Open Access Article
Open Access ArticleImpact of coagulation–ultrafiltration on long-term pipe biofilm dynamics in a full-scale chloraminated drinking water distribution system†
Kristjan
Pullerits
 ab,
Sandy
Chan‡
ab,
Jon
Ahlinder
c,
Alexander
Keucken
de,
Peter
Rådström
a and
Catherine J.
Paul
ab,
Sandy
Chan‡
ab,
Jon
Ahlinder
c,
Alexander
Keucken
de,
Peter
Rådström
a and
Catherine J.
Paul
 *ae
*ae
aApplied Microbiology, Department of Chemistry, Lund University, P.O. Box 124, SE-221 00 Lund, Sweden. E-mail: catherine.paul@tvrl.lth.se
bSweden Water Research AB, Ideon Science Park, Scheelevägen 15, SE-223 70 Lund, Sweden
cFOI, Swedish Defence Research Agency, Cementvägen 20, SE-906 21 Umeå, Sweden
dVatten & Miljö i Väst AB, P.O. Box 110, SE-311 22 Falkenberg, Sweden
eWater Resources Engineering, Department of Building and Environmental Technology, Lund University, P.O. Box 118, SE-221 00 Lund, Sweden
First published on 15th September 2020
Abstract
While pipe biofilms in DWDSs (drinking water distribution systems) are thought to affect the quality of distributed water, studies regarding the microbial processes are impeded by the difficulties in accessing biofilm undisturbed by DWDS maintenance. In this study, pipe sections were removed from a fully operational DWDS for biofilm sampling over two years and three months, and before and after start of ultrafiltration (UF) with coagulation treatment in the drinking water treatment plant (DWTP). Water (n = 31), surface biofilm (obtained by swabbing, n = 34) and deep pipe biofilm (obtained by scraping, n = 34) were analyzed with 16S rRNA gene amplicon sequencing; with flow cytometry, and chemical and natural organic matter (NOM) analysis as additional parameters for water quality. UF with coagulation decreased the total cell concentration in the DWDS bulk water from 6.0 × 105 ± 2.3 × 105 cells per ml to 6.0 × 103 ± 8.3 × 103 cells per ml, including fluctuations due to seasonal change, as well as decreasing most analyzed fractions of NOM. UF treatment of the water revealed that 75% ± 18% of the cells in the water originated from DWDS biofilm, confirmed by SourceTracker analysis, with the rest of the cells likely released from biofilm on DWTP storage tanks. Following UF start, the ASVs (amplicon sequence variants) in the deep pipe biofilm decreased, and Evenness and Shannon diversity indices decreased, reflecting the community's response to the new environment created by the altered water quality. The pipe biofilm community was dominated by ASVs classified as Nitrosomonadaceae, Nitrospira, Hyphomicrobium and Sphingomonas, with relative abundances ranging from 5–78%, and also included ASVs of genus Mycobacterium, genus Legionella and order Legionellales. This community composition, together with the observation that turnover of nitrogen compounds was unchanged by UF start, indicate that nitrification in the DWDS was localized to the pipe biofilm.
Water impactPipe biofilm in a full-scale DWDS did not result in any significant biofilm detachment following installation of coagulation and ultrafiltration in the treatment plant. The bacterial community did alter, with nitrifiers adapting to maintain turn-over of nitrogen compounds at pre-UF levels. Since removal of cells by UF didn't impact nitrification, this was localized to the biofilm in this system. |
1. Introduction
The surface of pipes in drinking water distribution systems (DWDSs) are colonized by microorganisms as biofilm. It has been estimated that >95% of the biomass in DWDSs is found in pipe biofilm and loose deposits1,2 and biofilm may include up to 108 bacteria per cm2.3 Pipe biofilm is of great importance and has been linked to the drinking water quality by altering aesthetics of the bulk water,4 enhancing nitrification in chloraminated systems,5 serving as a reservoir for persistent opportunistic pathogens such as Legionella and Mycobacteria6 and influencing corrosion.7Due to their location, sampling of pipe biofilm in a DWDS is challenging. To address this difficulty, studies have investigated different types of biofilms in the DWDS that are more accessible, including biofilm on water meters8 and coupons recovered from pilot- and full-scale systems,9,10 as well as estimations of the biofilm community by sampling water at different locations in the DWDS.11,12 Grab samples of pipe biofilm, obtained for research purposes or during pipe maintenance, have also been investigated.1,8,13–15
Maintaining disinfectant residuals such as chloramine to repress growth of problematic organisms in the DWDS is common practice. This use is however debatable since resistant microorganisms can be selected16 and, in combination with organic matter, cause undesirable disinfection byproducts to form.17 The use of chloramine generates ammonia in the DWDS by excess ammonia, from chloramine formation, and chloramine decay, which can stimulate growth of nitrifying bacteria and subsequent undesirable production of nitrite and nitrate.18 The ammonia is converted to nitrite by AOB (ammonia oxidizing bacteria) or AOA (ammonia oxidizing archaea); this nitrite can then support continued decay of monochloramine, and growth of NOB (nitrate oxidizing bacteria), producing both nitrate and growth of biofilm.18 Bacteria in chloraminated drinking water included a community with diverse approaches to nitrogen metabolism dominated by Nitrosomonas (AOB), and Nitrospira (NOB) and accompanied by heterotrophs such as Sphingomonas19 and these taxa were recently identified as actively conducting nitrification in diverse pipe biofilms.13
When a drinking water treatment plant (DWTP) in Varberg, Sweden, installed an ultrafiltration (UF) facility with two-stage filtration and in-line coagulation at the primary membrane stage,20 the new finished water (FW) contained virtually no bacteria, and less and different natural organic matter (NOM).11 The impact of this change on the bacterial community was examined by collecting water, swabbing the surface of the biofilm, and scraping to remove deep biofilm from multiple sections of pipe in series and excavated from the operational DWDS. In this drinking water system, we previously reported which bacteria were entering the water phase from the biofilm11 and how seasonal variations and residence time affect the water quality.21 In this study we examine how installation of the coagulation and ultrafiltration processes in the DWTP impacted the DWDS biofilm. The nature of the water quality in the DWDS was characterized in detail in order to determine if these changes could be linked to specific roles for the bacteria in the pipe biofilm, and we show that the bacterial biofilm alone can impact both biotic and abiotic aspects of water quality.
2. Methods
2.1 Study site and sampling
Samples were collected from the DWDS and DWTP (Kvarnagården) in Varberg, Sweden operated by Vatten & Miljö i Väst AB (VIVAB). Treatment consisted of pH adjustment, rapid sand filtration, storage tank 1 with addition of monochloramine, followed by storage tank 2 and 3 and UV disinfection (for more details see ref. 20). In November 2016, UF was added, hereafter defined as UF start, resulting in a treatment chain of rapid sand filtration, storage tank 1, coagulation and UF, pH adjustment, storage tank 2 with addition of monochloramine, storage tank 3 and UV disinfection (Fig. 1A). The monochloramine residual concentration in the finished water (FW) was between 0.13 and 0.21 mg L−1 as total chlorine. UF feed water was used for pH adjustment until March 2017 after which UF permeate water was used.11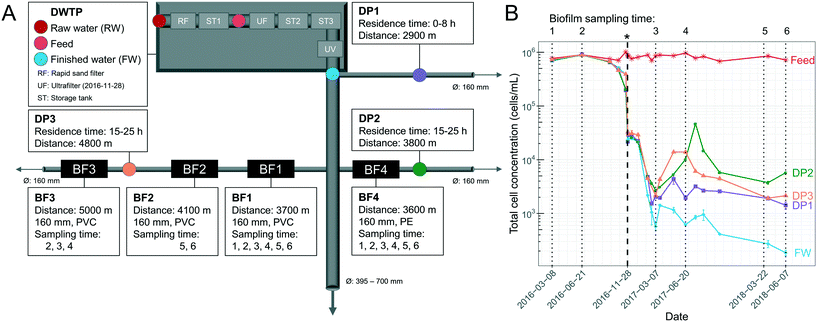 | ||
| Fig. 1 Overview of sampling locations and total cell concentrations (TCC) over time. (A) Schematic illustration of the drinking water treatment plant (DWTP) process steps, and distribution points (DPs) in the DWDS, indicating locations where water was sampled, as well as biofilm (BF) points, where pipe sections were excavated for biofilm sampling. Ø indicates pipe diameter, arrows indicate water flow direction, and pipe material was polyethylene (PE) or polyvinyl chloride (PVC). The ultrafilter (UF) started November 28, 2016. (B) TCC in the water, determined by flow cytometry, and indicating in time when biofilm was sampled (dotted lines) and the UF start (*, bold, dashed line). Error bars show standard deviations of technical triplicates. Before the UF start, raw-water (RW) is labelled “Feed”. Parts of this data are also shown in Chan et al. (2019)11 and Schleich et al. (2019).21 | ||
Pipe biofilm (BF) was sampled in four locations in the DWDS on six occasions over 27 months (Fig. 1A). Biofilms BF1, BF2 and BF3 were sampled from a 25–30 year-old PVC pipe, while BF4 was sampled from a 15 year-old PE pipe. Before UF start, BF1, BF2, BF3 and BF4 were sampled two (n = 8), none, one (n = 4) and two (n = 8) occasions, respectively. After UF start BF1, BF2, BF3 and BF4 were sampled four (n = 16), two (n = 8), two (n = 8) and four (n = 16) occasions, respectively. All sampled pipes had a diameter of 160 mm. Biofilm was collected from multiple adjacent sections, 5–10 m upstream of each previous sampling location; and with swabbing through 360° in the field and with duplicate cotton swabs (Fig. S1A†) for surface biofilm samples; a strategy supported by both Neu et al.22 and Liu et al.15 Following surface sampling, sections of at least 70 cm excavated pipes were sealed with parafilm and transported to the lab. Deep biofilm was then sampled from 50 cm of the bottom half of each section (total area: 0.13 m2) of excavated pipe (Fig. S1B†) within one hour, using a flame-sterilized custom-made metal scraper (Fig. S2†). Biofilm suspension was homogenized and collected using a 1 mL pipette with cut-off tips to avoid clogging the tips and suspensions were aliquoted into 1.5 mL Eppendorf tubes and duplicates used for sequencing. Cotton swabs and scraped biofilm were stored at −20 °C until DNA extraction.
Water was collected in the DWTP and from three distribution points (DPs; DP1, DP2 and DP3) in the DWDS (Fig. 1A) for conventional water quality analysis (n = 1, Eurofins Environment Testing Sweden AB, Linköping), NOM characterization (n = 1, LC-OCD-OND (liquid chromatography-organic carbon detection-organic nitrogen detection) at DOC-Labor (Germany)), flow cytometry (FCM) (n = 1, technical triplicates) and DNA sequencing (n = 1 and n = 2 for FW and DP3). FCM samples were collected in 15 mL Falcon tubes and when applicable, chlorine residuals were quenched with 1% (v/v) sodium thiosulfate (20 g L−1). Samples were stored on ice or at 4 °C for less than 12 hours before analysis. Water for sequencing was collected in sterilized borosilicate bottles (1 L before UF start, and feed/RW, 5 L after UF start), filtered onto 0.22 μm pore size filters (Merck, Germany) and stored at −20 °C until DNA extraction. Raw data of flow cytometry-, conventional- and LC OCD-analyses are available through figshare (https://doi.org/10.6084/m9.figshare.12555353).
2.2 FCM analysis
FCM analysis was conducted on raw water, feed to the UF, FW and three DPs according to Prest et al.23 and Gatza et al.24 using a BD Accuri C6 flow cytometer (Becton Dickinson, Belgium) with a 50 mW laser at a wavelength of 488 nm. Total cell concentrations (TCC) were measured by staining bacteria in water samples with SYBR Green I at a final concentration of 1× in triplicate, final volume of 500 μL. Intact cell concentrations (ICC) were measured by staining bacteria with SYBR Green I (1× final concentration, 500 μL final volume) and propidium iodide (0.3 mM final concentration, 500 μL final volume) in triplicate. Stained samples were incubated at 37 °C for 15 min and 50–250 μL was analyzed (to ensure >1000 cells per analysis) using a threshold of 500 arbitrary units in green fluorescence. FCS files were analyzed using FlowJo (Tree Star Inc., USA) and gated identically on green fluorescence (533 ± 30 nm) and red fluorescence (>670 nm). High nucleic acid (HNA) bacteria were measured by a cut-off in green fluorescence at >2 × 104 arbitrary units.23 The contribution of cells from the pipe biofilm (BFcells) was calculated using eqn (1) where cellsDP is the TCC at the distribution point and cellsFW the TCC in the Finished Water:| BFcells = cellsDP − cellsFW | (1) |
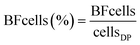 | (2) |
2.3 Bacterial community analysis
All DNA was extracted using the FastDNA Spin Kit for soil (MP Biomedicals, USA) following the manufacturer's instructions. Sodium phosphate was added to Lysing Matrix E tubes and either the cut-off tip of the cotton swabs; 120 μL of the scraped biofilm; or, filter papers cut into strips, were added to the tubes. Empty filters and unused cotton swabs were extracted as negative controls. DNA was stored at −20 °C. The V3-V4 region of the 16S rRNA gene was amplified with primers 341F (5-CCTACGGGNGGCWGCAG-3) and 785R (5-GACTACHVGGGTATCTAATCC-3).25 The PCR reaction (25 μL) included 12.6 μL MilliQ-water, 10 μL 5 Prime Hot MasterMix (Quantabio, USA), 0.4 μL (20 mg mL−1) bovine serum albumin, 0.5 μL (10 μM) forward and reverse primers and 1 μL template DNA or 5 μL template DNA in low biomass samples. PCR settings were 94 °C for 3 min and 35 cycles of 94 °C for 45 s, 50 °C for 1 min, 72 °C for 1.5 min and a final step of 72 °C for 10 min. Three PCR reactions were performed for each sample, pooled together, visualized by agarose gel, with DNA concentration quantified using a Qubit 2.0 dsDNA BR Assay Kit (Thermo Fisher Scientific, USA). Some primer dimers were observed following amplification from low biomass samples (FW, after UF start), although these were minimal in the amplicons from biofilm. Fifty ng of each pooled amplicon were pooled together and purified using Select-a-Size DNA clean and concentrator (Zymo Research, catalog #4080) and quantified using Qubit. Purified amplicon concentrations were adjusted to 2 nM, the library was denatured and diluted according to manufacturer's instruction (Illumina, USA), 10% PhiX was added to the sequencing run and amplicons sequenced using the MiSeq Reagent Kit v3 (600-cycles) (Illumina) according to the manufacturer's instructions.2.4 Bioinformatics and statistics
Raw sequencing reads were demultiplexed with deML26 and processed in QIIME2,27 version 2018.8.0. Reads were truncated at 280 and 215 bp for forward and reverse reads, respectively, and classified with the Greengenes database.28 Additional analyses were done with the Phyloseq package29 in R.30 Samples were visualized in a PCoA plot using Bray Curtis dissimilarity. Negative controls clustered with low biomass samples (FW, after UF start) having low read counts. Thus, samples with fewer than 2600 reads and all FW samples after UF start (eight negative controls and nine FW samples) were not included in further data analyses. Singletons and ASVs at a frequency less than 0.005% of the total number of reads31 and ASVs occurring in less than four samples were removed, resulting in 289 ASVs in 99 samples. Each ASV was identified with a number from 1–289 and letter to indicate its taxonomy when available (g = genus, f = family, o = order and c = class). Reads were normalized to relative abundances. R packages used for visualizations and calculations were; ggplot2,32 ggnomics,33 microbiome34 and VennDiagram.35 R script and plots from Kantor et al.36 were used as inspiration in some figures. Species diversity indices were calculated using Phyloseq (Shannon Index and observed ASVs) and the microbiome package (Pielou's measure) with non-normalized reads as suggested by McMurdie and Holmes37 since these indices are based on observations and not fractions. Differential abundance analysis was conducted with DESeq2,38 using non-normalized reads (Padjusted < 0.05). A Bayesian signature based microbial source tracking method (SourceTracker39) was utilized using default settings with the exception of a rarefaction depth set to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The source library included 68 biofilm communities, five raw-water communities and seven negative control communities (to account for possible contamination40).
000. The source library included 68 biofilm communities, five raw-water communities and seven negative control communities (to account for possible contamination40).
Pearson correlation (paired t-test) was calculated with the cor.test function, and the Welch t-test, were done in R. Locally weighted least squares (loess) regression was done using geom_smooth function in ggplot2 with the span parameter at 0.9. DNA sequences are available at the NCBI sequence read archive with the project accession number: PRJNA622401.
3. Results
3.1 Impact of UF start on water quality
Most water quality parameters responded predictably, based on expected changes introduced by UF start (Fig. S16† and Chan et al., (2019)11). The UF with coagulation removed 32% ± 4.4 of the dissolved organic carbon (DOC), from 2700 ± 300 ppb-C (feed) to 1800 ± 140 ppb-C (permeate) (Fig. S17†). Following UF start, DOC, chromatographic dissolved organic carbon (CDOC), chemical oxygen demand (COD-Mn), absorbance and color decreased in the DPs (Table 1). Throughout the study the UF feed water or raw water had a TCC of 820![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 90
000 ± 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (± standard deviation) (Fig. 1B). Before UF start, the finished water (FW) and water from the DWDS (DP1, DP2, DP3) had similar mean TCC, at 630
000 (± standard deviation) (Fig. 1B). Before UF start, the finished water (FW) and water from the DWDS (DP1, DP2, DP3) had similar mean TCC, at 630![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 200
000 ± 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL and 600
000 cells per mL and 600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 230
000 ± 230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL, with minimal contribution of cells from pipe biofilm (−0.13% ± 0.33; eqn (2)). After UF start, most cells in the FW originated from UF feed water used for pH regulation11 while after March 2017, UF permeate was used for pH regulation, and the TCC in the FW was 900 ± 560 cells per mL, likely originating from biofilm in DWTP storage tanks. Each DP had water with TCC of an overall mean of 6000 ± 8300 cells per mL (DP1: 2600 ± 1100, DP2: 9700 ± 13
000 cells per mL, with minimal contribution of cells from pipe biofilm (−0.13% ± 0.33; eqn (2)). After UF start, most cells in the FW originated from UF feed water used for pH regulation11 while after March 2017, UF permeate was used for pH regulation, and the TCC in the FW was 900 ± 560 cells per mL, likely originating from biofilm in DWTP storage tanks. Each DP had water with TCC of an overall mean of 6000 ± 8300 cells per mL (DP1: 2600 ± 1100, DP2: 9700 ± 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and DP3: 5800 ± 4500 cells per mL) and the contribution of cells from the DWDS pipe biofilm was an average of 75% ± 18 (64% ± 20, 81% ± 16 and 80% ± 14 at DP1, DP2 and DP3, respectively), reaching 98% at DP2 when the water temperature was elevated (Fig. S3†). While comparisons were limited, there was no difference in the number of cells entering the water, during the transition phase (sampling points 3 to 4; 4600 ± 4600 cells per mL; eqn (1)) compared to the later sampling times (sampling points 5 to 6; 2600 ± 1600 cells per mL) (P > 0.05, Welch t-test).
000 and DP3: 5800 ± 4500 cells per mL) and the contribution of cells from the DWDS pipe biofilm was an average of 75% ± 18 (64% ± 20, 81% ± 16 and 80% ± 14 at DP1, DP2 and DP3, respectively), reaching 98% at DP2 when the water temperature was elevated (Fig. S3†). While comparisons were limited, there was no difference in the number of cells entering the water, during the transition phase (sampling points 3 to 4; 4600 ± 4600 cells per mL; eqn (1)) compared to the later sampling times (sampling points 5 to 6; 2600 ± 1600 cells per mL) (P > 0.05, Welch t-test).
| DOC (ppb-C) | CDOC (ppb-C) | COD-Mn (mg O2 per L) | Absorbance 254 nm (A.U) | Color 410 nm (mg Pt per L) | |
|---|---|---|---|---|---|
| Before (n = 9) | 2500 ± 170 | 2400 ± 130 | 2.2 ± 0.12 | 0.41 ± 0.032 | 12 ± 1.6 |
| After (n = 24) | 1800 ± 170 | 1700 ± 120 | 1.3 ± 0.11 | 0.19 ± 0.048 | 0.38 ± 1.7 |
Observations of absolute counts of HNA cells (Fig. S4A†) and intact cells (Fig. S4B†) were similar to that of the TCC (Fig. 1B). From March 2017 to June 2018 the mean % HNA in the FW was 47% ± 10, and increased with distribution (65% ± 9, 71% ± 8 and 60% ± 6 in water from DP1, DP2 and DP3 respectively; Fig. S5A†). While the % HNA always increased at the DPs, the % ICC difference changed with water temperature (Fig. S5B†): % ICC difference decreased at the DPs from March 2017 to June/July 2017 and then increased, compared to the FW, until March 2018.
3.2 The pipe biofilm and water communities
The bacterial communities in water and biofilm were investigated with 16S rRNA gene amplicon sequencing. The number of observed biofilm ASVs were 20 ± 3.4, 19 ± 2.3, 42 ± 7.0 and 41 ± 7.8 at BF1, BF2, BF3 and BF4, respectively (Fig. S6†). Principal coordinates analysis (PCoA), based on Bray Curtis-dissimilarity, showed that communities at BF3 and BF4 were more similar than those at BF1 and BF2 (Fig. S7A†). The ASVs from the same pipe section (BF1, BF2, BF3), changed with distance from the treatment plant (Fig. 2B). The relative abundance of Nitrosomonadaceae decreased between BF1 to BF3, whereas that of Nitrospira increased with distance (Fig. 2 and S8†) which was also observed with differential abundance analysis (Fig. S9†). At BF1, ASVs classified as Nitrosomonadaceae, Hyphomicrobium and Sphingomonas dominated in relative abundance; at 64% ± 10, 23% ± 10 and 10% ± 12, respectively (Fig. 2A). The community at BF2 was also dominated by Nitrosomonadaceae, at 42% ± 6, while the ASV classified as Nitrospira increased to 31% ± 14, and relative abundances of Hyphomicrobium and Sphingomonas were comparable to those at BF1, at 17% ± 12 and 6% ± 2, respectively. Biofilms at BF3 and BF4 were dominated by the ASV classified as Nitrospira, at 78% ± 5 (BF3) and 75% ± 12 (BF4), relative abundance of Hyphomicrobium was less, at 5% ± 3 (BF3) and 6% ± 3 (BF4), and, relative abundance of Sphingomonas was similar to that at BF1 and BF2, at 6% ± 2 (BF3) and 6% ± 2 (BF4).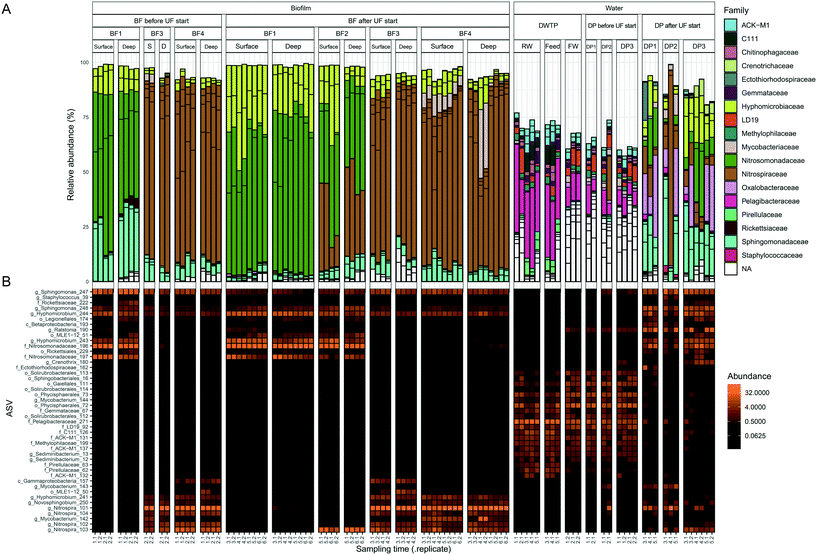 | ||
| Fig. 2 Bacterial community composition in biofilm and water. Samples are ordered in rows and grouped together based on sample type. ASVs present at >3% in one sample, representing 45 ASVs, are shown in both panels. Each column is one biological replicate. (A) Bar plot showing relative abundance at family level where each ASV is a bar separated by a black line. (B) Heatmap of ASVs with the most specified taxonomy when available, g = genus, f = family, o = order and c = class. Abbreviations; S: surface and D: deep. Sampling time indicates the date of sampling (Fig. 1B). | ||
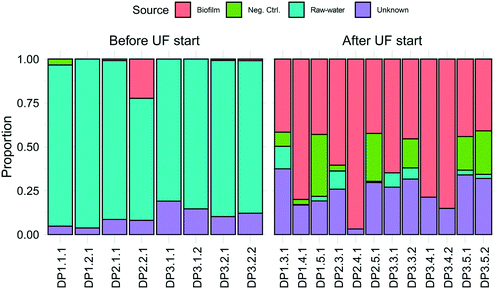
Observed ASVs in water from the DWTP were 79 ± 13, and 94 ± 13 at the DPs before UF start, decreasing to 45 ± 17 after UF start (Fig. S6†). Before UF start, ASVs in the water at the DPs were similar to the DWTP (Fig. S7B† and 2B), and distinct from those in biofilm. Following UF start, ASVs in the distributed water resembled those in the biofilm, including Nitrosomonadaceae, Nitrospira, Hyphomicrobium and Sphingomonas (Fig. 2B). Before UF start, the raw water contributed 86% ± 8 of the bacterial community at DPs (Fig. 3) while following UF start, this decreased to 4% ± 4 and the water community largely consisted of bacteria originating from biofilm (60% ± 20), estimated by SourceTracker. The taxa with highest assignment probability in the DPs within the biofilm source after UF start were Nitrosomonadaceae, Nitrospira, Hyphomicrobium and Sphingomonas (data not shown).
Mycobacterium_142, was present in 20/36 samples from BF3 and BF4 (Fig. S10†), ranging from 0.43% to 31%, while Mycobacterium_144 was present in the RW, feed and FW and water from the DPs before UF start. Following UF start, Mycobacterium_143 was observed in the water from DP1 and DP2. One ASV classified as the order Legionellales (Legionellales_174) was present in 47/68 biofilm samples (Fig. S11†), at 0.019% to 1.4% relative abundance, and was also present in 9/12 water samples from DPs after UF start (relative abundance 0.31% to 3.8%) (Fig. 2B). A second ASV classified to genus level as Legionella_175 was present in four samples from BF1.
The core biofilm community contained six ASVs present at all four BFs, regardless of sampling occasion: two ASVs classified as Sphingomonas, two ASVs classified as Hyphomicrobium, one ASV classified as Nitrosomonadaceae and one ASV classified as Rickettsiaceae (Fig. 4 and Table S1†). The biofilm communities at BF3 and BF4 shared the most ASVs, at 30.
 | ||
| Fig. 4 Identification of the core biofilm community. ASVs shared among the biofilm communities at different sampling points are shown in the different sections of the Venn diagram. ASV core communities for each biofilm represent ASVs >0.1% relative abundance in a minimum of 4 samples. The six ASVs shared by all four biofilms are shown with the most specified taxonomy available, g = genus and f = family. See ESI† Table S1 for all other shared ASVs. BF1: n = 24, BF2: n = 8, BF3: n = 12 and BF4: n = 24. | ||
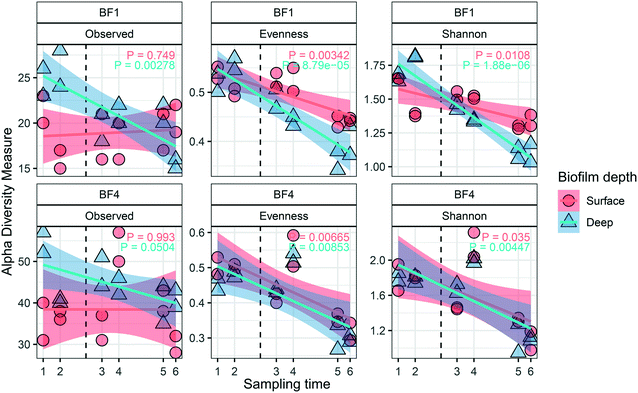
3.3 Dynamics in pipe biofilm community in response to UF start
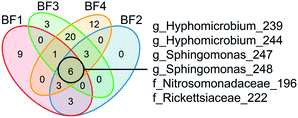
Dynamics in the biofilm community as a response to UF start were examined over time, and between surface and deep locations in the biofilm. Only BF1 and BF4 were sampled sufficiently often for statistically robust comparison. In deep biofilm at BF1 and BF4, observed ASVs decreased with time (Fig. 5, P < 0.05 and P = 0.0504, paired t-test), reaching numbers similar to those in their surface biofilm by the end of the study. The number of observed ASVs in the surface biofilm was similar over time at both locations (P > 0.7, paired t-test) while Shannon and Evenness indices decreased with time in surface and deep biofilm at both locations (P < 0.05, paired t-test). The biofilm communities at BF1 and BF4 responded in a similar way to UF start: changing from their original composition before UF start (Fig. S12†); moving through a transitional community shortly after UF start, to cluster together in a distinct post-UF community.Changes in relative abundance of ASVs over time (Fig. 6), and in surface and deep biofilm, defined the biofilm communities before UF (sampling time 1 and 2), during transition (sampling time 3 and 4), and after UF start (sampling time 5 and 6). At BF1, after UF start, Nitrosomonadaceae_196 increased in relative abundance, from 41% ± 8.5 to 65% ± 5.6 (surface and deep grouped together, P < 0.001, one-way ANOVA), whereas Nitrosomonadaceae_197 decreased in relative abundance from 17% ± 7.0 to 3.4% ± 1.8 (P < 0.05, one-way ANOVA). Hyphomicrobium_244 and Hyphomicrobium_243 both increased after UF start (7.0% ± 2.9 to 12% ± 2.4 and 4.4% ± 1.2 to 14% ± 3.7 (P < 0.05 and P < 0.05, one-way ANOVA), before and after UF start respectively) while Sphingomonas_247 decreased from a relative abundance of 26% ± 7.7 to 0.46% ± 0.33. Sphingomonas_248 had a relative abundance of 1.9% ± 0.97 before UF and decreased to 0.64 ± 0.23% in the transition state, before returning to 1.8% ± 0.81 relative abundance after UF start.
 | ||
| Fig. 6 Dynamics of the predominant relative abundant ASVs (>1.2% in one sample) in surface (n = 2 at each sampling time) and deep (n = 2 at each sampling time) biofilm over time. The change in relative abundance for individual ASVs from BF1 and BF4 are ordered from greatest relative abundance in top left panel, decreasing to bottom right. The vertical dashed line indicates the UF start. Shade areas in orange, white and green represent time periods defined as before UF (sampling time 1 and 2), transition (sampling time 3 and 4) and after UF (sampling time 5 and 6), respectively. ASVs are shown with the most specific taxonomy when available, g = genus, f = family, o = order and c = class. Blue and red lines show locally weighted least squares (loess) regression for each biofilm depth and the transparent areas are 95% confidence intervals for each loess regression. See ESI† Fig. S13 and S14 for full data sets. | ||
At BF4, and after UF start, Nitrospira_101 increased from 50% ± 6.4 to 76% ± 4.3 (P < 0.001, one way ANOVA), whereas three other ASVs in this genera decreased (Nitrospira_103 from 20% ± 6.4 to 2.3% ± 1.6; Nitrospira_102 from 5.6% ± 1.3 to 1.6% ± 0.87; and, Nitrospira_104 from 4.4% ± 1.1 to 1.0% ± 0.60; all P < 0.001, one-way ANOVA). Mycobacterium_142 increased from 0% to 3.3% ± 2.7, following UF start, reaching a maximum relative abundance of 31% ± 1.0 in the deep biofilm during transition (sampling time 4) with ASV Mycobacterium 141 showing a similar trend. As at BF1, Hyphomicrobium_244 increased in relative abundance at BF4 after UF start, from 1.4% ± 0.54 to 2.8% ± 1.4 (P < 0.05, one-way ANOVA). These observations were comparable to those obtained with differential abundance analysis (Fig. S15†).
3.4 Nitrification in the DWDS
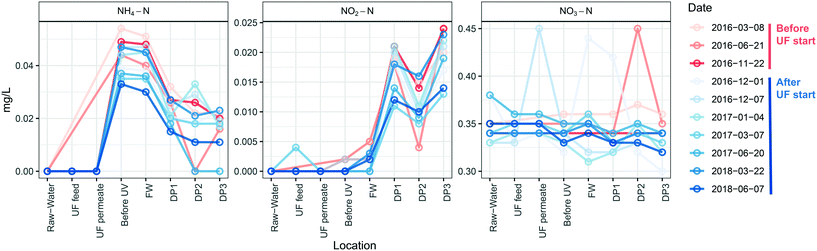
UF status did not impact concentrations of nitrogen species at different locations (Fig. 7) and therefore was not influenced by the number of cells in the distributed water. Ammonium nitrogen in the water decreased between the DWTP and the DPs by an average concentration of 0.025 ± 0.0045 mg L−1 before, and 0.021 ± 0.003 mg L−1 after UF start (P = 0.17, Welch t-test) with nitrite nitrogen showing an opposite trend and increasing as water passed through the DWDS, by 0.014 ± 0.0023 mg L−1 before, and 0.014 ± 0.0012 mg L−1 after the UF start (P = 0.87, Welch t-test). Nitrate nitrogen did not change during distribution of water before UF start (0.017 ± 0.017 mg L−1, P > 0.37, Welch t-test), and decreased only slightly after UF start (0.019 ± 0.017 mg L−1 (P < 0.05, Welch t-test). Ammonium nitrogen and nitrite nitrogen were below the detection limit for DPs further away from the DWTP than DP3 (data not shown).4. Discussion
This study describes the dynamics of the bacterial community in pipe biofilm and the associated drinking water over a 27 month period in a full scale DWDS. During this time the DWTP was upgraded to combine coagulation with UF treatment, and led to an immediate change in water quality, significantly altering the bacteria and NOM in the DWDS. By sampling multiple pipe sections, and with many fewer cells in the distributed water, the connection between these changes in water quality on the resident biofilm community over this period could be directly assessed and attributed to contact with the pipe biofilm, in both the immediate weeks following the UF start,11 and over the extended time period in the current study.4.1 Interaction of the biofilm with the distributed water
No large disturbance or sudden changes in the number of bacteria released from the biofilm were observed following UF start. The numbers of bacteria at the DPs were low, and changed mainly with season, echoing results in other studies of this DWDS21 with SourceTracker analysis confirming that the majority of these cells originated from the pipe biofilm, and not the DWTP. Significant impacts have been observed when the “balance of forces” required to maintain stability in the quality of distributed water have been altered by, for example, a change of source water, or physical flow characteristics.41 In the present study, while a great number of cells were removed from the water, with some impact on organic matter, other operational parameters such as chlorination41 were not changed: and hence the nature of the disturbance can explain why dramatic changes observed in other DWDS during transition were not observed.The identity of bacteria in the DPs was consistent with those released from biofilm directly after UF start,11 including more cells defined as HNA at the DPs than in the FW. While some cells may have increased in number during distribution due to growth, including growth of cells released from the biofilm, the doubling time of planktonic bacteria in oligotrophic drinking water has been estimated as 2.31 days (ref. 42) and together with the greatly reduced number of cells (which can also serve as a source of nutrition43 and the water temperature during the study period, the increase in cell number as the water was distributed can be largely attributed to those entering from the biofilm. The water community in both this and the previous study included abundant Nitrospira and Sphingomonadaceae and this agrees with the observation that these have previously been identified as HNA bacteria (>0.4 μm)44 as well as identification of these taxa as abundant members of the biofilms in this study. A previous description of this DWDS (also following UF start) noted a shift to LNA bacteria in the water during distribution, although this study examined water with significantly longer retention times (up to ∼170 h).21 As water is distributed, monochloramine is depleted, other chemical changes occur and there is increasing numbers of bacteria in this system released from the pipe biofilm in proportion to distance, suggesting that at more distal locations, the community in the biofilm is not similar to that observed in this study.
4.2 Changes in the biofilm community driven by nitrification
While pipe material and age in the branch of the DWDS where BF1–BF3 originated were identical, the observations in the biofilm community suggest that water residence time and changed water chemistry (due to upstream biofilm metabolism, see below) began to shift the community after 400 m, with a complete community change after 1300 m. Influence of hydraulic parameters on the community was also evident when comparing BF3 and BF4. Biofilms formed in contact with drinking water under lower flow rates have higher biomass, DNA concentration and total number of cells,45 suggesting that water flowing at a reduced rate at BF4 may have had contact with a similar amount of biomass, and subsequent nitrification, as that at BF3, and supporting similar communities at these locations. The pipe section with BF4 serves fewer consumers and thus while equally distant as BF1 from the DWTP, the water at BF4 likely had contact with the biofilm similar to that at BF3, and supported by the higher TCC in the water at DP2 than DP3. That flow is an influencing factor on the community at BF4 is further supported by observations that both nitrate concentration and the presence of Mycobacterium have been inversely correlated to flow.10As the presence of NOM facilitates decomposition of monochloramine into ammonia and nitrate;46 and increased concentrations of bacteria would remove monochloramine,47 UF start would increase the relative concentration of monochloramine in contact with the biofilm. In contrast, analysis of the nitrogen compounds showed no change at the different DPs following UF start, and nitrification was unaffected by removal of the cells in the water phase, strongly suggesting that nitrification takes place exclusively in the biofilm of this system. A significant role for the pipe biofilm in nitrogen transformation is supported by the observation that nitrifiers Nitrosomonadaceae and Nitrospira, together with Hyphomicrobium and Sphingomonas, accounted for 5–78% of the mean relative abundance in the biofilm. The role for chemoautotrophic nitrifiers is well established, metagenomics evidence has described participation of Sphingomonas in nitrogen cycling in drinking water.19 The core community here across all locations was comprised of only genera Hyphomicrobium and Sphingomonas and families Nitrosomonadaceae and Rickettsiaceae and coupled with the high relative abundance of Nitrosomonadaceae and Nitrospira, suggests that the biofilm in this study survives largely via ammonia and nitrite oxidation. The relative abundance of family Nitrosomonadaceae was higher in BF1, compared to BF3 in the same pipe section, and genus Nitrospira gradually predominated as distance, and water residence time, increased from the treatment plant, at sampling locations BF3 and BF4. As nitrification is the conversion of ammonium to nitrite by AOB (Nitrosomonadaceae), continued by nitrite to nitrate conversion by NOB (Nitrospira), it seems the biofilm converted the majority of the ammonium to nitrite in the distance from BF1 to BF3, and supported by high nitrite concentrations at DP3. Longer water residence time, such as that at BF4, would slow inflow of fresh ammonia, permitting complete nitrification to a greater extent at a shorter distance, and supporting the high relative abundance of Nitrospira at BF4. Only the biofilm at BF2 showed a mix of both AOB and NOB, showing that certain locations may support balanced mutualistic symbiosis48 and; communities at different locations will be determined by the delivery of nitrogen compounds in the flowing water, although the presence of commomox Nitrospira within the Nitrospira ASVs identified here cannot be ruled out.49 The water at DP2 downstream from BF4 showed lower nitrite concentrations than at DP3, preceeding the abundant Nitrospira at BF3, and ammonia and nitrite were below the detection limit in DPs with longer residence times (data not shown), further supporting that conversion of nitrite to nitrate relies on abundant Nitrospira in the biofilm.
Changes in the relative abundance and spatial distribution of ASVs for nitrifiers following UF start indicate that the biofilm community may buffer changes in relative ammonium and nitrate concentrations by changing the abundance of its members: after 1.5 years the changes in the community were apparent in ASVs, but not at family level (Fig. 2). This indicates that the biofilm maintained functionality50 while adapting to the new water quality.8,14 The communities at BF1 and BF4, and those in the water following UF start, decreased in evenness and Shannon diversity with time, indicating selection of bacteria (ASVs) for the new environment, and possibly reflecting the relative increase in monochloramine concentration as lower diversity has been proposed to correlate with higher disinfectant concentration.13 ASVs capable of oxidizing ammonia (Nitrosomonadaceae 197, BF1) decreased in relative abundance following UF start, however, other Nitrosomonadaceae taxa (196) increased at the same sampling location, reflecting adaptation, and demonstrating functional redundancy for conversion of ammonia to nitrite. Relative abundance of Nitrospira also responded to UF start, with decreases in Nitrospira ASVs 102![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 103 and 104 at sampling point BF4, and increase in Nitrospira ASV 101, with no change in nitrate concentrations. In a study using batch reactors, ammonia concentrations only decreased in the reactor containing particulate matter obtained from filtration of drinking water, while all reactors maintained similar TCC in the bulk water phase, supporting that nitrification may require cells attached to surfaces.51 Large populations of nitrifiers were identified in tropical chloraminated pipe biofilms, and nitrification activity in biofilm corresponded to identified taxa;13 and, nitrogen biotransformation in chloraminated DWDS and reservoirs has been linked to diverse populations including nitrifiers and heterotrophic bacteria.19,52 This suggests that to maintain monochloramine residual, drinking water producers need to consider nitrification occurring in the biofilm and that, regardless of biological content in the distributed water, disinfection concentration will continue to be determined by location within the DWDS.17
103 and 104 at sampling point BF4, and increase in Nitrospira ASV 101, with no change in nitrate concentrations. In a study using batch reactors, ammonia concentrations only decreased in the reactor containing particulate matter obtained from filtration of drinking water, while all reactors maintained similar TCC in the bulk water phase, supporting that nitrification may require cells attached to surfaces.51 Large populations of nitrifiers were identified in tropical chloraminated pipe biofilms, and nitrification activity in biofilm corresponded to identified taxa;13 and, nitrogen biotransformation in chloraminated DWDS and reservoirs has been linked to diverse populations including nitrifiers and heterotrophic bacteria.19,52 This suggests that to maintain monochloramine residual, drinking water producers need to consider nitrification occurring in the biofilm and that, regardless of biological content in the distributed water, disinfection concentration will continue to be determined by location within the DWDS.17
Sphingomonas and Hyphomicrobium have been observed in high relative abundances in pipe biofilm from real and model DWDS pipe materials exposed to a variety of residual disinfection types1,10,14,53 and have recently been linked to heterotrophic metabolism of nitrogen species.19 In the current study, relative abundance of Sphingomonas ASV 247 was most affected by UF start at BF1, where a decreased NOM content, together with higher flow than at BF4, may have pushed past the limits of oligotrophy tolerable for Sphingomonas. An increasing abundance of Sphingomonas has been associated with decreased disinfectant residual19 so this could also reflect a relative increase in monochloramine after UF start, and abundance of Sphingomonas in the water downstream of BF1 at DP3, and after UF start, suggests shedding of this ASV from BF1, due to the new environment. The changes in relative abundance of Sphinogomonas at BF1 may also be linked to the shift in ASV identity of the Nitrosomonas representative at BF1, as a metabolic link between these taxa, with Nitrosomonas producing tyrosine to be degraded by Sphingomonas, has been proposed.19 Depending on the metabolism of individual ASVs, changes in identity and relative abundance of Sphingomonas and Nitrosomonas at BF1 may also explain why ASVs for Hyphomicrobium increased following UF start, due to a lack of competition from Sphingomonas, and altered availability of C1 compounds and nitrite. The flexibility of Hyphomicrobium species to utilize C1-compounds likely gives them an advantage in oligotrophic environments like pipes, particularly those distributing ultrafiltered water, as well as via denitrification in monochloraminated DWDS.53
4.3 Impact of treatment change on taxa associated with opportunistic pathogens
ASVs classified as Mycobacterium and Legionellales were detected in biofilm and water, and family Rickettsiaceae was identified as a member of the core community. Individual species within these classifications (and others, including Sphingomonas) may harbor opportunistic pathogens. While this supports previous suggestions that biofilm could be a reservoir for these bacteria,6 it is important to consider that the approach used in this study is neither able to determine which bacteria are alive or dead, nor represent absolute quantification of any taxa resolved to species identity. Mycobacterium has been observed in chloraminated DWDS biofilms54,55 and bulk water in chlorinated and chloraminated DWDSs56 although risk of infection has not been associated with chloraminated DWDS.57Mycobacterium is known to be resistant to many disinfection methods,58 including ultraviolet (UV) disinfection,59 which the DWTP in this study is using. Legionella has been detected in pipe biofilms60 and also in bulk water both in chlorinated and chloraminated DWDSs,56 due in part to their ability to live within protozoa.61 Dynamics in Mycobacterium and Legionellales showed elevated relative abundances during summer (Fig. 6) likely due to increased temperatures. At BF1, the relative abundance of ASV Legionellales 174 was higher in the deep biofilm compared to the surface, but following UF start, the relative abundance of this ASV was similar in both surface and deep biofilm, and in lower abundance (relative to the deep biofilm). This was also observed at BF1, although this ASV was overall, less abundant (Fig. S14†). Rickettsiales (ASV 229) also decreased in relative abundance in the deep biofilm. This suggests that introduction of UF treatment may cause some members of these taxa to either disappear from the surface biofilm, or relocate from the deep to the surface and detection of these same ASVs in the water following UF start suggests they are released into the drinking water. While the same was not observed for Mycobacterium, these changes in location may not be specific to taxa including opportunistic pathogens, but may be part of a general remodeling of the deep biofilm following UF start. Overall, the number of observed ASVs decreased with time in the deep biofilm, reaching the same levels of abundance as in the surface biofilm, in both BF1 and BF4 (Fig. 4).5. Future outlook and conclusions
While biostability is currently defined as maintaining a defined microbial water quality until the drinking water reaches consumers,62 and “biologically stable water does not promote the growth of microorganisms during its distribution”,63 the current study, and others, suggest that this definition must be reconsidered. The concept of biostability could be revised to reflect natural dynamics in the microbial community which do not compromise public health.64 This new definition may be especially appropriate when there are few cells in the water and a diverse pipe biofilm, influenced by a number of operational parameters, governs the microbial water quality throughout the DWDS. This definition is needed in the context of DWTP with advanced treatment chains, such as observed in the present study, as well as in the context of water re-use, where produced water can be virtually cell-free. This will require significant efforts to understand how to monitor microbial dynamics with moving baselines influenced by season, residence times and other variables, while ensuring no impact on consumer satisfaction and safety.In addition, this study showed:
• UF with coagulation decreased the total cell concentration in the DWDS bulk water and altered the bacterial community composition in the pipe biofilm, but this change did not result in any significant biofilm detachment.
• Disinfection using monochloramine supports a high relative abundance of nitrification bacteria in this pipe biofilm, including Nitrosomonadaceae and Nitrospira. These taxa adapted to the change instigated by UF start, while retaining function, demonstrating functional redundancy in situ by a diverse nitrifier community.
• The majority of nitrification in this DWDS was performed by the pipe biofilm both before and after installation of UF.
• Taxa that include possible opportunistic pathogens were detected in the pipe biofilm and their location and prevalence in the biofilm may be influenced by change to distribution of ultrafiltered water, however additional quantitative investigations are required to assess any changes in risk to the consumer.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors want to thank Jennie Lindgren and Moshe Habagil for assistance in sampling, Emelie Salomonsson for assistance with sequencing, and Marcel Ollila who designed and made the biofilm scraper. The study was funded by Sweden Water Research AB, DRICKS center for drinking water research, Swedish Water and Wastewater Association (SVU), public joint-stock utility Vatten & Miljö i Väst AB (VIVAB).References
- G. Liu, G. L. Bakker, S. Li, J. H. G. Vreeburg, J. Q. J. C. Verberk, G. J. Medema, W. T. Liu and J. C. Van Dijk, Pyrosequencing reveals bacterial communities in unchlorinated drinking water distribution system: An integral study of bulk water, suspended solids, loose deposits, and pipe wall biofilm, Environ. Sci. Technol., 2014, 48, 5467–5476 Search PubMed.
- H. C. Flemming, S. L. Percival and J. T. Walker, Contamination potential of biofilms in water distribution systems, Water Sci. Technol.: Water Supply, 2002, 2, 271–280 Search PubMed.
- C. R. Proctor and F. Hammes, Drinking water microbiology — from measurement to management, Curr. Opin. Biotechnol., 2015, 33, 87–94 Search PubMed.
- K. Fish, A. M. Osborn and J. B. Boxall, Biofilm structures (EPS and bacterial communities) in drinking water distribution systems are conditioned by hydraulics and influence discolouration, Sci. Total Environ., 2017, 593–594, 571–580 Search PubMed.
- B. A. Carrico, F. A. Digiano, N. G. Love, P. Vikesland, K. Chandran, M. Fiss and A. Zaklikowski, Effectiveness of switching disinfectants for nitrification control, J. - Am. Water Works Assoc., 2014, 100(10), 104–115 Search PubMed.
- J. Wingender and H. C. Flemming, Biofilms in drinking water and their role as reservoir for pathogens, Int. J. Hyg. Environ. Health, 2011, 214, 417–423 Search PubMed.
- N. Kip and J. A. van Veen, The dual role of microbes in corrosion, ISME J., 2015, 9, 542–551 Search PubMed.
- K. Lührig, B. Canbäck, C. J. Paul, T. Johansson, K. M. Persson and P. Rådström, Bacterial Community Analysis of Drinking Water Biofilms in Southern Sweden, Microbes Environ., 2015, 30, 99–107 Search PubMed.
- P. Deines, R. Sekar, P. S. Husband, J. B. Boxall, A. M. Osborn and C. A. Biggs, A new coupon design for simultaneous analysis of in situ microbial biofilm formation and community structure in drinking water distribution systems, Appl. Microbiol. Biotechnol., 2010, 87, 749–756 Search PubMed.
- I. Douterelo, M. Jackson, C. Solomon and J. Boxall, Spatial and temporal analogies in microbial communities in natural drinking water biofilms, Sci. Total Environ., 2017, 581–582, 277–288 Search PubMed.
- S. Chan, K. Pullerits, A. Keucken, K. M. Persson, C. J. Paul and P. Rådström, Bacterial release from pipe biofilm in a full-scale drinking water distribution system, npj Biofilms Microbiomes, 2019, 5, 9 Search PubMed.
- I. Douterelo, B. E. Dutilh, K. Arkhipova, C. Calero and S. Husband, Microbial diversity, ecological networks and functional traits associated to materials used in drinking water distribution systems, Water Res., 2020, 173, 115586 Search PubMed.
- M. C. Cruz, Y. Woo, H. C. Flemming and S. Wuertz, Nitrifying niche differentiation in biofilms from full-scale chloraminated drinking water distribution system, Water Res., 2020, 176, 115738 Search PubMed.
- M. Waak, R. M. Hozalski, C. Hallé and T. M. Lapara, Comparison of the microbiomes of two drinking water distribution systems - With and without residual chloramine disinfection, Microbiome, 2019, 7(1), 1–14 Search PubMed.
- G. Liu, Y. Zhang, X. Liu, F. Hammes, W. Liu, G. Medema and P. Wessels, 360-Degree Distribution of Bio fi lm Quantity and Community in an Operational Unchlorinated Drinking Water Distribution Pipe, Environ. Sci. Technol., 2020, 54(9), 5619–5628 Search PubMed.
- T. H. Chiao, T. M. Clancy, A. Pinto, C. Xi and L. Raskin, Differential resistance of drinking water bacterial populations to monochloramine disinfection, Environ. Sci. Technol., 2014, 48, 4038–4047 Search PubMed.
- R. A. Li, J. A. McDonald, A. Sathasivan and S. J. Khan, Disinfectant residual stability leading to disinfectant decay and by-product formation in drinking water distribution systems: A systematic review, Water Res., 2019, 153, 335–348 Search PubMed.
- J. M. Regan, G. W. Harrington, H. Baribeau, R. De Leon and D. R. Noguera, Diversity of nitrifying bacteria in full-scale chloraminated distribution systems, Water Res., 2003, 37, 197–205 Search PubMed.
- S. C. Potgieter, Z. Dai, S. N. Venter, M. Sigudu and A. J. Pinto, Microbial Nitrogen Metabolism in Chloraminated Drinking Water Reservoirs, mSphere, 2020, 5(2) DOI:10.1128/mSphere.00274-20.
- A. Keucken, G. Heinicke, K. M. Persson and S. J. Köhler, Combined coagulation and ultrafiltration process to counteract increasing NOM in brown surface water, Water, 2017, 9(9), 697 Search PubMed.
- C. Schleich, S. Chan, K. Pullerits, M. D. Besmer, C. J. Paul, P. Rådström and A. Keucken, Mapping Dynamics of Bacterial Communities in a Full-Scale Drinking Water Distribution System Using Flow Cytometry, Water, 2019, 11, 2137 Search PubMed.
- L. Neu, C. R. Proctor, J. C. Walser and F. Hammes, Small-scale heterogeneity in drinking water biofilms, Front. Microbiol., 2019, 10, 1–14 Search PubMed.
- E. I. Prest, F. Hammes, S. Kötzsch, M. C. M. van Loosdrecht and J. S. Vrouwenvelder, Monitoring microbiological changes in drinking water systems using a fast and reproducible flow cytometric method, Water Res., 2013, 47, 7131–7142 Search PubMed.
- E. Gatza, F. Hammes and E. Prest, Assessing Water Quality with the BD Accuri™ C6 Flow Cytometer White Paper, BD Biosciences, 2013 Search PubMed.
- A. Klindworth, E. Pruesse, T. Schweer, J. Peplies, C. Quast, M. Horn and F. O. Glockner, Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies, Nucleic Acids Res., 2013, 41, e1 Search PubMed.
- G. Renaud, U. Stenzel, T. Maricic, V. Wiebe and J. Kelso, deML: robust demultiplexing of Illumina sequences using a likelihood-based approach, Bioinformatics, 2015, 31, 770–772 Search PubMed.
- E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, Y. Bai, J. E. Bisanz, K. Bittinger, A. Brejnrod, C. J. Brislawn, C. T. Brown, B. J. Callahan, A. M. Caraballo-Rodríguez, J. Chase, E. K. Cope, R. Da Silva, C. Diener, P. C. Dorrestein, G. M. Douglas, D. M. Durall, C. Duvallet, C. F. Edwardson, M. Ernst, M. Estaki, J. Fouquier, J. M. Gauglitz, S. M. Gibbons, D. L. Gibson, A. Gonzalez, K. Gorlick, J. Guo, B. Hillmann, S. Holmes, H. Holste, C. Huttenhower, G. A. Huttley, S. Janssen, A. K. Jarmusch, L. Jiang, B. D. Kaehler, K. Bin Kang, C. R. Keefe, P. Keim, S. T. Kelley, D. Knights, I. Koester, T. Kosciolek, J. Kreps, M. G. I. Langille, J. Lee, R. Ley, Y.-X. Liu, E. Loftfield, C. Lozupone, M. Maher, C. Marotz, B. D. Martin, D. McDonald, L. J. McIver, A. V. Melnik, J. L. Metcalf, S. C. Morgan, J. T. Morton, A. T. Naimey, J. A. Navas-Molina, L. F. Nothias, S. B. Orchanian, T. Pearson, S. L. Peoples, D. Petras, M. L. Preuss, E. Pruesse, L. B. Rasmussen, A. Rivers, M. S. Robeson, P. Rosenthal, N. Segata, M. Shaffer, A. Shiffer, R. Sinha, S. J. Song, J. R. Spear, A. D. Swafford, L. R. Thompson, P. J. Torres, P. Trinh, A. Tripathi, P. J. Turnbaugh, S. Ul-Hasan, J. J. J. van der Hooft, F. Vargas, Y. Vázquez-Baeza, E. Vogtmann, M. von Hippel, W. Walters, Y. Wan, M. Wang, J. Warren, K. C. Weber, C. H. D. Williamson, A. D. Willis, Z. Z. Xu, J. R. Zaneveld, Y. Zhang, Q. Zhu, R. Knight and J. G. Caporaso, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, 37, 852–857 Search PubMed.
- T. Z. Desantis, P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu and G. L. Andersen, Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB, Appl. Environ. Microbiol., 2006, 72, 5069–5072 Search PubMed.
- P. J. McMurdie and S. Holmes, phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data, PLoS One, 2013, 8, e61217 Search PubMed.
- R Core Team, 2018 Search PubMed.
- N. A. Bokulich, S. Subramanian, J. J. Faith, D. Gevers, J. I. Gordon, R. Knight, D. A. Mills and J. G. Caporaso, Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing, Nat. Methods, 2013, 10, 57–59 Search PubMed.
- H. Wickham, ggplot2: Elegant Graphics for Data Analysis, Springer-Verlag New York, 2016 Search PubMed.
- T. van den Brand, ggnomics: ggnomics. R package version 0.1.1, 2019 Search PubMed.
- L. Lahti and S. Shetty, microbiome R package, 2019 Search PubMed.
- H. Chen, VennDiagram: Generate High-Resolution Venn and Euler Plots, R package version 1.6.20 Search PubMed.
- R. S. Kantor, S. E. Miller and K. L. Nelson, The water microbiome through a pilot scale advanced treatment facility for direct potable reuse, Front. Microbiol., 2019, 10, 1–15 Search PubMed.
- P. J. McMurdie and S. Holmes, Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible, PLoS Comput. Biol., 2014, 10, e1003531 Search PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15, 550 Search PubMed.
- D. Knights, J. Kuczynski, E. S. Charlson, J. Zaneveld, M. C. Mozer, R. G. Collman, F. D. Bushman, R. Knight and S. T. Kelley, Bayesian community-wide culture-independent microbial source tracking, Nat. Methods, 2011, 8, 761–765 Search PubMed.
- M. Hägglund, S. Bäckman, A. Macellaro, P. Lindgren, E. Borgmästars, K. Jacobsson, R. Dryselius, P. Stenberg, A. Sjödin, M. Forsman and J. Ahlinder, Accounting for Bacterial Overlap Between Raw Water Communities and Contaminating Sources Improves the Accuracy of Signature-Based Microbial Source Tracking, Front. Microbiol., 2018, 9, 2364 Search PubMed.
- G. Liu, Y. Zhang, W. J. Knibbe, C. Feng, W. Liu, G. Medema and W. van der Meer, Potential impacts of changing supply-water quality on drinking water distribution: A review, Water Res., 2017, 116, 135–148 Search PubMed.
- R. Boe-Hansen, H. J. Albrechtsen, E. Arvin and C. Jørgensen, Bulk water phase and biofilm growth in drinking water at low nutrient conditions, Water Res., 2002, 36, 4477–4486 Search PubMed.
- I. Chatzigiannidou, R. Props and N. Boon, Drinking water bacterial communities exhibit specific and selective necrotrophic growth, npj Clean Water, 2018, 1, 22 Search PubMed.
- C. R. Proctor, M. D. Besmer, T. Langenegger, K. Beck, J.-C. Walser, M. Ackermann, H. Bürgmann and F. Hammes, Phylogenetic clustering of small low nucleic acid-content bacteria across diverse freshwater ecosystems, ISME J., 2018, 12(5), 1344–1359 Search PubMed.
- M. W. Cowle, G. Webster, A. O. Babatunde, B. N. Bockelmann-Evans and A. J. Weightman, Impact of flow hydrodynamics and pipe material properties on biofilm development within drinking water systems, Environ. Technol., 2019, 0, 1–13 Search PubMed.
- P. J. Vikesland, K. Ozekin and R. L. Valentine, Effect of natural organic matter on monochloramine decomposition: Pathway elucidation through the use of mass and redox balances, Environ. Sci. Technol., 1998, 32, 1409–1416 Search PubMed.
- A. Wilczak, L. L. Hoover and H. Hubert Lai, Effects of treatment changes on chloramine demand and decay, J. - Am. Water Works Assoc., 2003, 95, 94–106 Search PubMed.
- H. Daims, S. Lücker and M. Wagner, A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria, Trends Microbiol., 2016, 24, 699–712 Search PubMed.
- Y. Wang, L. Ma, Y. Mao, X. Jiang, Y. Xia, K. Yu, B. Li and T. Zhang, Comammox in drinking water systems, Water Res., 2017, 116, 332–341 Search PubMed.
- S. D. Allison and J. B. H. Martiny, Resistance, resilience, and redundancy in microbial communities, In the Light of Evolution, 2009, vol. 2, pp. 149–166 Search PubMed.
- E. Sawade, P. Monis, D. Cook and M. Drikas, Is nitrification the only cause of microbiologically induced chloramine decay?, Water Res., 2016, 88, 904–911 Search PubMed.
- S. Potgieter, A. Pinto, M. Sigudu, H. du Preez, E. Ncube and S. Venter, Long-term spatial and temporal microbial community dynamics in a large-scale drinking water distribution system with multiple disinfectant regimes, Water Res., 2018, 139, 406–419 Search PubMed.
- R. Liu, J. Zhu, Z. Yu, D. R. Joshi, H. Zhang, W. Lin and M. Yang, Molecular analysis of long-term biofilm formation on PVC and cast iron surfaces in drinking water distribution system, J. Environ. Sci., 2014, 26, 865–874 Search PubMed.
- M. Waak, T. M. Lapara, C. Hallé and R. M. Hozalski, Nontuberculous Mycobacteria in Two Drinking Water Distribution Systems and the Role of Residual Disinfection, Environ. Sci. Technol., 2019, 53, 8563–8573 Search PubMed.
- C. K. Gomez-Smith, T. M. Lapara and R. M. Hozalski, Sulfate reducing bacteria and mycobacteria dominate the biofilm communities in a chloraminated drinking water distribution system, Environ. Sci. Technol., 2015, 49, 8432–8440 Search PubMed.
- M. J. Donohue, S. Vesper, J. Mistry and J. M. Donohue, Impact of Chlorine and Chloramine on the Detection and Quantification of Legionella pneumophila and Mycobacterium Species, Appl. Environ. Microbiol., 2019, 85, 1–11 Search PubMed.
- N. Kotlarz, L. Raskin, M. Zimbric, J. Errickson, J. J. LiPuma and L. J. Caverly, Retrospective Analysis of Nontuberculous Mycobacterial Infection and Monochloramine Disinfection of Municipal Drinking Water in Michigan, mSphere, 2019, 4, e00160 Search PubMed.
- R. H. Taylor, J. O. Falkinham Iii, C. D. Norton and M. W. Lechevallier, Chlorine, Chloramine, Chlorine Dioxide, and Ozone Susceptibility of Mycobacterium avium, Appl. Environ. Microbiol., 2000, 66(4), 1702–1705 Search PubMed.
- K. Pullerits, J. Ahlinder, L. Holmer, E. Salomonsson, C. Öhrman, K. Jacobsson, R. Dryselius, M. Forsman, C. J. Paul and P. Rådström, Impact of UV irradiation at full scale on bacterial communities in drinking water, npj Clean Water, 5, 3, 1–10 Search PubMed.
- M. Waak, T. M. LaPara, C. Hallé and R. M. Hozalski, Occurrence of Legionella spp. in Water-Main Biofilms from Two Drinking Water Distribution Systems, Environ. Sci. Technol., 2018, 52, 7630–7639 Search PubMed.
- M. Steinert, U. Hentschel and J. Hacker, Legionella pneumophila: An aquatic microbe goes astray, FEMS Microbiol. Rev., 2002, 26, 149–162 Search PubMed.
- E. Prest, F. Hammes, M. C. M. van Loosdrecht and J. S. Vrouwenvelder, Front. Microbiol., 2016, 7, 45 Search PubMed.
- B. E. Rittmann and V. L. Snoeyink, Achieving biologically stable drinking water, J. - Am. Water Works Assoc., 1984, 76, 106–114 Search PubMed.
- J. El-Chakhtoura, E. Prest, P. Saikaly, M. Van Loosdrecht, F. Hammes and H. Vrouwenvelder, Dynamics of bacterial communities before and after distribution in a full-scale drinking water network, Water Res., 2015, 74, 180–190 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ew00622j |
| ‡ Present address: Sydvatten AB, Hyllie Stationstorg 21, SE-215 32 Malmö, Sweden. |
| This journal is © The Royal Society of Chemistry 2020 |