Open Access Article
Open Access ArticleGenetically encodable biosensors for Ras activity
Ryan
Weeks
 ab,
Sohum
Mehta
ab,
Sohum
Mehta
 b and
Jin
Zhang
b and
Jin
Zhang
 *abc
*abc
aDepartment of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA 92093, USA. E-mail: jzhang32@ucsd.edu; Tel: +1 (858) 246-0602
bDepartment of Pharmacology, University of California, San Diego, La Jolla, CA 92093, USA
cDepartment of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA
First published on 7th February 2024
Abstract
Genetically encoded Ras biosensors have been instrumental in illuminating the spatiotemporal dynamics of Ras activity since the beginning of the imaging revolution of the early 21st century. In general, these sensors employ Ras sensing units coupled with fluorescent proteins. These biosensors have not only helped elucidate Ras signalling dynamics at the plasma membrane but also revealed novel roles for Ras signalling within subcellular compartments such as the Golgi apparatus. In this review, we discuss the different classes of biosensors used to measure Ras activity and discuss their importance in uncovering new roles for Ras activity in cellular signalling and behavior.
Introduction
Background on Ras signalling studies
Ras is a small GTPase that regulates growth and proliferation. As a GTPase, Ras binds the guanine nucleotides GDP and GTP, and its activity is governed by the structural consequences of these bound nucleotide states, with GTP-bound Ras structurally poised for high-affinity interaction with downstream Raf kinases, activating the Raf-MEK-ERK mitogen activated kinase (MAPK) pathway.1–3 Ras nucleotide binding is primarily regulated by guanine exchange factors (GEFs) and GTPase activating proteins (GAPs).2 Ras signalling activity has long been understood to be localized at the plasma membrane. However, mounting evidence has shown complex spatiotemporal regulation of Ras activity, with distinct functional outcomes of Ras signals emanating from non-canonical subcellular compartments.4–6Ras signalling has been investigated for many decades, since its initial discovery as an oncogene and subsequent characterization as a regulator of eukaryotic growth and proliferation.7 Following the cloning of Aequorea victoria green fluorescent protein (GFP)8 and the advent of genetically encoded fluorescence, imaging studies into Ras began challenging the predominant view that Ras is exclusively present at the plasma membrane.9 When fluorescently labelled Ras, or Ras hyper-variable regions (HVR's), were expressed in cells, plasma membrane localization was apparent; however, fluorescence signals were also observed at endomembranes such as the endoplasmic reticulum (ER) and Golgi apparatus.10,11 Yet while these studies described novel localization of Ras protein, the imaging methodologies precluded assessment of Ras activity. In this review, we discuss the development of new molecular tools that address the need for spatiotemporally precise measurements of Ras activity in living cells, highlighting innovations that resulted in striking observations such as key roles for Ras signalling in cellular motility and chemotaxis, as well as demonstration of Ras activity at endomembrane compartments and novel functional roles for this subcellular activity.
Historically, cellular Ras activity was first measured using 32P radionucleotide labelling combined with Ras-immunoprecipitation and thin layer chromatography.12 But with the advent of GTPase pulldown assays,13 lysate pulldowns have been a preferred method for Ras activity measurements.14 These pulldown assays make use of a Ras-binding domain (RBD) derived from the primary Ras effector Raf1, which is used as a bait to capture active Ras in cell lysates. The enriched pool of active Ras is then quantitated using antibody-based techniques such as Western blotting or enzyme-linked immunoassay (ELISA). While these methods have traditionally been considered the standard for whole-cell Ras activity assays, subcellular measurements require additional fractionation steps, which may reduce signal and/or suffer from cross-contamination. Genetically encoded fluorescent biosensors have emerged as key tools that enable studies of the spatiotemporal regulation of signalling proteins with high precision and high resolution. Fluorescent biosensors are molecular tools designed to engage with components of cellular signalling pathways, be they small-molecule second messengers or signalling enzymes, and produce a measurable optical readout. As such, it is useful to characterize two key features of the fluorescent biosensor, one being the “sensing unit”, which directly interacts with the target signalling molecule, and the “reporting unit”, which produces the optical readout in conjunction with the sensing unit. As we discuss below, a protein domain commonly used in the context of Ras biosensor development is the aforementioned Raf1 RBD, which selectively binds active Ras. However, the fluorescent reporting unit can vary quite a lot, and over the years many types of design approaches have sought to couple RBD–Ras binding to different optical readouts for Ras activity.
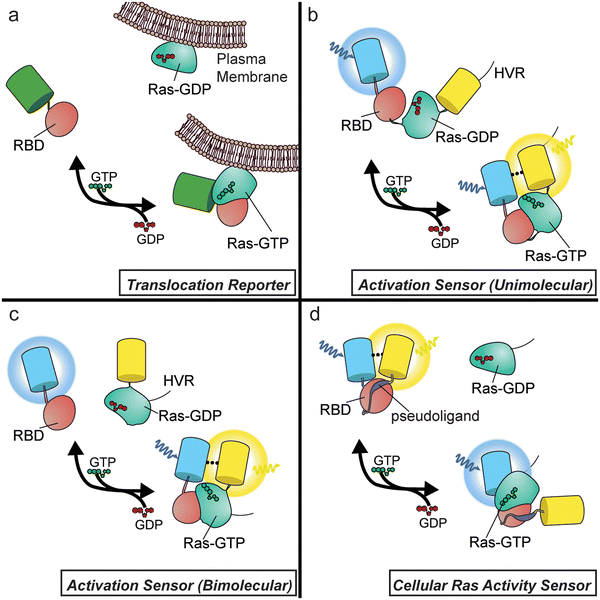
To date, Ras biosensors designs fall into three main categories (Fig. 1): (1) translocation reporters, (2) activation biosensors containing modified Ras, and (3) biosensors for sensing cellular Ras activity.
Translocation reporters
These sensors are typically constructed by fusing the Raf1 RBD, or a modified RBD, to a fluorescent protein (FP), whereby translocation of the fluorescence signal provides a readout of Ras activity. To measure Ras activity with this system, one must track the distribution of the sensor within the cell, and to provide quantitation, the ratio of fluorescence intensity within the cytosol versus the plasma membrane (or another compartment) can be calculated. Early studies using a simple GFP-tagged RBD were crucial in pointing to the Golgi as a putative location for Ras signalling.15,16 Since the wild-type (wt) RBD may result in little to no observable translocation for endogenous Ras, enhancement of the Ras-binding affinity is sometimes needed to obtain a signal for endogenous reporting. Strategies to enhance affinity include the use of tandem RBDs17 or the use of the Ras cysteine-rich domain (CRD).18 While this straightforward design allows sensing of endogenous Ras activity, reliable quantitation using these probes is challenging, as intensity changes caused by cell spreading, flattening, and membrane edge ruffling are common following growth-factor stimulation. Membrane ruffles, for instance, produce high-intensity regions irrespective of probe translocation events, complicating the optical readout of these tools.19,20Activation sensors (unimolecular)
Instead of relying on RBD translocation, other Ras biosensors feature a sensing unit design where the RBD is directly fused to a modified form of Ras. The resulting chimera functions as a molecular switch, with Ras activation by cellular GEFs resulting in an intramolecular interaction with the paired RBD, altering the conformation of the sensing unit. This conformational change is often used to modulate the distance and orientation of a pair of FPs capable of undergoing Förster Resonance Energy Transfer (FRET), which involves the radiationless transfer of excited-state energy between a donor and acceptor fluorophore with compatible emission and excitation spectra (e.g., CFP and YFP). Because FRET decreases donor fluorescence while increasing acceptor fluorescence, signals from FRET-based biosensors are often recorded as the ratio of sensitized acceptor emission (i.e., acceptor fluorescence following donor excitation) over donor-direct emission. This emission-ratiometric readout helps overcome many issues associated with imaging translocation-based reporters. The first Ras sensor to feature this design was a unimolecular biosensor known as Ras-RAICHU, in which the catalytic domain of Ras and the Raf1 RBD are sandwiched between CFP and YFP. Notably, Ras in this design is truncated to remove the hyper-variable region (HVR) responsible for proper localization of this protein within the cell, which is instead fused to the C-terminus of YFP. Many FRET-based GTPase sensors, including additional Ras-family,21,22 Rab-family,23 and Rho-family24,25 GTPase sensors, have since been developed based on this design, as reviewed previously.26Activation sensors (bimolecular)
An alternative strategy for constructing Ras sensors based on this Ras/RBD molecular switch is to use a bimolecular design. These sensors function similarly as the above unimolecular design, except that the Ras and Ras-binding RBD components are expressed as separate polypeptides fused to the corresponding parts of the reporting unit. This has been a common strategy for Ras sensor design, and was in fact the first design used for FRET-based small GTPase sensors, employed for Rac1.27 While many bimolecular Ras sensors contain FPs to provide a FRET-based readout,28,29 another popular strategy replaces the donor FP with luciferase, which will undergo bioluminescence resonance energy transfer (BRET) to an acceptor FP.30–32 More recently, the use of dimerization-dependent FPs, which are engineered FP pairs that show an increase in fluorescence intensity upon dimerization, has yielded intensiometric, single-color Ras sensors.33Biosensors for sensing cellular Ras activity
RAICHU and similar Ras sensors have been extremely useful in shedding light on the spatiotemporal regulation of Ras signalling. However, these designs preclude the detection of endogenous Ras activity, given that the sensors contain active Ras and rely on cellular GEFs to modulate the Ras–RBD interaction within the sensing unit, essentially behaving as GEF/GAP sensors. We believe that a sensor design capable of reporting on endogenous Ras activity would provide more direct insights into the spatiotemporal regulation of Ras activity.We recently sought to meet the need for endogenous Ras sensing by developing a Ras Activity Reporter (RasAR) capable of detecting cellular Ras activity.34 To do so, we engineered a molecular switch containing the Raf1 RBD fused to a short peptide derived from the RBD-interacting region of Ras, which is sandwiched between CFP and YFP. In the absence of Ras activity, the short peptide acts as a pseudoligand that binds the intramolecular RBD, whereas the pseudoligand peptide is displaced by the intermolecular interaction between the RBD and active cellular Ras, leading to a conformational change that alters the FRET efficiency. While this sensor was designed with the intention of measuring endogenous Ras activity, Ras overexpression proved necessary for these experiments, which thus share a similar limitation to previously discussed activation sensor designs. Nevertheless, RasAR features a unimolecular, FRET-based design, which allowed us to probe all three Ras isoforms, in addition to Y32F and Y32,64F mutants of HRasWT. RasAR was additionally amenable to subcellular targeting, revealing compartment-specific differences in Src-mediated regulation of HRas signaling under EGF stimulation.
While endogenous Ras detection was not feasible with RasAR, we suspect this to be largely attributable to insufficient affinity of the wild-type RBD. Enhanced affinity may thus be an important prerequisite for effectively sensing endogenous Ras. However, as we discuss further in the Summary and Future Directions section, a successful design would likely have to balance sensitivity for endogenous Ras with potential complications related to buffering of downstream signaling.
Biosensor applications
The proliferation of molecular tools for live-cell imaging of biochemical activities has spurred numerous investigations into the spatiotemporal dynamics of Ras activity over the past 25 years. While many studies naturally focused on the plasma membrane, given the established role for Ras at this location,9 more recent work is revealing key roles for Ras signalling at endomembrane compartments, as well as novel modes of Ras signalling regulation at both the cell surface and intracellular membranes.Ras signalling at the plasma membrane
Ras localization is determined by the C-terminal HVR, which contains sites for post-translational lipid modifications that increase the affinity of Ras for membranes, with the plasma membrane being the canonical site of Ras signalling. This tendency was first observed in Ras biosensor experiments using Ras-RAICHU. In EGF-stimulated COS-1 cells, RAICHU showed elevated FRET signals in membrane ruffles at the cell periphery. Ras activity was also shown to be depleted in regions of cell–cell contact.35 In PC12 cells, Ras was shown to be activated at the cell periphery upon NGF stimulation, and under prolonged differentiation stimulation, Ras was observed to exhibit sustained activity in the neurite extensions of differentiating PC12 cells. Additionally, Ras activity was shown to be distinctly localized compared with Rap1, whose activity was monitored using Rap-RAICHU.35This new model37 was further expanded in follow-up experiments using the same translocation-based methodology in Dictyostelium.38–40 For instance, folic acid was shown to induce Ras activity in response to directional cues dependent on Gβ-protein expression, whereas non-directional Ras activity in the context of cell motility was Gβ-protein independent.38 And, additionally, identification of the RasGAP DdNF1 in regulating Dictyostelium chemotaxis was assisted with translocation reporters.39 Researchers have continued to build upon these efforts by pursuing more detailed, modelling-focused studies into cell motility and chemotaxis,40 often making use of Ras biosensors.41–44
The PI3K-dependent amplification of Ras activity was also observed in NIH 3T3 fibroblasts. Specifically, Thevathasan et al. used the FKBP-FRB dimerization system to activate PI3K, wherein the FKBP-fused regulatory p85 subunit of PI3K is recruited to plasma membrane-localized FRB upon rapamycin addition, leading to membrane PI3K activity. Using RAICHU-HRas and this inducible PI3K activation probe, they observed increased HRas activity at the edge of protruding membranes upon rapamycin-induced PI3K activation.45 Furthermore, ablating HRas using siRNA abrogated the protrusion behaviour of these cells, further bolstering the role of PI3K in amplifying Ras activity during mammalian cellular protrusions.45 HRas and PI3K feedback was also observed in the migratory behaviour of epithelial cells, with the same signalling logic involving HRas, PI3K and F-actin in the process of intercalation and tube elongation,46 in which cells migrate to the surface layer of a tissue in 3D culture (intercalation) through a process involving prolonged cellular protrusions (tube elongation). When RBD-GFP was expressed in 3D-cultured tissue organoids, Ras activity was observed to accumulate within anterior protrusions of the migrating cells. This observation mirrored results for PI3K activity localization measured using PH-domain translocation probes. Furthermore, coexpressing RBD-GFP with the F-actin marker LifeAct within organoids revealed the colocalization of Ras activity with F-actin within the anterior protrusions. These probes were instrumental in analysing the key signalling molecules Ras, PI3K and F-actin in developing a model of cell migration within tissue culture in the process of radial intercalation.46
Recently, a novel set of in vivo imaging-compatible, bimolecular GTPase sensors, including for Ras, have been developed utilizing dimerization dependent fluorescent proteins.33 These sensors exhibit high dynamic ranges and were shown to be compatible with in vivo imaging, enabling Ras activity measurements in the brains of live mice. This was accomplished through Adeno-associated viral (AAV) transduction of the Ras biosensor to perform dynamic Ras activity measurements and even multiplexed imaging of Ras and Ca2+ dynamics through coexpression of GCaMP6.
Ras signalling at the Golgi apparatus
While Ras activity has long been known to be localized at the plasma membrane,9 mounting evidence from studies using fluorescent biosensors suggests that Ras also actively signals from endomembranes.10,11 Indeed, the advent of fluorescent biosensors has spurred numerous studies of Ras activity at endomembrane compartments such as the Golgi apparatus. Seminal studies from the Philips group demonstrated the presence of Golgi Ras activity using the RBD-GFP translocation sensor.15,16 In COS7 cells expressing HRas and RBD-GFP, treatment with EGF was observed to induce fast and transient Ras activity at the plasma membrane, along with slow and more sustained activity at the Golgi membrane.15 While this observation was in the context of HRas overexpression, the authors were also able to investigate endogenous Ras activity using a “bystander FRET” method. Specifically, Chiu et al. coexpressed plasma membrane- and Golgi-targeted FRET acceptors with the RBD-CFP translocation biosensor, such that increased FRET would indicate endogenous Ras activity within either compartment. They further showed that PP2 treatment to abolish Src activity abrogated RBD-GFP probe translocation to the Golgi apparatus, which was further corroborated by deletion of Src family kinase at the gene level.47 Further investigation into the regulation of Golgi Ras by Bivona and colleagues suggested the existence of a pathway in which phospholipase C induces Golgi Ras activity via RasGRP1 in a calcium-dependent manner. This pathway was substantiated in experiments using Jurkat T-cells and PC12 cells.16 In addition, the authors also observed that sustained Ras activity at the Golgi promoted PC12 cell differentiation. Extending upon these findings in immune cells, the authors coexpressed NRas with an RBD-GFP translocation probe and were able to additionally detect Ras activity at the Golgi in Jurkat T-cells stimulated by crosslinking with anti-CD3 and anti-CD28 antisera.48Expanding on these studies of Golgi Ras activity was the demonstration of the precise role of the Ras HVR in endomembrane activity. In particular, a series of biosensor-driven studies from the Bastiaens group revealed that the “Acylation Cycle” of HRas and NRas localization within the cell plays a key role in Ras Golgi activity.28,49,50 This model links H/NRas localization to the Golgi through the trapping of palmitoylated Ras isoforms by Golgi-prominent Acyl-palmitoyl transferases. As HRas and NRas show dual and single HVR palmitoylation, respectively, their dynamics should differ, which should have an impact on their corresponding Golgi activity. Using a bimolecular FRET approach consisting of YFP-tagged Ras and an RBD fused to two RFPs, the authors were able to track the spatiotemporal dynamics of Ras activity at the Golgi. Consistent with their expected localization dynamics, with NRas predicted to cycle faster through the Golgi compartment, Golgi NRas activity was observed to accumulate more quickly compared with HRas upon EGF stimulation. Additionally, the authors were able to confer NRas-like activity dynamics on HRas by blocking dual palmitoylation with an HRas C184S mutant. Given these observations of Golgi Ras activity, the authors concluded that activity in this compartment is likely an “echo” of activated Ras at the plasma membrane.28 However, later modelling work suggested the need for some endomembrane or cytosolic GEF to maintain the pool of active Golgi Ras, with additional experimental work using targeted dominant-negative mutants suggesting the possibility of an ER-localized GEF.50
Despite the impact of these findings, Golgi Ras activity has remained controversial. For example, using high-avidity sensors containing multiple RBD copies fused to GFP, Augsten et al. were able to detect endogenous Ras activity at the plasma membrane, but not the Golgi, in COS7, PC12, and Jurkat cells.17 Further investigation in T-cells by Rubio and colleagues, who incorporated tandem copies of EGFP into multi-RBD probes to enhance signal at low probe concentrations, again pointed to the plasma membrane as the sole location of Ras activity.51 While scepticism of overexpression studies is a valid concern, these conflicting results may be due to low signal, coupled with insufficient quantitative capacity of translocation sensors.
Recent work from the Crespo laboratory has revealed a novel mode of Golgi Ras activity regulation in MCF-7 cells.52 Using RBD-GFP as a translocation probe for Ras activity in the context of HRas expression, Casar et al. differentially stimulated MCF-7 cells with either EGF or TGF-β. While the proliferative stimulus of EGF induced only a transient spike of HRas activity at the plasma membrane, apoptotic TGF-β stimulation induced RBD-GFP translocation to the Golgi compartment. The authors further demonstrated a role for Golgi Ras activity in promoting RalGEF activation, leading to PTPRk-dependent antagonism of ERK activity.
More recently, we used our new FRET-based Ras sensor, RasAR, which omits full-length Ras from the sensing unit and enables direct measurements of cellular Ras activity, to probe the effect of Src inhibition on EGF-stimulated HRasWT activity.34 In COS7 cells coexpressing RasAR and HRasWT, treatment with the Src inhibitors PP2 or SrcI1 yielded enhanced RasAR response compared with the control (i.e., no Src inhibition), suggesting an inhibitory role for Src on Ras signalling. Notably, this effect was only observed when we targeted RasAR to the Golgi membrane and not with the plasma membrane-targeted RasAR. To investigate whether the mechanism of Src-mediated inhibition involves direct phosphorylation of Ras, as proposed previously,53,54 we coexpressed RasAR with an HRas Y32,64F double mutant that lacks both putative Src phosphorylation sites, and found that Src inhibitor pretreatment no longer had an effect on the EGF-stimulated Golgi-RasAR response in these cells. These studies highlight the crucial role of Ras biosensors in uncovering Ras signalling mechanisms and revealing non-canonical sites of activity, and continued efforts will greatly advance our understanding of this highly important signalling protein.
Ras biosensors in inhibitor studies
Multiple cancers are characterized by Ras mutations. These mutations primarily involve residues G12 or Q61 and lock the GTPase into a constitutively active state. Targeting mutant Ras has been a priority in cancer therapy for decades.55,56 Due to its lack of allosteric binding pockets and picomolar affinity for guanine nucleotides, Ras has long been considered “undruggable”. Nevertheless, pioneering work in characterizing the switch-II pocket has led to the development of KRasG12C inhibitors.57–60 This has spurred considerable excitement and renewed research into anti-Ras therapy and FDA-approved breakthrough therapy for the treatment of patients with locally advanced or metastatic non-small cell lung cancer with KrasG12C mutation.61 While this is an important step in treating cancers with mutated Ras, KRasG12C only makes up a fraction of oncogenic Ras mutations. Developing live-cell assays based on genetically encoded biosensors is highly desirable to allow robust assessment of Ras inhibition in the native cellular context with simultaneous assessment of drug permeability and stability within this environment.Notably, the translocation sensor Venus-RBDCRD, combined with high-throughput image analysis, was used to analyse the paradoxical effects of Ras pathway inhibitors on Ras activity. Crucially, the authors demonstrated that Ras pathway inhibition may lead to paradoxical Ras activation via the release of negative feedback mechanisms.62 Another technique was developed using full-length CRAF/BRAF fused to Renilla Luciferase II and Ras fused to GFP to produce a change in BRET upon Ras–Raf interaction.63 This study sought to explain the paradoxical activation of Ras by first-generation ATP-competitive Raf inhibitors. Using these live-cell assays for Ras–Raf interaction, coupled with similarly designed reporters for monitoring Raf-dimerization, the authors proposed a model in which these first-generation inhibitors induce Raf–Ras binding through disruption of the Raf auto-inhibitory state, coupled with BRaf/CRaf-heterodimerization facilitated by Ras dimers/nanoclusters, thereby increasing Ras activation.63
We also recently demonstrated the use of our RasAR sensor to investigate the dynamics of Ras inhibition in COS7 cells overexpressing KRasG12C. Interestingly, when we treated these cells with G12C inhibitors, we observed very slow inhibitory kinetics, requiring approximately 5 hours to achieve maximal inhibition. This slow inhibitory effect may be related to the targeting mechanism of first-generation KRasG12C inhibitors, which bind to the switch-II region of inactive, GDP-bound Ras. Since this state is very infrequently sampled in the case of constitutively active Ras mutants, the kinetics of inhibition are likely to slowed by this requirement.34 A number of groups are also pursuing the use of BRET-based sensor designs to study Ras inhibitors.30–32 For example, Bery and Rabbitts paired Ras fused to the luciferase RLuc8 with GFP-fused Ras-effector binding domains from PI3Kα, PI3Kγ, CRaf, or RalGDS, enabling them to monitor Ras activation and signalling along multiple downstream signalling axes. They used this system to identify a novel small-molecule inhibitor of Ras signalling,31 as well as to characterize a set of drugs identified from SPR screening.32
Summary and future directions
Ras biosensors have been very useful in elucidating the spatiotemporal dynamics of Ras activity. In the context of chemotactic signalling, Ras translocation reporters have been frequently used to detect endogenous Ras activity. These probes have been instrumental in discovering the upstream role Ras plays in directing the chemotaxis machinery, as well as in understanding the feedback mechanisms among Ras, PI3K and F-actin. Furthermore, these sensors have been useful in developing effective models for cell motility and chemotaxis. In the context of Ras signalling from intracellular hubs such as the Golgi apparatus, Ras biosensors have similarly been key tools, from initial studies using translocation reporters to recent work using Golgi-targeted ratiometric FRET reporters. Nevertheless, these studies have relied on Ras overexpression, which is a contentious methodology in this field. While previous efforts have yielded probes that can engage endogenous Ras, often with the use of tandem RBDs, these sensors have to date been limited to translocation-based designs. For instance, ground-breaking new research has identified active Ras within cytoplasmic granules formed by EML4-ALK oncogenic fusion kinases often associated with lung and other cancer subtypes.64 This work relied on high-avidity translocation probes51 to visualize Ras activity within these granules.To date, very few groups have reported biosensor designs with sensitivity for endogenous Ras activity that couple Ras binding to a quantitative output not dependent on translocation. Multiple factors contribute to this ongoing challenge. To start, the mechanism of endogenous detection with RBD-based designs requires reversible interaction with a limited pool of active GTPase molecules.65 To address this, the RBDs used in these sensors often employ enhanced affinity or avidity in order to outcompete endogenous effectors of Ras binding.18,51 However, the difficulty of effective Ras engagement is not the sole concern. For RBD-based sensors, interaction with endogenous Ras risks competitively inhibiting downstream Raf kinases and other Ras effectors. This effect may be further complicated by a shared engagement of RasGAPs and effectors for the effector-binding interface of Ras, possibly promoting RasGTP levels while simultaneously buffering downstream signals. Nevertheless, endogenous Ras activity measurements are an interest among Ras biologists, and translocation sensors have been developed to provide such measurements. We believe that this technology may be improved upon, with the development of biosensors that translate endogenous Ras binding into quantitative optical readouts in a reporting unit.
There have been few reports of biosensor-based Ras activity measurements in vivo. The sole prominent report of in vivo measurements employed AAV-based transduction33 of selected tissues in adult mice. Previous work reporting that transgenic expression of FRET-based Ras biosensors resulted in infertile specimens66 also suggests a potential for interference with endogenous signaling in vivo. However, this limitation may be overcome with the development of ultra-bright, red-shifted Ras biosensors. Sensors with red-shifted fluorescence spectra offer improved signal-to-noise ratio (SNR) in vivo due to the efficient tissue penetration of long-wavelength light. Similarly, ultra-bright sensors provide good SNR even at low expression levels, thus reducing concerns of downstream buffering, and decreasing the risk of interference with endogenous signalling pathways.
The development of a robust Ras biosensing platform will be instrumental in determining the spatiotemporal dynamics of Ras activity at the Golgi apparatus and other subcellular compartments. This endeavour likely necessitates the engineering of high-dynamic-range biosensors and sensing units capable of high-affinity Ras engagement and minimal pathway interference. But while most Ras biosensor designs utilize RBD-based approaches to detect Ras activity, it should be noted that this is not strictly necessary, as some sensors have utilized FRET between BODIPY-labelled GTP analogues and FP-tagged Ras.67 Further development in the Ras biosensor space will allow multiplexing of Ras to investigate crosstalk with other signalling pathways and open the possibility for robust Ras activity measurements in vivo. Continuing development of Ras biosensor capabilities will push key discoveries into the spatiotemporal dynamics of this pivotal signalling molecule.
Author contributions
R. W., S. M., and J. Z. coordinated, wrote, revised, and edited the text. R. W. designed the figures.Conflicts of interest
This review was done in the context of scientific collaboration with NIBR Global Scholars Program (NGSP), a funding program by Novartis.Acknowledgements
This work was supported by R35 CA197622, R01 DK073368, R01 DE030497, R01 HL162302, R01 CA262815, RF1 MH126707, and support from the NIBR Global Scholars Program (to J. Z).Notes and references
- D. R. Lowy and B. M. Willumsen, Function and Regulation of Ras, Annu. Rev. Biochem., 1993, 62, 851–891 CrossRef CAS PubMed.
- S. Lu, H. Jang, S. Muratcioglu, A. Gursoy, O. Keskin, R. Nussinov and J. Zhang, Ras Conformational Ensembles, Allostery, and Signaling, Chem. Rev., 2016, 116(11), 6607–6665 CrossRef CAS PubMed.
- R. J. A. Grand and D. Owen, The biochemistry of ras p21, Biochem. J., 1991, 279(3), 609–631 CrossRef CAS PubMed.
- S. P. Mo, J. M. Coulson and I. A. Prior, RAS variant signalling, Biochem. Soc. Trans., 2018, 46(5), 1325–1332 CrossRef CAS PubMed.
- I. A. Prior and J. F. Hancock, Ras trafficking, localization and compartmentalized signalling, Semin. Cell Dev. Biol., 2012, 23(2), 145–153 CrossRef CAS PubMed.
- E. C. Greenwald, S. Mehta and J. Zhang, Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks, Chem. Rev., 2018, 118(24), 11707–11794 CrossRef CAS PubMed.
- F. McCormick, Chapter One – A brief history of RAS and the RAS Initiative, in Advances in Cancer Research. 153, ed. J. P. O’Bryan and G. A. Piazza, Academic Press, 2022. pp.1–27 Search PubMed.
- M. Chalfie, Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, Green Fluorescent Protein as a Marker for Gene Expression, Science, 1994, 263(5148), 802–805 CrossRef CAS PubMed.
- M. C. Willingham, I. Pastan, T. Y. Shih and E. M. Scolnick, Localization of the src gene product of the Harvey strain of MSV to plasma membrane of transformed cells by electron microscopic immunocytochemistry, Cell, 1980, 19(4), 1005–1014 CrossRef CAS PubMed.
- E. Choy, V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov and M. R. Philips, Endomembrane trafficking of ras: The CAAX motif targets proteins to the ER and Golgi, Cell, 1999, 98(1), 69–80 CrossRef CAS PubMed.
- A. Apolloni, I. A. Prior, M. Lindsay, R. G. Parton and J. F. Hancock, H-ras but Not K-ras Traffics to the Plasma Membrane through the Exocytic Pathway, Mol. Cell. Biol., 2000, 20(7), 2475–2487 CrossRef CAS PubMed.
- T. Satoh, M. Endo, S. Nakamura and Y. Kaziro, Analysis of guanine nucleotide bound to ras protein in PC12 cells, FEBS Lett., 1988, 236(1), 185–189 CrossRef CAS PubMed.
- B. Franke, J. W. N. Akkerman and J. L. Bos, Rapid Ca2+-mediated activation of Rap1 in human platelets, EMBO J., 1997, 16(2), 252–259 CrossRef CAS PubMed.
- M. J. Baker and I. Rubio, Active GTPase Pulldown Protocol, Methods Mol. Biol., 2021, 2262, 117–135 CrossRef CAS PubMed.
- V. K. Chiu, T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Johnson, A. D. Cox and M. R. Philips, Ras signalling on the endoplasmic reticulum and the Golgi, Nat. Cell Biol., 2002, 4(5), 343–350 CrossRef CAS PubMed.
- T. G. Bivona, I. Pérez de Castro, I. M. Ahearn, T. M. Grana, V. K. Chiu, P. J. Lockyer, P. J. Cullen, A. Pellicer, A. D. Cox and M. R. Philips, Phospholipase Cγ activates Ras on the Golgi apparatus by means of RasGRP1, Nature, 2003, 424(6949), 694–698 CrossRef CAS PubMed.
- M. Augsten, R. Pusch, C. Biskup, K. Rennert, U. Wittig, K. Beyer, A. Blume, R. Wetzker, K. Friedrich and I. Rubio, Live-cell imaging of endogenous Ras-GTP illustrates predominant Ras activation at the plasma membrane, EMBO Rep., 2006, 7(1), 46–51 CrossRef CAS PubMed.
- J. D. McLaurin and O. D. Weiner, Multiple sources of signal amplification within the B-cell Ras/MAPK pathway, Mol. Biol. Cell, 2019, 30(13), 1610–1620 CrossRef CAS PubMed.
- S. Okumoto, A. Jones and W. B. Frommer, Quantitative imaging with fluorescent biosensors, Annu. Rev. Plant Biol., 2012, 63, 663–706 CrossRef CAS PubMed.
- T. G. Bivona, S. Quatela and M. R. Philips, Analysis of Ras Activation in Living Cells with GFP-RBD, Methods Enzymol., 2006, 407(1998), 128–143 CAS.
- A. Takaya, Y. Ohba, K. Kurokawa and M. Matsuda, RalA Activation at Nascent Lamellipodia of Epidermal Growth Factor-stimulated Cos7 Cells and Migrating Madin-Darby Canine Kidney Cells, Mol. Biol. Cell, 2004, 15(June), 2549–2557 CrossRef CAS PubMed.
- A. Takaya, T. Kamio, M. Masuda, M. Naoki, H. Sawa, M. Sato, K. Nagashima, A. Mizutani, A. Matsuno, E. Kiyokawa and M. Matsuda, R-Ras Regulates Exocytosis by Rgl2/Rlf-mediated Activation of RalA on Endosomes, Mol. Biol. Cell, 2007, 18(May), 1850–1860 CrossRef CAS PubMed.
- M. Kitano, M. Nakaya, T. Nakamura, S. Nagata and M. Matsuda, Imaging of Rab5 activity identifies essential regulators for phagosome maturation, Nature, 2008, 453(7192), 241–245 CrossRef CAS PubMed.
- R. E. Itoh, K. Kurokawa, Y. Ohba, H. Yoshizaki, N. Mochizuki and M. Matsuda, Activation of Rac and Cdc42 Video Imaged by Fluorescent Resonance Energy Transfer-Based Single-Molecule Probes in the Membrane of Living Cells, Mol. Cell. Biol., 2002, 22(18), 6582–6591 CrossRef CAS PubMed.
- O. Pertz, L. Hodgson, R. L. Klemke and K. M. Hahn, Spatiotemporal dynamics of RhoA activity in migrating cells, Nature, 2006, 440(7087), 1069–1072 CrossRef CAS PubMed.
- K. Aoki and M. Matsuda, Visualization of small GTPase activity with fluorescence resonance energy transfer-based biosensors, Nat. Protoc., 2009, 4(11), 1623–1631 CrossRef CAS PubMed.
- V. S. Kraynov, C. Chamberlain, G. M. Bokoch, M. A. Schwartz, S. Slabaugh and K. M. Hahn, Localized Rac activation dynamics visualized in living cells, Science, 2000, 290(5490), 333–337 CrossRef CAS PubMed.
- O. Rocks, A. Peyker, M. Kahms, P. J. Verveer, C. Koerner, M. Lumbierres, J. Kuhlmann, H. Waldmann, A. Wittinghofer and P. I. H. Bastiaens, An acylation cycle regulates localization and activity of palmitoylated ras isoforms, Science, 2005, 307(5716), 1746–1752 CrossRef CAS PubMed.
- M. Fivaz, S. Bandara, T. Inoue and T. Meyer, Robust Neuronal Symmetry Breaking by Ras-Triggered Local Positive Feedback, Curr. Biol., 2008, 18(1), 44–50 CrossRef CAS PubMed.
- N. Bery and T. H. Rabbitts, Bioluminescence Resonance Energy Transfer 2 (BRET2)-Based RAS Biosensors to Characterize RAS Inhibitors, Curr. Protoc. Cell Biol., 2019, 83(1), 1–22 Search PubMed.
- N. Bery, A. Cruz-Migoni, C. Bataille Jr., C. E. Quevedo, H. Tulmin, A. Miller, A. Russell, S. E. Phillips, S. B. Carr and T. H. Rabbitts, BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein–protein interactions, eLife, 2018, 7, 1–28 Search PubMed.
- C. E. Quevedo, A. Cruz-Migoni, N. Bery, A. Miller, T. Tanaka, D. Petch, C. J. R. Bataille, L. Y. W. Lee, P. S. Fallon, H. Tulmin, M. T. Ehebauer, N. Fernandez-Fuentes, A. J. Russell, S. B. Carr, S. E. V. Phillips and T. H. Rabbitts, Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment, Nat. Commun., 2018, 9, 3169 CrossRef PubMed.
- J. Kim, S. Lee, K. Jung, W. C. Oh, N. Kim, S. Son, Y. J. Jo, H. B. Kwon and W. Do Heo, Intensiometric biosensors visualize the activity of multiple small GTPases in vivo, Nat. Commun., 2019, 10, 211 CrossRef PubMed.
- R. Weeks, X. Zhou, T. L. Yuan and J. Zhang, Fluorescent Biosensor for Measuring Ras Activity in Living Cells, J. Am. Chem. Soc., 2022, 144(38), 17432–17440 CrossRef CAS PubMed.
- N. Mochizuki, S. Yamashita, K. Kurokawa, Y. Ohba, T. Nagai, A. Miyawaki and M. Matsuda, Spatio-temporal images of growth-factor-induced activation of Ras and Rap1, Nature, 2001, 411(6841), 1065–1068 CrossRef CAS PubMed.
- A. T. Sasaki, C. Chun, K. Takeda and R. A. Firtel, Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement, J. Cell Biol., 2004, 167(3), 505–518 CrossRef CAS PubMed.
- P. G. Charest and R. A. Firtel, Feedback signaling controls leading-edge formation during chemotaxis, Curr. Opin. Genet. Dev., 2006, 16(4), 339–347 CrossRef CAS PubMed.
- A. T. Sasaki, C. Janetopoulos, S. Lee, P. G. Charest, K. Takeda, L. W. Sundheimer, R. Meili, P. N. Devreotes and R. A. Firtel, G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility, J. Cell Biol., 2007, 178(2), 185–191 CrossRef CAS PubMed.
- S. Zhang, P. G. Charest and R. A. Firtel, Spatiotemporal Regulation of Ras Activity Provides Directional Sensing, Curr. Biol., 2008, 18(20), 1587–1593 CrossRef CAS PubMed.
- K. Takeda, D. Shao, M. Adler, P. G. Charest, W. F. Loomis, H. Levine, A. Groisman, W. J. Rappel and R. A. Firtel, Cell biology: Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway, Sci. Signaling, 2012, 5(205), 29–31 CrossRef PubMed.
- Y. Xiong, C. H. Huang, P. A. Iglesias and P. N. Devreotes, Cells navigate with a local-excitation, global-inhibition-biased excitable network, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(40), 17079–17086 CrossRef CAS PubMed.
- C. H. Huang, M. Tang, C. Shi, P. A. Iglesias and P. N. Devreotes, An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration, Nat. Cell Biol., 2013, 15(11), 1307–1316 CrossRef CAS PubMed.
- Y. Miao, S. Bhattacharya, M. Edwards, H. Cai, T. Inoue, P. A. Iglesias and P. N. Devreotes, Altering the threshold of an excitable signal transduction network changes cell migratory modes, Nat. Cell Biol., 2017, 19(4), 329–340 CrossRef CAS PubMed.
- J. Kuhn, Y. Lin and P. N. Devreotes, Using Live-Cell Imaging and Synthetic Biology to Probe Directed Migration in Dictyostelium, Front. Cell Dev. Biol., 2021, 9, 740205 CrossRef PubMed.
- J. V. Thevathasan, E. Tan, H. Zheng, Y. C. Lin, Y. Li, T. Inoue and M. Fivaz, The small GTPase HRas shapes local PI3K signals through positive feedback and regulates persistent membrane extension in migrating fibroblasts, Mol. Biol. Cell, 2013, 24(14), 2228–2237 CrossRef CAS PubMed.
- N. M. Neumann, M. C. Perrone, J. H. Veldhuis, R. J. Huebner, H. Zhan, P. N. Devreotes, G. W. Brodland and A. J. Ewald, Coordination of Receptor Tyrosine Kinase Signaling and Interfacial Tension Dynamics Drives Radial Intercalation and Tube Elongation, Dev. Cell, 2018, 45(1), 67–82 CrossRef CAS PubMed.
- S. Q. Zhang, W. Yang, M. I. Kontaridis, T. G. Bivona, G. Wen, T. Araki, J. Luo, J. A. Thompson, B. L. Schraven, M. R. Philips and B. G. Neel, Shp2 Regulates Src Family Kinase Activity and Ras/Erk Activation by Controlling Csk Recruitment signaling strategy. Ligand binding activates intrinsic (RTK) or associated (integrin) PTK activity, resulting in the recruitment and phosphorylation of substra, Mol. Cell, 2004, 13, 341–355 CrossRef CAS PubMed.
- I. Perez de Castro, T. G. Bivona, M. R. Philips and A. Pellicer, Ras Activation in Jurkat T cells following Low-Grade Stimulation of the T-Cell Receptor Is Specific to N-Ras and Occurs Only on the Golgi Apparatus, Mol. Cell. Biol., 2004, 24(8), 3485–3496 CrossRef PubMed.
- M. Schmick, A. Kraemer and P. I. H. Bastiaens, Ras moves to stay in place, Trends Cell Biol., 2015, 25(4), 190–197 CrossRef CAS PubMed.
- A. Lorentzen, A. Kinkhabwala, O. Rocks, N. Vartak and P. I. H. Bastiaens, Regulation of Ras localization by acylation enables a mode of intracellular signal propagation, Sci. Signaling, 2010, 3(140), 1–9 CrossRef PubMed.
- I. Rubio, S. Grund, S.-P. Song, C. Biskup, S. Bandemer, M. Fricke, M. Förster, A. Graziani, U. Wittig and S. Kliche, TCR-Induced Activation of Ras Proceeds at the Plasma Membrane and Requires Palmitoylation of N-Ras, J. Immunol., 2010, 185(6), 3536–3543 CrossRef CAS PubMed.
- B. Casar, A. P. Badrock, I. Jiménez, I. Arozarena, P. Colón-Bolea, L. F. Lorenzo-Martín, I. Barinaga-Rementería, J. Barriuso, V. Cappitelli, D. J. Donoghue, X. R. Bustelo, A. Hurlstone and P. Crespo, RAS at the Golgi antagonizes malignant transformation through PTPRκ-mediated inhibition of ERK activation, Nat. Commun., 2018, 9(1), 1–17 CrossRef CAS PubMed.
- S. Bunda, P. Heir, T. Srikumar, J. D. Cook, K. Burrell, Y. Kano, J. E. Lee, G. Zadeh, B. Raught and M. Ohh, Src promotes GTPase activity of Ras via tyrosine 32 phosphorylation, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(36), E3785–E3794 CrossRef CAS PubMed.
- Y. Kano, T. Gebregiworgis, C. B. Marshall, N. Radulovich, B. P. K. Poon, J. St-Germain, J. D. Cook, I. Valencia-Sama, B. M. M. Grant, S. G. Herrera, J. Miao, B. Raught, M. S. Irwin, J. E. Lee, J. J. Yeh, Z. Y. Zhang, M. S. Tsao, M. Ikura and M. Ohh, Tyrosyl phosphorylation of KRAS stalls GTPase cycle via alteration of switch I and II conformation, Nat. Commun., 2019, 10, 224, DOI:10.1038/s41467-018-08115-8.
- D. K. Simanshu, D. V. Nissley and F. McCormick, RAS Proteins and Their Regulators in Human Disease, Cell, 2017, 170(1), 17–33 CrossRef CAS PubMed.
- C. A. Stalnecker and C. J. Der, RAS, wanted dead or alive: Advances in targeting RAS mutant cancers, Sci. Signaling, 2020, 13(624), 1–7 CrossRef PubMed.
- P. Lito, M. Solomon, L. S. Li, R. Hansen and N. Rosen, Cancer therapeutics: Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism, Science, 2016, 351(6273), 604–608 CrossRef CAS PubMed.
- M. P. Patricelli, M. R. Janes, L. S. Li, R. Hansen, U. Peters, L. V. Kessler, Y. Chen, J. M. Kucharski, J. Feng, T. Ely, J. H. Chen, S. J. Firdaus, A. Babbar, P. Ren and Y. Liu, Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state, Cancer Discovery, 2016, 6(3), 316–329 CrossRef CAS PubMed.
- M. R. Janes, J. Zhang, L. S. Li, R. Hansen, U. Peters, X. Guo, Y. Chen, A. Babbar, S. J. Firdaus, L. Darjania, J. Feng, J. H. Chen, S. Li, S. Li, Y. O. Long, C. Thach, Y. Liu, A. Zarieh, T. Ely, J. M. Kucharski, L. V. Kessler, T. Wu, K. Yu, Y. Wang, Y. Yao, X. Deng, P. P. Zarrinkar, D. Brehmer, D. Dhanak, M. V. Lorenzi, D. Hu-Lowe, M. P. Patricelli, P. Ren and Y. Liu, Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor, Cell, 2018, 172(3), 578–589 CrossRef CAS PubMed.
- J. M. Ostrem, U. Peters, M. L. Sos, J. A. Wells and K. M. Shokat, K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions, Nature, 2013, 503(7477), 548–551 CrossRef CAS PubMed.
- H. A. Blair, Sotorasib: First Approval, Drugs, 2021, 81(13), 1573–1579 CrossRef CAS PubMed.
- D. J. Anderson, J. K. Durieux, K. Song, R. Alvarado, P. K. Jackson, G. Hatzivassiliou and M. J. C. Ludlam, Live-cell microscopy reveals small molecule inhibitor effects on MAPK pathway dynamics, PLoS One, 2011, 6(8), e22607 CrossRef CAS PubMed.
- T. Jin, H. Lavoie, M. Sahmi, M. David, C. Hilt, A. Hammell and M. Therrien, RAF inhibitors promote RAS-RAF interaction by allosterically disrupting RAF autoinhibition, Nat. Commun., 2017, 8, 1211 CrossRef PubMed.
- A. Tulpule, J. Guan, D. S. Neel, H. R. Allegakoen, Y. P. Lin, D. Brown, Y. T. Chou, A. Heslin, N. Chatterjee, S. Perati, S. Menon, T. A. Nguyen, J. Debnath, A. D. Ramirez, X. Shi, B. Yang, S. Feng, S. Makhija, B. Huang and T. G. Bivona, Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules, Cell, 2021, 184(10), 2649–2664 CrossRef CAS PubMed.
- A. F. Overbeck, T. R. Brtva, A. D. Cox, S. M. Graham, S. Y. Huff, R. Khorsavi-Far, L. A. Quilliam, P. A. Solski and C. J. Der, Guanine nucleotide exchange factors: Activators of Ras superfamily proteins, Mol. Reprod. Dev., 1995, 42, 468–476 CrossRef CAS PubMed.
- Y. Kamioka, K. Sumiyama, R. Mizuno and M. Matsuda Live imaging of transgenic mice expressing FRET biosensors. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS. 2013:125-8.
- H. Murakoshi, R. Iino, T. Kobayashi, T. Fujiwara, C. Ohshima, A. Yoshimura and A. Kusumi, Single-molecule imaging analysis of Ras activation in living cells, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(19), 7317–7322 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |