Open Access Article
Open Access ArticleAn antifouling and antiviral superhydrophobic elastomer formed by 3D printing and a peptide-based coating†
Tan
Hu
 ab,
Noa
Trink
ab,
Noa
Trink
 ab,
Shlomo
Magdassi‡
ab,
Shlomo
Magdassi‡
 *ab and
Meital
Reches‡
*ab and
Meital
Reches‡
 *ab
*ab
aInstitute of Chemistry and The Center for Nanoscience and Nanotechnology, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel. E-mail: magdassi@mail.huji.ac.il; meital.reches@mail.huji.ac.il
bSingapore-HUJ Alliance for Research and Enterprise (SHARE), Smart Grippers for Soft Robotics (SGSR), Campus for Research Excellence and Technological Enterprise (CREATE), Singapore, 138602, Singapore
First published on 13th August 2024
Abstract
Elastomers are used in various applications, including medical devices, conveyor belts that move food items through production, and soft grippers that handle objects such as plants, vegetables, and fruits. The undesirable contamination of the surface of these elastomers by microorganisms, dust, and sand harms their performance and service life. Herein, we describe the fabrication of an antifouling and antiviral superhydrophobic elastomer by combining 3D printing and a peptide-based coating. Superhydrophobicity is accomplished by an array of printed micropillars with multi-scale roughness due to embedded hydrophobic nanoparticles fabricated by digital light processing (DLP) 3D printing. The non-fluorinated silica particles embedded in the 3D pillars impart suitable roughness and surface energy to the printed elastomer. The resulting elastomer is superhydrophobic with a water contact angle of ∼158° and a rolling angle of ∼7°. The printed elastomers were coated by a short peptide that self-assembles onto the coating to provide this elastomer with antifouling properties. This coating reduces the number of bacteria on the elastomer and provides it with antiviral activity. Importantly, the coating does not alter the superhydrophobic properties of the elastomer and is mechanically stable. Overall, our work provides a new method for fabricating superhydrophobic antifouling elastomers.
Introduction
Elastomers are polymers possessing low modulus and high elasticity that can rapidly recover their original state upon release of the external stress.1,2 Due to their unique properties, they have been utilized in numerous applications, such as aerospace and automotive industries, consumer goods, construction, agriculture, and medicine.3–8 For some applications, including food preparation surfaces, biomedical devices, and soft grippers, the elastomers should avoid microbial, viral, and other contamination. The undesirable contamination by inorganic compounds such as dust and organic matter and microorganisms such as bacteria, fungi, and viruses harm their function and should be prevented.9–12Fouling, the undesirable accumulation of inorganic and organic matter on a surface, is related to surface energy, which is controlled by surface chemistry and topography.13 One approach to prevent fouling is by lowering the surface energy of a surface so it would be superhydrophobic, with a water contact angle higher than 150° and a rolling angle below 10°. Fabricating superhydrophobic elastomers usually relies on combining hierarchical surface structure and low surface energy materials.14 Studies in this field focus on generating micro-scale roughness followed by chemical surface modification.1,3,5,15,16 In addition, silicone- and fluor-based polymers can be used to impart superhydrophobicity.17 Yu et al. reported that combining commercial silicone elastomer (Eco flex) with micron-sized silica particles fabricated a robust superhydrophobic elastomer surface.15 Zheng et al. created highly durable superhydrophobic coatings obtained by a fluoropropyl methylsiloxane-dimethylsiloxane multi-block copolymer containing methacryloxyl group (MAc(PTFMS-alt-PDMS)) on elastomer.15 However, most work to generate the superhydrophobicity for elastomer surfaces involves the introduction of fluorinated compounds that are toxic and universally identified as environmental contaminants.16,18,19 Therefore, it is urgent to develop the superhydrophobic coating on elastomer substrates without fluorinated compounds. 3D printing enables making objects with desirable functionalities at pre-designed locations on the object, such as generating a roughness that results in superhydrophobicity. Our previous work developed an ink composition that contains acrylic monomers. Using the DLP technology, an array of micro-pillars was printed to create a surface roughness at the micro-scale level. The roughness of the surface, which led to superhydrophobicity, was accomplished by the chemical composition (embedded hydrophobic nanoparticles) and the structural design (micro-pillars).8,20
Another approach to prevent fouling is making a hydrophilic surface through chemical grafting of amphiphilic molecules on the surface of the polymer.9,10,14,21–25 This approach is commonly used for organic and biomaterials.26–28 Specifically to prevent adhesion and contaminations by bacteria and other microorganisms, chemical modification of the polymer surfaces can be done using quaternary ammonium salt17 or by adding a polymer such as polyethyleneimine, natural extracts (i.e., essential oils) and metallic particles (i.e., copper, silver, and zinc).22,29–32
Reches and her co-workers designed a tripeptide that self-assembles into an antifouling coating.14,33,34 This peptide approach has several advantages since peptides are non-toxic, environmentally friendly, can spontaneously form a coating on various substrates,35–38 including metals, oxides, and polymers, and can be manufactured on a large scale.14,24,34,35,38–42
Here, we achieved both superhydrophobicity and antifouling properties of objects composed of an elastomer by combining additive manufacturing, which is a fabrication by 3D printing, and peptide chemistry. The elastomer composition reported in this research is used as a model for demonstrating a proof of concept and is composed mainly of urethan acrylate, epoxy aliphatic acrylate, and lauryl acrylate. This composition results in polyurethanes, which are commonly used polymers, in a variety of fields. This includes microfluidic for blood-contacting applications,43 surgical drapes,44 catheters,45,46 and food packaging.47 The additive manufacturing imparts the substrate with superhydrophobicity while the peptide coating provides the antifouling and antiviral activity. We applied a tripeptide, DOPA-Phe(4F)-Phe(4F)-OMe, on a 3D-printed superhydrophobic elastomer as an antifouling coating. The coating maintained superhydrophobicity with a water contact angle of ∼158° and a rolling angle of ∼7° after coating and endowed the antifouling and antiviral properties (80% decrease in E. coli and S. epidermidis). Our findings provide a promising approach to design and developing the superhydrophobic antifouling elastomers by the combination of 3D printing and peptide chemistry.
Results and discussion
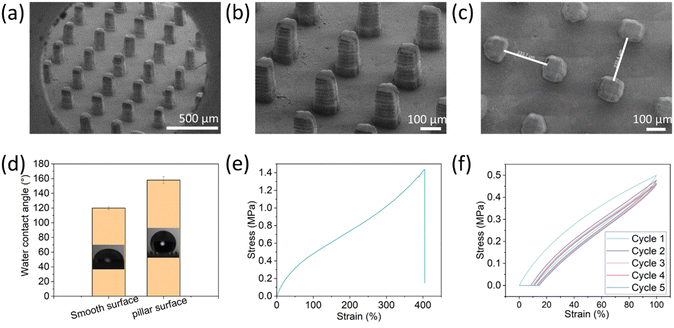
To generate the superhydrophobic surface, pillars at various dimensions and spacings were printed. The printing ink composition contains non-fluorinated monomers based on urethane acrylate with dispersed hydrophobic fumed silica (HFS) to introduce roughness and surface energy to the printed surfaces. Using the DLP method, an array of microscale pillars was printed to create the surface patterning at the scale of tens of micrometers. A micro-scale roughness of the surface was accomplished by the embedded hydrophobic nanoparticles. Overall, the superhydrophobicity was achieved by combining structural design (printed micro-pillars) and roughness by material properties (embedded particles).The resulting printed pillars were analyzed by SEM (Fig. 1). Their dimensions are 70 µm × 250 µm × 250 µm at the x, y, and z, respectively. As found earlier, for rigid polymeric substrates, these dimensions can provide the surface with superhydrophobic properties.26 Interestingly, the individual printed layers are visible at the Z-axis, and this structure may also contribute to the surface roughness at the Z-axis. The contact angle of the substrates with the printed pillars was 158° ± 5° compared to 120° ± 2° for a planar surface printed with the same composition (without pillars) (Fig. 1d). Moreover, the rolling angle of the 3D-printed pillar substrate was 7° ± 0.8°, while the rolling angle of the smooth surface was >50°. These values fit the definition of superhydrophobic surfaces (water contact angle larger than 150° and rolling angle less than 10°). These results indicate that the fabricated surface is superhydrophobic due to the microscale printed pillars that lead to air voids upon contact with water droplets, known as Cassie state wetting. Next, the mechanical properties of the printed elastomer were evaluated. As shown in Fig. 1e, the elastomer exhibited a strain-hardening behavior due to the limited extensibility of polymer chains,7 and the maximum strain was nearly 400%. We also evaluated the mechanical behavior with cyclic curves (5 times) to obtain the energy dissipation characteristics. During the cyclic loading and unloading process, the stress on reloading is observed to be lower than that on the initial loading for the same strain, resulting in a stress-softening phenomenon known as the Mullins effect.48 This effect contributes to a significant hysteresis loss in the first cycle, which is the ratio of dissipated energy to loaded energy, represented by the area of the curve and the integration of the loading curve, respectively. The hysteresis loss of the first cycle was about 26%.
To provide the superhydrophobic surface with antifouling properties, the printed surface was coated with the tripeptide DOPA-Phe(4F)-Phe(4F). The DOPA functional group can provide adhesion to any substrate.14,33,34 Phe(4F) residue can provide the self-assembly and antifouling properties.14 We have demonstrated that this tripeptide spontaneously self-assembles into a coating on various substrates at a low concentration (Fig. 2).38 This was done by immersing the 3D-printed elastomer substrate into a peptide solution at a concentration of 2 mg mL−1.38 After overnight incubation at room temperature, the substrate was washed with methanol to remove unattached peptide molecules and dried with nitrogen.
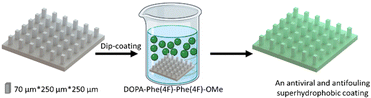 | ||
| Fig. 2 The scheme illustrates the coating process of the pillars by the peptide DOPA-Phe(4F)-Phe(4F)-OMe. | ||
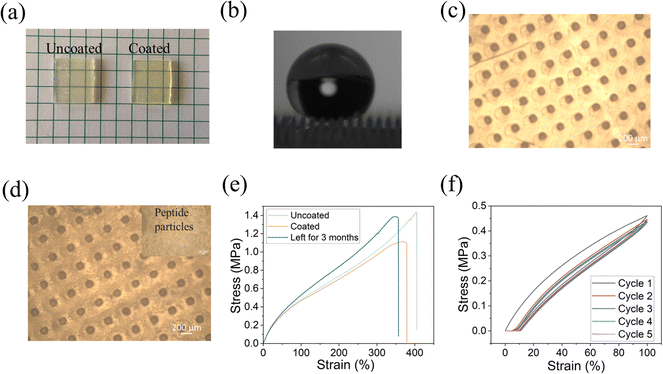
To confirm the attachment of the peptide to the printed substrate, we analyzed the surfaces by energy-dispersive X-ray spectroscopy (EDS) and optical microscopy. As shown in Fig. 3a, there was no significant visual change in morphology after coating the surface with the peptide. This is in agreement with our previous work regarding other substrates in which DOPA-Phe(4F)-Phe(4F)-Ome self-assembled into a transparent layer.14,35,36,38,41 From EDS analysis, we noted that the fluorine signal appeared all over the coated surface, while there was no fluorine signal on the uncoated surface (Fig. 3b). This suggests the formation of a coating on the surface by the self-assembly of the peptide DOPA-Phe(4F)-Phe(4F)-OMe. We used an optical microscope to study the morphology of the uncoated and coated elastomer substrates. We found that the peptide particles were attached to the 3D-printed substrate after coating (Fig. 3b and c), suggesting that the peptide bound on the elastomers. Importantly, the peptide coating did not change the structures of the pillars. To examine the elastic properties of the coated printed elastomer surface, we used the MTS criterion model 43 static mechanical tester to get stress–strain curves of uncoated and coated elastomers. The coated elastomer showed a similar maximum strain (nearly 400%) and hysteresis loss of the first cycle (∼25%) when compared to the uncoated one (Fig. 3e and f). Therefore, we inferred that the peptide coating did not alter the elastomer properties.
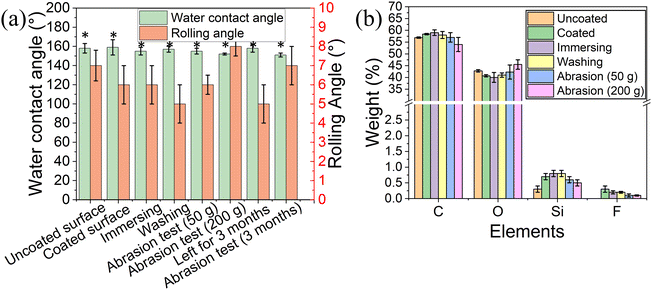
The water contact angle of the coated surface was 159° ± 8° and a rolling angle of 6° ± 1° (Fig. 4a), similar to the values obtained for the uncoated 3D printed elastomer substrate. This indicates that the peptide coating did not change the superhydrophobic characteristics of the surface, as the peptide only formed thin film on the surface.38 Our previous work demonstrated that the peptide DOPA-Phe(4F)-Phe(4F)-OMe can coat various substrates and increase the surface water contact angle.38 However, in this study, the surface superhydrophobicity was generated by the suitable roughness and surface energy. The peptide coating did not change the roughness of the elastomer substrates. These results revealed that the peptide can coat the substrates with a thin film that did not alter the superhydrophobicity of the substrate.
To determine the stability of the peptide coating on the 3D-printed surface, we immersed the coated elastomer substrate in distilled water for five minutes. Fig. 4a shows that the water contact angle and rolling angle did not decrease after immersing treatment (p > 0.05). Importantly, the fluorine signal could still be detected after the immersion in the water bath. These results suggest that the peptide coating on the 3D-printed elastomer substrate is of high stability. In addition, the water contact angle and rolling angle did not change upon washing the coated surfaces with distilled water three times indicating that the peptide coating is attached to the elastomer. (Fig. 4a).
It has been reported that the abrasion test is one of the most critical indicators for the stability of superhydrophobic coatings and soft materials. For this test, the coated elastomer substrate was placed against sandpaper (2000 grit size) with 50 g and 200 g weight on it and then was moved straight for ∼10 cm, denoted as one cycle.20 This test recorded the water contact angle, rolling angle, EDS analysis, and mechanical test after 10 cycles. As shown in Fig. 4a, the water contact angle and rolling angle did not alter significantly (p > 0.05), suggesting the high stability of the peptide coating. EDS analysis also demonstrated that the fluorine atoms could be detected after the abrasion test (Fig. 4b). This confirms that the peptide coating is still attached to the elastomer substrate after the abrasion test. The long-term storage stability of the superhydrophobic coating determined their practical application. We performed the tensile test and abrasion test after storage for 3 months. As shown in Fig. 3e, the coated elastomer after long-term storage had a similar maximum strain (nearly ∼350% strain) to the fresh elastomers. Moreover, we noted that the water contact angle (158° ± 4°) did not significantly change when compared to that of the fresh-coated elastomer. After 10 cycles of abrasion tests (200 g), the coated elastomer after storage for 3 months was still superhydrophobic (water contact angle > 150° and rolling angle < 10°) (Fig. 4a). These findings indicate that the long-term storage did not affect the coating stability and elastomer properties.
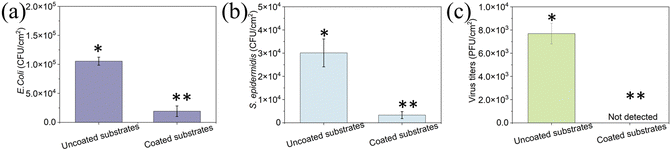
To determine the antifouling performance of the coating, we studied the attachment and biofilm formation of two bacterial strains: the Gram-negative E. coli and the Gram-positive S. epidermidis. Uncoated and coated surfaces were immersed in 107 CFU mL−1 bacterial suspension and incubated for 24 h to allow biofilm formation. A concentration of 1.1 × 105 CFU cm−2 on the uncoated surface was detected, while only 1.9 × 104 CFU cm−2E. coli was detected on the peptide-coated elastomer substrates. There was a decrease of ∼80% in E. coli on the coated surface compared to the uncoated elastomer substrate (p < 0.05) (Fig. 5a). Similarly, there was a significant reduction (80%) in S. epidermidis on the coated surface (p < 0.05) (Fig. 5b). These results indicate that the peptide coating could resist bacterial attachment and biofouling, showing antifouling activity in agreement with our previous findings that the tripeptide can form a coating on various substrates and present its ability to disrupt the biofouling process.38
To determine the antiviral activity of the uncoated and coated elastomer substrates, we incubated ∼106 PFU mL−1 T4 bacteriophages on each surface. T4 bacteriophage is a DNA-based virus that infects E. coli and causes them to burst. Fig. 5c presents the results of the inactivation of T4 bacteriophage for uncoated and coated elastomer substrates. The virus titers for uncoated elastomer substrates were 7.7 × 103 PFU cm−2. It was noted that no virus was detected on the coated elastomer substrates, indicating that it reduces the viral titers by 99.9% when compared to bare glass (Fig. 5c). Our previous work demonstrated that the antiviral activity for DOPA-Phe(4F)-Phe(4F)-OMe was generated by fluorinated phenylalanine and self-assemblies.49,50 DOPA is not essential for antiviral activity but is necessary when the peptide interacts with the 3D-printed elastomer substrates. In addition, we already proved in previous reports that the peptide-based coating is highly stable and can perform even after weeks in the presence of bacteria and even in biological fluids like saliva.36,51
Experimental
Materials
A bifunctional aliphatic urethane di-acrylate (AUD, Ebecryl 8413) and a monofunctional epoxy aliphatic acrylate (EAA, Ebecryl 113) were purchased from Allnex. A monofunctional monomer lauryl acrylate (SR335) was kindly provided by Sartomer-Arkema (Colombes Cedex, France). A photoinitiator diphenyl (2,4,6-trimethylbenzoyl) phosphine oxide (TPO) and acrylic acid (99%, stabilized, extra pure), were purchased from BASF (Ludwigshafen, Germany) and Acros organics respectively. Hydrophobic fumed silica particles, HFS (whose surface has been treated with dimethyldichlorosilane, TS-610), were kindly given by Cabot Specialty Chemicals Inc. Double distilled water was used for measuring the contact angle in all the experiments. Escherichia coli (ATCC# 25922) and Staphylococcus epidermidis (ATTC # 35984) bacteria were purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). Agar and lysogeny broth (LB) were purchased from Merck (New Jersey, USA) and Becton Dickinson (New Jersey, USA), respectively. Tryptic soy broth (TSB) for microbiology was purchased from Sigma Aldrich (St. Louis, Missouri, USA). E. coli strain B (Migula) Castellani and Chalmers (ATCC 11303) and E. coli bacteriophage T4 (ATCC 113030-B4) were purchased from ATCC (ATCC, Manassas, Virginia, USA).Printing composition
A typical 10 g composition for DLP printing was prepared as follows: 2.27 g of EA and 2.27 g of SR335 were mixed, followed by adding 0.20 g of TPO and then mixed in a sonication bath (Elmasonic P 30H) at 60 °C with 100% power at 80 kHz for 20 min. Afterward, 4.46 g of AUD was added to the mixture, followed by ∼10 min of mechanical mixing using DISPERMAT CV (Reichshof). Subsequently, 0.50 g of HFS particles were added to the mixture, followed by ∼10 min of mechanical mixing using DISPERMAT CV to obtain a homogenous dispersion. Then, 0.30 g of acrylic acid was added to the dispersion, followed by manual mixing. The dispersion underwent 5 min of defoaming using a planetary centrifugal mixer (Thinky) to obtain a clear dispersion without air bubbles.DLP 3D printing
Computer-aided design (CAD) files of arrays of square pillars with varying wall widths (x), spacings (y), and heights (z) were designed by Autodesk Inventor Professional 2023 software. These designs were converted to standard tessellation language (STL) or Stereolithography files. The STL files were further processed by the Asiga Max X35 UV (Asiga, Australia) 3D printer software, which sliced the design model into 2D cross-sections. The slicing is done according to a defined thickness of a single layer. The surfaces were printed using a light intensity of 25 mW cm−2 of the UV source at 385 nm. The initial layer, the burn layer, was exposed for 10 seconds, while all subsequent layers were irradiated for 1.2 seconds (Table S1, ESI†). Each printed layer of the pillar array had a thickness of 25 µm. To remove any uncured material trapped within the printed pillars, the printed surfaces were immersed in acetone for ∼10 min and dried by air pressure. Afterward, the printed surfaces underwent post-curing using a UV light (365 nm) for 15 min. To ensure gradual drying and complete post-curing, the surfaces were placed in an oven set at 60 °C for at least 1 h.SEM analysis for the printed elastomers
Images of the 3D printed pillars were acquired with an extra high-resolution scanning electron microscope (XHR-SEM) Magellan 400 L. Iridium was sputtered on the samples to avoid charging effects. The structures were mounted on a 4-axis motorized eccentric stage, and the instrument was operated in a low vacuum mode. Imaging was performed at an accelerating voltage of 2.0 kV and different magnifications (300×, 1300×, and 2500×). Elemental analysis was also performed by an energy dispersive X-ray detector (EDS, Quanta 200 FEG, FEI Company) attached to the SEM.Tensile test
The stress–strain curves of uncoated and coated substrates (1 cm × 1 cm) were acquired with MTS criterion model 43 (MTS Systems Corporation, Eden Prairie, MN, USA) static mechanical tester using a standard ISO procedure at a strain rate of 10 min−1. Cyclic tensile test measurements (5 times) were performed by stretching the samples to a limiting strain of 100% at the same strain rate. The hysteresis area was evaluated as the area difference between the loading and unloading curves.Abrasion test
The durability of the samples was evaluated by conducting a sandpaper abrasion test according to our previous work related to 3D-printed superhydrophobic elastomers.20 In brief, the sample was placed against a sandpaper (2000 grit size) with 200 g of weight. The sample was moved in a straight manner for ∼10 cm, and this was denoted as one cycle. 10 cycles were performed and the contact angle and rolling angle were recorded after these cycles.Preparation of peptide-based coatings
The peptide DOPA-Phe(4F)-Phe(4F)-OMe was dissolved in pure methanol to a concentration of 2 mg mL−1. Subsequently, the 3D polymeric objects were fully immersed in the peptide solution (horizontal) and were left overnight at room temperature. Then, the samples were rinsed extensively with methanol to remove excess peptides and were dried under nitrogen.Water contact angle measurements
Water contact angle and rolling angle were measured by Theta Lite optical tensiometer (Attension, Finland) using a sessile drop with a drop volume of 8 µL. Each experimental measurement had five repeats, and the reported angles were averaged.Rolling angle measurements
The rolling angle was measured by placing a drop of 8 µL distilled water on the printed surface, which has been fixed to a tiltable plate. Then, the plate was rotated slowly until the drop started to move. The corresponding angle was measured on a scale with a precision of about 0.5° and the angle from which the droplet started to move is referred to as the’rolling angle’. Five measurements were performed across two printed samples for each set of printed objects.Stability assay of the peptide-based elastomer coating
To examine the coating stability, the elastomer substrates coated by the peptide were immersed into TDW for 5 min or washed three times with 2 mL of TDW and then dried at room temperature according to our previous work.50 Moreover, peptide coatings were also tested by abrasion test as described above. The long-term stability was also performed after storage for 3 months. Then, the water contact angle, rolling angle measurements, and energy-dispersive X-ray spectroscopy (EDS) were performed.Antifouling assay
Uncoated and coated substrates (22 mm × 13 mm) were incubated overnight at 37 °C with either E. coli or S. epidermidis at a concentration of 107 CFU mL−1 in lysogeny broth (LB broth). Then, the surface was washed with PBS three times to remove non-adherent bacteria. Surfaces were transferred into a test tube containing 3 mL PBS. The test tubes were sonicated for 3 min and vortexed for 15 seconds to detach the bacteria from the substrates. Serial dilutions were made and seeded on the agar plates. The agar plates were incubated at 37 °C. Following an overnight incubation the number if colonies was recorded.Antiviral activity against bacteriophage T4
Ten decimal serial dilutions of the virus suspension were prepared using lysogeny broth (LB). Briefly, a drop of 16 µL of 1.0 × 106 PFU mL−1 bacteriophage T4 was placed on the uncoated and coated elastomer substrates (22 mm × 13 mm). Next, the phages were incubated under humid conditions at room temperature (25 °C) in a dark room for 24 h. After incubation, the phages were harvested by shaking with 2 mL of soya casein digest lecithin polysorbate (SCDLP) broth for 15 min to stop the incubation. The bacteriophage T4 in SCDLP broth was 10-fold diluted by LB. Subsequently, the samples and bacteria were mixed with 0.6% agarose. Then, the mixture was spread out on 1.5% LB agar to form a double agar layer. The plate was incubated at 37 °C for 18 h to form the plaques. For each sample, nine repeats were performed to assess the antiviral activity.Conclusions
This work demonstrates that the peptide DOPA-Phe(4F)-Phe(4F)-OMe coating on the 3D-printed superhydrophobic elastomer substrate results in antifouling and antiviral activity. In this study, for the 3D printing, we used a combination of monomers that results in polyurethane, as a model system to represent elastomers. Polyurethane is widely used in a variety of applications, and obviously a real application should be tailored to the specific product, and meet the requirements of this product, such as FDA approval for medical devices and food contact (leaching, degradation products, cell toxicity, etc.). Importantly, the peptide coating did not alter the superhydrophobicity of the printed elastomer as the water contact angle (159° ± 8°) and rolling angle (6° ± 1°) after coating did not decrease. Moreover, EDS analysis revealed that the peptide was tightly attached to the pillars-structured elastomer substrates. It was found that the peptide coating was stable even after performing an abrasion test. After coating, the printed structures showed excellent antifouling and antiviral activity. Antifouling results showed an 80% decrease in Gram-positive and Gram-negative bacteria compared to the uncoated surfaces. In addition, no viruses could be detected on the coated surfaces. After coating, the superhydrophobicity was still supported by the micropillars embedded with hydrophobic nanoparticles rather than the peptide itself. The peptide monolayer did not change the superhydrophobicity but provided the antifouling and antiviral properties. Our findings provide a new approach for making superhydrophobic elastomers with antifouling and antiviral functionalities. This approach of combining structuring for superhydrophobicity and peptide-coating can be applied wherever substrates are needed to reduce the risk of viral transmission and microbial adhesion for soft materials.Author contributions
The manuscript was written through the contributions of all authors. Meital Reches and Shlomo Magdassi: conceptualization, resources, visualization, validation, supervision, project administration, writing – reviewing and editing. Tan Hu and Noa Trink: methodology, data curation, writing – original draft preparation, formal analysis, visualization, writing – reviewing, and editing.Data availability
All data is available upon request for the corresponding authors.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This research was supported by grants from the National Research Foundation, Prime Minister's Office, Singapore, under its Campus of Research Excellence and Technological Enterprise (CREATE) program (grant 370184510).Notes and references
- D. Feldman, J. Macromol. Sci., Part A: Pure Appl. Chem., 2012, 49, 784–793 CrossRef CAS.
- M. Maiti, M. Bhattacharya and A. K. Bhowmick, Rubber Chem. Technol., 2008, 81, 384–469 CrossRef CAS.
- P. Hu, Y. Zhang, S. Zhou, T. Chen, D. Wang, T. Liu, Y. Wang, J. Chen, Z. Wang, J. Xu and J. Fu, Chem. Eng. J., 2023, 464, 142543 CrossRef CAS.
- S. Tang, J. Li, R. Wang, J. Zhang, Y. Lu, G.-H. Hu, Z. Wang and L. Zhang, SusMat, 2022, 2, 2–33 CrossRef CAS.
- M. M. Duran, G. Moro, Y. Zhang and A. Islam, Adv. Ind. Manuf. Eng., 2023, 7, 100125 Search PubMed.
- H. Yuk, B. Lu, S. Lin, K. Qu, J. Xu, J. Luo and X. Zhao, Nat. Commun., 2020, 11, 1604 CrossRef CAS PubMed.
- P. Huang, H. Fu, M. W. M. Tan, Y. Jiang and P. S. Lee, Adv. Mater. Technol., 2023, 2301642 Search PubMed.
- D. K. Patel, A. H. Sakhaei, M. Layani, B. Zhang, Q. Ge and S. Magdassi, Adv. Mater., 2017, 29, 1606000 CrossRef PubMed.
- R. Deng, T. Shen, H. Chen, J. Lu, H.-C. Yang and W. Li, J. Mater. Chem. A, 2020, 8, 7536–7547 RSC.
- W. Yao, L. Wu, L. Sun, B. Jiang and F. Pan, Prog. Org. Coat., 2022, 166, 106806 CrossRef CAS.
- J. H. Waite and M. L. Tanzer, Science, 1981, 212, 1038–1040 CrossRef CAS PubMed.
- D. Wang, Q. Sun, M. J. Hokkanen, C. Zhang, F.-Y. Lin, Q. Liu, S.-P. Zhu, T. Zhou, Q. Chang, B. He, Q. Zhou, L. Chen, Z. Wang, R. H. A. Ras and X. Deng, Nature, 2020, 582, 55–59 CrossRef CAS PubMed.
- A. M. C. Maan, A. H. Hofman, W. M. de Vos and M. Kamperman, Adv. Funct. Mater., 2020, 30, 2000936 CrossRef CAS.
- A. Dolid, L. C. Gomes, F. J. Mergulhão and M. Reches, Colloids Surf., B, 2020, 196, 111365 CrossRef CAS PubMed.
- J. Ju, X. Yao, X. Hou, Q. Liu, Y. S. Zhang and A. Khademhosseini, J. Mater. Chem. A, 2017, 5, 16273–16280 RSC.
- N. Zheng, X. Zhang, X. Min, J. Liu, W. Li and X. Ji, React. Funct. Polym., 2019, 143, 104329 CrossRef CAS.
- S. Dobretsov and J. C. Thomason, Biofouling, 2011, 27, 869–880 CrossRef CAS PubMed.
- K. Lv, J. Yu, K. Deng, J. Sun, Y. Zhao, D. Du and M. Li, J. Hazard. Mater., 2010, 173, 539–543 CrossRef CAS PubMed.
- V. C. Malshe and N. S. Sangaj, Prog. Org. Coat., 2005, 53, 207–211 CrossRef CAS.
- G. Kaur, A. Marmur and S. Magdassi, Addit. Manuf., 2020, 36, 101669 CAS.
- H.-C. Flemming, Appl. Microbiol. Biotechnol., 2002, 59, 629–640 CrossRef CAS PubMed.
- S. Behzadinasab, A. Chin, M. Hosseini, L. Poon and W. A. Ducker, ACS Appl. Mater. Interfaces, 2020, 12, 34723–34727 CrossRef CAS PubMed.
- N. Ashkenasy, W. S. Horne and M. R. Ghadiri, Small, 2006, 2, 99–102 CrossRef CAS PubMed.
- T. Hu, M. Kaganovich, Z. Shpilt, A. Pramanik, O. Agazani, S. Pan, E. Tshuva and M. Reches, Adv. Mater. Interfaces, 2023, 2202161 CrossRef CAS.
- I. Tagliaro, S. Seccia, B. Pellegrini, S. Bertini and C. Antonini, Carbohydr. Polym., 2023, 302, 120424 CrossRef CAS PubMed.
- A. Hooda, M. S. Goyat, J. K. Pandey, A. Kumar and R. Gupta, Prog. Org. Coat., 2020, 142, 105557 CrossRef CAS.
- I. S. Bayer, Adv. Mater. Interfaces, 2020, 7, 2000095 CrossRef CAS.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426–430 CrossRef CAS PubMed.
- D. Rahmawati, M. Chandra, S. Santoso and M. G. Puteri, AIP Conf. Proc., 1803, 2017, 20037 Search PubMed.
- M. Sarraf, A. Dabbagh, B. A. Razak, R. Mahmoodian, B. Nasiri-Tabrizi, H. R. M. Hosseini, S. Saber-Samandari, N. H. A. Kasim, H. Abdullah and N. L. Sukiman, Surf. Coat. Technol., 2018, 349, 1008–1017 CrossRef CAS.
- Z. Sun and K. Ken Ostrikov, Sustainable Mater. Technol., 2020, 25, e00203 CrossRef CAS.
- A. Kuroki, J. Tay, G. H. Lee and Y. Y. Yang, Adv. Healthcare Mater., 2021, 10, 2101113 CrossRef CAS PubMed.
- A. Dolid and M. Reches, J. Pept. Sci., 2019, 25, e3212 CrossRef CAS PubMed.
- S. L. Gaw, G. Sakala, S. Nir, A. Saha, Z. J. Xu, P. S. Lee and M. Reches, Biomacromolecules, 2018, 19, 3620–3627 CrossRef CAS PubMed.
- S. Nir, D. Zanuy, T. Zada, O. Agazani, C. Aleman, D. E. Shalev and M. Reches, Nanoscale, 2019, 11, 8752–8759 RSC.
- A. Friedlander, S. Nir, M. Reches and M. Shemesh, Front. Microbiol., 2019, 10, 1405 CrossRef PubMed.
- S. Yuran, A. Dolid and M. Reches, ACS Biomater. Sci. Eng., 2018, 4, 4051–4061 CrossRef CAS PubMed.
- S. Maity, S. Nir, T. Zada and M. Reches, Chem. Commun., 2014, 50, 11154–11157 RSC.
- H. Acar, S. Srivastava, E. J. Chung, M. R. Schnorenberg, J. C. Barrett, J. L. LaBelle and M. Tirrell, Adv. Drug Delivery Rev., 2017, 110–111, 65–79 CrossRef CAS PubMed.
- W.-Y. Chen, H.-Y. Chang, J.-K. Lu, Y.-C. Huang, S. G. Harroun, Y.-T. Tseng, Y.-J. Li, C.-C. Huang and H.-T. Chang, Adv. Funct. Mater., 2015, 25, 7189–7199 CrossRef CAS.
- G. P. Sakala and M. Reches, Adv. Mater. Interfaces, 2018, 5, 1800073 CrossRef.
- T. Hu, M. Kaganovich, Z. Shpilt, A. Pramanik, O. Agazani, S. Pan, E. Tshuva and M. Reches, Adv. Mater. Interfaces, 2023, 10, 2202161 CrossRef CAS.
- W.-I. Wu, K. N. Sask, J. L. Brash and P. R. Selvaganapathy, Lab Chip, 2012, 12, 960–970 RSC.
- B. Rogina-Car, S. Kovačević, S. Đorđević and D. Đorđević, Polymers, 2020, 12, 642 CrossRef CAS PubMed.
- P. Francois, P. Vaudaux, N. Nurdin, H. J. Mathieu, P. Descouts and D. P. Lew, Biomaterials, 1996, 17, 667–678 CrossRef CAS PubMed.
- A. Das and P. Mahanwar, Adv. Ind. Eng. Polym. Res., 2020, 3, 93–101 Search PubMed.
- D. Turan, Food Eng. Rev., 2021, 13, 54–65 CrossRef CAS.
- J. Diani, B. Fayolle and P. Gilormini, Eur. Polym. J., 2009, 45, 601–612 CrossRef CAS.
- A. S. Skwarecki, M. G. Nowak and M. J. Milewska, ChemMedChem, 2021, 16, 3106–3135 CrossRef CAS PubMed.
- T. Hu, O. Agazani, S. Nir, M. Cohen, S. Pan and M. Reches, ACS Appl. Mater. Interfaces, 2021, 13, 48469–48477 CrossRef CAS PubMed.
- D. Fang, S. Yuran, M. Reches, R. Catunda, L. Levin and M. Febbraio, J. Periodontal Res., 2020, 55, 503–510 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma00620h |
| ‡ Shlomo Magdassi and Meital Reches are the corresponding authors. |
| This journal is © The Royal Society of Chemistry 2024 |