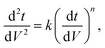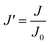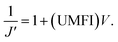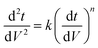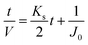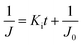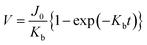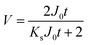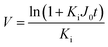Open Access Article
Open Access ArticleUltra-thin membrane filter with a uniformly arrayed nanopore structure for nanoscale separation of extracellular vesicles without cake formation†
Daesan
Kim
 a,
Jaehyuk
Lee
b,
Boyoung
Kim
b,
Yujin
Shin
b,
Jinhong
Park
cd,
Uijoo
Kim
ae,
Minbaek
Lee
cd,
Sang Bum
Kim
f and
Sunghoon
Kim
*ae
a,
Jaehyuk
Lee
b,
Boyoung
Kim
b,
Yujin
Shin
b,
Jinhong
Park
cd,
Uijoo
Kim
ae,
Minbaek
Lee
cd,
Sang Bum
Kim
f and
Sunghoon
Kim
*ae
aMedicinal Bioconvergence Research Center, Institute for Artificial Intelligence and Biomedical Research, Gangnam Severance Hospital, Yonsei University, Incheon 21983, Korea. E-mail: sunghoonkim@yonsei.ac.kr
bR&D Center, Metapore Co., Ltd, Advanced Institutes of Convergence Technology, 8F, Suwon 16229, Korea
cDepartment of Physics, Inha University, Incheon 22212, Korea
dThe Institute for Basic Science, Inha University, Incheon 22212, Korea
eCollege of Pharmacy, College of Medicine, Interdisciplinary Biomedical Center, Gangnam Severance Hospital, Yonsei University, Incheon 21983, Korea
fCollege of Pharmacy, Sahmyook University, Seoul 01795, Korea
First published on 22nd November 2022
Abstract
Extracellular vesicles (EVs) have emerged as vehicles that mediate diverse cell–cell communication. However, in-depth understanding of these vesicles is hampered by a lack of a reliable isolation method to separate different types of EVs with high levels of integrity and purity. Here, we developed a nanoporous and ultra-thin membrane structure (NUTS) that warrants the size-based isolation of EVs without cake formation, minimizing the sample loss during the filtration process. By utilizing the micro-electro-mechanical systems (MEMS) technique, we could also control the pore size in nanoscale. We validated the performance of this membrane to separate EVs according to their size range.
Introduction
Extracellular vesicles (EVs) are involved in various intercellular signaling pathways. Since they can deliver a wide range of cellular factors among cells, they attract much interest for their potential as a tool for diagnosis and therapeutic agents and even cosmetics.1–4 EVs are widely used as a general term for vesicles derived from cells. According to physical characteristics, biochemical composition, or cell of origin, EVs can be classified into different types, such as ectosomes, exosomes, apoptotic bodies, and oncosomes. Despite a significant potential to provide new biological information and biomedical applications, EVs are heterogeneous in size, function, and content to be defined as a single entity. Thus, a desirable method that can separate EVs to near homogeneity should be developed. Among many types of EVs, exosomes have a size range from 40 to 160 nm.1,5–7 Until recent decades, density-gradient or non-gradient ultracentrifugation was the most common method used to separate EVs from various types of biological fluids (e.g., cell-cultured media, plasma, and serum). Although other methods, such as differential precipitation, immune-affinity, magnetic beads, microfluidic separation, column chromatography, and ultrafiltration are also available,1,6,8–12 none of them seem to provide a desirable method to homogeneously separate exosomes with high purity, integrity, and yield. In fact, they often lose a part of their original properties and morphology in most isolation processes and thus may generate misleading results.13,14To investigate the exact biological function of exosomes, they should be isolated without affecting their intrinsic functional and morphological characteristics. In this study, to isolate exosomes without physical damage, we designed a unique structure of the membrane, which is the core of a filter, with a cylindrical pore structure and ultra-thin layer, and tested whether it can isolate undamaged exosomes with a high degree of purity and high yield of recovery.
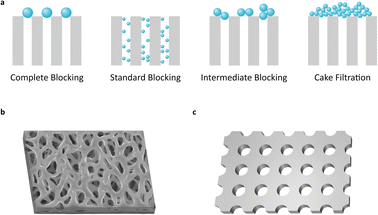
In a filtration process, membrane fouling phenomena are affected by various factors, such as pore blocking, cake formation, particle adsorption, and polarization of solution concentration.15,16 Among them, pore blocking causes major problems to filtration performance. However, it is a very complicated phenomenon in sub-micro scale filtration. The mechanism of pore blocking behavior has been debated from theoretical and practical perspectives. The most classical blocking filtration law, proposed by Hermans and Bredée,17 is described as four distinct mechanisms: complete blocking, standard blocking, intermediate blocking, and cake filtration (Fig. 1a).
Most previous filter membranes use fabric mesh or polymeric heterogeneous structures (Fig. 1b and S1†). Pore sizes are often determined as the mean size of the pore diameter on the membrane surface.18 However, the pore sizes of previous membranes show uncertain diameters and large distribution due to their irregular structure, raising a crucial problem to defining the pore diameter and cutoff size of the membrane.
While the membranes contain irregular porous structures, analytic models have been established based on ideal structures with regular and static morphology.15–17,19 The gap between theoretical and actual conditions can make a difference in predicting the performance of the filter and in designing the filtration system. Moreover, on a sub-micro scale, inexact experiment design often leads to completely unexpected results.
Commercial membrane filters mostly comprise irregular pore size and heterogeneous morphology in a single type of membrane (Fig. 1b). Meanwhile, the nanoporous and ultra-thin membrane structure (NUTS), fabricated in this study, has a homogeneously arranged pore structure, uniform pore size, and narrow distribution of size (Fig. 1c). This difference in core filtration structures may result in different blocking mechanisms and superior filtration performance. We then analyzed the filtration behavior of the NUTS matching with a pore blocking mechanism. Results indicate a distinct behavior and filtration performance. Moreover, cake formation is affected by various factors of filtration conditions (e.g., particle size distribution, flow rate, porosity, packing structure, and friction coefficient).20 In particular, membrane characteristics play a crucial role in cake formation. The NUTS has a relatively low friction force than a commercial filter membrane due to its uniform and flat surface. Additionally, particles deposited onto a flat surface have the disadvantage of forming a cake layer in terms of packing structures.
The outstanding characteristics of the NUTS suggested great potentials for various applications, especially the cutoff size resolution of filtration. For example, the filter assembled from the NUTS could achieve an accurate cutoff size and high-reliability results. For these extraordinary properties, micro-electro-mechanical system (MEMS) processes were used to fabricate the exact structure of the NUTS. Therefore, an MEMS can provide a precise specification standard for this filter fabrication.
Results and discussion
Design of the filter device using the NUTS
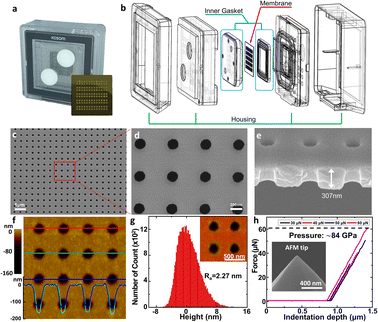
The NUTS comprises silicon composite to be utilized for the MEMS process. The fabrication method of the NUTS is briefly described in Fig. S2.† The silicon dioxide layer was grown on the bare silicon wafer. After the oxidation process, the chemical vapor deposition (CVD) process was carried out for a low-stress silicon nitride (LSN) layer for high-quality membrane development. The pore patterning on the LSN layer was achieved using the photolithography method. Then, the ion-coupled plasma (ICP) etching process vanished the unnecessary region. Back-side patterning was also implemented by using the photolithography process to remove the undesired layer for the development of an ultra-thin membrane structure. The residual oxidation layer was removed by using a buffered oxide etch (BOE) solution for the pore control process. Finally, the plasma-enhanced chemical vapor deposition (PE-CVD) was utilized for the development of a precisely designed pore size.The NUTS could be used for both direct filtration and tangential flow filtration (TFF). Direct filtration has a critical disadvantage in that slug is inevitably stuck on the membrane, reducing the filtration rate. In contrast, TFF can eliminate part of the slug or cake layer by the fluidic force perpendicular to the direction of the pores on the membrane.21–24
We designed a customized fluidic channel to use the NUTS as a filter for the application in the TFF system (Fig. 2a and b). In this system, the fluidic channel was designed to function as a gasket to prevent solution leakage and membrane damage. The assembled filter was first tested in nitrogen gas (N2) flow conditions for stability. The membrane was able to withstand input pressure of up to 200 kPa with a linear response to the output pressure (Fig. S3a†), indicating no membrane damage and no leakage during the process.
Physical characteristics of the NUTS
We observed the physical characteristics of the NUTS that was designed for the development of a novel concept filter. The NUTS comprises silicon composites, and as mentioned previously, the LSN and SiO2 composite was used for extraordinary physical characteristics of the membrane. The size of the chip is approximately 2 cm × 2 cm (Fig. 2a) and consists of 130 windows, each having a few hundreds of millions of evenly arranged nanopores. Pore structures have nanoscale morphology while appearing identical to a flat surface in an optical image (Fig. S4†). Scanning electron microscopy (SEM) was used to measure the pore structure and other features of the membrane (Fig. 2c–e). Clearly, the pores are arrayed with uniformity on the surface, and their diameter is about 187 ± 7.56 nm (Fig. S5†). The section profile of the membrane shows that the membrane has uniform thickness and pores have cylindrical structures. Since the upper pore diameter is narrower than the bottom pore diameter, it could have a positive effect to maintain the filtration performance by declining the pore blocking events. Moreover, due to the ultra-thin membrane, the filter could show much higher performance than filters with similar porosity.25,26Our NUTS was also measured using atomic force microscopy (AFM) to observe more detailed surface characteristics. Fig. 2f presents a topographic image with line profiles of the fabricated NUTS. In the line profiles of the topographic image, the fabricated pores were found to have a gradient slope around pores. The top SiO2/Si3N4 surface of the NUTS exhibited an average roughness of approximately 2.27 nm, which was sufficiently smooth for the filtration of bio-fluids without cake formation (Fig. 2g and S6†). Such morphological features could guide target materials to be filtrated through the NUTS. To examine the structural durability of the NUTS, a force–distance (FD) curve was measured at the surface of the SiO2/Si3N4 area by using the diamond tip (Fig. 2h). Various indentation forces were applied to the NUTS, and the typical indentation depth was about 0.5 μm. Note that, up to the indentation force of approximately 60 μN, the surface morphology of the NUTS was maintained before and after the FD measurement (Fig. S7a†). Upon converting force to local pressure, the NUTS can endure the local pressure of about 84 GPa without any mechanical damage. The penetration test was also conducted (Fig. S7b†). The NUTS was damaged locally at the points where a high indentation force, of over 84 GPa, was applied (Fig. S7c†). This implies that the pore shape or size on the membrane will not be affected by fluidic pressure, especially much lower than ∼84 GPa. Therefore, the cutoff size could be kept consistent because the pore structure remains unchanged during the filtration process. Unlike conventional membrane filters, one of the most remarkable features of this membrane is the flat surface morphology with the guided entrance. The filter cake comprises accumulated particles on a membrane surface during the filtration process.20,27 An interaction that establishes energetic stability is the particle adsorption on the surface. Moreover, the biomolecule structure is unfolded or denatured by hydrophobic interactions, which is an irreversible process. Meanwhile, molecular adsorption on a hydrophilic surface is a reversible process. Surface morphology influences particle deposition and filter cake formation. This is because the membrane surface with high roughness parameter has a higher chance of interacting with surrounding particles. Therefore, making a filter cake on a membrane with homogeneous morphology is difficult, such as the membrane in this study. Due to this feature, cake formation is rarely expected on the membrane surface.
Early stage filtration behavior using the NUTS
Since pore blocking of particles would result from a complex combination of different blocking mechanisms, the exact phenomena of whole physical and chemical pore blocking events is difficult to dissect, especially in long-term experiments. We thus conducted the experiment to observe the change of filtration behaviors in the early stage of the total filtration process.19,28For the typical pore blocking behaviors of filtration under constant pressure, it could be described as the following differential equation:28,29
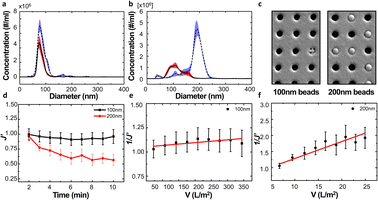
We examined the particle distribution of the solution before and after filtration, with different sizes of source solutions. Fig. 3a and b correspond to 100 nm and 200 nm bead solutions, respectively. The particle distributions of initial and filtrate solutions were plotted for both samples. The particle concentrations of the initial solution and filtrate containing 100 nm beads were measured as 2.37 × 108 and 1.24 × 108 ea ml−1, respectively. While the particle concentration was slightly decreased after the filtration process, the main peaks of the initial solution and filtrate show similar values of 77.4 and 74.3 nm, respectively (Fig. 3a).
Meanwhile, the peak size of the initial sample and filtrate containing 200 nm beads had drastically changed from 192.6 nm to 102.1 nm because the larger particles could not pass through the membrane of the filter. This clear difference indicates that the filtration process via the NUTS could separate and eliminate the particles with diameters larger than that of the pores (Fig. 3b).
During filtration, the different particle sizes resulted in varying pore blocking behavior. The tendency of filtrate flux change shows a clear difference in the results of 100 nm and 200 nm solutions (Fig. 3d).
The filtrate flux is affected by various factors of a membrane or solution, and one of the most important factors is transmembrane pressure (TMP). The effect of TMP is a complex and sensitive issue, and a filtration system could have a critical TMP condition in many cases.30–32 However, this filtration system was operated at TMP in the range under 1 kPa because the inlet flow was modulated at low-level pressure (Fig. S3b and c†).
In low-pressure membrane filtration, the simplified and modified Hermia's model could be utilized to determine the total fouling degree, which is the unified membrane fouling index (UMFI).31,33 The UMFI could show the accurate tendency of the pore blocking behavior using the following equation:
The UMFI value of 100 nm filtrate flux was about 2.9253 × 10−4, which implied that pore blocking occurred infrequently in this condition. In contrast, the UMFI value of 200 nm filtrate flux was about 0.0525, which was 179 times higher than that of the 100 nm filtrate result (Fig. 3e and f). This result showed that the particle size in the sample solution is a critical factor in the filtration performance. The SEM image of the membrane after the filtration showed a clear difference (Fig. 3c). Most of the pores in the membrane remained open after 100 nm beads filtration, while part of the pores in the 200 nm membrane was blocked. This suggests that blocking phenomena occurred in the filtration using 200 nm beads. Moreover, the image showed that cake formation did not occur. These results imply that we could design the filtering system with the exact cutoff size.
The filtrate flux of TFF is generally governed by cake formation on the membrane because the cake layer is constantly maintained for the filtration process.33 Among the four pore blocking mechanisms, the filtration performance is mainly affected by cake formation in the long-term stage of the filtration process. In the early stage of the filtration process, other blocking mechanisms have affected the filtration performance in a complex manner. Except for cake filtration, other pore blocking mechanisms are related to the membrane pore and particle sizes in the solution. If the particle size is larger than the pore size, complete and intermediate blocking mechanisms have mainly affected the filtration performance. However, the pore blockage by particles smaller than pore size could be explained via a standard blocking mechanism. Previous studies have attempted to explain the change of filtration performance by combining cake filtration with another dominant blocking mechanism.27,34
In this study, however, we attempted to minimize the effect of cake filtration to overcome drawbacks caused by cake formation. Cake formation has been considered an inevitable phenomenon in the previous membrane filter system because the morphology of the membrane has an irregular and entangled structure even though cake formation is one of the most significant factors affecting filtration performance.
Analysis of proportional pore blocking behaviors
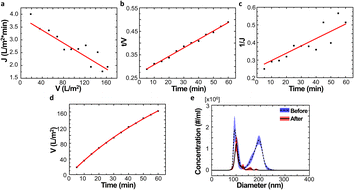
To simplify the complex problem in pore blocking, we employed the multiple linear models modified by Iritani for the analysis of the filtration result.15,16 Various pore blocking parameters were calculated using simple linear models and utilized into a single linearly combined model.34 Using the methodological way, we could find the proportional weight of each pore blocking mechanism to the total degradation of filter performance. To examine the complex effect of the pore blocking mechanism, the filtration was carried out using the mixture solution of 100 nm and 200 nm beads for 60 min.Fig. 4a–c show the linear relationship of resistance parameters of the pore blocking mechanism and filtration factors, J, V, and t. Each resistance parameter was calculated using the equation given in Table 1 linear expression term.15,16,28 The J0 value was used to calculate the average value of each linear pore blocking mechanism, as it was the common parameter of the whole mechanism. The values of Kb, Ks, and Ki were 0.0126, 0.0036, and 0.0041, respectively, which were calculated by a linear relationship. Importantly, the resistance parameter of cake filtration, Kc, was not used in this analysis because cake formation did not occur (Fig. 3c). Since the fitted filtration data using the cake filtration mechanism did not show a reasonable value, we used the other three pore blocking mechanisms, except for cake filtration.
In this study, we assumed that each pore blocking behavior was a coincident, but independent event. Therefore, we depicted the accumulated filtrate volume as follows:
| V(t)total = qbV(t)b + qsV(t)s + qiV(t)i + ε(where qb + qs + qi = 1, qb, qs, qi > 0). |
The solution before and after the filtration was measured by nanoparticle tracking analyzer (NTA) to verify the separation result of the particles (Fig. 4e). The particle distribution of the non-filtrate solution showed two major peaks at 94.5 nm and 194.5 nm with similar concentrations (blue line). In contrast, the filtrate solution showed a single major peak at 100.5 nm. The peak value of the red line was slightly changed by 6 nm compared to the blue line. Importantly, the particle distribution of the filtrate solution does not show the particles larger than 200 nm diameter. This indicates that the particles could be completely separated by a single filtration process via the NUTS.
Filtration result of the biologic sample
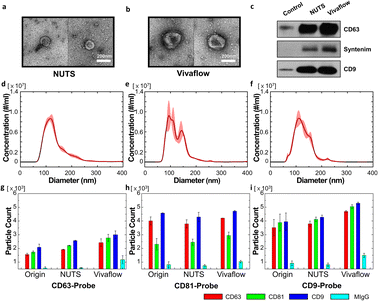
Our filter and its membrane, the NUTS, can have an accurate cutoff size and remain the same size, as there is no cake formation during the filtration process. This could be an advantage for isolating EVs from various biological samples, such as cell-cultured media, serum, and other body fluids. As a proof of concept, we filtrated cell-cultured media of HCT116 cells using our NUTS filter system and a Vivaflow 50 (Sartorius, Germany) for comparison.First, we took the EV images of the filtrates that passed through the NUTS and Vivaflow using the negative staining method of TEM (Fig. 5a and b, respectively). EVs in both filtrates showed the cup-shaped morphology, which is expected to result from the structural characteristics of empty space inside the EVs,35 although EVs in the Vivaflow filtrate showed larger sizes compared to those in the NUTS filtrate. It may be doubtful whether the cup-shaped EVs were caused by an external force during the isolation process. To prevent physical damage to the biological sample, note that no centrifugation method was used in these filtration processes. Therefore, the morphology of EVs suggests that EVs were physically undamaged, not being squashed or slanted to one side during the separation process.
To analyze the filtering characteristics, we first compared the filtering efficiency. The collection ratio of the filtrated samples was calculated in terms of particle number for the filtration time of 20 min considering biological stability. The collection ratio of both filters showed similar values: the NUTS was 37.96% and Vivaflow was 42.29%. Considering the dead volume of the tubing and filter itself, no significant difference exists in the filtering efficiency of both methods.
Thereafter, we analyzed the pore blocking behavior. The tendency of the filtrate flux of the bio-sample was slightly different from that of bead solutions (Fig. S8 and S9†). The main reason for the pore clogging was the larger particles compared to bead solutions' pore diameter. However, the bio-sample has various size distributions, unlike the bead solution. In this experiment, small particles have a higher portion than large particles, which shows the phenomenon close to standard blocking. A similar analytic method for bead solution was applied to the bio-sample. The proportions of complete, standard, and intermediate blocking were 13.02%, 65.17%, and 21.81%, respectively, indicating that standard blocking occurred five times more than complete blocking.
Exosomes are sometimes defined as harboring biomarkers that are often used for verification. CD63 and CD9 are well-known exosome membrane markers,1,4,7 and syntenin is a protein involved in biogenesis of exosomes.36,37 We thus detected CD63, CD9, and syntenin protein via the western blot (WB). After counting the number of vesicles in the sample using NTA for the filtration results of both filters and control, the amount of protein present in the same number of vesicles was measured by WB. In the WB results, the exosome marker proteins were better detected in both filtered samples compared to the control. Furthermore, we validated the presence of apoptotic bodies in the sample to confirm the sample quality. However, there was no signal detected by the markers for apoptotic bodies (Fig. S10†).
Then, we compared the particle size distribution of the filtrates obtained from both filters. All conditions were the same following the details in the Materials and methods section. The particle distribution of the original sample showed the single peak with the mean and mode of 137 nm and 112.8 nm (Fig. 5d). The value of D90 was 198.9 nm, indicating that about 10% of particles had larger diameters than 200 nm. The filtration results of Vivaflow and the NUTS are shown in Fig. 5e and f. Notably, most of the particles were smaller than 200 nm, which is the pore diameter of used filters and the mean pore diameters were similar, 126.8 nm for the NUTS and 124.1 nm for Vivaflow. However, the mode of the NUTS filtrate showed 100.4 nm, about 10 nm smaller than the Vivaflow filtrate (113.6 nm). The NUTS filtrate shows a sharper peak at a smaller size than that of Vivaflow, implying that the NUTS filtrate has a larger portion of smaller particles than the Vivaflow filtrate.
The filtrate samples were examined using ExoView to analyze the characteristics of the obtained EVs (Fig. 5g–i). The ExoView platform uses specific marker proteins, namely, CD63, CD9, and CD81, which are well-known exosome marker proteins.1,3,38 There is no particular difference in the Exoview result of the EV separation methods. However, MIgG, which is nonspecific binding, has slightly higher levels in Vivaflow than in others. In the overall results, the population of CD9 was higher, while CD63 was lower than other antibodies. Importantly, the CD81 level of the CD81-probe antibody was relatively low than other data. The plausible explanation is that the lower density of CD81 in single vesicles, compared to CD63 and CD9, results in multiple binding events rarely with CD81-probe antibodies.
Materials and methods
Membrane fabrication
Thermal oxidation was performed on 6 inch bare silicon wafers 〈100〉 (P06P, Nanopia, Korea) to grow a silicon dioxide layer with the thickness of about 200 nm using a furnace (Alpha-808DN, TEL, Japan). The low-pressure chemical vapor deposition (LP-CVD) (PF-D82, P&TECH, Korea) was then carried out on top of it for the development of a low-stress silicon nitride (LSN) layer, which has a high-quality property due to its lattice structure. The positive photoresist (Dongjin Semichem, Korea) was coated on the LSN layer for the photolithography process (PAS5500, ASML, Netherlands). After photolithography, the LSN layer was etched by the ion-coupled plasma (ICP) etching process (Plasma Pro 100 Cobra, Oxford Instruments, England). Subsequently, the PR was removed, and the first front-side pattern was achieved.Back-side patterning was implemented for the development of an ultra-thin layer of the NUTS. The PR was coated for back-side patterning of the photolithography process (MA200compact, SUSS Micro Tec, Germany). The patterned area was first etched via the ICP etching process for the LSN and SiO2 layers. On the open silicon wafer region, deep trenches (340 μm depth) were created via deep reactive-ion etching (DRIE) (Multiplex Lite ASE-SR, STS, England), which is a highly anisotropic etching process. The residual region of the silicon wafer was etched through a wet etching process using a potassium hydroxide solution. The silicon oxide layer, which functions as a barrier on the wet etching process, was eliminated by a buffered oxide etch (BOE) solution. The next step was the plasma-enhanced chemical vapor deposition (PE-CVD) (VL-LA-PECVD, Unaxis, USA) process for the development of the silicon dioxide layer in the free-standing membrane surface. Through this process, we could control the pore diameters on the membrane surface to obtain the designed sizes with high reliability. Finally, oxygen plasma treatment was performed for two cycles using PR Asher (YES-CV200RFS, Yield Engineering System, USA) to ash the organic residues. In addition, the plasma treatment altered the hydrophobic surface to a hydrophilic surface (Fig. S11†).
Filter assembly
A filter for the TFF system was constructed using the custom-made housing and the inner gasket with the fluidic channel (Fig. 2b and S12†). The housing (polycarbonate) and inner gasket (silicone) were ultrasonically cleaned with 70% ethanol for 10 min and then completely dried. Finally, the dried gasket and the nanopore membrane were combined and mounted on the housing (Fig. 2b).Optical and SEM measurement
The surface of the NUTS was observed by a 50× optical microscope (Nikon E400). The pore structure and section profile of the membrane were observed using high-resolution field emission scanning electron microscopes (FE-SEM: MERLIN, Carl Zeiss, and JSM-7401F, JEOL) to measure the pore diameter and membrane thickness.AFM measurement
The membrane morphology was measured by an atomic force microscope (AFM: XE-7, Park Systems). In detail, a non-contact mode of AFM was utilized with a cantilever (NCHR, Park Systems). The scanning rate, size, force constant, and resonance frequency were 1 Hz, 2.2 × 2.2 μm2, 42 N m−1, and 330 kHz, respectively. Force–distance (FD) and penetration tests were performed through AFM. To prevent any undesired effects, such as tip distortion, wear, and fracture under high force conditions, the diamond tip having a four-sided pyramid shape (PR-NM-RC, Probes), with a constant force of approximately 350 N m−1 was employed.Filtration conditions and equipment
Before injecting the sample solution, a buffer solution was filled into the channel to prevent excessive pressure on the membrane. The particle concentration in the solution was adjusted to about 108 ea ml−1, which was a similar value to the concentration of EVs in the biological fluid. It had the same condition at each experiment and differed only in terms of the particle size. The experiment was repeated three times under the inlet flow rate of 30 ml min−1 using the customized syringe pump, which could maintain the constant flow.NTA measurement
Vesicles and polystyrene beads were measured through a nanoparticle tracking analysis (NanoSight NS300, Malvern, USA). Each sample was loaded into a 1 ml syringe and measured three times to obtain the average value and deviation. Extracellular vesicle samples and 200 and 100 nm polystyrene beads were measured with setpoints of camera levels 13, 13 and, 8, respectively. The setpoint of detect threshold was fixed at 3 for all sample measurements.EV enrichment condition
Human colorectal cancer cell line, HCT-116 cells (Korean Cell Line Bank) were cultured in RPMI 1640 containing 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin to 80% confluency. After cultivation, the supernatants and washed cells twice were removed using PBS. Serum-free media were added on the dish and incubated for 72 h. The media under the starvation condition were collected and centrifuged under ×500g for 10 min at 4 °C to remove cells and conducted again ×1500g for 15 min at 4 °C to remove debris. After centrifugation, the supernatants were carefully collected and used for the filtration.TEM measurement
To obtain negatively stained EM images, 5 μl of each purified sample was applied to carbon-coated grids, which were glow-discharged (Harrick Plasma, USA) for 1 min in air. The grids were then negatively stained using 1% uranyl acetate.39The prepared grids were observed on a Tecnai 10 transmission electron microscope (TEM) equipped with a lanthanum hexaboride (LaB6, FEI) cathode operating at 100 kV. Images were recorded using a 2k × 2k UltraScan CCD camera (Gatan) at the magnification of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (1.0 nm per pixel).
000 (1.0 nm per pixel).
Western blotting
The filtrate sample was concentrated by ultrafiltration. Each sample was transferred to Amicon ultra 15 centrifugal filter units (100 kDa, Merck), and centrifugation was performed at 2000 rpm for 2 min at 4 °C. After measuring the number of vesicles in the samples via NTA, we prepared 2 × 107 particles per sample for SDS-PAGE. The samples were boiled at 100 °C for 5 min with the 5× sample buffer and loaded them to 10% acrylamide gels and separated the proteins by SDS-PAGE. Then, the separated proteins in the gel were transferred onto 0.45 μm PVDF (polyvinylidene fluoride) membrane and subjected to immunoblotting with CD63 (MX-49.129.5, Santa Cruz), CD9 (EPR2949, Abcam), and syntenin (EPR8102, Abcam) antibody.ExoView analysis
EVs were characterized by ExoView R100 (NanoView, Bioscience, USA) based on previous studies and manufacturer's instructions.40,41 Briefly, ExoView Tetraspanin chip with the array of detection antibodies (CD63, CD81, and CD9) and a negative control (MIgG) was used for the analysis experiments. Each sample solution of 35 μl was dropped on the chip and incubated at room temperature overnight. After treatment, the samples were scanned using the ExoView R100, and data were analyzed by ExoView Analyzer 3.0. The analysis conditions for the cutoff intensity values CD63, CD9, and CD81 were adjusted to 350, 350, and 830, respectively, as the min values. The max value was fixed to 5000 for all antibodies. Additional data are presented in Fig. S13.†Conclusions
The NUTS was fabricated using MEMS technology for the development of a filter system with accurate and quantitative performance. It has uniform size nanopores and a homogeneous array pattern on the membrane. It showed outstanding physical characteristics despite that the membrane is ultra-thin and has a flat surface. The NUTS was applied to the TFF system, and its filtration results were distinct from that of general filtering experiments. The unique feature of the membrane significantly reduced standard blocking and cake formation during the filtration process, while complete and intermediate blocking was dominantly affected in terms of filtration performance. The complete blocking phenomenon is directly affected by the relation between pore and particle size. Considering that the pore size of the NUTS could be easily controlled during the fabrication process using the MEMS technique, the cutoff size of the filter could be also easily controlled by controlling the pore size as well. This indicates that NUTS can be easily customized according to researchers' needs.As a proof of concept for biological applications, filtration was performed using cell-cultured media with enrichment of EVs. The existence of filtrated EVs was first confirmed by TEM images from the filtrate passed through the two types of filters. The WB data for specific exosome marker proteins confirmed the identity of EVs obtained through filtration. Although the particle distribution was similar between the two filtrates, the NUTS filtrate shows a higher population of smaller particles than the Vivaflow filtrate. Interestingly, the components of the filtrates showed some differences in the profile of tetraspanins that were analyzed using the ExoView platform, perhaps resulting from the size-dependent variation of EV components. The NUTS could be easily modulated for the cutoff size of the filter. In a further study, we plan to design various pore sizes of the NUTS to investigate EV characteristics for different size ranges. This study would help elucidate information regarding the relationships and components of EVs according to their size ranges. Moreover, this tool could be utilized to distinguish the property of different clinical samples, such as urine, plasma, and serum.
Author contributions
All authors have contributions in writing the manuscript and have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Yonsei University Research Fund of 2021-22-0293, 2021-22-0291, and 2021-22-0061; by a grant from the National Research Foundation of Korea (NRF) funded by the Korea Government (MSIT) (No. NRF-2021R1A3B1076605, 2021R1F1A1060166, and 2022R1C1C1013481); and by the Technology Development Program (S2854312) funded by the Ministry of SMEs and Startups (MSS, Korea).Notes and references
- R. Kalluri and V. S. LeBleu, Science, 2020, 367, 640 CrossRef PubMed
.
- C. Ciardiello, L. Cavallini, C. Spinelli, J. L. Yang, M. Reis-Sobreiro, P. de Candia, V. R. Minciacchi and D. Di Vizio, Int. J. Mol. Sci., 2016, 17, 175 CrossRef PubMed
.
- S. B. Kim, H. R. Kim, M. C. Park, S. Cho, P. C. Goughnour, D. Han, I. Yoon, Y. Kim, T. Kang, E. Song, P. Kim, H. Choi, J. Y. Mun, C. Song, S. Lee, H. S. Jung and S. Kim, J. Cell Biol., 2017, 216, 2201–2216 CrossRef CAS PubMed
.
- J. D. McBride, L. Rodriguez-Menocal and E. V. Badiavas, J. Invest. Dermatol., 2017, 137, 1622–1629 CrossRef CAS PubMed
.
- J. Kowal, G. Arras, M. Colombo, M. Jouve, J. P. Morath, B. Primdal-Bengtson, F. Dingli, D. Loew, M. Tkach and C. Thery, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E968–E977 CrossRef CAS PubMed
.
- H. Y. Zhang, D. Freitas, H. S. Kim, K. Fabijanic, Z. Li, H. Y. Chen, M. T. Mark, H. Molina, A. B. Martin, L. Bojmar, J. Fang, S. Rampersaud, A. Hoshino, I. Matei, C. M. Kenific, M. Nakajima, A. P. Mutvei, P. Sansone, W. Buehring, H. J. Wang, J. P. Jimenez, L. Cohen-Gould, N. Paknejad, M. Brendel, K. Manova-Todorova, A. Magalhaes, J. A. Ferreira, H. Osorio, A. M. Silva, A. Massey, J. R. Cubillos-Ruiz, G. Galletti, P. Giannakakou, A. M. Cuervo, J. Blenis, R. Schwartz, M. S. Brady, H. Peinado, J. Bromberg, H. Matsui, C. A. Reis and D. Lyden, Nat. Cell Biol., 2018, 20, 332 CrossRef CAS PubMed
.
- C. Thery, K. W. Witwer, E. Aikawa, M. J. Alcaraz, J. D. Anderson, R. Andriantsitohaina, A. Antoniou, T. Arab, F. Archer, G. K. Atkin-Smith, D. C. Ayre, J. M. Bach, D. Bachurski, H. Baharvand, L. Balaj, S. Baldacchino, N. N. Bauer, A. A. Baxter, M. Bebawy, C. Beckham, A. B. Zavec, A. Benmoussa, A. C. Berardi, P. Bergese, E. Bielska, C. Blenkiron, S. Bobis-Wozowicz, E. Boilard, W. Boireau, A. Bongiovanni, F. E. Borras, S. Bosch, C. M. Boulanger, X. Breakefield, A. M. Breglio, M. A. Brennan, D. R. Brigstock, A. Brisson, M. L. D. Broekman, J. F. Bromberg, P. Bryl-Gorecka, S. Buch, A. H. Buck, D. Burger, S. Busatto, D. Buschmann, B. Bussolati, E. I. Buzas, J. B. Byrd, G. Camussi, D. R. F. Carter, S. Caruso, L. W. Chamley, Y. T. Chang, C. C. Chen, S. Chen, L. Cheng, A. R. Chin, A. Clayton, S. P. Clerici, A. Cocks, E. Cocucci, R. J. Coffey, A. Cordeiro-da-Silva, Y. Couch, F. A. W. Coumans, B. Coyle, R. Crescitelli, M. F. Criado, C. D'Souza-Schorey, S. Das, A. D. Chaudhuri, P. de Candia, E. F. De Santana, O. De Wever, H. A. del Portillo, T. Demaret, S. Deville, A. Devitt, B. Dhondt, D. Di Vizio, L. C. Dieterich, V. Dolo, A. P. D. Rubio, M. Dominici, M. R. Dourado, T. A. P. Driedonks, F. V. Duarte, H. M. Duncan, R. M. Eichenberger, K. Ekstrom, S. E. L. Andaloussi, C. Elie-Caille, U. Erdbrugger, J. M. Falcon-Perez, F. Fatima, J. E. Fish, M. Flores-Bellver, A. Forsonits, A. Frelet-Barrand, F. Fricke, G. Fuhrmann, S. Gabrielsson, A. Gamez-Valero, C. Gardiner, K. Gartner, R. Gaudin, Y. S. Gho, B. Giebel, C. Gilbert, M. Gimona, I. Giusti, D. C. I. Goberdhan, A. Gorgens, S. M. Gorski, D. W. Greening, J. C. Gross, A. Gualerzi, G. N. Gupta, D. Gustafson, A. Handberg, R. A. Haraszti, P. Harrison, H. Hegyesi, A. Hendrix, A. F. Hill, F. H. Hochberg, K. F. Hoffmann, B. Holder, H. Holthofer, B. Hosseinkhani, G. K. Hu, Y. Y. Huang, V. Huber, S. Hunt, A. G. E. Ibrahim, T. Ikezu, J. M. Inal, M. Isin, A. Ivanova, H. K. Jackson, S. Jacobsen, S. M. Jay, M. Jayachandran, G. Jenster, L. Z. Jiang, S. M. Johnson, J. C. Jones, A. Jong, T. Jovanovic-Talisman, S. Jung, R. Kalluri, S. Kano, S. Kaur, Y. Kawamura, E. T. Keller, D. Khamari, E. Khomyakova, A. Khvorova, P. Kierulf, K. P. Kim, T. Kislinger, M. Klingeborn, D. J. Klinke, M. Kornek, M. M. Kosanovic, A. F. Kovacs, E. M. Kramer-Albers, S. Krasemann, M. Krause, I. V. Kurochkin, G. D. Kusuma, S. Kuypers, S. Laitinen, S. M. Langevin, L. R. Languino, J. Lannigan, C. Lasser, L. C. Laurent, G. Lavieu, E. Lazaro-Ibanez, S. Le Lay, M. S. Lee, Y. X. F. Lee, D. S. Lemos, M. Lenassi, A. Leszczynska, I. T. S. Li, K. Liao, S. F. Libregts, E. Ligeti, R. Lim, S. K. Lim, A. Line, K. Linnemannstons, A. Llorente, C. A. Lombard, M. J. Lorenowicz, A. M. Lorincz, J. Lotvall, J. Lovett, M. C. Lowry, X. Loyer, Q. Lu, B. Lukomska, T. R. Lunavat, S. L. N. Maas, H. Malhi, A. Marcilla, J. Mariani, J. Mariscal, E. S. Martens-Uzunova, L. Martin-Jaular, M. C. Martinez, V. R. Martins, M. Mathieu, S. Mathivanan, M. Maugeri, L. K. McGinnis, M. J. McVey, D. G. Meckes, K. L. Meehan, I. Mertens, V. R. Minciacchi, A. Moller, M. M. Jorgensen, A. Morales-Kastresana, J. Morhayim, F. Mullier, M. Muraca, L. Musante, V. Mussack, D. C. Muth, K. H. Myburgh, T. Najrana, M. Nawaz, I. Nazarenko, P. Nejsum, C. Neri, T. Neri, R. Nieuwland, L. Nimrichter, J. P. Nolan, E. N. M. Nolte-'t Hoen, N. N. Hooten, L. O'Driscoll, T. O'Grady, A. O'Loghlen, T. Ochiya, M. Olivier, A. Ortiz, L. A. Ortiz, X. Osteikoetxea, O. Ostegaard, M. Ostrowski, J. Park, D. M. Pegtel, H. Peinado, F. Perut, M. W. Pfaffl, D. G. Phinney, B. C. H. Pieters, R. C. Pink, D. S. Pisetsky, E. P. von Strandmann, I. Polakovicova, I. K. H. Poon, B. H. Powell, I. Prada, L. Pulliam, P. Quesenberry, A. Radeghieri, R. L. Raffai, S. Raimondo, J. Rak, M. I. Ramirez, G. Raposo, M. S. Rayyan, N. Regev-Rudzki, F. L. Ricklefs, P. D. Robbins, D. D. Roberts, S. C. Rodrigues, E. Rohde, S. Rome, K. M. A. Rouschop, A. Rughetti, A. E. Russell, P. Saa, S. Sahoo, E. Salas-Huenuleo, C. Sanchez, J. A. Saugstad, M. J. Saul, R. M. Schiffelers, R. Schneider, T. H. Schoyen, A. Scott, E. Shahaj, S. Sharma, O. Shatnyeva, F. Shekari, G. V. Shelke, A. K. Shetty, K. Shiba, P. R. M. Siljander, A. M. Silva, A. Skowronek, O. L. Snyder, R. P. Soares, B. W. Sodar, C. Soekmadji, J. Sotillo, P. D. Stahl, W. Stoorvogel, S. L. Stott, E. F. Strasser, S. Swift, H. Tahara, M. Tewari, K. Timms, S. Tiwari, R. Tixeira, M. Tkach, W. S. Toh, R. Tomasini, A. C. Torrecilhas, J. P. Tosar, V. Toxavidis, L. Urbanelli, P. Vader, B. W. M. van Balkom, S. G. van der Grein, J. Van Deun, M. J. C. van Herwijnen, K. Van Keuren-Jensen, G. van Niel, M. E. van Royen, A. J. van Wijnen, M. H. Vasconcelos, I. J. Vechetti, T. D. Veit, L. J. Vella, E. Velot, F. J. Verweij, B. Vestad, J. L. Vinas, T. Visnovitz, K. V. Vukman, J. Wahlgren, D. C. Watson, M. H. M. Wauben, A. Weaver, J. P. Webber, V. Weber, A. M. Wehman, D. J. Weiss, J. A. Welsh, S. Wendt, A. M. Wheelock, Z. Wiener, L. Witte, J. Wolfram, A. Xagorari, P. Xander, J. Xu, X. M. Yan, M. Yanez-Mo, H. Yin, Y. Yuana, V. Zappulli, J. Zarubova, V. Zekas, J. Y. Zhang, Z. Z. Zhao, L. Zheng, A. R. Zheutlin, A. M. Zickler, P. Zimmermann, A. M. Zivkovic, D. Zocco and E. K. Zuba-Surma, J. Extracell. Vesicles, 2018, 7, 1535750 CrossRef PubMed
.
- K. Boriachek, M. N. Islam, A. Möller, C. Salomon, N.-T. Nguyen, M. S. A. Hossain, Y. Yamauchi and M. J. A. Shiddiky, Small, 2018, 14, 1702153 CrossRef PubMed
.
- M. X. Wu, Y. S. Ouyang, Z. Y. Wang, R. Zhang, P. H. Huang, C. Y. Chen, H. Li, P. Li, D. Quinn, M. Dao, S. Suresh, Y. Sadovsky and T. J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 10584–10589 CrossRef CAS PubMed
.
- B. H. Wunsch, J. T. Smith, S. M. Gifford, C. Wang, M. Brink, R. L. Bruce, R. H. Austin, G. Stolovitzky and Y. Astier, Nat. Nanotechnol., 2016, 11, 936–940 CrossRef CAS PubMed
.
- D. B. Yang, W. H. Zhang, H. Y. Zhang, F. Q. Zhang, L. M. Chen, L. X. Ma, L. M. Larcher, S. X. Chen, N. Liu, Q. X. Zhao, P. H. L. Tran, C. Y. Chen, R. N. Veedu and T. Wang, Theranostics, 2020, 10, 3684–3707 CrossRef CAS PubMed
.
- W. Wang, J. Luo and S. Wang, Adv. Healthcare Mater., 2018, 7, 1800484 CrossRef PubMed
.
- L. Zhu, H. T. Sun, S. Wang, S. L. Huang, Y. Zheng, C. Q. Wang, B. Y. Hu, W. Qin, T. T. Zou, Y. Fu, X. T. Shen, W. W. Zhu, Y. Geng, L. Lu, H. L. Jia, L. X. Qin and Q. Z. Dong, J. Hematol. Oncol., 2020, 13, 152 CrossRef PubMed
.
- R. J. Lobb, M. Becker, S. W. Wen, C. S. Wong, A. P. Wiegmans, A. Leimgruber and A. Moller, J. Extracell. Vesicles, 2015, 4, 27031 CrossRef PubMed
.
- E. Iritani, Drying Technol., 2013, 31, 146–162 CrossRef
.
- E. Iritani and N. Katagiri, KONA Powder Part. J., 2016, 33, 179–202 CrossRef
.
- P. H. Hermans and H. L. Bredée, J. Soc. Chem. Ind., 1936, 55T, 1–4 Search PubMed
.
- S.-i. Nakao, J. Membr. Sci., 1994, 96, 131–165 CrossRef CAS
.
- C. C. Ho and A. L. Zydney, J. Membr. Sci., 1999, 155, 261–275 CrossRef CAS
.
- W.-M. Lu and K.-J. Hwang, AIChE J., 1995, 41, 1443–1455 CrossRef CAS
.
- M. R. Mackley and N. E. Sherman, Chem. Eng. Sci., 1992, 47, 3067–3084 CrossRef CAS
.
- K. J. Hwang, Y. H. Yu and W. M. Lu, J. Membr. Sci., 2001, 194, 229–243 CrossRef CAS
.
- W. M. Lu, K. J. Hwang and S. C. Ju, Chem. Eng. Sci., 1993, 48, 863–872 CrossRef CAS
.
- W. Wang, X. D. Jia and G. A. Davies, Chem. Eng. J. Biochem. Eng. J., 1995, 60, 55–62 CrossRef CAS
.
- J. R. Phillips and B. F. Dodge, AIChE J., 1963, 9, 93–98 CrossRef CAS
.
- G. Firpo, E. Angeli, L. Repetto and U. Valbusa, J. Membr. Sci., 2015, 481, 1–8 CrossRef CAS
.
- M. Kim, B. Sankararao, S. Lee and C. Yoo, Ind. Eng. Chem. Res., 2013, 52, 17198–17205 CrossRef CAS
.
- F. L. Wang and V. V. Tarabara, J. Colloid Interface Sci., 2008, 328, 464–469 CrossRef CAS PubMed
.
- J. Hermia, Trans. Inst. Chem. Eng., 1982, 60, 183–187 CAS
.
- D. J. Miller, S. Kasemset, D. R. Paul and B. D. Freeman, J. Membr. Sci., 2014, 454, 505–515 CrossRef CAS
.
- S. H. Jiang, Y. L. Zhang, F. C. Zhao, Z. J. Yu, X. F. Zhou and H. Q. Chu, Algal Res., 2018, 35, 613–623 CrossRef
.
- D. J. Karst, K. Ramer, E. H. Hughes, C. P. Jiang, P. J. Jacobs and F. G. Mitchelson, Biotechnol. Prog., 2020, 36, e3040 CrossRef CAS PubMed
.
- H. Huang, T. A. Young and J. G. Jacangelo, Environ. Sci. Technol., 2008, 42, 714–720 CrossRef CAS PubMed
.
- H. Xu, K. Xiao, J. L. Yu, B. Huang, X. M. Wang, S. Liang, C. H. Wei, X. H. Wen and X. Huang, Membranes, 2020, 10, 171 CrossRef CAS PubMed
.
- M. K. Jung and J. Y. Mun, J. Visualized Exp., 2018, 56482 Search PubMed
.
- M. F. Baietti, Z. Zhang, E. Mortier, A. Melchior, G. Degeest, A. Geeraerts, Y. Ivarsson, F. Depoortere, C. Coomans, E. Vermeiren, P. Zimmermann and G. David, Nat. Cell Biol., 2012, 14, 677–685 CrossRef CAS PubMed
.
- C. Thery, L. Zitvogel and S. Amigorena, Nat. Rev. Immunol., 2002, 2, 569–579 CrossRef CAS PubMed
.
- K. M. McAndrews and R. Kalluri, Mol. Cancer, 2019, 18, 52 CrossRef PubMed
.
- H. S. Jung, N. Billington, K. Thirumurugan, B. Salzameda, C. R. Cremo, J. M. Chalovich, P. D. Chantler and P. J. Knight, J. Mol. Biol., 2011, 408, 863–878 CrossRef CAS PubMed
.
- H. H. Jung, J. Y. Kim, J. E. Lim and Y. H. Im, Sci. Rep., 2020, 10, 14069 CrossRef PubMed
.
- R. Crescitelli, C. Lasser and J. Lotvall, Nat. Protoc., 2021, 16, 1548 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: SEM images of a commercial filter membrane, schematic diagram of fabrication of NMUS, relationships of pressure with various factors, optical image of a NMUS surface, distribution of pore diameter in single membrane, roughness of NMUS via AFM and partial fraction of NMUS by AFM indentation, SEM image of the membrane after bio-sample filtration, blocking analysis of bio-sample filtration, western blot data for the apoptotic body markers, contact angle measurement of the membrane surface, design of fluidic channel, microarray images of Exoview measurement. See DOI: https://doi.org/10.1039/d2na00227b |
| This journal is © The Royal Society of Chemistry 2023 |