Open Access Article
Open Access ArticleCombinatorial extracellular matrix cues with mechanical strain induce differential effects on myogenesis in vitro†
Alex H. P.
Chan‡
 abc,
Ishita
Jain‡
abc,
Beu P.
Oropeza
abc,
Tony
Zhou
ab,
Brandon
Nelsen
d,
Nicholas A.
Geisse
d and
Ngan F.
Huang
abc,
Ishita
Jain‡
abc,
Beu P.
Oropeza
abc,
Tony
Zhou
ab,
Brandon
Nelsen
d,
Nicholas A.
Geisse
d and
Ngan F.
Huang
 *abce
*abce
aStanford Cardiovascular Institute, Stanford, CA, USA. E-mail: ngantina@stanford.edu; Tel: +1 650-849-0559
bDepartment of Cardiothoracic Surgery, Stanford University, Stanford, CA, USA
cCenter for Tissue Regeneration, Repair and Restoration, Veterans Affairs Palo Alto Health Care System, Palo Alto, CA, USA
dCuri Bio, Inc., Seattle, WA, USA
eDepartment of Chemical Engineering, Stanford University, Stanford, CA 94305, USA
First published on 21st July 2023
Abstract
Skeletal muscle regeneration remains a clinical unmet need for volumetric muscle loss and atrophy where muscle function cannot be restored to prior capacity. Current experimental approaches do not account for the complex microenvironmental factors that modulate myogenesis. In this study we developed a biomimetic tissue chip platform to systematically study the combined effects of the extracellular matrix (ECM) microenvironment and mechanical strain on myogenesis of murine myoblasts. Using stretchable tissue chips composed of collagen I (C), fibronectin (F) and laminin (L), as well as their combinations thereof, we tested the addition of mechanical strain regimens on myogenesis at the transcriptomic and translational levels. Our results show that ECMs have a significant effect on myotube formation in C2C12 murine myoblasts. Under static conditions, laminin substrates induced the longest myotubes, whereas fibronectin produced the widest myotubes. Combinatorial ECMs showed non-intuitive effects on myotube formation. Genome-wide analysis revealed the upregulation in actin cytoskeletal related genes that are suggestive of myogenesis. When mechanical strain was introduced to C + F + L combinatorial ECM substrates in the form of constant or intermittent uniaxial strain at low (5%) and high (15%) levels, we observed synergistic enhancements in myotube width, along with transcriptomic upregulation in myosin heavy chain genes. Together, these studies highlight the complex role of microenvironmental factors such as ECM interactions and strain on myotube formation and the underlying signaling pathways.
1. Introduction
Skeletal muscle plays an important role in bearing weight and controlling movement of the body. Although skeletal muscle has high regenerative capacity, the ability to restore full muscle function is permanently impaired by traumatic injuries like volumetric muscle loss.1 Clinical treatments to muscle injuries include the grafting of autologous muscle flaps or scar tissue debridement.2–4 However, these therapies are often associated with significant donor site morbidity due to impaired endogenous regeneration and revascularization capacity. Current experimental approaches to induce muscle regeneration after traumatic injuries include tissue engineering, biological scaffolds, cell transplantation, rehabilitative exercise, and pro-regenerative soluble factors,5–12 although clinical translation is lagging.Muscle regeneration is a complex process in which satellite cells activate into myogenic precursors that proliferate, followed by cell cycle withdrawal and myoblast fusion to form multi-nucleated myotubes.13 This process is orchestrated by numerous signaling pathways, including insulin-like growth factor (IGF-1), notch, prostaglandin E2, and peroxisome proliferator activated receptor gamma (PPARG).14–16 It is well-recognized that the skeletal muscle microenvironment plays a critical role in skeletal muscle regeneration. These microenvironmental factors include the extracellular matrix (ECM) protein composition, biophysical patterning, and substrate stiffness.17 The ECM is important to normal muscle function. Skeletal muscle ECM is composed of a combination of proteins, including laminin, collagen I, collagen I, elastin and proteoglycans.18 Therefore, combinatorial ECMs may better reflect the native muscle ECM environment. Since these factors interact directly with muscle precursor cells, which give rise to multi-nucleated myofibers, it is generally agreed that these microenvironmental factors can modulate myogenesis.19 Additionally, owing to the dynamic nature of skeletal muscle, studying myogenesis in the presence of strain can reflect rehabilitative strategies to promote muscle regeneration. Rehabilitative exercise has previously been shown to improve muscle regeneration in preclinical models of muscle injury.20 Similarly, strain effects on myogenesis have been studied in vitro21,22 in which it was shown that strain mediated alignment of collagen I fibers enhanced the myogenesis of myoblasts, thereby recapitulating in vivo microenvironment.23
To study muscle myogenesis in an improved biomimetic microenvironment, we developed a stretchable tissue chip technology platform composed of customized combinatorial ECMs and strain regimens. Using this platform, we tested the effects of both intermittent and constant strain regimens at both low and high strain magnitude, in conjunction with combinatorial ECMs, on in vitro myogenesis. Our in vitro and transcriptomic results reveal complex interactions between ECM components and strain regimens on the myogenesis process.
2. Experimental
2.1. Cell culture
Murine myoblasts (C2C12, passages 4-7, ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. Cells were sub-cultured when they reached 80% confluency.2.2. ECM surface functionalization of stretchable tissue chips
24-Well uniaxial stretch chambers with aligned nanopatterns in the direction of strain (Curi Bio, USA) were disinfected with 70% ethanol and then modified with poly D-lysine (0.1 mg mL−1) for 16 hours at 37 °C to render them surface-reactive. Next, collagen I (C, Corning), fibronectin (F, Sigma) laminin (L, Life Technologies), and their combinations thereof were incubated at a total concentration of 10 μg mL−1 for 16 hours at 37 °C at equimolar ratios.2.3. Cell seeding
Murine myoblasts were seeded into 24-well stretch chambers having aligned nanopatterns parallel to the direction of strain (Curi Bio, USA) at density of 3 × 104 cells per cm2. Cells were grown to confluency before induction of myogenesis by switching to differentiation medium (DMEM with 2% horse serum and 1% penicillin/streptomycin) and simultaneously initiating mechanical stimulation.2.4. Mechanical stimulation
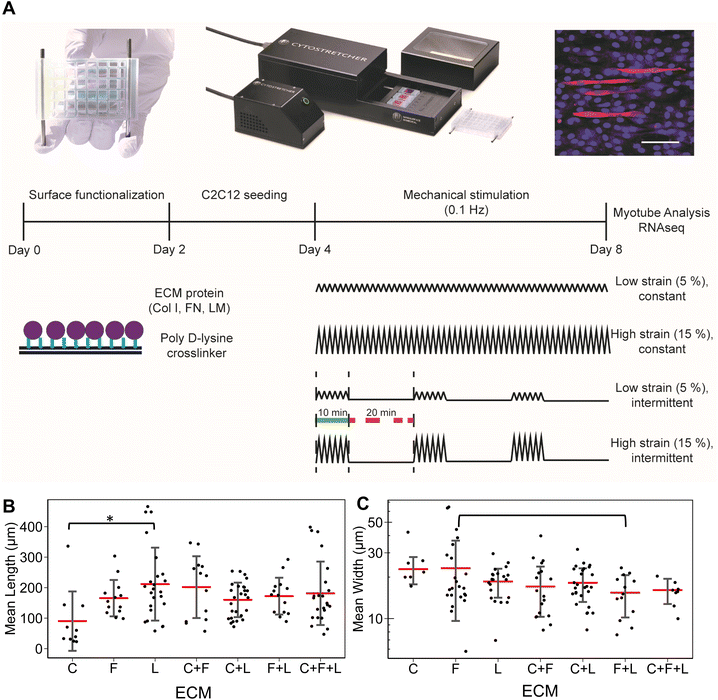
Uniaxial strain stimulation was applied to the tissue chips using the Cytostretcher (Curi Bio) as shown in Fig. 1A. The constant strain regime consisted of linearly applied low (5%) or high (15%) strain at 0.1 Hz. The intermittent strain regime consisted of repeating cycles of stimulation (10 minutes at 0.1 Hz followed by 20 minutes of rest). As a basis for comparison, the control samples were cultured under normal static conditions. Mechanical stimulation was applied for a total of 4 days, in which the differentiation media was changed on day 2 of stimulation.2.5. Immunofluorescence staining of myotube formation
After 4 days of differentiation, the cells were fixed with formalin (10%) for 15 minutes at room temperature and immunofluorescently stained for myosin heavy chain (Sigma), followed by incubation with secondary antibody conjugated with Alexa Fluor 594 (Fisher Scientific) and counter staining for nuclei with Hoechst 33342 (Fisher Scientific). Fluorescence images were captured at 10× magnification with an inverted confocal microscope (Zeiss LSM 710). For each independent experiment (n = 3), 3 images were acquired for image analysis of myotube formation.2.6. Image analysis
For in vitro studies, the lengths and widths of myotubes were measured from 3 images acquired for each independent experiment in each treatment group. Myotube length and widths were measured manually using ImageJ. Only myotube fully visible within the images were quantified.2.7. RNA isolation
A subset of samples was processed for RNA isolation to study the effects of combinatorial ECMs and strain on transcriptional modification. Medium-sized stretch chambers (12 × 12 mm, Curi Bio) with parallel-aligned microgrooves were functionalized with poly D-lysine (0.1 mg mL−1) for 16 hours before coating of collagen I, laminin, or C + L + F as described in section 2.2. Cell seeding was performed according to section 2.3. Upon reaching 90% confluency, these cell-seeded chambers underwent constant high strain (15%) in differentiation media on the Cytostretcher. After 4 days of strain stimulation, the cells were lysed and the RNA was extracted using GeneJET RNA isolation kit (Fisher Scientific).2.8 RNA sequencing
RNA sequencing was performed by Novogene Corporation. In brief, messenger RNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first strand cDNA was synthesized using random hexamer primers, followed by the second strand cDNA synthesis using either dUTP for directional library or dTTP for non-directional library. The library was checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries will be pooled and sequenced on Illumina platforms, according to effective library concentration and data amount.Original image data file from high-throughput Illumina sequencing platform was transformed to sequenced reads (Raw Data) by CASAVA base recognition (Base Calling). Raw data are stored in FASTQ (fq) format files which contain sequences of reads and corresponding base quality. Raw data was filtered by removing reads with adapter contamination, with uncertain nucleotides constituting more than 10 per cent of either read (n > 10%) and with low-quality nucleotides (base quality less than 5) constituting more than 50 per cent of the read. This was followed by alignment of reads using HISAT2 to the reference with the following considerations: (1) the whole sequence was aligned to a single exon, (2) the sequencing sequence was piecewise aligned to two exons of the genome, and (3) the sequencing sequence was segmented and aligned to more than three (including three) exons of the genome, and (4) the fragments per kilobase of transcript sequence per millions base pairs sequenced (FPKM) was quantified for each aligned read to estimate gene expression levels, which takes the effects into consideration of both sequencing depth and gene length on counting of fragments.
For RNAseq data analysis, the FPKM counts were normalized, and log transformed. Surrogate variable analysis was done using RUVseq package in R, and removeBatchEffect function was used to remove the surrogate variables found. Differential accessibility analysis was done using lmFit function of package limma in R. Specific comparisons (strain/ECM) were statistically tested in the model using contrasts.fit function. Adjusted P value (Padj) and fold change was calculated for each region and comparison. Specific differential gene expression lists were made for specific comparisons to enable enrichment analysis on EnrichR website. For gene set enrichment analysis (GSEA) analysis, the java desktop application was used, with instructions from the Wiki page was utilized. The bar plots were made using ggplot package and pheatmap package was used to create all the heatmaps in the figures.
2.9 Quantitative PCR (qPCR)
First, reverse transcription on isolated RNA sample was performed using the Superscript II reverse transcriptase (Invitrogen) and cDNA was synthesized according to the manufacturer's instruction and previous publications.24 Primers used for Taqman qPCR consisted of MyoD, Myh1, Myh3 and GAPDH (all from Applied Biosystems, Foster City, CA). The qPCR was performed on a 7900 real-time PCR system (Applied Biosystems) for 40 cycles. The data were analyzed and processed by the ΔΔCt method, normalized to GAPDH housekeeping gene, and then quantified as normalized relative fold changes with respect to collagen I under static conditions (n = 2–3).2.10 Statistical analysis
For in vitro studies, the lengths and widths of myotubes from 3 independent experiments were shown as mean ± standard deviation. Statistical analysis was performed by analysis of variance with Tukey post hoc test, where statistical significance is accepted at P < 0.05. For RNA sequencing, a linear model was computed for the gene expression as function of the ECM and mechanical stimulus (3 biological replicate each). The differences between each treatment group were calculated using established methods in the contrasts.fit function in limma package R. The statistics for the differential gene expression is calculated using eBayes functions in which analysis of variance (ANOVA) with Tukey's post hoc test was performed for all comparisons. Statistical significance was accepted at Padj < 0.05.3. Results
3.1 Combinatorial ECMs under static conditions differentially modulate myotube length and width
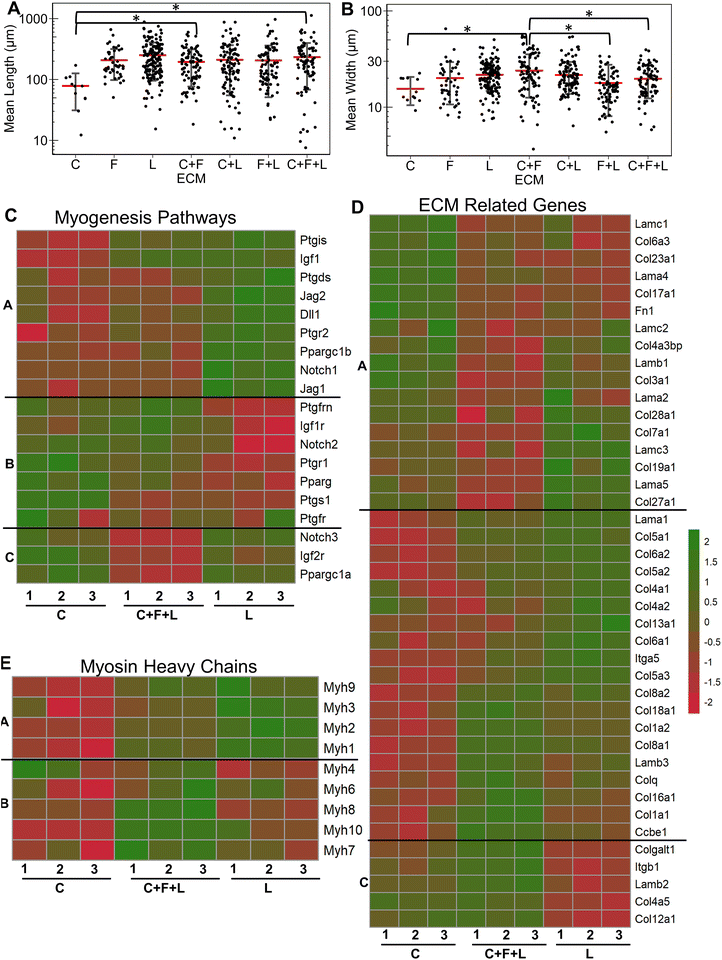
To study the effects of combinatorial ECMs on myogenesis, we first examined myotube formation over 4 days on stretch chambers that were surface-modified by collagen I (C), fibronectin (F), laminin (L); their two-factor combinations (C + F, C + L, F + L); and their three-factor combination (C + F + L) (Fig. 1A and ESI Fig. 1†). The average myotube length and width using myosin heavy chain immunostaining was quantified. These morphological parameters were selected based on prior publications that establish a correlation between morphological features and myotube formation.25,26 We first characterized the myogenesis of myoblasts in the presence of combinatorial microenvironments. Laminin exhibited the highest average myotube length (211 ± 119 μm) among all ECM compositions, reflecting >2-fold higher length compared to collagen I (90 ± 97 μm, P = 0.008) as represented in bar plot in Fig. 1B. However, myoblasts cultured on laminin showed no significant difference in myotube width compared to other ECMs (Fig. 1C). Interestingly, fibronectin + laminin (F + L) resulted in lowest average myotube width in myoblasts (15 ± 5.2 μm), especially compared to fibronectin alone, which supported the highest myotube width compared to all other ECM combinations (23 ± 13 μm, P = 0.06). This non-intuitive finding may be a result of inhibitory effects, as has been observed in combinatorial ECM interactions in other cellular contexts.27,28 It was also observed that different ECMs resulted in highest myotube length and width, pointing towards different molecular mechanisms being affected by different ECM combinations.3.2 RNA sequencing analysis reveals combinatorial ECM effects on myogenic differentiation of under static condition
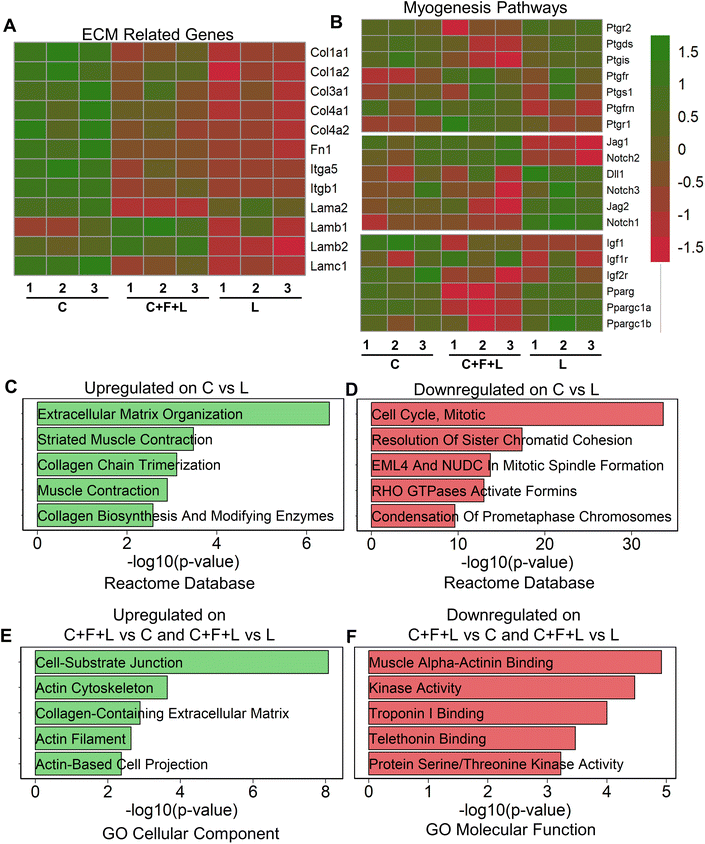
To understand regulatory mechanisms underlying differential effect of ECM combinations on average myotube length and width, we performed bulk RNA sequencing of myoblasts that were cultured on a subset of the ECM conditions. To study the molecular underpinnings in the presence of combinatorial vs. single-factor ECMs, we selected the three-factor ECM (C + F + L) combination, compared to the single-factor collagen I (C) and laminin (L) constituents that have distinct effects on myogenesis. Collagen I significantly exhibited upregulation of ECM related genes, specifically those relating to collagens, fibronectins, laminins, and integrins, including Col1a1, Col1a2, Itgb1, Itgb5 and Fn1 (Fig. 2A). This result suggests a higher ECM production in myoblasts on collagen I, compared to that on laminin or C + F + L under static conditions. Furthermore, gene enrichment analysis revealed the upregulation in the categories associated with ECM organization (Col1a1, Mfap5 (a glycoprotein in myofibrils in ECM), Itga11) and muscle contraction (Myl1, Tnnt1, Tcap (sarcomere assembly)) gene sets on collagen I compared to laminin (Fig. 2C). In agreement with higher myogenesis on collagen I, the gene set enrichment analysis showed downregulation of categories associated with cell cycle, compared to laminin, as withdrawal from cell cycle is required for myogenesis (Fig. 2D).29Since prostaglandin E2, notch, and PPARG (peroxisome proliferator activated receptor gamma) are also important myogenesis pathway genes, we observed that collagen I shows significant upregulation in a subset of genes from these pathways such as Pparg, Ppargc1a, Igf1, Ptgis, Jag1 compared to C + F + L under static conditions.30–33To substantiate the in vitro finding that laminin supported the formation of the longer myotubes, compared to collagen (Fig. 1B), we show that laminin substrates upregulated multiple genes in the prostaglandin E2, notch signaling and Pparg signaling pathways, signifying a unique muscle regeneration signature associated with laminin's role in modulating myogenesis (Fig. 2B). However, it is important to note that a different subset of myogenesis-related genes were upregulated on laminin substrates compared to both collagen I and C + F + L under static conditions. The C + F + L ECM combination specifically exhibited significantly higher expression of laminin b1 and b2 compared to both collagen I and laminin, while also demonstrated an overall higher expression of ECM related genes compared to only laminin, including Col1a1, Col1a2, and Fn1 (Fig. 2A).
Upregulated genes on C + F + L ECM combination, compared to either laminin or collagen I, included the categories of actin cytoskeleton and cell–substrate junction (Actn1, Fnlb, Vcam1) gene sets from the GO cellular component database (Fig. 2E). This finding suggests that the C + F + L ECM combination supported more interaction with the underlying substrate, compared to laminin and collagen I alone. Downregulated genes on C + F + L, compared to collagen I or laminin corresponded to muscle alpha-actinin binding, troponin binding pointing towards decreased expression of muscle contraction and assembly genes (Fig. 2F). Additionally, myogenesis pathway genes such as Ptgs1, Igfr1 and notch signaling receptor Notch2 were specifically upregulated on C + F + L (Fig. 2B). Together, these data suggested that single-component and combinatorial ECMs had a unique molecular signature and effect in modulating myogenesis.
Since sarcomeric structure is an important functional characteristic of myotube function, we quantified genes from sarcomere geneset in the gene ontology database in our transcriptomic analysis (ESI Fig. 2†). We discovered three distinct gene profiles, each enriched in a specific ECM combination. Specifically, genes in cluster A were upregulated in C + F + L combination under static conditions and corresponded to gene such as Calm1, Calm3 and Actn1, which are an integral part of the protein subunit for calcium signaling in sarcomeres.34 Additionally, cluster B corresponded to genes upregulated on either single-factor ECMs under static condition, containing genes such Ttn and Tnnt1, which are responsible for muscle contraction in sarcomeres.35 These findings suggest that sarcomeric genes were distinctively different on the C + F + L combination, compared to the single factor components.
3.3 Combined effects of strain with C + F + L on myogenesis
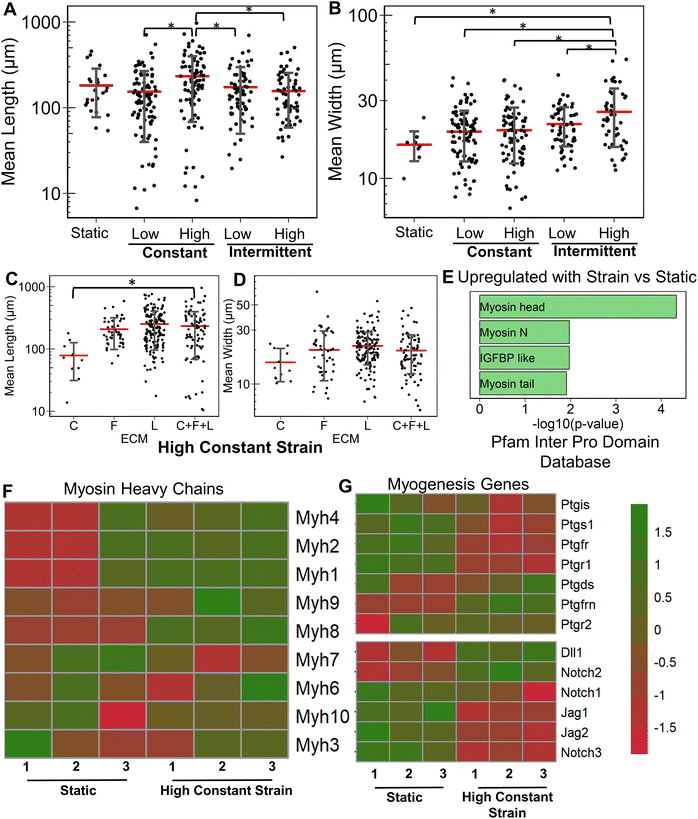
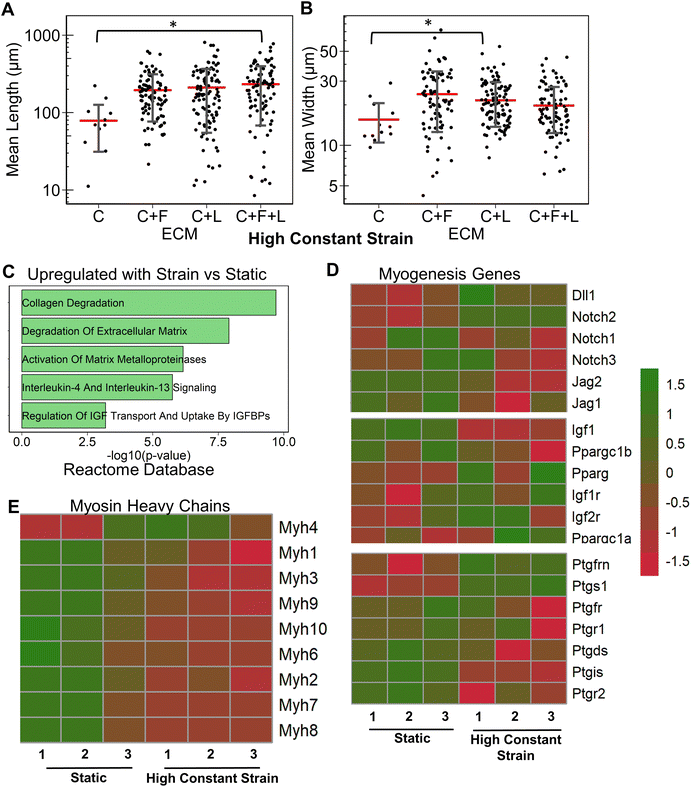
Having studied the effect of ECMs on the myogenic differentiation of C2C12 cells under static conditions, we next sought to understand the combined role of ECM composition and strain on the myogenic differentiation. Two levels of uniaxial strain (high or low, Fig. 1A) were applied to the cells cultured on all ECM combinations. Furthermore, these two levels of uniaxial strain were applied either constantly or intermittently. It was observed that high constant strain to myoblasts on C + F + L ECM combination resulted in the highest average myotube length, which was significantly higher compared to other strain regimens (Fig. 3A). Specifically, high constant strain on C2C12 myoblasts on C + F + L ECM exhibited a 50% increase compared to both high intermittent strain (P = 0.003) and low constant strain (P = 0.0005), and a 34% increase compared to low intermittent strain (P = 0.03; Fig. 3A). On the other hand, the highest myotube width was observed with high intermittent strain for C2C12 cells cultured on C + F + L. Specifically, an increase of 56% in myotube width was observed with high intermittent strain, compared to the static condition (P = 0.006; Fig. 3B). These findings show that high constant strain increased the average myotube length whereas high intermittent strain significantly increased average myotube width.Compared to the combinatorial environment of C + F + L, the individual components alone did not have a significant difference in both the myotube length and width with the high constant strain application, with the exception of a significantly lower myotube length on collagen I compared to C + F + L ECM (Fig. 3C and D). Genome-wide analysis of myoblasts cultured on C + F + L ECM with and without high constant strain demonstrated higher expression of multiple myosin heavy chains with the application of strain. In particular, Myh4 (slow), Myh1 (fast), Myh2 (fast) and Myh8 (fetal muscle) were found to be significantly upregulated with the strain on the C + F + L ECM (Fig. 3F). Additionally, upregulated genes C + F + L ECM with high constant strain compared to static conditions were enriched in myosin related protein domains (Myo1e, Myo5a) in the Pfam InterPro domains database (Fig. 3E). It was also observed that multiple muscle regeneration related growth factors and pathways such Igf1, Pparg, notch signaling was also upregulated with the application of strain on C + F + L (Fig. 3G). Overall, strain application to the differentiating myoblasts cultured on the C + F + L ECM combination enhanced both myotube width and length by upregulating certain myosin heavy chain expression and muscle-regeneration related pathways such as IGF-1 and notch.
3.4 Combined effects of strain and laminin on myogenesis
Next, we evaluated the effects of strain on laminin substrates. On laminin alone, a significantly higher myotube length was observed under high constant strain and low intermittent strain (P < 0.01), but not under low constant strain or high intermittent strain (Fig. 4A and ESI Fig. 3†). We also observed that all strain regimens increased the average width of myotubes, compared to on static conditions (P < 0.05, Fig. 4B). Interestingly, combination of C + L did not significantly affect the myotube width and length with application of high constant strain (Fig. 4C), even though under static conditions a significant decrease was observed in the myotube length with on C + L (P = 0.03) (Fig. 1A). A significant decrease in both myotube length and width was observed with pairing F + L under high constant strain condition, which was also observed under static conditions (Fig. 1A). Interestingly, this decrease in myotube length and width is not observed with the C + F + L ECM combinations, suggesting a non-intuitive influence of F + L compared to C + F + L on myogenic differentiation of myoblasts (Fig. 4C and D).At the transcriptional level, the expression of skeletal myosin heavy chain-associated genes was upregulated with the application of high constant strain on laminin. These genes included isoforms Myh3 (embryonic, associated with regenerating muscle), Myh1 (type IIa, fast) and Myh2 (type IIb, fast), signifying increased muscle functionality of differentiating myoblasts cultured on laminin with high constant strain, compared to on static conditions (Fig. 4E). Furthermore, gene set enrichment analysis revealed significantly higher enrichment in the category of skeletal myofibril assembly in myoblasts on laminin with high constant strain, compared to static conditions (enrichment score 0.7; P = 0.048) (Fig. 4F). Some of the upregulated genes in this category included cytoskeletal markers such as Tcap, Ttn, and Tmod1.
Additionally, we validated gene expression findings from RNAseq using quantitative PCR (ESI Fig. 4†). Similar to the RNAseq, upregulation of two myosin heavy chains, Myh1 and Myh3, was observed on laminin in the presence of high constant strain. Additionally, an early myogenesis marker, MyoD, was found to be upregulated on laminin in the presence of strain, signifying the synergistic effect of mechanical stimulation and combinatorial ECM combination on early myogenesis. Together, these findings suggest that C2C12 myoblasts cultured on laminin and under specific strain regime increased myotube width and length as well as resulted in overexpression of multiple myogenesis related genes.
3.5 Combined effects of strain and collagen I on myogenesis
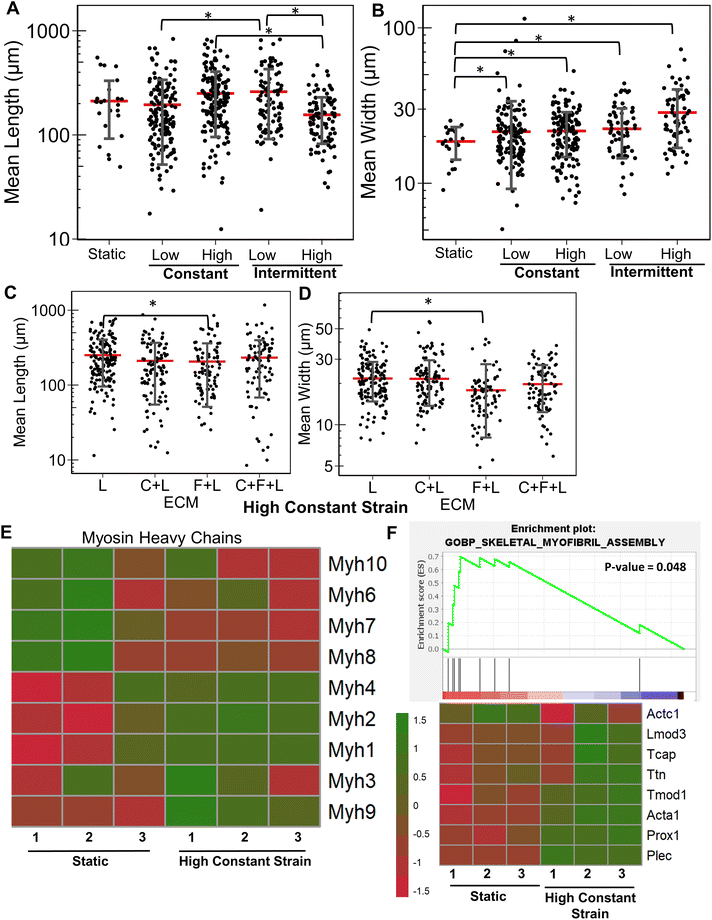
Similarly, we examined the strain effects on collagen I. A significant reduction in myotube length and width was observed with application of specific strain regimens (ESI Fig. 1†). With the application of high constant strain, a significant 26% increase in average myotube length was observed with C + F + L ECM compared to collagen I alone (Fig. 5A). However, it was C + L ECM combinations that had a significant increase in myotube width compared to collagen I alone (Fig. 5B). At the genome-wide level, upregulated genes on collagen I with high constant strain were enriched in categories related to ECM degradation and collagen degradation, compared to on static conditions (Fig. 5C). Furthermore, contrary to the strain effects on C + F + L vs. laminin (Fig. 4), strain on collagen I substrates led to a significant decrease in expression multiple myosin heavy chains (Fig. 5E). Additionally, upregulation of Ptgs1, Notch2 on strain was observed, concomitant with a downregulation of Igf1, Jag1, Jag2, Ppargc1b was also quantified (Fig. 5D). Overall, application of different strains to differentiating myoblasts on collagen I did not improve myogenic potential, and instead multiple myogenic genes were downregulated.3.6 Combinatorial ECMs under high constant strain conditions differentially regulate myogenesis
Having established the effects of different strain regimens, compared to static conditions, we then performed a comparative analysis of myogenesis in ECM compositions under high constant strain. Comparatively, the myotube length was significantly higher on C + F and C + F + L, compared to collagen I alone (P < 0.03, Fig. 6A). The myotube width was significantly higher on C + F, when compared to C + F + L (P = 0.02), F + L (P = 0.00009), or collagen I alone (P = 0.02) (Fig. 6B). A notable finding was that the ECM effect on myotube length was amplified by the application of high constant strain. For example, laminin exhibited a 3.2-fold increase in myotube length compared to collagen I under high constant strain (Fig. 6A), compared to only a 2.3-fold increase with static conditions (Fig. 1A). Similarly, under high constant strain, C + F + L also exhibited a significant 2.9-fold increase in myotube length, compared to collagen I (Fig. 6A), whereas there was no significant difference between the two ECM compositions under static conditions (Fig. 1A).Transcriptome analysis revealed various molecular signatures unique to each ECM composition in response to the high constant strain application. Three gene clusters were found in myogenesis pathway genes, whereas two distinct clusters were found in ECM genes (Fig. 6C). Specifically, cluster A in myogenesis pathways genes corresponded to upregulation of Igf1, Jag2, Dll1, Ppargc1b on laminin, along with a significant downregulation on collagen I. In contrast, cluster B corresponded to downregulation Notch2, Ptgs1, Igf1r on laminin and upregulation on collagen I. Cluster C in the myogenesis pathways genes corresponded to specific downregulation of genes on C + F + L ECM combination (Fig. 6C). With respect to ECM-related genes, the effects of high constant strain could be distinguished by three distinct clusters corresponding to genes downregulated on C + F + L (Fn1, Lamb1, Lama1), genes upregulated on both C + F + L and L (multiple isoforms of Col5 and Col6) and also the genes upregulated on C + F + L only (Col4a5, Lamb2, Itgb1) (Fig. 6D). Quantification of myosin heavy chains genes revealed upregulation of Myh1, Myh2 and Myh3 on laminin compared to collagen I (cluster A), whereas C + F + L led to the upregulation of Myh8 and Myh10 compared to both collagen I alone and laminin alone (cluster B) (Fig. 6E). These transcriptional signatures suggest that high constant strain effects on myogenesis show partial benefits in the setting of combinatorial ECMs, compared to single-component ECMs. Importantly, these findings also highlight the complexity of cell–ECM interactions that often lead to non-intuitive findings in combinatorial ECMs, compared to single ECMs.
3. Discussion
In this study, we investigated the effects of combinatorial ECM proteins and different regimens of uniaxial stretching on myogenesis. The salient findings are that: (1) ECM compositions had a significant effect on myotube formation. Under static conditions, laminin was observed to produce the longest myotubes, whereas fibronectin produced the widest myotubes; (2) ECM combinations often produced non-intuitive effects compared to that of the single component ECMs that comprise them; (3) the benefits of combined strain and ECM cues are complex. Myotube formation findings showed that different strain regimes on laminin and C + F + L substrates led to enhanced myotube length and width, whereas no improvement on collagen I was observed with the addition of strain; and (4) genome-wide analysis of strain and ECM composition demonstrated potential benefits of combinatorial ECMs over single-component ECMs in the modulating of myogenesis.We did not observe synergistic effects of combinatorial ECMs in relations to in vitro myotube morphology. However, when comparing transcriptional expression, we observed differential effects on the expression of combinatorial ECMs such as C + F + L, compared to collagen I alone. For example, cells cultured on collagen I alone in static conditions showed a significant downregulation of cell cycle related genes consistent with cell cycle withdrawal that precedes myotube formation (Fig. 2D), whereas cells on C + F + L substrates were enriched in actin cytoskeletal-related categories (Fig. 2E). These synergistic effects are compounded with mechanical stimulation, where myosin heavy chain gene expression levels were upregulated, compared to static culture alone (Fig. 3E). Together, our data suggests that combinatorial ECM microenvironment in concert with mechanical stimulation provide synergistic benefits to the myogenic process.
It is established that ECM proteins provide important biochemical cues for myoblast proliferation, attachment and differentiation.36 In our assessment of ECM effects, we studied three ECM components of skeletal muscle tissue, which included collagen I, laminin and fibronectin. Our results showed that laminin produced the longest myotube and this agreed with Duffy et al.,37 whereas Ding et al. observed fibronectin to be superior.38 Ito et al. observed that collagen IV resulted in the longest myotube.39 There are some conflicting data pertaining to the most myogenic ECM within the context of a singular component. In this study we have also included combination of the three ECM to investigate how multiple ECM interact to produce synergistic effects on myogenesis. We did not observe the additive or synergistic effect when combining two single-component ECMs. This could be due to a range of reasons such as interference of molecular binding sites of the ECM competing for the cellular receptors or steric interactions within ECMs resulting in a net negative number of total binding sites for cell-binding receptors. Investigating the molecular level using RNAseq, we were able to discern differences between singular component ECM (collagen and laminin) with the combination of three ECMs (C + F + L). This highlights the nuances in early myogenesis that are not observed in mature myotube that express myosin heavy chain by immunofluorescence staining.
Mechanical stimulation also promotes differentiation and maturation of myoblast into myotubes. Uniaxial stretching was achieved using the Cytostretcher system from Curi Bio. This system has been used for studies involving chondrocytes and peripapillary scleral fibroblasts.40–43 Strain and frequency were chosen to corroborate with previous studies as strains of more than 20% decreases myogenesis.44 Multiple studies have shown that mechanical stimulation increases myoblast differentiation and myotube formation.22,45–47 These tend to employ general molecular techniques to observe the regulation of known signaling pathways associated with muscle regeneration.48 Here, we used RNAseq to determine transcriptomic changes with respect to combinatorial ECMs in conjunction with strain regimens.
One interesting finding is the upregulation of growth factor-associated pathways was only in the C + F + L group while undergoing constant high straining. This suggests that combinatorial ECMs with strain provide beneficial effects to the myogenesis process. This data falls in line with other studies using decellularized ECM from muscle related tissue or Matrigel basement membrane matrix, which was shown to improve myogenesis.49 The synergistic effects of combinatorial ECM with mechanical stimulation could be potentially explained by mechanotransduction related pathways. Uniaxial stretching may also increase the presentation of ECM binding sites for cell interactions, as this phenomenon has been observed in fibronectin and collagen I.50,51
A limitation of this study includes the small number of ECMs studied, in order to study the interaction of ECM composition with strain regimens. We selected collagen I in this study as it is the most abundant collagen I isoform, although collagen IV is the second most abundant isoform and may also exert important signaling function. Future studies should expand on the number of ECM proteins to be investigated.39 Additionally, topography is also an important consideration. In this study, the cells were cultured on micropatterned substrates that confine the cells along the direction of strain. We and others have shown that micropatterning enables myoblasts to organize their cytoskeleton and form myotubes along the direction of the microgrooves.26,52,53 There have also been significant developments in incorporating spatiotemporal control of multiple biomechanical factors in directing cellular fate.54–57 Additional parameters such as three-dimensionality, substrate stiffness and viscoelasticity could be incorporated to develop a more biomimetic system that better reflects the complexity of the muscle microenvironment.58,59
In conclusion, we have developed a stretchable tissue chip system to probe the combined effects of ECM composition and strain on myogenesis. Using this platform, we could systematically study the effects of ECM combinations along with strain regimes on myotube formation and the associated transcriptional signatures. Our findings illustrate the complexity by which ECM and strain cues modulate myogenesis in myoblasts. This stretchable tissue chip system has broad applications towards other cell types and for modeling in vitro diseases.
Data availability
All data from this project are available from the corresponding author upon reasonable request.Author contributions
Conceptualization, A. H. P. C. and N. F. H.; methodology, A. H. P. C., I. J., B. N., N. G., and N. F. H.; investigation, A. H. P. C., I. J.; B. P. O., T. Z., resources, N. F. H.; data curation, A. H. P. C., I. J., and N. H.; writing – original draft preparation, A. H. P. C., I. J., and N. H.; writing – review and editing, A. H. P. C., I. J., B. N., N. G., B. P. O., and N. H.; visualization, A. H. P. C., I. J., and N. H.; supervision, N. F. H.; project administration, N. F. H.; funding acquisition, A. H. P. C., B. P. O., and N. F. H. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
B. D. and N. G. are employees of Curi Bio that manufactures the Cytostretcher equipment. Curi Bio, Inc. had no role in the design of in vitro or RNA sequencing studies.Acknowledgements
This work was supported in part by grants to NFH from the US National Institutes of Health (R01 HL127113 and R01 HL142718), the US Department of Veterans Affairs (1I01BX004259 and RX001222), the National Science Foundation (1829534 and 2227614), the American Heart Association (20IPA35360085 and 20IPA35310731), and the Tobacco Related-Disease Research Program (T33IP6580). This research received funding from the Alliance for Regenerative Rehabilitation Research & Training (AR3T), which is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institute of Neurological Disorders and Stroke (NINDS), and National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under award number P2CHD086843. NG was supported by a grant from the US National Institutes of Health (R43 HL146043). AHPC was supported by a postdoctoral fellowship from the Tobacco Related Disease Research Program (T30FT0860). BPO was supported in part by a diversity supplement from the US National Institutes of Health (3R01HL151997-03S1). NFH is a recipient of a Research Career Scientist award (IK6 BX006309) from the Department of Veterans Affairs.References
- K. Garg, C. L. Ward, B. J. Hurtgen, J. M. Wilken, D. J. Stinner, J. C. Wenke, J. G. Owens and B. T. Corona, Volumetric muscle loss: persistent functional deficits beyond frank loss of tissue, J. Orthop. Res., 2015, 33, 40–46 CrossRef PubMed.
- S. H. Lin, D. C. Chuang, Y. Hattori and H. C. Chen, Traumatic major muscle loss in the upper extremity: reconstruction using functioning free muscle transplantation, J. Reconstr. Microsurg., 2004, 20, 227–235 CrossRef PubMed.
- C. H. Lin, Y. T. Lin, J. T. Yeh and C. T. Chen, Free functioning muscle transfer for lower extremity posttraumatic composite structure and functional defect, Plast. Reconstr. Surg., 2007, 119, 2118–2126 CrossRef CAS PubMed.
- M. Klinkenberg, S. Fischer, T. Kremer, F. Hernekamp, M. Lehnhardt and A. Daigeler, Comparison of anterolateral thigh, lateral arm, and parascapular free flaps with regard to donor-site morbidity and aesthetic and functional outcomes, Plast. Reconstr. Surg., 2013, 131, 293–302 CrossRef CAS PubMed.
- M. Shayan and N. F. Huang, Pre-Clinical Cell Therapeutic Approaches for Repair of Volumetric Muscle Loss, Bioengineering, 2020, 7, 97 CrossRef CAS.
- B. M. Sicari, J. P. Rubin, C. L. Dearth, M. T. Wolf, F. Ambrosio and M. Boninger, et al., An acellular biologic scaffold promotes skeletal muscle formation in mice and humans with volumetric muscle loss, Sci. Transl. Med., 2014, 6, 234ra258 Search PubMed.
- B. T. Corona, M. A. Machingal, T. Criswell, M. Vadhavkar, A. C. Dannahower, C. Bergman, W. Zhao and G. J. Christ, Further development of a tissue engineered muscle repair construct in vitro for enhanced functional recovery following implantation in vivo in a murine model of volumetric muscle loss injury, Tissue Eng., Part A, 2012, 18, 1213–1228 CrossRef CAS PubMed.
- C. A. Alcazar, C. Hu, T. A. Rando, N. F. Huang and K. H. Nakayama, Transplantation of insulin-like growth factor-1 laden scaffolds combined with exercise promotes neuroregeneration and angiogenesis in a preclinical muscle injury model, Biomater. Sci., 2020, 8, 5376–5389 RSC.
- I. Eugenis, D. Wu, C. Hu, G. Chiang, N. F. Huang and T. A. Rando, Scalable macroporous hydrogels enhance stem cell treatment of volumetric muscle loss, Biomaterials, 2022, 290, 121818 CrossRef CAS PubMed.
- N. E. Gentile, K. M. Stearns, E. H. Brown, J. P. Rubin, M. L. Boninger, C. L. Dearth, F. Ambrosio and S. F. Badylak, Targeted rehabilitation after extracellular matrix scaffold transplantation for the treatment of volumetric muscle loss, Am. J. Phys. Med. Rehabil., 2014, 93, S79–S87 CrossRef PubMed.
- C. Hu, B. Ayan, G. Chiang, A. H. P. Chan, T. A. Rando and N. F. Huang, Comparative Effects of Basic Fibroblast Growth Factor Delivery or Voluntary Exercise on Muscle Regeneration after Volumetric Muscle Loss, Bioengineering, 2022, 9, 37 CrossRef CAS PubMed.
- K. H. Nakayama, M. Quarta, P. Paine, C. Alcazar, I. Karakikes and V. Garcia, et al., Treatment of volumetric muscle loss in mice using nanofibrillar scaffolds enhances vascular organization and integration, Commun. Biol., 2019, 2, 170 CrossRef PubMed.
- C. F. Bentzinger, Y. X. Wang and M. A. Rudnicki, Building muscle: molecular regulation of myogenesis, Cold Spring Harbor Perspect. Biol., 2012, 4, a008342 Search PubMed.
- A. Philippou, E. Papageorgiou, G. Bogdanis, A. Halapas, A. Sourla, M. Maridaki, N. Pissimissis and M. Koutsilieris, Expression of IGF-1 isoforms after exercise-induced muscle damage in humans: characterization of the MGF E peptide actions in vitro, In Vivo, 2009, 23, 567–575 CAS.
- M. F. Buas and T. Kadesch, Regulation of skeletal myogenesis by Notch, Exp. Cell Res., 2010, 316, 3028–3033 CrossRef CAS PubMed.
- T. Varga, R. Mounier, A. Patsalos, P. Gogolák, M. Peloquin and A. Horvath, et al., Macrophage PPARγ, a Lipid Activated Transcription Factor Controls the Growth Factor GDF3 and Skeletal Muscle Regeneration, Immunity, 2016, 45, 1038–1051 CrossRef CAS PubMed.
- A. Brougham-Cook, I. Jain, D. A. Kukla, F. Masood, H. Kimmel, H. Ryoo, S. R. Khetani and G. H. Underhill, High throughput interrogation of human liver stellate cells reveals microenvironmental regulation of phenotype, Acta Biomater., 2022, 138, 240–253 CrossRef CAS PubMed.
- R. Csapo, M. Gumpenberger and B. Wessner, Skeletal Muscle Extracellular Matrix - What Do We Know About Its Composition, Regulation, and Physiological Roles? A Narrative Review, Front. Physiol., 2020, 11, 253 CrossRef PubMed.
- W. Zhang, Y. Liu and H. Zhang, Extracellular matrix: an important regulator of cell functions and skeletal muscle development, Cell Biosci., 2021, 11, 65 CrossRef PubMed.
- J. Chen, R. Zhou, Y. Feng and L. Cheng, Molecular mechanisms of exercise contributing to tissue regeneration, Signal Transduction Targeted Ther., 2022, 7, 383 CrossRef.
- S. Bansai, T. Morikura, H. Onoe and S. Miyata, Effect of Cyclic Stretch on Tissue Maturation in Myoblast-Laden Hydrogel Fibers, Micromachines, 2019, 10, 399 CrossRef PubMed.
- Y. Wang, J. Song, X. Liu, J. Liu, Q. Zhang, X. Yan, X. Yuan and D. Ren, Multiple Effects of Mechanical Stretch on Myogenic Progenitor Cells, Stem Cells Dev., 2020, 29, 336–352 CrossRef PubMed.
- N. Shi, Y. Li, L. Chang, G. Zhao, G. Jin, Y. Lyu, G. M. Genin, Y. Ma and F. Xu, A 3D, Magnetically Actuated, Aligned Collagen Fiber Hydrogel Platform Recapitulates Physical Microenvironment of Myoblasts for Enhancing Myogenesis, Small Methods, 2021, 5, e2100276 CrossRef PubMed.
- N. F. Huang, F. Fleissner, J. Sun and J. P. Cooke, Role of nitric oxide signaling in endothelial differentiation of embryonic stem cells, Stem Cells Dev., 2010, 19, 1617–1626 CrossRef CAS PubMed.
- K. Ishikawa, K. Yoshida, K. Kanie, K. Omori and R. Kato, Morphology-Based Analysis of Myoblasts for Prediction of Myotube Formation, SLAS Discovery, 2019, 24, 47–56 CrossRef CAS PubMed.
- N. F. Huang, S. Patel, R. G. Thakar, J. Wu, B. S. Hsiao, B. Chu, R. J. Lee and S. Li, Myotube assembly on nanofibrous and micropatterned polymers, Nano Lett., 2006, 6, 537–542 CrossRef CAS PubMed.
- L. Hou, J. Coller, V. Natu, T. J. Hastie and N. F. Huang, Combinatorial extracellular matrix microenvironments promote survival and phenotype of human induced pluripotent stem cell-derived endothelial cells in hypoxia, Acta Biomater., 2016, 44, 188–199 CrossRef CAS PubMed.
- L. Hou, J. J. Kim, M. Wanjare, B. Patlolla, J. Coller, V. Natu, T. J. Hastie and N. F. Huang, Combinatorial Extracellular Matrix Microenvironments for Probing Endothelial Differentiation of Human Pluripotent Stem Cells, Sci. Rep., 2017, 7, 6551 CrossRef PubMed.
- A. M. Bennett and N. K. Tonks, Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases, Science, 1997, 278, 1288–1291 CrossRef CAS PubMed.
- A. T. V. Ho, A. R. Palla, M. R. Blake, N. D. Yucel, Y. X. Wang and K. E. G. Magnusson, et al., Prostaglandin E2 is essential for efficacious skeletal muscle stem-cell function, augmenting regeneration and strength, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 6675–6684 CrossRef CAS PubMed.
- C. Mo, R. Zhao, J. Vallejo, O. Igwe, L. Bonewald, L. Wetmore and M. Brotto, Prostaglandin E2 promotes proliferation of skeletal muscle myoblasts via EP4 receptor activation, Cell Cycle, 2015, 14, 1507–1516 CrossRef CAS PubMed.
- D. Luo, V. M. Renault and T. A. Rando, The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis, Semin. Cell Dev. Biol., 2005, 16, 612–622 CrossRef CAS PubMed.
- G. Dammone, S. Karaz, L. Lukjanenko, C. Winkler, F. Sizzano and G. Jacot, et al., PPARgamma Controls Ectopic Adipogenesis and Cross-Talks with Myogenesis During Skeletal Muscle Regeneration, Int. J. Mol. Sci., 2018, 19, 33 CrossRef PubMed.
- M. Rasmussen and J. P. Jin, Troponin Variants as Markers of Skeletal Muscle Health and Diseases, Front. Physiol., 2021, 12, 747214 CrossRef PubMed.
- M. Marcello, V. Cetrangolo, M. Savarese and B. Udd, Use of animal models to understand titin physiology and pathology, J. Cell. Mol. Med., 2022, 26, 5103–5112 CrossRef CAS PubMed.
- M. Kjaer, Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading, Physiol. Rev., 2004, 84, 649–698 CrossRef CAS PubMed.
- R. M. Duffy, Y. Sun and A. W. Feinberg, Understanding the Role of ECM Protein Composition and Geometric Micropatterning for Engineering Human Skeletal Muscle, Ann. Biomed. Eng., 2016, 44, 2076–2089 CrossRef.
- R. Ding, M. Horie, S. Nagasaka, S. Ohsumi, K. Shimizu, H. Honda, E. Nagamori, H. Fujita and T. Kawamoto, Effect of cell-extracellular matrix interaction on myogenic characteristics and artificial skeletal muscle tissue, J. Biosci. Bioeng., 2020, 130, 98–105 CrossRef CAS.
- A. Ito, M. Yamamoto, K. Ikeda, M. Sato, Y. Kawabe and M. Kamihira, Effects of type IV collagen on myogenic characteristics of IGF-I gene-engineered myoblasts, J. Biosci. Bioeng., 2015, 119, 596–603 CrossRef CAS PubMed.
- H. A. Abusharkh, A. H. Mallah, M. M. Amr, J. Mendenhall, B. A. Gozen, E. M. Tingstad, N. I. Abu-Lail and B. J. Van Wie, Enhanced matrix production by cocultivated human stem cells and chondrocytes under concurrent mechanical strain, In Vitro Cell. Dev. Biol.: Anim., 2021, 57, 631–640 CrossRef CAS PubMed.
- H. A. Abusharkh, O. M. Reynolds, J. Mendenhall, B. A. Gozen, E. Tingstad, V. Idone, N. I. Abu-Lail and B. J. Van Wie, Combining stretching and gallic acid to decrease inflammation indices and promote extracellular matrix production in osteoarthritic human articular chondrocytes, Exp. Cell Res., 2021, 408, 112841 CrossRef CAS PubMed.
- A. Chow, L. McCrea, E. Kimball, J. Schaub, H. Quigley and I. Pitha, Dasatinib inhibits peripapillary scleral myofibroblast differentiation, Exp. Eye Res., 2020, 194, 107999 CrossRef CAS PubMed.
- J. Szeto, A. Chow, L. McCrea, A. Mozzer, T. D. Nguyen, H. A. Quigley and I. Pitha, Regional Differences and Physiologic Behaviors in Peripapillary Scleral Fibroblasts, Invest. Ophthalmol. Visual Sci., 2021, 62, 27 CrossRef CAS.
- Y. Ma, S. Fu, L. Lu and X. Wang, Role of androgen receptor on cyclic mechanical stretch-regulated proliferation of C2C12 myoblasts and its upstream signals: IGF-1-mediated PI3K/Akt and MAPKs pathways, Mol. Cell. Endocrinol., 2017, 450, 83–93 CrossRef CAS PubMed.
- A. M. Collinsworth, C. E. Torgan, S. N. Nagda, R. J. Rajalingam, W. E. Kraus and G. A. Truskey, Orientation and length of mammalian skeletal myocytes in response to a unidirectional stretch, Cell Tissue Res., 2000, 302, 243–251 CrossRef CAS PubMed.
- A. Kumar, R. Murphy, P. Robinson, L. Wei and A. M. Boriek, Cyclic mechanical strain inhibits skeletal myogenesis through activation of focal adhesion kinase, Rac-1 GTPase, and NF-kappaB transcription factor, FASEB J., 2004, 18, 1524–1535 CrossRef CAS.
- T. Matsumoto, J. Sasaki, E. Alsberg, H. Egusa, H. Yatani and T. Sohmura, Three-dimensional cell and tissue patterning in a strained fibrin gel system, PLoS One, 2007, 2, e1211 CrossRef.
- Y. Da, Y. Mou, M. Wang, X. Yuan, F. Yan, W. Lan and F. Zhang, Mechanical stress promotes biological functions of C2C12 myoblasts by activating PI3K/AKT/mTOR signaling pathway, Mol. Med. Rep., 2020, 21, 470–477 CAS.
- F. Carton, D. Di Francesco, L. Fusaro, E. Zanella, C. Apostolo, F. Oltolina, D. Cotella, M. Prat and F. Boccafoschi, Myogenic Potential of Extracellular Matrix Derived from Decellularized Bovine Pericardium, Int. J. Mol. Sci., 2021, 22, 9406 CrossRef CAS.
- K. E. Kubow, R. Vukmirovic, L. Zhe, E. Klotzsch, M. L. Smith, D. Gourdon, S. Luna and V. Vogel, Mechanical forces regulate the interactions of fibronectin and collagen I in extracellular matrix, Nat. Commun., 2015, 6, 8026 CrossRef CAS PubMed.
- J. M. Szymanski, E. N. Sevcik, K. Zhang and A. W. Feinberg, Stretch-dependent changes in molecular conformation in fibronectin nanofibers, Biomater. Sci., 2017, 5, 1629–1639 RSC.
- P.-Y. Wang, W.-T. Li, J. Yu and W.-B. Tsai, Modulation of osteogenic, adipogenic and myogenic differentiation of mesenchymal stem cells by submicron grooved topography, J. Mater. Sci.: Mater. Med., 2012, 23, 3015–3028 CrossRef CAS PubMed.
- H. Gao, J. Xiao, Y. Wei, H. Wang, H. Wan and S. Liu, Regulation of Myogenic Differentiation by Topologically Microgrooved Surfaces for Skeletal Muscle Tissue Engineering, ACS Omega, 2021, 6, 20931–20940 CrossRef CAS PubMed.
- Y. Ma, M. Lin, G. Huang, Y. Li, S. Wang, G. Bai, T. J. Lu and F. Xu, 3D Spatiotemporal Mechanical Microenvironment: A Hydrogel-Based Platform for Guiding Stem Cell Fate, Adv. Mater., 2018, 30, e1705911 CrossRef PubMed.
- I. Jain, I. C. Berg, A. Acharya, M. Blaauw, N. Gosstola, P. Perez-Pinera and G. H. Underhill, Delineating cooperative effects of Notch and biomechanical signals on patterned liver differentiation, Commun. Biol., 2022, 5, 1073 CrossRef CAS PubMed.
- S. G. Zambuto, I. Jain, K. B. H. Clancy, G. H. Underhill and B. A. C. Harley, Role of Extracellular Matrix Biomolecules on Endometrial Epithelial Cell Attachment and Cytokeratin 18 Expression on Gelatin Hydrogels, ACS Biomater. Sci. Eng., 2022, 8, 3819–3830 CrossRef CAS PubMed.
- K. B. Kaylan, I. C. Berg, M. J. Biehl, A. Brougham-Cook, I. Jain and S. M. Jamil, et al., Spatial patterning of liver progenitor cell differentiation mediated by cellular contractility and Notch signaling, eLife, 2018, 7, e38536 CrossRef PubMed.
- C. Mueller, M. Trujillo-Miranda, M. Maier, D. E. Heath, A. J. O'Connor and S. Salehi, Effects of External Stimulators on Engineered Skeletal Muscle Tissue Maturation, Adv. Mater. Interfaces, 2021, 8, 2001167 CrossRef.
- Y. Ma, T. Han, Q. Yang, J. Wang, B. Feng, Y. Jia, Z. Wei and F. Xu, Viscoelastic Cell Microenvironment: Hydrogel-Based Strategy for Recapitulating Dynamic ECM Mechanics, Adv. Funct. Mater., 2021, 31, 2100848 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3bm00448a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |