Open Access Article
Open Access ArticleNasopharyngeal carcinoma related miRNA detection through a DSN enzyme assisted tetrahedral probe for more accurate prognosis†
Li Zhanga and
Zhiping Lou *b
*b
aDepartment of Otolaryngology, Zhuji Affiliated Hospital of Shaoxing University, Zhuji City, Zhejiang Province 311800, China
bDepartment of Otolaryngology, Zhuji Affiliated Hospital of Shaoxing University, Zhuji People's Hospital, 9 Jianmin Road, Taozhu Street, Zhuji City, Zhejiang Province 311800, China. E-mail: 624341450@qq.com; Fax: +86-057581782153
First published on 18th March 2021
Abstract
MicroRNAs (miRNAs) play a pivotal role in cellular functions and in the development and progression of cancer. Precise quantification of miRNAs from different clinical samples is a challenging but necessary endeavor that is largely neglected by many emerging fluorescence technologies. Herein, we propose here a novel miRNA detection method through a designed tetrahedral probe and DSN enzyme for attached signal amplification. The highlights of the method are calculated as: (i) the tetrahedral probe induced a much higher transfection efficiency than normal probes; (ii) the DSN enzyme-based cleavage ensures amplification and thus leads to a high detection sensitivity. In all, we believe that the proposed method could greatly improve the sensitivity of intracellular miRNA imaging methods and contribute to the fundamental research and prognosis of cancer.
Introduction
MicroRNAs (miRNAs) play pivotal roles in development, cellular differentiation, proliferation, cell cycle control and cell death, and have been implicated in a variety of human diseases, including cancer.1,2 A growing body of evidence has implied that miRNAs are closely related with the management of chemotherapy efficacy in diverse diseases. Nasopharyngeal carcinoma (NPC) is a tumor type implicated with Epstein–Barr virus (EBV) exposure, diet and genetic factors as its aetiology and arises from the epithelial cells that line the nasopharynx.3,4 A growing body of studies have demonstrated that abnormal expression of miRNAs is closely related with pathogenesis process of NPC.5,6 Among all the miRNAs reported, miR3188 is one of the most original miRNAs discovered and are important in both reducing cell-cycle transition and proliferation as well as significantly prolonging the survival time of tumor-bearing mice as well as sensitizes cells to 5-FU.7,8 Therefore, accurate and sensitive identification and quantification of both in vitro and intracellular miR3188 will contribute to the fundamental research and prognosis of NPC.The most classic method for miRNA detection is qrt-PCR, which is widely applied for in vitro miRNA detection from clinical samples with a detection limit down to aM level.9 Even though, qrt-PCR is also criticized from some instinct drawbacks, including complicated primer design, temperature cycle, heavy labor, high costs and the most importantly its incapability for in situ miRNA imaging, that greatly limited its further application with different requirements.10 Accurate and sensitive in situ miRNA detection in living cells is pivotal for the basically biomedical researches and disease diagnosis.11,12 However, the development of in situ miRNA imaging methods are faced with a great challenge because of their low expression level and complex environments. The most developed in situ miRNA fluorescence imaging is mostly with a one-to-one pattern that could not for accurate and sensitive monitoring of intracellular miRNAs.13,14 Furthermore, the unsatisfactory uptake efficiency of designed molecular beacons by parent cells adds difficulties to the miRNA imaging methods and lead to a low sensitivity. Therefore, it is in high demand to develop an accurate and sensitive miRNA imaging methods with a favorable molecular beacon uptake efficiency. In recent years, duplex-specific nuclease (DSN)-mediated signal amplification has attracted great interest because of its good enzyme stability and high catalytic activity. For example, Huiting Lu proposed an ultrasensitive and multiple miRNA detection strategy through the combination of three-dimensional tetrahedron-structured probes-modified gold nanoparticle probes and DSN-assisted signal amplification with a low limit of detection.15 Kai Zhang also successfully detected target miRNA from complicated biological samples in a sensitive way through two stages DSN-assisted target recycling signal amplification. Therefore, DSN enzyme has attracted abundant attention in the establishments of in vitro miRNA detection methods. However, few of the former methods have investigated the feasibility of DSN enzyme for intracellular imaging.
Herein, we propose here a novel in situ miRNA imaging methods through the integration of both tetrahedral induced efficient intake of molecular beacons and DSN enzymes-based signal amplification. In the proposed method, molecular beacon is designed with a tetrahedral structure so as to improve the uptake efficiency by target cells. Furthermore, tetrahedral structure probe could be recognized and then displaced by target intracellular miRNA, and thus keep the fluorescence moiety get away from corresponding quenching moiety. As a result, the obtained fluorescence is related with the amounts of intracellular miRNA. With the assistance of DSN enzymes, the DNA sequences that is complementary with target miRNA is degraded and thus the released miRNA could attend a next amplification cycle. Therefore, we believe that the proposed method will greatly improve the sensitivity of intracellular miRNA imaging methods and contribute to the fundamental research and prognosis of cancers.
Result and discussions
The principle of the proposed method for intracellular miRNA imaging
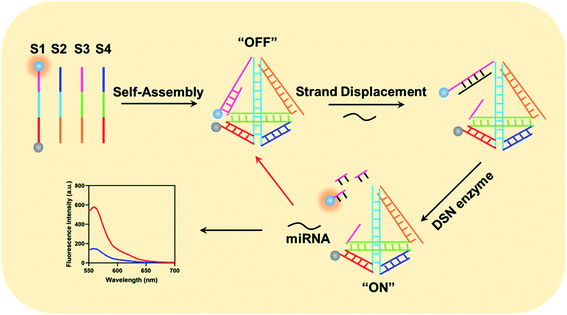
The working mechanism of the method is illustrated in Scheme 1. In the proposed method, we have carefully designed four DNA sequences: S1, S2, S3 and S4. In details, we have labeled a fluorescence moiety Cy3 (Sulfo-Cyanine3) and corresponding quenching moiety (BHQ-2, Black Hole Quencher-2). Furthermore, the 5′ terminal of S1 could specially hybridize with both part of S3 and miRNA. After gradient cooling of the S1–S4 mixture from 90 °C to room temperature, the four DNA sequences could auto-assembled into a tetrahedral structure. Notably, the 5′-terminal part of S3 probe could only partially hybridize with corresponding part in S1 and the molecular beacon is on its “OFF” state due to the fluorescence is quenched by BHQ-2. When target miRNA existed, it could specially recognize the uncomplemented region in the 5′ terminal of S1 probe and form a NA–RNA complex. Afterwards, the DSN enzyme could recognize the DNA–RNA complex in the tetrahedral beacons and specially degrade the DNA sequences in the complex. Eventually, Cy3 gets apart from BHQ-2 and generated fluorescence intensity, which is in positive correlation with the amounts of intracellular miRNA. Besides, the released miRNA could then enter the next amplification cycle to continuously generate fluorescence signals.Investigation of the designed tetrahedral probes for intracellular miRNA imaging
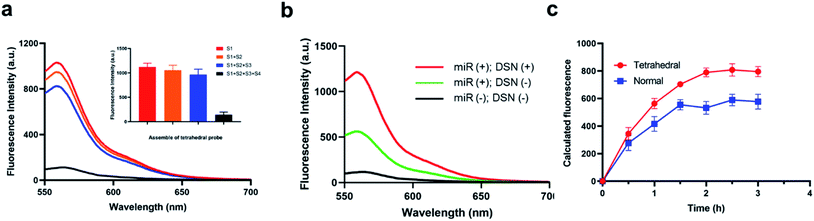
It is widely reported that the tetrahedral structure probe could induce a higher uptake efficiency and thus lead to a higher sensitivity. Therefore, we firstly studied the self-assemble of the designed tetrahedral probes via PAGE gel electrophoresis. As shown in Fig. S1,† the S1 DNA sequences is about 50 bp in the lane 1. When mixed with S2, the S1 + S2 complex is about 100 bp with a lower mobility (lane 2). Meanwhile, the mobility of S1 + S2 + S3 complex and S1 + S2 + S3 + S4 complex gradually decrease in lane 3 and lane 4. Therefore, the tetrahedral is successfully assembled. Besides the PAGE gel electrophoresis analysis, we have also studied the fluorescence-quenching effect in the self-assemble process through a fluorescence assay. In the original stated of S1 alone, we have observed a high fluorescence intensity about 789 a.u., and the fluorescence intensity showed no obvious decrease when S1 mixed with S2 and S3. When the four DNA sequences existed simultaneously, the fluorescence intensity have greatly decreased, indicating that only the successful assemble of the tetrahedral structure could initiate the quench of Cy3 fluorescence (Fig. 1a). We then investigated the feasibility of the method for miRNA detection through a fluorescence assay. As shown in Fig. 1b, a significantly high enhanced fluorescence responses were observed in the presence of target miRNA and active DSN enzymes. The sensing system with the active DSN enzymes could provide an approximate 2.5 times higher fluorescence signals than that is without active DSN enzymes, indicating that the participant of active DSN enzyme could provide an attached signal amplification which is consistent with its principles. Afterwards, we compared the uptake efficiency of tetrahedral structure probe and normal probes by target cells. Eventually, we have observed a gradually increased fluorescence intensity in both the tetrahedral probe group and the normal probe group, indicating both the two probes could enter target cancer cells (Fig. 1c). Notably, when incubated with target cancer cells for more than 30 minutes, the tetrahedral group exhibited a gradually higher fluorescence intensity than that in the normal probe group, suggesting that the tetrahedral could provide a much higher transmembrane efficiency than normal probes. The possible high transmembrane process of the tetrahedral probe may derived from its stable structure. At last, we have studied the cytotoxicity of the designed tetrahedral probes in different concentration and did not observe obvious cytotoxicity (Fig. S2).† Furthermore, we have also investigated the stability of the designed tetrahedral probe through detection of fluorescence intensity of tetrahedral probe in different experimental conditions (BSA, PBS buffer, RPMI-1640). From the result in Fig. S3,† there was no significant fluorescence signals enhancements when tetrahedral probe incubated with different experimental conditions, indicating a favorable stability of the probe and potential capability for intracellular application.Optimization of the experimental conditions
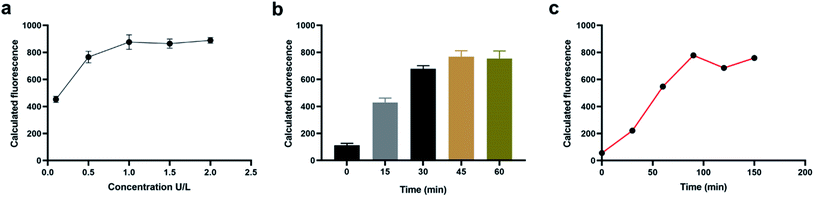
In order to obtained a better detection performance, we have then optimized the experimental conditions including the concentration of DSN enzymes, the incubation time of DSN enzymes and the incubation time of tetrahedral probes. In the experiment exploring the concentration of DSN enzymes used in the sensing system, we have diluted the purchased DSN enzymes into different concentration gradient and detected the fluorescence intensity of the whole systems with 100 nM miRNA. From the result in Fig. 2a, we have obtained a gradually increased fluorescence signals with the concentrations of DSN enzymes ranging from 0.1 U L−1 to 1 U L−1 and observed no more increasements with the concentration more than 1 U L−1, indicating the concentration 1 U L−1 could provide an optimized detection performance. We then studied the incubation time of DSN enzymes. As shown in Fig. 2b, then sensing system could provide a most significant fluorescence increments when incubated with DSN enzyme for about 45 min. Therefore, 45 min is selected as the most appropriate incubation time. With the same procedure, the incubation time of tetrahedral probe is eventually determined 90 min (Fig. 2c).Sensitivity and specificity of the sensing system for in vitro miRNA detection
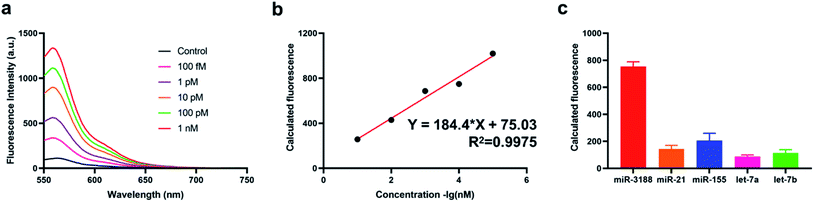
Under the optimized experimental conditions, we have then studied the sensitivity of the method through applying it for target miRNA detection with different concentrations ranging from 100 fM to 1 nM. The fluorescence intensity gradually increased with the concentration of target miRNA increased from 100 fM to 1 nM (Fig. 3a) and is positively correlated with the amounts of target miRNA. Eventually, the obtained correlation equation was determined Y = 184.4 × lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + 75.03 (C refers to the concentration of target miRNAs) with a correlation coefficient R2 = 0.9975 (Fig. 3b), indicating that the method could provide a sensitive response to the quantification of target miRNA. The limit of detection (LOD) of the proposed method was also calculated to be 25.7 fM through triple deviation method. Furthermore, we verified the selectivity of the method through applying it for the detection of different miRNAs including miR-21, miR-155, Let-7a, Let-7b. As a result, we have obtained a significantly enhanced fluorescence intensity for the detection of target miRNA compared with the other four miRNAs (Fig. 3c), indicating that the method also showed a favorable specificity for different miRNA detection, which could meet the clinical application requirements.
C + 75.03 (C refers to the concentration of target miRNAs) with a correlation coefficient R2 = 0.9975 (Fig. 3b), indicating that the method could provide a sensitive response to the quantification of target miRNA. The limit of detection (LOD) of the proposed method was also calculated to be 25.7 fM through triple deviation method. Furthermore, we verified the selectivity of the method through applying it for the detection of different miRNAs including miR-21, miR-155, Let-7a, Let-7b. As a result, we have obtained a significantly enhanced fluorescence intensity for the detection of target miRNA compared with the other four miRNAs (Fig. 3c), indicating that the method also showed a favorable specificity for different miRNA detection, which could meet the clinical application requirements.
Application of the method for in situ miRNA imaging
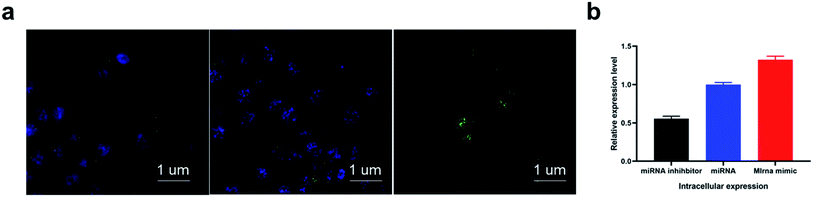
Considering the good biocompatibility, high transfection efficiency of the designed tetrahedral probe, we eventually applied the proposed sensing system for in situ miRNA imaging. From the imaging result in Fig. 4, the CNE-1 cells treated with miRNA inhibitors, which could depress the expression of target miRNA, showed negligible fluorescence, while the cells treated with miRNA mimic showed a much higher fluorescence response compared with the normal cell and inhibitors treated cells, indicating that the method could accurately report the dynamic variation of intracellular miRNAs. Confocal result with lower magnification in Fig. S4.†Conclusions
Herein, we report a novel intracellular miRNA imaging method implementing tetrahedral probe and cleavage characteristics of DSN enzymes and demonstrated its high sensitivity and specificity through a series of experiments. In the research, we have compared the tetrahedral structure probes with normal structure probes and also verified the high transfection efficiency of the designed tetrahedral probes. We find that the sensing system is amenable to detection of oncogenic miRNAs in CNE-1 cells with high sensitivity and successfully monitored the expression change of oncogenic miRNAs in cancer cells, providing a useful complementary tool for cancer clinical diagnosis and prognosis. We have then compared the in vitro detection capability of the method with some former developed methods (Table 1) and summarized the advantages as: (i) capability of both in vitro and in situ miRNA detection; (ii) a relatively high transmembrane efficiency derived from the tetrahedral structure probe; (iii) a comparable sensitivity with most of the former methods. This study not only has reference value in biomedical research but also has potential value in clinical analysis and product development applications such as the development of in vitro diagnostic reagents.| Title | Sensitivity | In vitro/in situ | Complexity | Ref. |
|---|---|---|---|---|
| a RCA: rolling circle amplification; UCNPs: upconversion nanoparticles. | ||||
| The method | fM | Both | Exo-III | |
| CRISPR-Cas12a | fM | In vitro | Cas12a enzyme; RCA related enzymes | |
| UCNPs based | fM | In situ | UCNPs | |
| Qrt-PCR | fM | In vitro | Equipment; primers; DNA polymerase | |
Conflicts of interest
There are no conflicts to declare.References
- L. Chen, L. Heikkinen, C. Wang, Y. Yang, H. Sun and G. Wong, Trends in the development of miRNA bioinformatics tools, Briefings Bioinf., 2019, 20(5), 1836–1852 CrossRef CAS PubMed.
- C. Foye, I. K. Yan, W. David, N. Shukla, Y. Habboush, L. Chase, K. Ryland, V. Kesari and T. Patel, Comparison of miRNA quantitation by Nanostring in serum and plasma samples, PLoS One, 2017, 12(12), e0189165 CrossRef PubMed.
- W. K. J. Lam and J. Y. K. Chan, Recent advances in the management of nasopharyngeal carcinoma, F1000Research, 2018, 7, Rev-1829 Search PubMed.
- H. M. Lee, K. S. Okuda, F. E. Gonzalez and V. Patel, Current Perspectives on Nasopharyngeal Carcinoma, Adv. Exp. Med. Biol., 2019, 1164, 11–34 CrossRef CAS PubMed.
- J. Yang, D. Zhu, S. Liu, M. Shao, Y. Liu, A. Li, Y. Lv, M. Huang, D. Lou and Q. Fan, Curcumin enhances radiosensitization of nasopharyngeal carcinoma by regulating circRNA network, Mol. Carcinog., 2020, 59(2), 202–214 CrossRef CAS PubMed.
- H. Zhao, A. Chang, J. Ling, W. Zhou, H. Ye and X. Zhuo, Construction and analysis of miRNA-mRNA regulatory networks in the radioresistance of nasopharyngeal carcinoma, 3 Biotech, 2020, 10(12), 511 CrossRef PubMed.
- M. Zhao, R. Luo, Y. Liu, L. Gao, Z. Fu, Q. Fu, X. Luo, Y. Chen, X. Deng, Z. Liang, X. Li, C. Cheng, Z. Liu and W. Fang, miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN, Nat. Commun., 2016, 7, 11309 CrossRef CAS PubMed.
- X. Zou, D. Zhu, H. Zhang, S. Zhang, X. Zhou, X. He, J. Zhu and W. Zhu, MicroRNA expression profiling analysis in serum for nasopharyngeal carcinoma diagnosis, Gene, 2020, 727, 144243 CrossRef PubMed.
- S. M. Majd, A. Salimi and F. Ghasemi, An ultrasensitive detection of miRNA-155 in breast cancer via direct hybridization assay using two-dimensional molybdenum disulfide field-effect transistor biosensor, Biosens. Bioelectron., 2018, 105, 6–13 CrossRef CAS PubMed.
- J. Jin, S. Vaud, A. M. Zhelkovsky, J. Posfai and L. A. McReynolds, Sensitive and specific miRNA detection method using SplintR Ligase, Nucleic Acids Res., 2016, 44(13), e116 CrossRef PubMed.
- S. Liu, H. Fang, C. Sun, N. Wang and J. Li, Highly sensitive and multiplexed miRNA analysis based on digitally encoded silica microparticles coupled with RCA-based cascade amplification, Analyst, 2018, 143(21), 5137–5144 RSC.
- A. Avino, C. S. Huertas, L. M. Lechuga and R. Eritja, Sensitive and label-free detection of miRNA-145 by triplex formation, Anal. Bioanal. Chem., 2016, 408(3), 885–893 CrossRef CAS PubMed.
- X. Zhao, L. Zhang, W. Gao, X. Yu, W. Gu, W. Fu and Y. Luo, Spatiotemporally Controllable MicroRNA Imaging in Living Cells via a Near-Infrared Light-Activated Nanoprobe, ACS Appl. Mater. Interfaces, 2020, 12(32), 35958–35966 CrossRef CAS PubMed.
- P. Svoboda, A toolbox for miRNA analysis, FEBS Lett., 2015, 589(14), 1694–1701 CrossRef CAS PubMed.
- H. Lu, K. Guo, Y. Cao, F. Yang, D. Wang, L. Dou, Y. Liu and H. Dong, Cancer Cell Membrane Vesicle for Multiplex MicroRNA Imaging in Living Cells, Anal. Chem., 2020, 92(2), 1850–1855 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00539a |
| This journal is © The Royal Society of Chemistry 2021 |