10, 23 CrossRef CAS.
- B. M. Gyori, G. Venkatachalam, P. S. Thiagarajan, D. Hsu and M.-V. Clement, Redox Biol., 2014, 2, 457–465 CrossRef CAS PubMed.
- R. Paulraj and J. Behari, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2006, 596, 76–80 CrossRef CAS PubMed.
- J. Liang, W. Li, J. Chen, X. Huang, Y. Liu, X. Zhang, W. Shu, B. Lei and H. Zhang, ACS Appl. Bio Mater., 2021, 4, 6937–6945 CrossRef CAS.
- H. Li, J. Huang, Y. Song, M. Zhang, H. Wang, F. Lu, H. Huang, Y. Liu, X. Dai, Z. Gu, Z. Yang, R. Zhou and Z. Kang, ACS Appl. Mater. Interfaces, 2018, 10, 26936–26946 CrossRef CAS.
- X. Zhang, X. Bai, X. Deng, K. Peng, Z. Zheng, J. Xiao, R. Zhang, Z. Huang, J. Huang, M. Chen and S. Weng, Carbon, 2023, 213, 118229 CrossRef CAS.
- P. Li, S. Liu, W. Cao, G. Zhang, X. Yang, X. Gong and X. Xing, Chem. Commun., 2020, 56, 2316–2319 RSC.
- Q. Luo, K. Qin, F. Liu, X. Zheng, Y. Ding, C. Zhang, M. Xu, X. Liu and Y. Wei, Analyst, 2021, 146, 1965–1972 RSC.
- H. Wang, M. Zhang, Y. Ma, B. Wang, M. Shao, H. Huang, Y. Liu and Z. Kang, J. Mater. Chem. B, 2020, 8, 2666–2672 RSC.
- Y.-J. Li, S. G. Harroun, Y.-C. Su, C.-F. Huang, B. Unnikrishnan, H.-J. Lin, C.-H. Lin and C.-C. Huang, Adv. Healthcare Mater., 2016, 5, 2545–2554 CrossRef CAS PubMed.
- N. Prathap, P. Balla, M. S. Shivakumar, G. Periyasami, P. Karuppiah, K. Ramasamy and S. Venkatesan, Sci. Rep., 2023, 13, 9676 CrossRef CAS PubMed.
- X. Hao, L. Huang, C. Zhao, S. Chen, W. Lin, Y. Lin, L. Zhang, A. Sun, C. Miao, X. Lin, M. Chen and S. Weng, Mater. Sci. Eng., C, 2021, 123, 111971 CrossRef CAS PubMed.
- L. Hui, J. Huang, G. Chen, Y. Zhu and L. Yang, ACS Appl. Mater. Interfaces, 2016, 8, 20–25 CrossRef CAS.
- R. Knoblauch, A. Harvey, E. Ra, K. M. Greenberg, J. Lau, E. Hawkins and C. D. Geddes, Nanoscale, 2021, 13, 85–99 RSC.
- C. Tang, C. Liu, Y. Han, Q. Guo, W. Ouyang, H. Feng, M. Wang and F. Xu, Adv. Healthcare Mater., 2019, 8, 1801534 CrossRef.
- P. Li, F. Han, W. Cao, G. Zhang, J. Li, J. Zhou, X. Gong, G. Turnbull, W. Shu, L. Xia, B. Fang, X. Xing and B. Li, Appl. Mater. Today, 2020, 19, 100601 CrossRef.
- Y. Lu, L. Li, M. Li, Z. Lin, L. Wang, Y. Zhang, Q. Yin, H. Xia and G. Han, Bioconjugate Chem., 2018, 29, 2982–2993 CrossRef CAS PubMed.
- W. Bing, H. Sun, Z. Yan, J. Ren and X. Qu, Small, 2016, 12, 4713–4718 CrossRef CAS PubMed.
- C. Zhao, X. Wang, L. Yu, L. Wu, X. Hao, Q. Liu, L. Lin, Z. Huang, Z. Ruan, S. Weng, A. Liu and X. Lin, Acta Biomater., 2022, 138, 528–544 CrossRef CAS.
- J. Yang, G. Gao, X. Zhang, Y.-H. Ma, X. Chen and F.-G. Wu, Carbon, 2019, 146, 827–839 CrossRef CAS.
- N. A. Travlou, D. A. Giannakoudakis, M. Algarra, A. M. Labella, E. Rodríguez-Castellón and T. J. Bandosz, Carbon, 2018, 135, 104–111 CrossRef CAS.
- H. Wang, Z. Song, J. Gu, S. Li, Y. Wu and H. Han, ACS Biomater. Sci. Eng., 2019, 5, 4739–4749 CrossRef CAS PubMed.
- M. Yu, X. Guo, H. Lu, P. Li, R. Huang, C. Xu, X. Gong, Y. Xiao and X. Xing, Carbon, 2022, 199, 395–406 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb02374e |
| This journal is © The Royal Society of Chemistry 2024 |