A three-dimensional DNA walker amplified FRET sensor for detection of telomerase activity based on the MnO2 nanosheet-upconversion nanoparticle sensing platform†
Mingdi
Xu
 a,
Junyang
Zhuang
a,
Junyang
Zhuang
 *b,
Xiaoyu
Jiang
a,
Xinzhong
Liu
a and
Dianping
Tang
*b,
Xiaoyu
Jiang
a,
Xinzhong
Liu
a and
Dianping
Tang
 *c
*c
aCollege of Ecological Environment and Urban Construction, Fujian University of Technology, Fuzhou 350108, P. R. China
bDepartment of Pharmaceutical Analysis, The Higher Educational Key Laboratory for Nano Biomedical Technology of Fujian Province, School of Pharmacy, Fujian Medical University, Fuzhou 350122, China. E-mail: Junyang.Zhuang@hotmail.com
cKey Laboratory of Analysis and Detection for Food Safety (Ministry of Education & Fujian Province), Institute of Nanomedicine and Nanobiosensing, Department of Chemistry, Fuzhou University, Fuzhou 350108, P. R. China. E-mail: dianping.tang@fzu.edu.cn
First published on 26th July 2019
Abstract
A novel fluorescent sensing platform for telomerase activity assay was developed by coupling a three-dimensional (3D) DNA walker with the MnO2 nanosheet-upconversion nanoparticle (UCNPs) complex-based fluorescence resonance energy-transfer (FRET) system.
Upconversion nanoparticles (UCNPs), which can convert low-energy near-infrared (NIR) photons into high-energy visible light, have attracted huge attention in the field of bioassay and biomedicine.1 Besides their outstanding biological compatibility, UCNPs have several inherent advantages over traditional dyes such as high photostability, low autofluorescence, large anti-Stokes shift, high signal-to-noise ratio and predominant luminescence.2
Manganese dioxide (MnO2) nanosheet, a two-dimensional transition metal oxide, has been proved to be an efficient and broad-spectrum fluorescence quencher on account of its broad absorption spectrum and large surface area.3 Numerous fluorescent biosensors based on the fluorescence resonance energy (FRET) technique have been established by employing MnO2 nanosheets as fluorescence quenchers.4
Commonly, these FRET sensors require suitable fluoroprobes as the energy donor. Among them, the MnO2-UCNP system is regarded as one of the most promising FRET platforms, because the UCNP system causes much lower background interference of scattering light and lower autofluorescence from ultraviolet-visible (UV-Vis) excitation compared with the traditional down-conversion fluoroprobes (e.g. organic dyes and quantum dots).5 For instance, Deng et al. demonstrated the rapid detection of intracellular glutathione based on the MnO2-UCNP FRET system;6 Yuan et al. successfully applied the MnO2-UCNP system in both ochratoxin A (OTA) and cathepsin D (Cat D) detection in complicated matrixes.7 Therefore, it is of great importance to further explore the potential of the MnO2-UCNP system in bioassay.
Signal amplification is the core part of many biosensors, which determines their sensitivity and feasibility for the detection of biomolecule species at very low abundance. In this regard, several isothermal nucleic acid amplification techniques, such as strand displacement amplification, rolling circle amplification and hybridization chain reactions,8 have been extensively studied and chosen as the signal amplification components of biosensors due to their excellent amplification ability and designability.
Recently, DNA walker, a new emerging DNA machine moving along the pre-determined DNA tracks, has provided a fascinating alternative approach for amplifying the sensing signal in biosensors.9 In these biosensors, the DNA walker as the signal amplifier is usually triggered by target molecules and powered by some specific reactions such as toehold-mediated strand displacement,10 DNAzyme-mediated strand cleavage11 and nuclease-mediated hydrolysis.12 By adopting suitable optical or electrochemical signal tags, an enhanced detection signal is produced following the implementation of the DNA walking behavior.10,13 In particular, the tracks for DNA walking laid on three-dimensional (3D) landscapes are of higher mobility and loading efficiency compared with those on one-dimensional or two-dimensional landscapes, so that the 3D landscapes (especially nanoparticles) are more suitable for amplifying the sensing signal.14 For example, Le et al. developed a 3D walking machine driven by Mn2+-specific DNAzyme to enable the autonomous DNA walking on gold nanoparticles and monitoring of miRNA in living cells.15 At present, the application of DNA walker in biosensors is still in its infancy, so more efforts should be devoted to this field.
Telomerase, as a ribonucleoprotein complex, catalyzes the addition of telomeric repeats (TTAGGG)n onto the 3′-end of human chromosomes via reverse transcription by using its intrinsic RNA as a template.16 Recent studies have revealed that the telomerase activity could be considered to be a useful biomarker for early diagnosis, prognosis, and understanding of the pathogenesis of cancer.17 Herein, we report on a novel 3D DNA walker-amplified fluorescence method for the telomerase activity assay by utilizing the MnO2-UCNP FRET system as the fluorescent signal readout.
The DNA walker is initiated by the telomerase-catalyzed extension reaction and operated on a 3D track anchored on magnetic beads (MBs). The spontaneous walking of DNA is powered by the cleavage reaction catalyzed by Mn2+-dependent DNAzyme, which is followed by the release of a trigger DNA strand. Furthermore, the trigger DNA strand will break the MnO2-UCNP based FRET system and recover the fluorescence of UCNPs to indicate the activity of telomerase. Owing to the powerful signal amplification ability of the DNA walker, the telomerase activity can be monitored with high sensitivity.
In the developed sensing platform (Scheme 1), magnetic beads (MBs) are simultaneously conjugated with a DNA walker scaffold and a substrate strand, forming DNA walker–MBs. The DNA walker scaffold consists of a walker strand and a locking strand. The walker strand includes two regions: the swing arm (containing 45 repetitive T bases) and the sequence of Mn2+-dependent DNAzyme. The locking strand hybridizes partially with the DNAzyme so that the enzyme activity of DNAzyme is “locked”. The substrate strand (as the track for DNA walker) consists of three regions: an immobilization region (to facilitate its immobilization on MBs), the cleavage site for DNAzyme and an extra segment named the trigger strand. To sense the telomerase activity, a telomerase substrate (TS) primer is employed in this design. In the presence of telomerase extracted from HeLa cells and dNTPs, the telomerase catalyzes the continuous addition of TTAGGG repeat units to the 3′-end of the TS primer, producing a longer strand (named the extension strand). The extension strand then hybridizes with the locking strand via a strand-displacement reaction, causing the release of the locking strand from the DNAzyme region and the activation of DNAzyme activity on MBs. The unlocked DNAzyme then hybridizes with its substrate sequence in the substrate strand and cleaves the substrate strand from the cleavage site in the presence of Mn2+ as the cofactor, resulting in the breakage of the substrate strand and the release of the trigger strand. The cleavage reaction provides the energy needed for the DNAzyme to move from one substrate strand to the next, resulting in its autonomous walking along the MBs. It is worth noting that the swing arm with a long length is customized to endow the DNAzyme with high flexibility. Significantly, each walking step of DNAzyme is accompanied by the release of the trigger strand.
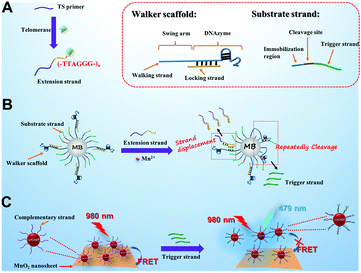 | ||
| Scheme 1 A schematic illustration of the developed 3D DNA walker amplified FRET sensing platform for telomerase activity assay based on the MnO2-UCNP system. | ||
On the other hand, a MnO2-UCNP FRET system is constructed as the readout signal for the DNA walking event. In detail, the NaYF4:Yb,Tm UCNPs prepared by a hydrothermal method are conjugated with a single stranded DNA (named the complementary strand) and assembled on the surface of MnO2 nanosheets via the affinity between single stranded DNA and MnO2 nanosheets,7 resulting in energy transfer from UCNPs to MnO2 and quenching of upconversion luminescence. The sequence of the complementary strand is complementary with that of the trigger strand. Once the trigger strand is introduced into the FRET system, it hybridizes with the complementary strand and a double-stranded DNA structure is formed. Since the affinity between double-stranded DNA and MnO2 nanosheets is very weak, the formation of double-stranded DNA will cause the separation of UCNPs from MnO2 nanosheets. In this situation, the FRET system is broken and the fluorescence of UCNPs is expected to be recovered, enabling the quantification of telomerase activity.
The morphology and size of the as-prepared NaYF4:Yb,Tm UCNPs were characterized by transmission electron microscopy (TEM). As shown in Fig. 1A, the NaYF4:Yb,Tm UCNPs were of a uniform spherical structure with an average diameter of 32 ± 2.3 nm. The presence of Na, Yb, F, Tm and Y elements in the NaYF4:Yb,Tm UCNPs was also confirmed by energy dispersive X-ray spectroscopy and (EDS) mapping (Fig. 1B).
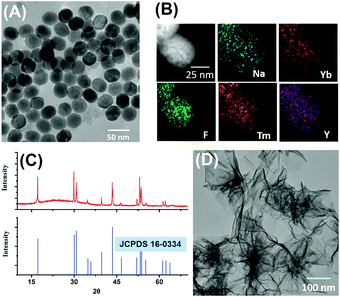 | ||
| Fig. 1 TEM (A), EDS mapping (B) and X-ray diffraction (XRD) patterns (C) of NaYF4:Yb,Tm UCNPs; (D) TEM of MnO2 nanosheets. | ||
In addition, the crystalline pattern of the NaYF4:Yb,Tm UCNPs was investigated by powder X-ray diffraction. As shown in Fig. 1C, the XRD crystalline peaks of NaYF4:Yb,Tm UCNPs were consistent with the standard data (JCPDS No. 16-0034) of the pure hexagonal-phase NaYF4. Next, the morphology and size of the as-prepared MnO2 nanosheets were characterized by TEM and dynamic laser scattering (DLS). As shown in Fig. 1D, the synthesized MnO2 nanomaterials were planar sheet-like, indicating that the nanosheets were successfully formed. The average size of the MnO2 nanosheets measured by DLS was about 198 nm (Fig. S2, ESI†).
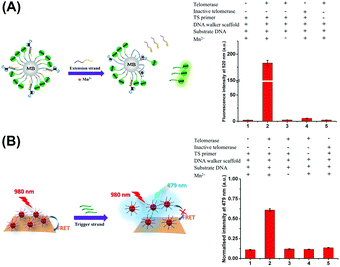
To verify the successful implementation of the DNA walking event, the substrate strand on the DNA walker–MBs was conjugated with FAM as the fluorescent reporter. Once the DNA walking event was initiated, the substrate strand would be cleaved to release the trigger DNA strand into the solution phase. By recording the fluorescence signal of FAM conjugated on the trigger DNA strand in solution, the implementation of the DNA walking event could be monitored.
As shown in Fig. 2A, almost no fluorescence signal of FAM at 518 nm was detected in the solution phase when the DNA walker–MBs were incubated with the TS primer in the absence of telomerase extracts (column 1), indicating that no substrate strand or trigger strand was released from MBs. In contrast, a strong fluorescence signal of FAM was detected with the introduction of telomerase extracts (column 2), manifesting the occurrence of the DNA walking event and the accompanying release of the trigger DNA strand. Importantly, the lack of Mn2+ ion (column 3) and the TS primer (column 5) disabled the DNA walking event, so that no fluorescence signals were detected. This result indicated that the DNA walking event was powered by the DNAzyme using Mn2+ as the cofactor and initiated by the extension strand produced on the TS primer. In addition, the telomerase extracts inactivated at 95 °C for 10 min also could not produce the fluorescence signal of FAM (column 4). These results clearly demonstrated the successful implementation of DNA walking on the surface of MBs.
The feasibility of the developed sensing platform (consisting of DNA walker–MBs and the MnO2-UCNP FRET system) for the telomerase activity assay was further confirmed. The fluorescence emission of NaYF4:Yb,Tm UCNPs at 479 nm was employed as the detection signal. The results are displayed in Fig. 2B. It was observed that the introduction of telomerase extracts into the sensing platform caused a significant increase in the fluorescence signal (columns 1 and 2). In the absence of Mn2+ or the TS primer, no obvious fluorescence signal was observed (columns 3 and 4). Moreover, the heat inactivation telomerase also could not produce an increasing fluorescence signal (column 5). These observations were similar to the phenomenon described above. It is noteworthy that the sequence of the trigger strand was specific to the disintegration of the MnO2-UCNP system and the recovery of the fluorescence signal (Fig. S4, ESI†). Based on these results, we might make a conclusion that the 3D DNA walker-based FRET sensing platform could be used for telomerase activity monitoring.
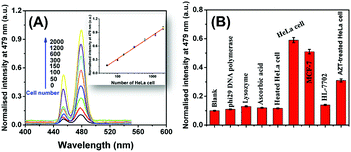
Under optimal conditions (Fig. S5, ESI†), the sensitivity of the developed sensing platform was studied by assaying telomerase extracts from HeLa cells with different concentrations. As shown in Fig. 3A, the fluorescence intensity increased with the increasing number of HeLa cells, and the fluorescence intensity was proportional to the numbers of HeLa cells in a linear range from 50 to 2000 (the inset in Fig. 3A). The linear regression equation was y = 0.518![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) x − 0.718 (y: the fluorescence intensity, x: the number of HeLa cells, and R2 = 0.9955). The detection limit calculated as three times the signal-to-noise ratio was estimated to be 23 HeLa cells.
x − 0.718 (y: the fluorescence intensity, x: the number of HeLa cells, and R2 = 0.9955). The detection limit calculated as three times the signal-to-noise ratio was estimated to be 23 HeLa cells.
Inspiringly, the sensitivity of the developed sensing platform is comparable or even superior to many previously reported strategies, such as the quartz crystal microbalance method based on gold nanoparticle induced signal amplification,18 portable pH meter based assay method,19 photoelectrochemical method,20 colorimetric method based on primer-modified gold nanoparticles,21 electrochemical method using PtNP-encapsulated MOFs as electrocatalytic tracers,22 molecular beacon-based DNA machines,23 hemin–graphene conjugate-based colorimetric method24 and energy-transfer-based photocathodic assay strategy25. However, the sensitivity of the developed method toward telomerase has not yet reached the single-cell level, which has been achieved in previous methods.26 In addition, the analytical performance of the sensing platform toward telomerase using FAM as the signal tag (as shown in Fig. 2A) was also investigated [Fig. S6, ESI†].
The selectivity of the developed sensing platform was evaluated by challenging it against several biomolecules such as phi29 DNA polymerase (20 U), lysozyme (20 U), ascorbic acid (0.2 mM) and heated HeLa cells (350 cells). Moreover, the response of the sensing platform toward telomerase extracts from MCF-7 (350 cells) and HL-7702 cell lines (350 cells) was also estimated. As shown in Fig. 3B, the phi29 DNA polymerase, lysozyme, ascorbic acid and heated HeLa cells caused no change in the fluorescence intensity. In addition, the HeLa cell extracts and MCF-7 cell extracts caused a significant increase in the fluorescence intensity, while the HL-7702 cell extracts resulted in low fluorescence intensity nearly to the background level. These results indicated that the fluorescence response was specifically dependent on the telomerase activity. The difference between the fluorescence response toward HeLa cell extracts and MCF-7 cell extracts could be explained by their different telomerase expression levels. On the other hand, it was reported that the telomerase expression levels in somatic cells were very low that the HL-7702 cell extracts did not cause significant fluorescence response.27 Significantly, the HeLa cell extracts treated with 50 mM zidovudine (AZT, a telomerase inhibitor) produced a relatively weaker fluorescence signal than the pure HeLa cell extracts. These results demonstrate that the 3D DNA walking-based sensing platform is of high specificity and generality for the telomerase activity assay and holds great potential in further clinical application. The feasibility of the developed sensing platform in complicated biological matrixes was also investigated by conducting a recovery experiment in spiked human urine samples (Table S2, ESI†).
In conclusion, a sensitive sensing platform for the telomerase activity assay was successfully developed by combining a 3D DNA walker-based signal amplification strategy with the MnO2-UCNP FRET system. The DNA walking event was triggered by a telomerase-catalyzed extension reaction. Powered by the cleavage reaction catalyzed by Mn2+-dependent DNAzyme, the DNA walker containing a DNAzyme sequence could spontaneously move along the substrate DNA-based track which was laid on magnetic beads. The DNA walking event was accompanied by the release of a trigger DNA strand, which was then used to break the MnO2-UCNP FRET system and recover the fluorescence emission of UCNPs. Due to the excellent signal amplification ability of the DNA walking strategy and the MnO2-UCNP FRET system, a high sensitivity was achieved and the detection limit could reach 23 HeLa cells. Moreover, the developed sensing platform was successfully applied in spiked human urine sample assays, with satisfactory recoveries ranging from 92.3% to 105.7%. Thus, the 3D DNA walker-based FRET sensing method can serve as a promising non-invasive diagnostic tool to screen bladder cancers in clinical applications.
This work was supported by the National Natural Science Foundation of China (21705092), the Natural Science Foundation of Fujian Province (2018J01442), the Educational Research Project of Young and Middle-aged Teachers in Fujian Province (JT180322 and JAT170487), the Foundation for High-Level Talents in Quanzhou City (2017Z029) and the Foundation in Fujian University of Technology (GY-Z18042).
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) G. Chen, H. Qiu, P. N. Prasad and X. Chen, Chem. Rev., 2014, 114, 5161 CrossRef CAS PubMed; (b) Q. Liu, T. Yang, W. Feng and F. Li, J. Am. Chem. Soc., 2012, 134, 5390 CrossRef CAS PubMed.
- (a) L. M. Wiesholler, C. Genslein, A. Schroter and T. Hirsch, Anal. Chem., 2018, 90, 14247 CrossRef CAS PubMed; (b) H. He, B. Liu, S. Wen, J. Liao, G. Lin, J. Zhou and D. Jin, Anal. Chem., 2018, 90, 12356 CrossRef CAS PubMed.
- (a) C. Peng, H. Xing, X. Fan, Y. Xue, J. Li and E. Wang, Anal. Chem., 2019, 91, 5762 CrossRef CAS PubMed; (b) X. L. Zhang, C. Zheng, S. S. Guo, J. Li, H. H. Yang and G. Chen, Anal. Chem., 2014, 86, 3426 CrossRef CAS PubMed.
- (a) X. Yan, Y. Song, C. Zhu, H. Li, D. Du, X. Su and Y. Lin, Anal. Chem., 2018, 90, 2618 CrossRef CAS PubMed; (b) T. Xiao, J. Sun, J. Zhao, S. Wang, G. Liu and X. Yang, ACS Appl. Mater. Interfaces, 2018, 10, 6560 CrossRef CAS PubMed.
- J. Chen, H. Meng, Y. Tian, R. Yang, D. Du, Z. Li, L. Qu and Y. Lin, Nanoscale Horiz., 2019, 4, 321 RSC.
- R. Deng, X. Xie, M. Vendrell, Y. T. Chang and X. Liu, J. Am. Chem. Soc., 2011, 133, 20168 CrossRef CAS PubMed.
- Y. Yuan, S. Wu, F. Shu and Z. Liu, Chem. Commun., 2014, 50, 1095 RSC.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491 CrossRef CAS PubMed.
- X. Qu, D. Zhu, G. Yao, S. Su, J. Chao, H. Liu, X. Zuo, L. Wang, J. Shi, L. Wang, W. Huang, H. Pei and C. Fan, Angew. Chem., Int. Ed., 2017, 56, 1855 CrossRef CAS PubMed.
- L. Wang, R. Deng and J. Li, Chem. Sci., 2015, 6, 6777 RSC.
- J. Chen, A. Zuehlke, B. Deng, H. Peng, X. Hou and H. Zhang, Anal. Chem., 2017, 89, 12888 CrossRef CAS PubMed.
- Y. Ji, L. Zhang, L. Zhu, J. Lei, J. Wu and H. Ju, Biosens. Bioelectron., 2017, 96, 201 CrossRef CAS PubMed.
- J. Zhang, L. L. Wang, M. F. Hou, Y. K. Xia, W. H. He, A. Yan, Y. P. Weng, L. P. Zeng and J. H. Chen, Biosens. Bioelectron., 2018, 102, 33 CrossRef CAS PubMed.
- (a) W. Li, L. Wang and W. Jiang, Chem. Commun., 2017, 53, 5527 RSC; (b) C. Jung, P. B. Allen and A. D. Ellington, Nat. Nanotechnol., 2015, 11, 157 CrossRef PubMed.
- H. Peng, X. F. Li, H. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- J. Lei, B. Han, S. Lv, Y. Li, J. Tang, Y. Mao and J. Zhuang, Electrochem. Commun., 2018, 92, 43 CrossRef CAS.
- H. R. Zhang, M. S. Wu, J. J. Xu and H. Y. Chen, Anal. Chem., 2014, 86, 3834 CrossRef CAS PubMed.
- H. Yang, Y. Li, D. Wang, Y. Liu, W. Wei, Y. Zhang, S. Liu and P. Li, Chem. Commun., 2019, 55, 5994 RSC.
- L. Wang, C. Chen, H. Huang, D. Huang, F. Luo, B. Qiu, L. Guo, Z. Lin and H. Yang, Biosens. Bioelectron., 2018, 121, 153 CrossRef CAS PubMed.
- S. Liu, S. Zhao, W. Tu, X. Wang, X. Wang, J. Bao, Y. Wang, M. Han and Z. Dai, Chem. – Eur. J., 2018, 24, 3677 CrossRef CAS PubMed.
- L. Zhang, S. Zhang, W. Pan, Q. Liang and X. Song, Biosens. Bioelectron., 2016, 77, 144 CrossRef CAS PubMed.
- P. Ling, J. Lei, L. Jia and H. Ju, Chem. Commun., 2016, 52, 1226 RSC.
- K. Li, L. Wang, X. Xu and W. Jiang, Talanta, 2017, 167, 645 CrossRef CAS PubMed.
- X. Xu, M. Wei, Y. Liu, X. Liu, W. Wei, Y. Zhang and S. Liu, Biosens. Bioelectron., 2017, 87, 600 CrossRef CAS PubMed.
- L. Zhang, X. M. Shi, Y. T. Xu, G. C. Fan, Y. Y. Liang, C. Wang and W. W. Zhao, Anal. Chem., 2019, 91, 6403 CAS.
- (a) H. H. Wang, H. Wang, C. H. Liu, X. R. Duan and Z. P. Li, Chem. Sci., 2016, 7, 4945 RSC; (b) L. J. Wang, Y. Zhang and C. Y. Zhang, Anal. Chem., 2013, 85, 11509 CrossRef CAS PubMed.
- L. Wang, C. Chen, H. Huang, D. Huang, F. Luo, B. Qiu, L. Guo, Z. Lin and H. Yang, Biosens. Bioelectron., 2018, 121, 153 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cc05387e |
| This journal is © The Royal Society of Chemistry 2019 |