Open Access Article
Open Access ArticleRecent advances in the discovery, biosynthesis, and therapeutic potential of isocoumarins derived from fungi: a comprehensive update†
Mohamed A. Tammam
 a,
Mariam I. Gamal El-Din
a,
Mariam I. Gamal El-Din
 b,
Amira Abood
b,
Amira Abood
 cd and
Amr El-Demerdash
cd and
Amr El-Demerdash
 *ef
*ef
aDepartment of Biochemistry, Faculty of Agriculture, Fayoum University, Fayoum, 63514, Egypt
bDepartment of Pharmacognosy, Faculty of Pharmacy, Ain-Shams University, Cairo 11566, Egypt
cChemistry of Natural and Microbial Products Department, National Research Center, Dokki, Cairo, Egypt
dSchool of Bioscience, University of Kent, Canterbury, UK
eOrganic Chemistry Division, Department of Chemistry, Faculty of Sciences, Mansoura University, Mansoura, 35516, Egypt. E-mail: a_eldemerdash83@mans.edu.eg
fDepartment of Biochemistry and Metabolism, John Innes Centre, Norwich Research Park, Norwich, NR4 7UH, UK. E-mail: Amr.El-Demerdash@jic.ac.uk
First published on 10th March 2023
Abstract
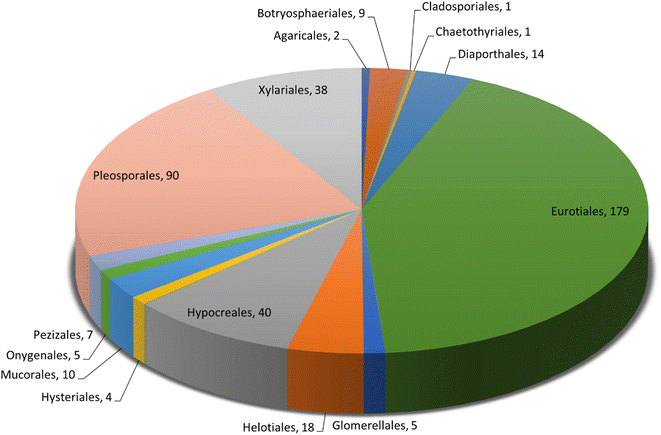
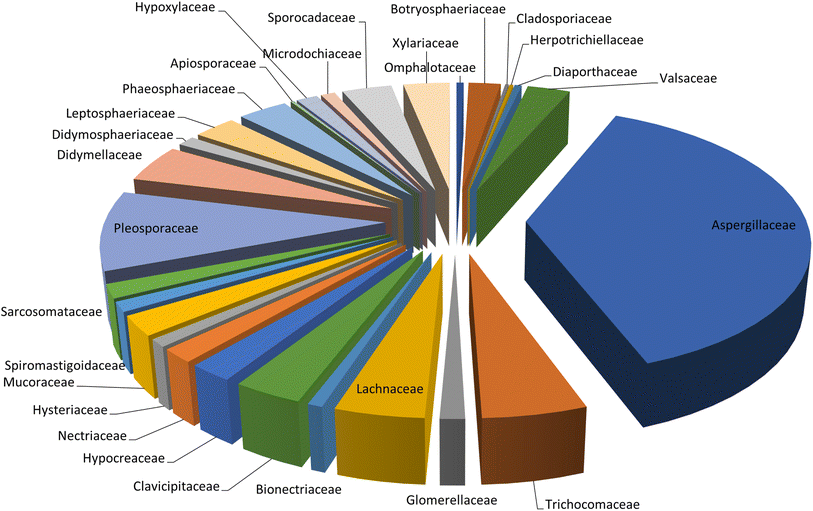
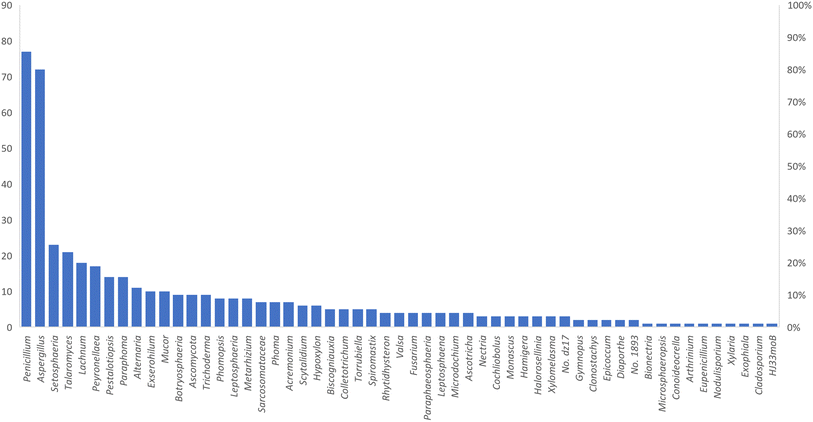
Microorganisms still remain the main hotspots in the global drug discovery avenue. In particular, fungi are highly prolific producers of vast structurally diverse specialized secondary metabolites, which have displayed a myriad of biomedical potentials. Intriguingly, isocoumarins is one distinctive class of fungal natural products polyketides, which demonstrated numerous remarkable biological and pharmacological activities. This review article provides a comprehensive state-of-the-art over the period 2000–2022 about the discovery, isolation, classifications, and therapeutic potentials of isocoumarins exclusively reported from fungi. Indeed, a comprehensive list of 351 structurally diverse isocoumarins were documented and classified according to their fungal sources [16 order/28 family/55 genera] where they have been originally discovered along with their reported pharmacological activities wherever applicable. Also, recent insights around their proposed and experimentally proven biosynthetic pathways are also briefly discussed.
1. Introduction
Natural products have long been recognized as a crucial mine for drug discovery. Indeed, they have been traditionally used for centuries for their biological significance that have been extended through a wide era of therapeutic fields including inflammatory disorders, cancer immunodeficiency, infectious, hepatic, cardiovascular, renal, and skin diseases.1,2 Besides, natural products are always a preferable source of bioactive drugs on account of their safety profile associated with their therapeutic potency compared to conventional synthetic drugs.3,4 Terrestrial plants, despite being a rich source of secondary metabolites and a major fundamental for traditional folk medicine for thousands of years, their overuse made them susceptible to overharvesting, depletion, and extinction of many rare species of high medicinal value.5 Hence, microorganisms, especially fungi, have recently received remarkably growing attention for discovering an enormous scaffold of natural products of indispensable medicinal values.6,7 Fungi constitute a broadly diverse class of eukaryotes, which inhabit a wide range of ecosystems including soil, air, and water, in addition to the fungal endophytes that dwell within their hosts of terrestrial and marine habitats.8,9 They are recognized as an abundant reservoir of bioactive metabolites that have demonstrated stunning therapeutic potentials for both human and animals.7,10 Dating back to 1929, Alexander Fleming explored the antibacterial activity of the mould Penicillium rubens, naming it Penicillin, which was the first of a series of antimicrobial agents isolated from fungi, marking the emergence of the golden era of antibiotics discovery.11 In addition, further β-lactam antibiotics were discovered from different fungi, causing substantial changes in the global health and the world pharmaceutical industry.12,13 Moreover, various fungal metabolites were identified and approved as commercial drugs with different biological activities, including the immunosuppressant cyclosporine, statins, the inhibitors of cholesterol synthesis, the antifungal, Griseofulvin, and kojic acid, the tyrosinase inhibitor, in addition to chemotherapeutic agents, such as, vincristine, paclitaxel, and camptothecin.13–15 Secondary metabolites isolated and identified from fungi are sorted to different chemical classes including peptides, steroids, quinones, terpenes, alkaloids, and isocoumarins.4,16–18 Isocoumarins constitute a distinguished class of secondary metabolites widely abundant in fungi, bacteria, and terrestrial plants. Chemically, as their name implies, isocoumarins are characterized by their inverted α-pyrone lactone nucleus, with substituted or unsubstituted 3-phenyl ring attached on the lactone ring, usually demonstrating 3-alkyl substitution (C1–C7), and possible oxygenation at 6 and 8 positions (Scheme 1).19,20 Their variant chemical substitution patterns account for their great chemical diversity that influence their wide array of biological and pharmacological activities. Isocoumarins were reported to possess antimicrobial, antifungal, insecticidal, antioxidant, anticancer, anti-inflammatory, and antidiabetic activities.21–25 Besides, isocoumarins constitute key intermediates in the synthesis of important heterocyclic compounds, viz., isochromenes, isoquinolines, and isocarbostyrils.26 Consequently, isocoumarins have recently gained great attention in the medicinal, synthetic, and drug discovery research fields. Various captivating reviews addressed natural isocoumarins, namely, the reviews by Saeed A., et al., which focused on the chemical structural diversity among isocoumarins and their associated pharmacological activities reported before 2016.27,28 Besides, Saddiqa et al. reviewed the isolated isocoumarins from natural sources before 2017 with highlights on their bioactivities and chemical synthesis.29 Noor et al. reported the isocoumarins isolated from endophytic fungi between 2019–2020 stressing on their chemistry, biosynthesis, and their pharmacological activities.31 Meanwhile, the recent review by Shabir et al., reported the natural isocoumarins isolated in the period of 2016–2020, focusing on their chemistry and their bioactivities.31 As a part of our ongoing research on biologically active natural products32–34 and with emphasis on pharmacologically active fungal natural products (FNPs),4,16,35–37 herein, we comprehensively present an up-to-date literature review for the period 2000–2022 on the chemical diversity and biological activities reported for isocoumarins isolated exclusively from different fungal strains. Indeed, the review systematically documents the distribution of a list of 351 isocoumarins among the various fungal genera, their chemical diversities along with their therapeutic potentialities. Furthermore, insights into their biogenesis are briefly discussed.2. General biosynthetic pathway of fungal isocoumarin
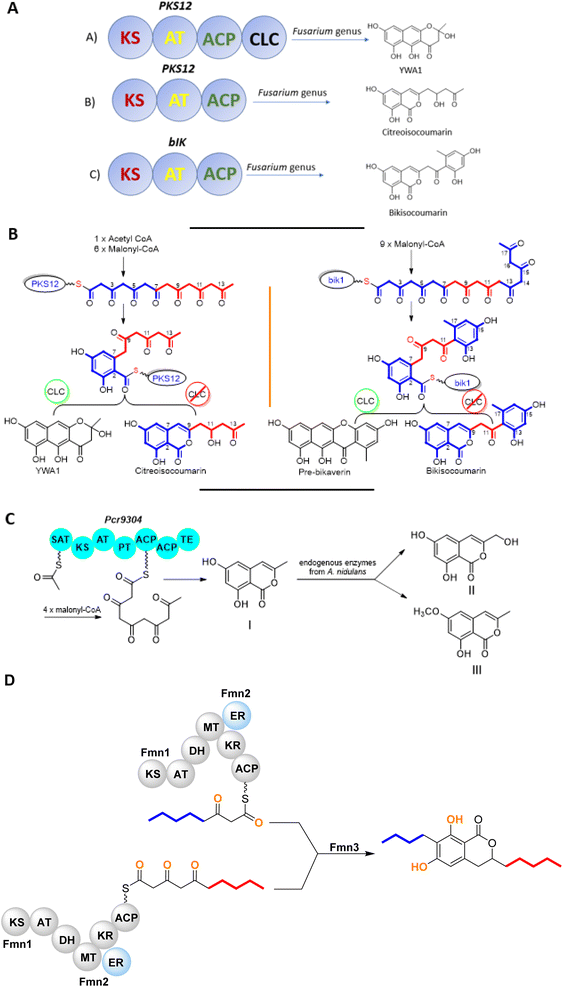
The chemistry of isocoumarin has been extensively studied since the 1950 or before, as reported by Barry.38 Isocoumarin is a well-known polyketide that had been biosynthesized by the polyketide synthase (PKS) pathway.39,40 The biosynthetic pathway of isocoumarin derivates had been studied by Birch's group.41,42 Their study investigated the biosynthesis of canescin from Penicillium canescens using stable isotopes (13C labelled) and NMR spectroscopy to determine the sites of incorporation. They pointed out that the isocoumarin portion of the canescin molecule was generated through the acetate/malonate pathway.42,43 Indeed, Birch et al., discovered that canescin's C-10 and C-14 were generated from methionine. Further study by Lewis's group confirmed canescin biosynthesis using the feeding experiment of [13C,2H3] methionine that was administered to Aspergillus malignus using batch-wise method over 5 days of incubation.43 Advances in bioinformatic analysis, genetic transformation experiments, and the progress in genome sequencing revealed that isocoumarins in Fusarium graminearum had been synthesized by polyketide synthase 12 (PKS12), a transcription factor (aurR1), and several tailoring enzymes.44–47 This type of PKS domain architecture consists of a β-ketoacyl synthase (KS), an acetyltransferase (AT), an acyl carrier protein (ACP), and a claisen-type cyclase (CLC), as shown in Fig. 1A. The biosynthetic pathway could be considered as a unified gate to generate different natural compounds by the modification of the PKS domain architecture. Enzymatically, the biosynthetic cascade begins with the condensation of one acetyl-coenzyme A (CoA) molecule and six malonyl-CoA molecules, which is mediated by PKS12 with the active CLC domain to furnish the naphthopyrone YWA1.48 However, the absence of the CLC domain tragically led to the biosynthesis of citreoisocoumarin.49,50 This demonstrates that similar PKS domain architectures in different species can lead to the biosynthesis of various polyketide derivatives.51 In addition, bikisocoumarin is another type of isocoumarin that biosynthesized by similar PKS named bIK45 which shared a similar architecture domain to previously mentioned PKS12. Nine malonyl-CoA molecules were employed to form pre-bikaverin with the presence of active CLC domain52,53 (Fig. 1B) while bikisocoumarin was only obtained with the deletion of CLC domain. Furthermore, Ma et al., 200852 was able to produce bikisocoumarin by heterologous expression in E. coli of a mutated bIK gene. Moreover, a successful heterologous expression insights has been done to elucidate the biosynthesis of some isocoumarins.54 Three isocoumarins derivatives (I–III) were accumulated as a result of the expression of the non-reducing polyketide synthase (NR-PKS) gene from Penicillium crustosum in Aspergillus nidulans. The domain architecture of this isocoumarin synthase consists of the SAT-KS-AT-PT-ACP-ACP-TE domain. Meanwhile, Xiang et al., in 2020 elucidated the structure of the three generated isocoumarins by 1H-NMR analysis.54 They concluded that compounds II–III are modification products obtained by endogenous host enzymes during the heterologous expression (Fig. 1C).In addition, fusamarins (FMN) is another type of dihydroisocoumarins, which is isolated from the plant-pathogenic fungus Fusarium mangiferae. Although it was reported 50 years ago, the biosynthetic pathway has not been revealed yet. Indeed, Atanasoff-Kardjalieff et al.,55 managed to investigate the gene cluster involved in fusamarins biosynthesis. They showed that the FmPKS8 biosynthetic gene cluster (FMN BGC) leads the biosynthesis, which is composed of FmPKS8 (FmFMN1), followed by FmFMN2, FmFMN3, and FmFMN4. Interestingly, PKS8 exhibits the characteristic domains of a highly reducing (HR)-PKS containing a dehydratase (DH), intrinsic S-adenosyl-methionine (SAM)-dependent methyltransferase (CMet), and keto reductase (KR) domain. They observed that the C-Met domain is not functional in the FMN BGC as the isolated metabolites were not C-methylated; it is conventional for HR-PKS to have the inactive CMet domain.56 On the other hand, FmFmn3 possesses a peptidase domain with α/β hydrolase fold, FmFmn2 has an ER domain, as well as domains with putative alcohol dehydrogenase activity (ADH). The isolated FMN's structural characteristics imply that two different carbon chains are fused during their production. It was hypothesized that (FmFMN1) produces two distinct polyketides, a tetra and a pentaketide, with changing numbers of double bonds dependent on the selective activities of the trans-acting ER FmFmn2 as only one PKS is expressed inside the FMN BGC (Fig. 2).
3. Chemistry and pharmacological potentials of isocoumarins isolated from fungi
3.1. Fungi of the order Agaricales
3.1.1.1. Genus Gymnopus (Marasmiaceae). Two chlorinated isocoumarin derivatives previously unreported, namely, gymnopalynes A (1) and B (2) (Fig. 2), were obtained from the fungus Gymnopus sp., isolated from the basidiomycete collected from the rain forest of Thailand. Compounds 1–2 were tested for their antimicrobial activity by assessing their MIC against several bacterial and fungal strains (oxytetracycline hydrochloride, gentamicin, and nystatin were used as positive controls), which exhibited weak to moderate activity against some of the examined strains. In addition, compounds 1–2 showed cytotoxic effect toward the mouse fibroblast cell line L-929 with IC50 values of 3.7 and 14.0 μM, respectively.57
3.2. Fungi of the order Botryosphaeriales
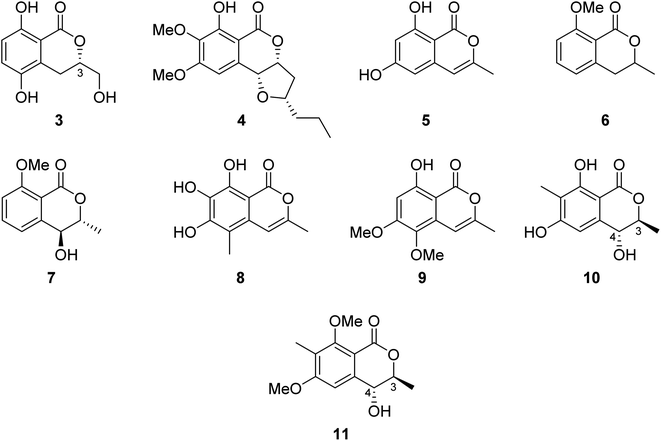
3.2.1.1. Fungi of the genus Botryosphaeria. The chemical examination of the EtOAc extract of the marine-derived fungus Botryosphaeria sp. KcF6 isolated from the fruit of the mangrove K. candel, collected from the bay of Daya, China, led to the isolation of the previously unreported 3S-5,8-dihydroxy-3-hydroxymethyldihydroisocoumarin (3) as well as other four previously reported isocoumarin derivatives, namely, monocerin (4), 3-methyl-6,8-dihydroxyisocoumarin (5), 8-methoxymellein (6), and trans-4-hydroxymellein (7) (Fig. 3). Though none of the isolated compound showed any cytotoxic effect against the studied cancer cell lines (K562, MCF-7, A549, U937, HeLa, DU145, HL60, BGC823, MOLT-4, and H1975), compound 3 showed antiinflammation properties through the inhibition of COX-2 activities with IC50 = 6.51 μM.58 Other four previously unreported isocoumarin derivatives, namely, botryospyrones A–D (8–11) (Fig. 3), were obtained from the EtOAc extract of the marine endophytic fungus Botryosphaeria ramosa L29 isolated from the leaf of Myoporum bontioides, collected from the mangrove of Leizhou Peninsula, China. The antifungal properties of compounds 8–10 against three phytopathogenic fungi, i.e., Fusarium oxysporum, Penicillium italicum, and Fusarium graminearum, were tested (triadimefon was used as the positive control). They showed antifungal activity ranging from weak to strong with MIC values ranging from 900 to 105.8 μM, except for compound 9, which showed no activity toward Penicillium italicum with MIC value >900.0 μM. Also, it is worth mentioning that compound 11 was not examined as it was obtained in a very minute amount.59
3.3. Fungi of the order Cladosporiales
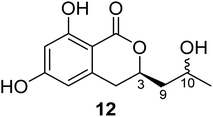
3.3.1.1. Fungi of the genus Cladosporium. Chemical investigation of the marine sponge-derived fungus Cladosporium sp. SCSIO41007 isolated from the sponge Callyspongia sp., collected from the sea near to the province of Guangdong, China, led to the isolation of the previously unreported dihydroisocoumarin derivatives, namely, (3R)-3-(2-hydroxypropyl)-6,8-dihydroxy-3,4-dihydroisocoumarin (12) (Fig. 4). It is worth mentioning that compound 12 was not assessed for any biological activity.60
3.4. Fungi of the order Chaetothyriales
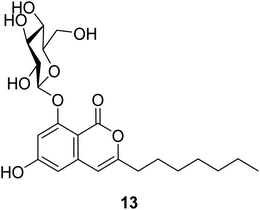
3.4.1.1. Fungi of the genus Exophiala. The previously unreported exophiarin (13) (Fig. 5) was isolated from the EtOAc extract of the soil derived fungus Exophiala sp. obtained from a dumped organic waste collected from Kaziranga, Assam. Compound 13 exhibited moderate activity in the glucose uptake activity when tested in vitro, using Rosiglitazone as a positive control.61
3.5. Fungi of the order Diaporthales
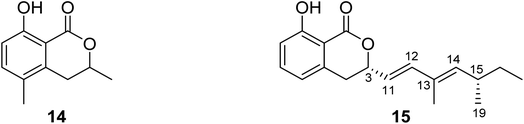
3.5.1.1. Fungi of the genus Diaporthe. (−)3,4-Dihydro-8-hydroxy-3,5-dimethyl-isocoumarin (14) (Fig. 6), also known as (−)5-methylmellein, a previously reported phytotoxic isocoumarin derivative, was isolated from the EtOAc extract of the pathogenic fungus Diaporthe eres obtained from Hedera helix infected leaf collected from Oxford, Mississippi. Compound 14 was examined for its phytotoxicity against Agrostis stolonifera (bentgrass) and Lactuca sativa (lettuce), and it was found to be more phytotoxic toward bentgrass than lettuce with IC50 ∼100 μM. It is worth mentioning that compound 14 is well known to be more active on monocots than dicots.62 A previously unreported dihydroisocoumarin derivative Diaporone A (15) (Fig. 6) was obtained from the EtOAc extract of the endophytic fungus Diaporthe sp., isolated from Pteroceltis tatarinowii Maxim collected from Nanjing, China. Compound 15 was examined for its antimicrobial activity against several bacterial strains including B. subtilis (ATCC 6633), Staphylococcus aureus (CGMCC 1.2465), Streptococcus pneumoniae (CGMCC 1.1692), Escherichia coli (CGMCC 1.2340), and the fungal strains Saccharomyces cerevisiae (ATCC 18824) and Candida albicans (CGMCC 2.2086) using gentamycin as a positive control, showing a moderate antibacterial activity toward B. subtilis with an MIC value of 66.7 μM. In addition, compound 15 was tested for its cytotoxic activity against a panel of human cancer cell lines including human glioma cell lines (SH-SY5Y), cervical epithelial cells (HeLa), human colon cancer cells (HCT116), human hepatocellular carcinoma cells (HepG2), human lung cancer cells (A549), and human breast cancer cells (MCF7). It was found to be a weak cytotoxic agent against HeLa cell lines with IC50 = 97.4 μM.63
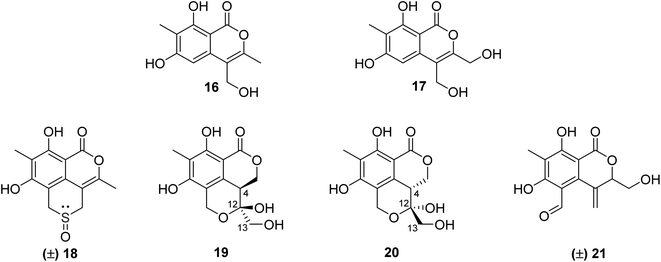
3.5.2.1. Fungi of the genus Phomopsis. The chemical examination of the EtOAc extract of the endophytic fungus Phomopsis prunorum isolated from the leaves of Hypericum ascyron, collected in Hubei, China, led to the isolation of two previously unreported isocoumarins, namely, phomoisocoumarins C–D (16–17) (Fig. 7). Compounds 16–17 were tested for their antibacterial activity toward a list of plant pathogenic bacterial strains including Pseudomonas syringae pv. Lachrymans, Xanthomonas citri pv. phaseoli var. fuscans, and the pathogenic bacteria E. coli, as well as the marine-derived bacteria Vibrio parahaemolyticus and Vibrio anguillarum using microplate assay and streptomycin as the positive control. They exhibited weak to moderate inhibition effect against P. syringae pv. Lachrymans with MIC values of 31.2 and 15.6 μg mL−1, respectively, and toward X. citri pv. phaseoli var. fuscans with MIC value of 31.2 μg mL−1.64 Chemical investigations of the organic extract of the endophytic fungus Phomopsis prunorum (F4-3) obtained from Hypericum ascyron leaves collected in Hubei, China, led to the isolation of three pairs of enantiomeric isocoumarin derivatives, including the previously unreported (±)-prunomarin A (18), (+)-pestalactone B (19) along with its known enantiomer (−)-pestalactone B (20), together with the known enantiomers pestalactone C (+)-(21) and oxoisochromane (−)-(21) (Fig. 7). Only compound 18 exhibited anti-inflammatory activity by the inhibition of nitric oxide (NO) production in the lipopolysaccharide (LPS)-stimulated mouse macrophage RAW 264.7 with IC50 = 84.2 μM, and the rest of the isolated compounds were found to be inactive.65
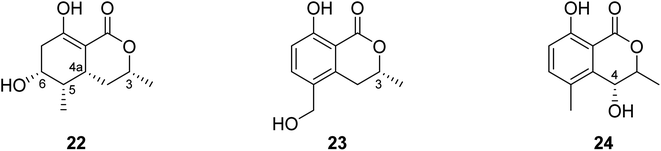
3.5.2.2. Fungi of the genus Valsa. (3R,4aR,5S,6R)-6-Hydroxy-5-methylramulosin (22), a previously unreported isocoumarin along with three other known derivatives, including the previously mentioned (−)5-methylmellein (14), (−)-5-hydroxymethylmellein (23), and (−)-(3R,4R)-cis-4-hydroxy-5-methylmellein (24) (Fig. 8), were isolated from the marine alga-derived fungus Valsa ceratosperma obtained from the green alga Codium fragile (SURINGAR) HARIOT, collected from the Japan sea at the bay of Toyama. While compounds 14, 23, and 24 showed no cytotoxic activity against HeLa cell lines, compound 22 displayed 65% inhibitory activity against HeLa cells growth at a concentration of 50 μg mL−1.66
3.6. Fungi of the order Eurotiales
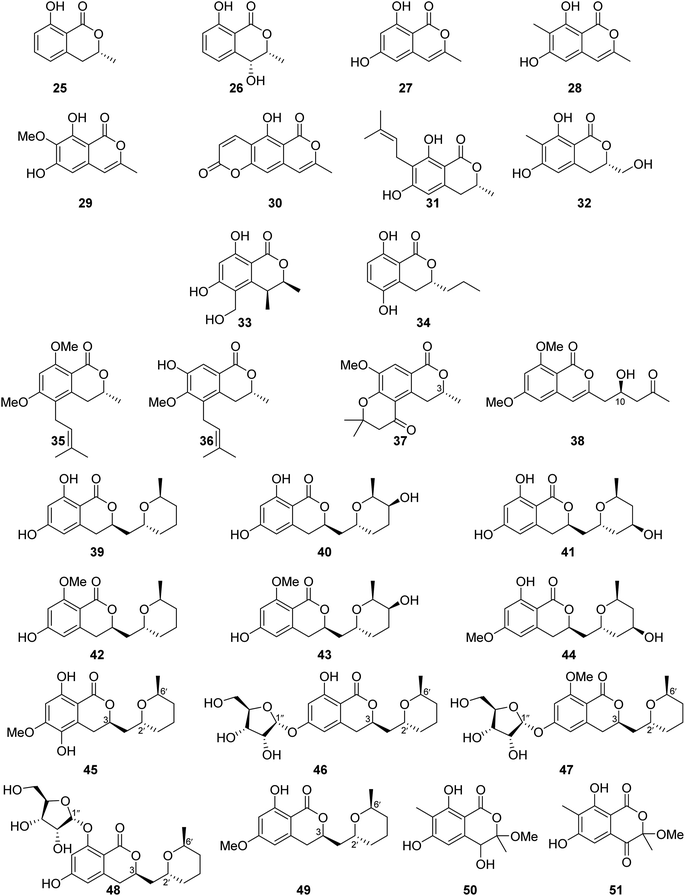
3.6.1.1. Fungi of the genus Aspergillus. The chemical investigation of the marine-derived fungus Aspergillus sp. associated with the ascidian Eudistoma vannamei, obtained from Northeast Brazil, led to the isolation of two previously reported isocoumarins, namely, mullein (25) and cis-4-hydroxymellein (26), along with the previously mentioned trans-4-hydroxymellein (7) (Fig. 9). All the isolated compounds exhibited no cytotoxicity once they were examined against two tumor cell lines HCT-8 and MDA-MB-435.67 Other four isocoumarins were isolated from the EtOAc extract of the marine-derived fungus Aspergillus similanensis sp. nov. KUFA 0013, obtained from the marine sponge Rhabdermia sp., collected in Thailand, from the Similan Islands coral reef, including the previously reported 6,8-dihydroxy-3-methylisocoumarin (27) and reticulol (29), along with previously unreported derivatives 6,8-dihydroxy-3,7-dimethylisocoumarin (28) and 5-hydroxy-8-methyl-2H,6H-pyrano[3,4-g]chromen-2,6-dione (30), which is also known as similanpyrone A (Fig. 9). Compounds 27–30 showed neither antifungal nor antibacterial when tested against C. albicans ATCC 10231, Gram-negative bacteria (E. coli ATCC 25922 and P. aeruginosa ATCC 27853), and Gram-positive bacteria (S. aureus ATCC 25923 and B. subtilis ATCC 6633).68 In addition, four previously reported isocoumarin derivatives, namely, angelicoin A (31), periplanetin D (32), (3S,4S)-dihydroascochin (33), and phomolactone B (34) together with three previously unreported congeners, including versicoumarins A (37), B (35), and C (36) (Fig. 9), were obtained from the endophytic fungus of Aspergillus versicolor isolated from P. marmorata stearn rhizome, collected from Yunnan, China. Compounds 31–37 were examined for their antiviral activity (anti-TMV activity) using the half-leaf method and ningnanmycin as a positive control. They exhibited an inhibition activity in the range from 11.5 to 28.6%. Furthermore, it is worth mentioning that compound versicoumarins A (37) exhibited the highest inhibitory activity of 28.6%. In addition, they were also tested for their cytotoxicity against MCF7, NB4, PC3, SHSY5Y, and A549 cancer cell lines using the MTT-assay and taxol as the positive control. Compounds 31–36 exhibited moderate cytotoxic effect against the examined cell lines with IC50 <10 μM; however, compound versicoumarins A (37) displayed strong cytotoxic effect toward MCF7 and A549 tumor cell lines with IC50 values of 4.0 and 3.8 μM, respectively.69 (S)-(−)-6,8-Di-O-methylcitreoisocoumarin (38) (Fig. 9), a previously unreported isocoumarin derivative, was isolated from the marine-derived fungus Aspergillus flavus OUCMDZ-2205 isolated from the prawn, Penaeus vannamei, collected in China from the sea area of Lianyungang. Compound 38 was not examined for any relevant biological activity.70 The chemical examination of the algicolous-derived endophytic fungus Aspergillus sp. F00785 isolated from the alga, Enteromorpha prolifera, collected in China from the Saltern of Jinjiang, afforded nine asperentin derivatives, including the previously reported asperentin (39), which is also known as cladosporin, 5′-hydroxyasperentin (40), 4′-hydroxyasperentin (41), asperentin-8-methyl ether (42), 5′-hydroxyasperentin-8-methyl ether (43), and 4′-hydroxyasperentin-6-methyl ether (44), together with three previously undescribed derivatives, i.e., 5-hydroxyl-6-O-methylasperentin (45), 6-O-α-D-ribosylasperentin (46), and 6-O-α-D-ribosyl-8-O-methylasperentin (47) (Fig. 9). Compounds 39–47 were examined for their antifungal effect against three crop pathogenic fungi, B. cinerea Pers, C. gleosporioides Penz., and C. gleosporioides (Penz.) Sacc., using the filter-paper disk method and as a positive control amphotericin has been used. Compounds 41–47 exhibited no antifungal effect against the examined fungi, but compound 39 displayed strong inhibition effect against C. gleosporioides Penz. at a concentration of 5 μg mL−1 with an inhibition zone of 19.7 mm.71 Six dihydroisocoumarins derivatives including the previously mentioned asperentin (39), 5′-hydroxyasperentin (40), 4′-hydroxyasperentin (41), asperentin-8-methyl ether (42), along with two previously undescribed derivatives, including cladosporin 8-O-α-ribofuranoside (48), and asperentin 6-O-methyl ether (49) (Fig. 9), were obtained from EtOAc extract of the marine-derived fungi Aspergillus sp. SF-5974 and Aspergillus sp. SF-5976, isolated from an unidentified red algae, collected from the Ross Sea at a depth of 300 m. Compounds 39–42 and 48–49, were tested for their antineuro–inflammatory activity, and they displayed inhibition activity toward prostaglandin E2 (PGE2) and LPS-induced nitric oxide (NO) production through inducible NO synthase (iNOS) and cyclooxygenase-2 (COX-2) expression, with IC50 values ranging from 10 to 80 μM.72 The chemical examination of the MeOH extract of the mangrove-derived fungus Aspergillus sp. 16-5B isolated from Sonneratia apetala leaves, collected in China, in Hainan Island, led to the isolation of two previously unidentified isocoumarin derivatives as a racemic mixture of the possible enantiomer. i.e., (±) 50 and (±) 51 (Fig. 9). Compounds 50–51 were tested for their ability to inhibit the enzyme α-glucosidase (using acarbose as a positive control); while compound 51 was inactive, compound 50 displayed inhibition activity with IC50 values of >200 and 90.4 μM, respectively.73
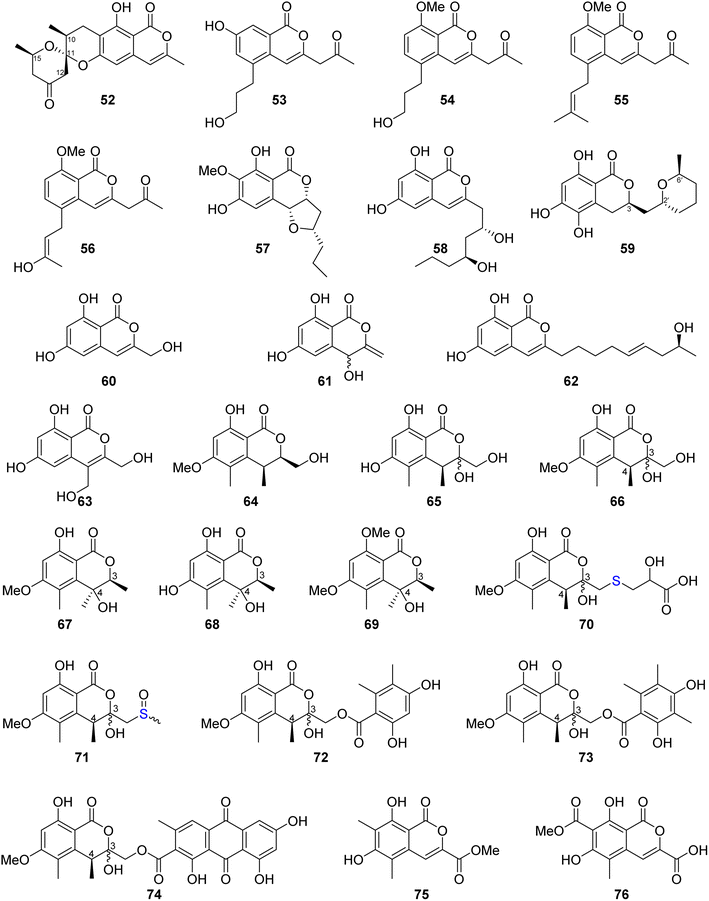
Similanpyrone C (52) (Fig. 10), a previously undescribed isocoumarin derivative, was isolated from the marine associated fungus Aspergillus similanensis KUFA 0013, obtained from the unidentified marine sponge. Compound 52 was not tested for any biological activity as it was obtained in a very minute amount.74 The chemical investigation of the endophytic fungus Aspergillus oryzae organic extract obtained from the rhizome of P. polyphylla var. yunnanensis, collected in Yunnan, China afforded six isocoumarin derivatives, including the previously unreported oryzaeins A (53) and B (54), together with the known derivatives, tabaisocoumarin A (55), caudacoumarin C (56), exserolide D (57), and exserolide F (58) (Fig. 10). Intriguingly, the presence of the unusual 2-oxopropyl group and a rare 3-hydroxypropyl group shows privileged compounds 53–54 to be the first examples of an isocoumarin possessing these unusual structural features. Only compounds 53–54 were examined for their antiviral effect toward tobacco mosaic virus (anti-TMV) using the half-leaf method and ningnanmycin as a positive control, showing an inhibition rate of 28.4 and 30.6, respectively, at a concentration of 20 μM. Moreover, they were tested for their cytotoxic ability against NB4, A549, SHSY5Y, PC3, and MCF7 cancer cell lines by the MTT method, exhibiting weak to moderate cytotoxic effect with IC50 values in the range of 2.8–8.8 μM.75 Asperentin B (59) (Fig. 10), a previously undescribed isocoumarin derivative of the asperentin-type, and the previously mention asperentin (39) were isolated from the marine-derived fungus Aspergillus sydowii obtained from the deep Mediterranean sea sediment (2769 m). Asperentin B (59) was found to display potent inhibitory activity against PTP1B enzyme with an IC50 value of 2.05 μM, when compared with suramin as a positive control that exhibited an IC50 value of 11.85 μM. In addition, 59 exhibited weak inhibitory activity against Propionibacterim acnes with an inhibition rate of 57% at a concentration of 100 μM. Moreover, compound 59 displayed no antimicrobial effect when tested against X. campestris, S. tritici, C. albicans, B. subtilis, and S. lentus at a concentration of 100 μM. Furthermore, 59 showed no cytotoxic effect at a concentration of 50 μM when tested against the HepG2 and HT29 tumor cell lines. On the other hand, while compound 39 showed no inhibition effect against the activity of PTP1B enzyme as well as exhibited no cytotoxic effect against HepG2 and HT29, it inhibited the growth of X. campestris, S. tritici, B. subtilis, T. mentagrophytes, and S. lentus with an inhibition rate in the range of 83–100% when compared with the positive control clotrimazole, which also displayed a very weak antifungal activity.76 The previously mentioned 6,8-dihydroxy-3-methylisocoumarin (27), along with four previously reported isocoumarin derivatives, namely, 6,8-dihydroxy-3-hydroxymethylisocoumarin (60), 4,6-dihydroxy-3,9-dehydromellein (61), fusariumin (62), and penicimarin F (63) (Fig. 10), were obtained from the EtOAc extract of the fermentation product of the endophytic fungus Aspergillus versicolor, obtained from the rhizome of Paris polyphylla var. yunnanensis, collected in Yunnan, China. No relevant biological activity was reported for 60–63.77 The chemical investigation of the fungus Aspergillus banksianus obtained from Banksia integrifolia collected from Australia in New South Wales led to the isolation of three previously described isocoumarin derivatives, namely, clearanol I (64), dothideomynone A (65), and banksialactone A (66), together with ten previously undescribed derivatives, including eight isocoumarins, namely, banksialactones B–I (67–74), as well as two new isocoumarins, named banksiamarins A–B (75–76) (Fig. 10). Compounds 64–76 were examined for their antibacterial activity against B. subtilis (ATCC6633), E. coli (ATCC25922), C. albicans (ATCC 10231), and S. cerevisiae (ATCC 9763), as well as for their antiprotozoal ability against T. fetus (KV-1) and for their cytotoxicity against the NS-1 cancer cell line using ampicillin, clotrimazole, mebendazole, and 5-fluorouracil, respectively, as positive controls. Though compounds 64–65, 67–71, and 75–76 displayed no activity in any of the investigated biological activity, compounds 72–74 exhibited weak to moderate activity on all the aforementioned biological assays.78
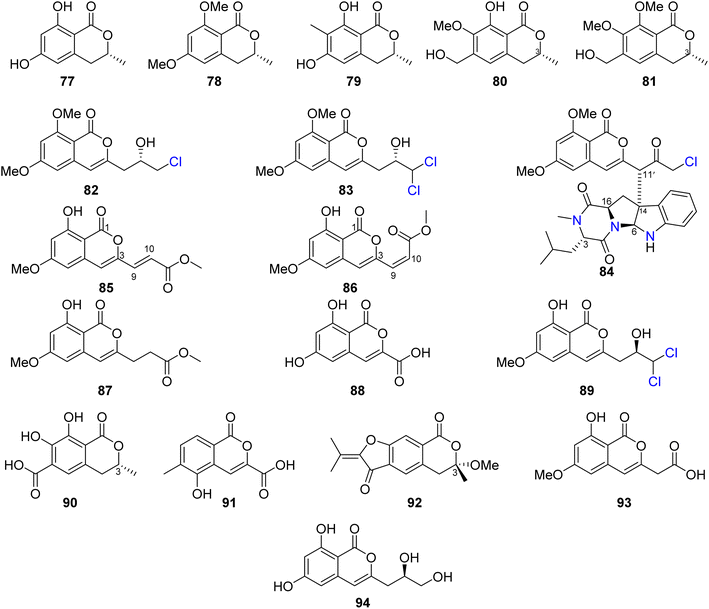
Three previously reported isocoumarins (R)-6-hydroxymellein (77), 6,8-dimethoxy-3-methyl-3,4-dihydro-1H-isochromen-1-one (78), and periplanetin B (79), along with the previously undescribed derivatives (3R)-methyl-8-hydroxy-6-(hydroxymethyl)-7-methoxydihydroisocoumarin (80) and (3R)-methyl-7,8-dimethoxy 6-(hydroxymethyl)dihydroisocoumarin (81) (Fig. 11), were isolated from the endophytic fungus Aspergillus versicolor derived from the Nicotiana tabacum rhizome, collected from Yunnan, China. Compounds 80–81 were tested for their antiviral activity against the TMV using the half leaf method and ningnanmycin as a positive control, showing an inhibition effect with an inhibition rate 21.8 and 18.6%, respectively.79 The chemical examination of the terrestrially-derived fungus Aspergillus sp. CPCC 400810, an endolichenic fungus obtained from Cetrelia sp., collected in Yunnan, China from the mount of Laojun, led to the isolation of the two previously identified isocoumarin derivatives 8-methyl-11-chlorodiaporthin (82) and 8-methyl-11,11-dichlorodiaporthin (83), along with one previously unreported isocoumarindole A (84), a hybrid molecule, featuring a polyketide-nonribosomal peptide (PKS-NRPS) biogenesis pathway (Fig. 11). Compound 84 displayed moderate antifungal effect toward C. albicans using caspofungin as a positive control, with MIC value of 32.0 μg mL−1. In addition, it exhibited potent cytotoxicity against MIA-PaCa-2 and AsPC-1 with IC50 values of 1.63 and 5.53 μM, respectively.80 Three previously undescribed isocoumarins, namely, apergisocoumrins A–C (85–87), along with the previously identified 8-dihydroxyisocoumarin-3-carboxylic acid (88) and dichlorodiaportin (89) (Fig. 11), were isolated from the mangrove-derived endophytic fungus Aspergillus sp. HN15-5D isolated from A. ilicifolius fresh leaves, collected from Hainan Island, China. Compounds 85–89 were tested for their cytotoxicity (using epirubicin as the positive control) against MDA-MB-435, HepG2, HCT116, H460, and MCF10A cancer cell lines. While 87–89 showed no cytotoxic activity against the examined tumor cell lines, 85 exhibited potent cytotoxic activity against MDA-MB-435, HepG2, H460, and MCF10A with IC50 value in the range of 5.08–43.7 μM; however, 86 displayed moderate cytotoxicity against MCF10A and MDA-MB-435 cancer cell lines with IC50 values of 21.4 and 4.98, respectively. Furthermore, 85–89 were tested for their antibacterial ability against S. aureus, S. epidermidis, E. coli, K. pneumoniae, and B. subtilis; only 89 displayed moderate antibacterial activity against with MIC values of 25 μg mL−1.81 Two previously unreported isocoumarins, namely, aspergillspins F–G (90–91) (Fig. 11), were reported from the gorgonian-derived fungus Aspergillus sp. SCSIO 41501 isolated from the soft coral Melitodes squamata collected from the South China Sea. Compounds 90–91 displayed neither cytotoxicity when examined against HL60, HepG2, and MCF-7 cancer cell lines using MTT methods nor antibacterial effect toward B. subtilis and E. coli using the standard disc diffusion.82 Asperisocoumarin G (92) (Fig. 11), a previously undescribed isocoumarin, was isolated from MeOH extract of the marine derived fungus Aspergillus sp. 085![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 242 isolated from the mangrove plant collected in China. Compound 92 exhibited moderate inhibition effect against α-glucosidase activity when compared with clinical acarbose as a positive control. In addition, it showed no antibacterial activity against S. aureus ATCC 6538, B. Subtilis ATCC 6633, E. coli ATCC 8739, P. Aeruginosa ATCC 9027, and Salmonella ATCC 14028.83 The chemical examination of the marine-derived fungus Aspergillus falconensis isolated from the marine sediment collected from Red Sea, Egypt from the Canyon at Dahab at 25 m depth led to the isolation of the previously mentioned dichlorodiaportin (89) along with the previously synthesized derivative 2-(8-hydroxy-6-methoxyisochromen-3′-yl)acetic acid (93) and the previously described desmethyldiaportinol (94). In silico studies of compounds 89 and 93–94 on human cyclin-dependent kinase 2 revealed a certain degree of stability in the active sites of CDK-2 (ΔG = −20.32, −22.30, and −20.46, respectively).84
242 isolated from the mangrove plant collected in China. Compound 92 exhibited moderate inhibition effect against α-glucosidase activity when compared with clinical acarbose as a positive control. In addition, it showed no antibacterial activity against S. aureus ATCC 6538, B. Subtilis ATCC 6633, E. coli ATCC 8739, P. Aeruginosa ATCC 9027, and Salmonella ATCC 14028.83 The chemical examination of the marine-derived fungus Aspergillus falconensis isolated from the marine sediment collected from Red Sea, Egypt from the Canyon at Dahab at 25 m depth led to the isolation of the previously mentioned dichlorodiaportin (89) along with the previously synthesized derivative 2-(8-hydroxy-6-methoxyisochromen-3′-yl)acetic acid (93) and the previously described desmethyldiaportinol (94). In silico studies of compounds 89 and 93–94 on human cyclin-dependent kinase 2 revealed a certain degree of stability in the active sites of CDK-2 (ΔG = −20.32, −22.30, and −20.46, respectively).84
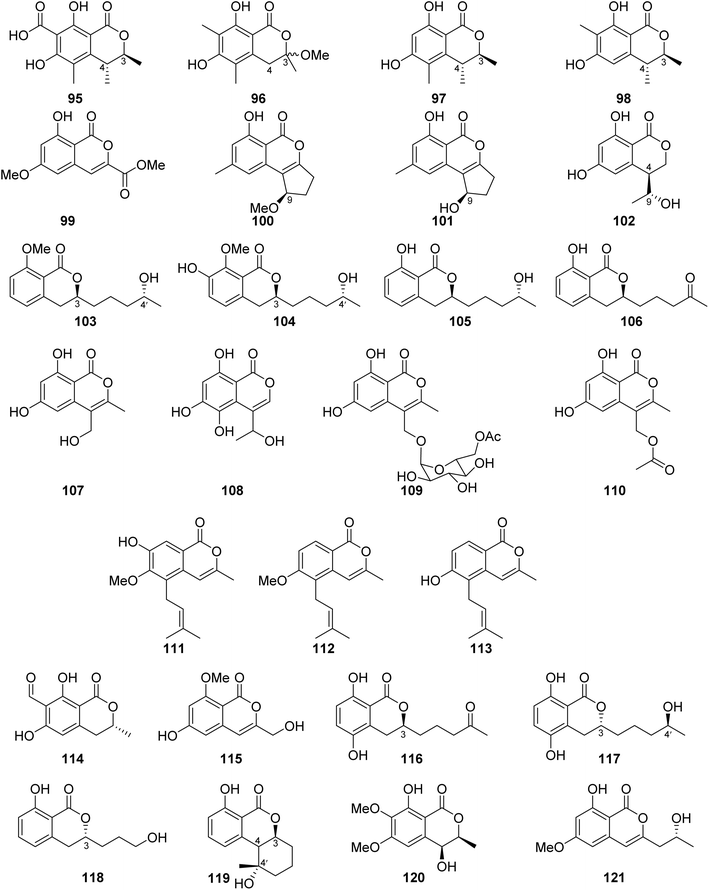
3.6.1.2. Fungi of the genus Penicillium. Dihydrocitrinone (95) (Fig. 12), a previously described isocoumarin, was reported from the marine-derived fungus Penicillium notatum B-52 isolated from the sediments, collected from the Lake of Qinghai, China. In addition, the chemical investigation of the marine-derived fungus P. stoloniferum QY2-10 isolated from an unidentified sea squirt collected from the bay of Jiaozhou, China, led to the isolation of two previously unreported derivatives, namely, stoloniferols A–B (96–97) (Fig. 12). Compounds 95–97 displayed no activity when tested for their cytotoxicity by the MTT method against P388, BEL-7402, A-549, and HL-60 tumor cell lines.85 Cis-4-hydroxymellein (26), a previously mentioned derivative (Fig. 9), was isolated from the marine-derived fungus P. sp., obtained from the green alga Ulva pertusa collected from the island of Beijing, Korea. No biological activity was reported for this compound.86 In addition, the chemical investigation of the marine derived endophytic fungus P. sp., 091
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 402 isolated from the roots of the mangrove Bruguiera sexangular Linn collected in China, Qinglan Port, Hainan led to the isolation of one previously unreported derivative, (3R*,4S*)-6,8-dihydroxy-3,4,7-trimethylisocoumarin (98), along with the known isocoumarin derivative, (3R,4S)-6,8-dihydroxy-3,4,5-trimethylisocoumarin (97) (Fig. 12). Compound 97 was not tested for any relevant bioactivity, while compound 98 displayed mild cytotoxic activity against K562 cancer cell line with IC50 = 18.9 μg mL−1.87 Penicilisorin (99) (Fig. 12), a previously undescribed isocoumarin, was isolated from the terrestrial endophytic fungus P. sclerotiorum PSUA13, isolated from G. Atroviridis leaves, collected in Thailand from Yala Province. Due to the minute quantity of 99, it was not subjected for any relevant biological activity.88 A previously unidentified citrinolactone D (100) along with the previously described citrinolactone B (101) (Fig. 12) was isolated from the marine sediment-derived fungus P. sp., ML226, isolated from the sediment of the mangrove region, Long Hai, China.
402 isolated from the roots of the mangrove Bruguiera sexangular Linn collected in China, Qinglan Port, Hainan led to the isolation of one previously unreported derivative, (3R*,4S*)-6,8-dihydroxy-3,4,7-trimethylisocoumarin (98), along with the known isocoumarin derivative, (3R,4S)-6,8-dihydroxy-3,4,5-trimethylisocoumarin (97) (Fig. 12). Compound 97 was not tested for any relevant bioactivity, while compound 98 displayed mild cytotoxic activity against K562 cancer cell line with IC50 = 18.9 μg mL−1.87 Penicilisorin (99) (Fig. 12), a previously undescribed isocoumarin, was isolated from the terrestrial endophytic fungus P. sclerotiorum PSUA13, isolated from G. Atroviridis leaves, collected in Thailand from Yala Province. Due to the minute quantity of 99, it was not subjected for any relevant biological activity.88 A previously unidentified citrinolactone D (100) along with the previously described citrinolactone B (101) (Fig. 12) was isolated from the marine sediment-derived fungus P. sp., ML226, isolated from the sediment of the mangrove region, Long Hai, China.
Compound 100 was tested for its cytotoxicity against the tumor HeLa and HepG-2 cell line (using the MTT method and cis-platinum as positive control) as well as evaluated for its antimicrobial activity against S. aureus (CMCC26003), E. coli (CMCC44103), C. albicans (AS2.538), and A. niger (ACCC30005) using the paper diffusion method. Compound 100 displayed neither cytotoxicity nor antimicrobial activities.89 The chemical examination of the sponge-derived fungus P. sp., (MWZ14-4), isolated from an unidentified sponge collected from the coral reef of the South China Sea, afforded ten isocoumarin derivatives, including five previously unreported hydroisocoumarins, namely, penicimarins A–C (102–104), penicimarins D–E (109–110), along with known congeners aspergillumarins B (105) and A (106), sescandelin B (107), 5,6,8-trihydroxy-4-(1′-hydroxyethyl)isocoumarin (108) (Fig. 12), and previously mentioned derivative penicimarin F (63) (Fig. 10). Compounds 63 and 102–110 were tested for their antibacterial activity against the terrestrial pathogenic bacteria E. coli, S. aureus, S. albus, B. subtilis, B. cereus, M. tetragenus, and K. rhizophila, as well as the marine-derived pathogenic bacteria V. parahemolyticus and V. anguillarum (using the conventional broth dilution method and ciprofloxacin as a positive control). Among all of them, 108 displayed the most potent activity against B. cereus and V. parahemolyticus, with MIC values of 6.25 μM. Furthermore, 63 and 102–110 displayed no cytotoxicity against HeLa, A549, K562, and HL-60 when examined by the MTT method.90
Terrecoumarins A–C (111–113), three previously unreported isocoumarins along with known ones including, periplanetin A (114), 6-hydroxy-3-hydroxymethyl-8-methoxyisocoumarin (115) (Fig. 12), periplanetin D (32) (Fig. 9), and 6,8-dihydroxy-3-hydroxymethylisocoumarin (60) (Fig. 10), were reported from the terrestrial-derived fungus P. oxalicum 0403 isolated from Nicotiana sanderae leaves collected in Yunnan Province, China. Compounds 32, 60, and 111–115 were tested for their antiviral activity against tobacco mosaic virus (using ningnamycin as a positive control).
Only 111 displayed strong anti-TMV activity with an inhibition rate of 25.4%, while all the other compounds displayed weak antiviral effect with an inhibition rate in the range of 11.3–18.9%.91 In addition, the chemical examination of the EtOAc extract of the marine sediment-derived fungus P. citrinum, isolated from the marine sediment collected in China, from the Island of Langqi, afforded a previously mentioned (3R,4S)-6,8-dihydroxy-3,4,5-trimethylisocoumarin (97) (Fig. 12). No relevant biological activity was reported for 97 obtained from this species.92 Furthermore, the chemical investigation of the EtOAc of the marine-derived fungus P. sp., (KY620115), isolated from the hydrothermal vent sediment, collected in Taiwan, from the Island Kueishantao, led to the isolation of six isocoumarin derivatives, including two previously reported analogues, aspergillumarins B (105) and A (106) together with four previously unreported derivatives, penicillisocoumarins A–D (116–119) (Fig. 12).
All the isolated compounds displayed no cytotoxicity against HepG2, SMMC-7721, and Bel-7402 cancer cell lines. However, 116–117 and 119 exhibited weak antibacterial activity against E. coli, with MIC value of 32 μg mL−1.93 Chen et al., 201794 recorded the chemical examination of the marine-derived fungus P. chrysogenum SCSIO 41001 obtained from the deep-sea sediment collected in the Indian Ocean, which led to the isolation of three known compounds, stoloniferol A (96), 4-hydroxykigelin (120), and diaporthin (121) (Fig. 12). Though Xin et al., 200785 suggested before that stoloniferol A (96) was reported as one pure compound, Chen et al., 2017 94succeeded in its isolation in the form of two enantiomers such as stoloniferol A (96), i.e., R-(−)-stoloniferol A and S-(+)-stoloniferol A (96). Compounds 96 and 120–121 displayed no activity when examined for their antibacterial, cytotoxic, antiviral, and anti-inflammatory (COX-2) activities.
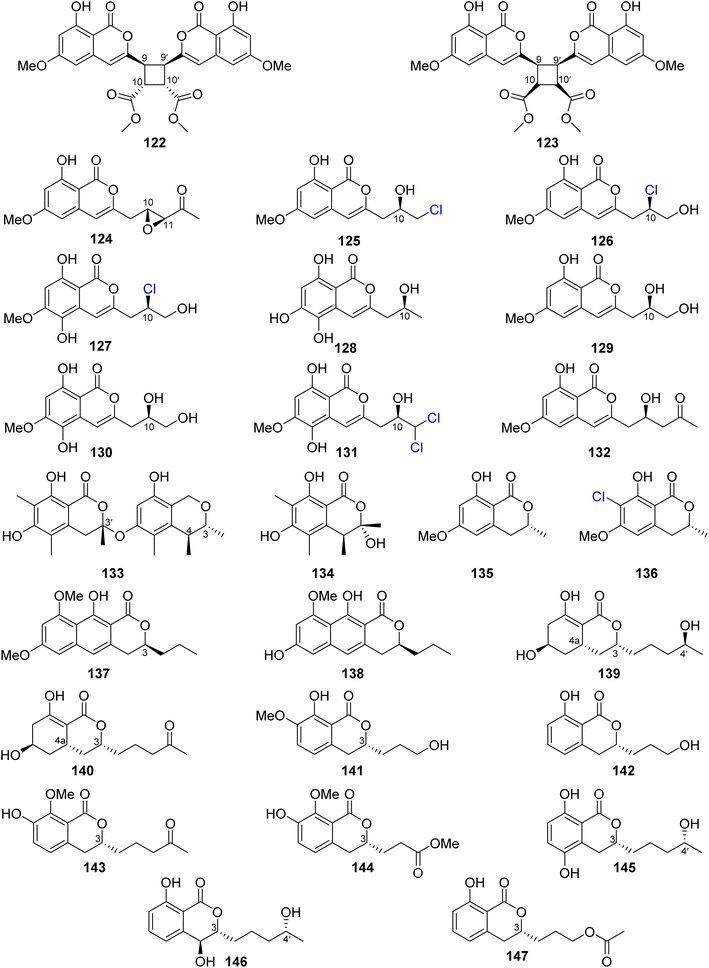
Dichlorodiaportin (89) (Fig. 11) and diaporthin (121) (Fig. 12), two previously reported derivatives, along with previously undescribed peniisocoumarins A–J (122–131), together with the previously reported (+)-6-O-methylcitreoisocoumarin (132) (Fig. 13), were obtained from the EtOAc extract of the mangrove-derived fungus P. commune QQF-3, isolated from Kandelia candel collected in Guangdong municipality, China.
Compounds 89 and 121–132 were tested for their α-glucosidase inhibition activity using acarbose as a positive control. Indeed, 89 and 126–127 displayed mild inhibitory effect with IC50 values of 102.4, 110.3, and 158.4 μM, respectively. Moreover, 124, 128, and 130–131 were found to be more potent than the positive control acarbose with IC50 values of 38.1, 40.5, 78.1, 45.1, and 478.4 μM, respectively. In addition, they were evaluated for their inhibition effect against MptpB, using oleanolic acid and p-nitrophenyl phosphate as a positive control/substrate. Compound 128 displayed a moderate inhibitory activity with an IC50 value of 20.7 μM, but the remaining compounds exhibited weak or no activity. Moreover, none of the isolated compounds exhibited cytotoxicity against A549, HepG2, HeLa, MCF-7, and HEK293T tumor cell lines using the MTT method.95 The chemical examination of the EtOAc of the marine-derived fungus P. piltunense KMM 4668, isolated from the subaqueous soil collected in Russia from the Island of Sakhalin, led to the isolation of the previously mentioned asperentin, also known as cladosporin (39), 5′-hydroxyasperentin (40) (Fig. 9). Compound 39 displayed cytotoxicity against the 22Rv1 cancer cell line with a high selectivity index. In addition, it displayed anti-inflammatory effect by decreasing the NO production by 24.1% in LPS-stimulated macrophages.96
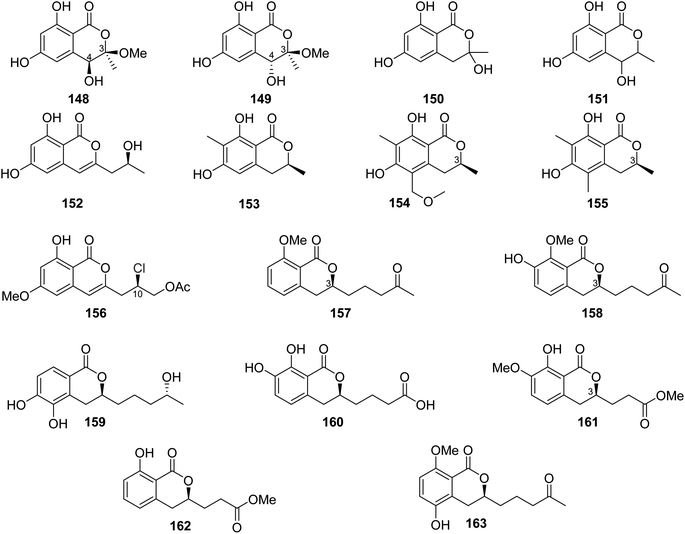
A previously undescribed penicitol D (133), along with four known derivatives, stoloniferol B (97) (Fig. 12), (3S,4S)-sclerotinin A (134), (3R)-6-methoxymellein (135), and (3R)-6-methoxy-7-chloromellein (136) (Fig. 13), were isolated from the deep sea-derived fungus P. citrinum NLG-S01-P1. Compounds 97 and 133–136 were tested for their antibacterial activity using continuous dilution in 96-well plates method against V. rotiferianus (MCCC E385), MRSA (ATCC 43300, CGMCC 1.12409), V. campbellii (MCCC E333), and V. vulnificus (MCCC E1758). Among them, 133 exhibited a potent antibacterial effect toward MRSA (ATCC 43300, CGMCC 1.12409), and 134 showed relatively stronger effect in comparison with the other compounds. In addition, 97 and 133–136 displayed weak cytotoxic effect when tested for their cytotoxicity against A549 and HeLa tumor cell lines (using the Cell Counting Kit-8 (CCK-8) (DOJINDO) method and as a positive control, doxorubicin was used).97
A chemical investigation of the sponge-derived fungus P. sp., XWS02F62, isolated from the marine sponge Callyspongia sp., collected in Guangdong, China, afforded one previously unrecorded isocoumarin, named 7-O-methylpenicitor A (137), along with previously reported aspergillumarins B (105) and A (106), and penicitor A (138) (Fig. 13). Compounds 105, 106, and 137–138 were examined for their cytotoxicity against MDA-MB-231, 143B, C4–2B, MGC803, and A549 cancer cell lines. Only 138 exhibited a moderate inhibitory activity against MDA-MB-231 and C4–2B tumor cells with inhibition rates of 31.3% and 25.7% at a concentration of 5 μM, respectively.98 Three previously mentioned derivatives, penicimarin C (104), aspergillumarin A (106), and (R)-3-(3-hydroxypropyl)-8-hydroxy-3,4-dihydroisocoumarin (142) (Fig. 12), along with eight new derivatives, peniciisocoumarins A (139), B (140), C (141), D (143), E (144), F (145), G (147), and H (146) together with the previously described (Fig. 13), were obtained from the marine-derived fungus P. sp., TGM112, isolated from the mangrove plant B. sexangula var. rhynchopetala, collected in China, from the South China Sea.
Compounds 139, 140, 144, and 146 displayed insecticidal effect against H. armigera Hubner (using azadirachtin as a positive control) with IC50 values of 200, 200, 100, and 100 μg mL−1 respectively. Moreover, none of the isolated compound exhibited cytotoxicity toward A549, HeLa, and HepG2 tumor cell lines. Furthermore, they displayed no antibacterial ability against E. coli (ATCC 25922), S. aureus (ATCC 25923), Methicillin-resistant S. aureus MRSA (ATCC 33591), Bacillus cereus (ATCC 11778), V. parahaemolyticus (ATCC 17802), and V. alginolyticus (ATCC 17749), using the microplate assay method and ciprofloxacin as the positive control. In addition, they showed no anti-inflammatory properties as they displayed no inhibitory activity against nitric oxide (NO) production in lipopolysaccharide (LPS)-induced RAW 246.7 mouse macrophages.99
The chemical analysis of the EtOAc extract of the mangrove-derived endophytic fungus P. coffeae MA-314 isolated from Laguncularia racemosa leaves collected in China from the Island of Haninan afforded five previously reported known derivatives including 6,8-dihydroxy-3-methylisocoumarin (27), 4,6-dihydroxy-3,9-dehydromellein (61), 3-methoxy-6,8-dihydroxy-3-methyl-3,4-dihydroisocoumarin (150), cis-4,6-dihydroxymellein (151), and O-demethyldiaporthin (152) along with two previously unreported enantiomers, penicoffrazins B (148) and C (149) (Fig. 14). The isolated compounds were tested for their antioxidant activity by measuring their ability to scavenge the DPPH free radical, and butylated hydroxytoluene (BHT) was used as a positive control. Only 150 displayed weak antioxidant effect with IC50 = 159 μM; however, the remaining compounds exhibited almost no antioxidant activity with IC50 >900 μM.100 A previously described isocoumarin, monaschromone (153), along with three previously non-recorded derivatives, namely, (S)-6,8-dihydroxy-5-(methoxymethyl)-3,7-dimethylisochroman-1-one (154), (S)-6,8-dihydroxy-3,5,7-trimethyl-isochroman-1-one (155), and (R)-2-chloro-3-(8-hydroxy-6-methoxy-1-oxo-1H-isochromen-3-yl) propyl acetate (156) (Fig. 14), was isolated from the MeOH extract of the marine endophytic fungus P. sp., YYSJ-3, isolated from Heritiera littoralis steam, collected from Guangdong Province, China. Compounds 153–156 were examined for their inhibitory activity toward α-glucosidase activity. Compound 153 displayed no effect, but 154–155 exhibited weak inhibition activity with IC50 values of 309.6 and 237.4 μmol L−1, respectively. In addition, among them, 156 was the most potent enzyme inhibitor with an IC50 value of 100.6 μmol L−1.101 Phytochemical examination of the marine-derived fungus P. sp., XR046, obtained from the soil collected from the area of Xinren coal area in the province of Guizhou, China, led to the isolation of five known derivatives, including penicimarins B–C (103–104), penicillisocoumarin A (116) (Figure 12), 5,6-dihydroxy-3R-(4S-hydroxypentyl)-isochroman-1-one (159), and 3R-(7,8-dihydroxy-1-oxoisochroman-3-yl)propanoic acid (160) (Fig. 14), along with two previously undescribed congeners, namely, 3R-8-methoxy-3-(4-oxo-pentyl) isochroman-1-one (157) and 3R-7-hydroxy-8-methoxy-3-(4-oxopentyl) isochroman-1-one (158) (Fig. 14). Compounds 103–104 and 157–160 were tested for their antimicrobial activity against C. albicans ATCC 5314, S. epidermidis, E. coli ATCC 25922, S. aureus ATCC 25923, and B. subtilis ATCC 6633. While 103–104 and 157–160 displayed mild antifungal effect against C. albicans ATCC 5314, 103–104 and 157–159 displayed weak antibacterial ability against S. epidermidis. Moreover, only 159–160 showed weak antibacterial activity toward B. subtilis ATCC 6633. In addition, among all the tested compounds, only 160 displayed weak antibacterial activity against E. coli ATCC 25922. The remaining isolated metabolites were inactive against S. aureus ATCC 25923.102 A previously undescribed penicimarin N (161) along with three previously reported derivatives, penicimarins I–H (162–163) (Fig. 14) and aspergillumarin A (106) (Fig. 12), was obtained from the EtOAc extract of the mangrove-derived fungus P. sp., TGM112 isolated from B. sexangula var. rhynchopetala, collected South China Sea, China. Compounds 106 and 161–163 were evaluated for their antioxidant activity (using Trolox as a positive control), antibacterial activity against S. aureus, methicillin-resistant S. aureus, S. albus, V. alginolyticus, and V. parahemolyticus as well as α-glucosidase initiatory activities. While 163 showed weak antioxidant ability, 161 displayed strong antioxidant effect with IC50 values of 9.0 and 0.1 mM, respectively. None of the examined metabolites exhibited antibacterial activity against the bacterial strains under investigation. Among the tested isocoumarin derivatives, 161 displayed moderate enzyme inhibitory effect toward α-glucosidase.103
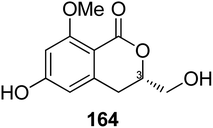
3.6.1.3. Fungi of the genus Eupenicillium. A chemical study of the marine-derived fungus Eupenicillium sp. 6A–9 obtained from the inner part of the marine sponge Plakortis simplex, collected from the Island of Yongxing, China, afforded the previously unreported isocoumarin derivative eupenicillin A (164) (Fig. 15). Compound 164 displayed no antibacterial activity against S. aureus ATCC 25923, methicillin-resistant S. aureus (MRSA) ATCC4330, and A. baumannii ATCC19606, but it exhibited moderate cytotoxic effect against MCF7 with an IC50 value of 53.48 μM.104
3.6.1.4. Fungi of the genus Hamigera. A previously described derivative, 8-methyl-11,11-dichlorodiaporthin (83) (Fig. 11), alongwith with two previously unidentified chlorinated congeners, (9R*)-8-methyl-9,11-dichlorodiaporthin (165) and (9S*)-8-methyl-9,11-dichlorodiaporthin (166) (Fig. 16), was isolated from the soil-derived fungus Hamigera fusca NRRL 35721, obtained from a soil sample treated with phenol collected from a banana tree in the Island of Grande Comore. Compounds 83 and 165–166 were tested for their cytotoxic activity against CCD25sk, SHSY5, MiaPaca-2, MCF-7, HepG2, A2058, and A549 cancer cell lines colorimetrically using the MTT assay and doxorubicin as a positive control. Compound 83 displayed moderate cytotoxicity toward the CCD25sk tumor cell line with a CC50 value of 27.0 μM. Furthermore, 83 and 165–166 exhibited moderate cytotoxicity against SHSY5y cells with the CC50 values ranging from 19.4 to 36.2 μM.105
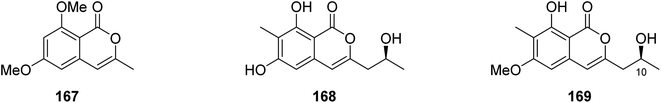
3.6.1.5. Fungi of the genus Monascus. A chemical inspection of the marine-derived fungus Monascus ruber BB5, isolated from the shellfish Meretrix collected in Yangjiang, from the Island of Hailing, China, led to the isolation of two previously described derivatives 6,8-dimethoxy-3-methylisocoumarin (167) and lunatinin (6,8-dihydroxy)-3-(2-hydroxypropyl)-7-methyl-1H-isochromen-1-one (168), together with one previously unreported derivative (S)-8-hydroxy-3-(2-hydroxypropyl)-6-methoxy-7-methyl-1H-isochromen-1-one, also known as monarubin B (169) (Fig. 17).
Compounds 167–168 were examined for their cytotoxic activity toward CNE1, CNE2, SUNE1, HONE1, HepG2, and QGY7701 cancer cell lines (using the MTT colorimetric assay and hirsutanol A as a positive control). Though all the isolated compounds displayed no cytotoxicity against CNE1 and HONE1, 168–169 showed weak cytotoxic effect toward CNE2 with IC50 values of 85.66 and 75.70 μM, respectively. Furthermore, they exhibited weak to potent cytotoxicity against the rest of the examined cell lines with IC50 values in the range of 0.71–72.07 μM.106
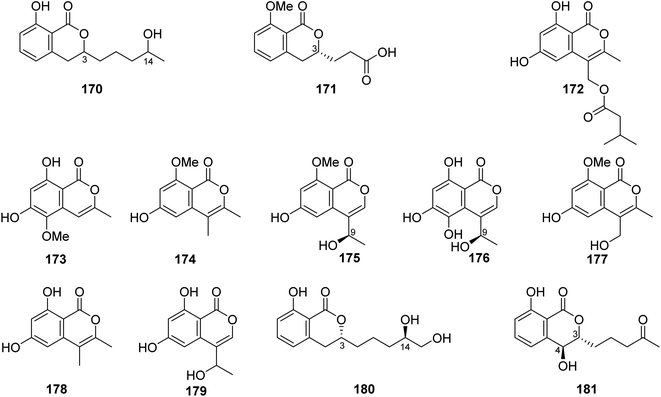
3.6.2.1. Fungi of the genus Talaromyces. A previously unreported isocoumarin derivative, named 8-hydroxy-3-(4-hydroxypentyl)-3,4-dihydroisocoumarin (170) (Fig. 18), was discovered from the plant-derived fungus Talaromyces verruculosus isolated from Stellera chamaejasme L. rhizosphere soil, collected from the Mountains of Qinling in the Province of Shaanxi, China. Compound 170 showed potent antibacterial ability against E. coli and S. aureus with MIC values of 2.5 and 5.0 μg mL−1, respectively. Furthermore, 170 displayed weak antifungal activity against A. solani, V. mali, C. lunata, and B. berengeriana with the growth inhibition percentage ranging from 87.2 to 97.3%.107 A chemical investigation of the marine sponge-derived fungus T. tratensis, obtained from the marine sponge Mycale sp., collected from the coral reefs of the Island of Samae San, Thailand, led to the isolation of the unreported previously tratenopyrone (171) (Fig. 18). Compound 171 displayed neither antibacterial nor anti-quorum sensing activities. In addition, it showed no cytotoxic and fungicidal activities.108 Five previously unreported isocoumarin derivatives including 172, 173, 6-hydroxy-8-methoxy-3,4-dimethylisocoumarin (174), S-(−)-5-hydroxy-8-methoxy-4-(10-hydroxyethyl)-isocoumarin (175), and 180, along with ten previously described compounds, including S-(−)-5,6,8-trihydroxy-4-(10-hydroxyethyl)isocoumarin (176), 6-hydroxy-4-hydroxymethyl-8-methoxy-3-methyl-isocoumarin (177), 3,4-dimethyl-6,8-dihydroxyisocoumarin (178) and sescandelin (179) (Fig. 18), penicimarins B (103), C (104), aspergillumarins B (105), A (106), sescandelin B (107), and 5,6-dihydroxy-3R-(4S-hydroxypentyl)-isochroman-1-one (159), were isolated from the marine endophytic fungus T. amestolkiae, isolated from the mangrove leaf of Kandelia obovate, collected from the Guangdong Province, China. Compounds 103–107, 159, and 172–180 exhibited moderate to weak inhibitory activities toward the enzyme α-glucosidase with IC50 ranging from 17.2 to 585.7 μM. Moreover, all the isolated compounds displayed no antibacterial activity when tested against S. aureus, S. epidermidis, E. coli, K. pneumoniae, and B. subtilis.23 A chemical investigation of the methanolic extract of the mangrove endophytic fungus T. sp., SCNU-F0041, obtained from Kandelia leaf, collected from Guangdong Province, China, afforded three previously mentioned aspergillumarins B (105), A (106) (Fig. 12), and 3R-(7,8-dihydroxy-1-oxoisochroman-3-yl) propanoic acid (160) (Fig. 14), along with a previously undescribed isocoumarin derivative aspergillumarin C (181) (Fig. 18). Compound 181 displayed no inhibitory effect toward AChE (acetylcholinesterase).109
3.7. Fungi of the order Glomerellales
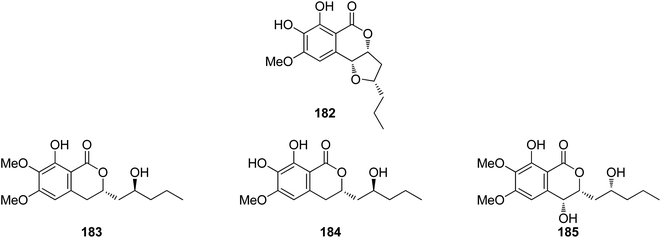
3.7.1.1. Fungi of the genus Colletotrichum. A previously mentioned derivative 4 (Fig. 3), along with other four previously reported isocoumarin derivatives, namely, monocerin demethylated (182), fusarentin-6,7-dimethyl ether (183), fusarentin-6-methyl ether (184), and fusarentin derivative (185) (Fig. 19), were obtained from the endophytic fungus Colletotrichum sp., CRI535-02, isolated from Piper ornatum, collected from the Province of Surat Thani. Compounds 4 and 182–185 were tested for their cytotoxicity against the HepG2, HuCCA-1, and A549 tumor cell lines (using the MTT assay) as well as toward the MOLT-3 cancer cell line (using the XTT assay). Though all the isolated compounds exhibited weak cytotoxicity or no activity against the examined cell lines, 182 displayed potent anticancer ability toward HepG2 with an IC50 value of 23.7 μM when compared with the positive control etoposide (IC50 = 15.8 μM). In addition, the potential cancer chemo-preventive characteristics of the isolated compounds were examined by determining their antioxidant activity and their inhibition activity against aromatase (CYP19). Indeed, among the isolated compounds, 182 and 184 displayed moderate DPPH free radical scavenging activities with IC50 values of 23.4 and 16.4 μM, respectively. In addition, 182 and 184 showed significant inhibitory activity against the formation of the superoxide anion radical with IC50 values of 52.6 and 4.3 μM, respectively. Moreover, none of the isolated compounds suppressed superoxide anion generation induced by 12-O-tetradecanoylphorbol-13-acetate (TPA) in differentiated HL-60 human promyelocytic leukemia cells. Furthermore, compounds 4, 182, and 183 displayed potent ORAC antioxidant properties with ORAC units in the range of 10.8–14.4 ORAC units.110
3.8. Fungi of the order Helotiales
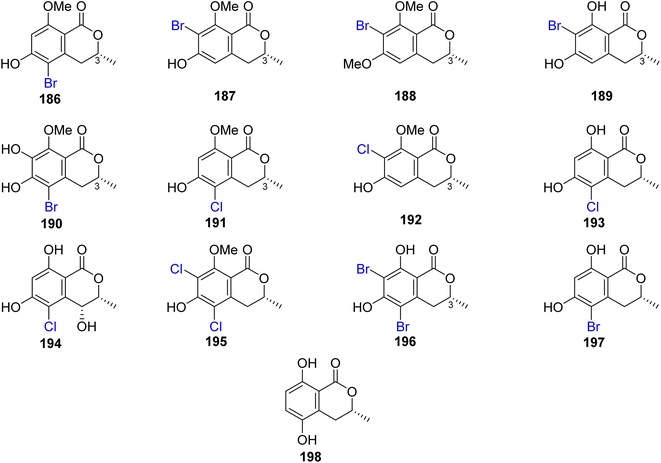
3.8.1.1. Fungi of the genus Lachnum. The chemical examination of the endophytic fungus Lachnum palmae isolated from Przewalskia tangutica Maxim. fresh tissue, collected Linzhou, China, led to the isolation of the previously mentioned derivatives, including trans-4-hydroxymellein (7), mullein (25), cis-4-hydroxymellein (26), (R)-6-hydroxymellein (77), and (3R)-6-methoxymellein (135), along with seven previously unreported palmaerones A–G (186–192), together with six previously described compounds, namely, (R)-5-cholro-6-hydroxymellein (193), (3R,4R)-5-cholro-4,6-dihydroxymellein (194), palmaerins A (195), B (196), D (197), and (R)-5-hydroxymellein (198) (Fig. 20). Compounds 186–192 were examined for their antimicrobial properties against three fungal strains, namely, C. neoformans, Penicillium sp., and C. albicans and two bacterial strains B. subilis and S. aureus (using the broth microdilution method, and amphotericin B and kanamycin as a positive controls). Generally, the brominated derivatives were found to be more potent as antimicrobial agents than the chlorinated congeners. Among them, 190 displayed the most powerful antimicrobial effect against the examined fungal and bacterial strains with MIC value ranging from 10 to 55 μg mL−1. In addition, 186–192 were tested for their anti-inflammatory activity (SMT; 2-methyl-2-thiopseudourea sulphate was used as a positive control). Compounds 186 and 190 exhibited mild no inhibitory activity with IC50 values of 26.3 and 38.7 μM, respectively. Further, only 190 displayed a weak cytotoxicity toward HepG2 with an IC50 value of 42.8 μM.111
3.9. Fungi of the order Hypocreales
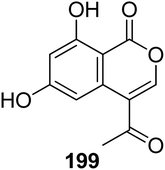
3.9.1.1. Fungi of the genus Bionectria. A previously described isocoumarin derivative AGI-7 (199) (Fig. 21) was isolated from the fungus Bionectria sp. (MSX 47401), isolated from leaf litter and leaves collected from a humid mountain forest. Compound 199 displayed no cytotoxicity against NCI-H460, MCF-7, and SF-268 cancer cell lines.112
3.9.1.2. Fungi of the genus Clonostachys. Chemical examination of the EtOAc extract of the marine-derived fungus Clonostachys sp. (AP4.1) isolated from the marine sponge Axinella polypoides, collected at a depth of 30 m from İlyosta–Ayvalık, Turkey, led to the isolation of two previously mentioned isocoumarin derivatives, dichlorodiaportin (89) and peniisocoumarin D (125). Compound 125 displayed no cytotoxicity against the L5178Y cancer cell line.113
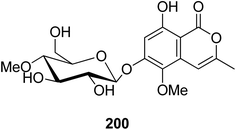
3.9.2.1. Fungi of the genus Conoideocrella. A previously unreported isocoumarin glycoside (200) (Fig. 22) was isolated from the organic extract of the insect pathogenic fungus Conoideocrella tenuis BCC 18627, obtained from Hemiptera scale, collected in Nakhon Nayok Province from the national park of Khao Yai. In addition, as a result of its limited abundance, 200 was not subjected to any relevant biological activity test.114
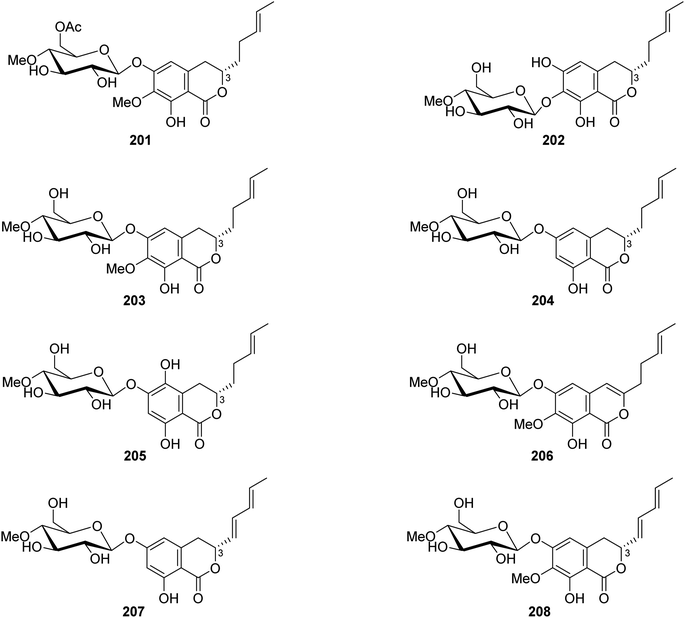
3.9.2.2. Fungi of the genus Metarhizium. The chemical exploration of the soil-derived fungus Metarhizium anisopliae (No. DTH12-10), isolated from a soil sample collected in China from the Province of Hunan, led to the isolation of eight previously undescribed isocoumarin glycosides (Fig. 23), namely, (3S)-6-O-(4′-O-methyl-6′-acetyl-β-D-glucopyranoside)-7-O-methyl-8-hydroxyl-3-[(3E)penta-3-enyl]-3,4-dihydroisocoumarin (201), (3S)-7-O-(4′-O-methyl-β-D-glucopyranoside)-6,8-dihydroxyl-3-[(3E)penta-3-enyl]-3,4-dihydroisocoumarin (202), (3S)-6-O-(4′-O-methyl-β-D-glucopyranoside)-7-O-methyl-8-hydroxyl-3-[(3E)-pent-3-enyl]-3,4-dihydroisocoumarin (203), (3S)-6-O-(4′-O-methyl-β-D-glucopyranoside)-8-hydroxyl-3-[(3E)-pent-3-enyl]-3,4-dihydroisocoumarin (204), (3S)-6-O-(4′-O-methyl-β-D-glucopyranoside)-5,8-dihydroxyl-3-[(3E)-pent-3-enyl]-3,4-dihydroisocoumarin (205), 6-O-(4′-O-methyl-β-D-glucopyranoside)-7-O-methyl-8-hydroxyl-3-[(3E)-penta-3-enyl]-isocoumarin (206), (3R)-6-O-(4′-O-methyl-β-D-glucopyranoside)-8-hydroxyl-3-[(1E,3E)-penta-1,3-dienyl]-dihydroisocoumarin (207), and (3R)-6-O-(4′-O-methyl-β-D-glucopyranoside)-7-O-methyl-8-hydroxyl-3-[(1E,3E)-penta-1,3-dienyl]-dihydroisocoumarin (208). Compounds 201–208 were tested for their biofilm and virulence factor secretion inhibition activities of toward P. aeruginosa strain PAOA (clinical isolates). Among them, 201 displayed antibacterial effect in comparison with (Z)-4-bromo-5-(bromomethylene)-2(5H)-furanone (BF as positive control).115
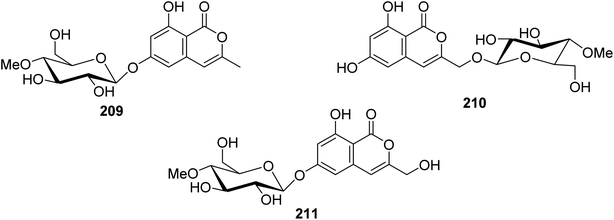
3.9.2.3. Fungi of the genus Torrubiella. Two previously described isocoumarin derivatives, including 6,8-dihydroxy-3-methylisocoumarin (27) and 6,8-dihydroxy-3-hydroxymethylisocoumarin (60), along with three previously undescribed isocoumarin glucosides 209–211 (Fig. 24), were obtained from the fungus Torrubiella tenuis BCC 12732, isolated from the scale of Homoptera, collected from the province of Chiang Mai, Thailand. Compounds 27, 60, and 209–211 were tested for their antimalarial, anti-mycobacterium, antiviral (against Herpes simplex virus-1), and cytotoxic activities against KB, MCF-7, NCI-H187, and Vero cells. Though all the isolated compounds displayed no biological activity, 60 displayed mild growth inhibitory activity against Mycobacterium tuberculosis H37Ra and Herpes simplex virus-1 with IC50 values of 25 and 50 μg mL−1, respectively.116
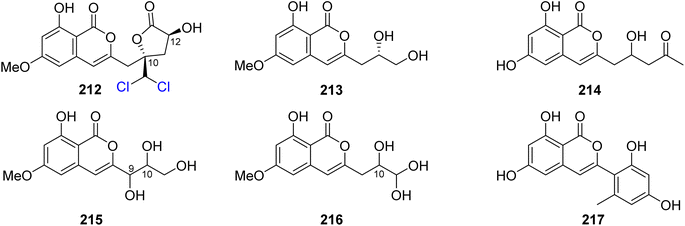
3.9.3.1. Fungi of the genus Trichoderma. Dichlorodiaportinolide (212), a previously undescribed derivative along with three previously reported congeners, including diaportinol (213), dichlorodiaportin (89), and diaporthin (121) (Fig. 25), was obtained from the endophytic fungus Trichoderma sp., 09, isolated from Myoporum bontioides A. Gray roots, collected from Guangdong Province, China. Compounds 121 and 213 were not tested for any relevant biological activity due to their minute available quantities; however, 89 and 212 were tested for their antifungal activity against a list of phytopathogenic fungi, including C. musae, F. graminearum, P. italic, and R. solani (using the broth dilution method and carbendazim as a positive control). Compound 212 displayed strong to mild antifungal properties against R. solani and C. musae, with MIC values of 6.25 and 25 μg mL−1, respectively. In addition, 89 displayed weak antifungal activity against R. solani and C. musae, with MIC values of 150 μg mL−1.117 A chemical investigation of the marine-derived fungus T. sp., HPQJ-34, isolated from the marine sponge Hymeniacidon perleve, collected from the Island of Dongji, China, led to the isolation of two previously described derivatives, including citreoisocoumarin (214) (Fig. 25) and (+)-6-O-methylcitreoisocoumarin (132). Compounds 132 and 214 were not subjected to any relevant biological activity test.118 In addition, two previously unreported isocoumarin derivatives (215) and (216) (Fig. 25), were recovered from the EtOAc extract of the endophyte fungus T. harzianum isolated from the fresh leaves of F. elastic. The antimicrobial activity of 215–216 was tested against B. subtilis, C. albicans, E. coli, P. aeruginosa, and S. aureus (using the well diffusion method, and chloramphenicol and fluconazole as positive controls). Compounds 215–216 showed moderate antibacterial effect against E. coli with MIC values of 32 μg mL−1.119 A chemical exploration of the marine-derived fungus T. citrinoviride A-WH-20-3, isolated from the inner tissue marine alga Laurencia okamurai, collected from the coast of Weihai, China, led to the isolation of a previously undescribed trichophenol A (217) (Fig. 25). Compound 217 displayed anti-microalgal activity against the marine phytoplankton Heterosigma akashiwo, Prorocentrum donghaiense, Karlodinium veneficum, and Chattonella marina with IC50 values of 9.1, 5.9, 20, and 4.4 μg mL−1, respectively. Furthermore, it displayed antibacterial activity against the marine-derived pathogenic bacterial strains including P. citrea, V. splendidus, V. harveyi, V. anguillarum, and V. parahaemolyticus with an inhibition zone of 21, 7.5, 7.0, 8.0, and 7.0 mm, respectively, at a concentration of 50 μg per disk.120
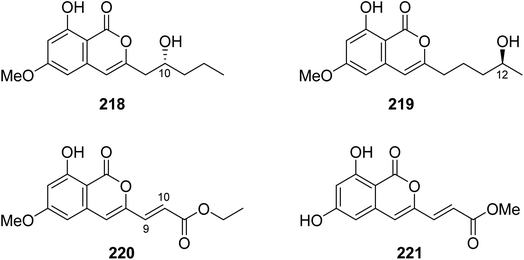
3.9.4.1. Fungi of the genus Fusarium. Four previously unreported isocoumarin derivatives, including fusarimarins A–C (218–220), along with a previously described aspergisocoumrin A (221) (Fig. 26), were isolated from the mangrove-derived fungus Fusarium sp., 2ST2, isolated from the leaves of Kandelia candel, collected from the South China Sea, China. Compounds 219–220 were tested for their cytotoxicity against A549, HELA, KYSE150, PC-3, and MDA-B-435 tumor cell lines (using the MTT method and DDP as a positive control). Only 221 displayed significant cytotoxicity toward A549 and MDA-B-435, with IC50 values of 6.2 and 2.8 μM, respectively. In addition, 220 showed weak cytotoxicity against MDA-B-435, with an IC50 value of 30.5 μM.121
3.9.4.2. Fungi of the genus Nectria. 3,4-dimethyl-6,8-dihydroxyisocoumarin (178), a previously mentioned derivative, along with two previously undescribed congeners, namely, nectriapyrones A–B (222–223) (Fig. 27), was obtained from the endophytic fungus Nectria pseudotrichia 120-1NP, isolated from Gliricidia sepium stem, collected from the forest of Wanagama, Indonesia. Compounds 178 and 222–223 were tested for their cytotoxicity, phytotoxicity, and antimicrobial activity, but they showed no activity.122
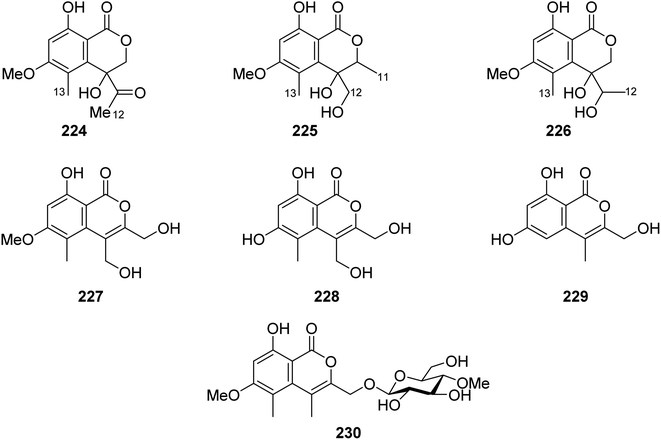
3.9.5.1. Fungi of the genus Acremonium. Phytochemical examination of the mangrove-derived fungus Acremonium sp., PSU-MA70, isolated from R. apiculata branch, collected from the Province of Satun, Thailand, afforded seven previously unreported isocoumarin derivatives, namely, acremonones B–H (224–230) (Fig. 28). Indeed, due to the lack of isolated quantities, only 227, was examined for its antifungal activity against C. albicans NCPF3153 and C. neoformans ATCC90113, but it showed no activity.123
3.10. Fungi of the order Hysteriales
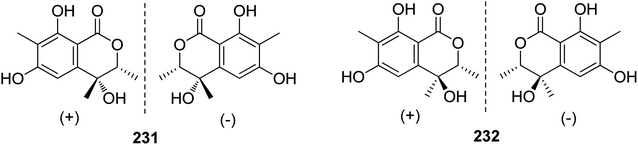
3.10.1.1. Fungi of the genus Rhytidhysteron. Two pairs of previously unreported enantiomers (±) 231 and (±) 232 (Fig. 29) were isolated from the fungus Rhytidhysteron sp., BZM-9. Compounds 231–232 displayed no cytotoxicity.124
3.11. Fungi of the order Mucorales
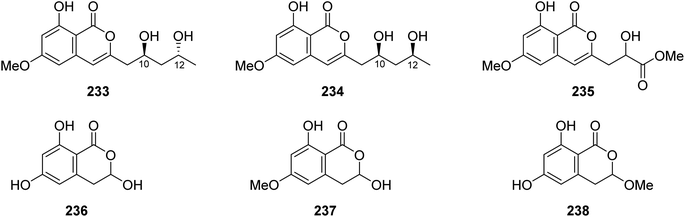
3.11.1.1. Fungi of the genus Mucor. Chemical inspection of the soil-derived fungus Mucor sp., (No. XJ07027-5), obtained from a mountainous soil collected from the region of Sinkiang Uyghur, China, led to the discovery of three previously unreported mucorisocoumarins A–C (233–235), along with seven previously described isocoumarin derivatives including 236–238 (Fig. 30), dichlorodiaportin (89), (+)-6-methylcitreoisocoumarin (132), O-demethyldiaporthin (152), and diaportinol (213). Compounds 89, 152, 213, and 233–23, were tested for their toxicity using a zebrafish model. Only 235 displayed toxicity toward the zebrafish embryos, while all the other compounds showed no toxicity.125
3.12. Fungi of the order Onygenales
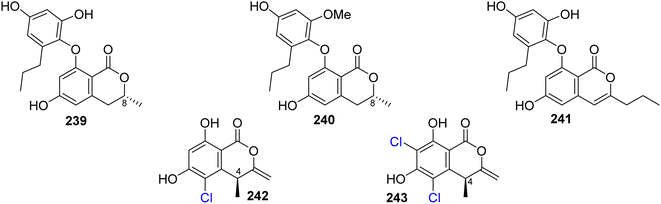
3.12.1.1. Fungi of the genus Spiromastix. Three previously unreported isocoumarin derivatives, namely, spiromastols G–I (239–241), (Fig. 31), were obtained from the EtOAc extract of the deep sea-derived fungus Spiromastix sp. MCCC 3A00308, isolated from the sediment of the deep ocean, collected from the South Atlantic Ocean. Compounds 239–240 were tested for their antibacterial activity against X. vesicatoria ATCC 11633, P. lachrymans ATCC11921, A. tumefaciens ATCC11158, R. solanacearum ATCC11696, B. thuringensis ATCC 10792, S. aureus ATCC 25923, and B. subtilis CMCC 63501 (using chloramphenicol as a positive control). While 239–240 displayed no effect against the examined bacterial strains, 241 showed mild antibacterial activity with MIC values ranging from 8 to 64 μg mL−1.126 A chemical examination of the marine-derived fungus S. sp. MCCC 3A00308, isolated from the sediment collected from the South Atlantic Ocean at a depth of 2869 m, led to the isolation of two previously undescribed chlorinated derivatives namely, spiromastimelleins A–B (242–243) (Fig. 31). Compounds 242–243 were evaluated for their antibacterial activity against the Gram-positive bacteria (S. aureus ATCC 25923, B. thuringiensis ATCC 10792, and B. subtilis CMCC 63501) and the Gram-negative bacterium (E. coli ATCC 25922) (using chloramphenicol as a positive control). While both compounds showed no activity against E. coli ATCC 25922, they displayed significant effect against S. aureus ATCC 25923, B. thuringiensis ATCC 10792, and B. subtilis CMCC 63501, with MIC values ranging from 4 to 32 μg mL−1.127
3.13. Fungi of the order Pezizales
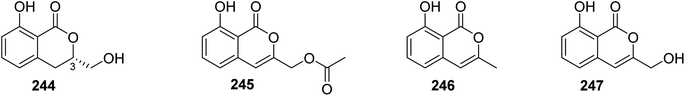
3.13.1.1. Fungi of the genus Sarcosomataceae. Three previously mentioned derivatives, including methoxymellein (6), trans-4-hydroxymellein (7), and mullein (25), along with two previously unreported analogues, namely, (3R)-3-hydroxymethyl-8-hydroxyl-3,4-dihydroisoucoumarin (244) and 3-acetoxyl-8-hydroxyl-isocoumarin (245), beside two previously reported congeners, 3-methyl-8-hydroxyisocoumarin (246) and 8-hydroxy-3-(hydroxymethyl)-lH-2-benzopyran-1-one (247) (Fig. 32), were isolated from the EtOAc extract of the endophytic fungus Sarcosomataceae sp., NO. 49-14-2-1, isolated from E. nepalense, collected from the province of Panzhihua, Sichuan, China. The isolated compounds were not tested for any relevant biological activity.128
3.14. Fungi of the order Pleosporales
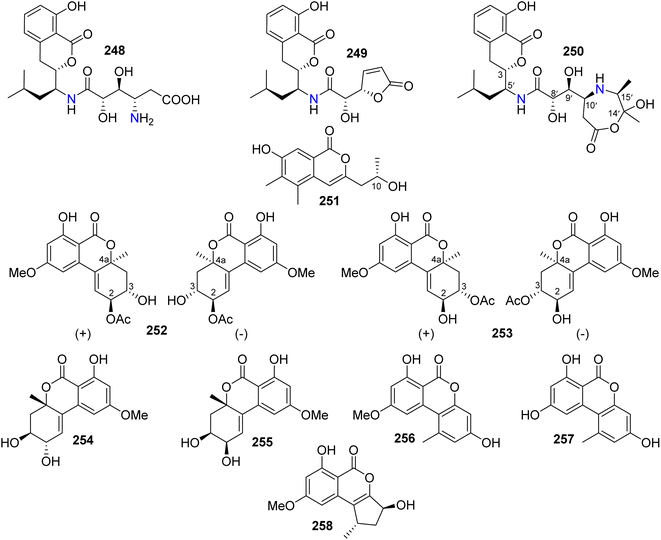
3.14.1.1. Fungi of the genus Alternaria. Three isocoumarin derivatives including two previously reported ones, namely, AI-77-B (248) and AI-77-F (249), alongwith a previously undescribed derivative Sg17-1-4 (250) (Fig. 33), were recorded from the marine-derived fungus Alternaria tenuis Sg17-1, isolated from an unidentified marine alga, collected from the island of Zhoushan, China. Compounds 248–250 were tested for their cytotoxicity against A375-S2 and HeLa cancer cell lines. Compound 248 displayed the strongest cytotoxicity followed by 250 with IC50 values of (0.1 and 0.02 mM) and (0.3 and 0.05 mM), respectively. In addition, 249 exhibited weak cytotoxicity against HeLa cells only with an IC50 value of 0.4 mM.129 The chemical examination of the endophytic fungus A. alternata, isolated from Camellia sinensis branches, collected from the Jiangsu Province, China, afforded three previously unreported derivatives, namely, (+)-(10R)-7-hydroxy-3-(2-hydroxy-propyl)-5,6-dimethyl-isochromen-1-one (251), altenuene-2-acetoxy ester (252), and altenuene-3-acetoxy ester (253), together with five previously described analogues including altenuene (254), 5′-epialtenuene (255), alternariol 9-methyl ether (256), alternariol (257), and phialophoriol (258) (Fig. 33). Compounds 251–258 were tested for their antimicrobial activity against S. aureus, B. subtilis, E. coli, C. albicans, and T. rubrum (using initial agar diffusion assay and as a positive control Penicillin and Fluconazole have been used). Among them, 257 exhibited the strongest antibacterial effect against B. subtilis with an MIC80 of 8.6 μg mL−1. In addition, 251–254 and 258 displayed weak to mild inhibition against the examined pathogenic microorganisms. Moreover, only 256 exhibited moderate cytotoxicity against the U2OS cancer cell line with an IC50 value of 28.3 μM.130
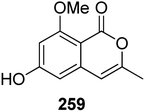
3.14.1.2. Fungi of the genus Cochliobolus. The phytochemical study of the marine-derived fungus Cochliobolus lunatus (TA26-46), isolated from the inner part tissue of zoanthid, collected from the coral reef of Weizhou, South China Sea, led to the isolation of two previously mentioned compounds 3-methyl-6,8-dihydroxyisocoumarin (5) and O-demethyldiaporthin (152), together with a previously described derivative, namely, 6-hydroxy-8-methoxy-3-methylisocoumarin (259) (Fig. 34). Compounds 5, 152, and 259 were tested for their antibacterial effect against S. aureus (ATCC 33591 and ATCC 43300), and for their inhibitory activity against acetylcholinesterase and topoisomerase I (Topo I) enzymes. They displayed neither antibacterial nor enzyme inhibition activities.131
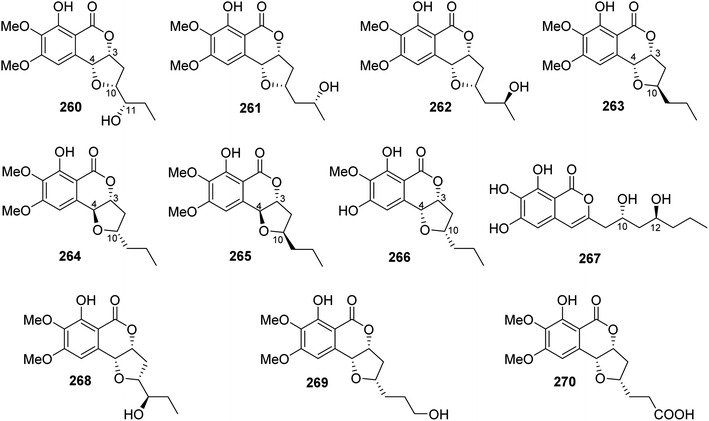
3.14.1.3. Fungi of the genus Exserohilum. The previously mentioned monocerin (4), together with a previously unreported derivative 11(R)-hydroxymonocerin (260), alongwith the previously described compound 12(R)-hydroxymonocerin (261) (Fig. 35), were isolated from the EtOAc extract of the endophytic fungus Exserohilum rostratum, derived from Stemona sp. leaves and roots, collected from the Province of Ayutthaya, Thailand. Compounds 4 and 260 displayed antimalarial activity against P. falciparum with IC50 values of 0.68 and 7.7 μM, respectively. In addition, they were evaluated for their cytotoxicity against BT474, CHAGO, Hep-G2, KATO-3, and SW620, but they showed no activity at a concentration of 20 μg mL−1.132 A chemical investigation of the endophytic fungus E. sp., isolated from A. truncatum Bunge leaves, collected from the mountain of Dongling, China, afforded three previously mentioned derivatives, namely, monocerin (4), 11(R)-hydroxymonocerin (260), and 12(R)-hydroxymonocerin (261), along with the previously reported 12(S)-hydroxymonocerin (262), together with five previously unreported isocoumarin derivatives, named exserolides A (263), B (264), C (265), D (266), and F (267) (Fig. 35). Compounds 4 and 260–267 were tested for their antibacterial and antifungal effects against E. coli (CGMCC 1.2340), B. subtilis (ATCC 6633), S. pneumoniae (CGMCC 1.1692), and S. aureus (CGMCC 1.2465), as well as fungus F. oxysporum (CGMCC 3.2830), an example of the plant pathogenic fungi (using ampicillin, gentamicin, and amphotericin B as positive controls, respectively). Compounds 261 and 265 showed moderate antifungal activity against F. oxysporum (CGMCC 3.2830) with MIC values of 20 μg mL−1. Meanwhile, 267 exhibited antibacterial effect against the tested bacterial strains with MIC values ranging from 5 to 20 μg mL−1.133 In addition, the previously mentioned derivatives including monocerin (4), 12(R)-hydroxymonocerin (261), 12(S)-hydroxymonocerin (262), exserolides B (264), and C (265), together with the previously described congeners, 11-hydroxymonocerin (268), exserolide I (269), and exserolide J (270) (Fig. 35), were recorded from the EtOAc extract of the marine-derived fungus E. sp. (CHNSCLM-0008), isolated from the zoanthid P. haddoni inner part tissues, collected from Weizhou coral reefs, South China Sea. Compounds 4 and 262 showed antimalarial activity with IC50 values of 1.13 and 11.7 μM, respectively; however 270 was found to be inactive.134
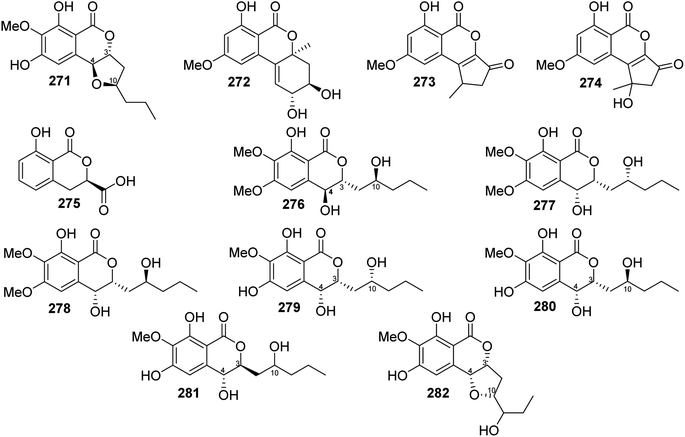
3.14.1.4. Fungi of the genus Setosphaeria. A phytochemical investigation of the marine-derived fungus Setosphaeria sp. SCSIO41009, isolated from the marine sponge Callyspongia sp., collected from the Guangdong Province, China, afforded seven previously mentioned derivatives, including 11(R)-hydroxymonocerin (260), 12(R)-hydroxymonocerin (261) exserolides B (264), C (265), D (266), 11-hydroxymonocerin (268), exserolides I (269), and J (270), together with a previously unreported exserolide K (271) (Fig. 36). All the isolated metabolites were tested for their antibacterial activity against S. aureus, antifungal activity toward F. oxysporum, C. gloeosporioides, C. acutatum, C. asianum, and P. oryza. Intriguingly, none of these compounds showed either antibacterial or antifungal activities against the examined microbial strains. In addition, the antioxidant activities of the purified compounds were evaluated. Only 266 exhibited moderate anti-radical activity with an IC50 value of 38 μM. Furthermore, these compounds were tested for their antiviral and MptpB inhibitory activities, but they showed no activity.135 Two previously mentioned derivatives, alternariol 9-methyl ether (256) and alternariol (257), along with four previously reported compounds, including isoaltenuene (272), 1-deoxyrubralactone (273), rubralactone (274), and phomasatin (275), together with a previously unreported setosphacohol A (276) (Fig. 36), were obtained from the fungus S. sp. (strain LGWB-2), isolated from H. axyridis, collected from the Hebei Province in Baoding, China. Compounds 256–257 and 272–276 were evaluated for their cytotoxicity against HeLa, A549, MCF-7, MGC-803, Huh-7, and H1975 cancer cell lines (using the MTT method and cisplatin as a positive control). While 272 and 274–276 displayed no cytotoxic activity against the tested cancer cell lines, 256–257 and 273 showed mild cytotoxicity with IC50 values ranging from 23.04 to 96.91 μg mL−1.136 A chemical examination of the S. rostrate LGWB-10, obtained from H. axyridis, collected from the Hebei Province, China, afforded two previously mentioned derivatives, 12(R)-hydroxymonocerin (261) and 12(S)-hydroxymonocerin (262), along with two previously described compounds, including (3R,4R)-4,8-dihydroxy-3-((R)-2-hydroxypentyl)-6,7-dimethoxyisochroman-1-one (277) and (3R,4R)-4,8-dihydroxy-3-((R)-2-hydroxypentyl)-6,7-dimethoxyisochroman-1-one (278) together with four previously undescribed natural products namely, setosphlides A–D (279–282) (Fig. 36). Compounds 279–282 displayed no cytotoxicity against MCF-7, MGC-803, HeLa, and Huh-7 with IC50 values >200 μM.137
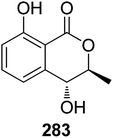
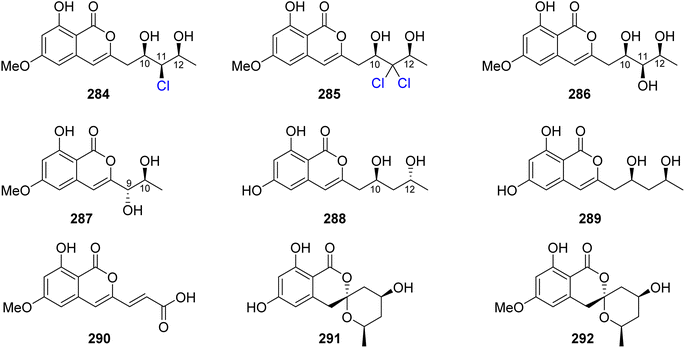
3.14.2.1. Fungi of the genus Epicoccum. A previously mentioned (R)-5-hydroxymellein (198), along with the previously reported (−)-(3R,4S)-4-hydroxymellein (283) (Fig. 37), were obtained from the EtOAc extract of the brown alga-derived fungus Epicoccum sp., isolated from Fucus vesiculous, collected from the coast of the North Sea, Germany. Compounds 198 and 283 were evaluated for their antioxidant activity (using the DPPH-radical scavenging and TBARS assays and BHT as a positive control). They displayed inhibitory activity in both the assays with an inhibition ratio of 28.4 and 9.6 at a concentration of 50.0 μg mL−1, and 30.4 and 18.4 at a concentration of 37.0 μg mL−1.138
3.14.2.2. Fungi of the genus Peyronellaea. The chemical study of the marine-derived fungus Peyronellaea glomerata XSB-01-15, obtained from the marine sponge Amphimedon sp., collected from the Island of Xisha, South China Sea, afforded five previously undescribed peyroisocoumarins A (284), B (285), C (286), D (287), and isocitreoisocoumarinol (288), along with four previously reported derivatives including, citreoisocoumarinol (289), LL-Z 1640-7 (290), demethylcitreoviranol (291), and citreoviranol (292) (Fig. 38), together with eight previously mentioned (+)-6-O-methylcitreoisocoumarin (132), O-demethyldiaporthin (152), diaportinol (213), citreoisocoumarin (214), mucorisocoumarins A (233), B (234), alternariol-9-methyl ether (256), and alternariol (257). All the isolated compounds were tested for their antibacterial activities against B. thuringiensis, S. aureus, P. lachrymans, X. vesicatoria, E. coli, A. tumefaciens, and R. solanacearum. Among the isolated metabolites, 257 displayed mild antibacterial activity toward S. aureus, A. tumefaciens, and R. solanacearum, 256 showed weak activity against A. tumefaciens, 152 and 299 also exhibited weak activity against R. solanacearum. Furthermore, all the isolated compounds showed ARE luciferase activity that was 0.88–2.95 folds more than the negative control (DMSO) when compared with the positive control tBHQ with an induction value 4.29 folds more than the solvent (DMSO).139
3.14.2.3. Fungi of the genus Phoma. A chemical study of the marine sponge-derived fungus Phoma sp. 135, isolated from the sponge Ectyplasia perox, collected from the Reef of Lauro Club, Dominica, led to the isolation of one previously mentioned derivative, (3R)-6-methoxy-7-chloromellein (136), and a previously unreported chlorinated metabolite, (3R,4S)-4-hydroxy-6-methoxy-7-chloromellein (293) (Fig. 39). Compounds 136 and 293 were not tested for any relevant biological assay.140 A previously mentioned phomasatin (275) was obtained from the EtOAc extract of the plant-derived fungus P. sp. YN02-P-3, isolated from the Sumbaviopsis J. J. Smith, collected from Yunnan, China. Compound 275 was evaluated for its cytotoxicity against HL-60, Molm 13, and PC-3 cancer cell lines (using 5-fluorouracil as a positive control), and it showed no activity.141 In addition, the chemical investigation of the marine-derived fungus P. sp. (TA07-1), isolated from the fresh tissues of the gorgonian Dichotella gemmacea (GX-WZ-2008003-4), collected from the coral reef of Weizhou, South China Sea, afforded four previously mentioned metabolites, including desmethyldiaportinol (94), diaportinol (213), citreoisocoumarin (214), and citreoisocoumarinol (289). The isolated compounds were tested for their antibacterial activity against S. albus, S. aureus, E. coli, V. parahaemolyticus, and V. anguillarum as well as for their lethal activity toward the brine shrimps, but none of them showed antibacterial or any lethal effect.142
3.14.3.1. Fungi of the genus Microsphaeropsis. A previously mentioned isocoumarin derivative 26 was isolated from the marine sponge derived fungus Microsphaeropsis sp. (strain number H5-50), isolated from the marine sponge Myxilla incrustans Jonnston (1842), collected around Helgoland, Germany. Compound 26 showed no antiplasmodial activity.143
3.14.3.2. Fungi of the genus Paraphaeosphaeria. A chemical study of the plant-derived fungus Paraphaeosphaeria sporulosa, isolated from Actinidia chinensis Planch stem, collected from the Sichuan Province, Cangxi, led to the isolation of two previously mentioned derivatives, acremonones E (227) and F (228) along with two previously undescribed isocoumarins, paraphaeones E (294) and F (295) (Fig. 40). Compounds 227–228 and 294–295 were evaluated for their antibacterial effect against P. syringae pv. Actinidiae (using streptomycin as a positive control). While 228 showed no activity, 227, 294, and 295 displayed moderate antibacterial activity with MIC values of 50, 25, and 50 μg mL−1, respectively.144
3.14.4.1. Fungi of the genus Leptosphaeria. Six previously mentioned derivatives, clearanol I (64), dothideomynone A (65), banksialactone A (66), 3,4-dimethyl-6,8-dihydroxyisocoumarin (178), acremonone F (228), and acremonone G (229), along with a previously unreported clearanol J (296) together with a previously described (R)-4,8-dihydroxy-6-methoxy-4,5-dimethyl-3-methyleneisochromen-1-one (297) (Fig. 41), were obtained from the deep sea-derived fungus Leptosphaeria sp. SCSIO 41005, isolated from the sediments of the Indian Ocean collected at a depth of 3614 m. All the obtained compounds were tested for their cytotoxicity and antiviral properties against K562, MCF-7, and SGC7901 cancer cell lines, and influenza A virus subtype H3N2, EV71, and HIV viruses. None of them showed either cytotoxicity or antiviral activities.145
3.14.4.2. Fungi of the genus Leptosphaena. A chemical exploration of the plant-derived fungus Leptosphaena maculans, isolated from Osmanthus fragrans, afforded three previously mentioned derivatives, monocerin (4), 12(R) hydroxymonocerin (261), and exserolide I (269) along with the previously unreported maculansline C (298) (Fig. 42). Compounds 4, 261, 269, and 298 were evaluated for their α-glucosidase inhibition effect, but none of them displayed any inhibitory activity.146
3.14.5.1. Fungi of the genus Paraphoma. Paraphamides A–C (299–301), three previously unreported isocoumarin derivatives, featuring a rare skeleton coupled with phenylethylamine moiety, along with another six previously unreported derivatives, namely, parapholactone (302), 7-hydroxyoospolactone (303), 7-methoxyoospolactone (304), 7-methoxy-9-hydroxyoospolactone (305), 10-acetoxy-9-hydroxyoospolactone 6-dehydroxysescandelin (306), and 6-dehydroxysescandelin (307), together with five previously described metabolites, including oospolactone (308), 8-O-methyloospolactone (309), 10-hydroxyoospolactone (310), 9,10-dihydroxyoospolactone (311), and oospoglycol (312) (Fig. 43), were discovered from the marine-derived fungus Paraphoma sp. CUGBMF180003, obtained from a mud, collected from the Shenzhen intertidal zones, China. All the isolated compounds were evaluated for their antibacterial activities toward C. albicans ATCC 10231 and S. aureus ATCC 25923. Among them, 302–303 displayed moderate antibacterial activity against S. aureus ATCC 25923 with MIC values of 12.5 μg mL−1.147
3.15. Fungi of the order Xylariales
3.15.1.1. Fungi of the genus Arthrinium. The chemical study of the marine-derived fungus Arthrinium sacchari, isolated from an identified marine sponge, collected from Atamishi coast, Japan, afforded the isolation of one previously unreported decarboxyhydroxycitrinone (313) (Fig. 44). Compound 313 exhibited weak cytotoxicity toward HUVECs and HUAECs cancer cell lines with IC50 values of 7.6 and 17.4 μM, respectively.148
3.15.2.1. Fungi of the genus Hypoxylon. Two previously mentioned (−)-(3R,4R)-cis-4-hydroxy-5-methylmellein (24) and phomasatin (275), along with two previously unreported (3R,4R)-4,8-dihydroxy-5-(hydroxymethyl)-3-methylisochroman-1-one (314) and (3R,4S)-4,8-dihydroxy-5-(hydroxymethyl)-3-methylisochroman-1-one (315), together with two previously described 3,5-dimethyl-8-hydroxy-7-methoxy-3,4-dihydroisocoumarin (316) and 3,5-dimethyl-8-methoxy-3,4-dihydroisocoumarin (317) (Fig. 45), were obtained from the EtOAc extract of the soil derived fungus Hypoxylon sp., isolated from a soil sample collected from the Province Heilongjiang Province, the mountain of Da Hinggan, China. It is worth mentioning that none of the obtained compounds was tested for any relevant biological activity.149
3.15.3.1. Fungi of the genus Microdochium. A chemical identification of the plant-derived fungus Microdochium bolleyi, isolated from Fagonia cretica, collected in Gomera, afforded four previously mentioned derivatives including, monocerin (4), 12(R)-hydroxymonocerin (261), 12(S) hydroxymonocerin (262), and (3R,4R)-4,8-dihydroxy-3-((R)-2-hydroxypentyl)-6,7-dimethoxyisochroman-1-one (277). The isolated metabolites were examined for their inhibitory activity against the fungal strain M. Violaceum, the bacterial strains E. coli and B. Megaterium, and against the algae C. fusca, (using the agar diffusion test and as a positive controls penicillin, tetracycline, nystatin, and actidione have been used). While 261 displayed anti-algal and antifungal activities, it showed no antibacterial activity. However, 4, 262, and 277 exhibited antifungal, antibacterial, and algal activity against the examined organisms.150
3.15.4.1. Fungi of the genus Pestalotiopsis. Two previously mentioned aspergillumarins B (105) and A (106), along with the previously unreported pestalotiorin (318) (Fig. 46), were obtained from the marine seagrass-derived fungus Pestalotiopsis sp. PSU-ES194, isolated from E. acoroides leaves, collected in Thailand. Compound 318 was examined for its anti-mycobacterial, antimalarial, and cytotoxic activities, but it showed no activity.151 In addition, two previously mentioned (−)-pestalactone B (20) and (+)-pestalactone C (21), along with three previously unreported derivatives, including pestalactone A (319), pestapyrones D (320), and E (321) (Fig. 46), were isolated from the EtOAc extract of the endophytic fungus Pestalotiopsis sp., obtained from P. frasery leaves, collected in Jiangsu, Nanjing, China. All the isolated compounds were tested for their antimicrobial effect against S. aureus ATCC 25923, B. subtilis ATCC 6633, E. coli ATCC 25922, P. aeruginosa ATCC 9027, and C. glabrata ATCC 90030. Among them, only 21 showed antifungal activity against C. glabrata ATCC 90030 with an MIC50 value of 3.49 μg mL−1.152 The chemical processing of the marine sponge-derived fungus P. heterocornis isolated from the marine sponge P. fusca collected in China from the Island of Xisha afforded two previously undescribed pestaloisocoumarins A–B (322–323), along with the previously reported gamahorin (324) (Fig. 46). Compounds 322–324 showed no cytotoxicity against BGC-823, H460, PC-3, and SMMC-7721 cancer cell lines. However, they exhibited antibacterial activity against S. aureus and B. subtilis with MIC values ranging from 25 to 100 μg mL−1. In addition, they were tested for their antifungal activity against C. albicans, C. parapsilosis, and C. neoformans, where 322 exhibited weak antifungal activity with MIC values of 100 μg mL−1. Moreover, 323 was found to be inactive against C. albicans and C. parapsilosis but exhibited weak effect against C. neoformans with an MIC value of 100 μg mL−1. Furthermore, 324 displayed weak antifungal ability toward C. parapsilosis and C. neoformans with MIC values of 100 μg mL−1 and was found to be inactive against C. albicans.153 A previously unreported pestalotiopisorin B (325), alongwith the co-isolated previously reported (R)-(−)-mellein methyl ether (326) (Fig. 46), were recorded from the mangrove-derived fungus Pestalotiopsis sp. HHL101, isolated from Rhizophora stylosa branches, collected from the Island of Hainan, China. Compound 325 was examined for its CN-inhibition activity, but it showed no activity. In addition, it displayed mild antibacterial effect against E. coli and P. aeruginosa. Moreover, it showed no cytotoxicity against HeLa, A549, and HepG2 cancer cell lines.154 A previously mentioned gamahorin (324), along with the previously undescribed microsporaline D (327) (Fig. 46), were reported from the marine-derived fungus P. microspora SC3082, isolated from Scaevola taccada (Gaertn.) Roxb leaves, collected from the island of Yongxing, China. Compound 327 was tested for its antifungal activity against C. albicans ATCC 10231 (using fluconazole as a positive control) but it showed no activity.155
3.15.5.1. Fungi of the genus Ascotricha. Two previously undescribed isocoumarin derivatives, namely, ascotrichols A–B (328–329), along with the two previously reported 3-(2-hydroxypropyl)-8-hydroxy-3,4-dihydroisocoumarin (330) and (R)-orthosporin (331) (Fig. 47), were reported from the ethanol extract of the marine mud-derived fungus Ascotricha sp. ZJ-M-5, isolated from the sea mud, collected from an unidentified location. It is worth mentioning that none of the isolated metabolites were subjected for any relevant biological activity test.156
3.15.5.2. Fungi of the genus Biscogniauxia. The previously described 6-methoxy-5-methyl mellein (332) (Fig. 48) was reported from the marine-derived fungus Biscogniauxia mediterranea strain LF657, isolated from the sediments collected in the Eastern Mediterranean Sea at a depth of 2800 m. Compound 332 displayed weak antifungal effect against the phytopathogenic fungi Septoria tritici and Trichophyton rubrum.157 In addition, a chemical examination of the plant endophytic fungus B. capnodes isolated from Averrhoa carambola L fruits, collected from Kandey, led to the isolation of the previously mentioned (−)-3,4-dihydro-8-hydroxy-3,5-dimethyl-isocoumarin (14) and reticulol (29), along with two previously described 6-O-methylreticulol (333) and 7-hydroxy-5-methylmellein (334) (Fig. 48). These compounds were tested for their antifungal properties against C. cladosporioides, antioxidant activity, their lethal activity against the brine shrimp, and their phytotoxicity. Only 29 displayed mild anti-radical effect with an IC50 value of 58 μg mL−1.158
3.15.5.3. Fungi of the genus Halorosellinia. A previously mentioned (R)-4,8-dihydroxy-6-methoxy-4,5-dimethyl-3-methyleneisochromen-1-one (297), along with two previously unreported halorosellins A–B (335–336) (Fig. 49), were recorded from the marine-derived fungus Halorosellinia oceanica, collected in the Province of Samutsongkram. Compounds 297, 335, and 336 displayed no antiviral, anti-mycobacterium, antimalarial, and cytotoxic effects.159
3.15.5.4. Fungi of the genus Nodulisporium. A chemical investigation of the algal-derived fungus Nodulisporium sp., isolated from an algal species inner tissue, collected from the island of Corfu, Greece, afforded the previously mentioned 7-hydroxy-5-methylmellein (334). Compound 334 was tested using the agar diffusion methods against the bacterial strains B. megaterium and E. coli, the fungal strains M. violaceum, E. rubrum, and M. microspora as well as the green microalga C. fusca, but it showed no activity. In addition, it showed no cytotoxicity when evaluated against a panel of 36 tumor cell lines.160
3.15.5.5. Fungi of the genus Xylaria. The previously undescribed xylariamalirin (337) (Fig. 50) was recorded from n-BuOH fraction of the endophytic fungus Xylaria mali isolated from Lepidagathis stenophylla C. B. Clarke ex Hayata stems, collected from southern Taiwan. Compound 337 was not evaluated for any relevant biological activity.161
3.16. Miscellaneous
3.16.4.1. Fungal strain No. 1893. The chemical examination of the marine-derived fungal strain No. 1893, isolated from the mangrove Kandelia candel, collected from the Sea coast of the South China Sea, afforded the previously mentioned (−)-3,4-dihydro-8-hydroxy-3,5-dimethyl-isocoumarin (14) and the previously described 5-carboxylmellein (350) (Fig. 54). No biological activity was reported for these metabolites.165
3.16.4.2. Fungal strain No. dz17. Two previously mentioned (–)-(3R,4R)-cis-4-hydroxy-5-methylmellein (24) and 5-carboxylmellein (350), along with the previously unreported 3,4-dihydro-6-methoxy-8-hydroxy-3,4,5-trimethyl-isocoumarin-7-carboxylic acid methyl ester (351) (Fig. 55), were recorded from the EtOAc extract of the marine-derived fungal strain No. dz17, obtained from a sample collected from the coast of South China Sea. Compound 351 exhibited cytotoxic effect toward HepG2 and Hep-2 tumor cell lines with IC50 values of 55 and 52 μg mL−1, respectively.166
3.16.4.3. Fungal strain HJ33moB. The chemical examination of the marine-derived fungal strain HJ33moB, isolated from an unidentified alga, afforded the previously mentioned 1-deoxyrubralactone (273). Compound 273 exhibited strong inhibition effect against families X and Y of eukaryotic pols.167
4. Conclusion and future prospectives
For decades, natural products have granted medicine an innumerable supply of safe and effective drugs in various medicinal fields. Fungi, the broad diverse class of eukaryotes, is characterized by its variable habitats extending between air, water, soil, and within other marine and terrestrial hosts. Moreover, fungi have been traditionally used in treating various ailments in different old nations, especially Asia. Indeed, notable attention has recently been directed toward fungi as a substantial robust reservoir of pharmacologically active metabolites of crucial value in drug discovery. Among the captivating classes of fungal secondary metabolites are isocoumarins. We hereby present a comprehensive up to date literature reviewing focused on isocoumarins isolated over the period 2000–2022 from different fungal natural sources. A total of 351 structurally diverse isocoumarins isolated from various fungal genera were documented in this current review. Their chemical diversities and biological potentials constituted the focal point of interest of the current review, beside presenting brief highlights on the isocoumarin biosynthetic pathways. Thoroughly, in this manuscript, the analysis of such massive data revealed the richness of the fungal order (Eurotiales) with isocoumarins, representing 51% of the reported isolated compounds (Fig. 56). Meanwhile, on the family level, family (Aspergillaceae) was the most represented fungal family comprising 38% of the reported isocoumarins, followed by (Pleosporaceae) and (Trichocomaceae), from which 10% and 5% of the recorded isocoumarins were reported, respectively (Fig. 57). Intriguingly, the gathered statistical data also revealed the significant contribution of the two fungal species, Penicillium and Aspergillus, which constitute the prevailing sources of the reported compounds (Fig. 58), where a total of 149 isocoumarins were solely isolated from these two fungal species, representing 42% out of the reported isocoumarins over the specified 23 years period of the review. With emphasis on the therapeutic potentials of the recorded isocoumarins, a broad array of multiple biological activities was documented, including anti-inflammatory, antioxidant, antimicrobial, cytotoxic, and enzyme inhibitory activities. In addition, it is worthy to mention that the cytotoxicity, antibacterial, and antifungal activities were the most profoundly studied biological assays. Moreover, such massive biological screening revealed the evaluation of 155 isocoumarins for their cytotoxic potentials against different cancer cell lines. Furthermore, a total number of 160 and 77 isocoumarins were evaluated for their antibacterial and antifungal activities, respectively (Fig. 58). Noteworthily, a considerable number of isocoumarins, for example, compounds 84, 108, 124, 128, 130–131, 134, and 156, demonstrated significant biomedical potentials, including antibacterial, cytotoxic, and antidiabetic, respectively, which exceeds the potency when compared to the standards. Moreover, asperentin (59) displays numerous potent pharmacological activities, including antifungal, antimicrobial, antiplasmodial, and recently, as inhibitors of the protein tyrosine phosphatase 1B. Actually, such massive linkage between the different chemical architectures and their associated biological significances sheds light on the considerable importance of fungi as a prolific reservoir of promising lead compounds, which are worth future exploration for pharmaceutical and industrial applications. In-depth study of the pharmacokinetic properties of the fungi-derived isocoumarins is highly recommended as a prerequisite step in enhancing the opportunities for discovering potential drug candidates for different preclinical and clinical trials (Fig. 59).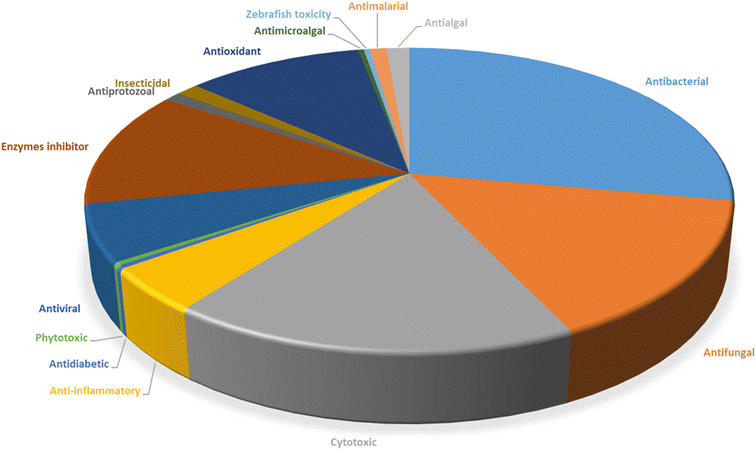 | ||
| Fig. 59 Total therapeutic activities of various isocoumarins isolated from different fungal species. | ||
Abbreviations
| A549 | Human lung cancer cells |
| AChE | Acetylcholinesterase |
| AsPC-1 | Xenograft model pancreatic cancer |
| BEL-7402 | Hepatoma cell line |
| BGC823 | Human gastric cancer cell line |
| BHT | Butylated hydroxytoluene |
| BT474 | Human breast cancer cell line |
| CCD25sk | Cellosaurus cancer cell line |
| CDK-2 | Human cyclin-dependent kinase 2 |
| CN | Ser/Thr phosphatase calcineurin |
| CNE1 | Cellosaurus CNE-1 |
| CNE2 | Cellosaurus CNE-2 |
| COX-2 | Cyclooxygenase-2 |
| CYP19 aromatase | Cytochrome P450 aromatase |
| DDP | cis-Diamminedichloroplatinum(II) |
| DEPT | Distortionless enhancement by polarization transfer |
| DMSO | Dimethyl sulfoxide |
| DPPH | 2,2-Diphenyl-1-picrylhydrazyl |
| DU145 | Prostate cancer cell line |
| EtOAc | Ethyl acetate |
| EV71 | Enterovirus 71 |
| H1975 | Human lung cancer cells |
| H2O2 | Hydrogen peroxide |
| H3N2 | Influenzavirus A |
| H460 | Human lung cancer cells |
| HCT116 | Human colon cancer cells |
| HCT-8 | Human colon cancer cells |
| HEK293T | Human embryonic kidney 293 cells |
| HeLa | Human cervical cancer cell line |
| Hep-2 | Human hepatocellular cancer cell lines |
| HepG2 | Human hepatocellular cancer cell lines |
| HIV | Human immunodeficiency virus |
| HL-60 | Human promyelocytic leukemia cells |
| HOAc | Acetic acid |
| HONE1 | Epithelial tumor cell lines |
| HT29 | Human colon adenocarcinoma cell line |
| HUAECs | Human amniotic epithelial cells |
| HuCCA-1 | Human cholangiocarcinoma cell line |
| Huh-7 | Human hepatocellular cancer cell lines |
| HUVECs | Human umbilical vein endothelial cells |
| IC50 | Inhibitory concentration that causes a 50% reduction in cell viability |
| iNOS | Inducible nitric oxide synthase |
| K562 | Human leukemia cancer cell line |
| KATO-3 | Human gastric cancer cell line |
| KB | Human epidermoid carcinoma, ATCC CCL-17 |
| KYSE150 | Esophageal adenocarcinoma cells |
| L5178Y | Mouse lymphoma |
| L-929 | Murine fibroblasts cancer cell line |
| LPS | Lipopolysaccharide |
| MCF10A | Human breast cancer cells |
| MCF-7 | Human breast cancer cells |
| MDA-MB-435 | Melanoma cell line |
| MeOH | Methanol |
| MGC-803 | Human gastric cancer cell line |
| MIA-PaCa-2 | Epithelial pancreatic cancer cell line |
| MIC | Minimum inhibitory concentration |
| Molm 13 | Acute myeloid leukemia |
| MOLT-3 | Human acute T lymphoblastic leukaemia |
| MOLT-4 | T lymphoblast cell line |
| MptpB | Mycobacterial protein tyrosine phosphatases A and B inhibitors |
| MTT assay | Colorimetric assay for assessing cell metabolic activity |
| NCI-H187 | Human small cell lung cancer, ATCC CRL-5804 |
| NCI-H460 | Human non-small-cell lung cancer cell line |
| NO | LPS-induced nitric oxide |
| NS-1 | Myeloma cancer cell line |
| ORAC | Oxygen Radical Absorbance Capacity |
| PC3 | Metastatic human prostate adenocarcinoma cancer cell line |
| PGE2 | Prostaglandin E2 |
| Psa | P. syringae pv. Actinidiae |
| PTP1B | Protein Tyrosine Phosphatase 1B |
| QGY7701 | Human hepatocellular cancer cell lines |
| RAW 264.7 | Macrophage-like, Abelson leukemia virus-transformed cell line derived from BALB/c mice |
| Rh2 (OCOCF3)4 | Rhodium;2,2,2-trifluoroacetic acid |
| SF-268 | Human CNS glioma cancer cell line |
| SGC7901 | Human gastric cancer cell line |
| SHSY5 | Thrice cloned subline of the neuroblastoma cell line |
| SH-SY5Y | Human glioma cell lines |
| SMMC-7721 | Human hepatocarcinoma |
| SW620 | Human colon cancer cells |
| TBARS | Thiobarbituric acid reactive substances |
| tBHQ | tert-Butylhydroquinone |
| TDDFT | Time Dependent DFT |
| TMV | Tobacco mosaic virus |
| Topo I | Topoisomerase I |
| TPA | 12-O-tetradecanoylphorbol-13-acetate |
| Vero cells | African green monkey kidney fibroblasts, ATCC CCL-81 |
| U2OS | Human Bone Osteosarcoma Epithelial Cells |
| U937 | Monocyte morphology |
| UV/vis | Ultraviolet-visible spectroscopy |
| XTT assay | Colorimetric method that uses the tetrazolium dye, 2,3-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2H)-tetrazolium-5-carboxanilide |
Author contributions
Conceptualization: Amr El-Demerdash. Validation: Amr El-Demerdash. Formal analysis: Mohamed A. Tammam and Amr El-Demerdash. Investigation: Mohamed A. Tammam, Mariam I. Gamal El-Din, Amira Abood and Amr El-Demerdash. Resources: Mohamed A. Tammam and Amr El-Demerdash. Data curation: Mohamed A. Tammam and Amr El-Demerdash. Writing original draft: Mohamed A. Tammam, Mariam I. Gamal El-Din, Amira Abood and Amr El-Demerdash. Writing-review & editing: Mohamed A. Tammam, Mariam I. Gamal El-Din, Amira Abood and Amr El-Demerdash.Conflicts of interest
The authors declare that they have no known competing commercial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
Amr El-Demerdash is immensely grateful to the John Innes Centre, Norwich Research Park, United Kingdom for the postdoctoral fellowship. Amr El-Demerdash is thankful to his home university, Mansoura University, Egypt for the unlimited support, inside and outside. Mohamed A. Tammam is humbly dedicating this work to the soul of his sister Dr Mai A. Tammam who passed away on 19 of March 2022, she was always kind supporter in all aspects of my life.References
- A. G. Atanasov, S. B. Zotchev, V. M. Dirsch, I. E. Orhan, M. Banach, J. M. Rollinger, D. Barreca, W. Weckwerth, R. Bauer, E. A. Bayer, M. Majeed, A. Bishayee, V. Bochkov, G. K. Bonn, N. Braidy, F. Bucar, A. Cifuentes, G. D'Onofrio, M. Bodkin, M. Diederich, A. T. Dinkova-Kostova, T. Efferth, K. el Bairi, N. Arkells, T. P. Fan, B. L. Fiebich, M. Freissmuth, M. I. Georgiev, S. Gibbons, K. M. Godfrey, C. W. Gruber, J. Heer, L. A. Huber, E. Ibanez, A. Kijjoa, A. K. Kiss, A. Lu, F. A. Macias, M. J. S. Miller, A. Mocan, R. Müller, F. Nicoletti, G. Perry, V. Pittalà, L. Rastrelli, M. Ristow, G. L. Russo, A. S. Silva, D. Schuster, H. Sheridan, K. Skalicka-Woźniak, L. Skaltsounis, E. Sobarzo-Sánchez, D. S. Bredt, H. Stuppner, A. Sureda, N. T. Tzvetkov, R. A. Vacca, B. B. Aggarwal, M. Battino, F. Giampieri, M. Wink, J. L. Wolfender, J. Xiao, A. W. K. Yeung, G. Lizard, M. A. Popp, M. Heinrich, I. Berindan-Neagoe, M. Stadler, M. Daglia, R. Verpoorte and C. T. Supuran, Nat. Rev. Drug Discovery, 2021, 20, 200–216 CrossRef CAS PubMed.
- M. I. Gamal El-Din, F. S. Youssef, M. L. Ashour, O. A. Eldahshan and A. N. B. Singab, Food Funct., 2020, 11, 1958–1965 RSC.
- S. Shams ul Hassan, H. zi Jin, T. Abu-Izneid, A. Rauf, M. Ishaq and H. A. R. Suleria, Biomed. Pharmacother., 2019, 109, 459–467 CrossRef CAS PubMed.
- M. A. Tammam, M. Sebak, C. Greco, A. Kijjoa and A. El-Demerdash, J. Mol. Struct., 2022, 1268, 133711 CrossRef CAS.
- F. Uzma, C. D. Mohan, C. N. Siddaiah and S. Chowdappa, Fungal Biol., 2019, 243–265 Search PubMed.
- A. Rani, K. C. Saini, F. Bast, S. Mehariya, S. K. Bhatia, R. Lavecchia and A. Zuorro, Molecules, 2021, 26, 1142 CrossRef CAS PubMed.
- G. F. Bills and J. B. Gloer, The fungal kingdom, 2017, pp. 1087–1119 Search PubMed.
- N. Rai, P. Kumari Keshri, A. Verma, S. C. Kamble, P. Mishra, S. Barik, S. Kumar Singh and V. Gautam, Mycology, 2021, 12, 139 CrossRef CAS PubMed.
- L. Boddy, The Fungi, 3rd edn, 2016, pp. 361–400 Search PubMed.
- A. Schueffler and T. Anke, Nat. Prod. Rep., 2014, 31, 1425–1448 RSC.
- D. J. Caruso, E. A. Palombo, S. E. Moulton and B. Zaferanloo, Microorganisms, 2022, 10, 1990 CrossRef CAS PubMed.
- A. H. Aly, A. Debbab and P. Proksch, Fungal Diversity, 2011, 50, 3–19 CrossRef.
- S. K. Deshmukh, V. Prakash and N. Ranjan, Front. Microbiol., 2018, 8, 2536 CrossRef PubMed.
- H. A. S. El-Nashar, M. I. Gamal El-Din, L. Hritcu and O. A. Eldahshan, Molecules, 2021, 26, 7546 CrossRef CAS PubMed.
- J. Ślusarczyk, E. Adamska and J. Czerwik-Marcinkowska, Nutrients, 2021, 13, 9 CrossRef PubMed.
- M. Sebak, F. Molham, C. Greco, M. A. Tammam, M. Sobeh and A. El-Demerdash, RSC Adv., 2022, 12, 24887–24921 RSC.
- J. Lenzi, T. M. Costa, M. D. Alberton, J. A. G. Goulart and L. B. B. Tavares, Appl. Microbiol. Biotechnol., 2018, 102, 5791–5810 CrossRef CAS PubMed.
- V. N. Ramachander Turaga, Bioactive Natural Products in Drug Discovery, 2020, pp. 713–730 Search PubMed.
- S. Pal, V. Chatare and M. Pal, Curr. Org. Chem., 2011, 15, 782–800 CrossRef CAS.
- P. Saikia and S. Gogoi, Adv. Synth. Catal., 2018, 360, 2063–2075 CrossRef CAS.
- K. Tianpanich, S. Prachya, S. Wiyakrutta, C. Mahidol, S. Ruchirawat and P. Kittakoop, J. Nat. Prod., 2011, 74, 79–81 CrossRef CAS PubMed.
- V. Das, P. P. Kaishap, G. Duarah, C. Chikkaputtaiah, H. P. Deka Boruah and M. Pal, Naunyn-Schmiedeb. Arch. Pharmacol., 2021, 394, 1437–1449 CrossRef CAS PubMed.
- S. Chen, Y. Liu, Z. Liu, R. Cai, Y. Lu, X. Huang and Z. She, RSC Adv., 2016, 6, 26412–26420 RSC.
- M. Kuramata, S. Fujioka, A. Shimada, T. Kawano and Y. Kimura, Biosci. Biotechnol. Biochem., 2007, 71, 499–503 CrossRef CAS PubMed.
- K. Krohn, U. Flörke, M. S. Rao, K. Steingröver, H. J. Aust, S. Draeger and B. Schulz, Nat. Prod. Lett., 2001, 15, 353–361 CrossRef CAS PubMed.
- S. Pal and M. Pal, Natural Occurrences, Synthetic Approaches and Pharmaceutical Applications, 2019, 978-0-12-815411-3 Search PubMed.
- A. Saeed, Eur. J. Med. Chem., 2016, 116, 290–317 CrossRef CAS PubMed.
- H. Hussain and I. R. Green, Expert Opin. Ther. Pat., 2017, 27, 1267–1275 CrossRef CAS PubMed.
- A. Saddiqa, M. Usman and O. Çakmak, Turk. J. Chem., 2017, 41, 153–178 CrossRef CAS.
- A. O. Noor, D. M. Almasri, A. A. Bagalagel, H. M. Abdallah, S. G. A. Mohamed, G. A. Mohamed and S. R. M. Ibrahim, Molecules, 2020, 25, 395 CrossRef CAS PubMed.
- G. Shabir, A. Saeed and H. R. El-Seedi, Phytochemistry, 2021, 181, 112568 CrossRef CAS PubMed.
- A. W. K. Yeung, A. El-Demerdash, I. Berindan-Neagoe, A. G. Atanasov and Y. S. Ho, Crit. Rev. Oncog., 2018, 23, 347–370 CrossRef PubMed.
- W. Yeung, N. Choudhary, D. Tewari, A. El-Demerdash, O. Horbanczuk, N. Das, V. Pirgozliev, M. Lucarini, A. Durazzo, E. Souto, A. Santini, H. Devkota, M. Uddin, J. Echeverria, D. Wang, R. Gan, M. Brncic, R. Kalfin, N. Tzvetkov, A. Jozwik, M. Solka, N. Strzalkowska, J. Horbanczuk and A. Atanasov, Anim. Sci. Pap. Rep., 2021, 39, 199–212 Search PubMed.
- A. el Demerdash, A. M. Dawidar, E. M. Keshk and M. Abdel-Mogib, Chem. Nat. Compd., 2012, 48, 646–648 CrossRef CAS.
- A. El-Demerdash, J. Fungus, 2018, 4, 130 CrossRef CAS PubMed.
- A. El-Demerdash, G. Genta-Jouve, M. Bärenstrauch, C. Kunz, E. Baudouin and S. Prado, Phytochemistry, 2019, 166, 112056 CrossRef CAS PubMed.
- A. El-Demerdash, C. Borde, G. Genta-Jouve, A. Escargueil and S. Prado, Nat. Prod. Res., 2022, 36, 1273–1281 CrossRef CAS PubMed.
- R. D. Barry, Chem. Rev., 1964, 64, 229–260 CrossRef CAS.
- C. Wu, H. Zhu, G. P. Van Wezel and Y. H. Choi, Metabolomics, 2016, 12, 90 CrossRef PubMed.
- R. Y. Song, X. B. Wang, G. P. Yin, R. H. Liu, L. Y. Kong and M. H. Yang, Fitoterapia, 2017, 122, 115–118 CrossRef CAS PubMed.
- A. J. Birch, J. H. Birkinshaw, P. Chaplen, L. Mo, A. H. Manchanda, A. Pelter and M. Riano-Martin, Aust. J. Chem., 1969, 22, 1933–1941 CrossRef CAS.
- A. J. Birch, L. Loh, A. Pelter, J. H. Birkinshaw, P. Chaplen, A. H. Manchanda and M. Riano-Martin, Tetrahedron Lett., 1965, 57, 29–32 CrossRef CAS PubMed.
- C. N. Lewis, J. Staunton and D. C. Sunter, J. Chem. Soc., Perkin trans., 1988, 1, 747–754 RSC.
- R. J. N. Frandsen, N. J. Nielsen, N. Maolanon, J. C. Sørensen, S. Olsson, J. Nielsen and H. Giese, Mol. Microbiol., 2006, 61, 1069–1080 CrossRef CAS PubMed.
- F. T. Hansen, J. L. Sørensen, H. Giese, T. E. Sondergaard and R. J. N. Frandsen, Int. J. Food Microbiol., 2012, 155, 128–136 CrossRef CAS PubMed.
- J. E. Kim, K. H. Han, J. Jin, H. Kim, J. C. Kim, S. H. Yun and Y. W. Lee, Appl. Environ. Microbiol., 2005, 71, 1701 CrossRef CAS PubMed.
- S. Malz, M. N. Grell, C. Thrane, F. J. Maier, P. Rosager, A. Felk, K. S. Albertsen, S. Salomon, L. Bohn, W. Schäfer and H. Giese, Fungal Genet. Biol., 2005, 42, 420–433 CrossRef CAS PubMed.
- R. J. N. Frandsen, C. Schütt, B. W. Lund, D. Staerk, J. Nielsen, S. Olsson and H. Giese, J. Biol. Chem., 2011, 286, 10419 CrossRef CAS PubMed.
- I. Fujii, A. Watanabe, U. Sankawa and Y. Ebizuka, Chem. Biol., 2001, 8, 189–197 CrossRef CAS PubMed.
- A. Watanabe, Y. Ono, I. Fujii, U. Sankawa, M. E. Mayorga, W. E. Timberlake and Y. Ebizuka, Tetrahedron Lett., 1998, 39, 7733–7736 CrossRef CAS.
- J. L. Sørensen, K. F. Nielsen and T. E. Sondergaard, Fungal Genet. Biol., 2012, 49, 613–618 CrossRef PubMed.
- S. M. Ma, J. Zhan, X. Xie, K. Watanabe, Y. Tang and W. Zhang, J. Am. Chem. Soc., 2008, 130, 38–39 CrossRef CAS PubMed.
- P. Wiemann, A. Willmann, M. Straeten, K. Kleigrewe, M. Beyer, H. U. Humpf and B. Tudzynski, Mol. Microbiol., 2009, 72, 931–946 CrossRef CAS PubMed.
- P. Xiang, L. Ludwig-Radtke, W. B. Yin and S. M. Li, Org. Biomol. Chem., 2020, 18, 4946–4948 RSC.
- A. K. Atanasoff-Kardjalieff, B. Seidl, K. Steinert, C. G. Daniliuc, R. Schuhmacher, H. U. Humpf, S. Kalinina and L. Studt-Reinhold, ChemBioChem, 2022, e202200342 Search PubMed.
- P. A. Storm, P. Pal, C. R. Huitt-Roehl and C. A. Townsend, ACS Chem. Biol., 2018, 13, 3043–3048 CrossRef CAS PubMed.
- B. Thongbai, F. Surup, K. Mohr, E. Kuhnert, K. D. Hyde and M. Stadler, J. Nat. Prod., 2013, 76, 2141–2144 CrossRef CAS PubMed.
- Z. Ju, X. Lin, X. Lu, Z. Tu, J. Wang, K. Kaliyaperumal, J. Liu, Y. Tian, S. Xu and Y. Liu, J. Antibiot., 2015, 68, 653–656 CrossRef CAS PubMed.
- Z. Wu, J. Chen, X. Zhang, Z. Chen, T. Li, Z. She, W. Ding and C. Li, Mar. Drugs, 2019, 17, 88 CrossRef CAS PubMed.
- X. Pang, X. Lin, J. Wang, R. Liang, Y. Tian, L. Salendra, X. Luo, X. Zhou, B. Yang, Z. Tu and Y. Liu, Steroids, 2018, 129, 41–46 CrossRef CAS PubMed.
- A. R. Gohil, S. K. Deshmukh, V. Bhattacharya, R. Lavhale, S. Verekar and A. S. Kate, Nat. Prod. Res., 2021, 35, 1573–1581 CrossRef CAS PubMed.
- K. M. Meepagala, W. E. Briscoe, N. Techen, R. D. Johnson, B. M. Clausen and S. O. Duke, Pest Manag. Sci., 2018, 74, 37–45 CrossRef CAS PubMed.
- L. Guo, S. Niu, S. Chen and L. Liu, J. Antibiot., 2019, 73(2), 116–119 CrossRef PubMed.
- H. R. Qu, W. W. Yang, X. Q. Zhang, Z. H. Lu, Z. S. Deng, Z. Y. Guo, F. Cao, K. Zou and P. Proksch, Phytochem. Lett., 2020, 37, 1–4 CrossRef CAS.
- X. Q. Zhang, Z. H. Lu, G. R. Xia, W. M. Song, Z. Y. Guo and P. Proksch, Tetrahedron Lett., 2021, 75, 153205 CrossRef CAS.
- A. A. El-Beih, H. Kato, T. Ohta and S. Tsukamoto, Chem. Pharm. Bull., 2007, 55, 953–954 CrossRef CAS PubMed.
- T. G. C. Montenegro, F. A. R. Rodrigues, P. C. Jimenez, A. L. Angelim, V. M. M. Melo, E. R. Filho, M. C. F. de Oliveira and L. V. Costa-Lotufo, Chem. Biodivers., 2012, 9, 2203–2209 CrossRef CAS PubMed.
- C. Prompanya, T. Dethoup, L. J. Bessa, M. M. M. Pinto, L. Gales, P. M. Costa, A. M. S. Silva and A. Kijjoa, Mar. Drugs, 2014, 12, 5160–5173 CrossRef CAS PubMed.
- Y. Q. Ye, C. F. Xia, J. X. Yang, Y. Qin, M. Zhou, X. M. Gao, G. Du, H. Y. Yang, X. M. Li and Q. F. Hu, Phytochem. Lett., 2014, 10, 215–218 CrossRef CAS.
- K. Sun, Y. Li, L. Guo, Y. Wang, P. Liu and W. Zhu, Mar. Drugs, 2014, 12, 3970–3981 CrossRef CAS PubMed.
- Q. Tang, K. Guo, X. Y. Li, X. Y. Zheng, X. J. Kong, Z. H. Zheng, Q. Y. Xu and X. Deng, Mar. Drugs, 2014, 12, 5993–6002 CrossRef CAS PubMed.
- D. C. Kim, T. H. Quang, N. T. T. Ngan, C. S. Yoon, J. H. Sohn, J. H. Yim, Y. Feng, Y. Che, Y. C. Kim and H. Oh, J. Nat. Prod., 2015, 78, 2948–2955 CrossRef CAS PubMed.
- Y. Liu, S. Chen, Z. Liu, Y. Lu, G. Xia, H. Liu, L. He and Z. She, Mar. Drugs, 2015, 13, 3091–3102 CrossRef CAS PubMed.
- C. Prompanya, C. Fernandes, S. Cravo, M. M. M. Pinto, T. Dethoup, A. M. S. Silva and A. Kijjoa, Mar. Drugs, 2015, 13, 1432–1450 CrossRef CAS PubMed.
- M. Zhou, K. Zhou, P. He, K. M. Wang, R. Z. Zhu, Y. de Wang, W. Dong, G. P. Li, H. Y. Yang, Y. Q. Ye, G. Du, X. M. Li and Q. F. Hu, Planta Med., 2016, 82, 414–417 CrossRef CAS PubMed.
- J. Wiese, H. Aldemir, R. Schmaljohann, T. A. M. Gulder, J. F. Imhoff and R. Kerr, Mar. Drugs, 2017, 15, 191 CrossRef PubMed.
- M. Zhou, J. Lou, Y. K. Li, Y. de Wang, K. Zhou, B. K. Ji, W. Dong, X. M. Gao, G. Du and Q. F. Hu, Arch. Pharm. Res., 2017, 40, 32–36 CrossRef CAS PubMed.
- N. K. Chaudhary, J. I. Pitt, E. Lacey, A. Crombie, D. Vuong, A. M. Piggott and P. Karuso, J. Nat. Prod., 2018, 81, 1517–1526 CrossRef CAS PubMed.
- Y. Q. Duan, L. Z. Dang, J. X. Jiang, Y. P. Zhang, N. J. Xiang, H. M. Yang, G. Du, H. Y. Yang and Q. Q. Li, Chem. Nat. Compd., 2018, 54, 249–252 CrossRef CAS.
- M. Chen, R. Wang, W. Zhao, L. Yu, C. Zhang, S. Chang, Y. Li, T. Zhang, J. Xing, M. Gan, F. Feng and S. Si, Org. Lett., 2019, 21, 1530–1533 CrossRef CAS PubMed.
- Y. Wu, S. Chen, H. Liu, X. Huang, Y. Liu, Y. Tao and Z. She, Arch. Pharm. Res., 2019, 42, 326–331 CrossRef CAS PubMed.
- X. Ma, X. Liang, Z. H. Huang and S. H. Qi, Nat. Prod. Res., 2020, 34, 1992–2000 CrossRef CAS PubMed.
- H. X. Guo, C. Y. Huang, Z. Y. Yan, T. Chen, K. Hong and Y. H. Long, Chin. J. Nat. Med., 2020, 18, 855–859 CAS.
- D. H. El-Kashef, F. S. Youssef, I. Reimche, N. Teusch, W. E. G. Müller, W. Lin, M. Frank, Z. Liu and P. Proksch, Bioorg. Med. Chem., 2021, 29, 115883 CrossRef CAS PubMed.
- Z. H. Xin, L. Tian, T. J. Zhu, W. L. Wang, L. Du, Y. C. Fang, Q. Q. Gu and W. M. Zhu, Arch. Pharmacal Res., 2007, 30, 816–819 CrossRef CAS PubMed.
- D. Zhang, X. Li, J. S. Kang, H. D. Choi, J. H. Jung and B. W. Son, J. Microbiol. Biotechnol., 2007, 17, 865–867 CAS.
- Z. Han, W. Mei, Y. Zhao, Y. Deng and H. Dai, Chem. Nat. Compd., 2009, 45, 805–807 CrossRef CAS.
- J. Arunpanichlert, V. Rukachaisirikul, Y. Sukpondma, S. Phongpaichit, S. Tewtrakul, N. Rungjindamai and J. Sakayaroj, Chem. Pharm. Bull., 2010, 58, 1033–1036 CrossRef CAS PubMed.
- M. L. Wang, C. H. Lu, Q. Y. Xu, S. Y. Song, Z. Y. Hu and Z. H. Zheng, Molecules, 2013, 18, 5723–5735 CrossRef CAS PubMed.
- J. Qi, C. L. Shao, Z. Y. Li, L. S. Gan, X. M. Fu, W. T. Bian, H. Y. Zhao and C. Y. Wang, J. Nat. Prod., 2013, 76, 571–579 CrossRef CAS PubMed.
- Q. Q. Li, L. Z. Dang, Y. P. Zhang, J. X. Jiang, C. M. Zhang, N. J. Xiang, H. Y. Yang, G. Du and Y. Q. Duan, J. Asian Nat. Prod. Res., 2015, 17, 876–881 CrossRef CAS PubMed.
- L. Chen, W. Liu, X. Hu, K. Huang, J.-L. Wu and Q.-Q. Zhang, Chem. Pharm. Bull., 2011, 59, 515–522 CrossRef CAS PubMed.
- C. Pan, Y. Shi, B. N. Auckloo, S. S. ul Hassan, N. Akhter, K. Wang, Y. Ye, C. T. Arthur Chen, X. Tao and B. Wu, Mar. Biotechnol., 2017, 19, 469–479 CrossRef CAS PubMed.
- S. Chen, J. Wang, Z. Wang, X. Lin, B. Zhao, K. Kaliaperumal, X. Liao, Z. Tu, J. Li, S. Xu and Y. Liu, Fitoterapia, 2017, 117, 71–78 CrossRef CAS PubMed.
- R. Cai, Y. Wu, S. Chen, H. Cui, Z. Liu, C. Li and Z. She, J. Nat. Prod., 2018, 81, 1376–1383 CrossRef CAS PubMed.
- S. S. Afiyatullov, O. I. Zhuravleva, A. S. Antonov, E. v. Leshchenko, M. v. Pivkin, Y. v. Khudyakova, V. A. Denisenko, E. A. Pislyagin, N. Y. Kim, D. v. Berdyshev, G. von Amsberg and S. A. Dyshlovoy, Mar. Drugs, 2019, 17, 647 CrossRef CAS PubMed.
- W. Wang, Y. Liao, B. Zhang, M. Gao, W. Ke, F. Li and Z. Shao, Mar. Drugs, 2019, 17, 46 CrossRef CAS PubMed.
- X. W. Luo, C. H. Gao, F. H. Han, X. Q. Chen, X. P. Lin, X. F. Zhou, J. J. Wang and Y. H. Liu, Magn. Reson. Chem., 2019, 57, 982–986 CrossRef CAS PubMed.
- M. Bai, C. J. Zheng, G. L. Huang, R. Q. Mei, B. Wang, Y. P. Luo, C. Zheng, Z. G. Niu and G. Y. Chen, J. Nat. Prod., 2019, 82, 1155–1164 CrossRef CAS PubMed.
- J. Cao, X. M. Li, X. Li, H. L. Li, L. H. Meng and B. G. Wang, Phytochem. Lett., 2019, 32, 1–5 CrossRef.
- P. Qiu, R. L. Cai, L. Li and Z. G. She, Chin. J. Nat. Med., 2020, 18, 256–260 CAS.
- G. B. Xu, F. Y. Yang, X. Y. Wu, R. Li, M. Zhou, B. Wang, X. S. Yang, T. T. Zhang and S. G. Liao, Nat. Prod. Res., 2021, 35, 1445–1451 CrossRef CAS PubMed.
- W. N. Zeng, J. Cai, B. Wang, L. Y. Chen, C. X. Pan, S. J. Chen, G. L. Huang and C. J. Zheng, J. Asian Nat. Prod. Res., 2022, 24, 679–684 CrossRef CAS PubMed.
- B. Bin Gu, Y. Wu, J. Tang, W. Hua Jiao, L. Li, F. Sun, S. P. Wang, F. Yang and H. W. Lin, Tetrahedron Lett., 2018, 59, 3345–3348 CrossRef.
- C. Almeida, I. Pérez-Victoria, V. González-Menéndez, N. de Pedro, J. Martín, G. Crespo, T. Mackenzie, B. Cautain, F. Reyes, F. Vicente and O. Genilloud, J. Nat. Prod., 2018, 81, 1488–1492 CrossRef CAS PubMed.
- Y. Q. Ran, W. J. Lan, Y. Qiu, Q. Guo, G. K. Feng, R. Deng, X. F. Zhu, H. J. Li and J. Dong, Mar. Drugs, 2020, 18, 100 CrossRef CAS PubMed.
- F. Miao, R. Yang, D. D. Chen, Y. Wang, B. F. Qin, X. J. Yang and L. Zhou, Molecules, 2012, 17, 14091–14098 CrossRef CAS PubMed.
- S. Buttachon, W. W. May Zin, T. Dethoup, L. Gales, J. A. Pereira, A. M. S. Silva and A. Kijjoa, Planta Med., 2016, 82, 888–896 CrossRef CAS PubMed.
- J. Li, C. Chen, T. Fang, L. Wu, W. Liu, J. Tang and Y. Long, Molecules, 2022, 27, 5766 CrossRef CAS PubMed.
- K. Tianpanich, S. Prachya, S. Wiyakrutta, C. Mahidol, S. Ruchirawat and P. Kittakoop, J. Nat. Prod., 2011, 74, 79–81 CrossRef CAS PubMed.
- M. Zhao, L. Y. Yuan, D. le Guo, Y. Ye, Z. M. Da-Wa, X. L. Wang, F. W. Ma, L. Chen, Y. C. Gu, L. S. Ding and Y. Zhou, Phytochemistry, 2018, 148, 97–103 CrossRef CAS PubMed.
- M. Figueroa, H. Raja, J. O. Falkinham, A. F. Adcock, D. J. Kroll, M. C. Wani, C. J. Pearce and N. H. Oberlies, J. Nat. Prod., 2013, 76, 1007–1015 CrossRef CAS PubMed.
- L. H. Meng, H. Q. Chen, I. Form, B. Konuklugil, P. Proksch and B. G. Wang, Nat. Prod. Commun., 2016, 11, 1293–1296 CrossRef PubMed.
- M. Isaka, S. Palasarn, S. Supothina, S. Komwijit and J. J. Luangsa-Ard, J. Nat. Prod., 2011, 74, 782–789 CrossRef CAS PubMed.
- J. F. Tian, P. J. Li, X. X. Li, P. H. Sun, H. Gao, X. Z. Liu, P. Huang, J. S. Tang and X. S. Yao, Bioorg. Med. Chem. Lett., 2016, 26, 1391–1396 CrossRef CAS PubMed.
- J. Kornsakulkarn, C. Thongpanchang, S. Lapanun and K. Srichomthong, J. Nat. Prod., 2009, 72, 1341–1343 CrossRef CAS PubMed.
- W. Li, J. Xu, F. Li, L. Xu and C. Li, Pharmacogn. Mag., 2016, 12, 259–261 CAS.
- F. Fang, J. Zhao, L. Ding, C. Huang, C. B. Naman, S. He, B. Wu, P. Zhu, Q. Luo, W. H. Gerwick, X. Yan, Q. Wang, Z. Zhang and W. Cui, Mar. Drugs, 2017, 15, 260 CrossRef PubMed.
- Z. Ding, T. Tao, L. Wang, Y. Zhao, H. Huang, D. Zhang, M. Liu, Z. Wang and J. Han, J. Microbiol. Biotechnol., 2019, 29, 731–738 CrossRef CAS PubMed.
- X. H. Liu, X. L. Hou, Y. P. Song, B. G. Wang and N. Y. Ji, Fitoterapia, 2020, 141, 104469 CrossRef CAS PubMed.
- Y. Chen, G. Wang, Y. Yuan, G. Zou, W. Yang, Q. Tan, W. Kang and Z. She, Front. Chem., 2022, 10, 57 Search PubMed.
- N. R. Ariefta, P. Kristiana, T. Aboshi, T. Murayama, K. Tawaraya, T. Koseki, N. Kurisawa, K. ichi Kimura and Y. Shiono, Fitoterapia, 2018, 127, 356–361 CrossRef CAS PubMed.
- V. Rukachaisirikul, A. Rodglin, Y. Sukpondma, S. Phongpaichit, J. Buatong and J. Sakayaroj, J. Nat. Prod., 2012, 75, 853–858 CrossRef CAS PubMed.
- S. Zhang, F. H. Kang, J. B. Tan, D. K. Chen, M. Kuang, W. X. Wang, K. P. Xu and Z. X. Zou, New J. Chem., 2021, 45, 12700–12704 RSC.
- C. C. Feng, G. D. Chen, Y. Q. Zhao, S. C. Xin, S. Li, J. S. Tang, X. X. Li, D. Hu, X. Z. Liu and H. Gao, Chem. Biodivers., 2014, 11, 1099–1108 CrossRef CAS PubMed.
- S. Niu, D. Liu, P. Proksch, Z. Shao and W. Lin, Mar. Drugs, 2015, 13, 2526–2540 CrossRef CAS PubMed.
- S. Niu, D. Liu, Z. Shao, J. Huang, A. Fan and W. Lin, RSC Adv., 2021, 11, 29661–29667 RSC.
- J. F. Tian, R. J. Yu, X. X. Li, H. Gao, D. Hu, L. D. Guo, J. S. Tang and X. S. Yao, J. Asian Nat. Prod. Res., 2015, 17, 550–558 CrossRef CAS PubMed.
- Y. F. Huang, L. H. Li, L. Tian, L. Qiao, H. M. Hua and Y. H. Pei, J. Antibiot., 2006, 59, 355–357 CrossRef CAS PubMed.
- Y. Wang, M. H. Yang, X. B. Wang, T. X. Li and L. Y. Kong, Fitoterapia, 2014, 99, 153–158 CrossRef CAS PubMed.
- J. S. Wu, X. H. Shi, Y. H. Zhang, J. Y. Yu, X. M. Fu, X. Li, K. X. Chen, Y. W. Guo, C. L. Shao and C. Y. Wang, Front. Chem., 2019, 7, 763 CrossRef CAS PubMed.
- R. Sappapan, D. Sommit, N. Ngamrojanavanich, S. Pengpreecha, S. Wiyakrutta, N. Sriubolmas and K. Pudhom, J. Nat. Prod., 2008, 71, 1657–1659 CrossRef CAS PubMed.
- R. Li, S. Chen, S. Niu, L. Guo, J. Yin and Y. Che, Fitoterapia, 2014, 96, 88–94 CrossRef CAS PubMed.
- L. Coronado, X. Q. Zhang, D. Dorta, N. Escala, L. M. Pineda, M. G. Ng, E. del Olmo, C. Y. Wang, Y. C. Gu, C. L. Shao and C. Spadafora, J. Nat. Prod., 2021, 84, 1434–1441 CrossRef CAS PubMed.
- X. Pang, X. Lin, J. Yang, X. Zhou, B. Yang, J. Wang and Y. Liu, J. Nat. Prod., 2018, 81, 1860–1868 CrossRef CAS PubMed.
- S. S. Liu, W. bin Gao, J. Kang, X. H. Yang, F. Cao, F. D. Kong, Y. X. Zhao and D. Q. Luo, Chem. Nat. Compd., 2020, 56, 799–802 CrossRef CAS.
- W. Gao, X. Wang, F. Chen, C. Li, F. Cao and D. Luo, Nat. Prod. Bioprospect., 2021, 11, 137–142 CrossRef CAS PubMed.
- A. Abdel-Lateff, K. M. Fisch, A. D. Wright and G. M. König, Planta Med., 2003, 69, 831–834 CrossRef CAS PubMed.
- Y. Zhao, D. Liu, P. Proksch, S. Yu and W. Lin, Chem. Biodivers., 2016, 13, 1186–1193 CrossRef CAS PubMed.
- M. F. Elsebai and H. A. Ghabbour, Tetrahedron Lett., 2016, 57, 354–356 CrossRef CAS.
- X. N. Sang, S. F. Chen, X. An, G. Chen, H. F. Wang and Y. H. Pei, J. Asian Nat. Prod. Res., 2017, 19, 436–443 CrossRef CAS PubMed.
- T. Shi, J. Qi, C. L. Shao, D. L. Zhao, X. M. Hou and C. Y. Wang, Mar. Drugs, 2017, 15, 146 CrossRef PubMed.
- A. D. Wright and N. Lang-Unnasch, Planta Med., 2005, 71, 964–966 CrossRef CAS PubMed.
- Q. Chen, J. J. Yu, J. He, T. Feng and J. K. Liu, Phytochemistry, 2022, 195, 113050 CrossRef CAS PubMed.
- X. Luo, X. Lin, L. Salendra, X. Pang, Y. Dai, B. Yang, J. Liu, J. Wang, X. Zhou and Y. Liu, Mar. Drugs, 2017, 15, 204 CrossRef PubMed.
- M. F. Liao, K. Wang, J. W. Ren, L. Liu, L. Cai, J. J. Han and H. W. Liu, J. Asian Nat. Prod. Res., 2019, 21, 939–946 CrossRef CAS PubMed.
- X. Xu, J. Li, K. Zhang, S. Wei, R. Lin, S. W. Polyak, N. Yang, F. Song, X. Xu, J. Li, K. Zhang, S. Wei, R. Lin, S. W. Polyak and N. Yang, Mar. Drugs, 2021, 19, 313 CrossRef CAS PubMed.
- M. Tsukada, M. Fukai, K. Miki, T. Shiraishi, T. Suzuki, K. Nishio, T. Sugita, M. Ishino, K. Kinoshita, K. Takahashi, M. Shiro and K. Koyama, J. Nat. Prod., 2011, 74, 1645–1649 CrossRef CAS PubMed.
- W. Zheng, Y. bin Ji, W. L. Li, J. Dong, N. Chen, X. F. Yu, J. Wu, D. Zhao and Z. Xiang, J. Asian Nat. Prod. Res., 2017, 19, 993–999 CrossRef CAS PubMed.
- W. Zhang, K. Krohn, S. Draeger and B. Schulz, J. Nat. Prod., 2008, 71, 1078–1081 CrossRef CAS PubMed.
- J. Arunpanichlert, V. Rukachaisirikul, S. Phongpaichit, O. Supaphon and J. Sakayaroj, Tetrahedron, 2015, 71, 882–888 CrossRef CAS.
- R. Y. Song, X. B. Wang, G. P. Yin, R. H. Liu, L. Y. Kong and M. H. Yang, Fitoterapia, 2017, 122, 115–118 CrossRef CAS PubMed.
- H. Lei, X. Lin, L. Han, J. Ma, Q. Ma, J. Zhong, Y. Liu, T. Sun, J. Wang and X. Huang, Mar. Drugs, 2017, 15, 69 CrossRef PubMed.
- Z. Xu, X. Wu, G. Li, Z. Feng and J. Xu, Nat. Prod. Res., 2020, 34, 1002–1007 CrossRef CAS PubMed.
- G. Liao, P. Wu, Z. Liu, J. Xue, H. Li and X. Wei, Nat. Prod. Res., 2020, 35, 3644–3651 CrossRef PubMed.
- Y. J. Dong, G. M. Hou, B. Lin, D. Y. Li and Z. L. Li, J. Asian Nat. Prod. Res., 2019, 21, 689–695 CrossRef CAS PubMed.
- B. Wu, J. Wiese, R. Schmaljohann and J. F. Imhoff, Mar. Drugs, 2016, 14, 204 CrossRef PubMed.
- L. Jayasinghe, T. Sritharan, N. Savitri Kumar, H. Araya and Y. Fujimoto, Nat. Prod. Commun., 2019, 5, 1–3 Search PubMed.
- M. Chinworrungsee, P. Kittakoop, M. Isaka, R. Chanphen, M. Tanticharoen and Y. Thebtaranonth, Int. J. Food Microbiol., 2002, 1(22), 2473–2476 Search PubMed.
- A. Pontius, I. Mohamed, A. Krick, S. Kehraus and G. M. König, J. Nat. Prod., 2008, 71, 272–274 CrossRef CAS PubMed.
- M. J. Cheng, M. der Wu, T. Aung, C. T. Chang, S. Y. Hsieh and J. J. Chen, Chem. Nat. Compd., 2020, 56, 221–223 CrossRef CAS.
- Y. Chen, Z. Liu, H. Liu, Y. Pan, J. Li, L. Liu and Z. She, Mar. Drugs, 2018, 16, 54 CrossRef PubMed.
- K. Krohn, H. Sohrab, H. J. Aust, S. Draeger and B. Schulz, Nat. Prod. Res., 2006, 18, 277–285 CrossRef PubMed.
- D. Lai, J. Li, S. Zhao, G. Gu, X. Gong, P. Proksch and L. Zhou, Nat. Prod. Res., 2021, 35, 4616–4620 CrossRef CAS PubMed.
- G. Chen, Y. Lin, L. L. P. Vrijmoed and W. F. Fong, Chem. Nat. Compd., 2006, 42, 138–141 CrossRef CAS.
- Z. Huang, C. Shao, Y. Chen, Z. She, Y. Lin and S. Zhou, Chem. Nat. Compd., 2007, 43, 655–658 CrossRef CAS.
- M. Naganuma, M. Nishida, K. Kuramochi, F. Sugawara, H. Yoshida and Y. Mizushina, Bioorg. Med. Chem., 2008, 16, 2939–2944 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra08245d |
| This journal is © The Royal Society of Chemistry 2023 |