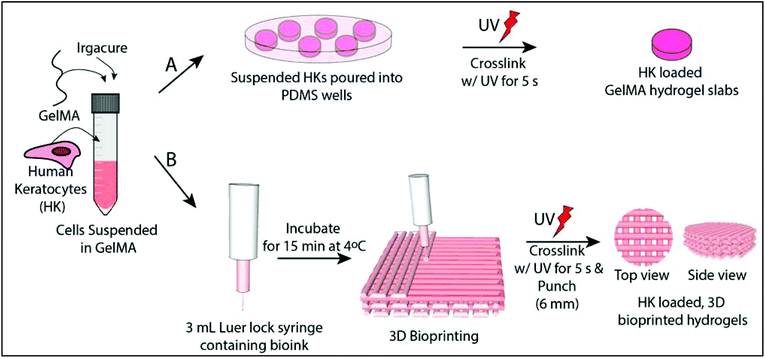
Cell loaded 3D bioprinted GelMA hydrogels for corneal stroma engineering†
Cemile
Kilic Bektas
 abc and
Vasif
Hasirci
abc and
Vasif
Hasirci
 *abcd
*abcd
aDepartment of Biological Sciences, Middle East Technical University (METU), Ankara, Turkey
bDepartment of Biotechnology, METU, Ankara, Turkey
cBIOMATEN, METU Center of Excellence in Biomaterials and Tissue Engineering, Ankara, Turkey
dAcıbadem Mehmet Ali Aydınlar University, Faculty of Engineering, Department of Medical Engineering, İstanbul, Turkey. E-mail: vasif.hasirci@acibadem.edu.tr
First published on 4th November 2019
Abstract
Tissue engineering aims to replace missing or damaged tissues and restore their functions. Three-dimensional (3D) printing has been gaining more attention because it enables the researchers to design and produce cell loaded constructs with predetermined shapes, sizes, and interior architecture. In the present study, a 3D bioprinted corneal stroma equivalent was designed to substitute for the native tissue. Reproducible outer and inner organization of the stroma was obtained by optimizing printing conditions such as the nozzle speed in the x–y direction and the spindle speed. 3D printed GelMA hydrogels were highly stable in PBS during three weeks of incubation (8% weight loss). Live–Dead cell viability assay showed 98% cell viability on day 21 indicating that printing conditions were suitable for keratocyte printing. Mechanical properties of the cell loaded 3D printed hydrogels increased 2-fold during this incubation period and approached those of the native cornea (ca. 20 kPa vs. 27 kPa, respectively). Expression of collagens types I and V, and proteoglycan (decorin) in keratocytes indicates maintenance of the phenotype in the hydrogels. Transparency of cell-loaded and cell-free hydrogels was over 80% (at 700 nm) during the three week culture period and comparable to that of the native cornea (85%) at the same wavelength. Thus, GelMA hydrogels bioprinted with keratocytes mimic the biological and physical properties of the corneal stroma with their excellent transparency, adequate mechanical strength, and high cell viability.
1. Introduction
The cornea, the outermost and transparent layer of the eye, is about 11–12 mm in diameter and 520 μm in thickness.1 It is the principal optical element of the eye which refracts about 65–75% of the incoming light. It also acts as a barrier against UV radiation and external and physical objects.2 The stroma is the thickest part of the cornea and constitutes 90% of the thickness. Collagen type I fibers are the main components of this layer and are organized parallel to each other in one layer and at nearly right angles to the fibers in adjacent layers, much like the fibers in plywood.3 This organization is critical for both the mechanical strength and the transparency of the cornea.4 The stroma is populated by sparsely distributed mesenchymal fibroblasts, keratocytes, which preserve the composition and the integrity of the stroma through continuous production of collagen, proteoglycan, and matrix metalloproteinases.5Corneal blindness caused by injuries or diseases affects the quality of life of millions of people. It is the third major cause of blindness after cataracts and glaucoma. Currently, transplantation is the major and long-term treatment; however, due to shortage of healthy corneas, only 1 in 70 patients is successful in getting one.6 Keratoprostheses (KPro) are partly synthetic artificial corneas for full thickness replacements. Boston KPro and Osteo-odonto KPro are the two commonly used products on the market.7 However, degradation of lamina, chronic inflammation, complex design, long surgical procedures, and calcification are some of their reported drawbacks.8 These imply that alternative, efficient approaches are needed to meet the demand and overcome the limitations of the current treatment methods.
Tissue engineering is a viable alternative to the abovementioned treatments. The complex organization, high transparency and mechanical property requirements make engineering of the corneal stroma a great challenge. To date, a number of scaffolds have been developed for corneal tissue engineering by either mimicking only one layer (epithelium, stroma, or endothelium), two layers (hemi-cornea; epithelium and stroma), or all layers of the cornea.9–12 These constructs were reported to be produced in various forms like foams, fibers, films, and decellularized cornea tissue.9,11,13–16 Scaffolds were constructed either using natural materials like silk, gelatin, collagen, and chitosan, or synthetic materials like polyglycolic acid (PGA) and poly(lactic acid-co-glycolic acid) (PLGA).10,17–21 Insufficient cellular population of the scaffolds, lack of micro-level organization of the corneal stroma, and low light transfer were reported in these studies. Although some of the replacements closely mimicked the physical/chemical organization of the stroma, only a few could manage to reach Phase I of clinical trials.22 Today there is still no engineered corneal tissue available for routine clinical use.
Gelatin is one of the most widely used natural materials because it is inexpensive, is available from a range of different sources, does not induce any antigenic response, and is suitable for attachment by cells due to its natural cell binding motifs like arginine–glycine–aspartic acid (RGD).23 Hydrogels composed of gelatin are highly attractive and widely studied in corneal tissue engineering.19,24 Hydrogels, hydrophilic polymeric networks, are appealing for corneal tissue engineering applications due to their similarity to the natural extracellular matrices, biocompatibility and structural integrity.25 Researchers have employed a variety of chemicals for the crosslinking of the gelatin like glutaraldehyde, carbodiimide, and NHS.26,27 However, these approaches have several disadvantages like inability to load cells during gel formation, long processing time, and the use of toxic or bioactive chemicals in crosslinking. Gel formation through photocrosslinking using UV or visible light, therefore, is a popular hydrogel forming technique due to the fast and controllable polymerization and the ability to load cells and other biological molecules into the hydrogel during crosslinking. Gelatin can be conveniently modified by methacrylation and form photocrosslinkable methacrylated gelatin (GelMA). Very short UV exposure (as short as 5 s) and the absence of organic molecules make loading of cells possible during crosslinking.16,28 GelMA has been used in many tissue engineering applications including meniscus,29 skin,30 and bone.31 Its suitability for corneal wound healing when used together with thiolated gelatin has also been reported.32 Our group has reported for the first time the use of GelMA hydrogels in corneal stroma engineering.16 The hydrogels were highly promising in terms of stability, biocompatibility, and transparency. However, they lacked the internal organization of a typical corneal stroma that is crucial for producing a satisfactory corneal stroma. In this study, we improved our product by introducing organization of the keratocytes within the hydrogel that highly mimics the organization in the native structure of the corneal stroma by employing 3D bioprinting. Thus, we achieved organized placement of the keratocytes within the hydrogels to form a construct that more closely mimics the natural tissue.
3D printing is commonly used to build complex structures, including scaffolds, by layer-by-layer deposition of biomaterials by using computer aided design (CAD) data sets.25,33 3D bioprinted corneal tissue equivalents have significant potential because of their transparency, high water content and the elasticity requirements of the tissue. These requirements of the tissue are provided by the hydrogels and complex organization of the tissue is mimicked by 3D bioprinting.
3D printing of corneal stroma constructs is very new and a few strategies have been reported by a small number of researchers.34,35 However, either the inability of the proposed model to mimic the natural organization of the corneal stroma or the high complexity of the system has limited the widespread use of this technique.
In this study, a highly transparent GelMA hydrogel was produced, preparation conditions were optimized, and hydrogels loaded with stromal keratocytes were printed using a simple pneumatic extrusion based bioprinter. The model consisted of parallel fibers within a layer, and adjacent layers at 90° to each other as in the native structure of the stroma. The organization of the stroma was mimicked for the first time by 3D bioprinting of GelMA hydrogels. High cell viability, transparency and adequate mechanical strength obtained with these hydrogels make the construct a strong alternative to the allografts, which are already in short supply.
2. Materials and methods
2.1 Methacrylated gelatin synthesis
Methacrylated gelatin (GelMA) was synthesized from the reaction of methacrylic anhydride (MA) (Sigma Aldrich, USA) and type A porcine skin gelatin (70–100 bloom, Sigma Aldrich, USA) as described previously.16 Briefly, 10% (w/v) gelatin solution was prepared in phosphate buffered saline (PBS, pH 7.4, 10 mM) at 60 °C. MA was added to the solution at 50 °C slowly to yield a 20% (v/v) final concentration. After 1 h, the reaction was stopped with warm PBS at 40 °C. Excess methacrylic acid and salts were removed by dialysis (SnakeSkin CO 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Hyclone, USA) against distilled water at 40 °C. The resulting solution was freeze dried (Labconco Freezone 6, USA) and stored at +4 °C until further use.
000, Hyclone, USA) against distilled water at 40 °C. The resulting solution was freeze dried (Labconco Freezone 6, USA) and stored at +4 °C until further use.
2.2 1H nucleic magnetic resonance (1H-NMR) of GelMA
Lyophilized GelMA and gelatin were dissolved in D2O (30 mg mL−1) at 40 °C. 1H NMR spectra were obtained using a Bruker DPX 400 spectrometer (Germany) at a 1H resonance frequency of 400 MHz. The degree of methacrylation was 70% as reported earlier.162.3 Preparation of GelMA hydrogel slabs
Hydrogel slabs were prepared on polydimethylsiloxane (PDMS) (Slygard 184, Dow Corning, USA) templates. PDMS templates were prepared by mixing a PDMS prepolymer and catalyst, pouring into glass Petri dishes and curing at 70 °C for 3 h. The resultant PDMS film was peeled off and small discs with different dimensions were prepared (h = 0.5 mm/r = 3.5 mm and h = 8 mm/r = 5 mm). 15% GelMA solution (GelMA15) (w/v, in PBS) was prepared in the presence of 0.5% Irgacure 2959 photoinitiator (2-(hydroxyl)-4-(2-hydroxyethoxy)-2-methylpropiophenone, Sigma Aldrich, USA) (w/v). The solution was poured into the PDMS templates, incubated for 15 min at 4 °C, and crosslinked for 5 s with OmniCure (S1500, Lumen Dynamics, Canada) (15 mW cm−2, at 365 nm) at a distance of 3 cm. The hydrogels were labeled as GelMA15-Slab.2.4 Preparation of 3D printed GelMA hydrogels
GelMA15 solution was prepared as mentioned above and loaded into a 3 mL Luer lock syringe compatible with a Bioscaffolder® (SYS-ENG, Salzgitter-Bad, Germany) 3D Printer low temperature dispense head, and incubated at 4 °C for 15 min. GelMA was printed at a movement speed of the nozzle in the x–y direction (Fxy) of 100, 200, or 300 mm min−1, a spindle speed (R/S) of 0.01, 0.02, or 0.03 dots per mm, and from a nozzle with a diameter of 0.26 mm (25 ga × ½ Luer stubs, Instech, USA). The layer thickness and the distance between two strands were set as 0.14 mm and 1 mm, respectively.After observation under a stereomicroscope, the following parameters were chosen for further studies: (1) Fxy 200 and R/S 0.01 (GelMA15-001), (2) Fxy 200 and R/S 0.2 (GelMA15-002), and (3) Fxy 300 and R/S 0.03 (GelMA15-003). Rectangular prism GelMA hydrogels (14 × 14 × 2 mm3 were prepared for mechanical tests, and 14 × 14 × 0.5 mm3 hydrogels were prepared for other studies (Table 1). Constructs were plotted according to a model prepared using Sketchup (Google Inc., USA) and loaded to the CAM (Computer Aided Manufacturing) software (Prim-CAM, Einsiedeln, Switzerland) of the Bioscaffolder®. Two dimensional (2D) layers were deposited perpendicular to each other at every layer. Models were crosslinked and stored as mentioned above.
| Spindle Speed (R/S) (dots per min) | Nozzle Speed (Fxy) (mm min−1) | Total number of layers | Abbreviation |
|---|---|---|---|
| 0.01 | 200 | 15 – mechanical tests | GelMA15-001 |
| 5 – in situ and in vitro tests | |||
| 0.02 | 200 | 10 layers-mechanical tests | GelMA15-002 |
| 3 layers- in situ and in vitro tests | |||
| 0.03 | 300 | 10 layers-mechanical tests | GelMA15-003 |
| 3 layers- in situ and in vitro tests |
2.5 Characterization of the hydrogels
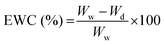 | (1) |
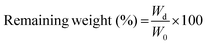 | (2) |
2.6 In vitro studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Thermo Fisher Scientific, USA), newborn calf serum (Thermo Fisher Scientific, USA) (10%), amphotericin B (Sigma Aldrich, USA) (1 μg mL−1), penicillin (100 UI mL−1), and streptomycin (Sigma Aldrich, USA) (100 μg mL−1), and Human basic fibroblast growth factor (bFGF, Cell Signaling Technology, Inc., USA) (10 ng mL−1)). Cells were cultured in tissue culture polystyrene (TCPS) flasks in a CO2 incubator (SanyoMCO-17 AIC, Japan) at 37 °C. Cells were trypsinized using trypsin-EDTA (0.25%, Sigma Aldrich, USA) at confluence and counted with a NucleoCounter (Chemo-Metec, Denmark). 1 × 106 cells per mL was taken in a separate centrifuge tube, centrifuged and suspended directly with GelMA solution prepared with growth medium. The gel precursors were exposed to UV as mentioned above (Fig. 1A). The hydrogel was washed several times with growth medium and incubated in the same medium at 37 °C in a CO2 incubator. The medium was changed every two days.
1, Thermo Fisher Scientific, USA), newborn calf serum (Thermo Fisher Scientific, USA) (10%), amphotericin B (Sigma Aldrich, USA) (1 μg mL−1), penicillin (100 UI mL−1), and streptomycin (Sigma Aldrich, USA) (100 μg mL−1), and Human basic fibroblast growth factor (bFGF, Cell Signaling Technology, Inc., USA) (10 ng mL−1)). Cells were cultured in tissue culture polystyrene (TCPS) flasks in a CO2 incubator (SanyoMCO-17 AIC, Japan) at 37 °C. Cells were trypsinized using trypsin-EDTA (0.25%, Sigma Aldrich, USA) at confluence and counted with a NucleoCounter (Chemo-Metec, Denmark). 1 × 106 cells per mL was taken in a separate centrifuge tube, centrifuged and suspended directly with GelMA solution prepared with growth medium. The gel precursors were exposed to UV as mentioned above (Fig. 1A). The hydrogel was washed several times with growth medium and incubated in the same medium at 37 °C in a CO2 incubator. The medium was changed every two days.
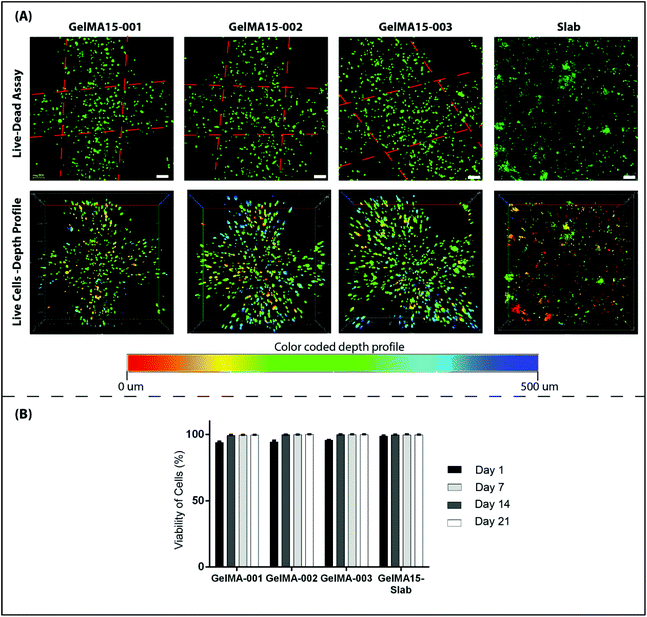
2.6.3.1 Cell viability. Cell viability was assessed by using a Live–Dead cell viability/cytotoxicity kit (Thermo Fisher Scientific, USA). Hydrogels were removed from the culture on days 1, 7, 14 and 21, and double stained with Calcein-AM (2 μM in PBS) and ethidium homodimer (EthD)-1 (4 μM) for 30 min at room temperature (RT). Hydrogels were examined under a Zeiss LSM 800 (Germany) Confocal Laser Scanning Microscope (CSLM). Semi-quantitative image analysis (n = 3) was performed using ImageJ NIH software to determine the viability of keratocytes (%) in the hydrogels according to the following equation:
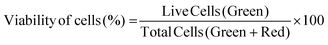 | (3) |
Metabolic activity of the cells was monitored using Alamar Blue® assay (Invitrogen Inc., USA). Samples (n = 3) were incubated in Alamar Blue solution (10% v/v in colorless growth medium) for 2 h. Reduction (%) of the dye was calculated according to the manufacturer's instructions using absorbance values obtained at 570 nm and 595 nm.
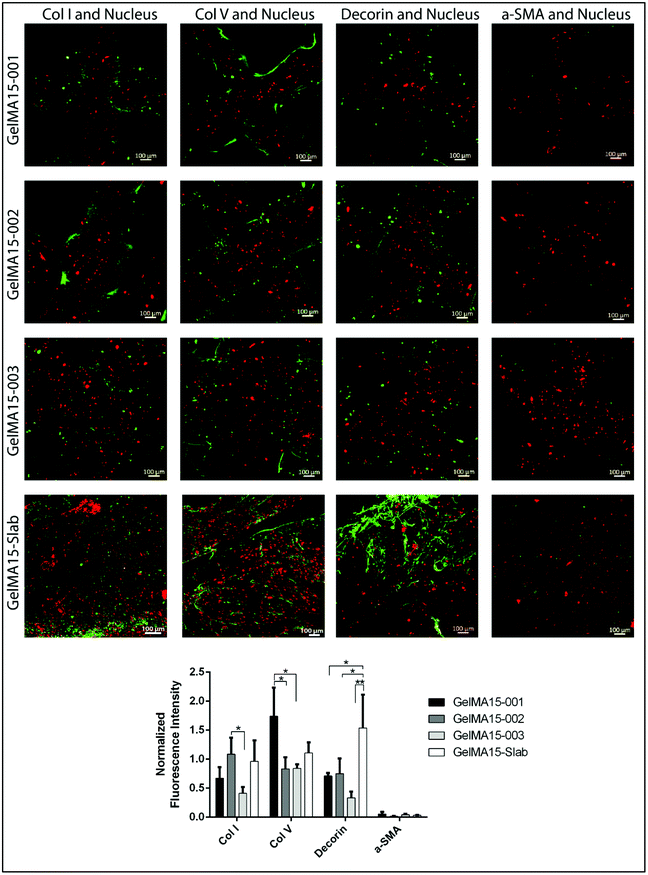
2.6.3.2 Immunofluorescence staining. Hydrogels were removed from the growth medium, HKs were fixed with 4% paraformaldehyde (PFA, Sigma Aldrich, USA) (w/v) for 30 min at RT, and their cell membranes were permeabilized with 1% Triton X-100 (AppliChem, USA) (v/v, in PBS) for 5 min. For blocking, samples were incubated in 1% bovine serum albumin (BSA, Sigma Aldrich, USA) (w/v, in PBS) at 37 °C for 1 h. Solutions of primary antibodies against collagen type I (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 v/v), collagen type V (1
100 v/v), collagen type V (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 v/v), decorin (4.8 μg mL−1), and α-SMA (1
100 v/v), decorin (4.8 μg mL−1), and α-SMA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 v/v) (all from Abcam Inc., MA) were prepared in 0.1% BSA (w/v, in PBS). Hydrogels were incubated with these primary antibodies overnight at 4 °C. The next day, samples were washed with PBS twice for 5 min and incubated with secondary antibodies (either anti-rabbit or anti-mouse Alexafluor 488 secondary antibody) (Invitrogen Inc., USA) (1
100 v/v) (all from Abcam Inc., MA) were prepared in 0.1% BSA (w/v, in PBS). Hydrogels were incubated with these primary antibodies overnight at 4 °C. The next day, samples were washed with PBS twice for 5 min and incubated with secondary antibodies (either anti-rabbit or anti-mouse Alexafluor 488 secondary antibody) (Invitrogen Inc., USA) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 v/v in 0.1% BSA in PBS) at 37 °C for 1 h. Hydrogels were then rinsed twice for 5 min and nuclei of the cells were stained with DRAQ5 (Thermo Fisher Scientific, USA) (1
100 v/v in 0.1% BSA in PBS) at 37 °C for 1 h. Hydrogels were then rinsed twice for 5 min and nuclei of the cells were stained with DRAQ5 (Thermo Fisher Scientific, USA) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 v/v in 0.1% BSA in PBS) for 1 h, at RT. Hydrogels were rinsed twice and stored in PBS until examination by CLSM.
1000 v/v in 0.1% BSA in PBS) for 1 h, at RT. Hydrogels were rinsed twice and stored in PBS until examination by CLSM.
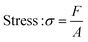 | (4) |
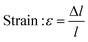 | (5) |
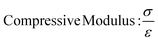 | (6) |
2.7 Statistical analysis
The GraphPad Prism program (Version 6.01) was used for statistical analysis. Two-way analysis of variance (ANOVA) with Tukey's post hoc test or Student's t test was used depending on the number of comparisons. Data are given as mean ± standard deviation (SD) (n = 3) and p ≤ 0.05 was reported as statistically significant.3. Results and discussion
3.1 Optimization of printing conditions
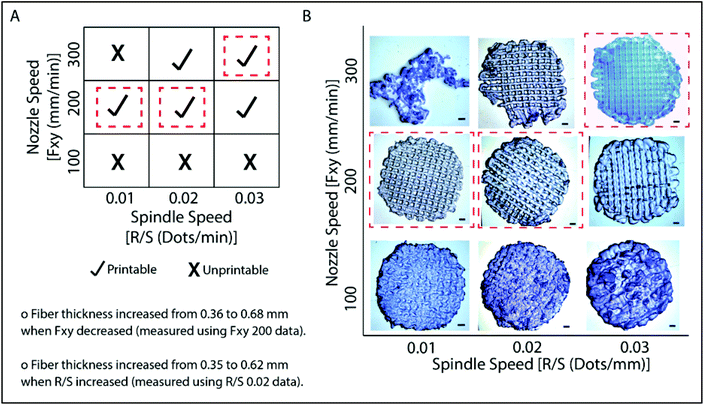
Models created by the Sketchup program were transferred as .stl files to the 3D printer, Bioscaffolder®. A low temperature dispense head was used and the fiber thickness was controlled with parameters such as the movement speed of the nozzle in the x–y direction (Fxy, mm min−1) and spindle speed (R/S, dots per min). The thickness of the fibers decreased as Fxy increased (Fig. 2). On the other hand, the fiber thickness increased as R/S increased. Printing conditions were optimized as presented in Fig. 2A. It was observed that the nozzle speed has an optimum at 200 mm min−1 when the spindle speed was set at 0.01 or 0.02 dots per min. Under these conditions, the gaps are well defined and the line thicknesses are quite consistent (Fig. 2B). Similarly, the model was properly printed at 300 mm min−1 nozzle speed and 0.03 dots per min spindle speed. These parameters were used in the following studies. Hydrogels which were prepared as slabs (section 2.3) were labeled as GelMA15-Slab. All 3D printed and slab hydrogels were stable after crosslinking. SEM images of the selected hydrogels clearly show the patterns and open pore structure of the scaffolds (Fig. S2†). Hydrogels retained a significant amount of water in their structures (ca. 90%), which is important to give cells space to proliferate.3.2 Degradation of hydrogels under in situ conditions
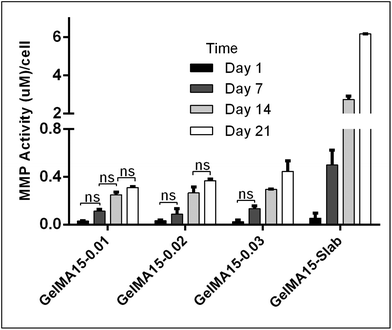
The in situ stability of the hydrogels was studied by incubating in PBS (pH 7.4, 10 mM, 0.5 mg mL−1 sodium azide) for three weeks. 3D printed and slab hydrogels were very stable and lost only 8% of their initial weights in 21 days (Fig. 3). Degradation rates of all hydrogels were similar and there was no statistically significant difference between them. The high stability of the hydrogels is most probably due to two reasons: (1) a high UV intensity was used in crosslinking, and (2) 3D printed hydrogels were incubated at 4 °C prior to crosslinking for physical gelation. The stability of the hydrogels, therefore, was enhanced due to dual crosslinking mechanisms which are physical (low temperature) and chemical (due to UV exposure). Other researchers reported similar results in terms of enhanced stability and mechanical properties due to physical gelation prior to chemical crosslinking.37–39 This phenomenon is known as the sol–gel transition which is induced thermally and a physical crosslinking takes place through noncovalent interactions.40 Most of the natural polymers like cellulose,41 agarose,42 and gelatin43 exhibit sol–gel behavior upon reduced temperature. Researchers suggest that crosslinking after these physical associations results in a more efficient covalent bond formation due to increased proximity of the polymer chains.38 In this study, physical gelation enhanced the production of the planned pattern and product; otherwise it is impossible to obtain printed architecture in a liquid phase.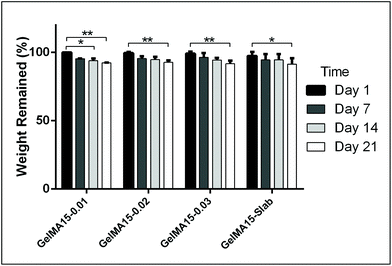 | ||
| Fig. 3 In situ stability test of 3D printed hydrogels (in PBS, pH 7.4, 10 mM) for 3 weeks. *p < 0.05, **p < 0.01, and ***p < 0.001. | ||
3.3 In vitro studies
Mesenchyme-derived fibroblasts, keratocytes, are sparsely distributed in the stroma and normally form an interconnected cellular network.5 However, HKs loaded in 3D printed hydrogels were mostly round in shape, most probably due to a tight network caused by physical crosslinking prior to UV exposure. This might limit the mobility of the cells and prevent cell interaction and elongation. Studies suggest that cells try to form stable contacts with the surrounding cells through their extensions and if this contact does not occur, they retract them.47 Cells in the hydrogels, therefore, might not interact with each other due to dense crosslinking and may not elongate. Similar results were reported by other researchers; for example gradually increasing concentration of GelMA hydrogels (from 5% to 30%) led to an increase in crosslink density and rigidity and thus to more round shaped cells.39 In several studies, researchers attempted to develop new methodologies like pre-crosslinking of the hydrogel solution (GelMA + gelatin or alginate) physically by thermal or ionic interactions to enhance printability of GelMA at low concentrations (≤5%).39,46 Alternatively, the cell density can be increased to obtain more interacting cells. A common seeding density of cells reported as 2 × 106 cells per mL by many studies35,48 can be used to enhance the interactions of the cells with each other. Morphology of the constructs was also shown to affect the behavior of cells. In a recent study, cells grown on curved constructs spread, elongated, produced an aligned extracellular matrix (ECM), and expressed corneal stromal cell markers without the need for any additional cues.49 The 3D printed construct proposed in our study, therefore, may be further improved by mimicking the curvature of the cornea either by printing on a curved template or crosslinking the printed structure while on a curved template for the construct.
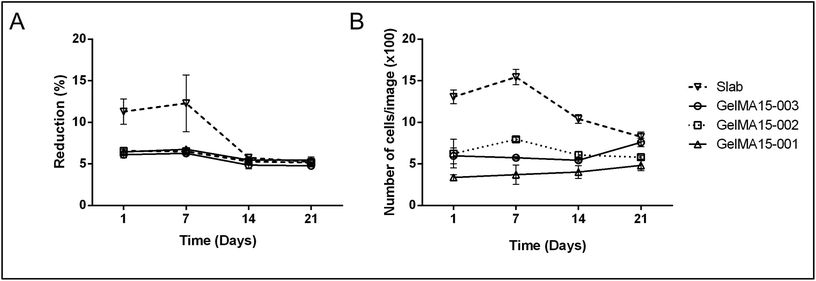 | ||
| Fig. 5 Proliferation of human keratocytes in 3D Printed and GelMA15-Slab hydrogels over 3 weeks. (A) Alamar Blue assay shows proliferation of cells in the hydrogels as indicated by the reduction % and (B) Live–Dead viability assay images (Fig. 4B and Fig. S3†) were used to count the number of cells per image (n = 3) with the NIH Image J program. | ||
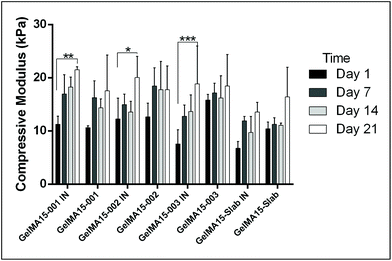
In this study, compressive tests were carried out to give an idea as to where the product stands in terms of mechanical strength compared to native cornea. Besides, compression tests for corneal constructs are a very widely reported and recommended technique in the literature37,57,58 since the cornea faces significant intraocular pressure in its native environment.
The expression of cornea-specific proteins and proteoglycans can be enhanced by increasing the loaded cell density to help cells to communicate with each other. Crosslinking conditions (photoinitiator concentration, UV duration and distance) can be optimized to obtain hydrogels with lower crosslinking density to enhance the growth medium and oxygen transport into the hydrogels.
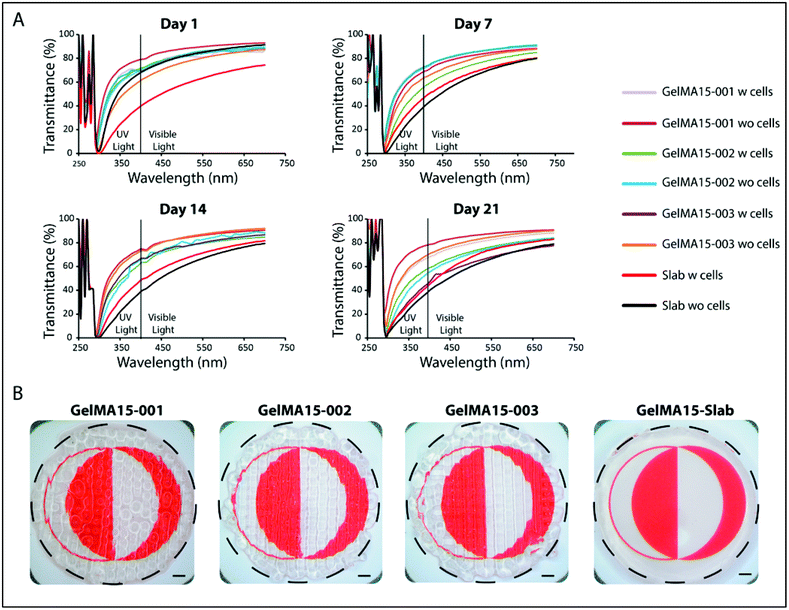
In the 290–400 nm range, transmittance was lower; this is the UVA and UVB regions and the transparency is comparable to that of the natural cornea (∼5% at UVB and 85% at 700 nm for a cornea of an 8 years old).62 However, transmittance at 300 nm (Table S1†) of slab hydrogels was lower than that of the 3D printed hydrogels (Table S1†). In conclusion, although light was scattered because of the patterns of 3D printed hydrogels, their transparency was still comparable with each other and with native cornea. The clarity of the 3D printed and slab hydrogels is seen in the stereomicrographs (Fig. 9B).
In this study, GelMA hydrogels bioprinted with human corneal keratocytes were shown to mimic the native structure of the corneal stroma with their excellent transparency, adequate mechanical strength and high cell viability. Our recent pilot in vivo study on rabbits has also proven the biocompatibility and biodegradability of the GelMA hydrogels63 which is an important indication of the potential of GelMA hydrogels for use in corneal stroma engineering applications.
4. Conclusion
In this study, highly transparent, biocompatible, and stable corneal stroma equivalents were produced by a rapid and effective 3D bioprinting method that mimics the micro-level organization of the stroma. In addition, high cell viability demonstrated the suitability of the printing process and conditions for the production of cell seeded corneal stroma. Synthesis of the specific collagens and proteoglycan by the seeded keratocytes indicated the maintenance of the cell phenotype during printing and under culture conditions. Optimization of the cell density in the hydrogels is necessary to promote the natural interaction and alignment of the cells. In vivo performance of the 3D printed hydrogels is also essential to show the compatibility of this biomaterial before clinical studies. These findings show great promise for the application of 3D bioprinting in corneal stroma engineering.Conflicts of interest
The authors have declared that there is no conflict of interest.Acknowledgements
The authors gratefully acknowledge BIOMATEN and the financial support from METU through BAP-01-08-2013-003 and BAP08-11-DPT-2011-K120350 projects and TUBITAK for the BIDEB2211-C Scholarship for C. K.References
- J. W. Ruberti and J. D. Zieske, Prog. Retinal Eye Res., 2008, 27, 549–577 CrossRef CAS PubMed.
- P. E. Ludwig and K. Sevensma, Anatomy, Head and Neck, Eye Cornea, 2019 Search PubMed.
- N. J. Fullwood, Structure, 2004, 12, 169–170 CrossRef CAS PubMed.
- J. Torbet, M. Malbouyres, N. Builles, V. Justin, M. Roulet, O. Damour, Å. Oldberg, F. Ruggiero and D. J. S. Hulmes, Biomaterials, 2007, 28, 4268–4276 CrossRef CAS PubMed.
- J. A. West-Mays and D. J. Dwivedi, Int. J. Biochem. Cell Biol., 2006, 38, 1625–1631 CrossRef CAS PubMed.
- P. Gain, R. Jullienne, Z. He, M. Aldossary, S. Acquart, F. Cognasse and G. Thuret, JAMA Ophthalmol., 2016, 134(2), 167–173 CrossRef PubMed.
- B. Salvador-Culla and E. P. Kolovou, J. Funct. Biomater., 2016, 7, 13 CrossRef PubMed.
- R. Viitala, V. Franklin, D. Green, C. Liu, A. Lloyd and B. Tighe, Acta Biomater., 2009, 5, 438–452 CrossRef CAS PubMed.
- N. E. Vrana, N. Builles, V. Justin, J. Bednarz, G. Pellegrini, B. Ferrari, O. Damour, D. J. S. Hulmes and V. Hasirci, Invest. Ophthalmol. Visual Sci., 2008, 49, 5325–5331 CrossRef PubMed.
- C. Kilic, A. Girotti, J. C. Rodriguez-Cabello and V. Hasirci, Biomater. Sci., 2014, 2, 318 RSC.
- A. Acun and V. Hasirci, J. Biomater. Sci., Polym. Ed., 2014, 25, 1110–1132 CrossRef CAS PubMed.
- S. Wang, C. E. Ghezzi, R. Gomes, R. E. Pollard, J. L. Funderburgh and D. L. Kaplan, Biomaterials, 2017, 112, 1–9 CrossRef CAS PubMed.
- M.-I. Huh, K.-P. Lee, J. Kim, S. Yi, B.-U. Park and H. K. Kim, J. Ophthalmol., 2018, 2018, 1–18 CrossRef PubMed.
- L. S. Wray and E. J. Orwin, Tissue Eng., Part A, 2009, 15(7), 1463–1472 CrossRef CAS PubMed.
- P. Zorlutuna, A. Tezcaner, I. Kiyat, A. Aydinli and V. Hasirci, J. Biomed. Mater. Res., Part A, 2006, 79, 104–113 CrossRef CAS PubMed.
- C. Kilic Bektas and V. Hasirci, J. Tissue Eng. Regener. Med., 2018, 12, e1899–e1910 CrossRef CAS PubMed.
- S. Hong, J. H. Yun, E.-S. Kim, J. S. Kim, H. Tchah and C. Hwang, Invest. Ophthalmol. Visual Sci., 2018, 59, 1475–1485 CrossRef CAS PubMed.
- E. S. Gil, B. B. Mandal, S.-H. Park, J. K. Marchant, F. G. Omenetto and D. L. Kaplan, Biomaterials, 2010, 31, 8953–8963 CrossRef CAS PubMed.
- T. Kawakita, K. Higa, S. Shimmura, J. Shimazaki and K. Tsubota, Invest. Ophthalmol. Visual Sci., 2014, 55, 5156 Search PubMed.
- M. Grolik, D. Kuźmicz, D. Dobrowolski, B. Wowra, E. Wylęgała, M. Nowakowska and K. Szczubiałka, J. Biomater. Tissue Eng., 2018, 8, 374–383 CrossRef.
- X. Hu, W. Lui, L. Cui, M. Wang and Y. Cao, Tissue Eng., 2005, 11, 1710–1717 CrossRef CAS PubMed.
- P. Fagerholm, N. S. Lagali, K. Merrett, W. B. Jackson, R. Munger, Y. Liu, J. W. Polarek, M. Söderqvist and M. Griffith, Sci. Transl. Med., 2010, 2, 46ra61 Search PubMed.
- A. I. Van Den Bulcke, B. Bogdanov, N. De Rooze, E. H. Schacht, M. Cornelissen and H. Berghmans, Biomacromolecules, 2000, 1, 31–38 CrossRef CAS PubMed.
- J. Rose, S. Pacelli, A. Haj, H. Dua, A. Hopkinson, L. White and F. Rose, Materials, 2014, 7, 3106–3135 CrossRef CAS PubMed.
- N. Hasirci, C. Kilic, A. Kömez, G. Bahcecioglu and V. Hasirci, in Gels Handbook, ed. M. R. Abidian, U. A. Gurkan and F. Edalat, World Scientific, 2016, vol. 2, pp. 1–52 Search PubMed.
- T. Mimura, S. Amano, S. Yokoo, S. Uchida, S. Yamagami, T. Usui, Y. Kimura and Y. Tabata, Mol. Vision, 2008, 14, 1819–1828 CAS.
- L. J. Luo, J. Y. Lai, S. F. Chou, Y. J. Hsueh and D. H. K. Ma, Acta Biomater., 2018, 65, 123–136 CrossRef CAS PubMed.
- G. Bahcecioglu, N. Hasirci, B. Bilgen and V. Hasirci, Biofabrication, 2019, 11, 25002 CrossRef CAS PubMed.
- G. Bahcecioglu, N. Hasirci, B. Bilgen and V. Hasirci, Int. J. Biol. Macromol., 2019, 122, 1152–1162 CrossRef CAS PubMed.
- G. Eke, N. Mangir, N. Hasirci, S. MacNeil and V. Hasirci, Biomaterials, 2017, 129, 188–198 CrossRef CAS PubMed.
- N. Celikkin, S. Mastrogiacomo, J. Jaroszewicz, X. F. Walboomers and W. Swieszkowski, J. Biomed. Mater. Res., Part A, 2018, 106, 201–209 CrossRef CAS PubMed.
- L. Li, C. Lu, L. Wang, M. Chen, J. White, X. Hao, K. M. McLean, H. Chen and T. C. Hughes, ACS Appl. Mater. Interfaces, 2018, 10, 13283–13292 CrossRef CAS PubMed.
- F. Rengier, A. Mehndiratta, H. von Tengg-Kobligk, C. M. Zechmann, R. Unterhinninghofen, H.-U. Kauczor and F. L. Giesel, Int. J. Comput. Assist. Radiol. Surg., 2010, 5, 335–341 CrossRef CAS PubMed.
- A. Sorkio, L. Koch, L. Koivusalo, A. Deiwick, S. Miettinen, B. Chichkov and H. Skottman, Biomaterials, 2018, 171, 57–71 CrossRef CAS PubMed.
- A. Isaacson, S. Swioklo and C. J. Connon, Exp. Eye Res., 2018, 173, 188–193 CrossRef CAS PubMed.
- B. A. Harley, J. H. Leung, E. C. Silva and L. J. Gibson, Acta Biomater., 2007, 3, 463–474 CrossRef CAS PubMed.
- M. Rizwan, G. S. L. Peh, H. P. Ang, N. C. Lwin, K. Adnan, J. S. Mehta, W. S. Tan and E. K. F. Yim, Biomaterials, 2017, 120, 139–154 CrossRef CAS PubMed.
- D. Hellio-Serughetti and M. Djabourov, Langmuir, 2006, 22, 8516–8522 CrossRef CAS PubMed.
- J. Yin, M. Yan, Y. Wang, J. Fu and H. Suo, ACS Appl. Mater. Interfaces, 2018, 10, 6849–6857 CrossRef CAS PubMed.
- Y. S. Zhang, K. Yue, J. Aleman, K. Mollazadeh-Moghaddam, S. M. Bakht, J. Yang, W. Jia, V. Dell'Erba, P. Assawes, S. R. Shin, M. R. Dokmeci, R. Oklu and A. Khademhosseini, Ann. Biomed. Eng., 2017, 45, 148–163 CrossRef PubMed.
- B. Jeong, S. W. Kim and Y. H. Bae, Adv. Drug Delivery Rev., 2012, 64, 154–162 CrossRef.
- Q. Chen, L. Zhu, C. Zhao, Q. Wang and J. Zheng, Adv. Mater., 2013, 25, 4171–4176 CrossRef CAS PubMed.
- H. B. Bohidar and S. S. Jena, J. Chem. Phys., 1993, 98, 8970–8977 CrossRef CAS.
- L. E. Bertassoni, J. C. Cardoso, V. Manoharan, A. L. Cristino, N. S. Bhise, W. A. Araujo, P. Zorlutuna, N. E. Vrana, A. M. Ghaemmaghami, M. R. Dokmeci and A. Khademhosseini, Biofabrication, 2014, 6, 024105 CrossRef PubMed.
- D. Joung, V. Truong, C. C. Neitzke, S.-Z. Guo, P. J. Walsh, J. R. Monat, F. Meng, S. H. Park, J. R. Dutton, A. M. Parr and M. C. McAlpine, Adv. Funct. Mater., 2018, 1801850 CrossRef.
- C. Colosi, S. R. Shin, V. Manoharan, S. Massa, M. Costantini, A. Barbetta, M. R. Dokmeci, M. Dentini and A. Khademhosseini, Adv. Mater., 2015, 28, 677–684 CrossRef PubMed.
- B. Hoffmann and C. Schäfer, Cell Adhes. Migr., 2010, 4, 190–193 CrossRef PubMed.
- D. B. Kolesky, R. L. Truby, A. S. Gladman, T. A. Busbee, K. A. Homan and J. A. Lewis, Adv. Mater., 2014, 26, 3124–3130 CrossRef CAS PubMed.
- R. M. Gouveia, E. Koudouna, J. Jester, F. Figueiredo and C. J. Connon, Adv. Biosyst., 2017, 1, 1700135 CrossRef.
- J. L. Battaglioli and R. D. Kamm, Invest. Ophthalmol. Visual Sci., 1984, 25, 59–65 CAS.
- S. J. Petsche, D. Chernyak, J. Martiz, M. E. Levenston and P. M. Pinsky, Invest. Ophthalmol. Visual Sci., 2012, 53, 873–880 CrossRef PubMed.
- P. Zorlutuna, N. Builles, O. Damour, A. Elsheikh and V. Hasirci, Biomaterials, 2007, 28, 3489–3496 CrossRef CAS PubMed.
- N. E. Vrana, A. Elsheikh, N. Builles, O. Damour and V. Hasirci, Biomaterials, 2007, 28, 4303–4310 CrossRef CAS PubMed.
- C. Kilic Bektas and V. Hasirci, J. Mater. Sci. Mater. Med., 2019 Search PubMed , (recently accepted for publication).
- T. S. Girton, T. R. Oegema, E. D. Grassl, B. C. Isenberg and R. T. Tranquillo, J. Biomech. Eng., 2000, 122, 216–223 CrossRef CAS PubMed.
- T. S. Girton, T. R. Oegema and R. T. Tranquillo, J. Biomed. Mater. Res., 1999, 46, 87–92 CrossRef CAS PubMed.
- M. A. Watsky, C. J. Doillon, M. Hakim, J. Wang, R. Munger, N. Laycock and M. Griffith, Invest. Ophthalmol. Visual Sci., 2002, 43, 2996 Search PubMed.
- S. R. Sloan, M. A. R. Garcia, Y. Khalifa and M. R. Buckley, Invest. Ophthalmol. Visual Sci., 2015, 56, 1108 CrossRef PubMed.
- J. V. Jester, P. A. Barry-Lane, H. D. Cavanagh and W. M. Petroll, Cornea, 1996, 15, 505–516 CrossRef CAS PubMed.
- G. P. Riley, R. L. Harrall, P. G. Watson, T. E. Cawston and B. L. Hazleman, Eye, 1995, 9(Pt 6), 703–718 CrossRef PubMed.
- J. M. Sivak and M. E. Fini, Prog. Retinal Eye Res., 2002, 21, 1–14 CrossRef CAS.
- J. D. Mallet and P. J. Rochette, Photochem. Photobiol. Sci., 2013, 12, 1310–1318 RSC.
- C. Kilic Bektas, A. Burcu, G. Gedikoglu, H. H. Telek, F. Ornek and V. Hasirci, J. Biomater. Sci., Polym. Ed., 2019, 30, 1803–1821 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9bm01236b |
| This journal is © The Royal Society of Chemistry 2020 |