Monitoring gestational diabetes at the point-of-care via dual glycated albumin lateral flow assays in conjunction with a handheld reader†
Sayali
Belsare
 *a,
Derek
Tseng
b,
Aydogan
Ozcan
*a,
Derek
Tseng
b,
Aydogan
Ozcan
 bc and
Gerard
Coté
ad
bc and
Gerard
Coté
ad
aDepartment of Biomedical Engineering, Texas A&M University, College Station, TX, USA. E-mail: sayali22@tamu.edu
bElectrical and Computer Engineering Department, University of California, Los Angeles, CA, USA
cBioengineering Department, University of California, Los Angeles, CA, USA
dTexas Engineering Experiment Station Centre for Remote Health Technologies and Systems, College Station, TX, USA
First published on 4th November 2022
Abstract
Chronic conditions like diabetes require monitoring of vital biomarkers over extended periods of time. Monitoring gestational diabetes mellitus (GDM) is crucial to avoid short- and long-term adverse effects on both mother and infant. Providing monitoring systems to patients at the point-of-care (POC) has the potential to help mitigate these effects. In this manuscript, we propose the use of a sensing system combining lateral flow assays (LFAs) with a handheld colorimetric reader for use in tracking the glycemic status of a GDM patient at the POC. Current strategies of GDM monitoring include glucose and HbA1c measurements. These are often too frequent or not frequent enough for effective monitoring. Hence, we have developed a sensor for an intermediate interval biomarker – glycated albumin (GA). Based on the half-life of the protein, GA is measured once every 2–3 weeks. Here we first present two lateral flow assays, one for GA and another for total serum albumin used in conjunction with a handheld reader to read the colorimetric signals. Both assays have a sandwich aptamer format and measure the target proteins in their physiologically relevant ranges. The GA assay has a dynamic range of 3–20 mg ml−1 and the serum albumin assay has a range of 20–50 mg ml−1 without any sample dilution. Both LFAs were then incorporated into a single dual assay cartridge such that both assays could run simultaneously and provide the % glycated albumin value from a single test. Thus, the dual assay cartridge plus reader system has the potential to provide an effective platform for measuring GA for tracking GDM at the POC.
Introduction
Gestational diabetes monitoring
Gestational diabetes is the third most prevalent type of diabetes mellitus after Type I and Type II diabetes.1 As the name suggests, it affects women during pregnancy. GDM most commonly develops between the 24th to 28th week of pregnancy1,2 and has a range of short and long-term adverse effects on both the pregnant woman and foetus. The short-term effects to the patient include pre-eclampsia, shoulder dystocia and increased risk of C-section.3 Short-term effects on the infant include neonatal hypoglycemia and neonatal jaundice. Further, type II diabetes may develop in both mother and infant in the long term.4 To avoid these effects, it is crucial to monitor GDM carefully during pregnancy up to a couple of months after the baby is born.Currently, the standard strategy to monitor GDM is direct glucose measurement and/or HbA1c measurement. Due to the rate of glucose metabolism, it is typically recommended that patients with diabetes measure it 4–5 times a day. Since GDM is primarily characterized by consistent hyperglycemia5 with rare hypoglycemic episodes, multiple glucose measurements may not always be necessary. Since the recommendation is multiple readings per day, traditional glucose meters would require multiple fingerpricks every day, which can result in non-compliance from the patients’ side. The other standardized biomarker for diabetes is glycated hemoglobin (HbA1c).6 Due to its longer half-life, HbA1c is measured only once in 2–3 months.7 In this case, the longer timeline is not ideal since the crucial monitoring period for GDM is about 6–7 months. Hence, specifically for GDM, an intermediate biomarker (interval: 2 weeks) would be advantageous for monitoring glycemic status. It would also be useful for monitoring the effects of therapeutic strategies like alterations in dietary regimens and exercise routines.8
One such intermediate interval biomarker is glycated albumin (GA).9,10 Free glucose in the bloodstream binds to serum albumin via non-enzymatic glycation reaction to form glycated albumin.11 It reflects glycemic excursions of postprandial plasma glucose along with average plasma glucose.12 Since glycated albumin is reported as a ratio of glycated to total albumin, unglycated protein also has to be measured in addition to the glycated version. In normal conditions, about 10 to 16% of serum albumin is glycated.10 However, in cases of prolonged hyperglycemia, up to 40% of albumin can be glycated.10 Since glycation of albumin as a phenomenon is not restricted to GDM, as a biomarker, it can be used to monitor type I and type II diabetes as well.13
Point-of-care monitoring with lateral flow assays
In the past two years, COVID-19 has demonstrated the importance of point-of-care (POC) tests. Testing at home is not just convenient for patients but also plays a major role in relieving the load on healthcare providers.14–16 Most commercially available at-home tests like Panbio rapid test (Abbott) or SD Biosensor rapid test (Roche) use the standard lateral flow assay (LFA) design.17 Lateral flow assays are one of the most established forms of paper based POC test platforms.18,19 Their compact size, portability, low-cost and ease-of-use make them ideal candidates for POC applications. To date, several LFAs have been developed for a range of applications like COVID-19, influenza, bacterial pathogens, heart failure etc.18,20–22The principal components of any LFA typically include recognition elements like antibodies23,24 for the capture of the target biomolecule onto the test strip and transduction elements like enzymes25,26 or nanoparticles21,23 to transduce the recognition event into a readable signal. Antibodies have been established as recognition elements in lateral flow assays owing to their high sensitivity and specificity.27,28 However, they also have drawbacks like high cost of production and thermal instability. On the other hand, aptamers, which are single-stranded DNA or RNA sequences, also have the potential to be used as recognition elements for LFAs due to their thermal stability, stability over a range of pH, low-cost and longer shelf life which make them more suited for use in point-of-care tests.29–31
The other main component of any biosensor is the transduction element. For qualitative POC tests, the signal generated by the transduction element can be observed visually to get a yes/no answer.32 However, for quantitative tests, the transduced signal must be measured and quantified.24,33 Several optical techniques like colorimetry,34 fluorescence35 and surface-enhanced Raman spectroscopy36 have been explored for lateral flow applications. The use of bare gold nanoparticles for colorimetric signal generation is a sensitive and cost-effective optical transduction technique since it does not require any dye and uses less sophisticated equipment for signal generation and measurement.37–40
Ikeda et al.,41 Testa et al.,42 and Ko et al.43 have published assays for glycated albumin detection using various enzymatic methodologies. Inoue et al.,44 and Bohli et al.45 have published enzyme and antibody-based sensors using electrochemical detection methods. Ki et al.46,47 have published colorimetric antibody-based assays to measure glycated and serum albumin. Apiwat et al.48 and Gosh et al.49 have published fluorescence-based detection systems with aptamers as the recognition elements. While successfully detecting and measuring glycated albumin, all these systems have certain limitations in terms of POC applications. Enzymes and antibodies have low thermal stability and hence require low-temperature storage for effective use. Transduction methods like electrochemistry are not ideal for POC application since the associated equipment is likely not compact or portable. Lastly, most of the above-mentioned assays require an additional dilution step for sensing due to high concentrations of the target proteins. Currently the only commercially available assay for GA is the Lucica GA-L assay.50 While this assay is a commercial standard, it is a lab-based enzymatic assay which uses clinical analysers.
In this manuscript, a sensing system has been developed for monitoring gestational diabetes at the point-of-care using glycated albumin as the target biomarker. The sensing system is comprised of a dual assay cartridge consisting of two aptamer-based sandwich assays on lateral flow platforms, for glycated and serum albumin (Fig. 1(a)). Initially both assays were developed separately, and their results have been reported. That was followed by incorporation of both assays into a single dual assay cartridge to run both assays simultaneously. The signal generated by the assays was read using a custom handheld colorimetric reader (Fig. 1(b)–(d)) following our earlier designs.51,52 The combination of lateral flow tests with a compact reader satisfies the RE-ASSURED criteria set by World Health Organization (WHO) for point-of-care testing.53 The combination of dual lateral flow assay cartridge and the handheld reader was designed specifically for simultaneous measurement of both forms of albumin (glycated and total) such that the % glycated albumin value can be obtained by the patient in 30 min at-home without diluting the samples.
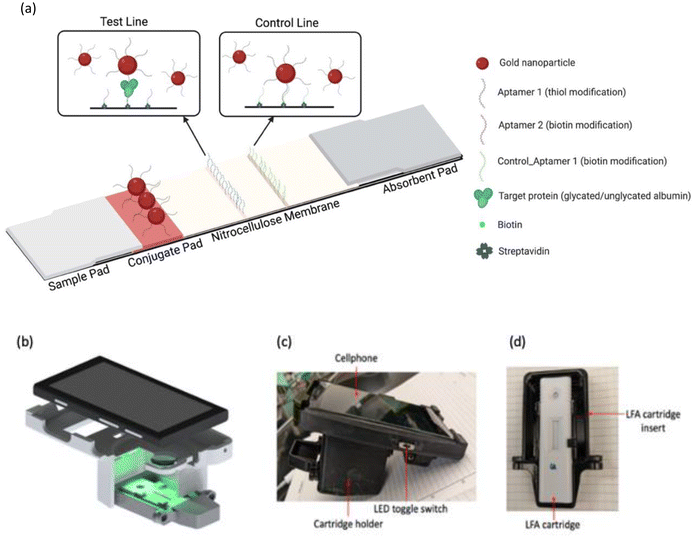 | ||
| Fig. 1 (a) Lateral flow assay schematic (created with BioRender.com); (b) handheld colorimetric reader (CAD design); (c) handheld colorimetric reader and (d) lateral flow assay cartridge holder. | ||
Materials and methods
Materials
Gold(III) chloride trihydrate (HAuCl4) and tri-sodium citrate dihydrate (Na3C5H5O7) for gold nanoparticle synthesis were purchased from MilliporeSigma (St. Louis, MO). Syringe filters (0.2 μm) were purchased from VWR (Pennsylvania, USA). All five aptamers were purchased from Integrated DNA Technologies (Iowa, USA). The oligonucleotide sequences are listed in the ESI (Table S1†). Tris (2-carboxyehtyl) phosphine hydrochloride (TCEP), sodium chloride (NaCl) and phosphate-buffered saline (PBS; pH 7.4) tablets were purchased from Sigma. Nanosep centrifugal filters were purchased from Pall Corporation (New York, USA). Serum albumin, glycated albumin and glucose were purchased from Sigma. Simulated blood serum (without albumin) was purchased from Biochemazone (Edmonton, Canada). Milli-Q deionized (DI) water (18.2 M Ωcm−1) was used in all experiments. Whatmann G4 chromatography paper and nitrocellulose membrane (FF170HP) were purchased from Cytiva (Marlborough, MA). Glass fiber membrane was purchased from EMD Millipore (Burlington, MA). Thick absorbent paper (Trans-blot filter paper) was purchased from BioRad (Hercules, CA). Lateral flow cartridges and backing cards were purchased from DCN Dx (San Diego, CA). ESun pla+filament was purchased from Amazon. UV-Visible spectrometry was conducted using a Tecan Infinite 200 Pro (Männedorf, Switzerland) plate reader.Zeta potential and dynamic light scattering measurements were conducted on a Zetasizer Nano ZS90 (Malvern, UK). Dual assay cartridges were printed on a Prusa i3 mk3s+printer. Colorimetric measurements were performed using a custom handheld reader developed by our team.
Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the glycated albumin assay and 2000
1 for the glycated albumin assay and 2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the serum albumin assay). The aptamer-nanoparticle binding was carried out for one hour in a shaker and then left on the benchtop overnight. Salt aging was conducted after incubation using 2 M NaCl.56 After salt addition was complete, particles were left on the benchtop overnight. The particles were then centrifuged at 10k rcf to remove the unbound aptamers and resuspended in 1× PBS. Particles were resuspended to a final OD 1 for the glycated albumin assay and OD 1.5 for the serum albumin assay. These particles were then stored at room temperature on the benchtop until further use.
1 for the serum albumin assay). The aptamer-nanoparticle binding was carried out for one hour in a shaker and then left on the benchtop overnight. Salt aging was conducted after incubation using 2 M NaCl.56 After salt addition was complete, particles were left on the benchtop overnight. The particles were then centrifuged at 10k rcf to remove the unbound aptamers and resuspended in 1× PBS. Particles were resuspended to a final OD 1 for the glycated albumin assay and OD 1.5 for the serum albumin assay. These particles were then stored at room temperature on the benchtop until further use.
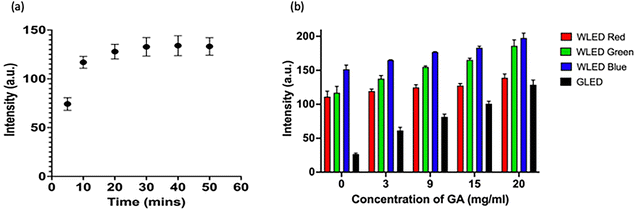
Determination of assay run time. The run time of the assay was the time interval measured starting from the addition of the sample to the development of a constant signal intensity at the test line. It determined by measuring the colorimetric signal intensity at different time points after sample addition: 5 min, 10 min, 20 min, 30 min, 40 min and 50 min.
Determination of colorimetric channel. The custom handheld reader was designed with two inbuilt LEDs: white and green. The ideal colorimetric channel was determined by capturing images with both white and green LED and analysing and comparing all colorimetric channels from both LEDs. The ideal channel was chosen to be the one that gave the highest signal sensitivity from lowest to highest target concentrations for both assays.
Five mins after sample addition, 50 μl of running buffer (1× PBS + 0.1% Tween-20) was added to push all residual conjugates from the conjugate layers and flow through the nitrocellulose membrane. Colorimetric intensities were measured 30 min after sample addition. Each sample was tested in triplicate.
Results and discussion
Nanoparticle characterization
Gold nanoparticles were synthesized by the seed growth method as described above. After synthesis was completed, the absorbance of the nanoparticles was measured using a plate reader. Using the absorbance spectrum, the size and concentration of the nanoparticles was calculated. The spectrum, as seen in Fig. 2(a), has an absorbance peak at 524 nm, which was used to calculate the size of the nanoparticles. These 40 nm nanoparticles were used in all experiments. After both aptamers were conjugated to separate sets of nanoparticle solutions, conjugation was confirmed using via absorbance measurements. An absorbance peak shift of 4 nm confirmed aptamer conjugation onto the nanoparticles for both assays. The peak shifts can be observed in Fig. 2(a). OD 1 particles were used for the glycated albumin assay, while OD 1.5 particles were used in the serum albumin assay owing to the higher concentration range. Nanoparticle size and conjugation was further confirmed by measuring the zeta potential and hydrodynamic size. As seen in Fig. 2(b), a 10 nm increase in hydrodynamic size was observed after conjugation and a reduction of zeta potential from −31 mV to −42 mV was also seen (Fig. 2(c)).Assay response
Once the assay parameters were finalized, both assays were run to obtain responses in the relevant concentration ranges. Since the assays are colorimetric, qualitative responses of both assays were obtained and can be visually observed in Fig. 4(a) and (b). Intensities of colorimetric signals at the test lines (first red line) increased as the concentration of the target protein increased. However, for monitoring purposes, a qualitative result provides insufficient information since knowledge of disease severity is required. It has been established that up to 16% glycated albumin indicates a healthy non-diabetic person.10 However, if the % GA value is much higher, it indicates the need for intervention from healthcare professionals by prescribing alternative dietary or exercise regimens. Quantitative results were obtained by conducting analyses of images obtained using the handheld reader. The quantitative responses (calibration curves) in (Fig 5(a) and (b)) were obtained by analysing images from the green LED, 30 min after sample addition. Each concentration was tested in triplicate. A dynamic range of 3–20 mg ml−1 was achieved for the glycated albumin assay and 20–50 mg ml−1 for the serum albumin assay. It was observed from the plots that both assays have a linear correlation with the concentrations of the target proteins. Regression parameters for both assays have been listed in the ESI (Table S2†). Based on these parameters, the LoD for the glycated albumin assay was calculated to be 0.8 mg ml−1 and for the serum albumin assay was 1.5 mg ml−1.
Assay response in serum
Simulated blood serum was used to evaluate the response of both assays in a complex biological matrix. The serum was purchased without any albumin to control the amount of glycated and serum albumin present. This enabled the creation of samples containing different concentrations of glycated albumin and serum albumin to test both assays. The results are listed in Table 1 below. It can be observed that the recoveries are in the range of 93% to 104%, which implies that serum components do not have a major effect on the assays. Serum dilution was not required for either assay due to the running buffer volume of 50 μl, which was instrumental in improving the flow and overcoming the higher complexity of the serum by preventing non-specific binding.| Sample | Doped (mg ml−1) | Recovered (mg ml−1) | Recovery (%) | SD |
|---|---|---|---|---|
| GA 1 | 8 | 8.25 | 103.15 | 4.87 |
| GA 2 | 20 | 19.48 | 97.4 | 2.7 |
| HSA 1 | 35 | 33 | 94.28 | 3.61 |
| HSA 2 | 50 | 46.89 | 93.78 | 4.04 |
Dual assay response
After both assays were optimized for the individual lateral flow configuration and tested in a complex biological medium, the final step was to incorporate them into a single cartridge. The dual assay cartridge was designed to enable the user to run both assays simultaneously and generate the % glycated albumin result in a single test instead of having to run two separate tests with two separate samples. The cartridge was designed in Solidworks and 3D printed in the lab. Fig. 6(a) illustrates the design of the cartridge. Fig. S1† includes top and bottom views of the cover and bottom of the cartridge. After designing and printing the cartridges, the symmetry of flow to both sides of the cartridge was confirmed by measuring and comparing signals for the same assay on both sides of the cartridge. It can be observed from Fig. 6(b) that signal intensities of assays on both sides of the cartridge were comparable implying that similar sample volumes flowed on both sides of the assays. Once flow symmetry was confirmed, dual assay response was tested via three samples with varying concentrations of glycated and serum albumin: S1 consisted of 3.5 mg ml−1 GA and 25 mg ml−1 HSA; S2 was comprised of 10 mg ml−1 GA and 30 mg ml−1 HSA; S3 was a mix of 20 mg ml−1 GA and 30 mg ml−1 HSA. The concentration levels were chosen such that the dynamic range of each assay was covered between the three samples. Fig. 7 demonstrates the dual assay response for all levels. Concentration levels of sample one (GA: 3.5 mg ml−1 and SA: 25 mg ml−1) were chosen to demonstrate that lowest concentrations of both assay dynamic ranges could be successfully detected in the dual assay system. Sample 1 was equivalent to 12% glycation which corresponded to a non-diabetic healthy person. The second sample was at the mid-point of the glycated albumin assay range. For the serum albumin assay, the total serum albumin concentrations combined was equivalent to the mid-point of its dynamic range.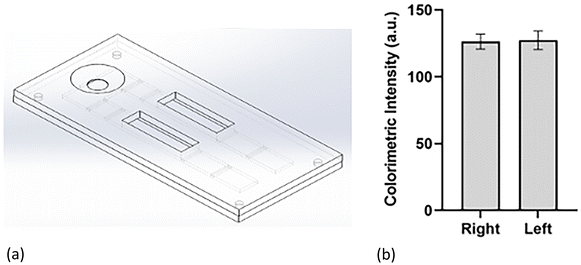 | ||
| Fig. 6 (a) Dual assay cartridge design; (b) symmetry of flow in both channels of dual assay cartridge. | ||
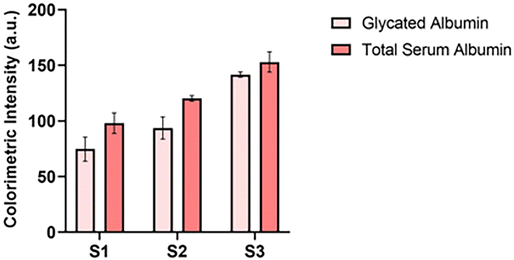 | ||
| Fig. 7 Dual assay response (S1: 3.5 mg ml−1 GA + 25 mg ml−1 HSA; S2: 10 mg ml−1 GA + 30 mg ml−1 HSA; S3: 20 mg ml−1 GA + 30 mg ml−1 HSA). | ||
This sample corresponded to 25% glycation which was comparable to mild to moderately severe diabetes. Finally, in the last sample, the concentration of glycated albumin was chosen to be the upper limit of the dynamic range and serum albumin concentration was chosen such that the summation of both concentrations was equivalent to the highest relevant concentration for serum albumin. It was a 40% glycation sample which corresponded to a severely diabetic condition. In sample 3, unglycated serum albumin concentration was maintained at the same concentration as sample 2 to demonstrate that only an increase in glycated albumin could also be detected by the serum albumin assay. It was observed that there was a 7–10% increase in signal intensities compared to individual assays run separately. However, this was expected since in case of the glycated albumin assay, non-specific binding from unglycated serum albumin to a small extent was anticipated. And in case of the serum albumin assay, aptamer binding to unglycated and glycated albumin was expected to differ to some extent resulting in minor differences in resultant signal intensities even at the same concentrations. In addition, all samples were doped in simulated blood serum which also contributed to the non-specific binding albeit to a small extent. However, it was noted that signal intensities for all concentration levels increased to a similar extent and hence the dual assay cartridge could in fact successfully be used to simultaneously measure glycated and total serum albumin to obtain the % glycated albumin value from a single sample and single test.
Conclusion
In this manuscript, two aptamer-based lateral flow assays were developed to measure glycated albumin and serum albumin in their relevant concentration ranges. The choice of aptamers was made over other recognition elements owing to their thermal stability, low cost, and relatively long shelf life. Both assays used gold nanoparticle-based colorimetry since it does not require any additional use of dyes which lowered the overall cost of the system. Both assays were immobilized onto lateral flow platforms, which afforded advantages like compact size, portability, and ease-of-use. A handheld colorimetric reader developed by the team was used in conjunction with lateral flow cartridges. When the sample was added to the cartridge, the target protein interacted and bound to the aptamer on the gold nanoparticles which were stored in the conjugate layer. The protein-particle complexes then flowed onto the nitrocellulose membrane, where they were captured at the test line. The number of particles at the test line was proportional to the concentration of the target protein in the sample, which was observed in the assay responses. The dynamic ranges for both assays were 3–20 mg ml−1 and 20–50 mg ml−1, which are both physiologically relevant, implying that sample dilution will not be required to run the assays. Finally, a dual assay cartridge was used to run both assays simultaneously to obtain the value of % glycated albumin from a single test. In conclusion, this research demonstrated that a dual lateral flow assay plus handheld colorimetric reader setup can effectively monitor glycated and serum albumin for ultimate use as a point-of-care sensor for monitoring gestational diabetes.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Science Foundation Precise Advanced Technologies and Health Systems for Underserved Populations (PATHS-UP) Engineering Research Center through award no. EEC-1648451. The authors would also like to acknowledge the discussions with Dr Samuel Mabbott on lateral flow assay optimization.References
- American Diabetes Association, Gestational Diabetes Mellitus, Diabetes Care, 2004, 27, s88–s90, DOI:10.2337/diacare.27.2007.S88.
- R. Kaaja and T. Rönnemaa, Gestational diabetes: Pathogenesis and consequences to mother and offspring, Rev Diabet. Stud., 2008, 5(4), 194–202, DOI:10.1900/RDS.2008.5.194.
- Y. Yogev, E. M. J. Xenakis and O. Langer, The association between preeclampsia and the severity of gestational diabetes: The impact of glycemic control, Am. J. Obstet. Gynecol., 2004, 191(5), 1655–1660, DOI:10.1016/j.ajog.2004.03.074.
- Maryland. Mild Traumatic Brain Injury (mTBI)/Concussion Policy and Management Plan. Published online 2015: 2–21.
- C. Kühl, P. J. Hornnes and O. Andersen, Etiology and Pathophysiology of gestational diabetes mellitus, Diabetes, 1985, 34(2), 66–70, DOI:10.1021/ma9704673.
- P. B. Renz, G. Cavagnolli, L. S. Weinert, S. P. Silveiro and J. L. Camargo, HbA1c test as a tool in the diagnosis of gestational diabetes mellitus, PLoS One, 2015, 10(8), 1–11, DOI:10.1371/journal.pone.0135989.
- S. I. Sherwani, H. A. Khan, A. Ekhzaimy, A. Masood and M. K. Sakharkar, Significance of HbA1c test in diagnosis and prognosis of diabetic patients, Biomarker Insights, 2016, 11, 95–104, DOI:10.4137/Bmi.s38440.
- Management of Diabetes in Pregnancy, Standards of medical care in Diabetes-2018, Diabetes Care, 2018, 41, S137–S143, DOI:10.2337/dc18-S013.
- Y. Huang, Y. Hu, Y. Ma and G. Ye, Glycated albumin is an optimal biomarker for gestational diabetes mellitus, Exp. Ther. Med., 2015, 10(6), 2145–2149, DOI:10.3892/etm.2015.2808.
- T. Kohzuma, T. Yamamoto, Y. Uematsu, Z. K. Shihabi and B. I. Freedman, Basic performance of an enzymatic method for glycated albumin and reference range determination, J. Diabetes Sci. Technol., 2011, 5(6), 1455–1462, DOI:10.1177/193229681100500619.
- I. Sirangelo and C. Iannuzzi, Understanding the role of protein glycation in the amyloid aggregation process, Int. J. Mol. Sci., 2021, 22(12), 1–20, DOI:10.3390/ijms22126609.
- M. Koga, Glycated albumin; clinical usefulness, Clin. Chim. Acta, 2014, 433, 96–104, DOI:10.1016/j.cca.2014.03.001.
- H. V. Roohk and A. R. Zaidi, A Review of Glycated Albumin as an Intermediate Glycation Index for Controlling Diabetes, J. Diabetes Sci. Technol., 2008, 2(6), 1114–1121, DOI:10.1177/193229680800200620.
- C. P. Price and A. St. John, Will COVID-19 be the coming of age for point-of-care testing?, BMJ Innov., 2021, 7(1), 3–5, DOI:10.1136/bmjinnov-2020-000466.
- M. J. O'Kane, Patient self-testing in chronic disease management, J. Appl. Lab. Med., 2020, 44(2), 81–87, DOI:10.1515/labmed-2019-0175.
- D. Dunlap, E. Ding and K. Abramo, et al., Point-of-care testing, your cardiologist, and affairs of the heart, Cardiovasc. Digit. Health J., 2021, 2(6), 331–335, DOI:10.1016/j.cvdhj.2021.10.004.
- V. M. Corman, V. C. Haage and T. Bleicker, et al., Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study, Lancet Microbe, 2021, 2(7), e311–e319, DOI:10.1016/S2666-5247(21)00056-2.
- Y. Zhou, Y. Wu, L. Ding, X. Huang and Y. Xiong, Point-of-care COVID-19 diagnostics powered by lateral flow assay, Trends Anal. Chem., 2021, 145, 116452 CrossRef CAS PubMed.
- M. Pohanka, Point-of-Care Diagnoses and Assays Based on Lateral Flow Test, Int. J. Anal. Chem., 2021, 6685619, DOI:10.1155/2021/6685619.
- T. T. Le, P. Chang, D. J. Benton, J. W. McCauley, M. Iqbal and A. E. G. Cass, Dual Recognition Element Lateral Flow Assay Toward Multiplex Strain Specific Influenza Virus Detection, Anal. Chem., 2017, 89(12), 6781–6786, DOI:10.1021/acs.analchem.7b01149.
- R. Wang, K. Kim and N. Choi, et al., Highly sensitive detection of high-risk bacterial pathogens using SERS-based lateral flow assay strips, Sens. Actuators, B, 2018, 270, 72–79, DOI:10.1016/j.snb.2018.04.162.
- M. You, M. Lin and Y. Gong, et al., Household Fluorescent Lateral Flow Strip Platform for Sensitive and Quantitative Prognosis of Heart Failure Using Dual-Color Upconversion Nanoparticles, ACS Nano, 2017, 11(6), 6261–6270, DOI:10.1021/acsnano.7b02466.
- D. Zhang, L. Huang and B. Liu, et al., Quantitative and ultrasensitive detection of multiplex cardiac biomarkers in lateral flow assay with core-shell SERS nanotags, Biosens. Bioelectron., 2018, 106, 204–211, DOI:10.1016/j.bios.2018.01.062.
- J. Deng, M. Yang, J. Wu, W. Zhang and X. Jiang, A Self-Contained Chemiluminescent Lateral Flow Assay for Point-of-Care Testing, Anal. Chem., 2018, 90(15), 9132–9137, DOI:10.1021/acs.analchem.8b01543.
- K. K. Fung, C. P. Y. Chan and R. Renneberg, Development of enzyme-based bar code-style lateral-flow assay for hydrogen peroxide determination, Anal. Chim. Acta, 2009, 634(1), 89–95, DOI:10.1016/j.aca.2008.11.064.
- D. Calabria, M. M. Calabretta and M. Zangheri, et al., Recent advancements in enzyme-based lateral flow immunoassays, Sensors, 2021, 21(10), 1–19, DOI:10.3390/s21103358.
- M. Sajid, A. N. Kawde and M. Daud, Designs, formats and applications of lateral flow assay: A literature review, J. Saudi Chem. Soc., 2015, 19(6), 689–705, DOI:10.1016/j.jscs.2014.09.001.
- K. M. Koczula and A. Gallotta, Lateral flow assays, Essays Biochem., 2016, 60(1), 111–120, DOI:10.1042/EBC20150012.
- A. Chen and S. Yang, Replacing antibodies with aptamers in lateral flow immunoassay, Biosens. Bioelectron., 2015, 71, 230–242, DOI:10.1016/j.bios.2015.04.041.
- S. Dalirirad and A. J. Steckl, Aptamer-based lateral flow assay for point of care cortisol detection in sweat, Sens. Actuators, B, 2019, 283, 79–86, DOI:10.1016/j.snb.2018.11.161.
- M. Jauset-Rubio, M. S. El-Shahawi, A. S. Bashammakh, A. O. Alyoubi and C. K. O′Sullivan, Advances in aptamers-based lateral flow assays, TrAC, Trends Anal. Chem., 2017, 97, 385–398, DOI:10.1016/j.trac.2017.10.010.
- A. Valdivia, I. Torres and D. Huntley, et al., Qualitative assessment of SARS-CoV-2-specific antibody avidity by lateral flow immunochromatographic IgG/IgM antibody assay, J. Med. Virol., 2021, 93(2), 1141–1144, DOI:10.1002/jmv.26344.
- D. Tu, A. Holderby and G. L. Coté, Aptamer-based surface-enhanced resonance Raman scattering assay on a paper fluidic platform for detection of cardiac troponin I, J. Biomed. Opt., 2020, 25(9), 097001, DOI:10.1117/1.JBO.25.9.097001.
- D. Kong, L. Liu, S. Song, Q. Zheng, X. Wu and H. Kuang, Rapid detection of tenuazonic acid in cereal and fruit juice using a lateral-flow immunochromatographic assay strip, Food Agric. Immunol., 2017, 28(6), 1293–1303, DOI:10.1080/09540105.2017.1337085.
- L. G. Lee, E. S. Nordman, M. D. Johnson and M. F. Oldham, A low-cost, high-performance system for fluorescence lateral flow assays, Biosensors, 2013, 3(4), 360–373, DOI:10.3390/bios3040360.
- S. Choi, J. Hwang, S. Lee, D. W. Lim, H. Joo and J. Choo, Quantitative analysis of thyroid-stimulating hormone (TSH) using SERS-based lateral flow immunoassay, Sens. Actuators, B, 2017, 240, 358–364, DOI:10.1016/j.snb.2016.08.178.
- R. Pan, Y. Jiang and L. Sun, et al., Gold nanoparticle-based enhanced lateral flow immunoassay for detection of Cronobacter sakazakii in powdered infant formula, J. Dairy Sci., 2018, 101(5), 3835–3843, DOI:10.3168/jds.2017-14265.
- T. Mahmoudi, B. Shirdel, B. Mansoori and B. Baradaran, Dual sensitivity enhancement in gold nanoparticle-based lateral flow immunoassay for visual detection of carcinoembryonic antigen, Anal. Sci. Adv., 2020, 1–12, DOI:10.1002/ansa.202000023.
- J. Liu, X. Hu, F. Cao, Y. Zhang, J. Lu and L. Zeng, A lateral flow strip based on gold nanoparticles to detect 6-monoacetylmorphine in oral fluid, R. Soc. Open Sci., 2018, 5(6), 1–11, DOI:10.1098/rsos.180288.
- Y. Panraksa, A. Apilux and S. Jampasa, et al., A facile one-step gold nanoparticles enhancement based on sequential patterned lateral flow immunoassay device for C-reactive protein detection, Sens. Actuators, B, 2020, 129241, DOI:10.1016/j.snb.2020.129241.
- K. Ikeda, Y. Sakamoto and Y. Kawasaki, et al., Determination of glycated albumin by enzyme-linked boronate immunoassay (ELBIA), Clin Chem., 1998, 44(2), 256–263 CrossRef CAS.
- R. Testa, E. Guerra, A. Rita, N. Gaetano di and G. Santini, Analytical Performances of an Enzymatic Assay for the Measurement of Glycated Albumin, J. Appl. Lab. Med., 2016, 01(02), 162–171, DOI:10.1373/jalm.2016.020446.
- E. Ko, V. K. Tran and Y. Geng, et al., Determination of glycated albumin using boronic acid-derived agarose beads on paper-based devices, Biomicrofluidics, 2018, 12(1), 1–8, DOI:10.1063/1.5021395.
- Y. Inoue, M. Inoue, M. Saito, H. Yoshikawa and E. Tamiya, Sensitive Detection of Glycated Albumin in Human Serum Albumin Using Electrochemiluminescence, Anal. Chem., 2017, 89(11), 5909–5915, DOI:10.1021/acs.analchem.7b00280.
- N. Bohli, O. Meilhac, P. Rondeau, S. Gueffrache, L. Mora and A. Abdelghani, A facile route to glycated albumin detection, Talanta, 2018, 184, 507–512, DOI:10.1016/j.talanta.2018.03.027.
- H. Ki, J. Oh, G. R. Han and M. G. Kim, Glycation ratio determination through simultaneous detection of human serum albumin and glycated albumin on an advanced lateral flow immunoassay sensor, Lab Chip, 2020, 20(4), 844–851, 10.1039/c9lc00967a.
- H. Ki, H. Jang and J. Oh, et al., Simultaneous Detection of Serum Glucose and Glycated Albumin on a Paper-Based Sensor for Acute Hyperglycemia and Diabetes Mellitus, Anal. Chem., 2020, 92(17), 11530–11534, DOI:10.1021/acs.analchem.0c02940.
- C. Apiwat, P. Luksirikul and P. Kankla, et al., Graphene based aptasensor for glycated albumin in diabetes mellitus diagnosis and monitoring, Biosens. Bioelectron., 2016, 82, 140–145, DOI:10.1016/j.bios.2016.04.015.
- S. Gosh, D. Datta, M. Cheema, M. Dutta and M. Stroscio, Aptasensor based optical detection of glycated albumin for diabetes mellitus diagnosis, Nanotechnology, 2017, 28, 435505–435516 CrossRef PubMed.
- T. Kouzuma, T. Usami, M. Yamakoshi, M. Takahashi and S. Imamura, An enzymatic method for the measurement of glycated albumin in biological samples, Clin. Chim. Acta, 2002, 324(1–2), 61–71, DOI:10.1016/S0009-8981(02)00207-3.
- O. Mudanyali, S. Dimitrov, U. Sikora, S. Padmanabhan, I. Navruz and A. Ozcan, Integrated rapid-diagnostic-test reader platform on a cellphone, Lab Chip, 2012, 12(15), 2678–2686, 10.1039/c2lc40235a.
- H. A. Joung, Z. S. Ballard and J. Wu, et al., Point-of-Care Serodiagnostic Test for Early-Stage Lyme Disease Using a Multiplexed Paper-Based Immunoassay and Machine Learning, ACS Nano., 2020, 14(1), 229–240, DOI:10.1021/acsnano.9b08151.
- K. J. Land, D. I. Boeras, X. S. Chen, A. R. Ramsay and R. W. Peeling, REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes, Nat. Microbiol., 2019, 4(1), 46–54, DOI:10.1038/s41564-018-0295-3.
- N. G. Bastús, J. Comenge and V. Puntes, Kinetically controlled seeded growth synthesis of citrate-stabilized gold nanoparticles of up to 200 nm: Size focusing versus ostwald ripening, Langmuir, 2011, 27(17), 11098–11105, DOI:10.1021/la201938u.
- S. Belsare and G. Coté, Development of paper-based colorimetric assays for monitoring gestational diabetes at the point-of-care, Proc. SPIE, 2021, 11651, 21, DOI:10.1117/12.2585542.
- B. Liu and J. Liu, Methods for preparing DNA-functionalized gold nanoparticles, a key reagent of bioanalytical chemistry, Anal. Methods, 2017, 9(18), 2633–2643, 10.1039/c7ay00368d.
- E. B. Bahadır and M. K. Sezgintürk, Lateral flow assays: Principles, designs and labels, TrAC, Trends Anal. Chem., 2016, 82, 286–306, DOI:10.1016/j.trac.2016.06.006.
- A. M. López-Marzo, J. Pons, D. A. Blake and A. Merkoçi, High sensitive gold-nanoparticle based lateral flow Immunodevice for Cd2+detection in drinking waters, Biosens. Bioelectron., 2013, 47, 190–198, DOI:10.1016/j.bios.2013.02.031.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an01238c |
| This journal is © The Royal Society of Chemistry 2022 |