 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microcalorimetry and fluorescence show stable peptide nucleic acid (PNA) duplexes in high organic content solvent mixtures†
Samuel
Núñez-Pertíñez
 ,
Thomas R.
Wilks
,
Thomas R.
Wilks
 * and
Rachel K.
O'Reilly
* and
Rachel K.
O'Reilly
 *
*
School of Chemistry, University of Birmingham, Edgbaston, Birmingham, B15 2TT, UK. E-mail: T.R.Wilks@bham.ac.uk; R.OReilly@bham.ac.uk
First published on 6th August 2019
Abstract
The selectivity of nucleic acid hybridisation can be exploited to template chemical reactions, enabling materials discovery by chemical evolution. However, to date the range of reactions that can be used has been limited to those that are compatible with aqueous media, since the addition of organic co-solvents can have a large impact on the stability of nucleic acid duplexes. Peptide nucleic acids (PNAs) are promising in this regard because previous studies have suggested they may be stable as duplexes in high organic content solvent mixtures. Here, we use micro-differential scanning calorimetry (micro-DSC) to confirm for the first time that double-stranded PNA (dsPNA) is stable in N,N-dimethylformamide (DMF)/water mixtures up to 95 vol% DMF. Using fluorescence, we corroborate these results and show that the isothermal annealing of PNA in high DMF content solution is also rapid. These findings suggest that PNA could enable the use of a range of water-sensitive chemistries in nucleic acid templating applications.
Nucleic acids have the remarkable ability to recognise complementary strands. This selective hybridisation has been exploited to template the synthesis of oligomers1,2 and for the production of combinatorial libraries3,4 in an analogous manner to the genetically controlled biosynthesis of proteins.5 Interestingly, this approach to organic synthesis has the potential to perform directed chemical evolution: whole libraries of DNA-tagged molecules can be screened in one pot, allowing the selection and identification of those that perform a particular function, such as catalysis or enzyme inhibition.6–10
One major limitation of this nucleic acid-templated synthesis (NATS) is the incompatibility of many chemical reactions with aqueous solution. Aqueous solution is required because nucleic acid duplexes are greatly destabilised by the addition of organic cosolvents.11 Overcoming this limitation would allow the use of a wider variety of chemical transformations, increasing the scope of NATS technology.
Peptide nucleic acids (PNAs) are DNA analogues in which the phosphate and sugar backbone is replaced by N-(2-aminoethyl)glycine linkages.12 PNAs have been used as molecular tags to encode the identity of molecules in combinatorial chemical libraries,13 and as templates in several examples of templated synthesis.14–16 PNAs are capable of selective recognition through hybridisation and, in contrast to native DNA and RNA, are soluble in a range of organic solvents such as 1,4-dioxane, dimethylformamide (DMF), N-methylpyrrolidone (NMP) and dimethyl sulfoxide (DMSO).15 Previous studies have also shown that double-stranded PNA (dsPNA) is stable in up to 70 vol% DMF/water mixtures.17 They are therefore promising candidates to allow extension of NATS into high organic content mixtures, or even pure organic solvents.
The duplex melting temperature (Tm) is the most commonly used proxy of the thermal stability of dsPNA, and is defined as the temperature at which half the possible duplexes have formed. There are three main techniques available to measure the Tm of nucleic acid duplexes (see Table 1): ultraviolet (UV) absorbance, fluorescence and calorimetry. The formation of dsPNA in hydrated organic solvent was studied by Sen and Nielsen through UV absorbance, and confirmed duplex stability up to 5018 and 70 vol%17 DMF. However, study of higher organic content mixtures was not possible because of DMF's interference with the UV absorbance measurements. This limitation also holds true for a wide range of organic solvents with high UV cut-offs. This problem can be avoided by the use of fluorescence to measure Tm, by tagging one nucleic acid strand with a fluorophore and the other with either a fluorescence quencher or Förster resonance energy transfer (FRET) pair and following the change in fluorescence as hybridisation brings the two into close proximity.19 However, this approach has proven effects on the Tm.20
| UV absorbance | Fluorescence | Calorimetry | |
|---|---|---|---|
| Sensitivity | Moderate | High | Low |
| Chemical modification | None required | Required | None required |
| Solvent range | Narrow | Wide | Wide |
Micro-differential scanning calorimetry (micro-DSC), meanwhile, has proved a useful tool to determine the thermal stability of macromolecular systems,21 allowing quantification of small heat flow changes in dilute solutions. In contrast to fluorescence, it does not require the chemical modification of the macromolecules and unlike UV absorbance is compatible with a wide range of solvents – the only requirement is that the process being studied involves heat exchange.
Here, micro-DSC has been used to study the thermal stability of dsPNA across the whole range of aqueous–organic solvent compositions without any post-synthetic modification of the strands. DMF was chosen as the organic solvent due to the reported stability of dsPNA in DMF/water mixtures,17,18 its high boiling point, miscibility with water and the fact that it would be a good solvent for PNA-templated synthesis in future applications. The findings were further corroborated by fluorescence measurements, based on fluorescence quenching, which also confirmed the rapid isothermal annealing of dsPNA in high organic content solvent mixtures.
In order to establish how comparable the previously reported values for the Tm of dsPNA in DMF determined by UV absorbance17 would be with those measured by micro-DSC, a comparative study was performed. The Tms of 10-mer dsDNA, PNA·DNA and dsPNA duplexes with the same base sequence, that had previously shown good stability in DMF/water mixtures up to 70 vol% DMF, were determined in aqueous solution using UV absorbance, fluorescence and micro-DSC, and good agreement was found between the three techniques (Fig. 1). Measurement of the Tm of the dsPNA duplex by fluorescence was not possible as we were unable to find a synthetically accessible dye/quencher pair that gave sufficient sensitivity in aqueous solution. Nevertheless, UV absorbance and micro-DSC identified the following stability trend in aqueous solution: dsPNA ≈ PNA·DNA > dsDNA. There was a maximum discrepancy between the Tms measured by different techniques of approximately 7 °C in dsDNA, 8 °C in PNA·DNA and 5 °C in dsPNA. We hypothesise that these differences arose from the different accuracies in the control of the heating flow for each instrumental set up. The higher concentrations required for micro-DSC measurements (due to the lower sensitivity of this technique) may also have played a role. For fluorescence measurements, the conjugation of a fluorophore and quencher may have had an additional impact on the Tm.20 Despite the differences in Tm determined through different techniques, micro-DSC and UV absorbance showed the same stability trend across the dsPNA, PNA·DNA, and dsDNA series. It was therefore concluded that micro-DSC would provide results that could be meaningfully compared with previously reported data obtained using UV absorbance.
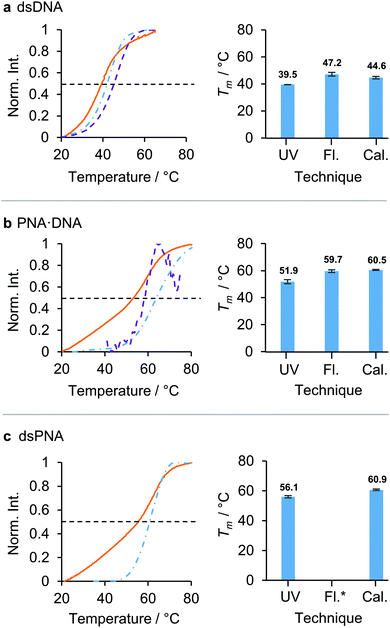 | ||
| Fig. 1 Comparison of three techniques for the measurement of the Tm of nucleic acid duplexes in aqueous solution: UV absorbance (UV, orange solid line), fluorescence (Fl., violet dashed line) and calorimetry (Cal., blue dashed-dotted line). The black dashed line indicates the 50% maximum intensity from which the Tms were calculated. The duplexes studied were: (a) dsDNA, (b) PNA·DNA, (c) dsPNA. The calculated melting temperatures for each duplex are displayed in the bar charts in the right hand column. *Fluorescence was not measurable. The normalised intensities (norm. int.) corresponded to: UV absorbance at 260 nm, fluorescence emission measured in counts and the numeric integral of the excess heat capacity. (dsDNA was excited at 495 nm and the fluorescence emission was measured at 512 nm. PNA·DNA was excited at 290 nm and the fluorescence emission was measured at 340 nm. Sigmoidal baselines were fitted and subtracted to produce the molar excess heat calorimetry signals.26 Sequences: [10]-DNA/PNA1 5′/N – TCA CTA GAT G – C/3′ [10]-DNA/PNA2 5′/N – CAT CTA GTG A – C/3′. For full sequences and modifications, see section 1 ESI.† For solvents, concentrations and heating rates see Table S3 ESI.†) | ||
We next moved on to study the stabilities of dsPNA duplexes in very high organic content solvent mixtures. Fig. 2 shows that for a short dsPNA duplex (10-mer) similar to the one studied previously by Sen and Nielsen17 micro-DSC confirmed that stability was retained up to 90 vol% DMF. Interestingly, in previous reports an approximately linear relationship between Tm and DMF vol% was identified and it was hypothesised that the Tm would be slightly above 50 °C in pure organic solvent. However, our results demonstrate that this is not the case. A representation of the Tm as a function of the molar percentage (mol%) of DMF rather than vol% illustrates why: because DMF and water have similar densities but different molecular weights, previous studies had only explored a relatively small range of DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water molar ratios, making extrapolation of the trend to high DMF content prone to inaccuracy (see Fig. S6 and S7 ESI†).
water molar ratios, making extrapolation of the trend to high DMF content prone to inaccuracy (see Fig. S6 and S7 ESI†).
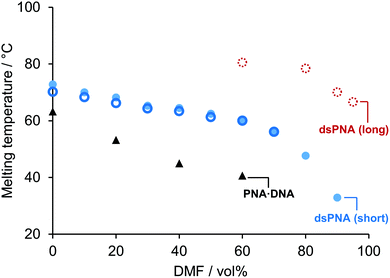 | ||
| Fig. 2 Evolution of the melting temperature (Tm) of dsPNA as a function of the volumetric DMF content. 15-mer dsPNA micro-DSC measurements (open dashed red circle), 10-mer dsPNA micro-DSC measurements (solid blue dot), 10-mer dsPNA UV absorbance reported data (open blue circle) and 10-mer PNA·DNA hybrid duplex micro-DSC measurements (black triangle). No transition was observed in 100 vol% DMF solutions (data not shown). (Sequences: [10]-DNA/PNA1 5′/N – TCA CTA GAT G – C/3′ [10]-DNA/PNA2 – 5′/N – CAT CTA GTG A – C/3′ [15]-PNA1 N – CGC CGT CAC TAG ATG – C [15]-PNA2 N – CAT CTA GTG ACG GCG – C.) For further details and standard error evaluation see section 4 ESI†. | ||
The stability of PNA·DNA hybrid duplexes was also studied by micro-DSC in different DMF/water mixtures. It was found that addition of DMF had a much greater impact on the stability of DNA·PNA duplexes than on dsPNA.
Our results led us to speculate that a longer PNA duplex might remain stable at even higher DMF vol%, because increasing the length of a nucleic acid duplex generally results in a greater Tm, at least for short sequences.22 In pure aqueous solutions, extending the length of unmodified PNA is not an option because strong inter-strand interactions render it insoluble,23 but we speculated that the addition of DMF would allow longer PNA strands to dissolve and hybridise in a reversible and well-controlled manner. These hypotheses were tested with a 15-mer dsPNA (see Fig. 2). As expected, the addition of five extra nucleobases increased the Tm in 90 vol% DMF from 33 ± 1.1 °C to 67.7 ± 0.3 °C. Furthermore, while the 10-mer dsPNA duplex was not observed in solutions with a DMF content higher than 90 vol%, it was found that the 15-mer sequence formed stable dsPNA in up to 95 vol% DMF. A previous study has demonstrated that reducing the concentration of water to 5 vol% enables the use of water-sensitive transformations such as the pyrrolidine catalysed aldol reaction to afford nucleic acid-templated products in high yield (up to 88%), illustrating the impact of our findings on future NATS applications.24
Nucleic acid duplexes are stabilised by a combination of hydrophobic, hydrogen bonding, π–π stacking, van der Waals and electrostatic interactions.25 The reduction in the Tm that we observed at lower molar water content indicated that the hydrophobic effect accounted for a considerable fraction of this stabilisation for dsPNA. It was previously proposed by Sen and Nielsen that as DMF content increases, the major contributor to duplex stability shifts towards hydrogen bonding.17 As DMF has the ability to accept hydrogen bonds, this suggests that alternative, non-hydrogen bonding solvents hold potential for further stabilisation of the duplex and could be identified in the future by micro-DSC.
Finally, to assess the potential of dsPNA for use in more complex multi-step templated syntheses,2 we investigated the isothermal annealing of PNA duplexes in 90 vol% DMF and aqueous solution (Fig. 3).
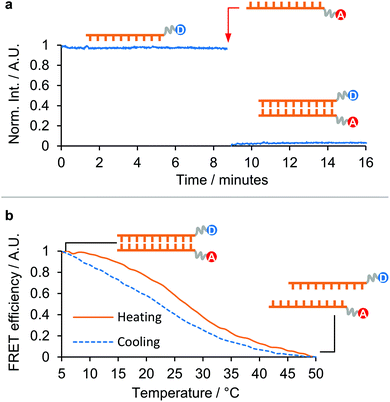 | ||
| Fig. 3 Isothermal annealing of a 10-mer dsPNA in 90 vol% DMF: (a) plot of the emission of the tryptophan-labelled PNA strand over time. The addition of the complementary quencher-labelled PNA strand is labelled with a red arrow. (b) Thermal denaturation of the isothermally annealed sample, and subsequent re-annealing upon cooling. (The donor moiety in [10]-PNA1-F was Tryptophan, the acceptor moiety in [10]-PNA2-Q was Dansyl. Tryptophan was excited at 290 nm and the emission was recorded at 340 nm. Dansyl emission was recorded at 550 nm.) For further details see section 5 ESI†. | ||
One 10-mer PNA strand was labelled with a tryptophan group at the N′ end and the fluorescence in solution recorded for several minutes to ensure a stable baseline. The fluorescence cuvette was then removed, the complementary strand labelled with a 5-(dimethylamino)naphthalene-sulfonamide (dansyl) group at the C′ was added, and the cuvette shaken to ensure thorough mixing, before being replaced in the instrument and measurements continued.
In 90 vol% DMF, annealing was found to be rapid (Fig. 3a), with full quenching observed by the time the cuvette had been replaced in the instrument following addition of the complementary strand (a delay of around 12 s). It was also found that the freshly dissolved PNA strands required a heating/cooling cycle in the desired solvent prior to isothermal annealing in order to display these fast kinetics, otherwise annealing took approximately 1.5 h to complete (see Fig. S14 ESI†). This was interpreted as the effect of kinetically trapped states present in the dry pellet that provided an additional barrier to dsPNA formation. The isothermally annealed PNA was subsequently thermally denatured and was observed to re-anneal upon cooling, demonstrating the full reversibility of the process (Fig. 3b). This supported the micro-DSC evidence of dsPNA formation in 90 vol% DMF and suggests that multistep PNA templated synthesis in organic solvents may be possible in the future.
In conclusion, it has been demonstrated that microcalorimetry allows reliable determination of the thermal stability of dsPNA in solvent mixtures where the widely employed UV absorbance method cannot be used as a result of the high UV cut-off of the solvent. This technique was subsequently used to study the stability of 10-mer and 15-mer dsPNA duplexes in 0 to 100 vol% DMF showing the formation of dsPNA in up to 95 vol% DMF. A remarkable stability in high organic solvent content was observed, which was directly dependent on the length of the PNA. Finally, it was shown that PNAs isothermally anneal rapidly in high DMF content solutions. Altogether, these findings highlight the potential of microcalorimetry to study duplex formation events in a wider range of solvent media and in the absence of a measurable spectrometric change. They also provide a good starting point for further applications of peptide nucleic acids as chemical templates in water-free media.
Conflicts of interest
There are no conflicts to declare.Notes and references
- M. L. McKee, P. J. Milnes, J. Bath, E. Stulz, A. J. Turberfield and R. K. O'Reilly, Angew. Chem., Int. Ed., 2010, 49, 7948–7951 CrossRef CAS PubMed.
- Y. He and D. R. Liu, J. Am. Chem. Soc., 2011, 133, 9972–9975 CrossRef CAS PubMed.
- M. H. Hansen, P. Blakskjaer, L. K. Petersen, T. H. Hansen, J. W. Hoøjfeldt, K. V. Gothelf and N. J. V. Hansen, J. Am. Chem. Soc., 2009, 131, 1322–1327 CrossRef CAS PubMed.
- W. Meng, R. A. Muscat, M. L. McKee, P. J. Milnes, A. H. El-Sagheer, J. Bath, B. G. Davis, T. Brown, R. K. O'Reilly and A. J. Turberfield, Nat. Chem., 2016, 8, 542–548 CrossRef CAS PubMed.
- X. Li and D. R. Liu, Angew. Chem., Int. Ed., 2004, 43, 4848–4870 CrossRef CAS PubMed.
- C. J. Krusemark, N. P. Tilmans, P. O. Brown and P. B. Harbury, PLoS One, 2016, 11, e0154765 CrossRef PubMed.
- Z. J. Gartner, B. N. Tse, R. Grubina, J. B. Doyon, T. M. Snyder and D. R. Liu, Science, 2004, 305, 1601–1605 CrossRef CAS PubMed.
- B. N. Tse, T. M. Snyder, Y. Shen and D. R. Liu, J. Am. Chem. Soc., 2008, 130, 15611–15626 CrossRef CAS PubMed.
- M. W. Kanan, M. M. Rozenman, K. Sakural, T. M. Snyder and D. R. Liu, Nature, 2004, 431, 545–549 CrossRef CAS PubMed.
- B. Seelig and J. W. Szostak, Nature, 2007, 448, 828–831 CrossRef CAS PubMed.
- D. D. Albergo and D. H. Turner, Biochemistry, 1981, 20, 1413–1418 CrossRef CAS PubMed.
- P. E. Nielsen, M. Egholm, R. H. Berg and O. Buchardt, Science, 1991, 254, 1497–1500 CrossRef CAS PubMed.
- C. Zambaldo, S. Barluenga and N. Winssinger, Curr. Opin. Chem. Biol., 2015, 26, 8–15 CrossRef CAS PubMed.
- S. Barluenga and N. Winssinger, Acc. Chem. Res., 2015, 48, 1319–1331 CrossRef CAS PubMed.
- A. Singhal, V. Bagnacani, R. Corradini and P. E. Nielsen, ACS Chem. Biol., 2014, 9, 2612–2620 CrossRef CAS PubMed.
- S. Ficht, A. Mattes and O. Seitz, J. Am. Chem. Soc., 2004, 126, 9970–9981 CrossRef CAS PubMed.
- A. Sen and P. E. Nielsen, Nucleic Acids Res., 2007, 35, 3367–3374 CrossRef CAS PubMed.
- A. Sen and P. E. Nielsen, Biophys. J., 2006, 90, 1329–1337 CrossRef CAS PubMed.
- L. E. Morrison, T. C. Halder and L. M. Stols, Anal. Biochem., 1989, 183, 231–244 CrossRef CAS PubMed.
- B. G. Moreira, Y. You, M. A. Behlke and R. Owczarzy, Biochem. Biophys. Res. Commun., 2005, 327, 473–484 CrossRef CAS PubMed.
- P. L. Privalov and A. I. Dragan, Biophys. Chem., 2007, 126, 16–24 CrossRef CAS PubMed.
- T. Ratilainen, A. Holmén, E. Tuite, P. E. Nielsen and B. Nordén, Biochemistry, 2000, 39, 7781–7791 CrossRef CAS PubMed.
- P. E. Nielsen, G. Haaima, A. Lohse and O. Buchardt, Angew. Chem., Int. Ed., 2004, 35, 1939–1942 CrossRef.
- M. M. Rozenman and D. R. Liu, ChemBioChem, 2006, 7, 253–256 CrossRef CAS PubMed.
- M. Sundaralingam and P. K. Ponnuswamy, Biochemistry, 2004, 43, 16467–16476 CrossRef CAS PubMed.
- M. C. Chakrabarti and F. P. Schwarz, Nucleic Acids Res., 1999, 27, 4801–4806 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methods, HPLC and LC-MS characterisation, UV absorbance, micro-DSC and fluorescence data. See DOI: 10.1039/c9ob01460h |
| This journal is © The Royal Society of Chemistry 2019 |
