Open Access Article
Open Access ArticleUsing membrane–water partition coefficients in a critical membrane burden approach to aid the identification of neutral and ionizable chemicals that induce acute toxicity below narcosis levels†
Steven T. J.
Droge
 *a,
Geoff
Hodges
*a,
Geoff
Hodges
 b,
Mark
Bonnell
b,
Mark
Bonnell
 c,
Steve
Gutsell
c,
Steve
Gutsell
 b,
Jayne
Roberts
b,
Jayne
Roberts
 b,
Alexandre
Teixeira
b,
Alexandre
Teixeira
 b and
Elin L.
Barrett
b and
Elin L.
Barrett
 b
b
aDepartment of Freshwater and Marine Ecology (FAME), Institute for Biodiversity and Ecosystem Dynamics (IBED), Universiteit van Amsterdam (UvA), Science Park 904, 1098XH Amsterdam, The Netherlands. E-mail: steven.droge@wur.nl; Tel: +31 317484410
bSafety and Environmental Assurance Centre, Unilever, Colworth Science Park, Sharnbrook, Bedfordshire, UK
cEnvironment and Climate Change Canada, Ecological Assessment Division, Science and Risk Assessment Directorate, Gatineau, Quebec, Canada
First published on 13th February 2023
Abstract
The risk assessment of thousands of chemicals used in our society benefits from adequate grouping of chemicals based on the mode and mechanism of toxic action (MoA). We measure the phospholipid membrane–water distribution ratio (DMLW) using a chromatographic assay (IAM-HPLC) for 121 neutral and ionized organic chemicals and screen other methods to derive DMLW. We use IAM-HPLC based DMLW as a chemical property to distinguish between baseline narcosis and specific MoA, for reported acute toxicity endpoints on two separate sets of chemicals. The first set comprised 94 chemicals of US EPA's acute fish toxicity database: 47 categorized as narcosis MoA, 27 with specific MoA, and 20 predominantly ionic chemicals with mostly unknown MoA. The narcosis MoA chemicals clustered around the median narcosis critical membrane burden (CMBnarc) of 140 mmol kg−1 lipid, with a lower limit of 14 mmol kg−1 lipid, including all chemicals labelled Narcosis_I and Narcosis_II. This maximum ‘toxic ratio’ (TR) between CMBnarc and the lower limit narcosis endpoint is thus 10. For 23/28 specific MoA chemicals a TR >10 was derived, indicative of a specific adverse effect pathway related to acute toxicity. For 10/12 cations categorized as “unsure amines”, the TR <10 suggests that these affect fish via narcosis MoA. The second set comprised 29 herbicides, including 17 dissociated acids, and evaluated the TR for acute toxic effect concentrations to likely sensitive aquatic plant species (green algae and macrophytes Lemna and Myriophyllum), and non-target animal species (invertebrates and fish). For 21/29 herbicides, a TR >10 indicated a specific toxic mode of action other than narcosis for at least one of these aquatic primary producers. Fish and invertebrate TRs were mostly <10, particularly for neutral herbicides, but for acidic herbicides a TR >10 indicated specific adverse effects in non-target animals. The established critical membrane approach to derive the TR provides for useful contribution to the weight of evidence to bin a chemical as having a narcosis MoA or less likely to have acute toxicity caused by a more specific adverse effect pathway. After proper calibration, the chromatographic assay provides consistent and efficient experimental input for both neutral and ionizable chemicals to this approach.
Environmental significanceTo prioritize more detailed risk assessment for certain chemicals of concern, it is important for risk assessors to identify chemicals that induce toxicity by an adverse effect pathway other than narcosis. Our study shows that the membrane lipid–water distribution ratio (DMLW) is a key descriptor for both neutral and ionizable organic chemicals. By measuring new DMLW values for 121 chemicals we derive the critical membrane burden (CMB) threshold for fish acute toxicity below which a specific mode of action other than narcosis drives toxicity and use this CMB approach on the response of different (non-)target species to a variety of herbicides. |
1. Introduction
(i) Mode of action analysis to support priority setting and risk assessment
With increasing regulatory and ethical drive to accelerate and strengthen progress on chemical management and at the same time reduce reliance on in vivo data to fill data gaps, there is now an increased emphasis on the use of alternative approaches and data sources to support decision making.1,2 The approach taken to chemical management varies from region to region. However, widely applicable methods such as chemical grouping are employed under many regulatory schemes such as for read-across and data-gap filling under the EU REACH regulation where ECHA developed a Read-Across Assessment Framework (RAAF).3 One of the critical components for successful chemical grouping is an understanding of both the modes and mechanisms of action (MoA and MechoA) of the target and analogue structures in addition to comparable structural and bioavailability properties. If a chemical category includes members with disparate modes and mechanisms of action (including metabolites of parent chemicals in the category, if possible), the justification of the read-across for hazard information becomes less certain and may lead to false positives or false negatives.The consideration of modes and mechanisms of action can also be very useful as one descriptor of hazards when prioritizing chemicals for further regulatory action. In Canada, for example, identifying organic chemicals with specific and non-specific modes and mechanisms of action was incorporated into version 1.0 of the Ecological Risk Classification (ERC1) approach2,4 used in 2016 by Environment and Climate Change Canada (ECCC) to reset priorities for 640 organic chemicals for phase three of the Chemicals Management Plan (CMP). ERC1 introduced the concept of data consensus weighting between critical body residue (CBR) derived toxicity ratios (TRs) and quantitative structure–activity relationship (QSAR) classification of MoA as one hazard descriptor for priority setting in ERC1. Specific modes of action were responsible for 40% of the high hazard classifications (i.e., not final risk classification) identified in 2016 by ECCC using ERC1.
Building on ERC1, version 2.0 of the ERC5 (ERC2) was developed in 2018 and is currently in use by ECCC for identifying chemicals of concern among 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 organic chemicals for post 2020 work planning reasons. ERC2 is a weight of evidence logical model relying on data consensus to determine the risk classification, risk confidence and risk scale of organic chemicals for further regulatory consideration. ERC2 takes the MechoA concept further than ERC1 by expanding the number and type of tissue residue and QSAR approaches used for identifying specific and non-specific modes of action. ERC2 also integrates both molecular initiating event information (MIE) and modes and mechanisms of action using the adverse outcome pathway (AOP) concept.6
200 organic chemicals for post 2020 work planning reasons. ERC2 is a weight of evidence logical model relying on data consensus to determine the risk classification, risk confidence and risk scale of organic chemicals for further regulatory consideration. ERC2 takes the MechoA concept further than ERC1 by expanding the number and type of tissue residue and QSAR approaches used for identifying specific and non-specific modes of action. ERC2 also integrates both molecular initiating event information (MIE) and modes and mechanisms of action using the adverse outcome pathway (AOP) concept.6
The degree of MoA consensus in ERC2 was also evaluated by comparing MoA classifications from the five MoA QSAR and five methods used to calculate tissue residue TRs associated with median lethality in fish2 in ERC2. The confidence score associated with the MoA classification in ERC2 is directly related to the degree of consensus between all methods above. The results, based on 929 organic chemicals with available averaged acute fish median lethality data gathered using the OECD QSAR Toolbox,7,8 revealed that 100% consensus between all ten methods was greater for non-specific (narcosis) chemicals (∼53%) than those with specific modes of action (∼29%). There was no large distinction in MoA classification among the five tissue residue methods and the authors suggest further curation of aquatic toxicity and water solubility data may improve the correlation among all methods.
A consensus MoA approach has also been incorporated into automated on-line tools for determining predicted no-effect concentrations (PNECs) for risk assessment using the Ecological Threshold of Toxicological Concern (Eco-TTC) and curated ENVIROTOX database tools.9–11 MechoA considerations have also been incorporated into the derivation of assessment (safety) factors for PNEC derivation used in risk assessments of new and existing substances by ECCC. The evaluation of modes and mechanisms of action is reviewed and documented to ensure that the selection of critical toxicity values used to derive a chronic PNEC are related to the mode and mechanisms of action.
A number of schemes exist to support the identification of the MoA of chemicals. Typically schemes such as those developed by Verhaar and subsequent updates,12–14 the EPA Mode of Action and Toxicity (MOAtox) database15 and the Acute Aquatic Toxicity MOA by OASIS (AAT OASIS) scheme,16 also incorporated into the US EPA inhouse expert system ASTER (ASsessment Tools for the Evaluation of Risk17) and the US EPA Toxicity Estimation Software Tool (TEST18), are easily accessible for such an application. The Verhaar and OASIS schemes also have been incorporated into the OECD QSAR toolbox as mechanistic profilers. More recently there has been a growing recognition that classifying chemicals using the mechanism of action can add more confidence in toxicity prediction.19 The MechoA scheme as one such example has recently been developed and available freely online is the KREATiS MechoA scheme20 to predict the toxicity mechanism based on the chemical structure.21,22 The recent scheme of Sapounidou et al.23,24 is another such example which follows an analogous approach to the MechoA scheme. However, it remains that such schemes have potential for discrepancies in assigned MoA and also have limitations in being able to classify the full chemical space.9 With the benefits of having a reliable understanding of MoA for supporting chemical prioritization and risk assessment in addition to data gap filling, there is, therefore, a continued need to develop consensus on MoA.10 The development of new and complementary approaches to support MoA is thus needed.
In the current study, we investigate the expansion of an existing approach considering the application of the critical body residue (CBR) and critical membrane burden (CMB) to aid the determination of MoA. Specifically, we consider the role of the membrane lipid–water distribution ratio (DMLW, as used for ionizable species, or KMLW for neutral species) to distinguish between chemicals that operate in the range of baseline toxicants from chemicals which are likely to induce toxicity via another MoA at a CMB lower than typical for narcosis. Since the cell membrane is not the target site for most specific adverse effect pathways (e.g., covalent interactions with DNA), the DMLW-approach is not intended to classify a chemical to a certain MoA. Our approach can be used to identify chemicals for which further information on MoA would be considered of high relevance for risk assessment, because they do not induce acute toxicity by baseline narcosis. The key aim is thus to define the lower limit CMB of polar and non-polar narcotic chemicals. DMLW can be derived using a range of experimental methods (e.g. unilamellar liposome vesicles25 or solid supported lipid membranes26,27), or in silico approaches, such as COSMOmic28,29 or molecular dynamics.30–32 Here, we consider the use of a chromatographic column retention approach to derive DMLW values for MoA determination. The overall aim was to generate two strategic DMLW data sets that would allow for MoA assessment with high quality toxicity data, extending previous efforts in multiple aspects in terms of both chemical space, DMLW data quality, and MoA domains. The first data set focuses on chemicals listed in the acute fish toxicity data from EPA's Fathead minnow (FHM) database. The second set involves herbicides for which toxicity data for several types of aquatic species are compared. Before discussing the experimental part and discuss the data interpretation, below we briefly introduce in Section (ii) the chromatographic approach and comparable studies that precede the current work, (iii) the link between DMLW and toxicity, (iv) alternative ways to derive DMLW, and (v) which toxicity data sets were used to evaluate the CMB-MoA analysis. The method section then starts with explaining which chemicals from those toxicity databases were selected to obtain the chromatographic retention data for.
(ii) Chromatographic retention measurements on phospholipid coated particles
Chromatographic retention time measurements allow for consistent experimental values of physico-chemical descriptors for a wide range of organic chemicals, because the retention capacity factors are proportional to the interaction energy with the column's coating material.33–42 The development of silica particles coated with an immobilized artificial phospholipid membrane for use in high performance liquid chromatography (IAM-HPLC)43–46 has enabled measurements of retention capacity factors (kIAM) that closely relate to descriptors that are valuable for drug development, toxicokinetic modeling and chemical risk assessment in general.47IAM-HPLC provides a cost-effective, high-throughput, consistent approach to experimentally derive indicative DMLW values directly from these kIAM measurements. Using kIAM obtained in (or extrapolated to) fully aqueous eluent (i.e., k0IAM), multiplication with a pre-determined medium/phospholipid volume ratio of the IAM-column (φ = 18.9 (ref. 45)) gives the partition coefficient between the bulk aqueous eluent and IAM phospholipid monolayer (KIAM), which should be analogous to DMLW:
| DMLW − KIAM = φ × k0IAM = 18.9 × k0IAM | (1) |
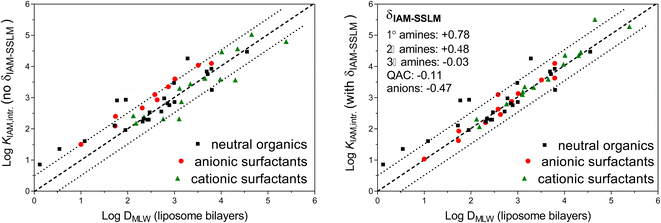
A review on liposomal partition coefficients showed a linear relationship between DMLW values and IAM-HPLC retention capacity factors (kIAM) for 24 neutral organic chemicals, as shown in Fig. 1 by black squares (kIAM already converted to KIAM partition coefficients by using eqn (1)).25 For most chemicals, the IAM-HPLC estimate was within a factor of ±3, but for some neutral chemicals with high H-bond donor capacities the KIAM overestimated DMLW by a factor of ∼10. Two studies, on anionic surfactants27 (including perfluorinated anions) and cationic surfactants26 (including quaternary ammonium chemicals), both determined KIAM values and DMLW (via solid supported lipid membranes), which extends the DMLW–KIAM comparison to also include ionic chemicals. The 19 cationic surfactants26 are shown in Fig. 1 as green triangles, and the 10 (fluorinated and non-fluorinated) anionic surfactants as red dots. Since the aim is to derive quantitative and precise DMLW values from kIAM for the CMB approach, and not work with relative chromatographic indices, we need to take into account that the silica particles used in the IAM-columns cause confounding electrostatic effects on the retention time of ionic chemicals. Only intrinsic IAM phospholipid-partition coefficients (KIAM,intr) are to be used for predominantly/fully ionized chemicals, which are corrected for this confounding attraction using empirically based or theoretically derived Boltzmann corrective factors.48 This Boltzmann correction is in more detail reviewed for dissociated acids and protonated bases in ESI Section S1.† While the neutral data in Fig. 1 were obtained from a wide variety of studies and different experimental approaches to derive DMLW and KIAM, the scatter between the series of KIAM and DMLW values obtained for ionic surfactants was obtained in a single research institute. Consistent empirical differences for KIAM and DMLW for specific types of ionic moieties on the surfactant data indicated that it may be necessary to adjust the IAM-HPLC results by several empirical increments (δIAM-SSLM) to improve the KIAM–DMLW consistency, as shown in the plot on the right in Fig. 1: −0.5 log units for all anionic chemicals, +0.8 log units for primary (1°) amine cations, +0.5 log units for secondary (2°) amine cations, −0.03 log units for tertiary (3°) amine cations, and −0.1 log units for quaternary ammonium cations (see also ESI-Table S3†). The reasons for these ionic-type specific increments to align the phospholipid-coating based KIAM data and phospholipid bilayers based DMLW data remain to be elucidated.
The primary goal of the current study was to use a measured DMLW value to predict the baseline toxicity to aquatic organisms for a large and diverse set of organic chemicals, and identify whether a chemical is not a baseline toxicant and very likely induces acute toxicity by a reactive or more specific MoA. Since IAM-HPLC allows for high-throughput and experimental consistency, the KIAM values obtained by this method are considered to be the best DMLW proxies to do so. Several studies have already demonstrated that kIAM values (i.e., IAM-HPLC retention capacity factors) strongly correlate with the acute effect concentrations of non-specific toxicants to aquatic organisms (LC50,Narcosis),49–54 for example tadpoles (Rana temporaria, from the data set on narcotics by Overton and Meyer) and the fathead minnow fish (Pimephales promelas). The set-up of these valuable kIAM-LC50,Narcosis comparative studies, however, are limited in several aspects, and we aimed to extend these aspects in this study. First, while the reported studies used kIAM retention capacity factors that were based on measurements with eluent that contained 40% acetonitrile, the current study aims to derive new IAM-retention data for a large number of strategic chemicals (regarding available toxicity data, ionization, and data scarcity) obtained at, or adequately extrapolated to, fully aqueous eluent. As a result, our IAM-HPLC data set allows for simple conversion of k0IAM (0% solvent, 100% water) to KIAM values that are direct proxies for DMLW, rather than using kIAM as a relative scaling index. Second, whereas the reported studies included only neutral chemicals, the current study aimed to include largely ionized organic bases (or permanently charged cations) and strong organic acids in toxicity evaluations. The KIAM–DMLW data set on ionic surfactants allows for a validation set in deriving the intrinsic KIAM values to relate to DMLW values for ionic species (Fig. 1). Third, while the reported studies included only chemicals with narcosis MoA, we aim to include chemicals with proven specific strongly toxic MoA alongside those with only narcosis MoA. This should exemplify the extent to which IAM-based DMLW values accurately distinguish specific MoA and narcosis MoA chemicals.
(iii) D MLW values predict baseline toxicity
Like all DMLW proxies, IAM-HPLC based retention capacity factors can provide a reasonable estimate of the dissolved concentration of a chemical that is toxic, because organic chemicals are toxic at a certain accumulated cellular concentration. For a wide variety of organic chemicals considered to only act through a non-specific “narcosis” MoA, the total critical body residue (CBRwet, in mmol kg−1 wet weight) is found to be in the range of 2–8 mmol kg−1 for a range of different aquatic organisms.55–57 Narcosis MoA is most often related to a critical accumulation level of molecules in cell membranes, which relates to the “baseline toxicity” for all chemicals. At the critical membrane burden (CMB, in mmol kg−1 phospholipid) basic cellular membrane functions become impaired.56 The relationship between CBRwet and CMB can be expressed as the ratio between the two driving partition coefficients.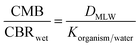 | (2) |
According to partition coefficients to different tissue phases, membranes being one specific component besides for example storage lipids, carbohydrates, (structural) proteins, and water,58–62 it was derived from a reviewed set of CBR data for narcosis chemicals that CMBnarc ranges between 80 and 250 mmol kg−1 membrane, with a geometric mean of 140 mmol kg−1.63
Using this narrow range of CMBnarc, dissolved concentrations that are acutely lethal to 50% of aquatic organisms due to narcosis (LC50,narc) can be back-calculated for any chemical using DMLW, as defined by the target lipid model by Di Toro et al. (2000):64
| LC50,narc (in mmol L−1 water) = CMBnarc (∼140 mmol kg−1 membrane)/DMLW (in L water per kg membrane) | (3) |
Chemicals that are considered to distinctively exert acute toxicity by a MoA other than narcosis are expected to have a critical membrane burden significantly below 140 mmol kg−1, and hence have an acute (lethal) effect concentration well below the DMLW-calculated LC50,narc. It is important for risk assessors to identify this level of endpoint specificity, as near baseline cytotoxic levels many specific cellular pathways will also be affected, as a so-called “cytotoxic burst”.65,66 The primary aim of the current study was to derive a minimum CMBnarc for as large a set of chemicals as possible classified as narcotics, with the cell membrane as the target site of action for narcosis, and a measured membrane–water partition coefficient. Any organic chemical is likely to induce acute toxicity due to baseline narcosis above this range, whereas chemicals that act via a specific mechanism of action are expected to display a CMB below this minimum CMBnarc and thus indicative of a specific MoA driving the observed acute toxicity. Note that the chronic MoA to aquatic organisms is not taken into account in this CMB-MoA approach. For most baseline toxicants the acute to chronic ratio (ACR) is small (∼10×), and the chronic CMBnarc is thus not much lower than the acute CMBnarc. This suggests that below the threshold of basic membrane disturbance the exposed organism may withstand this toxic pressure for a prolonged period, with a certain loss of energy used for maintenance. However, a small fraction of chemicals identified as baseline toxicants were found to have an acute to chronic ratio of more than 30, indicating that upon chronic exposure these chemicals may act via a specific MoA.67 It is therefore not possible to rely on the current selection of chemicals classified by acute effect studies.
A prediction of the baseline toxic concentration (LC50,narc) of any organic chemical is relevant for risk assessment. First, it easily translates to an initial approach to set maximal allowable concentrations for chemicals for which no toxicity data are available. Second, it could be a check whether the adverse effect concentration reported for a certain chemical is due to a specific MoA, or whether the adverse effect occurred at a level where baseline toxicity is expected (apparently specific effects may have occurred only as part of the cytotoxic burst).68 Third, most environmental pollution occurs as complex chemical mixtures, and in most cases chemicals with a specific MoA are dissolved at concentrations well below the level that induces a specific effect. However, any chemical in a mixture contributes to the accumulation of chemicals in membranes, and each chemical therefore contributes to the narcosis CMB of the total mixture in a (molar) concentration-additive way.69–71
The CMB approach is generally evaluated using the octanol–water partition coefficient (KOW) as a proxy for DMLW, using toxicity data sets for algae, daphnids, and fish.57,64 Baseline toxicity has also been expressed as a function of chemical activity, using the maximum water solubility (Sw) in relation to the LC50 as a metric to assess the MoA.72 There are relevant uncertainties related to using both the KOW and Sw that could lead to false classification of chemicals having or not having a specific MoA.73 This involves uncertainties surrounding the KOW and Sw values, but also the relevance of these values in relation to the DMLW values driving the actual CMB, as discussed below. The approach of the current study aims to obtain DMLW experimentally, which would by-pass several uncertainties relating to KOW and Sw and thereby assess MoA specificity with a higher level of confidence.
(iv) Different ways to derive KMLW/DMLW
There are multiple approaches of deriving the DMLW in eqn (2) and (3), either by experiment or calculations. Below, we first describe commonly used and more recent ways to calculate sorption affinity to phospholipid membranes (‘phospholipophilicity’). For many neutral organic chemicals, KMLW values are closely related to octanol–water partition coefficients,25 with a nearly 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 relationship based on a structurally diverse KMLW data set:
1 relationship based on a structurally diverse KMLW data set:Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KMLW = 1.01(±0.02) × log KMLW = 1.01(±0.02) × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW + 0.12(±0.07) (n = 156, SD = 0.426, R2 = 0.948) KOW + 0.12(±0.07) (n = 156, SD = 0.426, R2 = 0.948) | (4) |
Although accurate KOW values can be obtained according to standardized protocols, there can be wide margins in reported values for many chemicals. And although large experimental KOW data sets have been used to create a variety of commonly used predictive algorithms, predicted KOW values further contribute to uncertainty in derived KMLW values according to eqn (4). Octanol also does not necessarily reflect the specific interactive properties of neutral chemicals with phospholipids in a cell membrane. Therefore, a phospholipid-water specific poly-parameter linear free energy relationship (ppLFER) has been constructed (eqn (5)).25 This ppLFER uses five chemical descriptors that should adequately cover the contribution of different solute/system interactions: molecular volume (Vx), hydrogen-bond acidity (A) and basicity (B), hexadecane-air partition coefficients (L), and a parameter to account for excess polarizability (S).
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KMLW (25 °C) = 1.65(±0.17) × Vx + 0.55(±0.03) × L − 0.95(±0.09) × S − 0.05(±0.09) × A − 4.02(±0.10) × B + 0.48(±0.09) (n = 131, SD = 0.294, R2 = 0.977) KMLW (25 °C) = 1.65(±0.17) × Vx + 0.55(±0.03) × L − 0.95(±0.09) × S − 0.05(±0.09) × A − 4.02(±0.10) × B + 0.48(±0.09) (n = 131, SD = 0.294, R2 = 0.977) | (5) |
While Vx is calculated, the descriptors S, A, and B are recommended to be derived experimentally from consistent column retention studies, as collected in the UFZ LSER database (https://www.ufz.de/lserd), as to avoid stacking of estimation uncertainties for each descriptor. So instead of the more generic hydrophobicity descriptor KOW, more accurate values can be calculated via ppLFER if descriptors are available. This indeed requires the experimental ppLFER descriptors S, A, B and L to be available or to be newly derived, e.g. from multiple chromatographic retention data.
For ionizable organic chemicals (IOCs), however, neither KOW nor the ppLFER approach adequately takes into account the ionic interactions between charged sites in the phospholipid headgroup domain with charged groups in ionized organic chemicals. The octanol–water distribution ratio (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D) of the largely ionized form of a strong acid (pKa ≪ testing pH, e.g. <3 units) or strong base (pKa ≫ testing pH, e.g. >3 units) is mostly several orders of magnitude lower than the octanol–water partition coefficient (log
D) of the largely ionized form of a strong acid (pKa ≪ testing pH, e.g. <3 units) or strong base (pKa ≫ testing pH, e.g. >3 units) is mostly several orders of magnitude lower than the octanol–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) of the fully neutral acid or base, because the ionized form strongly prefers the readily polarizable aqueous phase.74,75 Moreover, the affinity of the ionic form for octanol strongly depends on the presence and concentration of counterions. However, the majority of cell membrane phospholipids contain a zwitterionic phosphatidylcholine headgroup. In anisotropically organized phospholipid layers of liposomes and cell membranes, the zwitterion moieties together form a partially hydrated headgroup ‘region’, which shields off the highly hydrophobic ‘region’ formed by the densely packed fatty acid tails from the bulk water phase. Due to strongly favorable ionic interactions with the zwitterionic headgroups, combined with partial embedding in the hydrophobic bilayer core, ionized forms of many organic bases and organic acids have a phospholipid membrane–water distribution ratio (DMLW,ion) only marginally lower than, or even equal to, the KMLW,N of their corresponding neutral forms.29,76–81
P) of the fully neutral acid or base, because the ionized form strongly prefers the readily polarizable aqueous phase.74,75 Moreover, the affinity of the ionic form for octanol strongly depends on the presence and concentration of counterions. However, the majority of cell membrane phospholipids contain a zwitterionic phosphatidylcholine headgroup. In anisotropically organized phospholipid layers of liposomes and cell membranes, the zwitterion moieties together form a partially hydrated headgroup ‘region’, which shields off the highly hydrophobic ‘region’ formed by the densely packed fatty acid tails from the bulk water phase. Due to strongly favorable ionic interactions with the zwitterionic headgroups, combined with partial embedding in the hydrophobic bilayer core, ionized forms of many organic bases and organic acids have a phospholipid membrane–water distribution ratio (DMLW,ion) only marginally lower than, or even equal to, the KMLW,N of their corresponding neutral forms.29,76–81
The affinity of ionic organic chemicals for phospholipid membranes can be estimated rather crudely for both predominantly charged acids and bases as a first approach.82
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW,ion = log DMLW,ion = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KMLW,N − 1 KMLW,N − 1 | (6) |
However, a single increment between the DMLW values of the ionic and neutral forms is overly simplistic, and can be further refined for different ion types. Based on rather small sample sizes, ion-type specific scaling factors (ΔMW) have been derived according to eqn 7
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW,ion = log DMLW,ion = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KMLW,N − ΔMW KMLW,N − ΔMW | (7) |
The ΔMW describes the average difference between log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW,ion and log
DMLW,ion and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KMLW,N: −0.75 for phenolates, −2 for all other anionic chemicals, −0.3 for primary amines, −0.5 for secondary amines, and −1.25 for tertiary amines and other cationic chemicals.80,81,83–85
KMLW,N: −0.75 for phenolates, −2 for all other anionic chemicals, −0.3 for primary amines, −0.5 for secondary amines, and −1.25 for tertiary amines and other cationic chemicals.80,81,83–85
After defining the DMLW of both ionic and neutral forms, the pH-dependent fractions of neutral forms (fN) and ionic forms (1 − fN), the overall distribution ratio at a certain pH (DMLW(pH)) can be calculated according to eqn (8):
| DMLW(pH) = fN × KMLW,N + (1 − fN) × DMLW,ion | (8) |
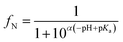 | (9) |
While the ΔMW-approach takes the specific ion-type differences into account, the KMLW,N is often still based on KOW values and eqn (4). For certain chemicals, particularly for IOCs, it becomes relevant that octanol is a bulk solvent, while phospholipid membranes are anisotropically structured. The charged moiety will favorably position in the headgroup region while the most hydrophobic molecular portion will extend into the core, and this position may strongly differ for the neutral form of the same IOC. Computational methods such as the COSMOmic and COSMOconf modules of the commercial software package COSMOlogic (3ds Dassault systèmes/BIOVIA) and molecular dynamics simulations can be of use in spatially oriented prediction of DMLW. COSMOmic combines quantum chemistry and thermodynamics and uses the three-dimensional (3D) structure of both the solute and the hydrated phospholipid membrane. The internal membrane potential and surface charge density distributions of ionogenic chemicals as well as the phospholipid structure can also be considered.26,29,86,87 Molecular dynamics simulations consider the conformation of, and interactions between, all compounds in the membrane, water and solute, and can also be used in the study of the impact of conformation on DMLW.
Experimental measurements on phospholipophilicity may be preferred over descriptor calculated values for chemicals of environmental concern that require higher tier level risk assessment, particularly for ionizable chemicals. Many pharmaceuticals and illicit drugs are ionizable chemicals,88 as well as a considerable fraction of high production volume chemicals for which chemical fate and hazard assessment needs to be more detailed.89 Using the neutral form of an acid or base to calculate the phospholipophilicity of the anionic or cationic species, e.g. viaeqn (4) + (6), or (5) + (6), entails considerable uncertainty. Although various batch tests with pure artificial phospholipids are possible,79,90–92 no widely recognized testing protocols, such as OECD or ASTM, are available. Chromatographic methods, however, are already standardized in OECD guideline 117 to determine octanol–water distribution ratios93 and OECD 121 to screen for soil organic carbon sorption affinities. Retention on a commercially available HPLC-column with an immobilized artificial membrane (IAM) facilitates consistent measurements of large numbers of chemicals, with simple HPLC systems pumping aqueous eluent into various detectors (RI, ELSD, UV, FLU, and MS/MS).43–46 The experimentally feasible range covers log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW 0–6, depending on detection limits for the strongest sorbing chemicals.94–96 Although the silica IAM packing can have confounding coulombic electrostatic effects on the retention of anions and cations, as discussed in Section S1,† this can be corrected for by empirical or modeled Boltzmann factors.27,48,97 Consistent IAM-based DMLW data sets have been derived for both cationic (>150 chemicals26,77,78,98,99) and anionic chemicals (>20 (ref. 27 and 100)).
DMLW 0–6, depending on detection limits for the strongest sorbing chemicals.94–96 Although the silica IAM packing can have confounding coulombic electrostatic effects on the retention of anions and cations, as discussed in Section S1,† this can be corrected for by empirical or modeled Boltzmann factors.27,48,97 Consistent IAM-based DMLW data sets have been derived for both cationic (>150 chemicals26,77,78,98,99) and anionic chemicals (>20 (ref. 27 and 100)).
(v) Rationale for selecting toxicity data and chemicals to derive KIAM
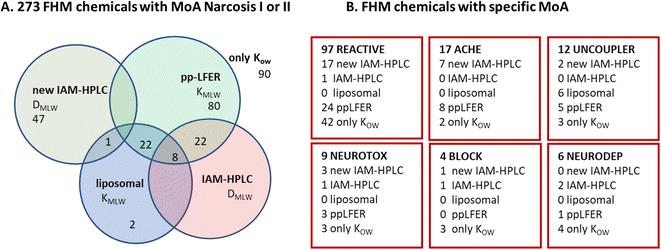
In order to evaluate whether the CMB-approach can adequately distinguish chemicals with different MoA based on KIAM values, the collection of toxicity data should be of high quality. Ideally, this experimental toxicity data are obtained using a standardized test protocol, measured exposure concentrations, and consistent handling of test procedures, and have minimum uncertainty margins due to differences between species of the same type, e.g. fish.The first MoA-evaluation approach we selected was to use a widely recognized consistent database of acute toxicity values on a single fish species, performed by a single institute with measured exposure concentrations. The United States Environmental Protection Agencies Fathead Minnow (FHM) database16 provides such MoA data for 616 organic chemicals. We selected 74 neutral chemicals for IAM-measurement: 47 narcotics, 27 with a specific MoA. One of the other criteria for selecting neutral chemicals was that we wanted to address the scarcity of physicochemical information for many chemicals in the FHM set, and selected only chemicals for which no other estimate of KMLW was available other than (estimates of) KOW, as discussed in the Methods section in more detail.
The second MoA-evaluation approach selected was to use chemicals that are designed to exert specific MoA on a certain type of species, while they are expected to have only baseline effects (non-specific MoA) for non-target species. For pesticides, standard toxicity tests on different representative species under strict testing protocols are mandatory in the regulatory process. Also, from the interest of (eco)toxicological assessment, typically pesticides have high quality toxicity data sets on diverse test organisms that allow for adequate comparisons. In this case, we performed IAM-measurements for 12 neutral herbicides as well as for a set of 17 strongly acidic herbicides, which under physiological pH exist for >99.9% as organic anions. For these herbicides, we used the CMBnarc approach to determine to what extent toxicity to ‘likely target’ organisms was more specific than toxicity to ‘non-target’ organisms. For example, algae are likely affected specifically by herbicides, whereas fish are hopefully only reacting non-specifically to herbicides at levels predicted by the CMBnarc approach.
2. Materials and methods
(i) Chemical selection
The chemical selection was primarily limited by the toxicity data set used for CMB evaluation, but furthermore based on the following criteria and goals: ease of purchase in adequate purity, ease of detection based on expected retention time and detector sensitivity, the goal of extending the kIAM database with new chemicals,94 the goal of including a substantial number of ionizable acids and bases for which also liposome based DMLW,ion data exist for comparison with kIAM, and the goal of covering a sufficient number of chemicals to compare narcosis I and II (set 1), to compare narcosis with several specific MoA (set 1), and herbicides (set 2) with a specific MoA expected for algae/plants but nonspecific for other aquatic organisms, and to extend the KIAM data for anionic chemicals (set 3). In total, this study determined new KIAM values for 121 chemicals.| Code | 47 narcosis FHM chemicals | CAS | FHM LC50 (mmol L−1) | FHM LC50 (mg L−1) | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW (EpiSuite) KOW (EpiSuite) |
New log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM KIAM |
Nr injections | Solvent range (%) | CMBIAM (mmol kg−1) |
|---|---|---|---|---|---|---|---|---|---|
a Coding of MoA of chemicals in the FHM database (set 1): N_I = Narcosis_I; N_II = Narcosis_II (polar narcosis); log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P as reported in the FHM database. P as reported in the FHM database.
|
|||||||||
| 1N_I-01 | 2-Hydroxyethyl ether | 111-46-6 | 709 | 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
−1.30 | −0.46 | 2 | 0 | 246 |
| 1N_I-02 | Triethylene glycol | 112-27-6 | 399 | 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
−1.24 | 0.01 | 1 | 0 | 408 |
| 1N_I-03 | 2-Methyl-2,4-pentanediol | 107-41-5 | 90.5 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
−0.67 | 0.75 | 1 | 0 | 509 |
| 1N_I-04 | Urethane | 51-79-6 | 58.8 | 5240 | −0.15 | 0.48 | 3 | 0 | 178 |
| 1N_I-05 | 3-Methyl-1-pentyn-3-ol | 77-75-8 | 12.4 | 1220 | 0.86 | 1.19 | 1 | 0 | 192 |
| 1N_I-06 | n-Phenyldiethanolamine | 120-07-0 | 4.06 | 735 | 0.44 | 1.84 | 2 | 0 | 281 |
| 1N_I-07 | 3-Methyl-3-pentanol | 77-74-7 | 6.58 | 672 | 1.53 | 1.43 | 5 | 0 | 177 |
| 1N_I-08 | 2,4,5-Trimethyloxazole | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662-84-4 662-84-4 |
4.04 | 449 | 1.79 | 1.78 | 3 | 0 | 243 |
| 1N_I-09 | cis-3-Hexen-1-ol | 928-96-1 | 3.80 | 381 | 1.34 | 1.58 | 2 | 0 | 144 |
| 1N_I-10 | 2-Methyl-3,3,4,4-tetrafluoro-2-butanol | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 553-26-2 553-26-2 |
3.64 | 582 | 1.03 | 1.53 | 1 | 0 | 123 |
| 1N_I-11 | trans-3-Hexen-1-ol | 928-97-2 | 2.71 | 271 | 1.34 | 1.62 | 2 | 0 | 113 |
| 1N_I-12 | 2-Phenoxyethanol | 122-99-6 | 2.49 | 344 | 1.16 | 1.70 | 5 | 0 | 125 |
| 1N_I-13 | 2,6-Dichlorobenzamide | 2008-58-4 | 2.47 | 469 | 1.25 | 1.51 | 3 | 0 | 80 |
| 1N_I-14 | 1-Ethynyl-cyclohexanol | 78-27-3 | 2.06 | 256 | 1.73 | 1.80 | 2 | 0 | 130 |
| 1N_I-15 | 3,4-Dimethyl-1-pentyn-3-ol | 1482-15-1 | 1.84 | 205 | 1.26 | 1.59 | 2 | 0 | 72 |
| 1N_I-16 | Diethyl benzylphosphonate | 1080-32-6 | 1.47 | 336 | 1.59 | 2.41 | 3 | 0 | 378 |
| 1N_I-17 | 2-(Bromomethyl)-tetrahydro-2h-pyran | 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 723-82-5 723-82-5 |
1.14 | 205 | 1.61 | 1.77 | 4 | 0 | 67 |
| 1N_I-18 | 2′,3′,4′-Trimethoxy-acetophenone | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 909-73-4 909-73-4 |
1.09 | 229 | 1.12 | 2.39 | 3 | 0 | 268 |
| 1N_I-19 | 2-Dimethylaminopyridine | 5683-33-0 | 1.04 | 127 | 1.43 | 1.96 | 3 | 0 | 95 |
| 1N_I-20 | 2-Amino-4-chloro-6-methylpyrimidine | 5600-21-5 | 1.02 | 147 | 1.13 | 1.76 | 3 | 0 | 59 |
| 1N_I-21 | 2-Phenyl-3-butyn-2-ol | 127-66-2 | 0.773 | 113 | 1.68 | 2.15 | 2 | 0 | 109 |
| 1N_I-22 | 2,5-Dimethylfuran | 625-86-5 | 0.740 | 71.1 | 2.62 | 2.05 | 4 | 0 | 83 |
| 1N_I-23 | 2,3-Dihydrobenzofuran | 496-16-2 | 0.680 | 81.7 | 2.14 | 2.18 | 4 | 0 | 113 |
| 1N_I-24 | 5-Ethyl-2-methylpyridine | 104-90-5 | 0.669 | 81.1 | 2.49 | 2.51 | 3 | 0 | 216 |
| 1N_I-25 | Benzyl-tert-butanol | 103-05-9 | 0.404 | 66.4 | 2.57 | 2.59 | 3 | 0 | 157 |
| 1N_I-26 | Benzyl sulfoxide | 621-08-9 | 0.348 | 80.1 | 1.96 | 2.73 | 3 | 0 | 187 |
| 1N_I-27 | 2-(n-Ethyl-m-toluidino)ethanol | 91-88-3 | 0.295 | 52.9 | 2.49 | 2.74 | 3 | 0 | 163 |
| 1N_I-28 | α,α,α-Trifluoro-o-tolunitrile | 447-60-9 | 0.247 | 42.2 | 2.46 | 2.54 | 3 | 0 | 86 |
| 1N_I-29 | α,α,α-4-Tetrafluoro-m-toluidine | 2357-47-3 | 0.168 | 30.1 | 2.62 | 2.64 | 4 | 0 | 73 |
| 1N_I-30 | α,α,α-4-Tetrafluoro-o-toluidine | 393-39-5 | 0.165 | 29.6 | 2.62 | 2.54 | 7 | 0 | 57 |
| 1N_I-31 | 4-Ethoxy-2-nitroaniline | 616-86-4 | 0.143 | 26.0 | 2.47 | 2.87 | 3 | 0 | 106 |
| 1N_I-32 | 4-(Diethylamino)-benzaldehyde | 120-21-8 | 0.135 | 23.9 | 2.94 | 3.10 | 6 | 0 | 170 |
| 1N_I-33 | 4-Phenylpyridine | 939-23-1 | 0.104 | 16.1 | 2.59 | 3.05 | 3 | 0 | 117 |
| 1N_I-34 | 1,1-Diphenyl-2-propyn-1-ol | 3923-52-2 | 0.053 | 11.1 | 2.71 | 3.34 | 3 | 0 | 116 |
| 1N_I-35 | Butyl phenyl ether | 1126-79-0 | 0.0384 | 5.8 | 3.65 | 3.64 | 7 | 15–30% | 168 |
| 1N_I-36 | 4-(Diethylamino)-salicylaldehyde | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 754-90-4 754-90-4 |
0.0277 | 5.4 | 3.34 | 3.35 | 2 | 0 | 62 |
| 1N_I-37 | Flavone | 525-82-6 | 0.0157 | 3.5 | 3.56 | 3.90 | 6 | 15–30% | 125 |
| 1N_I-38 | 2-Amino-4′-chloro-benzophenone | 2894-51-1 | 0.0092 | 2.1 | 3.95 | 4.21 | 6 | 15–30% | 149 |
| 1N_I-39 | Di-n-butylisophthalate | 3126-90-7 | 0.0032 | 0.9 | 5.53 | 4.67 | 9 | 20–30% | 150 |
| 1N_I-40 | 3-(4-tert-Butylphenoxy)-benzaldehyde | 79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 124-76-8 124-76-8 |
0.0015 | 0.4 | 5.93 | 5.41 | 4 | 20–30% | 386 |
| 1N_I-41 | 3-(3,4-Dichlorophenoxy)-benzaldehyde | 69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 770-23-6 770-23-6 |
0.0011 | 0.3 | 5.49 | 4.98 | 4 | 20–30% | 105 |
| Average | 160 | ||||||||
| St. dev. | 105 | ||||||||
| 1N_II-01 | 6-Chloro-2-pyridinol | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 879-02-0 879-02-0 |
1.65 | 214.0 | 1.78 | 1.34 | 3 | 0 | 36 |
| 1N_II-02 | 2-Amino-5-bromopyridine | 1072-97-5 | 1.02 | 177.0 | 1.39 | 2.12 | 3 | 0 | 134 |
| 1N_II-03 | 2-Chloro-4-methylaniline | 615-65-6 | 0.253 | 35.9 | 2.58 | 2.55 | 5 | 0 | 90 |
| 1N_II-04 | 4-Amino-2-nitrophenol | 119-34-6 | 0.235 | 36.2 | 0.96 | 1.74 | 6 | 0 | 13 |
| 1N_II-05 | 2-Chloro-4-nitroaniline | 121-87-9 | 0.109 | 18.9 | 2.17 | 2.84 | 4 | 0 | 75 |
| 1N_II-06 | 3-Trifluoromethyl-4-nitrophenol | 88-30-2 | 0.0441 | 9.1 | 3.00 | 3.28 | 3 | 0 | 84 |
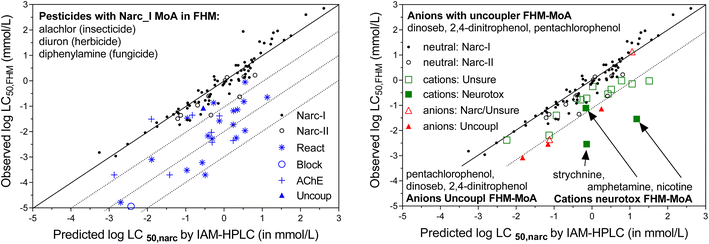
As shown in Fig. 2B, we also selected a set of 27 neutral FHM chemicals that were categorized by MoA, 17 with electrophile/pro-electrophile reactivity (‘REACTIVE’), 7 acetylcholinesterase inhibitors (‘ACHE’), 1 uncoupler of oxidative phosphorylation, and 1 that acts as a respiratory blocker/inhibitor (‘BLOCK’). The selection was aimed at minimizing overlap with the existing IAM-HPLC/liposomal database (see further details in ESI section S2, Table S1B†). These specific MoA FHM-chemicals are listed in Table 2, sorted per MoA by decreasing reported FHM LC50 in mmol L−1.
| Codea | 27 specific MoA FHM chemicals | CAS | FHM LC50 (mmol L−1) | LC50 (mg L−1) | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW (Epi-Suite) KOW (Epi-Suite) |
New log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM KIAM |
Nr inj. | Solvent range (%) | CMBIAM (mmol kg−1) |
|---|---|---|---|---|---|---|---|---|---|
| a Coding of MoA of chemicals in the FHM database (set 1): Ach = acetylcholinesterase inhibitors; Rea = electrophile/proelectrophile reactivity; Unc = uncouplers of oxidative phosphorylation (AUnc = acidic uncoupler); Blo = respiratory blocker/inhibitor; 1CNeu = cationic neurotoxin; CunsA = cationic unsure amine mode of action; AN = acidic narcosis chemical. b δ IAM-SSLM corrected. NA = not available. δIAM-SSLM corrections for 1°, 2°, and 3° amines: +0.78, +0.47, and −0.03, respectively, as in ref. 77. | |||||||||
| 1Ach-01 | Carbaryl | 63-25-2 | 0.0444 | 8.93 | 2.46 | 2.98 | 3 | 0 | 42.40 |
| 1Ach-02 | Propoxur | 114-26-1 | 0.0421 | 8.80 | 1.45 | 2.27 | 3 | 0 | 7.84 |
| 1Ach-03 | Diazinon | 333-41-5 | 0.0307 | 9.35 | 4.19 | 4.05 | 7 | 15–30% | 344.46 |
| 1Ach-04 | Aminocarb | 2032-59-9 | 0.0094 | 1.95 | 2.16 | 2.48 | 4 | 0 | 2.84 |
| 1Ach-05 | Aldicarb | 116-06-3 | 0.0045 | 0.86 | 1.12 | 1.91 | 4 | 0 | 0.37 |
| 1Ach-06 | Carbofuran | 1563-66-2 | 0.0038 | 0.84 | 2.32 | 2.46 | 4 | 0 | 1.10 |
| 1Ach-07 | EPN | 2104-64-5 | 0.0002 | 0.08 | 3.85 | 5.10 | 7 | 20–30% | 25.18 |
| 1Ach-08 | Azinphos-methyl | 86-50-0 | 0.0002 | 0.06 | 2.75 | 3.57 | 7 | 15–30% | 0.74 |
| 1Rea-01 | Butanal | 123-72-8 | 0.2219 | 16.00 | 0.88 | 1.02 | 1 | 0 | 2.32 |
| 1Rea-02 | 4-Fluoroaniline | 371-40-4 | 0.1521 | 16.90 | 1.15 | 1.63 | 3 | 0 | 6.49 |
| 1Rea-03 | 2,4-Dinitrotoluene | 121-14-2 | 0.1334 | 24.30 | 2.00 | 2.42 | 3 | 0 | 35.09 |
| 1Rea-04 | 4-Nitrobenzaldehyde | 555-16-8 | 0.0668 | 10.10 | 1.50 | 1.90 | 3 | 0 | 5.31 |
| 1Rea-06 | Benzaldehyde | 100-52-7 | 0.0717 | 7.61 | 1.48 | 1.80 | 3 | 0 | 4.52 |
| 1Rea-05 | 2,4,5-Tribromoimidazole | 2034-22-2 | 0.0261 | 7.96 | 1.96 | 2.27 | 3 | 0 | 4.86 |
| 1Rea-07 | 4-Dimethylamino-cinnamaldehyde | 6203-18-5 | 0.0337 | 5.90 | NA | 3.11 | 3 | 0 | 43.41 |
| 1Rea-08 | Salicylaldehyde | 90-02-8 | 0.0188 | 2.30 | 1.81 | 1.91 | 3 | 0 | 1.53 |
| 1Rea-09 | 4-Chlorocatechol | 2138-22-9 | 0.0109 | 1.58 | 1.97 | 1.60 | 1 | 0 | 0.43 |
| 1Rea-11 | 1-Benzoylacetone | 93-91-4 | 0.0068 | 1.10 | 1.05 | 2.57 | 3 | 0 | 2.53 |
| 1Rea-10 | Pentafluorobenzaldehyde | 653-37-2 | 0.0056 | 1.10 | 2.45 | 1.83 | 3 | 0 | 0.38 |
| 1Rea-12 | α,α,α-Trifluoro-m-tolualdehyde | 454-89-7 | 0.0053 | 0.92 | 2.47 | 2.46 | 3 | 0 | 1.53 |
| 1Rea-13 | 1,3,5-Trichloro-2,4-dinitrobenzene | 6284-83-9 | 0.0008 | 0.22 | 2.65 | 4.04 | 9 | 20–30% | 8.77 |
| 1Rea-14 | 2-Methyl-1,4-naphthoquinone | 58-27-5 | 0.0006 | 0.11 | 2.20 | 2.72 | 3 | 0 | 0.31 |
| 1Rea-15 | α-Bromo-2′,5′-dimethoxyacetophenone | 1204-21-3 | 0.0003 | 0.09 | 2.39 | 3.12 | 3 | 0 | 0.40 |
| 1Rea-16 | 1,3-Dichloro-4,6-dinitrobenzene | 3698-83-7 | 0.0002 | 0.05 | 2.49 | 2.64 | 4 | 0 | 0.09 |
| 1Rea-17 | N-Vinylcarbazole | 1484-13-5 | 0.0000166 | 0.0032 | NA | 4.85 | 7 | 20–30% | 1.18 |
| 1Blo-01 | Rotenone | 83-79-4 | 0.0000114 | 0.0045 | 4.10 | 4.58 | 6 | 20–30% | 0.43 |
| 5 Mostly anionic FHM chemicals | FHM LC50 (mmol L−1) | LC50 (mg L−1) | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW KOW |
New log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM KIAM |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAMb KIAMb |
CMBIAM (mmol kg−2) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
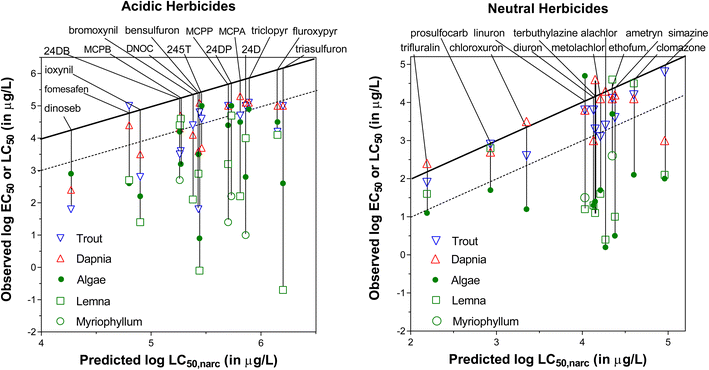
| 1AUns-01 | Salicylic acid | 54-21-7 | 13.5 | 2160 | 2.26 | 1.49 | 0.99 | 1 | 0 | 131.9 |
| 1AUnc-01 | 2,4-Dinitrophenol | 51-28-5 | 0.072 | 13.3 | 1.67 | 2.32 | 1.82 | 2 | 0 | 4.8 |
| 1AUnc-02 | Dinoseb | 88-85-7 | 0.0029 | 0.70 | 4.62 | 3.76 | 3.26 | 2 | 0 | 5.3 |
| 1AUnc-03 | Pentachlorophenol | 87-86-5 | 0.00083 | 0.22 | 5.12 | 4.45 | 3.95 | 7 | 15–30% | 7.4 |
| 1AN-01 | 2,3,4,6-Tetrachlorophenol | 58-90-2 | 0.0044 | 1.03 | 4.12 | 3.74 | 3.29 | 8 | 15–30% | 7.6 |
| 15 Mostly cationic FHM chemicals | FHM LC50 (mmol L−1) | LC50 (mg L−1) | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW KOW |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAMb (ref. 77) KIAMb (ref. 77) |
CMBIAM (mmol kg−2) | ||
|---|---|---|---|---|---|---|---|
| 1CNeu-01 | Amphetamine sulfate | 60-13-9 | 0.078 | 28.80 | 1.76 | 2.31 | 15.9 |
| 1CNeu-02 | Nicotine sulfate | 65-30-5 | 0.029 | 12.20 | 1.17 | 0.96 | 0.3 |
| 1CNeu-03 | Strychnine hemisulphate | 60-41-3 | 0.0029 | 1.11 | 1.93 | 2.28 | 0.6 |
| 1CUnsA-01 | Phenyltrimethylammonium methosulfate | 98-04-4 | 0.924 | 243 | NA | 0.64 | 4.0 |
| 1CUnsA-02 | Benzyltriethylammonium chloride | 56-37-1 | 0.952 | 161 | NA | 1.37 | 22.3 |
| 1CUnsA-03 | N,N-Dimethylbenzylamine | 103-83-3 | 0.707 | 37.8 | 1.95 | 1.08 | 8.5 |
| 1CUnsA-04 | Hexylamine | 111-26-2 | 0.559 | 56.6 | 2.06 | 2.12 | 73.7 |
| 1CUnsA-05 | N-Ethylbenzylamine | 14321-27-8 | 0.422 | 57.1 | 2.04 | 1.64 | 18.4 |
| 1CUnsA-06 | Benzylamine | 100-46-9 | 0.280 | 102 | 1.09 | 1.73 | 15.0 |
| 1CUnsA-07 | N-Heptylamine | 111-68-2 | 0.190 | 21.8 | 2.57 | 2.54 | 65.9 |
| 1CUnsA-08 | tert-Octylamine | 107-45-9 | 0.189 | 24.6 | 2.43 | 2.29 | 36.9 |
| 1CUnsA-09 | 1-Adamantanamine | 768-94-5 | 0.165 | 25.0 | 2.44 | 2.44 | 45.4 |
| 1CUnsA-10 | Octylamine | 111-86-4 | 0.040 | 5.19 | 3.04 | 3.08 | 48.1 |
| 1CUnsA-11 | Di-n-hexylamine | 143-16-8 | 0.0065 | 0.78 | 4.77 | 3.28 | 12.4 |
| 1CUnsA-12 | N-Decylamine | 2016-57-1 | 0.0042 | 1.03 | 4.10 | 4.40 | 105.5 |
A substantial number of chemicals in the FHM database are ionizable chemicals: forty nine are bases that are >95% ionized at tested pH (pKa > 8.5) and nineteen are acids with pKa < 6 (>95% ionized at physiological pH 7.4). MoA classification of most of the ionizable chemicals is equivocal. For chemical set 1, we selected fifteen strong bases/cations listed in the FHM database for which KIAM values were already available, i.e. three neurotoxic bases (amphetamine, strychnine, and nicotine)78 and twelve bases/cations of the type “UNSURE AMINES”.77 The five strongly dissociated acids selected from the FHM database for chemical set 1 had different MoA labels, including three phosphorylation uncouplers. The fifteen selected cations were assumed to be a relevant set of chemicals to evaluate the MoA approach based on the CMB for cations, with expectations that the “UNSURE AMINES” would be considered non-specific (narcosis) chemicals, as they lack specific functional moieties other than the charged amine group, while the three neurotoxic cations are expected to have a CMB significantly below the narcosis level. The set of anions was considered too small to evaluate the MoA approach based on the CMB, which is another reason we included the strongly acidic herbicides for chemical set 2 as part of this study. ESI Section S2† provides more details on the chemical selection procedure, and ESI Table S2† presents a list of the purity and suppliers of the chemicals purchased to create Chemical set 1.
| Code | Name (FHM set 1 code, LC50 in mg L−1, MoA) | CAS | Herbicide MoA | pKa | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW neutral ACD labs KOW neutral ACD labs |
New log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM (anion KIAM,intr) KIAM (anion KIAM,intr) |
New log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAMδIAM-SSLM corrected KIAMδIAM-SSLM corrected |
Nr inj. | Solvent range (%) | Toxicity endpoint source |
|---|---|---|---|---|---|---|---|---|---|---|
| a Coding of MoA of chemicals in Set 2: A = acid; N = neutral; 1 Lemna endpoint (or Myriophyllum endpoint between brackets) as listed in Table 5; 2 alg = EC50Raphidocelis subcapitata if no data on aquatic plants were retrieved; 3Oncorhynchys mykiss (rainbow trout) as listed in Table 5. 4 7d ErC50 at pH 7.8, reported as 0.000202 mg L−1 (nom) for the Frond number of Lemna gibba in EFSA J. 2015; 13(1):3958.5 14d ErC50, reported as 0.0008 mg L−1 for the Frond number of Lemna gibba in EFSA Scientific Report 2008; 178, 1. | ||||||||||
| Anionic herbicides | ||||||||||
| 2A-01 | Clopyralid | 1702-17-6 | Synth. auxin | 2.02 | 1.31 | 0.64 | 0.14 | 1 | 0 | EFSA scientific report, 2005, 50, 1–65 |
| 2A-02 | Fluroxypyr | 69377-81-7 | Synth. auxin | 2.22 | 3.16 | 1.90 | 1.40 | 3 | 0 | EFSA J., 2011, 9(3), 2091 |
| 2A-03 | Triclopyr | 55335-06-3 | Synth. auxin | 2.26 | 2.98 | 2.16 | 1.66 | 3 | 0 | EFSA scientific report, 2005, 56, 1–103 |
| 2A-04 | MCPA | 94-74-6 | Synth. auxin | 3.14 | 2.49 | 2.14 | 1.64 | 6 | 0 | EC (2008), Review report for the active substance MCPA (SANCO/4062/2001-final; pp. 1–62) |
| 2A-05 | 2,4-D | 94-75-7 | Synth. auxin | 2.6 | 2.59 | 2.13 | 1.63 | 3 | 0 | EFSA J. 2014; 12(9), 3812 |
| 2A-06 | MCPP | 93-65-2 | Synth. auxin | 3.19 | 2.84 | 2.28 | 1.78 | 6 | 0 | EFSA J. 2017; 15(5), 4832 |
| 2A-07 | 2,4-DP | 120-36-5 | Synth. auxin | 3.1 | 3.43 | 2.29 | 1.79 | 3 | 0 | EFSA J. 2018; 16(6), 5288 |
| 2A-08 | MCPB | 94-81-5 | Synth. auxin | 4.84 | 3.42 | 2.74 | 2.24 | 3 | 0 | EC (2005). Review report for the active substance MCPB (No. SANCO/4063/2001-final; pp. 1–42) |
| 2A-09 | 2,4-DB | 94-82-6 | Synth. auxin | 4.95 | 3.53 | 2.78 | 2.28 | 3 | 0 | EFSA J. 2016; 14(5), 4500 |
| 2A-10 | 2,4,5-T | 93-76-5 | Synth. auxin | 2.88 | 3.3 | 2.59 | 2.09 | 3 | 0 | US EPA ECOTOX/PPDB |
| 2A-11 | DNOC | 534-52-1 | Uncoupler | 4.31 | 2.13 | 2.65 | 2.15 | 3 | 0 | US EPA ECOTOX (EC (1998). Review report for the active substance DNOC (no. 7777/VI/98-rev. 3; pp. 1–3)) |
| 2A-12 | Dinoseb | 88-85-7 | Uncoupler | 4.62 | 3.60 | 3.76 | 3.26 | 2 | 0 | REACH registration dossier https://echa.europa.eu/nl/registration-dossier/-/registered-dossier/12446 |
| 2A-13 | Bromoxynil | 1689-84-5 | PS-II inh. | 4.09 | 2.95 | 2.70 | 2.20 | 2 | 0 | EFSA J., 2017; 15(6), 4790 |
| 2A-14 | Ioxynil | 1689-83-4 | PS-II inh. | 3.96 | 3.43 | 3.31 | 2.81 | 2 | 0 | EC (2004). Review report for the active substance ioxynil (no. SANCO/4349/2000 final; pp. 1–98) |
| 2A-15 | Triasulfuron | 82097-50-5 | AHAS inh. | 4.34 | 2.36 | 2.05 | 1.55 | 3 | 0 | EFSA J., 2015; 13(1), 3958 |
| 2A-16 | Bensulfuron-methyl | 83055-99-6 | AHAS inh. | 3.50 | 2.38 | 2.82 | 2.32 | 6 | 10–20% | EFSA scientific report, 2008, 178, 1–102 |
| 2A-17 | Fomesafen | 72178-02-0 | PROTOX | 2.42 | 2.94 | 3.48 | 2.98 | 6 | 0–20% | Australian pesticides and veterinary medicines authority (APVMA): public release summary on fomesafen in the product reflex herbicide, ISSN 1443–1335 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||||
| Neutral herbicides | ||||||||||
| 2N-01 | Alachlor | 15972-60-8 | Cell div. inh. (VLCFA) | 3.59 | 3.31 | 10 | 0–30% | EC (2007). Review report for the active substance alachlor (SANCO/4331/2000-final)//US EPA ECOTOX | ||
| 2N-02 | Metolachlor | 1418095-19-8 | Cell div. inh. (VLCFA) | 3.45 | 3.39 | 8 | 0–30% | Public consultation_S-Metolachlor_RAR_23_LoEP_ 2018-09-06.pdf | ||
| 2N-03 | Simazine | 122-34-9 | PS-II inh. | 1.78 | 2.49 | 3 | 0 | US EPA ECOTOX (EC (2003)), Review report for the active substance simazine SANCO/10495/2003-rev. Final) | ||
| 2N-04 | Ametryn | 834-12-8 | PS-II inh. | 2.60 | 3.14 | 3 | 0 | US EPA ECOTOX/REACH registration dossier https://echa.europa.eu/nl/registration-dossier/-/registered-dossier/2171/6/2/1 | ||
| 2N-05 | Terbuthylazin | 5915-41-3 | PS-II inh. | 2.48 | 3.36 | 3 | 0 | EFSA J. 2011; 9(1):1969/US EPA ECOTOX | ||
| 2N-06 | Diuron | 330-54-1 | PS-II inh. | 2.53 | 3.38 | 3 | 0 | EFSA scientific report, 2005, 25, 1–58 | ||
| 2N-07 | Linuron | 330-55-2 | PS-II inh. | 2.30 | 3.51 | 3 | 0 | EFSA J., 2016; 14(7), 4518 | ||
| 2N-08 | Chloroxuron | 1982-47-4 | PS-II inh. | 3.43 | 4.26 | 6 | 15–30% | US EPA ECOTOX | ||
| 2N-09 | Ethofumesate | 26225-79-6 | Lipid synth inh. | 2.34 | 3.25 | 3 | 0 | EFSA J., 2016, 14(1), 4374 | ||
| 2N-10 | Prosulfocarb | 52888-80-9 | Lipid synth inh. | 4.17 | 4.62 | 7 | 15–30% | EFSA scientific report, 2007, 111, 1–81 | ||
| 2N-11 | Clomazone | 81777-89-1 | Lycopene cyclase inh. | 2.93 | 2.93 | 14 | 0–30% | EFSA scientific report, 2007, 109, 1–73 | ||
| 2N-12 | Trifluralin | 1582-09-8 | Mitosis inh. | 4.60 | 5.48 | 4 | 20–30% | EFSA scientific report, 2009, 327, 1–111 | ||
| Code | Name (code in Set 1; FHM LC50 in mg L−1, MoA) | CAS | pKa | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW KOW |
New log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM KIAM |
Nr inj. | Solvent range (%) | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW,ion DMLW,ion |
δ IAM-liposome | New log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM KIAM |
|---|---|---|---|---|---|---|---|---|---|---|
| Anion KIAM,intr | Liposome | Log units | δ IAM-SSLM corrected | |||||||
| a Coding of MoA of chemicals in set 3: Ca = carboxylic acid; Su = sulfonate acid; Ph = phenolic acid. b Considered outlier (>2 time st. dev.), not included in the calculation of average δIAM-liposome. | ||||||||||
| 3Ca-01 | Salicylic acid (1AUns-01; 2160, UNSURE) | 54-21-7 | 2.75 | 2.26 | 1.49 | 1 | 0 | 1.03 | 0.46 | 0.99 |
| 3Ca-02 | 5-Phenylvaleric acid | 2270-20-4 | 4.88 | 2.70 | 1.97 | 1 | 0 | 1.66 | 0.31 | 1.47 |
| 3Ca-03 | Ibuprofen | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 687-27-1 687-27-1 |
4.45 | 3.50 | 2.78 | 1 | 0 | 1.81 | 0.97 | 2.28 |
| 3Ca-04 | Fenamic acid | 91-40-7 | 3.99 | 4.36 | 3.43 | 3 | 0 | 2.28 | 1.15 | 2.93 |
| 3Ca-05 | Diclofenac | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 307-86-5 307-86-5 |
3.99 | 4.5 | 3.76 | 6 | 0 | 2.64 | 1.12 | 3.26 |
| 3Ca-06 | Diflunisal | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 494-42-4 494-42-4 |
3.00 | 4.44 | 3.73 | 6 | 0 | 2.73 | 1.00 | 3.23 |
| 3Su-01 | 4-Octylbenzene-1-sulfonate | 6149-03-7 | <0 | 4.65 ACD | 4.81 | 2 | 0 | 3.63 | 1.11 | 4.31 |
| 3Su-02 | C10-2-LAS | NA | <0 | 5.53 ACD | 5.58 | 8 | 15–30% | 4.79 | 0.79 | 5.11 |
| 3Ph-01 | Warfarin | 81-81-2 | 4.9 | 2.7 | 3.06 | 5 | 20–30% | 1.40 | 1.66 | 2.56 |
| 3Ph-02 | Pentafluorophenol | 771-61-9 | 5.53 | 3.05 ACD | 2.20 | 1 | 0 | 1.74 | 0.46 | 1.70 |
| 3Ph-03 | 2,4-Dinitrophenol (not in set 1; 13.3, UNCOUPLER_1) | 51-28-5 | 3.94 | 1.67 | 2.36 | 1 | 0 | 1.90 | 0.46 | 1.86 |
| 3Ph-04 | Bromoxynil | 1689-84-5 | 4.09 | 2.8 | 2.70 | 2 | 0 | 2.10 | 0.78 | 2.20 |
| 3Ph-05 | 2-Methyl-4,6-dinitrophenol | 534-52-1 | 4.31 | 2.20 ACD | 2.77 | 2 | 0 | 2.35 | 0.46 | 2.27 |
| 3Ph-06 | 4-tert-Butyl-2,6-dinitrophenol | 4097-49-8 | 4.11 | 3.56 ACD | 3.38 | 1 | 0 | 3.23 | 0.18 | 2.88 |
| 3Ph-07 | Dinoseb (1AUnc-01; 0.7, UNCOUPLER_3) | 88-85-7 | 4.62 | 3.56 | 3.76 | 1 | 0 | 3.35 | 0.43 | 3.26 |
| 3Ph-08 | 2,3,4,6-Tetrachlorophenol (1AN-01; 1.03, NARCOSIS_I_3) | 58-90-2 | 5.40 | 4.12 | 3.79 | 2 | 0 | 3.46 | 0.28 | 3.29 |
| 3Ph-09 | 2,3,5,6-Tetrachlorophenol | 935-95-5 | 5.14 | 3.88 | 3.78 | 8 | 15–30% | 3.49 | 0.28 | 3.28 |
| 3Ph-10 | Pentachlorophenol (1AUnc-02; 0.22, UNCOUPLER_1) | 87-86-5 | 4.75 | 5.12 | 4.45 | 8 | 15–30% | 4.35 | 0.10 | 3.95 |
| Average | 0.60 | |||||||||
| St. dev. | 0.36 | |||||||||
(ii) Chromatographic conditions
A 100 × 4.6 mm IAM.PC.DD2 column (Regis Technologies, Inc., Morton Grove, IL USA), with an IAM.PC.DD2 10/300 guard cartridge, was operated with typical phosphate buffer saline (PBS, pH 7.4) as the eluent, at a flow rate of 1.0 mL min−1 (23 ± 2 °C). Eluting chemicals were detected by using a UV-diode array (Agilent 1100 system) at various wavelengths (207, 220, 254, and 278 nm) relative to the signal at 360 nm. This way, in all cases there was at least one wavelength that clearly detected the eluting peak of all of the chemicals tested on the IAM-HPLC system with a single detector setting. The PBS was composed of 10 mM buffer with 8.0 g L−1 NaCl (137 mM) and 0.2 g L−1 KCl (2.7 mM). Triplicate IAM-injections (20 μL) were run on the same day for most chemicals if fully aqueous eluent was used. 3-Nitroaniline was used as the neutral reference chemical to check the IAM column performance throughout the testing period. The negative peak apex signal of injected pure water (MilliQ, Millipore Merck) was used as a neutral non-retained tracer (t0) in UV-diode array detection. The peak apex of the eluted peak (tr) on both detectors was used to calculate retention capacity factors (kIAM) based on the ratio tr/(tr − t0). The intrinsic phospholipid sorption coefficient (KIAM,intr) was obtained by (i) multiplying kIAM by the solvent/sorbent phase ratio of 18.9 for the IAM.PC.DD2 column, and (ii) accounting for electrostatic repulsion by IAM surfaces at a certain pH with Boltzmann factors.27A series of at least 3 different eluent mixtures with acetonitrile (≤30%) were applied to chemicals with a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow >3 as a first indication (Fig. S2†). At least 3 measurements were performed on 3 different solvent mixtures for these chemicals. Linear trends between log
Kow >3 as a first indication (Fig. S2†). At least 3 measurements were performed on 3 different solvent mixtures for these chemicals. Linear trends between log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM and fraction solvent in the eluent mixtures were extrapolated to estimate log
KIAM and fraction solvent in the eluent mixtures were extrapolated to estimate log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM values with a fully aqueous eluent composition in MS Excel.
KIAM values with a fully aqueous eluent composition in MS Excel.
3. Results and discussion
The key purpose of this study was to evaluate the MoA-classification based on critical membrane burdens for chemical set 1 (fish toxicity with different MoA, using the FHM database) and chemical set 2 (chemicals with a herbicidal MoA, but evaluated for different species). The chromatographic method IAM-HPLC is used to obtain membrane-lipid/water distribution coefficients for the more than 100 chemicals selected (details on all IAM measurements in Table S4†).For 28 chemicals in set 1,2, or 3, where IAM-HPLC retention capacity was measured for a series of (water/acetonitrile) compositions, ESI-fig. S2† shows the extrapolated linear trendlines to derive the KIAM,intr at 100% water. For seven chemicals (2A-16, 2A-17, 2N-01, 2N-02, 2N-11, 3Ca-05, and 3Ca-06), a solvent range was determined as well as measurements at 0% solvent, confirming the linearity of the trendline in this solvent range. For most chemicals with a solvent range extrapolation, the 95% confidence limit for KIAM,intr at 0% solvent was <0.2 log units (details in Fig. S2†), particularly if multiple measurements were made per solvent composition. In some cases, with single measurements per solvent composition (3Ca-04 fenamic acid), or one deviating point (3Ca-05 diclofenac), and with a limited range of 20–30% solvent (1Ach-07 EPN), the 95% confidence limits around the extrapolated 100% aqueous medium KIAM,intr are actually too high to derive a reliable DMLW value for further interpretation. For all other chemicals measured in 100% aqueous medium, replicate IAM measurements demonstrate high consistency (<0.1 log unit deviations for KIAM,intr), and as such also single KIAM measurements for 15 chemicals in 100% aqueous buffer are considered sufficiently reliable to derive the DMLW value.
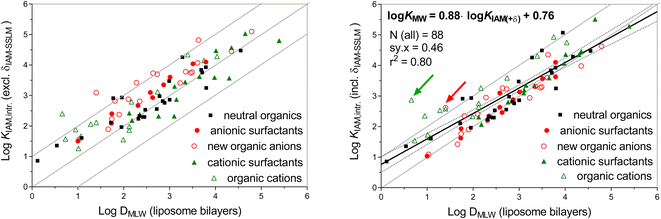
Since both set 1 and set 2 contain largely dissociated acids, for which the chromatographic method is used to determine the DMLW, the intrinsic KIAM (accounting for electrostatic repulsion from the IAM surface at neutral pH) obtained for the acids of chemical set 3 will be presented and discussed first. Whilst the alignment between liposomal DMLW and KIAM,intr values has been presented in other studies for neutral organics25 and organic cations,26,77,78 the current study provides data for a substantial set of organic anions in addition to the anionic surfactants27 that are already presented in Fig. 1. This collection should demonstrate the uncertainty margins with which IAM-HPLC can be used to derive DMLW for a wide chemical domain that includes both neutral and ionizable organic chemicals.
(i) Assessment of the alignment between IAM-retention factors and liposomal DMLW (chemical set 3)
Table 4 shows the IAM-HPLC results for the strong acids, with the number of injections included to derive KIAM,intr, and the solvent (acetonitrile) range applied. Nearly all acids were at least 99% dissociated at the tested pH 7 because of the very low pKa values (>3 units lower than the tested pH). The observed retention time is thus due to the interaction of the anionic form with the IAM surface. In Table 4, the IAM retention capacity factor (kIAM) is already converted to the IAM phospholipid–water partition coefficient (KIAM) using eqn (1). The logarithmic Boltzmann factor (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B) for pH 7.0 and eluent salinity of 0.1 M (log
B) for pH 7.0 and eluent salinity of 0.1 M (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BpH 7/0.1 M) is 0.5 (see ESI-Fig. S1†). The left plot in Fig. 3 shows the same surfactant data as shown in the left plot of Fig. 1, but now also includes the newly derived KIAM,intr data for organic anions (open red circles). In addition, data for organic cations other than the cationic surfactants in Fig. 1 are now shown (open green triangles), using the consistent KIAM,intr data set for organic cations78 and liposomal KMLW data collected elsewhere (see details in ESI-Table S3†).29,76 These data sets present the widest chemical space of the KIAM–KMLW comparison that is currently available.
BpH 7/0.1 M) is 0.5 (see ESI-Fig. S1†). The left plot in Fig. 3 shows the same surfactant data as shown in the left plot of Fig. 1, but now also includes the newly derived KIAM,intr data for organic anions (open red circles). In addition, data for organic cations other than the cationic surfactants in Fig. 1 are now shown (open green triangles), using the consistent KIAM,intr data set for organic cations78 and liposomal KMLW data collected elsewhere (see details in ESI-Table S3†).29,76 These data sets present the widest chemical space of the KIAM–KMLW comparison that is currently available.
The data for non-surfactant organic ions demonstrate more scatter than the surfactants. Obviously, surfactants have very simple hydrocarbon or fluorocarbon structures, and don't account for the influence of polar groups on the interaction difference between the IAM monolayer and bilayer liposomes. The empirical incremental δIAM-MLW correction of −0.47 log units derived from the different anionic surfactants was also applied to all corresponding types of organic anions. As shown in the right plot of Fig. 3, for all anions this indeed brings nearly all KIAM,intr values for anions closer to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 line with liposomal KMLW,anion values. Warfarin (3Ph-01, indicated by the red arrow in Fig. 4) is the organic anion with the highest deviating KIAM,intr (δIAM-MLW adjusted KIAM,intr still 1.2 log units above liposomal KMLW,anion. Warfarin also showed a higher KIAM compared to liposomal KMLW (pH 7.4) in another study, although the slightly different IAM.PC.DD column was used.101 It is not clear what features of warfarin are responsible for this deviation, although it has a very delocalised charge in comparison to the other carboxylic acids and phenolic acids in the selection. Leaving out warfarin as an atypical outlier, the average difference between KIAM,intr and KMLW,anion for 27 anionic compounds is 0.54 log units, so the final δIAM-MLW remains at −0.5. The root mean square error (RMSE) for all 27 δIAM-MLW adjusted anion log
1 line with liposomal KMLW,anion values. Warfarin (3Ph-01, indicated by the red arrow in Fig. 4) is the organic anion with the highest deviating KIAM,intr (δIAM-MLW adjusted KIAM,intr still 1.2 log units above liposomal KMLW,anion. Warfarin also showed a higher KIAM compared to liposomal KMLW (pH 7.4) in another study, although the slightly different IAM.PC.DD column was used.101 It is not clear what features of warfarin are responsible for this deviation, although it has a very delocalised charge in comparison to the other carboxylic acids and phenolic acids in the selection. Leaving out warfarin as an atypical outlier, the average difference between KIAM,intr and KMLW,anion for 27 anionic compounds is 0.54 log units, so the final δIAM-MLW remains at −0.5. The root mean square error (RMSE) for all 27 δIAM-MLW adjusted anion log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM,intr values compared to log
KIAM,intr values compared to log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KMLW,anion is 0.38. This indicates that there is about a factor of ±3 uncertainty when extrapolating IAM-HPLC measurements for anions (incl. δIAM-MLW) to liposomal DMLW.
KMLW,anion is 0.38. This indicates that there is about a factor of ±3 uncertainty when extrapolating IAM-HPLC measurements for anions (incl. δIAM-MLW) to liposomal DMLW.
For cations, δIAM-MLW was defined for various types of simple amine structures in another study.26 Surprisingly, the diverse set of organic cations does not seem to converge to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 line with the δIAM-MLW increments set by the cationic surfactants (Fig. 3B). Several adjusted KIAM,intr values even deviate by more than a log unit from DMLW data, and not one cation has an adjusted KIAM partition coefficient lower than the KMLW values. The most outlying cation is acebutolol (indicated by the green arrow in Fig. 4, log
1 line with the δIAM-MLW increments set by the cationic surfactants (Fig. 3B). Several adjusted KIAM,intr values even deviate by more than a log unit from DMLW data, and not one cation has an adjusted KIAM partition coefficient lower than the KMLW values. The most outlying cation is acebutolol (indicated by the green arrow in Fig. 4, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KMLW,ion 0.66, log
KMLW,ion 0.66, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM,intr 2.4, and log
KIAM,intr 2.4, and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM,intr + δIAM-MLW 2.9). For the majority of chemicals in the right plot of Fig. 4, the δIAM-MLW corrected KIAM,intr values are within a factor of 0.7–10 of the liposomal DMLW data, with a tendency to particularly overestimate DMLW for lower affinity chemicals. Using all data on neutral, anionic and cationic chemicals, the overall double-log linear trendline shows a standard deviation of the residuals (sy·x, the square root of the average squared deviation) of 0.46 log units:
KIAM,intr + δIAM-MLW 2.9). For the majority of chemicals in the right plot of Fig. 4, the δIAM-MLW corrected KIAM,intr values are within a factor of 0.7–10 of the liposomal DMLW data, with a tendency to particularly overestimate DMLW for lower affinity chemicals. Using all data on neutral, anionic and cationic chemicals, the overall double-log linear trendline shows a standard deviation of the residuals (sy·x, the square root of the average squared deviation) of 0.46 log units:
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW = 0.88 × (KIAM,intr + δIAM-MLW) + 0.76 DMLW = 0.88 × (KIAM,intr + δIAM-MLW) + 0.76 | (10) |
Eqn (10) may be used to further minimize the error margins between (KIAM,intr + δIAM-MLW) and DMLW for the wide variety of neutral and ionizable chemicals. However, for the current evaluation we only used δIAM-MLW corrective increments for anionic and cationic surfactant DMLW determination, no further corrections were applied for the KIAM of neutral chemicals.
(ii) Evaluation of the CMBnarc-approach based on chemicals with a narcosis MoA from the FHM database (chemical set 1)
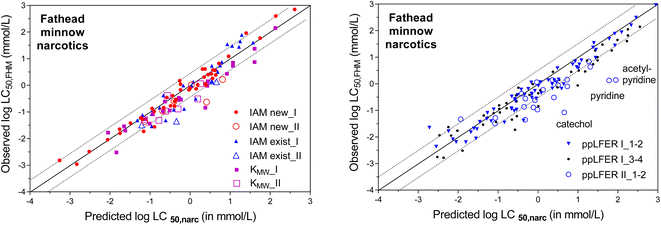
Neutral chemicals with a narcosis MoA are the easiest data set to start the evaluation of the CMBnarc-approach. Table 3 lists the measured IAM partition coefficients for FHM chemicals classified as having a narcosis MoA. The acutely toxic concentrations observed for narcotic chemicals to fathead minnow (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50,FHM), as classified in EPA's FHM database as Narcosis_I and Narcosis_II, are plotted against CMB predicted narcosis concentrations (log
LC50,FHM), as classified in EPA's FHM database as Narcosis_I and Narcosis_II, are plotted against CMB predicted narcosis concentrations (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50,narc), using eqn (1) with a CMBnarc of 140 mmol kg−1 as defined by Endo (2016)63 (Fig. 4). To calculate log
LC50,narc), using eqn (1) with a CMBnarc of 140 mmol kg−1 as defined by Endo (2016)63 (Fig. 4). To calculate log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC50,narc, different membrane–water partition coefficients were available as input values, we used:
LC50,narc, different membrane–water partition coefficients were available as input values, we used:
(i) 29 liposomal sorption coefficients (KMLW in the left plot of Fig. 4)
(ii) 30 existing IAM-HPLC partition coefficients (KIAM-exist in the left plot of Fig. 4)
(iii) 47 new IAM-HPLC based KIAM values obtained in this study (KIAM-new in the left plot of Fig. 4)
(iv) 138 ppLFER predicted KMLW values (right plot of Fig. 4).
As shown in the two plots of Fig. 4, most of the predicted toxic concentrations are within a factor of ±3 of the observed acutely toxic concentrations for fathead minnows. When using experimental membrane lipid–water distribution coefficients (Fig. 4 left), this is the case for 79% of the LC50,narc values based on liposomal KMLW values (97% within a factor of 10), and 84% of the LC50,narc values based on (existing and new) IAM-HPLC values (96% within a factor of 10). When using KMLW calculated with ppLFER descriptors (Fig. 4 right), this is the case for 71% of the LC50,narc values based on ppLFER calculated KMLW (93% within a factor of 10).
Fig. 4 demonstrates that the prediction of baseline toxic MoA based on IAM-HPLC derived partition coefficients is accurate for 96% of the tested neutral narcosis chemicals; only for 4% of the tested narcosis chemicals in the FHM set, the LC50 deviates by more than a factor of 10 from the LC50,narc. Fig. 4 also indicates that the previously derived CMB of 140 mmol kg−1 seems to apply equally to Narcosis_I and Narcosis_II classified chemicals, as discussed further below in section (iii). The ppLFER predictions show comparably successful predictions of acutely toxic concentrations, but this typically is only possible when experimental ppLFER descriptors are available.102 The three Narcosis_II classified chemicals that have a ppLFER calculated CMB <14 mmol kg−1 (catechol, pyridine and acetylpyridine) may even be re-classified as having a more specific mode of toxic action.
(iii) What is the lower CMB range for narcosis chemicals, and do Narcosis_II chemicals have a lower CMB than Narcosis_I chemicals (chemical set 1)?
The distinction between Narcosis_I and Narcosis_II chemicals (or non-polar narcosis and polar narcosis) stems from observed differences in respiratory–cardiovascular responses in fish acute toxicity syndromes (FATS), and the fact that lethal body burdens are generally lower for polar narcosis. When LC50 values are plotted against KOW values as the chemical descriptor of hydrophobicity, Narcosis_I and Narcosis_II chemicals typically are somewhat separated clusters.103 We examined this using only chemicals labeled in the FHM data set as Narcotics with confidence classes 1 and 2 used (highest confidence groups in EPA classification) and only for those chemicals for which experimentally derived KOW values were available (listed in EpiSUITE) to avoid co-influence of KOW-algorithm prediction biases. As listed in Table 5, multiplying KOW with LC50 indeed results in a higher CMBnarc for Narcosis_I chemicals (CMB = 169 mmol kg−1) compared to Narcosis_II chemicals (CMB = 29.1 mmol kg−1), on average a factor 5.8 difference. A comparable distinction based on KOW was observed in the study by Vaes et al.103 for guppy toxicity data; the recalculated average CMB for 8 Narcosis_I chemicals (342 mmol kg−1) and 10 Narcosis_II chemicals (75.6 mmol kg−1) according to eqn (2) indicates a difference of a factor of 4.5.| MoA class | Using experimental DMLW neutral/ions | Using ppLFER (neutral only) | Using experimental KOW neutral only | Using COSMOmic neutral/ion | |
|---|---|---|---|---|---|
| a The highest calculated CMB of 3832 was considered an outlier, see the text. | |||||
| Narcosis_I (EPA) | CMB mmol kg−1 | 155 | 132 | 169 | 397 |
| Classes 1 and 2 only | St. dev. | 119 | 202 | 138 | 1039 |
| (130 out of 225 classes 1–4) | Min–max | 30–568a | 13–1645 | 2–825 | 2–10599 |
| N used | 63 | 65 | 87 | 130 | |
| Narcosis_II (EPA) | CMB mmol kg−1 | 72 | 46 | 29 | 35 |
| Classes 1 and 2 only | St. dev. | 59 | 45 | 36 | 51 |
| (26 out of 36 classes 1–4) | Min–max | 13–247 | 2–152 | 0.9–97.4 | 3–275 |
| N used | 15 | 23 | 24 | 29 | |
| Specific toxic | CMB mmol kg−1 | 21 | 6 | 20 | n.a. |
| Mode of action | St. dev. | 64 | 11 | 66 | |
| Min–max | 0.09–341 | 0.24–32 | 0.08–285 | ||
| N used | 26 | 8 | 18 | ||
| Unsure amines | Average CMB mmol kg−1 | 45 | 179 | ||
| St. dev. | 45 | 244 | |||
| Min–max | 3–164 | 2–734 | |||
| N used | 12 | 12 | |||
| Neurotox amphetamine | CMB mmol kg−1 | 16 | 23 | ||
| Neurotox nicotine | CMB mmol kg−1 | 0.27 | 0.06 | ||
| Neurotox strychnine | CMB mmol kg−1 | 0.55 | 0.06 | ||
The current study involves a much larger data set on Narcosis_I and Narcosis_II chemicals than the study by Vaes et al.103 Instead of using IAM-HPLC partition coefficients to calculate LC50,narc for comparison with reported LC50 values, as done in the previous section (ii), the KIAM for narcosis FHM chemicals can also be used to re-calculate the CMBnarc. We can now do this for 82 Narcosis_I chemicals (64 classes 1, 2 and 18 class 3) from the FHM database for which KIAM is available. For one Narcosis_I chemical, 2-methyl-2-propanol, a very high CMBnarc of 3832 mmol kg−1 was calculated based on an IAM-HPLC based log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW of 1.65 (kIAM = 0.37 from ref. 104, and as such included in a review105 on IAM-HPLC capacity factors). This CMBnarc is more than 6 times higher than the second highest CMB value of 570 mmol kg−1 for Narcosis_I chemicals. The reported log
DMLW of 1.65 (kIAM = 0.37 from ref. 104, and as such included in a review105 on IAM-HPLC capacity factors). This CMBnarc is more than 6 times higher than the second highest CMB value of 570 mmol kg−1 for Narcosis_I chemicals. The reported log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW may be a considerable overestimation, because the pp-LFER-based log
DMLW may be a considerable overestimation, because the pp-LFER-based log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW estimate is 0.05, which would result in a 40 times lower CMB. The same IAM-HPLC study also reported an almost 3 times lower retention capacity factor for the more hydrophobic homolog 2-methyl-2-butanol, which would have a log
DMLW estimate is 0.05, which would result in a 40 times lower CMB. The same IAM-HPLC study also reported an almost 3 times lower retention capacity factor for the more hydrophobic homolog 2-methyl-2-butanol, which would have a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW of only 1.20. Without this outlier, a CMBnarc range of 19–570 mmol kg−1 (average 151 ± 114 s. d.), which closely compares to the average value of 140 mmol kg−1 derived recently.63 For Narcosis_II chemicals still only a limited set of 17 KIAM are available (15 classes 1, 2). This Narcosis_II set shows an average CMBnarc range of 13–247 mmol kg−1 (average 71 ± 56 s. d.). Based on this set of 98 chemicals (the one outlier excluded) with a defined narcosis MoA and measured DMLW, there is a significant distinction between the CMBnarc for Narcosis_I and Narcosis_II chemicals (using Graphpad Prism V9, unpaired t-test, p = 0.006, t = 2.813, and df = 96). This is consistent with a data compilation of several studies where internal whole body residues were measured and it was found that the range of polar narcosis overlaps with non-polar but goes lower in all data sets used. Still, a valuable distinction seems to be the lowest observed CMB of 13 mmol kg−1 for both types of narcosis chemicals.103 For simplicity, we set this limit to 14 mmol kg−1 from here on, as 10% of the average CMBnarc of 140 mmol kg−1.63 Chemicals with CMBnarc calculated using LC50/DMLW above 14 mmol kg−1 most likely do not exert lethal toxicity via a specific MoA, while chemicals with CMBnarc below 14 mmol kg−1 may be considered to have lethal adverse effects via some kind of specific or reactive MoA.
DMLW of only 1.20. Without this outlier, a CMBnarc range of 19–570 mmol kg−1 (average 151 ± 114 s. d.), which closely compares to the average value of 140 mmol kg−1 derived recently.63 For Narcosis_II chemicals still only a limited set of 17 KIAM are available (15 classes 1, 2). This Narcosis_II set shows an average CMBnarc range of 13–247 mmol kg−1 (average 71 ± 56 s. d.). Based on this set of 98 chemicals (the one outlier excluded) with a defined narcosis MoA and measured DMLW, there is a significant distinction between the CMBnarc for Narcosis_I and Narcosis_II chemicals (using Graphpad Prism V9, unpaired t-test, p = 0.006, t = 2.813, and df = 96). This is consistent with a data compilation of several studies where internal whole body residues were measured and it was found that the range of polar narcosis overlaps with non-polar but goes lower in all data sets used. Still, a valuable distinction seems to be the lowest observed CMB of 13 mmol kg−1 for both types of narcosis chemicals.103 For simplicity, we set this limit to 14 mmol kg−1 from here on, as 10% of the average CMBnarc of 140 mmol kg−1.63 Chemicals with CMBnarc calculated using LC50/DMLW above 14 mmol kg−1 most likely do not exert lethal toxicity via a specific MoA, while chemicals with CMBnarc below 14 mmol kg−1 may be considered to have lethal adverse effects via some kind of specific or reactive MoA.
Experimental DMPC membrane–water partition coefficients have been used as the chemical descriptor to plot LC50 values against.103 The difference between Narcosis_I and Narcosis_II chemicals in their analysis was still a factor 1.8 lower average CMBnarc for Narcosis_II chemicals, comparable to our evaluation. For the 8 Narcosis_I chemicals, the CMBnarc ranged between 33 and 513 mmol kg−1 (average 173 mmol kg−1), and for the 10 Narcosis_II chemicals, 11–174 mmol kg−1 (average 94 mmol kg−1). This set did not show a significant difference (p = 0.13, t = 1.555, and df = 17). As discussed in that study, the sorption affinities to the DMPC phospholipids (KDMPC) of Narcosis_II chemicals are relatively higher than to octanol, compared to Narcosis_I chemicals. The fact that Lethal Body Burden (LBB) values differ for Narcosis_I and Narcosis_II chemicals in this study may be related to the fact that these values relate to the whole body of the fish, including storage lipid, and the more polar Narcosis_II chemicals have a distinctly lower affinity for neutral storage lipids compared to polar phospholipids. The rationale behind a similar CMBnarc for all chemicals with a narcosis MoA is that the target site is the cell membrane, and that when normalized to this specific lipid pool all organic chemicals exert baseline toxicity at a comparable molar cell membrane loading.
In conclusion, the CMBnarc is on average about a factor 2 lower for more polar chemicals with a narcosis MoA, and based on experimental DMLW values, there is a significant difference in CMBnarc for Narcosis_I and Narcosis_II classified chemicals. The lower CMBnarc limit of 98 narcosis chemicals (classes 1–3) of the current study and the 18 narcosis chemicals from Vaes et al.103 is 11 mmol kg−1, or about 10% of the average CMBnarc of 140 mmol kg−1 phospholipid. If for a chemical the CMB is lower than 14 mmol kg−1, the toxic effect is thus very likely associated with an adverse effect pathway other than narcosis.
(iv) Using the IAM-based CMB to evaluate the classification of neutral FHM chemicals with a MoA other than narcosis (chemical set 1)
Table 2 lists the IAM partition coefficients for 28 chemicals with a specific MoA, and the 20 largely ionized chemicals, all selected from the FHM database. Fig. 5 plots all neutral FHM chemicals for which IAM-HPLC data are available, including the narcosis chemicals similar to those in Fig. 4 (black symbols), but now plotted together with the specific MoA chemicals selected from the FHM database (blue symbols in the left plot of Fig. 5). Each reported LC50 for Fathead minnow (LC50,FHM) is plotted against the calculated LC50,narc which is derived by dividing the CMBnarc of 140 mmol kg−1 by the chemical's DMLW obtained with IAM. The first dotted line next to the solid line in Fig. 5 marks a tenfold higher toxicity than expected based on the CMBnarc level of 140 mmol kg−1, i.e. a CMB of 14 mmol kg−1, which represents the lower limit of any evaluated narcosis chemical's CMB (13 mmol kg−1 derived in the discussion section above).Most of the tested FHM chemicals (23 out of 28) that were originally classified to have a specific MoA are on the right of this dividing line, i.e. these chemicals had lethally toxic effects occurring at membrane burdens below 14 mmol kg−1 (according to the IAM-based DMLW values). This confirms that these chemicals act via a MoA other than narcosis. It is interesting to see that several acetylcholinesterase inhibitors (AChE, indicated by + signs: diazinon, carbaryl, and EPN) are not as specifically (acutely) toxic to fish via the AChE mechanism as compared to other AChE chemicals, but have lethal membrane concentrations associated with a nonspecific narcosis effect. Although these chemicals may still adversely affect fish via the AChE mechanism, the potencies with regard to binding to the enzyme is rather low for these chemicals.
The AChE pesticide most toxic to fathead minnow fish is aldicarb (2.6 log units more toxic than LC50,narc). The maximum difference between the observed fish LC50,FHM and LC50,narc is 3.2 log units (i.e. an “excessive toxicity factor” of 1600) for the reactive chemical 1,3-dichloro-4,6-dinitro-benzene. The most toxic chemical tested in terms of dissolved concentrations was the respiratory blocker/inhibitor rotenone (0.01 μmol L−1), although the reactive chemical 1,3-dichloro-4,6-dinitro-benzene is toxic at the lowest calculated cell membrane concentration (0.1 mmol kg−1). Whether any organic chemical is likely to exerts a specific MoA at levels below or within the CMBnarc range is not readily derived by the current CMB approach. This is part of more detailed risk profile assessments, which may be done using the various MoA and MechoA tools available mentioned in the Introduction, or even based on the likelihood of interactions with the key initiating receptor using the chemical properties of the solute using a polyparameter approach.106
(v) Using the IAM-based CMB to evaluate whether largely ionized FHM chemicals act by a specific MoA (chemical set 1)
The right graph of Fig. 5 plots the largely cationic (green symbols) and anionic chemicals (red symbols) in the FHM data set for which IAM-HPLC data are available, alongside the neutral (black symbols) narcosis chemicals. The tested cationic amines classified as ‘unsure amines’ all have a molecular formula alike the cationic surfactants: CxHyN+.77 It is thus expected that the KIAM,intr values adjusted with δIAM-MLW increments (set by CxHyN+ cationic surfactants) give realistic KMLW values to apply to the CMBnarc approach. For most of these “Unsure MoA amines”, the observed LC50,FHM deviates less than a factor of 10 from the predicted LC50,narc – ranging on average a factor of 8.4 with a range between factors 1.3 and 35. Only the quaternary ammonium chemical phenyltrimethylammonium methosulfate (factor 35) and the tertiary amine N,N-dimethylbenzylamine (factor 16) have a more than tenfold lower LC50,FHM than LC50,narc. The average CMB for all 12 unsure amines is 38 mmol kg−1 phospholipid, suggesting that on average these cationic amines with a simple molecular formula of CxHyN+ are not toxic to fathead minnow fish by a MoA other than narcosis. Three FHM neurotoxic amines that are mostly cationic at neutral pH are strychnine (rat poison), amphetamine, and nicotine. Based on the adjusted KIAM,intr values, both strychnine and nicotine are calculated to have an LC50,FHM that is 250 and 520 times lower, respectively, than the predicted LC50,narc, clearly indicative of a specific MoA operating at lower levels than narcosis. Amphetamine, however, is calculated to be only 9 times more toxic than the predicted LC50,narc, positioning more in the range of the other ‘unsure amines’ and appears to have an acute lethal effect concentration based on narcosis for this fish species.The organic anion salicylic acid is classified in the FHM database as “unsure” MoA, but according to the CMB-approach this organic anion acts as a narcosis organic acid to fish. The acid 2,3,4,6-tetrachlorophenol (pKa 5.4) is classified as a Narcosis_I-3 chemical, but falls in line with the other uncoupler acids, acting at the same level as dinoseb (17 and 24 times more toxic than LC50,narc, respectively). This was already established for guppy fish data based on liposomal distribution coefficients for these same two acids.107 The four tested acidic uncouplers are lethally toxic to FHM at a level 17–24 times below the predicted LC50,narc, and thus also appear to act as toxicants by a specific MoA based on the IAM-HPLC derived KMLW values. Unfortunately, the number of largely dissociated acids in the FHM database with a narcosis MoA is limited, so we focused on herbicides to further evaluate the use of IAM-HPLC values for acidic chemicals to distinguish between specific MoA and baseline toxicity.
(vi) Chemical set 2: predicting baseline toxicity for herbicides
For 29 herbicides, divided in a set of 17 predominantly anionic and 12 nonionic chemicals, IAM-HPLC based DMLW values were measured, as listed in Table 2. We use the herbicide toxicity data as a second case study to (i) demonstrate how the CMBnarc approach can perform risk assessment on the specificity of adverse effects of chemicals to suspected sensitive organisms (i.e., herbicides affecting aquatic primary producers via a specific MoA), and to non-sensitive non-target organisms (i.e. only via baseline narcosis if the key molecular initiating event is not present), and (ii) whether IAM-HPLC is a useful tool in deriving the LC50,narc also for organic anions. The goal of this CMB evaluation with herbicides was not to prove whether herbicides are specifically toxic to various non-target organisms, as this would require inclusion of all available toxicity data (including chronic endpoints) and more detailed analysis of the possible adverse outcome pathways.As discussed above, in order to translate the IAM-HPLC based KIAM,intr values to DMLW, an empirical corrective increment δIAM-MLW of −0.47 for anions was applied to the values listed in Table 2. Using the critical membrane burden of 140 mmol kg−1 as derived for neutral chemicals, the baseline LC50,narc was calculated, in Table 6 shown in mg L−1 for comparison to the toxicity data.
| LC50,narc (mg L−1) (CMBnarc of 140 mmol kg−1) | Algal EC50b (mg L−1) | Algal TR | Macrophyte EC50 (mg L−1) Lemna sp.a (Myrio-phyllum) | Macrophyte TR | Invertebrate EC50c (mg L−1) | Invertebrate TR | Fish LC50d (mg L−1) | Fish TR | |
|---|---|---|---|---|---|---|---|---|---|
| a 7 d 50% biomass reduction for duckweed species. b 3 d 50% growth reduction of fresh water green algae Raphidocelis subcapitata (formerly Pseudokirchneriella subcapitata). c 2 d 50% immobilization of Daphnia magna. d 4 d 50% lethal effects on rainbow trout (O. mykiss). e SA: synthethic auxin; Unc: Uncoupler of phosphorylation; AHAS inh.: inhibits plant amino acid synthesis – acetohydroxyacid synthase AHAS; PROTOX: inhibits protoporphyrinogen oxidase (PROTOX), leading to irreversible cell membrane damage. PS-II: inhibitor for photosystem II. (VLCFA: very-long-chain fatty acid (inhibition of cell division); LSH: lipid synthesis inhibitor; LCI: lycopene cyclase inhibitor; MIT: mitosis inhibitor. f ✗ = no reliable ecotoxicity data retrieved. g Italic values: not from regulatory dossiers but through the US EPA ECOTOX database. | |||||||||
| Anionic herbicidese (MoA, as in Table 3) | |||||||||
| Clopyralid (SA) | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 297 297 |
30 | 643 | 89 | 217 | 130 | 148 | 53 | 364 |
| Fluroxypyr (SA) | 1417 | 35.3 | 40 | 12.3 | 115 | 100 | 14 | 14.3 | 99 |
| Triclopyr (SA) | 779 | 75.8 | 10 | ✗f | ✗ | 131 | 6 | 117 | 7 |
| MCPA (SA) | 647 | 32.9 | 20 | 0.152 | 4258 | 190 | 3 | 50 | 13 |
| 2,4-D (SA) | 717 | 0.68 | 1054 | 10.66(0.011) |
67(65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 169) 169)
|
134.2 | 5 | 100 | 7 |
| MCPP (SA) | 503 | 23.9 | 21 | 1.6(0.027) |
315(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 708) 708)
|
91 | 6 | 93 | 5 |
| 2,4-DP (SA) | 532 | 100 | 5 | 50(0.156) | 11(3411) | 46.6 | 11 | 46.6 | 11 |
| MCPB (SA) | 184 | 1.5 | 123 | 37 | 5 | 55 | 3 | 4.3 | 43 |
| 2,4-DB (SA) | 181 | 16.4 | 11 | 23.47(0.51) | 8(355) | 21.2 | 9 | 3 | 60 |
| 2,4,5-T (SA) | 286 | 100.8 | 3 | ✗ | ✗ | 5.0 | 57 | 38.2 | 7 |
| DNOC (Unc) | 267 | 3.4 | 79 | 0.81 | 330 | 3.67 | 73 | 0.066 | 4050 |
| Dinoseb (Unc) | 19 | 0.74 | 25 | ✗ | ✗ | 0.24 | 77 | 0.058 | 319 |
| Bromoxynil (PS-II) | 242 | 0.12 | 2018 | 0.118 | 2052 | 12.5 | 19 | 23 | 11 |
| Ioxynil (PS-II) | 80 | 0.15 | 533 | 0.027 | 2959 | 3.14 | 25 | 0.64 | 125 |
| Triasulfuron (AHAS) | 1586 | 0.39 | 4067 | 0.000202 |
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 852 852![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 263 263
|
100 | 16 | 100 | 16 |
| Bensulfuron-methyl (AHAS) | 274 | 0.0077 |
35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 558 558
|
0.0008 |
342![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 249 249
|
130 | 2 | 66 | 4 |
| Fomesafen (PROTOX) | 64 | 0.42 | 151 | 0.48 | 133 | 25 | 3 | >99 | 1 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Neutral herbicides (MoA) | |||||||||
| Alachlor (VLCFA) | 18 | 0.0016 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 279 279
|
0.0023 | 8043 | 18.4 | 1 | 2.24 | 8 |
| Metolachlor (VLCFA) | 16 | 0.056 | 289 | 0.0367 | 441 | 11.24 | 1.4 | 1.23 | 13 |
| Simazine (PS-II) | 91 | 0.10 | 914 | 0.14 | 653 | 1.10 | 83 | 60.2 | 2 |
| Ametryn (PS-II) | 23 | 0.0032 | 7205 | 0.009 | 2506 | 15 | 2 | 3.6 | 6 |
| Terbuthylazine (PS-II) | 14 | 0.028 | 501 | 0.0128 | 1097 | 36 | 0.4 | 2.2 | 6 |
| Diuron (PS-II) | 14 | 0.019 | 716 | 0.0183 | 743 | 1.1 | 12 | 6.7 | 2 |
| Linuron (PS-II) | 11 | 50.7 | 0.2 | 0.017(0.0317) | 634(340) | 5.81 | 2 | 6.7 | 2 |
| Chloroxuron (PS-II) | 2.2 | 0.0160 | 140 | ✗ | 2.95 | 0.8 | 0.43 | 5 | |
| Ethofumesate (LSI) | 23 | 4.7 | 5 | 42(0.38) | 0.5(59) | 13.5 | 2 | 11.9 | 2 |
| Prosulfocarb (LSI) | 0.8 | 0.049 | 17 | 0.69 | 1.2 | 0.51 | 2 | 11.9 | 1 |
| Clomazone (LCI) | 39 | 0.14 | 290 | 34 | 1.2 | 12.7 | 3 | 15.5 | 3 |
| Trifluralin (MIT) | 0.2 | 0.0122 | 13 | 0.0435 | 4 | 0.245 | 0.6 | 0.088 | 2 |
Dividing CMBnarc by DMLW, the LC50,narc was calculated as the aqueous concentration at which non-specific MoA was expected to result in a 50% (sub-)lethal effect (Table 6). Just as we did for the evaluation above for 1 fish species (fathead minnow), the LC50,narc serves as the benchmark value to compare the observed toxic concentrations for the four different aquatic organisms evaluated here.
Using the baseline CMBnarc approach, the specificity of an observed toxic effect is more easily expressed by the toxic ratio (TR) in eqn (11):
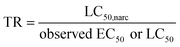 | (11) |
When including the margins of CMBnarc between 80 and 250 mmol kg−1, and residual uncertainty in the KIAM–DMLW relationship, a TR > 10 is a strong indicator of a chemical exerting adverse effects other than via a non-specific MoA. Since LC50,narc is based on the CMBnarc of 140 mmol kg−1, a TR > 10 is set as the cut-off concentration of the critical membrane burden of 14 mmol kg−1 below which chemicals induce toxicity through a specific MoA. This corresponds to the observation for narcosis chemicals in chemical set 1 with the US-EPA fathead minnow database, where out of 78 Narcosis_I and Narcosis_II chemicals the lowest CMB was 13. The CMBnarc is a fairly constant value across all kinds of organisms56,57,107 and the FHM evaluation above already showed adequate predictions for a broad range of narcosis chemicals in fish. Table 6 lists the TR values for the herbicide endpoints for different aquatic organisms relative to LC50,narc.
We assumed that all herbicides are toxic to aquatic plants via a specific MoA, but mostly act by narcosis (non-specific MoA) on non-target invertebrates and fish, except for uncouplers of oxidative phosphorylation (DNOC and dinoseb). Fig. 6 shows the range of toxicity endpoints for various aquatic species for each herbicide along the Y-axis, in relation to the expected baseline LC50,narc.
As expected, for 21 of the 25 herbicides with complete toxicity data on duckweed and green algae, the herbicides do seem to impair growth by a specific toxic MoA (TR > 10) for at least one of these two aquatic plant species (Table 6). When comparing the two plots of Fig. 6, it appears that most acidic herbicides are more toxic to duckweed than to green algae, while most neutral herbicides affect green algae at lower concentrations than duckweed.
For several anionic synthetic auxins, Lemna and algae appeared to be affected only by baseline toxicity, with TR of less than 10 for 2,4,5-T (algae), 2,4-DB (algae and Lemna), 2,4-DP (Lemna), MCPB (Lemna). Lemna does show a high specific effect for MCPA with a TR > 4000. Myriophyllum, however, appears to be a much more sensitive primary producer than Lemna and R. subcapitata, with a TR of 6.5 × 104 for 2,4-D.
For the herbicide 2,4,5-T, only an algal toxicity endpoint was retrieved, which showed a TR < 10. For 3 out of the 29 herbicides the TR was < 10 for green algae, but TR was > 10 for one or both of the aquatic plants. For the neutral herbicides targeting lipid synthesis (ethofumesate and prosulfocarb), microtubule assembly during mitosis (trifluralin), and lycopene cyclase to block provitamin A carotenoid synthesis (clomazone), duckweed appeared to be affected only through baseline toxicity (TR <10). For ethofumesate, however, Myriophyllum showed a higher TR of 59, and green algae appeared to be specifically affected by clomazone (TR of 290).
The herbicides most toxic to aquatic plants in our data set are the acidic N-sulfonylurea chemicals bensulfuron-methyl and triasulfuron. In target plants, these herbicides inhibit plant-essential amino acid synthesis (acetohydroxyacid synthase AHAS). Particularly duckweed growth is affected by this (or related) specific MoA, at a TR of 3.4 × 105 and 7.8 × 107, for bensulfuron-methyl and triasulfuron respectively. Triasulfuron has been banned for use in the EU since 2017, while bensulfuron-methyl is approved in several European countries, according to the University of Herfortshire's PPDB.
However, the CMB-approach also demonstrates that several herbicides affect non-target aquatic species other than aquatic plants via a specific MoA. Since some of the acidic herbicides were classified as having an uncoupler MoA already in the FHM database, as well as in PPDB (Table 5: DNOC, dinoseb), it was expected that some other acidic herbicides may also demonstrate specific toxicity to daphnids and fish. For 11 of the 17 acidic herbicides the TR was indeed higher than 10, ranging up to 4050. For some of these acidic herbicides fish were the most sensitive species, although no data for Lemna/Myriophyllym were available for both DNOC and dinoseb to evaluate over multiple aquatic plant species. The herbicides most toxic to fish were indeed the phosphorylation uncouplers DNOC and dinoseb, with respective TR values of 4050 and 319. Dinoseb is even considered highly toxic to birds, and while it was once widely used, it has therefore been banned as a pesticide in most countries. DNOC has no longer been approved in the EU and North America since 1991.
Simazine is the only neutral herbicide example in the current selection with a specific toxicity to daphnids, with a TR of 83, while it is not acutely toxic via a specific MoA to rainbow trout. The acids 2,4,5-T, DNOC and dinoseb also appear to affect daphnids by a specific toxicity, with a TR of 53, 77, and 73, as well as the photosystem inhibitors bromoxynil and ioxynil (TR of 19 and 25, resp.).
4. Synopsis
The CMBnarc approach for determining acute fathead minnow toxicity and acute effects of herbicides on non-target organisms shows that IAM-HPLC is able to predict baseline toxic concentrations and distinguish between chemicals that exert baseline narcosis and toxicity due to another more specific MoA, for neutral, cationic and anionic organic chemicals. A Toxic Ratio (TR) of observed toxic concentrations against the predicted LC50,narc of >10, corresponding to a CMB <14 mmol kg−1, is a good indication that the chemical exerts toxicity via a specific MoA for the purposes of screening chemicals as part of a consensus MoA approach. The fact that IAM-HPLC is a simple experimental assay makes it a valuable alternative to octanol–water based predictions for chemicals that fall outside the applicability domain of the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW–log
KOW–log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KMLW relationships. The limiting factor of IAM-HPLC is that the maximum apparent log
KMLW relationships. The limiting factor of IAM-HPLC is that the maximum apparent log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIAM that seems experimentally feasible is about 6, which translates to retention times of ∼24 h with an eluent flow rate of 1 mg min−1. At a higher retention capacity factor, higher solvent fractions are required, which may not linearly extrapolate to fully aqueous eluent, particularly for ionogenic chemicals. Still, a series of high solvent fractions in the eluent could provide for a rough value of components in complex (technical) mixtures, or provide a lower limit to the tested chemical of log
KIAM that seems experimentally feasible is about 6, which translates to retention times of ∼24 h with an eluent flow rate of 1 mg min−1. At a higher retention capacity factor, higher solvent fractions are required, which may not linearly extrapolate to fully aqueous eluent, particularly for ionogenic chemicals. Still, a series of high solvent fractions in the eluent could provide for a rough value of components in complex (technical) mixtures, or provide a lower limit to the tested chemical of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMLW ∼6, which would already indicate strong sorption to cell membranes, a potentially high bioaccumulation factor, and toxic baseline concentrations <0.1 mg L−1.
DMLW ∼6, which would already indicate strong sorption to cell membranes, a potentially high bioaccumulation factor, and toxic baseline concentrations <0.1 mg L−1.
Author contributions
The experimental work was performed by SD. The manuscript was written by SD with contributions of all authors. All authors have given approval to the final version of the manuscript. The conclusions expressed in the paper represent the expert judgement of the authors, but not necessarily the opinion of their affiliation.Conflicts of interest
There are no conflicts of interest to declare. The contribution by SD is based on his experimental work during employment at Utrecht University, which ended on September 30th, 2016, and at the University of Amsterdam, which ended on December 31st, 2019.References
- UN, Global Chemicals Outlook II, from Legacies to Innovative Solutions, United Nations Environment Programme, UN, 2019 Search PubMed.
- OECD, Prioritization of Chemicals Using the Integrated Approaches for Testing and Assessment (IATA)-based Ecological Risk Classification, Case Study Third Cycle, OECD, 2017 Search PubMed.
- ECHA, ECHA Read across Framework (ECHA-17-R-01-EN), European Chemicals Agency, ECHA, 2017 Search PubMed.
- ECCC – Environmental and Climate Change Canada, Science Approach Document: Ecological Risk Classification of Organic Substances, ECCC – Environment and Climate Change Canada, Gatineau (QC), 2016, https://www.ec.gc.ca/ese-ees/default.asp?lang=En&n=a96e2e98-1 Search PubMed.
- Environment and Climate Change Canada (ECCC), Science Approach Document - Ecological Risk Classification of Organic Substances Version 2.0 (ERC2, Government of Canada. ECCC, Gatineau, Quebec, 2022, https://www.canada.ca/en/environment-climate-change/services/evaluating-existingsubstances/science-approach-document-ecological-risk-classification-organic-substances-erc2.html Search PubMed.
- G. T. Ankley, R. S. Bennett, R. J. Erickson, D. J. Hoff, M. W. Hornung, R. D. Johnson, D. R. Mount, J. W. Nichols, C. L. Russom, P. K. Schmieder, J. A. Serrrano, J. E. Tietge and D. L. Villeneuve, Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment, Environ. Toxicol. Chem., 2010, 29, 730–741, DOI:10.1002/etc.34.
- J. M. Armitage, J. A. Arnot and M. Bonnell, Comparing Mode of Action (MOA) Classification Using Body Residues, Membrane Concentrations and Chemical Activity for Chemical Prioritization, SETAC North America 39th Annual Meeting. Society of Environmental Toxicology and Chemistry, Pensacola, Florida, 2018 Search PubMed.
- OECD, The OECD QSAR Toolbox, OECD, 2020, https://qsartoolbox.org Search PubMed.
- A. Kienzler, M. G. Barron, S. E. Belanger, A. Beasley and M. R. Embry, Mode of Action (MOA) assignment classifications for ecotoxicology: an evaluation of approaches, Environ. Sci. Technol., 2017, 51, 10203–10211, DOI:10.1021/acs.est.7b02337.
- A. Kienzler, K. A. Connors, M. Bonnell, M. G. Barron, A. Beasley, C. G. Inglis, T. J. Norberg-King, T. Martin, H. Sanderson, N. Vallotton, P. Wilson and M. R. Embry, Mode of action classifications in the EnviroTox Database: Development and implementation of a consensus MOA classification, Environ. Toxicol. Chem., 2019, 38, 2294–2304, DOI:10.1002/etc.4531.
- K. A. Connors, A. Beasley, M. G. Barron, S. E. Belanger, M. Bonnell, J. L. Brill, D. de Zwart, A. Kienzler, J. Krailler and M. R. Embry, Creation of a curated aquatic toxicology database: EnviroTox, Environ. Toxicol. Chem., 2019, 38, 1062–1073, DOI:10.1002/etc.4382.
- H. J. M. Verhaar, C. J. van Leeuwen and J. L. M. Hermens, Classifying environmental pollutants. 1. Structure-Aacivity-relationships for prediction of aquatic toxicity, Chemosphere, 1992, 25, 471–491, DOI:10.1016/0045-6535(92)90280-5.
- H. J. M. Verhaar, J. Solbé, J. Speksnijder, C. J. Van Leeuwen and J. L. M. Hermens, Classifying environmental pollutants: Part 3. External validation of the classification system, Chemosphere, 2000, 40, 875–883, DOI:10.1016/S0045-6535(99)00317-3.
- S. J. Enoch, M. Hewitt, M. T. D. Cronin, S. Azam and J. C. Madden, Classification of chemicals according to mechanism of aquatic toxicity: An evaluation of the implementation of the Verhaar scheme in Toxtree, Chemosphere, 2008, 73, 243–248, DOI:10.1016/j.chemosphere.2008.06.052.
- M. G. Barron, C. R. Lilavois and T. M. Martin, MOAtox: A comprehensive mode of action and acute aquatic toxicity database for predictive model development, Aquat. Toxicol., 2015, 161, 102–107, DOI:10.1016/j.aquatox.2015.02.001.
- C. L. Russom, S. P. Bradbury, S. J. Broderius, D. E. Hammermeister and R. A. Drummond, Predicting modes of toxic action from chemical structure: Acute toxicity in the fathead minnow (Pimephales Promelas), Environ. Toxicol. Chem., 1997, 16, 948–967 CrossRef CAS.
- U.S. EPA, User Guide for ASTER (ASsessment Tools for the Evaluation of Risk) Version 2.0, 2012 Search PubMed.
- U.S. EPA, User Guide for T.E.S.T. (Version 4.2) (Toxicity Estimation Software Tool): A Program to Estimate Toxicity from Molecular Structure, 2016 Search PubMed.
- P. C. Thomas, P. Bicherel and F. Bauer, How in silico and QSAR approaches can increase confidence in environmental hazard and risk assessment, Integr. Environ. Assess. Manage., 2019, 15, 40–50, DOI:10.1002/ieam.4108.
- KREATiS, (2020) iSafeRat® (in silico Algorithms For Environmental Risk And Toxicity) Online version v2.1https://isaferat.kreatis.eu, accessed July 2021.
- F. J. Bauer, P. C. Thomas, S. Y. Fouchard and S. J. M. Neunlist, A new classification algorithm based on mechanisms of action, Comput. Toxicol., 2018, 5, 8–15, DOI:10.1016/j.comtox.2017.11.001.
- F. J. Bauer, P. C. Thomas, S. Y. Fouchard and S. J. M. Neunlist, High-accuracy prediction of Mechanisms of Action using structural alerts, Comput. Toxicol., 2018, 7, 36–45, DOI:10.1016/j.comtox.2018.06.004.
- M. Sapounidou, D. Ebbrell, M. Bonnell, B. Campos, J. Firman, S. Gutsell, G. Hodges, J. Roberts and M. Cronin, Development of an enhanced mechanistically driven mode of action classification scheme for adverse effects on environmental species, Environ. Sci. Technol., 2021, 55, 1897–1907, DOI:10.1021/acs.est.0c06551.
- J. W. Firman, D. J. Ebbrell, F. J. Bauer, M. Sapounidou, G. Hodges, B. Campos, J. Roberts, S. Gutsell, P. C. Thomas, M. Bonnell and M. T. D. Cronin, Construction of an in silico structural profiling tool facilitating mechanistically grounded classification of aquatic toxicants, Environ. Sci. Technol., 2022, 56, 17805–17814, DOI:10.1021/acs.est.2c03736.
- S. Endo, B. I. Escher and K. U. Goss, Capacities of membrane lipids to accumulate neutral organic chemicals, Environ. Sci. Technol., 2011, 45, 5912–5921, DOI:10.1021/es200855w.
- N. Timmer and S. T. J. Droge, Sorption of cationic surfactants to artificial cell membranes: comparing phospholipid bilayers with monolayer coatings and molecular simulations, Environ. Sci. Technol., 2017, 51, 2890–2898, DOI:10.1021/acs.est.6b05662.
- S. T. J. Droge, Membrane-water partition coefficients to aid risk assessment of perfluoroalkyl anions and alkyl sulfates, Environ. Sci. Technol., 2019, 53, 760–770, DOI:10.1021/acs.est.8b05052.
- A. Klamt, U. Huniar, S. Spycher and J. Keldenich, COSMOmic: A Mechanistic Approach to the Calculation of Membrane-Water Partition Coefficients and Internal Distributions within Membranes and Micelles, J. Phys. Chem. B, 2008, 112, 12148–12157 CrossRef CAS PubMed.
- K. Bittermann, S. Spycher, S. Endo, L. Pohler, U. Huniar, K. U. Goss and A. Klamt, Prediction of phospholipid-water partition coefficients of ionic organic chemicals using the mechanistic model COSMOmic, J. Phys. Chem. B, 2014, 118, 14833–14842 CrossRef CAS PubMed.
- T. Ingram, S. Storm, L. Kloss, T. Mehling, S. Jakobtorweihen and I. Smirnova, Prediction of micelle/water and liposome/water partition coefficients based on molecular dynamics simulations, COSMO-RS, and COSMOmic, Langmuir, 2013, 29, 3527–3537 CrossRef CAS PubMed.
- S. Jakobtorweihen, A. C. Zuniga, T. Ingram, T. Gerlach, F. J. Keil and I. Smirnova, Predicting solute partitioning in lipid bilayers: Free energies and partition coefficients from molecular dynamics simulations and COSMOmic, J. Chem. Phys., 2014, 141, 045102 CrossRef CAS PubMed.
- T. D. Potter, E. L. Barrett and M. A. Miller, Automated Coarse-Grained Mapping Algorithm for the Martini Force Field and Benchmarks for Membrane-Water Partitioning, J. Chem. Theory Comput., 2021, 17, 5777–5791 CrossRef CAS PubMed.
- H. Könemann, R. Zelle, F. Busser and W. E. Hammers, Determination of log Poct values of chloro-substituted benzenes, toluenes and anilines by high-performance liquid chromatography on ODS-silica, J. Chromatogr. A, 1979, 178, 559–565, DOI:10.1016/S0021-9673(00)92516-0.
- B. McDuffie, Estimation of octanol/water partition coefficients for organic pollutants using reverse-phase HPLC, Chemosphere, 1981, 10, 73–83, DOI:10.1016/0045-6535(81)90171-5.
- C. V. Eadsforth and P. Moser, Assessment of reverse-phase chromatographic methods for determining partition coefficients, Chemosphere, 1983, 12, 1459–1475, DOI:10.1016/0045-6535(83)90076-0.
- C. Altomare, A. Carotti, G. Trapani and G. Liso, Estimation of partitioning parameters of nonionic surfactants using calculated descriptors of molecular size, polarity, and hydrogen bonding, J. Pharm. Sci., 1997, 86, 1417–1425 CrossRef CAS PubMed.
- M. H. Abraham, C. F. Poole and S. K. Poole, Classification of stationary phases and other materials by gas chromatography, J. Chromatogr. A, 1999, 842, 79–114 CrossRef CAS.
- A. Stenzel, S. Endo and K. U. Goss, Measurements and predictions of hexadecane/air partition coefficients for 387 environmentally relevant compounds, J. Chromatogr. A, 2012, 1220, 132–142 CrossRef CAS PubMed.
- A. Stenzel, K. U. Goss and S. Endo, Prediction of partition coefficients for complex environmental contaminants: Validation of COSMOtherm, ABSOLV, and SPARC, Environ. Toxicol. Chem., 2014, 33, 1537–1543, DOI:10.1002/etc.2587.
- G. Greco and T. Letzel, Main interactions and influences of the chromatographic parameters in HILIC separations, J. Chromatogr. Sci., 2013, 51, 684–693, DOI:10.1093/chromsci/bmt015.
- J. Hammer, J. J. Haftka, P. Scherpenisse, J. L. Hermens and P. W. de Voogt, Fragment-based approach to calculate hydrophobicity of anionic and nonionic surfactants derived from chromatographic retention on a C18 stationary phase, Environ. Toxicol. Chem., 2017, 36, 329–336, DOI:10.1002/etc.3564.
- J. Hammer, J. J. Haftka, P. Scherpenisse, J. L. Hermens and P. W. de Voogt, Investigating hydrophilic and electrostatic properties of surfactants using retention on two mixed-mode liquid chromatographic columns, J. Chromatogr. A, 2018, 1571, 185–192, DOI:10.1016/j.chroma.2018.08.024.
- D. Rhee, R. Markovich, W. G. Chae, X. Qiu and C. Pidgeon, Chromatographic surfaces prepared from lyso phosphatidylcholine ligands, Anal. Chim. Acta, 1994, 297, 377–386 CrossRef CAS.
- S. Ong, H. Liu, X. Qiu, G. Bhat and C. Pidgeon, Membrane partition coefficients chromatographically measured using immobilized artificial membrane surfaces, Anal. Chem., 1995, 67, 755–762 CrossRef CAS PubMed.
- S. Ong and D. Pidgeon, Thermodynamics of solute partitioning into immobilized artificial membranes, Anal. Chem., 1995, 67, 2119–2128 CrossRef CAS PubMed.
- S. Ong, H. Liu and C. Pidgeon, Immobilized-artificial-membrane chromatography: Measurements of membrane partition coefficient and predicting drug membrane permeability, J. Chromatogr. A, 1996, 728, 113–128 CrossRef CAS PubMed.
- F. Tsopelas, C. Stergiopoulos and A. Tsantili-Kakoulidou, Immobilized artificial membrane chromatography: From medicinal chemistry to environmental sciences, ADMET DMPK, 2018, 6, 225–241, DOI:10.5599/admet.553.
- S. T. J. Droge, Dealing with Confounding pH-Dependent Surface Charges in Immobilized Artificial Membrane HPLC Columns, Anal. Chem., 2016, 88, 960–967, DOI:10.1021/acs.analchem.5b03708.
- E. Lázaro, C. Ràfols, M. H. Abraham and M. Rosés, Chromatographic estimation of drug disposition properties by means of immobilized artificial membranes (IAM) and C18 columns, J. Med. Chem., 2006, 49, 4861–4870, DOI:10.1021/jm0602108.
- M. Hidalgo-Rodríguez, E. Fuguet, C. Ràfols and M. Rosés, Modeling nonspecific toxicity of organic compounds to the fathead minnow fish by means of chromatographic systems, Anal. Chem., 2012, 84, 3446–3452, DOI:10.1021/ac2034453.
- A. Fernández-Pumarega, S. Amézqueta, E. Fuguet and M. Rosés, Tadpole toxicity prediction using chromatographic systems, J. Chromatogr. A, 2015, 1418, 167–176, DOI:10.1016/j.chroma.2015.09.056.
- A. Fernández-Pumarega, S. Amézqueta, S. Farré, L. Muñoz-Pascual, M. H. Abraham, E. Fuguet and M. Rosés, Modeling Aquatic Toxicity through Chromatographic Systems, Anal. Chem., 2017, 89, 7996–8003, DOI:10.1021/acs.analchem.7b01301.
- C. Stergiopoulos, D. Makarouni, A. Tsantili-Kakoulidou, M. Ochsenkühn-Petropoulou and F. Tsopelas, Immobilized artificial membrane chromatography as a tool for the prediction of ecotoxicity of pesticides, Chemosphere, 2019, 224, 128–139, DOI:10.1016/j.chemosphere.2019.02.075.
- C. Stergiopoulos, F. Tsopelas, K. Valko and M. Ochsenkühn-Petropoulou, The use of biomimetic chromatography to predict acute aquatic toxicity of pharmaceutical compounds, Toxicol. Environ. Chem., 2022, 104, 1–19, DOI:10.1080/02772248.2021.2005065.
- A. P. Van Wezel and A. Opperhuizen, Narcosis due to environmental pollutants in aquatic organisms: Residue-based toxicity, mechanisms, and membrane burdens, Crit. Rev. Toxicol., 1995, 25, 255–279 CrossRef CAS PubMed.
- B. I. Escher, R. I. L. Eggen, U. Schreiber, Z. Schreiber, E. Vye, B. Wisner and R. P. Schwarzenbach, Baseline toxicity (narcosis) of organic chemicals determined by in vitro membrane potential measurements in energy-transducing membranes, Environ. Sci. Technol., 2002, 36, 1971–1979 CrossRef CAS PubMed.
- B. I. Escher and J. L. M. Hermens, Modes of action in ecotoxicology: Their role in body burdens, species sensitivity, QSARs, and mixture effects, Environ. Sci. Technol., 2002, 36, 4201–4217, DOI:10.1021/es015848h.
- S. Endo, T. N. Brown and K. U. Goss, General model for estimating partition coefficients to organisms and their tissues using the biological compositions and polyparameter linear free energy relationships, Environ. Sci. Technol., 2013, 47, 6630–6639 CrossRef CAS PubMed.
- A. Geisler, S. Endo and K. U. Goss, Partitioning of organic chemicals to storage lipids: Elucidating the dependence on fatty acid composition and temperature, Environ. Sci. Technol., 2012, 46, 9519–9524 CrossRef CAS PubMed.
- S. Endo and K. U. Goss, Serum albumin binding of structurally diverse neutral organic compounds: data and models, Chem. Res. Toxicol., 2011, 24, 2293–2301, DOI:10.1021/tx200431b.
- S. Endo, B. I. Escher and K. U. Goss, Capacities of membrane lipids to accumulate neutral organic chemicals, Environ. Sci. Technol., 2011, 45, 5912–5921, DOI:10.1021/es200855w.
- A. Geisler, S. Endo and K. U. Goss, Partitioning of polar and non-polar neutral organic chemicals into human and cow milk, Environ. Int., 2011, 37, 1253–1258 CrossRef CAS PubMed.
- S. Endo, Re-analysis of narcotic critical body residue data using the equilibrium distribution concept and refined partition coefficients, Environ. Sci.: Processes Impacts, 2016, 18, 1024–1029, 10.1039/c6em00180g.
- D. M. Di Toro, J. A. Mcgrath and D. J. Hansen, Technical basis for narcotic chemicals and polycyclic aromatic hydrocarbon criteria. I. Water and tissue, Environ. Toxicol. Chem., 2000, 19, 1951–1970, DOI:10.1002/etc.5620190803.
- R. Judson, K. Houck, M. Martin, A. M. Richard, T. B. Knudsen, I. Shah, S. Little, J. Wambaugh, R. W. Setzer, P. Kothya, J. Phuong, D. Filer, D. Smith, D. Reif, D. Rotroff, N. Kleinstreuer, N. Sipes, M. Xia, R. Huang, K. Crofton and R. S. Thomas, Analysis of the effects of cell stress and cytotoxicity on in vitro assay activity across a diverse chemical and assay space, Toxicol. Sci., 2016, 152, 323–339, DOI:10.1093/toxsci/kfw092.
- K. A. Fay, D. L. Villeneuve, J. Swintek, S. W. Edwards, M. D. Nelms, B. R. Blackwell and G. T. Ankley, Differentiating pathway-specific from nonspecific effects in high-throughput toxicity data: A foundation for prioritizing adverse outcome pathway development, Toxicol. Sci., 2018, 163, 500–515, DOI:10.1093/toxsci/kfy049.
- J. Ahlers, C. Riedhammer, M. Vogliano, R. Ebert, R. Kühne and G. Schüürmann, Acute to chronic ratios in aquatic toxicity - Variation across trophic levels and relationship with chemical structure, Environ. Toxicol. Chem., 2006, 25, 2937–2945, DOI:10.1897/05-701R.1.
- A. I. Okonski, D. B. MacDonald, K. Potter and M. Bonnell, Deriving predicted no-effect concentrations (PNECs) using a novel assessment factor method, Hum. Ecol. Risk Assess., 2021, 27, 1613–1635, DOI:10.1080/10807039.2020.1865788.
- J. Hermens, H. Canton, N. Steyger and R. Wegman, Joint effects of a mixture of 14 chemicals on mortality and inhibition of reproduction of Daphnia magna, Aquat. Toxicol., 1984, 5, 315–322, DOI:10.1016/0166-445X(84)90012-2.
- J. Y. M. Tang, S. McCarty, E. Glenn, P. A. Neale, M. S. J. Warne and B. I. Escher, Mixture effects of organic micropollutants present in water: Towards the development of effect-based water quality trigger values for baseline toxicity, Water Res., 2013, 47, 3300–3314, DOI:10.1016/j.watres.2013.03.011.
- P. A. Neale, F. D. L. Leusch and B. I. Escher, Applying mixture toxicity modelling to predict bacterial bioluminescence inhibition by non-specifically acting pharmaceuticals and specifically acting antibiotics, Chemosphere, 2017, 173, 387–394, DOI:10.1016/j.chemosphere.2017.01.018.
- P. Thomas, J. Dawick, M. Lampi, P. Lemaire, S. Presow, R. Van Egmond, J. A. Arnot, D. Mackay, P. Mayer and M. Galay Burgos, Application of the Activity Framework for Assessing Aquatic Ecotoxicology Data for Organic Chemicals, Environ. Sci. Technol., 2015, 49, 12289–12296, DOI:10.1021/acs.est.5b02873.
- K. U. Goss and S. Endo, Comment on "Application of the Activity Framework for Assessing Aquatic Ecotoxicology Data for Organic Chemicals, Environ. Sci. Technol., 2016, 50, 4139–4140, DOI:10.1021/acs.est.5b05534.
- C. A. Johnson and J. C. Westall, Effect of pH and potassium chloride concentration on the octanol-water distribution of methylanilines, Environ. Sci. Technol., 1990, 24, 1869–1875 CrossRef CAS.
- C. T. Jafvert, J. C. Westall, E. Grieder and R. P. Schwarzenbach, Distribution of hydrophobic ionogenic organic compounds between octanol and water: organic acids, Environ. Sci. Technol., 1990, 24, 1795–1803 CrossRef CAS.
- K. Bittermann, S. Spycher and K. -. Goss, Comparison of different models predicting the phospholipid-membrane water partition coefficients of charged compounds, Chemosphere, 2016, 144, 382–391, DOI:10.1016/j.chemosphere.2015.08.065.
- S. T. J. Droge, J. L. M. Hermens, J. Rabone, S. Gutsell and G. Hodges, Phospholipophilicity of CxHyN+ amines: Chromatographic descriptors and molecular simulations for understanding partitioning into membranes, Environ. Sci.: Processes Impacts, 2016, 18, 1011–1023, 10.1039/c6em00118a.
- S. T. J. Droge, J. L. M. Hermens, S. Gutsell, J. Rabone and G. Hodges, Predicting the phospholipophilicity of monoprotic positively charged amines, Environ. Sci.: Processes Impacts, 2017, 19, 307–323, 10.1039/c6em00615a.
- B. I. Escher, R. P. Schwarzenbach and J. C. Westall, Evaluation of liposome-water partitioning of organic acids and bases. 2. Comparison of experimental determination methods, Environ. Sci. Technol., 2000, 34, 3962–3968 CrossRef CAS.
- B. I. Escher, R. P. Schwarzenbach and J. C. Westall, Evaluation of liposome-water partitioning of organic acids and bases. 1. Development of a sorption model, Environ. Sci. Technol., 2000, 34, 3954–3961 CrossRef CAS.
- J. Neuwoehner, K. Fenner and B. I. Escher, Physiological modes of action of fluoxetine and its human metabolites in algae, Environ. Sci. Technol., 2009, 43, 6830–6837 CrossRef CAS PubMed.
- B. I. Escher, R. Baumgartner, M. Koller, K. Treyer, J. Lienert and C. S. McArdell, Environmental toxicology and risk assessment of pharmaceuticals from hospital wastewater, Water Res., 2011, 45, 75–92 CrossRef CAS PubMed.
- J. M. Armitage, J. A. Arnot, F. Wania and D. Mackay, Development and evaluation of a mechanistic bioconcentration model for ionogenic organic chemicals in fish, Environ. Toxicol. Chem., 2013, 32, 115–128 CrossRef CAS PubMed.
- B. I. Escher and R. P. Schwarzenbach, Partitioning of substituted phenols in liposome-water, biomembrane-water, and octanol-water systems, Environ. Sci. Technol., 1996, 30, 260–270 CrossRef CAS.
- S. Spycher, P. Smejtek, T. I. Netzeva and B. I. Escher, Toward a class-independent quantitative structure - Activity relationship model for uncouplers of oxidative phosphorylation, Chem. Res. Toxicol., 2008, 21, 911–927, DOI:10.1021/tx700391f.
- K. Bittermann and K. U. Goss, Assessing the toxicity of ionic liquids – Application of the critical membrane concentration approach, Chemosphere, 2017, 183, 410–418, DOI:10.1016/j.chemosphere.2017.05.097.
- B. I. Escher, A. Baumer, K. Bittermann, L. Henneberger, M. Konig, C. Kuhnert and N. Kluver, General baseline toxicity QSAR for nonpolar, polar and ionisable chemicals and their mixtures in the bioluminescence inhibition assay with Aliivibrio fischeri, Environ. Sci.: Processes Impacts, 2017, 19, 414–428, 10.1039/C6EM00692B.
- D. T. Manallack, The pKa distribution of drugs: application to drug discovery, Perspect. Med. Chem., 2007, 1, 25–38 Search PubMed.
- A. Franco, A. Ferranti, C. Davidsen and S. Trapp, An unexpected challenge: Ionizable compounds in the REACH chemical space, Int. J. Life Cycle Assess., 2010, 15, 321–325 CrossRef CAS.
- A. Loidl-Stahlhofen, T. Hartmann, M. Schottner, C. Rohring, H. Brodowsky, J. Schmitt and J. Keldenich, Multilamellar liposomes and solid-supported lipid membranes (TRANSIL): Screening of lipid-water partitioning toward a high-throughput scale, Pharm. Res., 2001, 18, 1782–1788 CrossRef CAS PubMed.
- A. Avdeef, K. J. Box, J. E. A. Comer, C. Hibbert and K. Y. Tam, pH-Metric logP 10. Determination of liposomal membrane-water partition coefficients of ionizable drugs, Pharm. Res., 1998, 15, 209–215 CrossRef CAS PubMed.
- H.-J. Lehmler, W. Xie, G. D. Bothun, P. M. Bummer and B. L. Knutson, Mixing of perfluorooctanesulfonic acid (PFOS) potassium salt with dipalmitoyl phosphatidylcholine (DPPC), Colloids Surf., B, 2006, 51, 25–29, DOI:10.1016/j.colsurfb.2006.05.013.
- OECD, T. No. 117: Partition Coefficient (n-octanol/water), HPLC Method. OECD Guidelines for the Testing of Chemicals, Section 1, OECD, Paris, France, 2004 Search PubMed.
- M. R. Ledbetter, S. Gutsell, G. Hodges, J. C. Madden, S. O'Connor and M. T. D. Cronin, Database of published retention factors for immobilized artificial membrane HPLC and an assessment of the effect of experimental variability, Environ. Toxicol. Chem., 2011, 30, 2701–2708 CrossRef CAS PubMed.
- M. R. Ledbetter, S. Gutsell, G. Hodges, J. C. Madden, P. H. Rowe and M. T. D. Cronin, Robustness of an immobilized artificial membrane high-performance liquid chromatography method for the determination of lipophilicity, J. Chem. Eng. Data, 2012, 57, 3696–3700 CrossRef CAS.
- M. R. Ledbetter, S. Gutsell, G. Hodges, S. O'Connor, J. C. Madden, P. H. Rowe and M. T. D. Cronin, Prediction of immobilised artificial membrane chromatography retention factors using theoretical molecular fragments and structural features, SAR QSAR Environ. Res., 2013, 24, 661–678 CrossRef CAS PubMed.
- Y.-X. Yang, Q. Zhang, Q. -. Li, Z. -. Xia, H. Chen, K. Zhou and F. -. Yang, pH-dependent surface electrostatic effects in retention on immobilized artificial membrane chromatography: Determination of the intrinsic phospholipid-water sorption coefficients of diverse analytes, J. Chromatogr. A, 2018, 1570, 172–182, DOI:10.1016/j.chroma.2018.07.081.
- J. Dolzonek, C. -. Cho, P. Stepnowski, M. Markiewicz, J. Thöming and S. Stolte, Membrane partitioning of ionic liquid cations, anions and ion pairs – Estimating the bioconcentration potential of organic ions, Environ. Pollut., 2017, 228, 378–389, DOI:10.1016/j.envpol.2017.04.079.
- F. Barbato, G. di Martino, L. Grumetto and M. I. La Rotonda, Can protonated [beta]-blockers interact with biomembranes stronger than neutral isolipophilic compounds?: A chromatographic study on three different phospholipid stationary phases (IAM-HPLC), Eur. J. Pharm. Sci., 2005, 25, 379–386 CrossRef CAS PubMed.
- L. Grumetto, C. Carpentiero, P. Di Vaio, F. Frecentese and F. Barbato, Lipophilic and polar interaction forces between acidic drugs and membrane phospholipids encoded in IAM-HPLC indexes: Their role in membrane partition and relationships with BBB permeation data, J. Pharm. Biomed. Anal., 2013, 75, 165–172 CrossRef CAS PubMed.
- C. Ottiger and H. Wunderli-Allenspach, Immobilized artificial membrane (IAM)-HPLC for partition studies of neutral and ionized acids and bases in comparison with the liposomal partition system, Pharm. Res., 1999, 16, 643–650 CrossRef CAS PubMed.
- S. Endo and K. U. Goss, Applications of polyparameter linear free energy relationships in environmental chemistry, Environ. Sci. Technol., 2014, 48, 12477–12491, DOI:10.1021/es503369t.
- W. H. J. Vaes, E. U. Ramos, H. J. M. Verhaar and J. L. M. Hermens, Acute toxicity of nonpolar versus polar narcosis: Is there a difference?, Environ. Toxicol. Chem., 1998, 17, 1380–1384 CrossRef CAS.
- A. Taillardat-Bertschinger, F. Barbato, M. T. Quercia, P. A. Carrupt, M. Reist, M. I. LaRotonda and B. Testa, Structural properties governing retention mechanisms on immobilized artificial membrane (IAM) HPLC columns, Helv. Chim. Acta, 2002, 85(2), 519–532, DOI:10.1002/1522-2675(200202)85:23.0.CO;2-Q.
- L. Sprunger, B. H. Blake-Taylor, A. Wairegi, W. E. Acree Jr and M. H. Abraham, Characterization of the retention behavior of organic and pharmaceutical drug molecules on an immobilized artificial membrane column with the Abraham model, J. Chromatogr. A, 2007, 1160, 235–245, DOI:10.1016/j.chroma.2007.05.051.
- K. S. Boone and D. M. Di Toro, Target site model: predicting Mode of Action and aquatic organism acute toxicity using Abraham parameters and feature-weighted k-nearest neighbors classification, Environ. Toxicol. Chem., 2019, 38, 375–386, DOI:10.1002/etc.4324.
- B. I. Escher and R. P. Schwarzenbach, Mechanistic studies on baseline toxicity and uncoupling of organic compounds as a basis for modeling effective membrane concentrations in aquatic organisms, Aquat. Sci., 2002, 64, 20–35 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Section S1 reports on the IAM assessment, Section S2 and Tables S1A and B on FHM chemical selection, Table S2 on chemical suppliers, Table S3 on incremental δIAM-MLW correction factors, Table S4 on IAM-HPLC measurement details, and Fig. S2 on solvent range extrapolations of KIAM. See DOI: https://doi.org/10.1039/d2em00391k |
| This journal is © The Royal Society of Chemistry 2023 |