DOI:
10.1039/D4NA00249K
(Paper)
Nanoscale Adv., 2024,
6, 5020-5030
Effects of silver-decorated PLGA nanoparticles on Staphylococcus epidermidis biofilms and evaluation of the detoxification limit of bacteria against these nanoparticles†
Received
25th March 2024
, Accepted 16th August 2024
First published on 31st August 2024
Abstract
Silver nanoparticles exert high antibacterial activity and thus have been used in polymeric formulations for drug delivery. In recent years, polymeric formulations containing silver nanoparticles have been found to be highly effective against biofilm infections, which are difficult to treat with drugs only. However, the antibacterial effects of silver nanoparticles in polymer composites depending on administration time are still unknown. In this study, we used transmission electron microscopy to investigate the antibacterial activity of silver decorated poly(DL-lactide-co-glycolide) (PLGA) (Ag PLGA) nanoparticles against Staphylococcus epidermidis biofilms at different treatment times. LIVE/DEAD assay result showed that approximately 85% of the bacteria in the biofilms was killed after 6 h of administering the Ag PLGA nanoparticles. The formulation comprising Ag PLGA nanoparticles was found to be highly effective and to exhibit low cytotoxicity. However, silver nanoparticles were ejected from the bacterial cells up to 4 h after treatment administration due to the self-protection properties of the bacteria. On the basis of the results, we propose a potential mechanism for the antibacterial activity of silver nanoparticles per treatment time, taking into account the detoxification activity of bacterial cells. This information can contribute not only to an understanding of foreign-body elimination but also to the design of effective formulations against biofilm infections.
1. Introduction
Biofilms and drug-resistant bacteria are serious problems, and developing pharmaceutical products to tackle these problems is crucial.1,2 Biofilms can cause infections around indwelling medical devices, such as catheters, shunts, grafts, intraocular lenses, valves, and cardiac pacemakers.3,4 Prevention efforts against nosocomial infections are also being made worldwide.5,6 A biofilm is an aggregation of bacteria encased in a thick layer of extracellular polymeric substances (EPSs), which can shield the bacteria from antimicrobial agents. We have previously defined this EPS layer, which envelops the bacterial cells and constitutes the biofilm, as the EPS film.7 Biofilms are also known to be associated with various diseases, such as periodontal disease, chronic osteomyelitis, chronic prostatitis, chronic rhinosinusitis, chronic otitis media, chronic wounds, recurrent urinary tract infections, and dental caries.8,9 Therefore, efficient treatment and formulation development are required. Recently, metal nanoparticles have attracted considerable attention in the field of drug formulation because of their high antimicrobial properties.10,11 In particular, the high antibacterial activity of silver nanoparticles is well known. The antibacterial properties of silver nanoparticles depend on the nanoparticle size, pH, medium, and capping agent.12 As a general mechanism, silver nanoparticles and silver ions can provide antibacterial efficacy by stimulating membrane impairment, reactive oxygen species (ROS) generation, oxidation and denaturation of proteins, dysfunction of mitochondria, damage to DNA, and obstruction of cell propagation.13,14 Some researchers have concentrated on silver nanoparticles synthesized with lasers or those that incorporate other metal species.15–17 Their focus has been on biofilms formed by strains such as Staphylococcus aureus, Streptococcus oralis, Porphyromonas gingivalis, and Enterococcus faecalis, among others. Torres-Mendieta et al.15 have demonstrated that silver nanoparticles doped with iron and cobalt can enhance antibacterial efficacy by disrupting biofilms when subjected to an external magnetic field. However, they did not focus on S. epidermidis biofilms. The mechanisms underlying the antibacterial efficacy and toxicity of silver nanoparticles remain unclear and have yet to be fully elucidated and visualized. Additionally, the toxicity of silver nanoparticles was a problem.18,19 Hence, drug delivery systems based on composites of silver and polymeric materials have been developed recently.20–23 Some researchers have tried to develop silver nanoparticles integrated with polymers without cytotoxicity.24–26 As a result, they have confirmed the high biocompatibility of their designed formulation, the high dispersibility of silver nanoparticles, and its significant antibacterial efficacy. On the other hand, Niidome et al. have reported that the antibacterial activity of silver nanoparticles against planktonic bacteria was reduced by compositing with PLGA.27 However, our target is mature biofilms, not planktonic bacteria. We also focused on the high biocompatibility of PLGA and its high adherence to biofilms and bacteria that we have revealed in our previous studies.28 We have developed submicron-sized silver decorated polymeric nanoparticles for drug delivery and revealed their high efficacy against Staphylococcus epidermidis (S. epidermidis) biofilms without any encapsulation of drugs.28,29 We are exploring the application of Ag PLGA nanoparticles in a paste formulation designed for local administration. The size of these particles ranges from 200 nm to several μm. By tailoring properties such as polymer size and cross-linking to a specific treatment site and intended application, we can finely tune the formulation's degradation rate and achieve controlled release of metal ions. Unlike conventional drug administration, nanoparticles can adhere to the surface of biofilms and penetrate the biofilms. The metal particles and drugs modified or encapsulated in the nanoparticle formulation can then act directly on the EPS film layer of biofilms and can also act on the biofilm-forming bacteria.28,30 In a previous study, we found ejection of silver nanoparticles from S. epidermidis. However, the foreign body exclusion system of bacteria remains unknown and it is also unclear how it relates to the antibacterial activity exhibited by silver nanoparticles. Moreover, the correlation between silver ion release and antibacterial activity was not clear.
In this study, we performed scanning electron microscopy and transmission electron microscopy. For transmission electron microscopy (TEM), we employed two types of TEM techniques, namely, conventional TEM and annular dark field (ADF) scanning transmission electron microscopy (STEM). Silver decorated poly(DL-lactide-co-glycolide) (PLGA) (Ag PLGA) nanoparticles were applied as the antibacterial preparation. PLGA is a biodegradable polymer that can degrade into CO2 and H2O in the human body.31,32 Therefore, we focused on PLGA with high biocompatibility properties to reduce cytotoxicity. Additionally, we prepared drug delivery carriers using modified PLGA to enhance antibacterial activity by adhering to biofilms. We are considering the treatment of biofilm infections using developed nanocarriers, where drug delivery alone is not effectively therapeutic, as an application focus. To obtain information on the behaviors of the Ag PLGA nanoparticles in bacterial cells, samples were prepared using the ultrathin section method. Using two types of TEM techniques, we imaged biofilms treated with Ag PLGA nanoparticles at different treatment times (from the time of treatment to 6 h after treatment administration). We successfully visualized the action of silver decorated polymeric particles on the bacterial cells by capturing changes in the cells over time. Concurrently, the ejection of silver nanoparticles from the bacterial cells due to the self-protection properties of the bacterial cells was observed over this timescale. We revealed the conditions under which the antibacterial effect of Ag PLGA nanoparticles on the bacteria is exerted and the conditions under which the ejection system of the bacteria can be exerted. In this study, we also investigated the correlation between the release rate of silver ions from Ag PLGA nanoparticles and their antibacterial activity.
2. Materials and methods
2.1. Materials
S. epidermidis (ATCC14990T) strain was applied as a sample. This strain was stored at −80 ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C and routinely grown at 37
°C and routinely grown at 37 ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C for 24 h in 0.5% CO2 in Tryptone Soy Broth (TSB) supplemented with 0.25% glucose (Becton, Dickinson and Company Co., USA). Biofilms were prepared using our established procedure.7 The absorbance of the S. epidermidis medium was adjusted to 0.2 using a UV/visible spectrophotometer at a wavelength of 520 nm (Ultrospec2100Pro, GE Healthcare Life Sciences Co., UK). Subsequently, the adjusted medium was incubated in 24 well plates at 37 °C for 24 h in 0.5% CO2 for the formation of the biofilms. For the preparation of Ag PLGA nanoparticles, PLGA (lactide
°C for 24 h in 0.5% CO2 in Tryptone Soy Broth (TSB) supplemented with 0.25% glucose (Becton, Dickinson and Company Co., USA). Biofilms were prepared using our established procedure.7 The absorbance of the S. epidermidis medium was adjusted to 0.2 using a UV/visible spectrophotometer at a wavelength of 520 nm (Ultrospec2100Pro, GE Healthcare Life Sciences Co., UK). Subsequently, the adjusted medium was incubated in 24 well plates at 37 °C for 24 h in 0.5% CO2 for the formation of the biofilms. For the preparation of Ag PLGA nanoparticles, PLGA (lactide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycolide of 75
glycolide of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25; 20
25; 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da; Kuraray Co., Japan), AgNO3 (Nacalai Tesque Inc., Japan) and NaBH4 (Kishida Chemical Co., Japan) were used. PLGA nanoparticles were prepared using the emulsion solvent diffusion method.30 The hydrophilic ionic liquid (1-butyl-3-methylimidazolium tetrafluoroborate [BMIM][BF4]) for the preparation of the scanning electron microscopy (SEM) sample was purchased from Kanto Chemical Co., Japan, and dried in a vacuum desiccator at 60 °C for 1 day. Its water content was below 128 ppm.
000 Da; Kuraray Co., Japan), AgNO3 (Nacalai Tesque Inc., Japan) and NaBH4 (Kishida Chemical Co., Japan) were used. PLGA nanoparticles were prepared using the emulsion solvent diffusion method.30 The hydrophilic ionic liquid (1-butyl-3-methylimidazolium tetrafluoroborate [BMIM][BF4]) for the preparation of the scanning electron microscopy (SEM) sample was purchased from Kanto Chemical Co., Japan, and dried in a vacuum desiccator at 60 °C for 1 day. Its water content was below 128 ppm.
2.2. Ag-decorated PLGA nanoparticles
Ag PLGA nanoparticles were prepared following a previously published procedure.28 Using dynamic light scattering (Zetasizer Nano ZS90, Malvern Instruments Ltd, Malvern, UK), we measured the size and zeta potential of the prepared nanoparticles. To understand silver release from PLGA nanoparticles, the following measurements were performed. Ag PLGA nanoparticles (1.0 mg) were mixed with ultrapure water (10 mL). Then, the solution was put in a warm water bath at 37 °C. Samples were collected at different time points (0 min, 5 min, 30 min, 2 h, 4 h, and 6 h). After being centrifuged, each supernatant was mixed with HNO3 for acidification. Using atomic absorption spectroscopy (280Z AA, Agilent Co., USA), we determined the silver concentration.
2.3. Imaging of samples
S. epidermidis biofilms were grown in a 24 well plate according to the method described in 2.1 Materials and then washed with purified water. Ag PLGA nanoparticle suspensions (1.0 mg mL−1) in TSB medium were added to each well. The bacterial cells were treated with Ag PLGA nanoparticles for 30 min, 2 h, 4 h, and 6 h at 37 °C under 0.5% CO2. It is difficult to observe all of the biofilms in the wells of a 24 well plate by SEM and TEM. Therefore, biofilms in the wells of the 24 well plate were sampled with a cell scraper to observe a portion of the thick biofilm present in each well. In the present study, we performed SEM and TEM. To observe large areas of bacteria treated with Ag PLGA nanoparticles, FE-SEM (JXA-8530FA; JEOL Co., Japan) with an accelerating voltage of 10.0 kV was applied. The sample preparation technique using an ionic liquid is described elsewhere.30 For TEM observations, we applied the ultrathin section method to prepare samples. We prepared TEM samples according to the following procedures. (1) Fixation with 2% glutaraldehyde in cacodylate buffer (pH 7.4) at 4 °C for 2 h, (2) postfixation with 1% osmium tetroxide in the same buffer at 4 °C for 1 h, (3) dehydration and embedding in Quetol-653 (Nisshin EM, Tokyo, Japan), (4) polymerization of the resin at 70 °C for 48 h, (5) preparation of ultrathin sections using an ultramicrotome (PT-X Power Tome, Boeckeler Instruments Inc., USA), and (6) counterstaining of the sections with uranyl acetate and lead citrate. Samples of bacterial cells immediately treated with Ag PLGA nanoparticles were also prepared. TEM (JEM-1400 Plus, JEOL Co., Japan) images were obtained at an accelerating voltage of 80.0 kV, while STEM (JEM-ARM200F, JEOL Co., Japan) images were obtained at an accelerating voltage of 200.0 kV. TEM images for Ag PLGA nanoparticles and PLGA nanoparticles were recorded using a TEM (JEM-ARM200F, JEOL Co., Japan) equipped with an EM-31160 cooling holder (JEOL Co., Japan). The sample was cooled from room temperature to 100 K using liquid nitrogen, and images were obtained with an accelerating voltage of 200.0 kV.
2.4. Antibacterial assay studies
Antibacterial assays were performed using a LIVE/DEAD BacLight bacterial viability kit (catalog number L-13152; Life Technologies Co., Japan). Samples were prepared as follows. Biofilms were grown in a 24 well plate, washed with purified water, and then treated with PLGA nanoparticles (1 mg), Ag PLGA nanoparticles (1 mg), and Ag nanoparticles (1 mg) for 2 h. Additionally, to understand the differences in dosing time, Ag PLGA nanoparticles (1 mg) were treated for 0 min, 30 min, 2 h, 4 h, 6 h, 8 h, 10 h, and 12 h. A detailed protocol for the antibacterial assay using the LIVE/DEAD BacLight bacterial viability kit is described elsewhere (Takahashi et al.,30 2015). The percentage of viable cells after various treatments was calculated (n = 3). After checking whether the variances of the two populations were equal with an F test,33 a two-sample T test34 with equal variances was applied.
2.5. Cytotoxicity studies
The human KB oral epithelial cell line (ATCC) was used to determine the cytotoxicity of PLGA nanoparticles, Ag PLGA nanoparticles, and Ag nanoparticles. The cell lines were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (Life Technologies) at 37 °C under 5% CO2. The cells were seeded onto a 96 well plate at a density of 2.0 × 105 cell per mL. After achieving confluent growth, the medium was replaced with 100 μL of fresh RPMI-1640 medium containing PLGA and Ag PLGA nanoparticles at varying concentrations (0.5, 1.0, and 2.5 mg mL−1). This experimental setup was designed to compare the cytotoxic effects of PLGA nanoparticles with those of Ag PLGA nanoparticles. Additionally, 100 μL of RPMI-1640 medium containing mixtures of Ag PLGA nanoparticles or silver nanoparticles was introduced into the wells, with silver nanoparticle concentrations adjusted to 12.5, 25, and 100 μg mL−1. In this case, the focus was on the concentration of silver nanoparticles rather than the weight of the formulation, in order to assess and compare the cytotoxic effects of Ag PLGA nanoparticles and silver nanoparticles based on their silver content. The 96 well plates were incubated for 6 h and the cytotoxicity was determined using a colorimetric assay involving 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). Detailed procedures are described in a previous study.28 All experiments were performed in duplicate and were repeated 3 times. The p-value is the probability of obtaining results as extreme as the observed results of a statistical hypothesis test, assuming that the null hypothesis is true.
3. Results and discussion
3.1. Physicochemical properties of Ag PLGA nanoparticles
Table 1 shows the physicochemical properties of the PLGA nanoparticles and Ag PLGA nanoparticles. The sizes of the PLGA nanoparticles and Ag PLGA nanoparticles were 221.7 nm and 283.1 nm, respectively. Due to the silver decoration, the Ag PLGA nanoparticles were larger than the PLGA nanoparticles. The zeta potential of the PLGA nanoparticles was −26.9 mV due to the dissociation of the carboxyl group of PLGA. On the other hand, the zeta potential of the Ag PLGA nanoparticles was −17.3 mV. The zeta potential of the Ag PLGA nanoparticles increased to −17.3 mV due to the neutralization of –COO− charges in lactic acid by Ag+, likely through the interaction between Ag+ and –COO− in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. Fig. 1a shows the release profile of silver ions from the PLGA nanoparticles, which was determined using a standard curve obtained with known concentrations of silver ions (Fig. S1†). After 6 h, the release of silver ions from the PLGA nanoparticles in ultrapure water was sustained at 34.2%. From 5 min to 30 min, silver ions were rapidly released from the PLGA nanoparticles (silver ion release rate: 0.3%/min). From 30 min to 1 h, 1 to 2 h, 2 to 4 h, and 4 to 6 h, silver ion release rates were 0.06%/min, 0.09%/min, 0.07%/min, and 0.09%/min, respectively. Fig. 1b shows a TEM image of the Ag PLGA nanoparticles and plain PLGA nanoparticles. The sizes of the PLGA nanoparticles and Ag PLGA nanoparticles were similar to those of physicochemical studies. Silver nanoparticles were successfully decorated onto PLGA nanoparticles.
5. Fig. 1a shows the release profile of silver ions from the PLGA nanoparticles, which was determined using a standard curve obtained with known concentrations of silver ions (Fig. S1†). After 6 h, the release of silver ions from the PLGA nanoparticles in ultrapure water was sustained at 34.2%. From 5 min to 30 min, silver ions were rapidly released from the PLGA nanoparticles (silver ion release rate: 0.3%/min). From 30 min to 1 h, 1 to 2 h, 2 to 4 h, and 4 to 6 h, silver ion release rates were 0.06%/min, 0.09%/min, 0.07%/min, and 0.09%/min, respectively. Fig. 1b shows a TEM image of the Ag PLGA nanoparticles and plain PLGA nanoparticles. The sizes of the PLGA nanoparticles and Ag PLGA nanoparticles were similar to those of physicochemical studies. Silver nanoparticles were successfully decorated onto PLGA nanoparticles.
Table 1 Particle size and zeta potential of the Ag PLGA NPs (n = 3)
| Condition |
PLGA |
Ag PLGA |
| Size |
221.67 ± 6.5 |
283.1 ± 9.8 |
| Zeta potential |
−26.9 ± 1.0 |
−17.3 ± 0.5 |
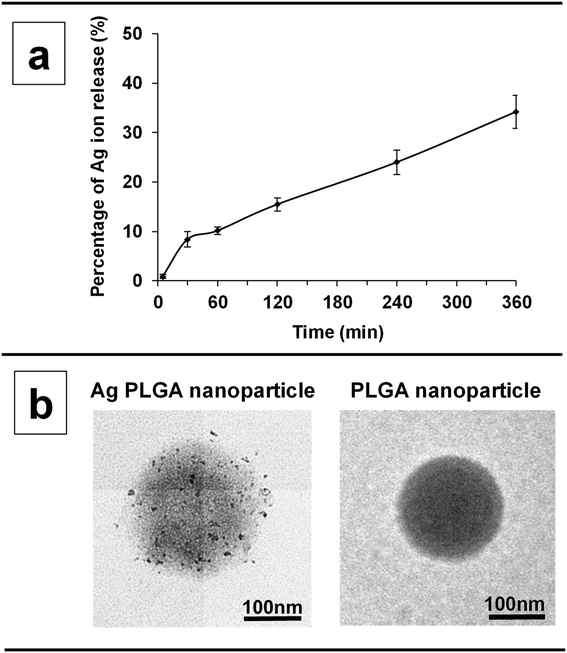 |
| | Fig. 1 (a) Percentage of silver ions released from Ag PLGA nanoparticles at 5, 30, 60, 120, 240, and 360 min. (b) TEM images of an Ag PLGA nanoparticle and PLGA nanoparticle. | |
3.2. Imaging of biofilms treated with Ag PLGA nanoparticles using SEM
SEM observation was performed to observe large areas of the S. epidermidis biofilms treated with Ag PLGA nanoparticles. The S. epidermidis bacterial cells are spherical as shown in Fig. 2a. Once a mature biofilm is formed, a three-dimensional (3D) structure of the biofilm can be observed (Fig. 2b). In our previous studies, we defined a 3D structured biofilm as an aggregation of bacteria covered by a thick EPS film.7,30 Even immediately after treatment with Ag PLGA nanoparticles, the presence of 3D-structured biofilms was still observed (Fig. 2c). A thick EPS film covered 1 μm sized spherical S. epidermidis bacteria. After 30 min of treatment with Ag PLGA nanoparticles, a wall-like EPS film covering individual bacteria was removed (Fig. 2d). After removal of the thick EPS film, we observed the aggregation of the S. epidermidis bacterial cells. After 2 h of treatment with Ag PLGA nanoparticles, Ag PLGA nanoparticles attached to bacterial cell aggregates were observed (Fig. 2e). In this state, large 3D-structured biofilms were not frequently observed. After 4 h of treatment with Ag PLGA nanoparticles, Ag PLGA nanoparticles penetrated into the aggregation of the bacterial cells (Fig. 2f). After 6 h of treatment with Ag PLGA nanoparticles, flat rather than spherical bacteria were observed (Fig. 2g). To further understand the interaction of each S. epidermidis bacterium with Ag PLGA nanoparticles, we performed TEM, as discussed in Section 3.3.
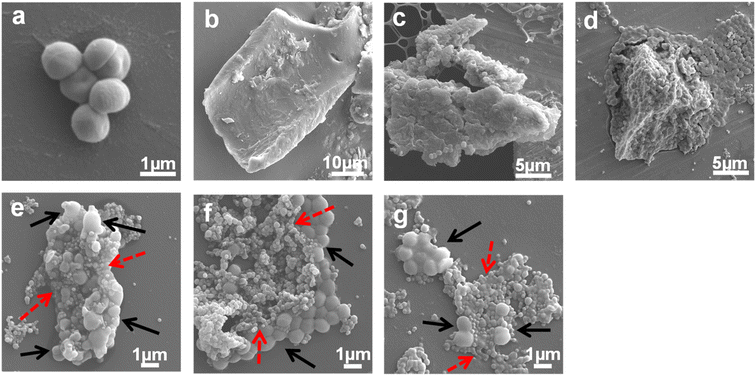 |
| | Fig. 2 SEM images of (a) the S. epidermidis bacterial cells, (b) biofilms without any treatment, and the biofilms treated with Ag PLGA nanoparticles immediately (c), and after (d) 30 min, (e) 2 h, (f) 4 h, and (g) 6 h. The black and dashed red arrows indicate bacterial cells and Ag PLGA nanoparticles. | |
3.3. Imaging of biofilms treated with Ag PLGA nanoparticles using TEM
3.3.1 TEM imaging of biofilms treated with Ag PLGA nanoparticles at various treatment times.
To investigate the activity of PLGA nanoparticles against S. epidermidis biofilms and the self-protection properties of S. epidermidis cells, TEM images were acquired. Fig. 3 shows the TEM images of S. epidermidis biofilms treated with Ag PLGA nanoparticles. At the time of treatment with Ag PLGA nanoparticles, damage to S. epidermidis bacterial cells was not observed (Fig. 3a and b). Spherical Ag PLGA nanoparticles were also observed (dashed red arrow in Fig. 3b). After 30 min of treatment, attachment of Ag PLGA nanoparticles to bacterial cells was observed (dashed red arrows in Fig. 3c). In this state, the shape of some Ag PLGA nanoparticles changed, from spheres to ellipsoids. Interestingly, some silver nanoparticles were observed close to the fibrils of bacterial cells (dashed black arrows in Fig. 3d). To clearly observe fibrils, we applied ADF-STEM imaging of the biofilms after treatment with Ag PLGA nanoparticles for 30 min (Fig. S2†). The benefits of ADF-STEM imaging are discussed in the subsequent section. Silver nanoparticles were found to be incorporated within the fibrils (Fig. S2†). Additionally, small silver nanoparticles were observed in the vicinity of the bacterial cells (red arrows in Fig. S2†). It is well known that fibrils play an important role in the multiplication of bacteria.35,36 Fibrils have been found on a variety of other Gram-negative bacteria and can mediate cell–cell contact among bacteria or between bacteria and eukaryotic host cells. Our previous report showed that tube-like fibrils recruit silver nanoparticles with changes in the size and shape of silver nanoparticles.28 We suggested that silver nanoparticles are ejected through ion efflux. Some reports have mentioned that many bacteria can produce large periplasmic efflux pumps to remove excess toxic metal ions.37–39 In a previous study, we only observed biofilms treated with Ag PLGA nanoparticles for 2 h. To elucidate the mechanism of the self-protection of bacterial cells, we obtained images of biofilms treated with Ag PLGA nanoparticles for different times. In the present study, the fibrils recruited silver nanoparticles after 2 h of treatment. Moreover, the shape of some Ag PLGA nanoparticles changed, and some silver nanoparticles aggregated (red arrow in Fig. 3e). Aggregation of silver nanoparticles was observed at the edge of Ag PLGA nanoparticles. Silver nanoparticles combined, and the combined silver particles were larger than the original silver nanoparticles. The size was approximately 10 to 50 nm (red arrows in Fig. 3f). In this state, the matrix of PLGA nanoparticles was drastically deformed. The results shown in Fig. 3d and S3† suggested that the fibrils attached to the Ag PLGA nanoparticles. The silver nanoparticles were then recruited into the fibril tubes, which promoted the aggregation of silver nanoparticles. In addition to the presence of Ag PLGA nanoparticles, fibrils are likely to be present between bacteria and silver nanoparticle aggregates. In this paper, we have not clarified the presence and location of fibrils; however we would like to address this in future studies. After 4 h of treatment, some parts of the bacterial cells were damaged (yellow arrows in Fig. 3g). Damage to the nucleoid and deformation of the cell membrane and wall were observed (yellow arrows in Fig. 3g). Aggregation of silver nanoparticles was observed (Fig. 3g) after 2 h of treatment. In this state, aggregates of Ag nanoparticles were larger than the aggregates observed after 2 h of treatment (red arrow in Fig. 3g). The size range was approximately 10 to 100 nm (red arrows in Fig. 3h). Bacterial cells seem to have the ability to produce large metal aggregates. We found that the behaviours of Ag PLGA nanoparticles against the bacterial cells of the biofilms and the self-protection of the bacterial cells occurred at the same time. After 6 h of treatment, the bacterial cells were damaged, especially the cell wall. The cell wall was broken and collapsed (yellow arrows in Fig. 3j). Some researchers have reported this phenomenon.40–43 After 6 h of treatment, the inner structure of the bacterial cells was also drastically damaged (Fig. 3j). Lysis of intracellular bacterial cells and aggregation of silver nanoparticles were observed (red arrows in Fig. 3i and j). These phenomena are consistent with the observations of previous studies. In this study, we observed aggregation of silver nanoparticles. Some researchers have reported that EPSs can promote the aggregation of metal particles. They found that the antibacterial activity of aggregated metal particles is less effective than that of nano-sized metal nanoparticles.44,45 Our results suggested that individual silver nanoparticles with high antibacterial activity form aggregates of silver particles with low antibacterial activity because of the detoxification effect of the bacteria. We discuss the role of fibrils by focusing on silver nanoparticles in Section 3.3.2.
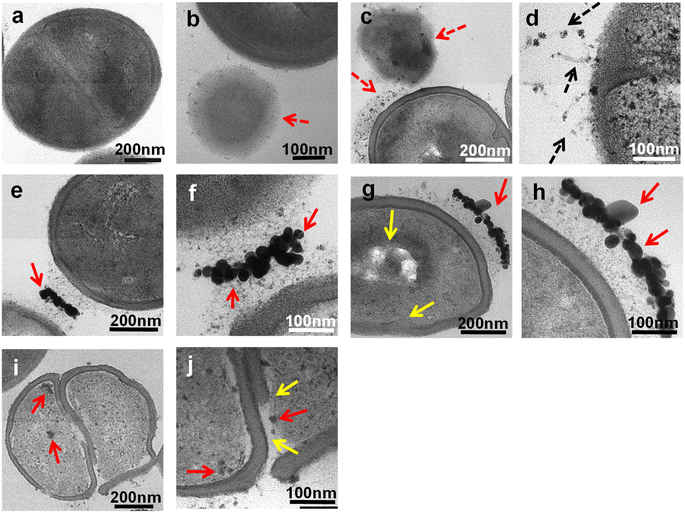 |
| | Fig. 3 TEM images of S. epidermidis biofilms treated with Ag PLGA nanoparticles (a and b) at the time of treatment, (c and d) after 30 min of treatment, (e and f) after 2 h of treatment, (g and h) after 4 h of treatment, and (i and j) after 6 h of treatment. The dashed red, dashed black, red, and yellow arrows indicate Ag PLGA nanoparticles, fibrils, silver nanoparticles, and damaged bacterial cells, respectively. | |
3.3.2 STEM imaging of biofilms treated with Ag PLGA nanoparticles at various treatment times.
We obtained ADF-STEM images to visualize silver nanoparticles clearly. ADF-STEM has some advantages. Z-Dependent high angle ADF images are formed by incoherently scattered electrons. It can take advantage of the Z difference, which is effective for observing metal nanoparticles clearly. After 30 min of treatment, silver nanoparticles were clearly observed in spherical PLGA nanoparticles (dashed red arrow in Fig. 4a). Interestingly, fibrils were attached to Ag PLGA nanoparticles (dashed arrow in Fig. 4a). It seems that fibrils collected only silver nanoparticles within Ag PLGA nanoparticles and sequestered silver nanoparticles from PLGA nanoparticles (red arrows in Fig. 4b). Our previous study indicated that the configuration and shape of fibrils can change easily.28 After 2 h of treatment, we found that some fibrils were similar to those after 30 min of treatment. In some parts, silver aggregation was observed (Fig. 4c and d). After 4 h of treatment, silver aggregation was mainly observed (Fig. 4e). Silver nanoparticles were found at the edge of PLGA nanoparticles. Moreover, the density of aggregates of silver nanoparticles was higher than that observed after 2 h of treatment. In this state, some silver nanoparticles were observed inside the bacterial cells (red arrow in Fig. 4f). The SAED pattern obtained from silver nanoparticles in Fig. 4f consisted of concentric diffraction rings (Fig. 4i). From the SAED pattern, the d-spacing derived from silver was mainly indexed. There are d-space values of 2.35 Å, 2.06 Å, 1.47 Å, and 1.28 Å that correspond to d111 Ag, d200 Ag, d220 Ag, and d311 Ag lattice fringes, respectively. After 6 h of treatment, the bacterial cells were collapsed, and aggregated silver nanoparticles were observed inside the bacterial cells (red arrows in Fig. 4h). In this state, silver nanoparticles were not aggregated in contrast to silver nanoparticles observed at 2 h and 4 h of treatment. The ejection of silver nanoparticles from the bacterial cells seemed difficult after 6 h of treatment. These results indicate that up to 4 h of treatment, the detoxification of bacterial cells was effective. However, after 6 h of treatment, the antibacterial activity of silver nanoparticles was more effective than the detoxification of the bacteria. According to previous reports and our present results, we found that both silver ions and silver nanoparticles contributed to the damage to the bacterial cells of the biofilms.
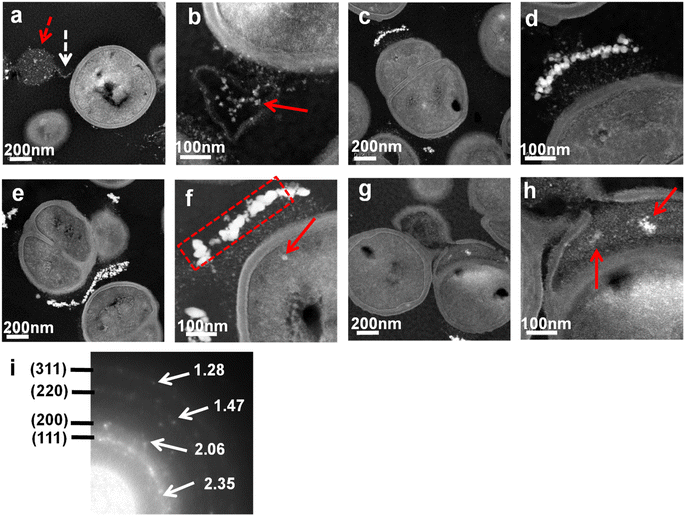 |
| | Fig. 4 ADF-STEM images of S. epidermidis biofilms treated with Ag PLGA nanoparticles for (a and b) 30 min, (c and d) 2 h, (e and f) 4 h, and (g and h) 6 h. (i) SAED pattern of Ag PLGA nanoparticles within the labeled red dotted square in (f). The dashed red, dashed white, and red arrows indicate Ag PLGA nanoparticles, fibrils, and silver nanoparticles. | |
3.4. Antibacterial activity of Ag PLGA nanoparticles against the biofilms
To investigate the antibacterial activity of Ag PLGA nanoparticles against the biofilms, LIVE/DEAD assays were performed. In this study, the biofilms were treated with different types of nanoparticles for different treatment times. The percentages of viable bacteria for the biofilms treated with PLGA nanoparticles, Ag PLGA nanoparticles, and silver nanoparticles for 2 h were 81.2%, 45.2%, and 57.2%, respectively (Fig. 5a). In this study, 1 mg of particles was used for each treatment. The percentage of viable bacteria decreased from 81.2% to 45.2% after silver nanoparticles were incorporated into the PLGA nanoparticles, indicating that silver nanoparticles worked as an antibacterial agent against the bacterial cells of the biofilms. However, the percentages of viable bacteria indicated that treatment with silver nanoparticles alone was less effective than treatment with Ag PLGA nanoparticles. Huangfu et al. reported that EPSs promote the aggregation of silver nanoparticles.46 They revealed that the aggregation of silver nanoparticles in the absence of EPSs follows Derjaguin–Landau–Verwey–Overbeek (DLVO) theory. In the case of Ag PLGA nanoparticles, silver nanoparticles were trapped by the PLGA nanoparticles, inside or on the surface. Therefore, the aggregation of silver nanoparticles was prevented by integrating silver nanoparticles with PLGA. The percentages of viable bacteria after the biofilm was treated with Ag PLGA nanoparticles for 30 min, 2 h, 4 h, 6 h, 8 h, 10 h, and 12 h were 93.0%, 46.4%, 31.1%, 14.2%, 6.3%, 1.4%, and 1.1%, respectively (Fig. 5b). After 10 h and 12 h of treatment, the bacterial cells of the biofilms were nearly dead. It is clear that the number of dead bacteria increased as the time of treatment increased. This result indicates that the antibacterial efficacy of Ag PLGA nanoparticles was high, even with the ejection of silver nanoparticles by the bacteria. Interestingly, there was a significant increase in the number of dead bacteria between 4 h and 6 h treatments. In particular, after 6 h of treatment with Ag PLGA nanoparticles, TEM and STEM images showed that most bacterial cells were damaged and collapsed (Fig. 3i, j, 4g and h). Silver ion release results showed that 24% and 34% of silver ions were released after 4 h and 6 h of Ag PLGA nanoparticle treatment, respectively. It is likely that when more than 25% of Ag ions are released from PLGA nanoparticles, the bacteria of the biofilms will be killed by the antibacterial activity of Ag PLGA nanoparticles without the ejection system of the bacteria being fully activated. The results of the LIVE/DEAD assays are consistent with the results of STEM and TEM observations. We confirmed from two LIVE/DEAD assays that Ag PLGA nanoparticles are effective against the bacterial cells of the biofilms.
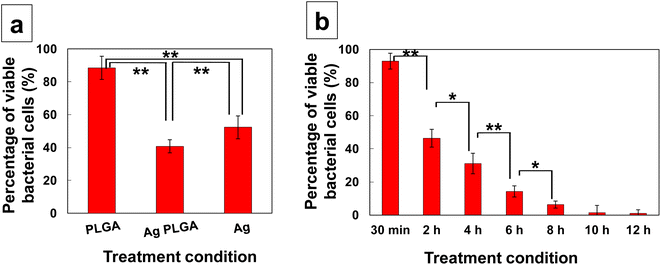 |
| | Fig. 5 (a) Percentage of viable bacterial cells after various treatments (PLGA nanoparticles, Ag PLGA nanoparticles, and silver nanoparticles) for 2 h. (b) Percentage of viable bacterial cells after treatment with Ag PLGA nanoparticles for 30 min, 2 h, 4 h, 6 h, 8 h, 10 h, and 12 h. Data are shown as mean ± SD (n = 3). **Significant difference between each treatment (p < 0.01). *Significant difference between each treatment (p < 0.05). | |
3.5. Cytotoxicity of Ag PLGA nanoparticles
In this study, we used MTT assay to compare the cytotoxicity of (1) PLGA nanoparticles and Ag PLGA nanoparticles as well as (2) silver nanoparticles and Ag PLGA nanoparticles. To compare the cytotoxicity of PLGA nanoparticles and Ag PLGA nanoparticles, PLGA nanoparticle concentrations of 0.5, 1.0, and 2.5 mg mL−1 were used. To compare the cytotoxicity of silver nanoparticles and Ag PLGA nanoparticles, silver nanoparticle concentrations of 12.5, 25, and 100 μg mL−1 were applied. PLGA nanoparticles and Ag PLGA nanoparticles in the dose range of 0.5 to 2.5 mg mL−1 did not significantly affect KB oral epithelial cell line viability (Fig. 6a). In contrast, silver nanoparticles at concentrations of 25 and 100 μg mL−1 were toxic to the KB oral epithelial cell line (Fig. 6b). Ag PLGA nanoparticles were not toxic to the KB oral epithelial cell line, even at the silver-nanoparticle concentration of 100 μg mL−1. By integrating silver nanoparticles with PLGA, the toxicity of silver nanoparticles was drastically reduced. Ag PLGA nanoparticles exhibit antibacterial activity against biofilm-forming bacteria with low cytotoxicity, indicating that Ag PLGA nanoparticles are applicable for the prevention of biofilm infections and removal of biofilms. In this study, PLGA with a molecular weight of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da was prepared using lactide and glycolide in a ratio of 75
000 Da was prepared using lactide and glycolide in a ratio of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25. In the future, we can change the sustained release profiles of silver nanoparticles by changing the composition of the polymer. It is also possible to design polymeric formulations with long-term sustained release of silver nanoparticles.
25. In the future, we can change the sustained release profiles of silver nanoparticles by changing the composition of the polymer. It is also possible to design polymeric formulations with long-term sustained release of silver nanoparticles.
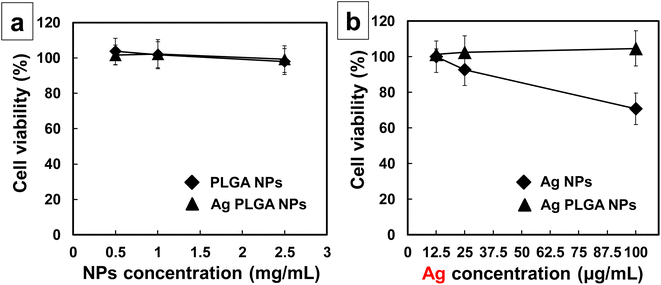 |
| | Fig. 6 Cytotoxic side effects of Ag PLGA nanoparticles at (a) different nanoparticle concentrations (0.5, 1.0, and 2.5 mg mL−1) or (b) different silver concentrations (12.5, 25, and 100 μg mL−1) in the KB oral epithelial cell line at 37 °C. All nanoparticles were incubated for 6 h. The viability of the treated KB oral epithelial cell line was determined using an MTT assay. Data are shown as mean ± SD (n = 3). | |
3.6. Mechanism of the self-protection of S. epidermidis bacterial cells against Ag PLGA nanoparticles and the antibacterial activity of Ag PLGA nanoparticles
Fig. 7 shows the correlation between silver ion release rate from Ag PLGA nanoparticles and S. epidermidis bacterial cell viability. It was indicated that a certain period of time is required for silver ions to act on bacteria and lead to their death after the release of silver ions. The EPS film layer of biofilms may have functioned as a barrier. After 2 h of Ag PLGA nanoparticle administration, a significant decrease in bacterial viability was observed. Thereafter, bacterial viability decreased continuously and steadily up to 6 h. The antibacterial activity of silver nanoparticles has been explained by some researchers. Silver nanoparticles can release silver ions, which exhibit bactericidal activity.12 Silver ions can interact with the nucleosides of nucleic acids to form complexes. Moreover, silver ions can adhere to the cytoplasm and cell wall of bacterial cells through electrostatic attraction and reactions with sulfur proteins. Subsequently, silver ions penetrate the bacterial cells and disrupt the bacterial casings.47 Respiratory enzymes are then inactivated and this triggers the production of reactive oxygen species (ROS),48 and the release of adenosine triphosphate (ATP) is inhibited. ROS such as superoxide radicals (˙O2−), hydroxyl radicals (˙OH), hydrogen peroxide (H2O2), and singlet oxygen (1O2) disrupt the cellular membrane and alter deoxyribonucleic acid (DNA). Silver ions can interact with the phosphorus and sulfur moieties of DNA, which damages DNA and disrupts DNA replication and cell proliferation. Silver ions can also inhibit protein synthesis by denaturing cytoplasmic ribosomal components.49 In addition to silver ions, silver nanoparticles can also exhibit antibacterial activity. Silver nanoparticles can denature cell membranes and penetrate bacterial cells, which changes the arrangement of cell membranes.50 Based on these findings and the results obtained, a potential mechanism can be proposed (Fig. 8). The mechanism of the antibacterial activity of Ag PLGA nanoparticles against S. epidermidis bacterial cells in biofilms, taking into account the detoxification effects of bacteria on silver nanoparticles, is as follows: (1) Ag PLGA nanoparticles initially attach to biofilms. (2) Silver ions and silver nanoparticles released from the Ag PLGA nanoparticles attack biofilms. (3) Ag PLGA nanoparticles attach to biofilm forming bacteria. (4) Silver nanoparticles are ejected from the bacterial cells via fibrils and large aggregates of silver nanoparticles are formed. (5) Continuously released silver ions and silver nanoparticles from the Ag PLGA nanoparticles force the collapse of bacterial cells.
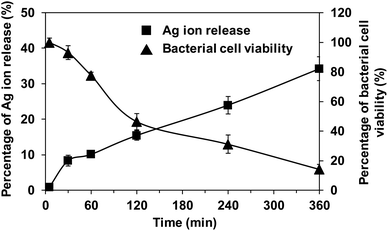 |
| | Fig. 7 Correlation between the silver ion release rate and bacterial cell viability. ■ and ▲ show the percentage of silver ion release and the percentage of bacterial cell viability, respectively. | |
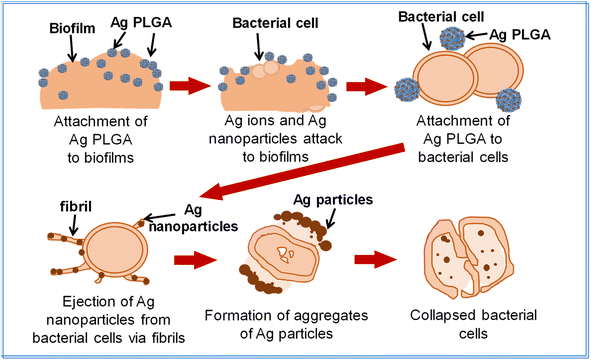 |
| | Fig. 8 Schematic image of the mechanism of the antibacterial activity of Ag PLGA nanoparticles against S. epidermidis biofilms. | |
4. Conclusions
In the present study, the detoxification of S. epidermidis cells and the activity of silver nanoparticle-decorated PLGA nanoparticles against bacteria of biofilms at different treatment times were investigated by acquiring images using two types of TEM techniques. On the basis of the results of this study, we proposed the antibacterial mechanism of Ag PLGA nanoparticles against S. epidermidis bacterial cells of biofilms. Additionally, the self-protection of the bacterial cells against Ag PLGA nanoparticles was clearly visualized. Ag PLGA nanoparticles showed high efficacy against thick EPS biofilms. After 6 h of treatment with Ag PLGA nanoparticles, over 85% of the bacterial cells of the biofilms were dead. The low cytotoxicity of Ag PLGA nanoparticles indicates their potential for application as a formulation. This study elucidated that a certain period of time is required for silver ions to exert antibacterial effects and cause bacterial cell death, based on the correlation between the silver ion release rate from Ag PLGA nanoparticles and bacterial cell viability. It is also possible to change the sustained release rate and antimicrobial effect by changing the shape and size of the silver nanoparticles and the composition of the polymeric base material. Formulations comprising Ag PLGA nanoparticles can be used not only against S. epidermidis but also against other bacteria, e.g., Gram-negative bacteria. In the future, we will design various types of metal decorated polymeric particles that are highly effective against multidrug-resistant bacteria and biofilms formed from multiple bacteria. We will also design formulations that are adaptable for both the prevention of biofilm infections and the treatment of biofilm infections. These findings from this study will contribute to research in the fields of pharmacy, bacteriology and biology.
Data availability
The data that support the findings of this study are available from the corresponding author, C. T., upon reasonable request.
Author contributions
C. T. conceived the research, designed the experiments, analysed the data, and prepared the manuscript draft. K. M. supported TEM experiments and finalized the manuscript. All authors have given approval to the final version of the manuscript.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
This study was partially supported by JSPS KAKENHI Grant Numbers JP17KK0178 and JP18K18388.
References
- J. L. Del Pozo, Expert Rev. Anti-Infect. Ther., 2018, 16, 51–65 CrossRef CAS.
- E. Christaki, M. Marcou and A. Tofarides, J. Mol. Evol., 2020, 88, 26–40 CrossRef CAS.
- J. P. O'Gara and H. Humphreys, J. Med. Microbiol., 2001, 50, 582–587 CrossRef.
- D. Mack, H. Rohde, L. G. Harris, A. P. Davies, M. A. Horstkotte and J. K. M. Knobloch, Int. J. Artif. Organs, 2006, 29, 343–359 CrossRef CAS.
- I. Francolini and G. Donelli, FEMS Immunol. Med. Microbiol., 2010, 59, 227–238 CrossRef CAS PubMed.
- N. K. D. Ragupathi, B. Veeraraghavan, E. Karunakaran and P. N. Monk, Front. Med., 2022, 9, 987011 CrossRef PubMed.
- C. Takahashi, G. Kalita, N. Ogawa, K. Moriguchi, M. Tanemura, Y. Kawashima and H. Yamamoto, Anal. Bioanal. Chem., 2015, 407, 1607–1613 CrossRef CAS.
- M. Burmolle, T. R. Thomsen, M. Fazli, I. Dige, L. Christensen, P. Homøe, M. Tvede, B. Nyvad, T. Tolker-Nielsen, M. Givskov and C. Moser, FEMS Immunol. Med. Microbiol., 2010, 59, 324–336 CrossRef.
- M. Elashiry, M. M. Meghil, R. M. Arce and C. W. Cutler, J. Periodontal Res., 2019, 54, 1 CrossRef CAS PubMed.
- A. M. Fayaz, K. Balaji K, M. Yadav, R. Girilal, P. T. Kalaichelvan and R. Venketesan, Nanomedicine, 2020, 6, 103–109 CrossRef.
- Y. N. Slavin, J. Asnis, U. O. Häfeli and H. Bach, J. Nanobiotechnol., 2017, 15, 65 CrossRef PubMed.
- R. A. Bapat, T. V. Chaubal, C. P. Joshi, P. R. Bapat, H. Choudhury, M. Pandey, B. Gorain and P. Kesharwani, Mater. Sci. Eng., C, 2018, 91, 881–898 CrossRef CAS.
- K. B. R. Ahmed, A. M. Nagy, R. P. Brown, Q. Zhang, S. G. Malghan and P. L. Goering, Toxicol. In Vitro, 2017, 38, 179–192 CrossRef.
- M. Ema, H. Okuda, M. Gamo and K. A. Honda, Reprod. Toxicol., 2017, 67, 149–164 CrossRef CAS PubMed.
- R. Torres-Mendieta, N. H. A. Nguyen, A. Guadagnini, J. Semerad, D. Łukowiec and P. Parma,
et al.
, Nanoscale, 2022, 14, 18143–18156 RSC.
- R. Pérez-Tanoira, M. Fernández-Arias, C. Potel, R. Carballo-Fernández, S. Pérez-Castro, M. Boutinguiza, M. Górgolas, F. Lusquiños and J. Pou, Int. J. Mol. Sci., 2022, 23, 12027 CrossRef.
- N. Heine, K. Doll-Nikutta, F. Stein, J. Jakobi, A. Ingendoh-Tsakmakidis, C. Rehbock, A. Winkel, S. Barcikowski and M. Stiesch, Sci. Rep., 2024, 14, 3405 CrossRef CAS.
- S. Lekamge, A. F. Miranda, A. Abraham, V. Li, R. Shukla, V. Bansal and D. Nugegoda, Front. Environ. Sci., 2018, 6, 152 CrossRef.
- Z. Ferdous and A. Nemmar, Int. J. Mol. Sci., 2020, 21, 2375 CrossRef CAS PubMed.
- L. Guo, W. Yuan, Z. Lu and C. M. Li, Colloids Surf., A, 2013, 439, 69–83 CrossRef CAS.
- M. Stevanović, I. Bračko, M. Milenković, N. Filipović, J. Nunić, M. Filipič and D. P. Uskoković, Acta Biomater., 2014, 10, 151–162 CrossRef PubMed.
- I. Tse, A. Jay, I. Na, S. Murphy, N. Niño-Martínez, G. A. Martínez-Castañon, J. Magrill and H. Bach, Materials, 2021, 14, 7681 CrossRef CAS.
- T. V. Mirolyubova, L. V. Redina, I. A. Chmutin and A. A. Kurnosova, Fibre Chem., 2023, 54, 303–308 CrossRef.
- K. Zou, Q. Liu, J. Chena and J. Du, Polym. Chem., 2014, 5, 405–411 RSC.
- C. Wu, G. Zhang, T. Xia, Z. Li, K. Zhao, Z. Deng, D. Guo and B. Peng, Mater. Sci. Eng., C, 2015, 55, 155–165 CrossRef CAS.
- F. Chen, J. Han, Z. Guo, C. Mu, C. Yu, Z. Ji, L. Sun, Y. Wang and J. Wang, Materials, 2023, 16, 3895 CrossRef CAS.
- R. Nawata, S. Maruyama, W. Xu and T. Niidome, Chem. Lett., 2024, 53, upae084 CrossRef.
- C. Takahashi C, N. Matsubara, Y. Akachi, N. Ogawa, G. Kalita, T. Asaka, M. Tanemura, Y. Kawashima and H. Yamamoto, Mater. Sci. Eng., C, 2017, 72, 143–149 CrossRef.
- C. Takahashi, T. Yamada, S. Yagi, T. Murai and S. Muto, Mater. Sci. Eng., C, 2021, 121, 111718 CrossRef CAS.
- C. Takahashi, N. Ogawa, Y. Kawashima and H. Yamamoto, Microscopy, 2015, 64, 169–180 CrossRef CAS PubMed.
- Y. Kawashima, H. Yamamoto, H. Takeuchi, T. Hino and T. Niwa, Eur. J. Pharm. Biopharm., 1998, 45, 41–48 CrossRef CAS PubMed.
- E. M. Elmowafy, M. Tiboni and M. E. Soliman, J. Pharm. Invest., 2019, 49, 347–380 CrossRef CAS.
-
G. W. Snedecor and W. G. Cochran, Statistical Methods, Iowa State University Press, 8th edn, 1989 Search PubMed.
- T. K. Kim, Korean J. Anesthesiol., 2015, 68, 540–546 CrossRef PubMed.
- M. A. Banner, J. G. Cunniffe, R. L. Macintosh, T. J. Foster, H. Rohde, D. Mack, E. Hoyes, J. Derrick, M. Upton and P. S. Handley, J. Bacteriol., 2007, 189, 2793–2804 CrossRef CAS PubMed.
- M. Bagheri, H. Nikolenko, S. Arasteh, N. Rezaei, M. Behzadi, M. Dathe and R. E. W. Hancock, ACS Appl. Mater. Interfaces, 2020, 12, 26852–26867 CrossRef CAS PubMed.
- S. Silver, FEMS Microbiol. Rev., 2003, 27, 341–353 CrossRef CAS PubMed.
- D. H. Nies, FEMS Microbiol. Rev., 2003, 27, 313–339 CrossRef CAS PubMed.
- K. N. Chacón, T. D. Mealman, M. M. McEvoy and N. J. Blackburn, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 15373–15378 CrossRef PubMed.
- P. D. Bragg and D. J. Rainnie, Can. J. Microbiol., 1974, 20, 883–889 CrossRef CAS PubMed.
- R. T. Belly and G. C. Kydd, Dev. Ind. Microbiol., 1982, 23, 567–577 Search PubMed.
- I. Sondi and B. Salopek-Sondi, J. Colloid Interface Sci., 2004, 275, 177–182 CrossRef CAS PubMed.
- W. K. Jung, H. C. Koo, K. W. Kim, S. Shin, S. H. Kim and Y. H. Park, Appl. Environ. Microbiol., 2008, 74, 2171–2178 CrossRef CAS PubMed.
- J. Hou, L. Miao, C. Wang, P. Wang, Y. Ao, J. Qian and S. Dai, J. Hazard. Mater., 2014, 276, 164–170 CrossRef CAS PubMed.
- N. Joshi, B. T. Ngwenya and C. E. French, J. Hazard. Mater., 2012, 241–242, 363–370 CrossRef CAS PubMed.
- W. Chen, J. Song, S. Jiang, Q. He, J. Ma and X. Huangfu, Front. Environ. Sci. Eng., 2021, 16, 16 CrossRef.
- S. Khorrami, A. Zarrabi, M. Khaleghi, M. Danaei and M. Mozafari, Int. J. Nanomed., 2018, 13, 8013–8024 CrossRef CAS.
- S. S. Das, S. Alkahtani, P. Bharadwaj, M. T. Ansari, M. D. ALKahtani, Z. Pang, M. S. Hasnain, A. K. Nayak and T. Aminabhavi, Int. J. Pharm., 2020, 585, 119556 CrossRef CAS.
- V. Pareek, R. Gupta and J. Panwar, Mater. Sci. Eng., C, 2018, 90, 739–749 CrossRef CAS.
- C. Liao, Y. Li and S. C. Tjong, Int. J. Mol. Sci., 2019, 20, 449 CrossRef.
|
| This journal is © The Royal Society of Chemistry 2024 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a and
Keiichi
Moriguchi
*a and
Keiichi
Moriguchi
 bc
bc
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C and routinely grown at 37
°C and routinely grown at 37 ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C for 24 h in 0.5% CO2 in Tryptone Soy Broth (TSB) supplemented with 0.25% glucose (Becton, Dickinson and Company Co., USA). Biofilms were prepared using our established procedure.7 The absorbance of the S. epidermidis medium was adjusted to 0.2 using a UV/visible spectrophotometer at a wavelength of 520 nm (Ultrospec2100Pro, GE Healthcare Life Sciences Co., UK). Subsequently, the adjusted medium was incubated in 24 well plates at 37 °C for 24 h in 0.5% CO2 for the formation of the biofilms. For the preparation of Ag PLGA nanoparticles, PLGA (lactide
°C for 24 h in 0.5% CO2 in Tryptone Soy Broth (TSB) supplemented with 0.25% glucose (Becton, Dickinson and Company Co., USA). Biofilms were prepared using our established procedure.7 The absorbance of the S. epidermidis medium was adjusted to 0.2 using a UV/visible spectrophotometer at a wavelength of 520 nm (Ultrospec2100Pro, GE Healthcare Life Sciences Co., UK). Subsequently, the adjusted medium was incubated in 24 well plates at 37 °C for 24 h in 0.5% CO2 for the formation of the biofilms. For the preparation of Ag PLGA nanoparticles, PLGA (lactide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycolide of 75
glycolide of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25; 20
25; 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da; Kuraray Co., Japan), AgNO3 (Nacalai Tesque Inc., Japan) and NaBH4 (Kishida Chemical Co., Japan) were used. PLGA nanoparticles were prepared using the emulsion solvent diffusion method.30 The hydrophilic ionic liquid (1-butyl-3-methylimidazolium tetrafluoroborate [BMIM][BF4]) for the preparation of the scanning electron microscopy (SEM) sample was purchased from Kanto Chemical Co., Japan, and dried in a vacuum desiccator at 60 °C for 1 day. Its water content was below 128 ppm.
000 Da; Kuraray Co., Japan), AgNO3 (Nacalai Tesque Inc., Japan) and NaBH4 (Kishida Chemical Co., Japan) were used. PLGA nanoparticles were prepared using the emulsion solvent diffusion method.30 The hydrophilic ionic liquid (1-butyl-3-methylimidazolium tetrafluoroborate [BMIM][BF4]) for the preparation of the scanning electron microscopy (SEM) sample was purchased from Kanto Chemical Co., Japan, and dried in a vacuum desiccator at 60 °C for 1 day. Its water content was below 128 ppm.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. Fig. 1a shows the release profile of silver ions from the PLGA nanoparticles, which was determined using a standard curve obtained with known concentrations of silver ions (Fig. S1†). After 6 h, the release of silver ions from the PLGA nanoparticles in ultrapure water was sustained at 34.2%. From 5 min to 30 min, silver ions were rapidly released from the PLGA nanoparticles (silver ion release rate: 0.3%/min). From 30 min to 1 h, 1 to 2 h, 2 to 4 h, and 4 to 6 h, silver ion release rates were 0.06%/min, 0.09%/min, 0.07%/min, and 0.09%/min, respectively. Fig. 1b shows a TEM image of the Ag PLGA nanoparticles and plain PLGA nanoparticles. The sizes of the PLGA nanoparticles and Ag PLGA nanoparticles were similar to those of physicochemical studies. Silver nanoparticles were successfully decorated onto PLGA nanoparticles.
5. Fig. 1a shows the release profile of silver ions from the PLGA nanoparticles, which was determined using a standard curve obtained with known concentrations of silver ions (Fig. S1†). After 6 h, the release of silver ions from the PLGA nanoparticles in ultrapure water was sustained at 34.2%. From 5 min to 30 min, silver ions were rapidly released from the PLGA nanoparticles (silver ion release rate: 0.3%/min). From 30 min to 1 h, 1 to 2 h, 2 to 4 h, and 4 to 6 h, silver ion release rates were 0.06%/min, 0.09%/min, 0.07%/min, and 0.09%/min, respectively. Fig. 1b shows a TEM image of the Ag PLGA nanoparticles and plain PLGA nanoparticles. The sizes of the PLGA nanoparticles and Ag PLGA nanoparticles were similar to those of physicochemical studies. Silver nanoparticles were successfully decorated onto PLGA nanoparticles.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da was prepared using lactide and glycolide in a ratio of 75
000 Da was prepared using lactide and glycolide in a ratio of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25. In the future, we can change the sustained release profiles of silver nanoparticles by changing the composition of the polymer. It is also possible to design polymeric formulations with long-term sustained release of silver nanoparticles.
25. In the future, we can change the sustained release profiles of silver nanoparticles by changing the composition of the polymer. It is also possible to design polymeric formulations with long-term sustained release of silver nanoparticles.







