Selective and rapid detection of mercury ion based on DNA assembly and nicking endonuclease-assisted signal amplification
Abstract
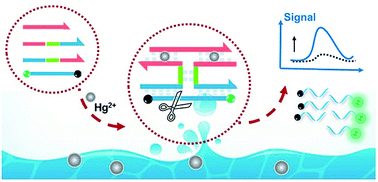
Sensitive and rapid detection of mercury ions (Hg2+) is of great significance for public health control. In this work, an innovative enzyme-assisted target recycling amplification strategy is proposed based on an I-shaped DNA assembly structure coupled with endonuclease-assisted isothermal amplification, termed ID–EIA. For this purpose, four DNA probes were designed, namely, a recognition probe, semi-enzyme-A, semi-enzyme-B, and fluorescein/quencher labeled reporting probe. In the absence of Hg2+, the four probes were free, while in the presence of Hg2+, an I-shaped DNA assembly structure was rapidly formed due to the specific Hg2+-mediated thymine–thymine base pairing (T-Hg2+–T). A nicking enzyme recognition locus appeared in the stem of the I-shaped junction, which allowed Nt.BstNBI to further cleave the reporting probe and generate fluorescence signal. Another reporting probe was then spontaneously hybridized with the remaining part of the junction and cleaved. The continuous cycle of hybridization and nicking resulted in significant signal amplification, enabling the sensitive detection of Hg2+ under a constant temperature condition. A linear range from 5 to 250 nM Hg2+ was achieved, with a detection limit of 1.7 nM (0.34 ppb). Furthermore, the analysis showed satisfactory selectivity and remarkable simplicity and could be accomplished facilely in one step within 1 h, which is much faster than other methods. The method was successfully implemented in tap water samples with satisfactory results, showing good practical value.
- This article is part of the themed collection: Analytical Methods Recent HOT articles


 Please wait while we load your content...
Please wait while we load your content...