Open Access Article
Open Access ArticleWastewater-based surveillance of COVID-19 and removal of SARS-CoV-2 RNA across a major wastewater treatment plant in San Antonio, Texas†
Haya
Al-Duroobi
a,
Kiran Kumar
Vadde
a,
Duc C.
Phan‡
a,
Sina V.
Moghadam
a,
Arash
Jafarzadeh
a,
Akanksha
Matta
b,
Marcio
Giacomoni
a and
Vikram
Kapoor
 *a
*a
aSchool of Civil & Environmental Engineering, and Construction Management, University of Texas at San Antonio, San Antonio, TX 78249, USA. E-mail: vikram.kapoor@utsa.edu
bDepartment of Chemistry, University of Texas at San Antonio, San Antonio, TX 78249, USA
First published on 15th March 2023
Abstract
Wastewater-based epidemiology (WBE) targeting SARS-CoV-2 RNA detection in municipal wastewater is considered a valuable tool for COVID-19 surveillance in a community. However, the persistence and removal of SARS-CoV-2 RNA in wastewater treatment plants (WWTPs) have not been well investigated. This study is aimed at detecting SARS-CoV-2 RNA in wastewater to correlate viral concentrations with clinical COVID-19 cases in the sewershed and determine whether the SARS-CoV-2 genetic material is detectable after treatment. Raw influent, primary effluent (after primary clarification), secondary effluent (after activated sludge treatment), and final effluent (after chlorination) samples were collected two times a week from the largest WWTP in San Antonio (Texas) during April to November 2021 and analyzed for SARS-CoV-2 RNA (N1 and N2 genes) concentrations using the reverse transcription droplet digital polymerase chain reaction (RT-ddPCR). SARS-CoV-2 RNA was detected in 98.5% (n = 34 weeks) of the raw influent samples and anticipated the trends of the COVID-19 outbreak. Furthermore, a higher correlation between viral concentrations and COVID-19 cases was observed for two days a week sampling frequency (ρ = 0.75, p <0.001) than one day per week (ρ = 0.60, p <0.001). Despite the high SARS-CoV-2 RNA concentrations in raw sewage, a significant amount of viral RNA was removed at primary and secondary clarifiers (removal efficiencies were 54% and 94%, respectively) and was undetectable in final effluents. These results demonstrate the performance of the WWTP in reducing the SARS-CoV-2 RNA concentration and further highlight the role of tertiary treatment and chlorination in eliminating SARS-CoV-2 RNA in receiving waters.
Environmental significanceThe wastewater-based surveillance technology has been targeting SARS-CoV-2 RNA in wastewater to monitor COVID-19 prevalence alongside clinical testing. However, very few studies have investigated the fate of SARS-CoV-2 RNA in wastewater treatment plants. Determining this vital information would be useful to better characterize the risk related to effluent discharges to the environment, sludge disposal and recycled water reuse. This comprehensive study aimed at detecting the SARS-CoV-2 RNA in raw influent samples to correlate viral concentrations with clinical COVID-19 cases and evaluating the persistence and removal of the SARS-CoV-2 genetic material along four stages of the activated sludge treatment process at a wastewater treatment plant in San Antonio, Texas. |
Introduction
The current COVID-19 pandemic caused by the novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) has accounted for more than 6.6 million deaths and 650 million confirmed cases worldwide as of December 2022.1 Due to the limited availability of clinical testing resources in the early stages of the pandemic and high levels of asymptomatic infected individuals (approximately 30% of all carriers), difficulties in tracking the transmission and assessing the true scale of COVID-19 burden in communities were reported.2,3 However, although SARS-CoV-2 is primarily a respiratory-transmitted pathogen that transmits through respiratory droplets or aerosols, the virus shedding in feces of both symptomatic and asymptomatic infected individuals enabled the detection of viral RNA in the untreated sewage or raw influents of wastewater treatment plants (WWTPs) and facilitated the wastewater-based epidemiology (WBE) of COVID-19 infections.4–7 Subsequently, as untreated sewage or raw influent testing is not impacted by asymptomatic infections or the availability of diagnostic testing resources, WBE has emerged as a valuable tool for the surveillance of SARS-CoV-2 transmission in sewershed areas, providing more insights about the infection trends of the community that are not captured by clinical testings.8–10 Currently, wastewater-based surveillance programs are employed globally to monitor COVID-19 trends in sewershed communities, and the priorities of these programs have evolved from general virus detection6 to the identification of emerging variants by sequencing the whole genome of the virus in wastewater.11,12 However, the detection of high SARS-CoV-2 RNA concentrations in raw influent samples around the world prompts public health concerns due to possible fecal-oral or aerosol transmission of the virus, yet only limited studies addressing the SARS-CoV-2 removal efficiencies in conventional and advanced WWTPs were carried out.13,14To date, there is minimal or no evidence supporting the potential transmission risk of COVID-19 through wastewater.15–17 For instance, few studies reported the possible risk of SARS-CoV-2 transmission via exposure to aerosols generated through untreated wastewater,18–22 while other studies stated that the evidence of virus infectivity via wastewater exposure is limited.23–26 For example, Gholipour et al.18 reported that the presence of SARS-CoV-2 RNA in the aerosols generated at WWTPs indicated an increased COVID-19 infection risk to wastewater workers, although the survival and fate of SARS-CoV-2 in wastewater were not assessed in this study. However, Bivins et al.23 reported that SARS-CoV-2 RNA was found to be significantly more persistent than infectious SARS-CoV-2, indicating that the environmental detection of RNA alone does not substantiate the risk of infection. Furthermore, a recent study performed on 134 wastewater workers from 59 wastewater treatment plants in Spain showed no increase in seroprevalence of IgG for SARS-CoV-2 (antibodies against COVID-19 infection), indicating that there is limited evidence of infection via wastewater.26 In another study it was reported that during the 2003 SARS epidemic in Hong Kong, wastewater aerosols containing coronaviruses (SARS-CoV-1) from a leaking sewage pipe in a residential apartment may have resulted in a cluster of community infections but there is neither epidemiological nor laboratory support for such mechanisms.27 Thus, there seems to be no evidence of infective viral particles in wastewater based on cell culture;16 furthermore, detection of SARS-CoV-2 RNA alone in wastewater does not suffice to characterize the risk of infection attributable to exposure to that wastewater.23
There is a critical need for further in-depth studies on SARS-CoV-2 and other enveloped viruses' persistence through wastewater treatment steps. Furthermore, the persistence of the SARS-CoV-2 genetic material in the treatment process impacts decision-making about the final discharge of treated effluent, sludge disposal and applications of recycled water reuse. Therefore, improving knowledge about the fate of SARS-CoV-2 RNA through wastewater primary, secondary and tertiary treatment stages and their removal performances in WWTPs would be useful to better characterize the risk related to discharges to the environment or wastewater reuse.14 Although cell culture-based methods are considered ideal for evaluating viral infectivity, infectious SARS-CoV-2 detection in wastewater has been rarely studied due to the complexity of isolation, detection sensitivity, and time consumption.23,28 However, PCR-based methods are commonly applied to detect SARS-CoV-2 RNA in treated effluents of WWTPs to investigate virus reduction and minimize the potential risk.5,29,30 It has been reported that viral loads are significantly reduced by conventional wastewater treatment processes; however, information on the reduction of SARS-CoV-2 RNA is still limited.13,30–34 Additional studies are required to demonstrate the role of WWTPs in removal of SARS-CoV-2 RNA in the effluents from secondary and/or tertiary treatments and to verify whether additional treatments are needed to protect the receiving environments.
In this regard, the present comprehensive study aimed at (1) detecting the SARS-CoV-2 RNA in raw influent samples to correlate viral concentrations with clinical COVID-19 cases and (2) evaluating the persistence of the SARS-CoV-2 genetic material throughout the activated sludge treatment process at the largest WWTP in San Antonio, Texas. In addition to quantifying viral loads in primary and secondary effluents, the final treated effluent was also evaluated to determine the effectiveness of the chlorine disinfection method. SARS-CoV-2 RNA quantification using the reverse transcription droplet digital polymerase chain reaction (RT-ddPCR) in raw influent samples collected from April 2021 to November 2021 captured the third COVID-19 wave in San Antonio. We used both raw and recovery-adjusted SARS-CoV-2 RNA concentrations to correlate with COVID-19 case data in the city. Also, we assessed the lag in clinical data compared to WBE data, using correlations with COVID-19 incidence reported by the City of San Antonio. Furthermore, sampling frequency was also evaluated to verify if sampling twice a week versus once a week does indeed enhance the WBE strategy for better estimates of community-wide incidence rates. The SARS-CoV-2 genetic material removal during the different treatment stages presented in this study provides valuable information to the water industry for ensuring the reduction of the virus in wastewater and may enhance public awareness and confidence in the potable reuse practice.
Materials and methods
Description of the wastewater treatment plant and sample collection
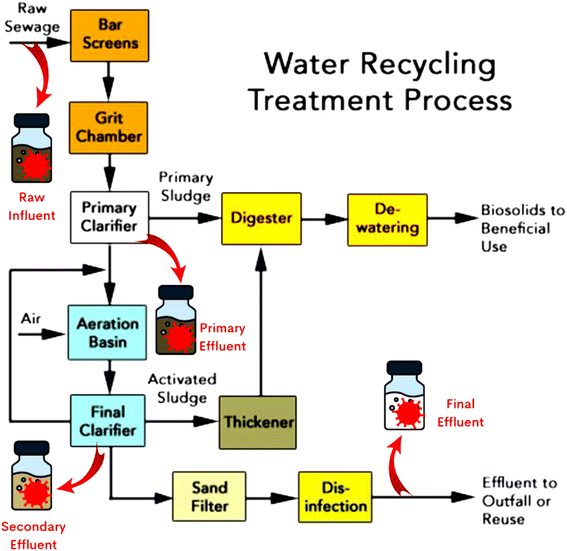
This study was conducted using samples collected from the Steven M. Clouse Water Recycling Center (WRC), which is the largest wastewater treatment plant in San Antonio, Texas. It serves approximately 399![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 wastewater customers and has a permitted flow of 125 million gallons per day (MGD). Wastewater samples were collected from four stages of the treatment process: raw influent at the plant's inlet, primary effluent, secondary effluent, and the final effluent before outfall (Fig. 1). Primary effluent samples were captured after the primary clarification stage where raw influent is routed through bar screens and grit chambers and undergoes primary clarification in which biosolids are settled and removed. Secondary effluent samples were collected after secondary biological treatment (activated sludge system) that involves water flowing into aeration basins and then final clarifiers. The tertiary treatment includes sand filtration, followed by 20 minutes of chlorine disinfection and dechlorination with sulfur dioxide. After the advanced (tertiary) treatment, final effluent samples were collected before the outfall.
000 wastewater customers and has a permitted flow of 125 million gallons per day (MGD). Wastewater samples were collected from four stages of the treatment process: raw influent at the plant's inlet, primary effluent, secondary effluent, and the final effluent before outfall (Fig. 1). Primary effluent samples were captured after the primary clarification stage where raw influent is routed through bar screens and grit chambers and undergoes primary clarification in which biosolids are settled and removed. Secondary effluent samples were collected after secondary biological treatment (activated sludge system) that involves water flowing into aeration basins and then final clarifiers. The tertiary treatment includes sand filtration, followed by 20 minutes of chlorine disinfection and dechlorination with sulfur dioxide. After the advanced (tertiary) treatment, final effluent samples were collected before the outfall.
24 hour composite samples were collected from each of the four treatment stages using ISCO 3700 autosamplers (Teledyne ISCO, Lincoln, NE). Raw influent samples were collected twice a week (Mondays and Wednesdays) from April 12, 2021 to November 29, 2021, for a total of 34 weeks (n = 67) to study the correlation between viral concentrations and clinical COVID-19 cases in the sewershed. Similarly, primary, secondary, and final effluent samples were collected twice a week from April 12 to July 26, 2021, for a total of 16 weeks (n = 31 for each type of effluent sample) to assess the virus removal efficiency between different stages of the treatment plant. After collection, samples were stored on ice and delivered to the laboratory at the University of Texas at San Antonio (UTSA). The samples were immediately concentrated upon delivery and frozen at −80 °C, followed by molecular analyses.
Virus concentration and nucleic acid extraction
The SARS-CoV-2 virus in the wastewater samples was concentrated using an adsorption–extraction method combined with high-speed centrifugation as described in previous studies.35–37 To determine the recovery efficiency of the virus concentration and nucleic acid extraction methods applied in this study, an attenuated vaccine strain of bovine coronavirus (Calf-Guard, Zoetis, Parsippany, NJ) was used as a surrogate viral RNA target. Prior to the sample concentration process, wastewater samples were directly spiked with 106 copies of bovine coronavirus (BCoV) per liter of wastewater. Subsequently, 200 mL of each sample was centrifuged at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min at 4 °C to pellet the solids, and the supernatant was passed through 0.45 μm pore-size, 47 mm diameter electronegative membranes (GN-6 Metricel Membrane Disc Filter, Pall Laboratory). Both the pellet and membrane were stored together in the same tube at −80 °C until nucleic acid extraction.
000g for 15 min at 4 °C to pellet the solids, and the supernatant was passed through 0.45 μm pore-size, 47 mm diameter electronegative membranes (GN-6 Metricel Membrane Disc Filter, Pall Laboratory). Both the pellet and membrane were stored together in the same tube at −80 °C until nucleic acid extraction.
Total viral RNA and DNA were extracted simultaneously from the samples using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) and in combination with the automated robot QIAcube Connect (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Briefly, the concentrated samples (filter membranes and pellets) were transferred into screw-cap microcentrifuge tubes containing DNase and RNase-free glass beads (PowerBead Tube, Qiagen, Hilden, Germany) and subjected to bead-beating for 3 min at maximum speed (PowerLyzer 24 Homogenizer, MO-BIO, Carlsbad, CA) in the presence of lysis buffer (Buffer RLT Plus; Qiagen, Hilden, Germany) containing β-mercaptoethanol (Sigma-Aldrich Co., St. Louis, MO). Samples were centrifuged at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130g for 10 min and the supernatant was used for the extraction of RNA and DNA separately with the AllPrep DNA/RNA Minikit, following the manufacturer's instructions. Finally, the extracted RNA and DNA were each eluted in 100 μL RNase-free water, respectively. The concentration and purity of RNA and DNA were determined by using a NanoDrop One Spectrophotometer (Thermo Scientific, Wilmington, DE). RNA extracts were stored at −80 °C and DNA extracts were stored at −20 °C until they were used in molecular assays.
130g for 10 min and the supernatant was used for the extraction of RNA and DNA separately with the AllPrep DNA/RNA Minikit, following the manufacturer's instructions. Finally, the extracted RNA and DNA were each eluted in 100 μL RNase-free water, respectively. The concentration and purity of RNA and DNA were determined by using a NanoDrop One Spectrophotometer (Thermo Scientific, Wilmington, DE). RNA extracts were stored at −80 °C and DNA extracts were stored at −20 °C until they were used in molecular assays.
RT-ddPCR
RNA extracts were used as templates for the RT-ddPCR assays using a Bio-Rad QX200™ Droplet Digital™ PCR System as per the procedures detailed in the ESI† (RT-ddPCR detailed procedure). SARS-CoV-2 RNA was measured in each sample using the CDC N1 and N2 assays (Table 1). Technical duplicates were run (two wells) and wells were merged for data analysis. For each plate, positive control and no-template control (NTC) were run in duplicate. Ten-fold dilutions of selected RNA extracts were used to test for PCR inhibition as described in the ESI (see RT-ddPCR detailed procedure and Table S6†). The 2019-nCoV_N_Positive Control containing the complete nucleocapsid gene of SARS-CoV-2 was purchased from Integrated DNA Technologies (IDT, Coralville, IA) and was used as a positive control in this study. The ddPCR data was analyzed using the QuantaSoft Analysis Pro (Bio-Rad) software and concentrations were calculated by either allowing the program to auto select a threshold or by manual calling when the program was not able to auto threshold. The required number of droplets for a sample with merged duplicate wells was at least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The limit of detection (LOD) and the limit of quantification (LOQ) were determined for the N1 and N2 assays (see ESI† – RT-ddPCR detailed procedure). Average concentrations per reaction were converted to copies per liter of wastewater using dimensional analysis (Table S1†). For virus recovery efficiency, BCoV was quantified using RNA extracts and a one-step RT-ddPCR assay targeting the bovine coronavirus gene (Table 1; see ESI† – RNA recovery efficiency). The RT-ddPCR data for wastewater samples are provided in Tables S2 and S3† and examples of the positive and negative results for each assay are listed in Table S4,† and a Minimum Information for Publication of Quantitative Digital PCR Experiments (dMIQE) checklist is shown in Table S5.†
000. The limit of detection (LOD) and the limit of quantification (LOQ) were determined for the N1 and N2 assays (see ESI† – RT-ddPCR detailed procedure). Average concentrations per reaction were converted to copies per liter of wastewater using dimensional analysis (Table S1†). For virus recovery efficiency, BCoV was quantified using RNA extracts and a one-step RT-ddPCR assay targeting the bovine coronavirus gene (Table 1; see ESI† – RNA recovery efficiency). The RT-ddPCR data for wastewater samples are provided in Tables S2 and S3† and examples of the positive and negative results for each assay are listed in Table S4,† and a Minimum Information for Publication of Quantitative Digital PCR Experiments (dMIQE) checklist is shown in Table S5.†
| Assay | Primer/probe | Reference |
|---|---|---|
| Bovine coronavirus (BCoV) | F: CTGGAAGTTGGTGGAGTT | Decaro et al. 2008 |
| R: ATTATCGGCCTAACATACATC | ||
| P: FAM-CCTTCATAT-ZEN-CTATACACATCAAGTTGTT-IBFQ | ||
| SARS-CoV-2 (CDC N1) | F: GACCCCAAAATCAGCGAAAT | Lu et al. 2020 |
| R: TCTGGTTACTGCCAGTTGAATCTG | ||
| P: FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ-1 | ||
| SARS-CoV-2 (CDC N2) | F: TTACAAACATTGGCCGCAAA | Lu et al. 2020 |
| R: GCGCGACATTCCGAAGAA | ||
| P: FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ-1 |
Clinical data
Clinical testing data for COVID-19 was obtained from the City of San Antonio's COVID-19 open data repository (https://cosacovid-cosagis.hub.arcgis.com/). The daily incident COVID-19 cases include COVID-19 cases reported on each date and the weekly average COVID-19 cases is the moving average of COVID-19 cases reported in the last 7 days.38Statistical analysis
All statistical analyses and graphs were completed using GraphPad Prism, version 9.3.1 (LaJolla, CA). The wastewater virus RNA concentrations were reported as copies per L of wastewater. Paired and unpaired t-tests were used to compare groups after confirming that the data were normally distributed. Spearman's rank correlation coefficients were used to determine the relationship between the SARS-CoV-2 RNA concentration in wastewater and the associated COVID-19 cases. Using the 7 day average of clinical COVID-19 cases reported on the wastewater sample collection date, the correlation between average weekly SARS-CoV-2 N1 and N2 concentrations in raw influent samples and the weekly moving average of reported COVID-19 cases in the sewershed area was determined. Correlations were also examined using SARS-CoV-2 wastewater concentrations adjusted to the BCoV recovery rate. In addition, correlations of SARS-CoV-2 N1 concentrations with non-lagged and 7 day lagged clinical data were estimated using Spearman's correlation. Similarly, Spearman's correlations were also carried out using single and two-day sampling frequency per week data to assess the ideal sampling frequency required to accomplish valid COVID-19 trend estimates.Results and discussion
Recovery of BCoV in wastewater samples
The recovery efficiency of virus concentration and nucleic acid extraction protocols applied in this study for each sample type was measured by spiking the BCoV vaccine as a surrogate viral RNA (Table 2). The raw influent samples had the highest mean recovery of 25.7 ± 12.5%, followed by the primary effluent with 20.2 ± 17.0% mean recovery. The secondary effluent had the lowest mean BCoV recovery (19.7 ± 18.6%) among the three sample types. Although the difference in the BCoV recovery percentage between raw influent and subsequent effluent samples was not statistically significant (ANOVA, p = 0.35), the higher recovery efficiency of BCoV RNA in raw influent samples collected prior to any treatment could be due to the presence of a larger number of suspended solids and biosolids which provides enhanced viral particle adsorption.31,39–41 However, the process control virus recovery efficiencies could differ among studies due to variations in characteristics of wastewater, the volume used for processing the samples, and the virus concentration methods applied.3,34,37,42 For instance, Randazzo et al. 202042 reported that the recovery efficiency of process controls viz PEDV (porcine coronavirus) and MgV (mengovirus) using an aluminum hydroxide adsorption–precipitation concentration method was 10 ± 3.5% and 10 ± 2.1% for raw influents and 3.3 ± 1.6% and 6.2 ± 1.0% for final effluents, respectively; while Polanco et al. 202334 reported a higher recovery (27.2%) of process controls using Φ6 phage (phi6 phage) in effluent samples after an ultraviolet advanced oxidation process.| Raw influent | Primary effluent | Secondary effluent | |
|---|---|---|---|
| BCoV recovery % | 25.7 (±12.5%) | 20.2 (±17.0%) | 19.7 (±18.6%) |
SARS-CoV-2 RNA detection in raw wastewater and comparison with clinical data
The concentrations of SARS-CoV-2 genetic targets N1 and N2 (copies per L) were measured in raw influent samples to monitor the trend of SARS-CoV-2 RNA in the largest sewershed of San Antonio, Texas. A total of 67 influent samples were collected between April and November 2021 with a twice weekly (every Monday and Wednesday) sampling frequency campaign. A total of 66 tested samples (98.5%) showed a positive detection for N1 with a concentration ranging from 4.5 × 102 and 2.8 × 104 copies per L of wastewater (quantifiable samples (QS) n = 63, ESI Table S2†). The N2 gene target was positively detected in 62 (93%) samples collected with a concentration ranging between 4.7 × 102 and 1.4 × 104 copies per L of wastewater (QS n = 52) (Fig. 2). The SARS-CoV-2 N1 and N2 concentrations showed a significant positive correlation with each other (Spearman's rank correlation coefficient ρ = 0.90; p <0.001). The results of the current study are consistent with those of previous studies which indicated that the SARS-CoV-2 N1 assay is more sensitive than the N2 assay and both are highly correlated.35,43,44 During the first three months of our study period, the average concentrations of both N1 and N2 gene targets were below 5000 copies per L. However, the trend in viral loads started to increase on July 5th and reached its peak on August 2nd, 2021, where the SARS-CoV-2 N1 and N2 concentrations in influent samples were 2.8 × 104 copies per L and 5.2 × 103 copies per L, respectively.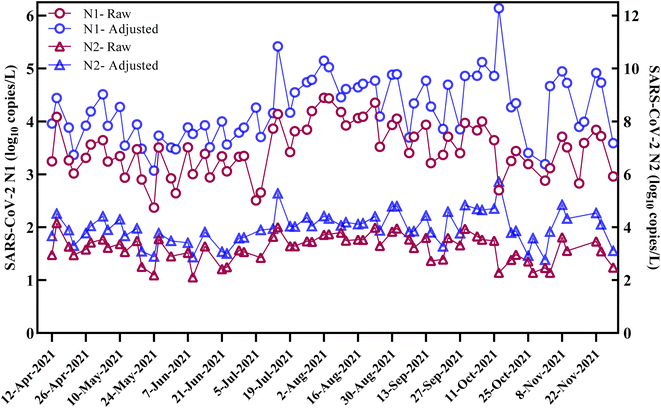 | ||
Fig. 2 Concentrations of SARS-CoV-2 N1 and N2 (log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) copies per L) in raw and BCoV recovery adjusted influent samples. copies per L) in raw and BCoV recovery adjusted influent samples. | ||
During this 34 week surveillance study from April 2021 to November 2021, the daily incident COVID-19 cases in San Antonio were relatively high in July and August, with the highest daily cases on August 14, 2021 (Fig. 3). We quantitatively compared SARS-CoV-2 RNA concentrations in wastewater with publicly reported data on COVID-19 cases. SARS-CoV-2 N1 and N2 levels exhibited both increasing and decreasing trends over the course of the study period (Fig. 3). Furthermore, these results suggest that recurring COVID-19 outbreaks in the sewershed contribute to the SARS-CoV-2 viral RNA loading into local WWTPs.45,46 Wastewater samples collected during the third wave of COVID-19 cases (July–August 2021) showed relatively higher SARS-CoV-2 RNA concentrations as compared to other periods in the study area.35,38 Notably, the SARS-COV-2 RNA levels were relatively lower from April–June 2021 and September–November 2021, which corresponded well with the initiation of vaccination in the study area and the decline in daily incident COVID-19 cases after the third wave, respectively.34
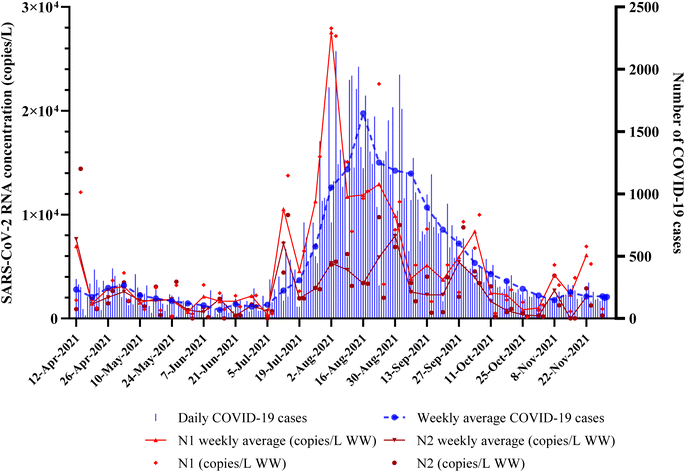 | ||
| Fig. 3 Correlation between reported COVID-19 cases in San Antonio and SARS-CoV-2 RNA concentrations (N1 and N2) measured in raw influent samples collected over 34 weeks (n = 67). | ||
Using the 7 day average of clinical COVID-19 cases reported on the wastewater sample collection date, the correlation between average weekly SARS-CoV-2 N1 and N2 concentrations in raw influent samples and the weekly moving average of reported COVID-19 cases in the sewershed area was determined. A significant positive correlation was observed between the weekly average COVID-19 cases and concentrations of N1 and N2 in raw wastewater (Spearman rank correlation coefficient ρ = 0.75 for N1; ρ = 0.64 for N2, p <0.001). However, when analyzed with recovery-adjusted data (Fig. 2), the correlation between SARS-CoV-2 RNA concentrations and weekly reported cases was reduced (ρ = 0.62 for N1 and ρ = 0.57 for N2, p <0.001). These findings are consistent with those of previous studies which indicated that normalization with the BCoV surrogate virus may reduce correlation with clinical data and may not be suitable for correcting SARS-CoV-2 RNA concentrations.3,35,47 As the SARS-CoV-2 N1 assay was able to detect targets in wastewater more consistently than the N2 assay and based on the above correlation results, the concentrations of SARS-CoV-2 N1 targets were selected for further correlation analysis.
Estimation of reporting lag time in clinical data
Our study was able to capture wastewater surveillance data from the third major COVID-19 pandemic wave (Delta variant) that ran from June 15, 2021, to August 31, 2021,.48,49 During this period, wastewater surveillance data collected in the first week of August had the peak weekly average reported concentrations for N1 (Fig. 3, Table S2†). However, the highest weekly average for COVID-19 cases was reported in the third week of August (Fig. 3), approximately one week after our highest SARS-CoV-2 RNA detection. To test the lag in clinical data reporting, the data of average weekly N1 concentrations were 7 days lagged and correlation analysis between lagged and non-lagged data was performed. The resulting analysis showed that the correlation between 7 day lagged N1 and weekly COVID-19 cases was slightly improved (ρ = 0.78, p <0.001), whereas correlation with same-day COVID-19 cases remained the same in the no-lag scenario as mentioned earlier (ρ = 0.75, p <0.001). The improved correlation for 3–14 days of lagged data was indicated in previous studies.50,51 For instance, a study conducted on a university campus at a subwatershed level50 and another larger scale wastewater monitoring project in Houston52 showed that lagged data correlated well with SARS-CoV-2 concentrations. Consequently, the wastewater viral load data collected in this study led to changes in the COVID-19 cases by one week, emphasizing the early-warning role that wastewater surveillance can play when coupled with clinical surveillance.Assessment of ideal sampling frequency for the wastewater-based surveillance program
This study was conducted with a sampling frequency of two sampling events per week (Mondays and Wednesdays). This sampling frequency is considered more frequent and over a longer period of time when compared to other similar studies.31,53 The frequency of sampling to enhance wastewater surveillance efforts has been one of the major research gaps that requires further probing. In order to assess whether an improvement in data correlation between viral loads and clinical cases was established with a 2 day versus 1 day per week sampling frequency, Spearman's correlation test was conducted (Table 3). Notably, the correlation between weekly average N1 concentrations and 7 day average cases was higher when sampling on two days (ρ = 0.75, p <0.001) rather than once a week (ρ = 0.60, p <0.001). However, the correlation between N1 and same-day COVID-19 case numbers was weak (ρ = 0.56, p <0.001). The CDC's guidelines on wastewater-based surveillance suggest a once per week sampling frequency for detecting the presence of SARS-CoV-2, and a minimum of 3 samples within a trend period of interest for surveillance purposes. Therefore, our results can be even more informative for establishing trends if the sample frequency is raised to three times per week. However, the higher sampling frequency can also be limited due to the laboratory capacity and supply chain shortages.54| Sampling frequency | Correlation between N1 & same-day COVID-19 cases | Correlation between average weekly N1 & 7 day average COVID-19 cases |
|---|---|---|
| Spearman's Rho (ρ) | Spearman's Rho (ρ) | |
| 2 days per week | 0.56 | 0.75 |
| Mondays ONLY | 0.50 | 0.60 |
| Wednesdays ONLY | 0.60 | 0.60 |
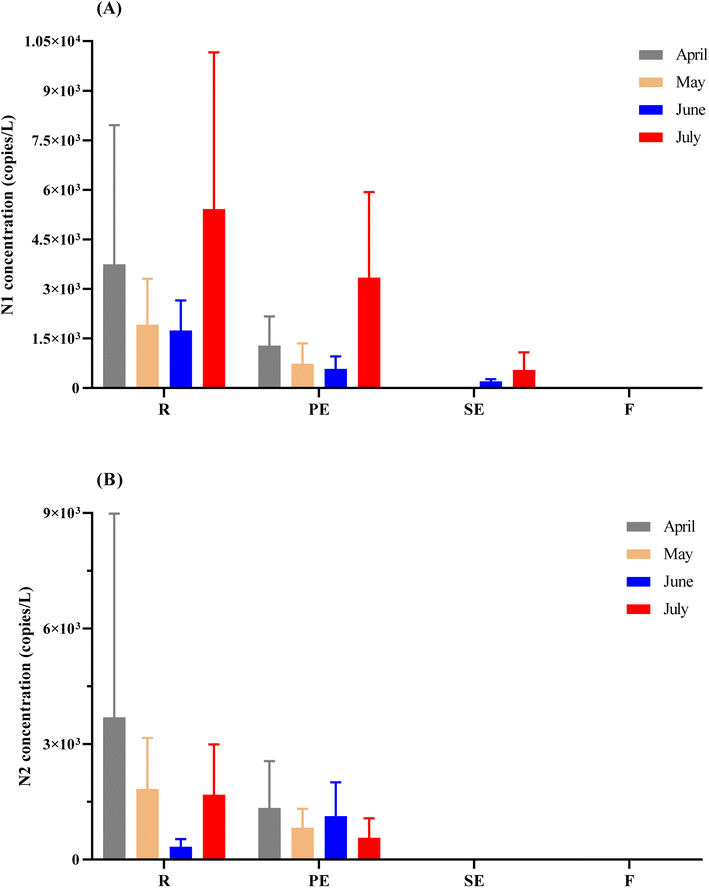
Removal of SARS-CoV-2 RNA by the activated sludge treatment process
In this study, the four stages of activated sludge treatment were sampled twice weekly over four months and samples (n = 31 for each type of sample) were analyzed to evaluate the persistence and removal of the SARS-CoV-2 genetic material. SARS-CoV-2 N1 gene targets were detected in all of the raw influent (R) and primary effluent (PE) samples, while N2 gene targets were detected in more than 95% of these sample types (Fig. 4A and B, ESI Table S3†). As for secondary effluent (SE) samples, SARS-CoV-2 N1 gene targets were detected only during June and July sampling campaigns (Fig. 4A, ESI Table S3†), and none of the samples were positive for the SARS-CoV-2 N2 gene target (Fig. 4B, ESI Table S3†). In the case of final effluents (F), all the samples tested were below the detection limit for SARS-CoV-2 N1 and N2 gene targets.The removal efficiency was evaluated using average SARS-CoV-2 N1 concentrations (n = 31) in copies per L of wastewater in the raw influent samples and subsequent effluent samples. In general, a 54% (0.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction) removal rate was achieved in the primary treatment stage, followed by 94% (1.2
log10 reduction) removal rate was achieved in the primary treatment stage, followed by 94% (1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction) in the secondary treatment stage, and 100% at the tertiary stage since no detections were observed in the final effluent samples. Evaluating the treatment efficiency between different stages showed that more than 87% (0.9
log10 reduction) in the secondary treatment stage, and 100% at the tertiary stage since no detections were observed in the final effluent samples. Evaluating the treatment efficiency between different stages showed that more than 87% (0.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction) of the viral load was removed between primary and secondary treatment. The higher levels of the N1 gene target were measured in July in the primary and secondary treatment stages. This result is related to the period when the viral RNA concentration measured in the raw influent started to increase (Fig. 4). Therefore, it can be anticipated that as the viral RNA loading increases in the raw influent, the removal of viral RNA by primary treatment starts to become more complex down the process train. The high viral RNA removal after primary and secondary clarification could be related to the hydrophobic envelope of SARS-CoV-2, which reduces the solubility of the virus in water and increases the probability of its adsorption to activated sludge and suspended solids.31,39,40 Therefore, collecting sludge samples spent after the primary and secondary treatment stages can verify the credibility of this result. Furthermore, evaluating the concentration of viral loads in the sludge samples can also identify the risk associated with handling and disposal of dewatered sludge. One of the limitations of the current study is that it did not include sludge analysis as we were not able to gain access to sludge at the treatment plant due to logistical and financial issues. Few previous studies reported high concentrations of SARS-CoV-2 RNA in primary, secondary, and thickened anaerobically treated sludge.13,39 It has been suggested that only after applying thermal hydrolysis SARS-CoV-2 RNA was non-detectable in sludge samples.32 Hence, the pre-treatment of dewatered sludge prior to disposal might be necessary to ensure the complete removal of SARS-CoV-2 RNA.
log10 reduction) of the viral load was removed between primary and secondary treatment. The higher levels of the N1 gene target were measured in July in the primary and secondary treatment stages. This result is related to the period when the viral RNA concentration measured in the raw influent started to increase (Fig. 4). Therefore, it can be anticipated that as the viral RNA loading increases in the raw influent, the removal of viral RNA by primary treatment starts to become more complex down the process train. The high viral RNA removal after primary and secondary clarification could be related to the hydrophobic envelope of SARS-CoV-2, which reduces the solubility of the virus in water and increases the probability of its adsorption to activated sludge and suspended solids.31,39,40 Therefore, collecting sludge samples spent after the primary and secondary treatment stages can verify the credibility of this result. Furthermore, evaluating the concentration of viral loads in the sludge samples can also identify the risk associated with handling and disposal of dewatered sludge. One of the limitations of the current study is that it did not include sludge analysis as we were not able to gain access to sludge at the treatment plant due to logistical and financial issues. Few previous studies reported high concentrations of SARS-CoV-2 RNA in primary, secondary, and thickened anaerobically treated sludge.13,39 It has been suggested that only after applying thermal hydrolysis SARS-CoV-2 RNA was non-detectable in sludge samples.32 Hence, the pre-treatment of dewatered sludge prior to disposal might be necessary to ensure the complete removal of SARS-CoV-2 RNA.
The removal efficiency after tertiary treatment was 100%, which suggests that biosolid settlement is crucial to achieving maximum removal prior to disinfection. Previous studies indicated that untreated wastewater sludge and Class B biosolids (mesophilic anaerobically digested biosolids) contained the genetic material from other coronaviruses in more than 80% of the samples.41 While this could be a concern, it is highly unlikely that infection can occur from Class B biosolids based on current evidence.24 The disinfection process used by the treatment plant in the current study is chlorination and it was reported that SARS-CoV-2 is extremely susceptible to chlorination degradation.31,44 The treatment plant in the current study does not have a requirement of residual chlorine. Although chlorination is effective in removing the virus, the application of adequate chlorine dosing is necessary to ensure the complete removal of SARS-CoV-2 traces.31 During periods of high levels of a community outbreak, wastewater treatment removal efficiency can be simplified by testing only treated effluent samples after tertiary treatment to confirm the absence of SARS-CoV-2 viral RNA.
Several studies indicated that the SARS-CoV-2 virus in feces can reach the WWTP in around 2–10 h after excretion and the time spent to reach may not be sufficient for the total inactivation of the virus by environmental factors, indicating the possible presence of viable and infectious viruses in the raw influents.25,28 In addition, previous studies reported that coronaviruses could be aerosolized at the pre-treatment stages or diffused with droplets, particularly through the pumping and movement of wastewater.27,34 A recent quantitative microbial risk assessment (QMRA) study conducted at the inlet of WWTPs indicated that workers involved in the manual cleaning of coarse screening may potentially be exposed to aerosols and could be at risk of infections;55 however, a recent epidemiological analysis found no increased risk of being affected by COVID-19 due to exposure to wastewaters contaminated with SARS-CoV-2 under the working conditions confronted by WWTPs' workers.26 High concentrations of SARS-CoV-2 RNA are normally detected in raw influent samples worldwide during the peak of outbreaks, including the current study. Therefore, further testing on viral survival and infectivity in the aerosols generated at the inlets of WWTPs and uncontrolled sewage discharge sites is necessary to better understand the risk of exposure.
Conclusion
Although WBE has emerged as a vital tool for the surveillance of SARS-CoV-2 transmission in sewershed areas, data regarding viral RNA quantification among different treatment stages of WWTPs are currently limited. The current comprehensive study applied both wastewater surveillance of SARS-CoV-2 RNA and evaluated the fate of SARS-CoV-2 RNA along the treatment process at a WWTP in San Antonio, Texas. SARS-CoV-2 RNA was detected in 98.5% (n = 34 weeks) of raw influents and captured the third wave of the COVID-19 outbreak. Correlation analysis of wastewater viral RNA loads with 7 day lagged reported COVID-19 cases showed that wastewater data are leading by a week, which confirms the early warning capabilities of WBE. A two days sampling frequency per week was found to be better than one day sampling frequency to ensure valid trend estimates. Regarding SARS-CoV-2 persistence in treatment stages, viral RNA was positively detected in all raw influent and primary effluent samples over four months. Temporal variability in detection with secondary effluents was exhibited, where heightened COVID-19 infectivity lowered removal efficiency by primary and secondary treatments. With most of the viral RNA adsorbed on biosolids, the average viral RNA removal efficiencies achieved were 54% at the primary treatment stage, 94% at the secondary treatment stage, and 100% at the tertiary treatment stage. As the pandemic continues to evolve, the current study presents important data regarding the SARS-CoV-2 RNA removal performance in different treatment stages of the WWTP, which is still rare in the literature.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This research was supported in part by the Health Collaborative and the City of San Antonio. We would like to thank the San Antonio Water System for providing access to wastewater samples. We also thank the NSF Research Coordination Network on Wastewater Surveillance of SARS-CoV-2 for providing guidance and resources that helped shape this study.References
- WHO, WHO Coronavirus (COVID-19) Dashboard, Coronavirus disease (COVID-19) Weekly Epidemiological Updates and Monthly Operational Updates, 2022, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Search PubMed.
- Q. Bi, J. Lessler, I. Eckerle, S. A. Lauer, L. Kaiser, N. Vuilleumier, D. A. Cummings, A. Flahault, D. Petrovic and I. Guessous, Insights into household transmission of SARS-CoV-2 from a population-based serological survey, Nat. Commun., 2021, 12, 1–8 CrossRef PubMed.
- S. Feng, A. Roguet, J. S. McClary-Gutierrez, R. J. Newton, N. Kloczko, J. G. Meiman and S. L. McLellan, Evaluation of Sampling, Analysis, and Normalization Methods for SARS-CoV-2 Concentrations in Wastewater to Assess COVID-19 Burdens in Wisconsin Communities, ACS ES&T Water, 2021, 1, 1955–1965 Search PubMed.
- R. Wölfel, V. M. Corman, W. Guggemos, M. Seilmaier, S. Zange, M. A. Müller, D. Niemeyer, T. C. Jones, P. Vollmar, C. Rothe, M. Hoelscher, T. Bleicker, S. Brünink, J. Schneider, R. Ehmann, K. Zwirglmaier, C. Drosten and C. Wendtner, Virological assessment of hospitalized patients with COVID-2019, Nature, 2020, 581, 465–469 CrossRef PubMed.
- S. P. Sherchan, S. Shahin, L. M. Ward, S. Tandukar, T. G. Aw, B. Schmitz, W. Ahmed and M. Kitajima, First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA, Sci. Total Environ., 2020, 743, 140621 CrossRef CAS PubMed.
- W. Ahmed, N. Angel, J. Edson, K. Bibby, A. Bivins, J. W. O'Brien, P. M. Choi, M. Kitajima, S. L. Simpson, J. Li, B. Tscharke, R. Verhagen, W. J. M. Smith, J. Zaugg, L. Dierens, P. Hugenholtz, K. V. Thomas and J. F. Mueller, First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community, Sci. Total Environ., 2020, 728, 138764 CrossRef CAS PubMed.
- G. Medema, F. Been, L. Heijnen and S. Petterson, Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: Opportunities and challenges, Curr. Opin. Environ. Sci. Health, 2020, 17, 49–71 CrossRef PubMed.
- W. Ahmed, B. Tscharke, P. M. Bertsch, K. Bibby, A. Bivins, P. Choi, L. Clarke, J. Dwyer, J. Edson, T. M. H. Nguyen, J. W. O'Brien, S. L. Simpson, P. Sherman, K. V. Thomas, R. Verhagen, J. Zaugg and J. F. Mueller, SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study, Sci. Total Environ., 2021, 761, 144216 CrossRef CAS PubMed.
- J. Weidhaas, Z. T. Aanderud, D. K. Roper, J. VanDerslice, E. B. Gaddis, J. Ostermiller, K. Hoffman, R. Jamal, P. Heck, Y. Zhang, K. Torgersen, J. V. Laan and N. LaCross, Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds, Sci. Total Environ., 2021, 775, 145790 CrossRef CAS PubMed.
- P. M. D'Aoust, E. Mercier, D. Montpetit, J.-J. Jia, I. Alexandrov, N. Neault, A. T. Baig, J. Mayne, X. Zhang, T. Alain, M.-A. Langlois, M. R. Servos, M. MacKenzie, D. Figeys, A. E. MacKenzie, T. E. Graber and R. Delatolla, Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence, Water Res., 2021, 188, 116560 CrossRef PubMed.
- V. Vo, A. Harrington, S. Afzal, K. Papp, C.-L. Chang, H. Baker, P. Aguilar, E. Buttery, M. A. Picker, C. Lockett, D. Gerrity, H.-Y. Kan and E. C. Oh, Identification of a rare SARS-CoV-2 XL hybrid variant in wastewater and the subsequent discovery of two infected individuals in Nevada, Sci. Total Environ., 2023, 858, 160024 CrossRef CAS PubMed.
- D. S. Smyth, M. Trujillo, D. A. Gregory, K. Cheung, A. Gao, M. Graham, Y. Guan, C. Guldenpfennig, I. Hoxie, S. Kannoly, N. Kubota, T. D. Lyddon, M. Markman, C. Rushford, K. M. San, G. Sompanya, F. Spagnolo, R. Suarez, E. Teixeiro, M. Daniels, M. C. Johnson and J. J. Dennehy, Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater, Nat. Commun., 2022, 13, 635 CrossRef CAS PubMed.
- R. Wang, M. Alamin, S. Tsuji, H. Hara-Yamamura, A. Hata, B. Zhao, M. Ihara and R. Honda, Removal performance of SARS-CoV-2 in wastewater treatment by membrane bioreactor, anaerobic-anoxic-oxic, and conventional activated sludge processes, Sci. Total Environ., 2022, 851, 158310 CrossRef CAS PubMed.
- P. Foladori, F. Cutrupi, M. Cadonna and S. Manara, Coronaviruses and SARS-CoV-2 in sewerage and their removal: Step by step in wastewater treatment plants, Environ. Res., 2022, 207, 112204 CrossRef CAS PubMed.
- M. Kumar, M. Alamin, K. Kuroda, K. Dhangar, A. Hata, H. Yamaguchi and R. Honda, Reply: Potential discharge, attenuation and exposure risk of SARS-CoV-2 in natural water bodies receiving treated wastewater, npj Clean Water, 2021, 4, 33 CrossRef CAS.
- S. Albert, A. Ruíz, J. Pemán, M. Salavert and P. Domingo-Calap, Lack of evidence for infectious SARS-CoV-2 in feces and sewage, Eur. J. Clin. Microbiol. Infect. Dis., 2021, 40, 2665–2667 CrossRef CAS PubMed.
- W. Ahmed, K. Bibby, P. M. D'Aoust, R. Delatolla, C. P. Gerba, C. N. Haas, K. A. Hamilton, J. Hewitt, T. R. Julian, D. Kaya, P. Monis, L. Moulin, C. Naughton, R. T. Noble, A. Shrestha, A. Tiwari, S. L. Simpson, S. Wurtzer and A. Bivins, Differentiating between the possibility and probability of SARS-CoV-2 transmission associated with wastewater: empirical evidence is needed to substantiate risk, FEMS Microbes, 2021, 2 Search PubMed.
- S. Gholipour, F. Mohammadi, M. Nikaeen, Z. Shamsizadeh, A. Khazeni, Z. Sahbaei, S. M. Mousavi, M. Ghobadian and H. Mirhendi, COVID-19 infection risk from exposure to aerosols of wastewater treatment plants, Chemosphere, 2021, 273, 129701 CrossRef CAS PubMed.
- M. Gormley, T. J. Aspray and D. A. Kelly, COVID-19: mitigating transmission via wastewater plumbing systems, Lancet Global Health, 2020, 8, e643 CrossRef PubMed.
- M. Kitajima, W. Ahmed, K. Bibby, A. Carducci, C. P. Gerba, K. A. Hamilton, E. Haramoto and J. B. Rose, SARS-CoV-2 in wastewater: State of the knowledge and research needs, Sci. Total Environ., 2020, 739, 139076 CrossRef CAS PubMed.
- V. Naddeo and H. Liu, Editorial Perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond?, Environ. Sci.: Water Res. Technol., 2020, 6, 1213–1216 RSC.
- J. D. Shutler, K. Zaraska, T. Holding, M. Machnik, K. Uppuluri, I. G. C. Ashton, Ł. Migdał and R. S. Dahiya, Rapid Assessment of SARS-CoV-2 Transmission Risk for Fecally Contaminated River Water, ACS ES&T Water, 2021, 1, 949–957 Search PubMed.
- A. Bivins, J. Greaves, R. Fischer, K. C. Yinda, W. Ahmed, M. Kitajima, V. J. Munster and K. Bibby, Persistence of SARS-CoV-2 in Water and Wastewater, Environ. Sci. Technol. Lett., 2020, 7, 937–942 CrossRef CAS.
- M. D. Sobsey, Absence of virological and epidemiological evidence that SARS-CoV-2 poses COVID-19 risks from environmental fecal waste, wastewater and water exposures, J. Water Health, 2021, 20, 126–138 CrossRef PubMed.
- I. D. Amoah, S. Kumari and F. Bux, Coronaviruses in wastewater processes: Source, fate and potential risks, Environ. Int., 2020, 143, 105962 CrossRef CAS PubMed.
- B. Muñoz-Palazon, P. R. Bouzas, J. González-López and M. Manzanera, Transmission of SARS-CoV-2 associated with wastewater treatment: a seroprevalence study, Int. J. Water Resour. Dev., 2022, 38, 928–937 CrossRef.
- L. S. Hung, The SARS epidemic in Hong Kong: what lessons have we learned?, J. R. Soc. Med., 2003, 96, 374–378 CrossRef PubMed.
- S. G. Rimoldi, F. Stefani, A. Gigantiello, S. Polesello, F. Comandatore, D. Mileto, M. Maresca, C. Longobardi, A. Mancon, F. Romeri, C. Pagani, F. Cappelli, C. Roscioli, L. Moja, M. R. Gismondo and F. Salerno, Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers, Sci. Total Environ., 2020, 744, 140911 CrossRef CAS PubMed.
- E. Haramoto, B. Malla, O. Thakali and M. Kitajima, First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan, Sci. Total Environ., 2020, 737, 140405 CrossRef CAS PubMed.
- S. Westhaus, F.-A. Weber, S. Schiwy, V. Linnemann, M. Brinkmann, M. Widera, C. Greve, A. Janke, H. Hollert, T. Wintgens and S. Ciesek, Detection of SARS-CoV-2 in raw and treated wastewater in Germany – Suitability for COVID-19 surveillance and potential transmission risks, Sci. Total Environ., 2021, 751, 141750 CrossRef CAS PubMed.
- H. Abu Ali, K. Yaniv, E. Bar-Zeev, S. Chaudhury, M. Shagan, S. Lakkakula, Z. Ronen, A. Kushmaro and O. Nir, Tracking SARS-CoV-2 RNA through the Wastewater Treatment Process, ACS ES&T Water, 2021, 1, 1161–1167 Search PubMed.
- A. Serra-Compte, S. Gonzalez, M. Arnaldos, S. Berlendis, S. Courtois, J. F. Loret, O. Schlosser, A. M. Yanez, E. Soria-Soria, M. Fittipaldi, G. Saucedo, A. Pinar-Mendez, M. Paraira, B. Galofre, J. M. Lema, S. Balboa, M. Mauricio-Iglesias, A. Bosch, R. M. Pinto, I. Bertrand, C. Gantzer, C. Montero and X. Litrico, Elimination of SARS-CoV-2 along wastewater and sludge treatment processes, Water Res., 2021, 202, 117435 CrossRef CAS PubMed.
- M. Pourakbar, A. Abdolahnejad, S. Raeghi, F. Ghayourdoost, R. Yousefi and A. Behnami, Comprehensive investigation of SARS-CoV-2 fate in wastewater and finding the virus transfer and destruction route through conventional activated sludge and sequencing batch reactor, Sci. Total Environ., 2022, 806, 151391 CrossRef CAS PubMed.
- J. A. Polanco, B. Schmitz, S. Choi, J. Safarik, S. Prasek and M. H. Plumlee, SARS-CoV-2 genetic material is removed during municipal wastewater treatment and is undetectable after advanced treatment, Sci. Total Environ., 2023, 859, 159575 CrossRef CAS PubMed.
- K. K. Vadde, H. Al-Duroobi, D. C. Phan, A. Jafarzadeh, S. V. Moghadam, A. Matta and V. Kapoor, Assessment of Concentration, Recovery, and Normalization of SARS-CoV-2 RNA from Two Wastewater Treatment Plants in Texas and Correlation with COVID-19 Cases in the Community, ACS ES&T Water, 2022, 2, 2060–2069 Search PubMed.
- W. Ahmed, A. Bivins, S. L. Simpson, W. J. Smith, S. Metcalfe, B. McMinn, E. M. Symonds and A. Korajkic, Comparative analysis of rapid concentration methods for the recovery of SARS-CoV-2 and quantification of human enteric viruses and a sewage-associated marker gene in untreated wastewater, Sci. Total Environ., 2021, 799, 149386 CrossRef CAS PubMed.
- W. Ahmed, P. M. Bertsch, A. Bivins, K. Bibby, A. Gathercole, E. Haramoto, P. Gyawali, A. Korajkic, B. R. McMinn and J. F. Mueller, Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater, Sci. Total Environ., 2020, 739, 139960 CrossRef CAS PubMed.
- H. Al-Duroobi, S. V. Moghadam, D. C. Phan, A. Jafarzadeh, A. Matta and V. Kapoor, Wastewater surveillance of SARS-CoV-2 corroborates heightened community infection during the initial peak of COVID-19 in Bexar County, Texas, FEMS Microbes, 2021, 2 Search PubMed.
- J. Peccia, A. Zulli, D. E. Brackney, N. D. Grubaugh, E. H. Kaplan, A. Casanovas-Massana, A. I. Ko, A. A. Malik, D. Wang and M. Wang, Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics, Nat. Biotechnol., 2020, 38, 1164–1167 CrossRef CAS PubMed.
- K. E. Graham, S. K. Loeb, M. K. Wolfe, D. Catoe, N. Sinnott-Armstrong, S. Kim, K. M. Yamahara, L. M. Sassoubre, L. M. Mendoza Grijalva and L. Roldan-Hernandez, SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed, Environ. Sci. Technol., 2020, 55, 488–498 CrossRef PubMed.
- K. Bibby and J. Peccia, Identification of viral pathogen diversity in sewage sludge by metagenome analysis, Environ. Sci. Technol., 2013, 47, 1945–1951 CrossRef CAS PubMed.
- W. Randazzo, P. Truchado, E. Cuevas-Ferrando, P. Simón, A. Allende and G. Sánchez, SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area, Water Res., 2020, 181, 115942 CrossRef CAS PubMed.
- T. Gonzalez, M. A. de la Rubia, K. P. Hincz, M. Comas-Lopez, L. Subirats, S. Fort and G. M. Sacha, Influence of COVID-19 confinement on students' performance in higher education, PLoS One, 2020, 15, e0239490 CrossRef CAS PubMed.
- B. Pecson, D. Gerrity, K. Bibby, J. E. Drewes, C. Gerba, R. Gersberg, R. Gonzalez, C. N. Haas, K. A. Hamilton and K. L. Nelson, Editorial perspectives: will SARS-CoV-2 reset public health requirements in the water industry? Integrating lessons of the past and emerging research, Environ. Sci.: Water Res. Technol., 2020, 6, 1761–1764 RSC.
- C. Duvallet, F. Wu, K. A. McElroy, M. Imakaev, N. Endo, A. Xiao, J. Zhang, R. Floyd-O’Sullivan, M. M. Powell, S. Mendola, S. T. Wilson, F. Cruz, T. Melman, C. L. Sathyanarayana, S. W. Olesen, T. B. Erickson, N. Ghaeli, P. Chai, E. J. Alm and M. Matus, Nationwide Trends in COVID-19 Cases and SARS-CoV-2 RNA Wastewater Concentrations in the United States, ACS ES&T Water, 2022, 2, 1899–1909 Search PubMed.
- C. S. McMahan, D. Lewis, J. A. Deaver, D. Dean, L. Rennert, C. A. Kalbaugh, L. Shi, D. Kriebel, D. Graves, S. C. Popat, T. Karanfil and D. L. Freedman, Predicting COVID-19 Infected Individuals in a Defined Population from Wastewater RNA Data, ACS ES&T Water, 2022, 2, 2225–2232 Search PubMed.
- Y. Ai, A. Davis, D. Jones, S. Lemeshow, H. Tu, F. He, P. Ru, X. Pan, Z. Bohrerova and J. Lee, Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States, Sci. Total Environ., 2021, 801, 149757 CrossRef CAS PubMed.
- N. L. Fahrenfeld, W. R. Morales Medina, S. D'Elia, A. S. Deshpande and G. Ehasz, Year-long wastewater monitoring for SARS-CoV-2 signals in combined and separate sanitary sewers, Water Environ. Res., 2022, 94, e10768 CrossRef CAS PubMed.
- P. Tandon, E. S. Leibner, A. Hackett, K. Maguire, N. Mashriqi and R. Kohli-Seth, The Third Wave: Comparing Seasonal Trends in COVID-19 Patient Data at a Large Hospital System in New York City, Critical Care Explorations, 2022, 4, e0653 CrossRef PubMed.
- A. Bivins and K. Bibby, Wastewater Surveillance during Mass COVID-19 Vaccination on a College Campus, Environ. Sci. Technol. Lett., 2021, 8, 792–798 CrossRef CAS.
- L. Hopkins, D. Persse, K. Caton, K. Ensor, R. Schneider, C. McCall and L. B. Stadler, Citywide wastewater SARS-CoV-2 levels strongly correlated with multiple disease surveillance indicators and outcomes over three COVID-19 waves, Sci. Total Environ., 2023, 855, 158967 CrossRef CAS PubMed.
- L. Hopkins, D. Persse, K. Caton, K. Ensor, R. Schneider, C. McCall and L. B. Stadler, Citywide wastewater SARS-CoV-2 levels strongly correlated with multiple disease surveillance indicators and outcomes over three COVID-19 waves, Sci. Total Environ., 2023, 855, 158967 CrossRef CAS PubMed.
- M. Kumar, K. Kuroda, A. K. Patel, N. Patel, P. Bhattacharya, M. Joshi and C. G. Joshi, Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with Upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques, Sci. Total Environ., 2021, 754, 142329 CrossRef CAS PubMed.
- CDC, Sampling Strategy: Where, How, and What to Sample, Developing a Wastewater Surveillance Sampling Strategy, accessed 10/11/2022, https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/developing-a-wastewater-surveillance-sampling-strategy.html#anchor_1602855374139 Search PubMed.
- R. N. Zaneti, V. Girardi, F. R. Spilki, K. Mena, A. P. C. Westphalen, E. R. da Costa Colares, A. G. Pozzebon and R. G. Etchepare, Quantitative microbial risk assessment of SARS-CoV-2 for workers in wastewater treatment plants, Sci. Total Environ., 2021, 754, 142163 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3va00015j |
| ‡ Present address: US Salinity Laboratory, USDA-ARS, Riverside, CA 92507, USA. |
| This journal is © The Royal Society of Chemistry 2023 |