Open Access Article
Open Access ArticleModulation of intestine development by fecal microbiota transplantation in suckling pigs†
H. Diao‡
ab,
H. L. Yan‡a,
Y. Xiao‡a,
B. Yua,
P. Zhenga,
J. He a,
J. Yua,
X. B. Mao
a,
J. Yua,
X. B. Mao *a and
D. W. Chen
*a and
D. W. Chen *a
*a
aInstitute of Animal Nutrition, Sichuan Agricultural University, Key Laboratory for Animal Disease-Resistance Nutrition of China Ministry of Education, Xinkang Road 46#, Ya'an, Sichuan Province 625014, People's Republic of China. E-mail: acatmxb2003@163.com; dwchen@sicau.edu.cn; Fax: +86-835-288-5106; Tel: +86-835-288-5106
bInstitute of Animal Nutrition, Sichuan Academy of Animal Science, No. 7 Niusha Road, Chengdu, Sichuan 610066, People's Republic of China
First published on 27th February 2018
Abstract
The present study was conducted to investigate the effects of early fecal microbiota transplantation on gut development in sucking piglets. A total of 24 3 day-old DLY sucking piglets (2.11 ± 0.15) kg were randomly divided into four groups (TMP, YMP, RMP and control group (CON)), which were transplanted with intact fecal microbiota of Tibetan pig (TP), Yorkshire pig (YP), Rongchang pig (RP), and without transplantation, respectively. The whole trial lasted for 56 d. The results are as follows: when compared with the YMP and RMP treatments, TMP and CON had a lower diarrhea index (P < 0.05), TMP and CON had higher GLP-2 and ANG4 mRNA abundances in the ileum (P < 0.05), and the TMP had a higher jejunal villus height: crypt depth and a higher colonic GLP-2 mRNA abundance (P < 0.05). Moreover, when compared with the YMP and RMP treatments, TMP had an enhanced DMT1 mRNA abundance in the duodenum (P < 0.05), TMP and CON had a greater lactase activity and a higher DMT1 mRNA abundance in the jejunum (P < 0.05), and CON had a higher γ-GT activity in the jejunum (P < 0.05). The jejunal Ca2+, Mg2+-ATPase activity in TMP was higher than that in CON, and the jejunal Na+, K+-ATPase activity in TMP was higher than that in the other three treatments (P < 0.05). Besides, when compared with the YMP and RMP treatments, TMP had a lower MDA content and a higher MUC1 mRNA abundance in the jejunum (P < 0.05); CON had a higher SOD activity in the jejunum (P < 0.05), whereas TMP and CON had a higher butyric acid concentration in the colon and a lower LPS content in the serum (P < 0.05). Finally, when compared with the TMP treatment, the other three treatments had an enhanced IL-10 mRNA abundance in the colon (P < 0.05), YMP and CON had higher counts of Escherichia coli in the colonic digesta (P < 0.05), and the CON had lower counts of Lactobacillus spp in the cecal and colonic digesta (P < 0.05). These data indicated that early transplantation of the fecal microbiota from the Yorkshire pigs and Rongchang pigs to DLY suckling piglets would destroy the gut microbiota balance and thus damage intestinal health.
Introduction
Many microorganisms inhabit the gastrointestinal tract, skin and mucosal surfaces of our body, with by far the largest number of microorganisms in the gut.1 The different regions of the host intestine harbor specific microbial communities altering in density and diversity, and whose composition are influenced by heredity, diet and environment.2 Indeed, it is clear that a complex and balanced environment for symbiosis between gut microbiota and host has been formed through long-term cooperative coevolution.3 In normal condition, the host provides essential developmental location for microbiota and, in turn, the microbiota contributes to many host physiological processes, including providing nutrients, modulating gastrointestinal development and shaping immune system.4,5Gut microbiota, known as animal second genome, are closely related to host gut development. The overall balance of the gut microbial communities is considerable in assuring homeostasis at the intestinal mucosa. Comparisons of conventional raised animals and germ free animals reveal the important role of gut microbiota in the structural development of the gastrointestinal tract.6–9 Germ free animals possess hypoplastic Peyer's patches, abnormal numbers of several immune cell types, as well as decreased numbers of mature isolated lymphoid follicles, along with reducing levels of secreted immunoglobulins.10–12 Moreover, normal gut microbiota are necessary for the expression of antimicrobial peptides in intestinal paneth cells.13 Germ-free mice colonized with B. thetaiotaomicron induce matrilysin expression in paneth cells, and the matrilysin is a matrix metalloproteinase that activates antimicrobial peptides.14 Besides, the cecum are greatly enlarged and the villus thickness are smaller in germ free animals, resulting in functional gastrointestinal disorders.9,15 Taken together, gut microbiota can play a key role in the intestinal development.
Currently, fecal microbiota transplant (FMT) is commonly used in the research of the relationship between gut microbes and the host.16,17 Studies on humans reveal that it is crucial to recover the normal microbial composition and cure such intestinal diseases, for example, inflammatory bowel disease and clostridium difficile enterocolitis, through transplanting fecal microbiota from healthy individual to those with these diseases.18–21 Compared with human, pig is a preferable animal model to study the role of environment to the microbiota development as a result of its relatively controllable diet, as well as growth environment. Moreover, it is a major task for current pig industry to improve disease resistance, and host gut characteristics plays a critical role in determining pig's resistance to diseases. One of our previous studies has shown that the microbiota composition differed among pig breeds (Tibetan pigs, Yorkshire pigs and Rongchang pigs), and the differences in gut-phenotypes among pig breeds would partially convey to germ-free recipient mice by fecal microbiota transplantation, especially intestinal morphology and enzyme activities.22 However, it is not clear whether gut microbiota mediated the intestinal development in conventional raised pig model via fecal microbiota transplantation. Besides, in actual physiology, the intestine of pigs is not sterile. Hence, the present study was conducted to investigate the effects of early fecal microbiota transplantation on gut development, digestion and barrier function in sucking piglets, which could provide the scientific basis for using fecal microbiota transplantation in pig production.
Materials and methods
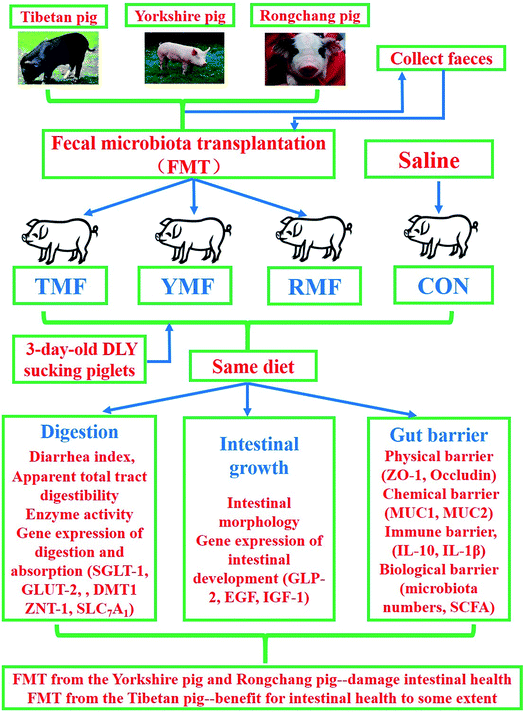
Animals, management, diets
The 5 Tibetan pigs, 5 Yorkshire pigs and 5 Rongchang pigs (12 weeks of age) provided by a reservation farm were used in this experiment as fecal donors. According to the standard for donor identification, all pigs did not have digestive disorders, and never received antibiotics and probiotics in the latest 8 weeks.23 The management of all the donor pigs were the same with our previous study.24 In brief, Tibetan pigs, Yorkshire pigs and Rongchang pigs were housed separately in individual metabolic cages in three environmentally controlled rooms for 8 weeks, in which pigs were allowed ad libitum access to water and diet.The new born pigs were segregated from their mothers 24 hours after birth and then were taught and fed with artificial milk for 2 days. A total of 24 3 day-old DLY (Duroc × Landrace × Yorkshire) sucking piglets (2.11 ± 0.15) kg were randomly allotted into four groups with six replicates per group and one pig per replicate, which were transplanted with intact fecal microbiota of Tibetan pigs, Yorkshire pigs, Rongchang pigs, and without transplantation, respectively. Four treatment groups were: (1) Tibetan microbiota-associated pigs (TFM), (2) Yorkshire microbiota -associated pigs (YMP), (3) Rongchang microbiota-associated pigs (RMP), and, (4) control group (CON). The whole trial lasted for 56 d and was divided into three periods, days 1–15, days 16–28 and days 29–56. All the suckling piglets were fed artificially with milk-substitutes in the first 15 days of trial, and then solid feed were used until the end of study. Ingredients and compositions of the diet in each period are presented in Tables 1, 2 and 3, respectively. There were no antibiotics in any diets. Pigs in each group were housed separately in three environmentally controlled rooms (room temperature maintained at 24–28 °C and relative humidity controlled at 60–70%) with ventilation and disinfection installations at the entrances.
| Items | TMP | YMP | RMP | CON | SEM | P-Value |
|---|---|---|---|---|---|---|
| a TMP, Tibetan porcine flora-associated pig; YMP, Yorkshire porcine flora-associated pig; RMP Rongchang porcine flora-associated pig.b Within a row, means without a common superscript differ (P < 0.05). | ||||||
| Pre-weaning | 1.110 | 1.098 | 0.793 | 0.732 | 0.176 | 0.319 |
| Post-weaning | 0.148b | 0.442a | 0.393a | 0.196b | 0.048 | 0.001 |
| Full period | 0.545 | 0.713 | 0.558 | 0.420 | 0.082 | 0.128 |
Fecal microbiota transplantation
In order to obtain representative fecal material for each breed, the fresh feces of all the donor pigs were collected separately after 12 hour fasting, and then the fecal samples of each breed were thoroughly mixed. Before they were pooled, all pigs fecal samples were collected individually, and analyzed for microbiota using 16S rRNA amplicon sequencing. The stool suspension was prepared as previously described by Zeng et al. (2013) and Diao et al. (2016).24,25 In brief, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (w/v) sterile saline was added into the mixed fresh feces, which was followed by mixing the suspension and passing stainless steel laboratory sieves (2.0, 1.0 and 0.5 mm, respectively). Piglets in the TMP, YMP and RMP groups were colonized with 10 mL fecal suspension derived from Tibetan pig, Yorkshire pig and Rongchang pig by intragastric administration daily for the first three days of the trial. During days 4–15, piglets received 10 mL fecal suspension every two days, and 20 mL during days 16–46 every five days. The CON group received the same amount of sterile saline at each inoculation time-point.
9 (w/v) sterile saline was added into the mixed fresh feces, which was followed by mixing the suspension and passing stainless steel laboratory sieves (2.0, 1.0 and 0.5 mm, respectively). Piglets in the TMP, YMP and RMP groups were colonized with 10 mL fecal suspension derived from Tibetan pig, Yorkshire pig and Rongchang pig by intragastric administration daily for the first three days of the trial. During days 4–15, piglets received 10 mL fecal suspension every two days, and 20 mL during days 16–46 every five days. The CON group received the same amount of sterile saline at each inoculation time-point.
Sample collection
Faeces of each piglet recipient was collected on days 53–56, and then added 10% hydrochloric acid for fixing excreta nitrogen. The fecal sample of each piglet was dried and used to measure the apparent total tract digestibility (ATTD) of dry matter (DM), calcium (Ca), phosphorous (P), crude ash, crude protein (CP) and ether extract (EE). All the piglets recipients were sacrificed on d 57 by using intravenously administrated dose of Zoletil 50 (Beijing PET Technology Co., LTD, Beijing, China, 10 mg kg−1 body weight) as anesthetics, and the blood samples were collected from the precaval vein into vacuum tubes and centrifuged (3000 rpm) for 10 min. The lengths and weights of the small intestine and large intestine were measured, and the sections of duodenum, jejunum, ileum and colon were immediately obtained and fixed in 4% paraformaldehyde solution. After that, collected the digesta of jejunum, ileum, cecum and colon for analysis of microbiota and metabolites. Samples of jejunal mucosa, duodenum, jejunum and ileum were immediately collected and stored at −80 °C for analysis of gene expressions and enzymes activities.Diarrhea index
The occurrence of diarrhea for each pig was visually assessed every morning and evening for 56 days of the trial based on the following scoring system: 0 = normal, firm feces, 1 = possible slight diarrhea, soft, formed feces, 2 = moderately diarrhea, definitely unformed, starchiness, moderately fluid feces, 3 = very watery and frothy diarrhea, severe fluid feces.26 Diarrhea index was calculated according to the formula where diarrhea index = the sum of diarrhea scores recorded in the morning and evening/observational days.Histology of intestine
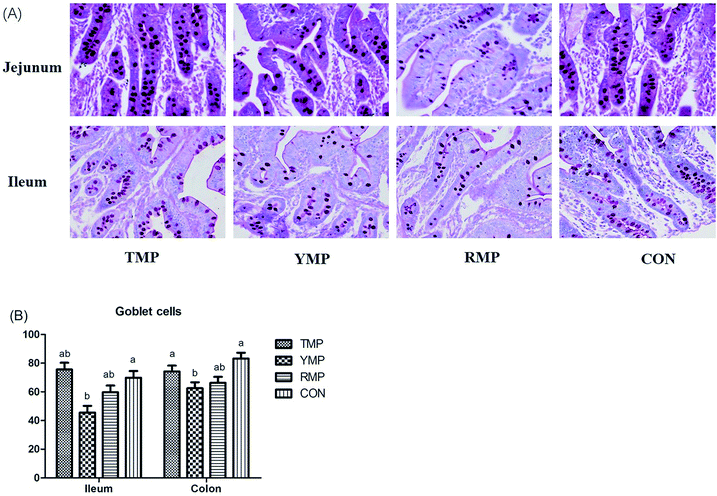
Samples of duodenum, jejunum and ileum were obtained and preserved in 4% paraformaldehyde solution, later dehydrated and embedded in paraffin for histological examination. Sections of 5 μm were cut and stained with hematoxylin and eosin (H&E) to determine the morphology of the intestinal samples. Ten well orientated sections of each sample were selected, photographed and detected villi height and crypts depth using the Olympus CK 40 (Olympus, Tokyo, Japan) at 40× magnification.27 Besides, the numbers of goblet cells in the ileum and colon were determined using alcian blue and periodic acid Schiff (AB–PAS) as described before.28 All the observations were measured by a single experimenter who was unknown to the treatments.Apparent total tract digestibility (ATTD)
The ATTD was determined as previously described by Diao et al. (2016).22 All feed and feces were analyzed for AIA (acid insoluble ash, method GB/T 23742), DM (method 930.15, AOAC, 1995), Ca (method 927.02, AOAC, 1995), EE (method 945.16, AOAC, 1995), crude ash (method 942.05, AOAC, 1995), P (method 995.11, AOAC, 1995), CP (method 990.03, AOAC, 1995) and GE. The GE was measured using bomb calorimetry (Parr Instrument 1563, Moline IL). The ATTD was calculated as (100 − A1F2/A2F1 × 100), where A1 represents the AIA content of the diet; F1 represents the nutrient content of the diet; A2 represents the AIA content of faeces; F2 represents the nutrient content of faeces.LPS and DAO concentrations, antioxidant capacity and the digestive and absorptive enzyme activities
The serum LPS (lipopolysaccharide) concentration was determined using pig Enzyme-linked Immunosorbent Assay Kits from R&D system (Minneapolis, MN) combined with a BioTek Synergy HT microplate reader (BioTek Instruments, Winooski, VT). The serum DOA (diamine oxidase) concentration was determined by a commercial kit (Nanjing Jiancheng Institute of Bioengineering, Nanjing, China) combined with a UV-VIS Spectrophotometer (UV1100, MAPADA, Shanghai, China) according to the manufacturer's protocol.The crude enzyme solutions of jejunal digesta and mucosa were prepared according to procedure previously described by Diao et al. (2015).29 Samples of jejunal digesta and mucosa were collected and homogenized in sterile saline (m/v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9). The homogenates were centrifuged at 3000 rpm at 4 °C for 15 min. After that, the supernatant was collected and used for subsequent analysis of the digestive and absorptive enzyme activities (lactase, sucrase, maltase, amylase, lipase, trypsin, γ-glutamyltransferase (γ-GT), Na+, K+-ATPase and Ca+, Mg+-ATPase), and the antioxidant capacity (total antioxidant capacity (T-AOC), superoxide dismutase (SOD) and methane dicarboxylic aldehyde (MDA)), which were measured by the kits (Nanjing Jiancheng Institute of Bioengineering, Nanjing, China) combined with a UV-VIS Spectrophotometer (UV1100, MAPADA, Shanghai, China).
9). The homogenates were centrifuged at 3000 rpm at 4 °C for 15 min. After that, the supernatant was collected and used for subsequent analysis of the digestive and absorptive enzyme activities (lactase, sucrase, maltase, amylase, lipase, trypsin, γ-glutamyltransferase (γ-GT), Na+, K+-ATPase and Ca+, Mg+-ATPase), and the antioxidant capacity (total antioxidant capacity (T-AOC), superoxide dismutase (SOD) and methane dicarboxylic aldehyde (MDA)), which were measured by the kits (Nanjing Jiancheng Institute of Bioengineering, Nanjing, China) combined with a UV-VIS Spectrophotometer (UV1100, MAPADA, Shanghai, China).
Total RNA extraction, reverse transcription reaction and real-time quantitative PCR
Total RNA was extracted from the frozen mucosa of duodenum, jejunum and ileum using the TRIzol reagent (Biotechnology Company, Dalian, China). Reverse transcription reaction was conducted by using a PrimeScript RT reagent kit (Biotechnology Company, Dalian, China), and then obtained cDNA of each sample. The genes related to intestinal development (EGF, epidermal growth factor; GLP-2, glucagon-like peptide-2; ANG4, angiogenin 4; IGF-1, insulinlike growth factor-1; IGF-1R, insulinlike growth factor-1 receptor), digestion and absorption (SGLT-1, sodium/glucose cotransporter 1; GLUT-2, glucose transporter type 2; ZNT-1, zinc transporters-1; DMT1, divalent metal transporter-1; SLC7A1, solute carrier family 7), and intestinal barrier (MUC1, mucin 1; MUC2, mucin 2; REGIIIγ, regeneration protein IIIγ; Occludin; ZO-1, zonula occludens 1; IL-10, interleukin-10; IL-1β, interleukin-1β) were detected by the QuantStudio™ Real-Time PCR System (Thermo Fisher Scientific, Shanghai, China) as described by Diao et al. (2016).22 Briefly, The PCR system was composed of 0.5 μL of forward and 0.5 μL of reverse primers (100 nM), 1 μL cDNA, 3 μL diethylpyrocarbonate-treated water and 5 μL 2× SYBR Premix Ex Taq (Biotechnology Company, Dalian, China). The PCR cycling conditions were as follows: 95 °C for 10 s, followed by forty cycles of 95 °C for 5 s, annealing at 60 °C for 10 s and 72 °C for 15 s. The melting curve conditions were 95 °C for 30 s, 55 °C for 60 s and 95 °C for 60 s, and this procedure was performed after each real-time quantitative PCR to check the purity and specificity of PCR products. The primers listed in Table S4† were synthesized commercially by Invitrogen (shanghai, China).Enumeration of Escherichia coli, Lactobacillus spp, Bifidobacterium spp, Bacillus spp, and total bacteria by PCR
Bacterial DNA was extracted from the frozen ileal, cecal and colonic digesta using a commercial stool DNA isolation kit (Omega Bio-Tek, Doraville, CA). Sequences of primers and probes (Table S5†) for Escherichia coli, Lactobacillus spp, Bifidobacterium spp, Bacillus spp and total bacteria were obtained from previous studies,30,31 and synthesized from Invitrogen (shanghai, China). The reaction system and quantitative real-time PCR conditions used in present study were referring to Qi, et al. (2011).30 The reaction system for detecting the total bacteria was composed of 12.5 μL SYBR Premix Ex Taq (2×), 9.5 μL double distilled water (ddH2O), 1 μL of forward and 1 μL of reverse primers (100 nM) and 1 μL DNA. The PCR conditions were as follows: 95 °C for 10 s, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 59 °C for 25 s and extension at 95 °C for 10 s. The reaction system for detecting Lactobacillus, Escherichia coli, Bacillus and Bifidobacterium was composed of 8 μL realMasterMix (2.5×), 7.7 μL ddH2O, 1 μL probe enhancer solution (20×), 1 μL of forward and 1 μL of reverse primers (100 nM), 0.3 μL probe (100 nM) and 1 μL DNA. The PCR conditions were as follows: 1 cycle of predenaturation at 95 °C for 10 s, followed by 50 cycles of denaturation at 95 °C for 5 s, annealing at 53–60 °C for 25 s and extension at 95 °C for 10 s.Short-chain fatty acid
The SCFAs (acetic acid, propionic acid and butyric acid) of the frozen colonic digesta samples were measured as previously described by Diao et al. (2014).32 In brief, the supernatants of colonic digesta samples were centrifuged at 500 × g for 15 min after adding 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 sterile distilled water, this was followed by transferring 2 mL supernatant into a sterile tube (centrifuged at 12
1 sterile distilled water, this was followed by transferring 2 mL supernatant into a sterile tube (centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min). Then 1 mL supernatant was transferred into a new sterile tube, and 0.2 mL 25% metaphosphoric acid was added and stood for 30 min (centrifuged at 12
000 × g for 10 min). Then 1 mL supernatant was transferred into a new sterile tube, and 0.2 mL 25% metaphosphoric acid was added and stood for 30 min (centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min). After that, 500 μL supernatant was transferred to another sterile tube, and 500 μL methanol was added (centrifuged at 12
000 × g for 10 min). After that, 500 μL supernatant was transferred to another sterile tube, and 500 μL methanol was added (centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min). Finally, the supernatant was transferred into a sterile tube and was stored at −20 °C until testing. The acetic acid, propionic acid and butyric acid were separated and quantified in a gas chromatographic system (VARIAN CP-3800, Varian, California, USA).
000 × g for 10 min). Finally, the supernatant was transferred into a sterile tube and was stored at −20 °C until testing. The acetic acid, propionic acid and butyric acid were separated and quantified in a gas chromatographic system (VARIAN CP-3800, Varian, California, USA).
Statistical analysis
Data were analyzed by ANOVA using the statistic software SAS 8.2 (SAS Inst. Inc., NC) where each pig was the statistical unit. All differences were considered significant at P < 0.05, and all the results were expressed as mean and SEM.Results
Diarrhea index
There were no significant differences in diarrhea index of pre-weaning and full period among the four treatments, nevertheless, the diarrhea index of pre-weaning and full period for pigs in the fecal microbiota transplantation groups (TMP, YMP and RMP) was higher than that in the CON group to various extent (P > 0.05, Table 1). However, the diarrhea index of post-weaning for pigs in the YMP and RMP groups was higher than that in the TMP and CON groups (P < 0.05).Intestinal morphology
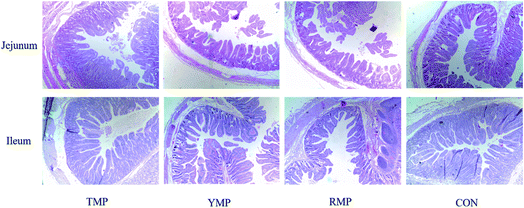
When compared to the TMP treatment, YMP had a higher crypt depth in the jejunum (P < 0.05, Table 2 and Fig. 1), whereas YMP and RMP had a lower villus height: crypt depth in the jejunum (P < 0.05). When compared to the RMP treatment, TMP had a higher villus height in the jejunum (P < 0.05), and CON had a higher villus height in the ileum (P < 0.05).| Items | TMP | YMP | RMP | CON | SEM | P-Value |
|---|---|---|---|---|---|---|
| a TMP, Tibetan porcine flora-associated pig; YMP, Yorkshire porcine flora-associated pig; RMP Rongchang porcine flora-associated pig; CON, control group, without fecal microbiota transplantation.b Within a row, means without a common superscript differ (P < 0.05). | ||||||
| Jejunums | ||||||
| Villus height (μm) | 546.800a | 523.720a,b | 491.790b | 521.360a,b | 13.186 | 0.077 |
| Crypt depth (μm) | 210.290b | 262.610a | 249.530a,b | 229.620a,b | 12.220 | 0.049 |
| Villus height: crypt depth | 2.631a | 2.0166b | 1.997b | 2.306a,b | 0.143 | 0.027 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Ileum | ||||||
| Villus height (μm) | 563.870a,b | 473.680a,b | 416.130b | 609.830a | 45.095 | 0.041 |
| Crypt depth (μm) | 229.960 | 202.680 | 175.720 | 244.020 | 27.375 | 0.344 |
| Villus height: crypt depth | 2.464 | 2.351 | 2.411 | 2.615 | 0.139 | 0.591 |
The relative mRNA expression of intestinal development-related genes
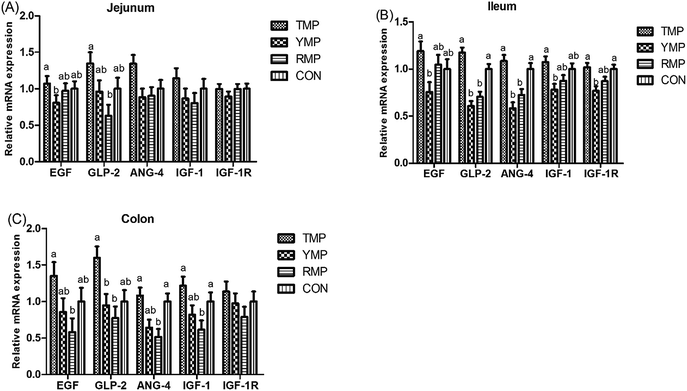
As shown in Fig. 2, compared with the RMP group, TMP had a higher GLP-2 mRNA abundance in the jejunum and a higher EGF mRNA abundance in the colon (P < 0.05). The colonic ANG4 and IGF-1 mRNA abundances in TMP and CON were higher than those in RMP (P < 0.05). Compared with the YMP group, TMP and CON had a higher IGF-1R mRNA abundance in the ileum (P < 0.05), and TMP had higher IGF-1 and EGF mRNA abundances in the ileum (P < 0.05). Compared with the YMP and RMP groups, TMP and CON had higher ANG4 and GLP-2 mRNA abundances in the ileum (P < 0.05), and TMP had a higher GLP-2 mRNA abundance in the colon (P < 0.05).Apparent total tract digestibility
When compared to the RMP group, the ATTD of CP was increased in YMP and CON (P < 0.05, Table 3), the ATTD of crude ash and Ca were improved in TMP, YMP and CON (P < 0.05), and the ATTD of DM and energy were increased in CON (P < 0.05). Moreover, the ATTD of P in TMP and the ATTD of EE in TMP and CON were enhanced as compared with RMP (P < 0.05). Besides, the ATTD of EE in CON was higher than that in YMP (P < 0.05), and the ATTD of Ca in TMP and CON was higher than that in YMP (P < 0.05).| Items | TMP | YMP | RMP | CON | SEM | P-Value |
|---|---|---|---|---|---|---|
| a TMP, Tibetan porcine flora-associated pig; YMP, Yorkshire porcine flora-associated pig; RMP Rongchang porcine flora-associated pig; CON, control group, without fecal microbiota transplantation.b Within a row, means without a common superscript differ (P < 0.05). | ||||||
| CP | 73.005a,b | 75.860a | 69.675b | 77.230a | 1.466 | 0.012 |
| DM | 83.9483a,b | 84.097a,b | 82.050b | 85.7933a | 0.632 | 0.007 |
| Crude ash | 55.997a | 51.962a | 46.367b | 53.667a | 1.273 | <0.001 |
| EE | 72.602a,b | 68.923b,c | 66.527c | 74.757a | 1.088 | <0.001 |
| Ca | 54.710a | 48.977b | 41.813c | 51.503a,b | 1.218 | <0.001 |
| P | 38.953a | 35.617a,b | 27.803b | 33.570a,b | 2.424 | 0.035 |
| Energy | 83.555a,b | 83.540a,b | 81.242b | 85.497a | 0.693 | 0.006 |
Digestive and absorptive enzyme activity in jejunum
The impacts of fecal microbiota transplantation on the digestive and absorptive enzyme activity in jejunum are shown in Table 4. Compared with the RMP and YMP treatments, TMP and CON had a higher lactase activity in the jejunal mucosa (P < 0.05), and CON had a higher γ-GT activity in the jejunal mucosa (P < 0.05). The sucrase activity of jejunal mucosa in TMP was higher than that in YMP (P < 0.05), and the maltase activity of jejunal mucosa in TMP was higher than that in RMP (P < 0.05). The Ca+, Mg+-ATP activity of jejunal mucosa in TMP was higher than that in CON (P < 0.05), whereas the Na+, K+-ATP activity of jejunal mucosa in TMP was higher than that in other three treatments (P < 0.05).| Items | TMP | YMP | RMP | CON | SEM | P-Value |
|---|---|---|---|---|---|---|
| a TMP, Tibetan porcine flora-associated pig; YMP, Yorkshire porcine flora-associated pig; RMP Rongchang porcine flora-associated pig; CON, control group, without fecal microbiota transplantation.b Within a row, means without a common superscript differ (P < 0.05). | ||||||
| Total protein (mgprot g−1) | 46.533 | 47.611 | 55.803 | 55.083 | 3.330 | 0.133 |
| Lactase (U mgprot−1) | 123.756a | 85.497b | 91.023b | 112.820a | 4.777 | <0.001 |
| Sucrase (U mgprot−1) | 63.314a | 50.130b | 61.021a,b | 60.896a,b | 2.841 | 0.022 |
| Maltase (U mgprot−1) | 498.880a | 449.560a,b | 422.150b | 483.340a,b | 16.733 | 0.024 |
| γ-GT (U gprot−1) | 42.502a,b | 32.741b | 31.307b | 49.154a | 3.085 | 0.003 |
| Na+, K+-ATPase (μmolPi per mgprot per hour) | 9.175a | 7.6634b | 7.871b | 7.651b | 0.277 | 0.004 |
| Ca2+, Mg2+-ATPase (μmolPi per mgprot per hour) | 7.670a | 6.770a,b | 6.984a,b | 6.506b | 0.240 | 0.022 |
| CK (U mgprot−1) | 2.048 | 1.890 | 1.691 | 1.803 | 0.102 | 0.134 |
Digestion and absorption-related genes in small intestine
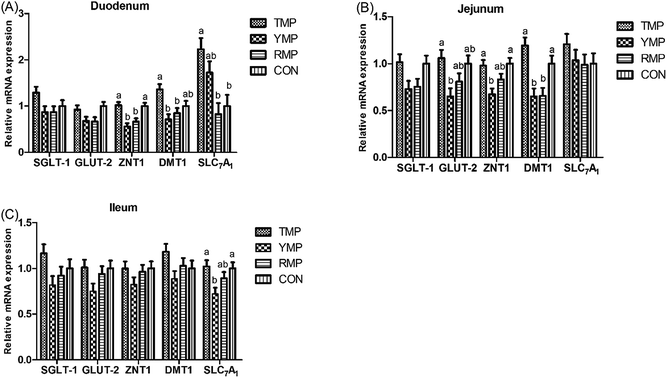
Fig. 3 presents the relative mRNA expressions of digestion and absorption-related genes in small intestine. Compared with the RMP and YMP treatments, TMP and CON had a higher ZNT-1 mRNA abundance in the duodenum and a higher DMT-1 mRNA abundance in the jejunum (P < 0.05), and TMP had a higher DMT-1 mRNA abundance in the duodenum (P < 0.05). The relative mRNA expression of duodenal SLC7A1 in TMP was higher than that in RMP and CON (P < 0.05). Compared with YMP, TMP and CON had a higher ZNT1 mRNA abundance in the jejunum and a higher SLC7A1 mRNA abundance in the ileum (P < 0.05), and TMP had a higher GLUT-2 mRNA abundance in the jejunum (P < 0.05).Jejunal antioxidant capacity
The effects of fecal microbiota transplantation on jejunal antioxidant capacity in piglets are shown in Table 5. Compared with the RMP and YMP treatments, TMP had a lower MDA content in the jejunum (P < 0.05), and CON had a higher SOD activity in the jejunum (P < 0.05). However, fecal microbiota transplantation failed to cause jejunal T-AOC capacity alterations (P > 0.05).| Items | TMP | YMP | RMP | CON | SEM | P-Value |
|---|---|---|---|---|---|---|
| a TMP, Tibetan porcine flora-associated pig; YMP, Yorkshire porcine flora-associated pig; RMP Rongchang porcine flora-associated pig; CON, control group, without fecal microbiota transplantation.b Within a row, means without a common superscript differ (P < 0.05). | ||||||
| MDA (nmol mgprot−1) | 1.016b | 1.556a | 1.446a | 1.339a,b | 0.090 | 0.004 |
| T-AOC (U mgprot−1) | 0.337 | 0.387 | 0.422 | 0.328 | 0.036 | 0.257 |
| SOD (U mgprot−1) | 126.493a,b | 109.062b | 106.192b | 133.097a | 5.834 | 0.013 |
Intestinal barrier function
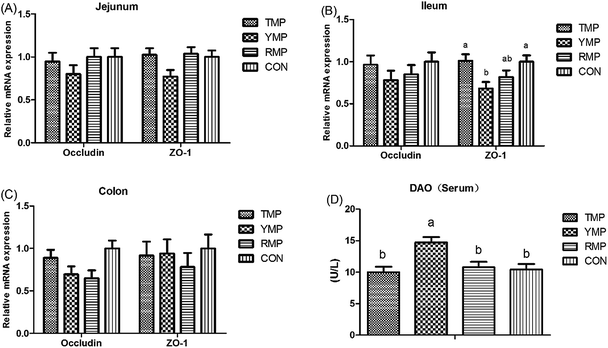
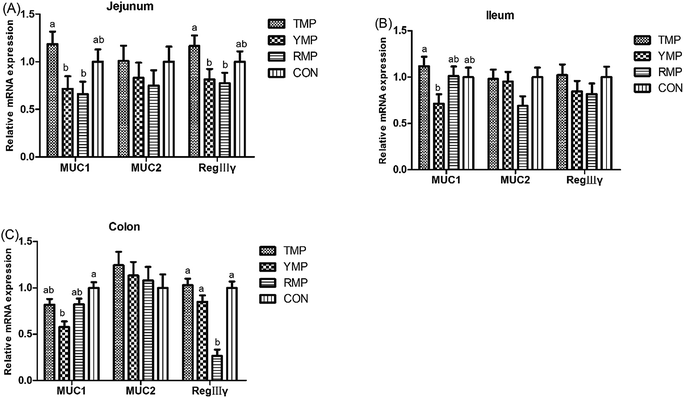
According to Fig. 4, we found the relative mRNA expression of ileal ZO-1 in TMP and CON was higher than that in YMP (P < 0.05), and the relative mRNA expression of colonic occludin in TMP and CON was higher than that in RMP (P < 0.05). Meanwhile, compared with other three treatments, YMP had a greater DAO concentration in the serum (P < 0.05).As shown in Fig. 5, compared with the RMP and YMP treatments, TMP had higher MUC1 and RegIIIγ mRNA abundances in the jejunum (P < 0.05). The relative mRNA expression of ileal MUC1 in TMP was higher than that in YMP (P < 0.05). The relative mRNA expression of colonic RegIIIγ in CON was higher than that in YMP (P < 0.05). Compared with YMP, TMP and CON had greater numbers of goblet cells in the colon (P < 0.05, Fig. 6), and CON had greater numbers of goblet cells in the ileum (P < 0.05).
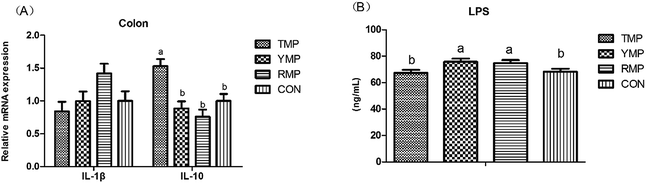
Fig. 7 depicts the effects of fecal microbiota transplant on the serum LPS concentration and the relative mRNA expression of colonic IL-1β and IL-10 in suckling pigs. Compared with other treatments, RMP had a higher IL-1β mRNA abundance in the colon (P < 0.05), and TMP had a higher IL-10 mRNA abundance in the colon (P < 0.05). The serum LPS concentration in TMP and CON was lower than that in YMP and RMP (P < 0.05).
According to Table 6, TMP had greater counts of Lactobacillus spp in the cecum and colon compared with CON (P < 0.05). The numbers of cecal Escherichia coli in TMP were lower than those in YMP and CON (P < 0.05), and the numbers of colonic Escherichia coli in TMP were lower than that in YMP (P < 0.05). We further measured the microbial metabolites in the colon for the four groups (Table 7). Specifically, the contents of colonic propionic acid and total short chain fatty acid in TMP and CON were higher than those in YMP (P < 0.05). The concentration of colonic butyric acid in TMP and CON was higher than that in RMP and YMP (P < 0.05). Besides, compared with RMP, TMP had a higher content of total short chain fatty acid in the colon (P < 0.05).
| Items | TMP | YMP | RMP | CON | SEM | P-Value |
|---|---|---|---|---|---|---|
| a TMP, Tibetan porcine flora-associated pig; YMP, Yorkshire porcine flora-associated pig; RMP Rongchang porcine flora-associated pig; CON, control group, without fecal microbiota transplantation.b Within a row, means without a common superscript differ (P < 0.05). | ||||||
| Cecum | ||||||
| Total bacteria | 11.422 | 11.475 | 11.395 | 11.503 | 0.061 | 0.600 |
| Bacillus spp | 9.898 | 9.913 | 9.761 | 9.777 | 0.061 | 0.214 |
| Lactobacillus spp | 8.965a | 8.516a,b | 8.383a,b | 8.345b | 0.155 | 0.047 |
| Escherichia coli | 8.157b | 8.932a | 8.6503a,b | 8.576a,b | 0.159 | 0.026 |
| Bifidobacterium spp | 8.006 | 8.092 | 8.161 | 8.021 | 0.144 | 0.865 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Colon | ||||||
| Total bacteria | 11.670 | 11.653 | 11.728 | 11.532 | 0.056 | 0.131 |
| Bacillus spp | 10.066 | 10.142 | 10.117 | 9.933 | 0.102 | 0.498 |
| Lactobacillus spp | 9.389a | 9.087a,b | 9.035a,b | 8.475b | 0.185 | 0.023 |
| Escherichia coli | 8.153b | 9.062a | 8.859a,b | 8.738a,b | 0.221 | 0.057 |
| Bifidobacterium spp | 7.931 | 7.682 | 7.808 | 7.629 | 0.076 | 0.054 |
| Items | TMP | YMP | RMP | CON | SEM | P-Value |
|---|---|---|---|---|---|---|
| a TMP, Tibetan porcine flora-associated pig; YMP, Yorkshire porcine flora-associated pig; RMP Rongchang porcine flora-associated pig; CON, control group, without fecal microbiota transplantation.b Within a row, means without a common superscript differ (P < 0.05). | ||||||
| Acetic acid | 53.41 | 50.659 | 51.86 | 55.828 | 1.845 | 0.265 |
| Propionic acid | 29.340a | 24.728b | 27.465a,b | 31.384a | 0.965 | 0.002 |
| Butyric acid | 16.544a | 12.957b | 12.863b | 16.973a | 0.866 | 0.005 |
| Total volatile fatty acid | 99.294a,b | 88.344c | 92.187b,c | 104.185a | 2.245 | <0.001 |
Discussion
Gut microbiota, the second genome, are closely related to host intestinal health.33 Studies of fecal microbiota transplantation from healthy individuals to those with inflammatory bowel disease and clostridium difficile enterocolitis have revealed that it is crucial to recover the normal microbial composition during curing these diseases.18–21 As there has been particularly tricky intestinal disorder problems in practical pig production, we can consider whether the pig intestinal health can be improved through fecal microbiota transplantation. Besides, studies on transplanting fecal microbiota to conventional raised pig are hardly researched. In addition, the numbers and species of gut microbiota are very few when pigs are born, indicating the microbiota are flexible when pigs are young. Therefore, the present study was conducted to investigate the effects of early fecal microbiota transplantation on gut development, digestion and barrier function in 3 day old sucking piglets.Although there was no statistical significance on the diarrhea index for pigs received the fecal microbiota from the Tibetan pigs, Yorkshire pigs, Rongchang pigs and control group in the pre-weaning period, the diarrhea index in the three transplantation groups was higher than that in the control group to various extent from the view point of numerical data, which suggested that inoculating fecal microbiota from Tibetan pigs, Yorkshire pigs and Rongchang pigs brings large stress to the new born piglets, and this may be attributed to complex components in the donor faeces, as well as the different composition and diversity of gut microbiota in donor and recipient with different physiological stages.34,35 However, the diarrhea index of post-weaning for pigs in the TMP and CON groups was lower than that in the YMP and RMP groups, indicating microbiota adaptation was existed in the intestines after pigs received the fecal microbiota from the Tibetan pigs, but not from the Yorkshire pigs and Rongchang pigs. Comparison of Tibetan pigs and ordinary pigs revealed Tibetan pigs had an increasing microbial diversity in intestine,36 which may be beneficial to recipient piglets after an adaptation period.
The body health is affected by the intestinal function, and the intestinal growth, digestion, absorption and barrier integrity functions compose gut development.37 In the present study, compared with the CON group, pigs received the fecal microbiota from the Yorkshire pigs and Rongchang pigs had lower GLP-2 and ANG4 mRNA abundances in the ileum, a reducing jejunal lactase activity, a decreasing DMT1 mRNA abundance in the jejunum, a lower butyric acid concentration in the colon whereas a higher LPS content in the serum. However, different results were found in pigs received the fecal microbiota from the Tibetan pigs, who had greater activities of jejunal Na+, K+-ATPase and Ca2+, Mg2+-ATPase, and higher counts of Lactobacillus spp in the cecal and colonic digesta as compared with the CON group. Meanwhile, there were no differences between TMP and CON in most of these indicators. Based on the foregoing view, we showed that early transplantation of the fecal microbiota from the Yorkshire pigs and Rongchang pigs to DLY suckling piglets had destructive effects on intestinal health, which could damage the intestinal growth, digestion, absorption and barrier integrity. Nevertheless, transplantation of the fecal microbiota derived from the Tibetan pigs would not harm the intestinal health, on the contrary, elevate the activities of absorption enzymes and the counts of beneficial bacteria in DLY suckling piglets, indicating the microbial diversity and predominant flora in the intestine of Tibetan pigs may act an important part to this phenomenon. A study using the method of denaturing gradient gel electrophoresis revealed one clone in faeces of Tibetan pigs was similar to Anaerobiospirillum succinici prouducens, an important genus of cellulose-degrading bacteria.36 Similarly, a cellulolytic bacterium was isolated from a Tibetan pig's intestine, and the cellulase production of this bacterium was high.38 16S rRNA gene sequencing in one of our previous studies has shown that the microbiota composition differed among Tibetan pigs, Yorkshire pigs and Rongchang pigs.24 Tibetan pigs had higher levels of bacteria from Elusimicrobia, Fibrobacteres and Spirochaetes, and all of the three phylum are capable of degrading fiber components, which contribute to enhance the hemicellulose digestibility.39–41 Moreover, our former study also revealed higher counts of Lactobacillus, Roseburia and Blautia were observed in Tibetan pigs.24 As is known to us, Blautia and Roseburia are major bacteria that produce acetic acid and butyrate, respectively,42,43 and Lactobacillus is found to be positively correlated with the cure of enteritis, which is beneficial to gut health.44 In our present study, pigs received the fecal microbiota from the Tibetan pigs had greater count of Lactobacillus spp in the cecum. It appears, therefore, that transplantation of the fecal microbiota derived from the Tibetan pigs may promote intestinal development.
Incomplete immune system in germ free animals, characterized by abnormal numbers of several immune cell types and immune cell products, deficient in local and systemic lymphoid structures, hypoplastic Peyer's patches, as well as decreased numbers of mature isolated lymphoid follicles, and reducing levels of secreted immunoglobulins A and immunoglobulins G, results in irregularities of cytokine levels and damages in body health.10–12 Lacking the expansion of CD4+ T-cell populations is one of the major immune deficiency exhibited by germ free animals, and this deficiency phenomenon can be reversed by colonizing with Bacteroides fragilis.45 Similarly, germ-free mice colonized with fecal microbiota derived from conventional mice induced the proliferation and differentiation of Th17 cells.46 In our present study, a higher IL-10 mRNA abundance in the colon was observed in pigs received the fecal microbiota from the Tibetan pigs, which may be associated with certain microbes in their intestine. Previous studies have demonstrated that Tibetan pigs had higher proportions of Lactobacillus and Parabacteroides, which are found to be positively correlated with the cure of enteritis through mediating cytokine levels.24,44,47,48 Besides, higher proportions of Roseburia and Blautia were also observed in Tibetan pigs, and the two genus of microbes can produce short chain fatty acids, regulating immune functions via their receptors (GPR41 and GPR43).42,43,49 As indicated above, the fecal microbiota of Tibetan pigs strongly affect immune function of suckling piglets, suggesting gut microbiota occupy a central role in the development of intestinal mucosal immunity system.
Conclusion
Early transplantation of the fecal microbiota from the Yorkshire pigs and Rongchang pigs to DLY suckling piglets would destroy the gut microbiota balance and thus damage intestinal health. However, transplantation of the fecal microbiota derived from the Tibetan pigs would not harm the intestinal health, on the contrary, promote absorption enzymes activities in DLY suckling piglets (Fig. 8).Funding
This study was financially supported by the National Natural Science Foundation of China (31672436), the earmarked fund for the China Agriculture Research System (CARS-35), the National Key Research and Development Program of China (2017YFD0500503), and the National High Technology Research and Development Program of China (2014AA022209).Authors' contributions
DWC and XBM conceived the study and designed the experiment. HD, HLY and YX performed the experiments, including chemical analysis, analyzed the experimental data, and wrote the manuscript. JY and BY verified the validity of the experimentation and checked the results. JH and PZ participated in its design and gave important intellectual advice for approval. All of the authors read and approved the final version of this manuscript.Consent for publication
Not applicable.Ethics approval and consent to participate
All experimental procedures and animal care were accomplished in accordance with the Guide for the Care and Use of Laboratory Animals provided by the Institutional Animal Care Advisory Committee for Sichuan Agricultural University. All animal protocols used in this study were approved by the Animal Care and Use Committee of Sichuan Agricultural University under permit number DKY-B20131704.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
The authors wish to thank the veterinarians XP Shi from College of Veterinary Medicine, Sichuan Agricultural University, for the elaborative animal examination and sampling. Thanks also go to Kevin Morgen who is a native English speaker for the modification of the manuscript.References
- T. Requena, P. Cotter, D. R. Shahar, C. R. Kleiveland, M. C. Martínez-Cuesta, C. Peláez and T. Lea, Trends Food Sci. Technol., 2013, 34, 44–53 CrossRef CAS.
- M. R. Hufeldt, D. S. Nielsen, F. K. Vogensen, T. Midtvedt and A. K. Hansen, Comp. Med., 2010, 60, 336–347 CAS.
- R. E. Ley, C. A. Lozupone, M. Hamady, R. Knight and J. I. Gordon, Nat. Rev. Microbiol., 2008, 6, 776–788 CrossRef CAS PubMed.
- L. V. Hooper and A. J. Macpherson, Nat. Rev. Immunol., 2015, 15, 329 CrossRef CAS.
- J. L. Round and S. K. Mazmanian, Nat. Rev. Immunol., 2009, 9, 313–323 CrossRef CAS PubMed.
- M. Banasaz, E. Norin, R. Holma and T. Midtvedt, Appl. Environ. Microbiol., 2002, 68, 3031–3034 CrossRef CAS PubMed.
- H. A. Gordon and E. Bruckner-Kardoss, Am. J. Physiol., 1961, 201, 175–178 CAS.
- A. M. O'Hara and F. Shanahan, EMBO Rep., 2006, 7, 688–693 CrossRef PubMed.
- B. Wostmann and E. Bruckner-Kardoss, Am. J. Physiol., 1959, 197, 1345–1346 CAS.
- H. Ishikawa, K. Tanaka, Y. Maeda, Y. Aiba, A. Hata, N. M. Tsuji, Y. Koga and T. Matsumoto, Clin. Exp. Immunol., 2008, 153, 127–135 CrossRef CAS PubMed.
- D. Bouskra, C. Brezillon, M. Berard, C. Werts, R. Varona, I. G. Boneca and G. Eberl, Nature, 2008, 456, 507–510 CrossRef CAS PubMed.
- A. J. Macpherson and N. L. Harris, Nat. Rev. Immunol., 2004, 4, 478–485 CrossRef CAS PubMed.
- M. Othman, R. Aguero and H. C. Lin, Curr. Opin. Gastroenterol., 2008, 24, 11–16 CrossRef PubMed.
- Y. S. Lopez-Boado, C. L. Wilson, L. V. Hooper, J. I. Gordon, S. J. Hultgren and W. C. Parks, J. Cell Biol., 2000, 148, 1305–1315 CrossRef CAS PubMed.
- A. M. O'Hara and F. Shanahan, EMBO Rep., 2006, 7, 688–693 CrossRef PubMed.
- E. G. Pamer, Mucosal Immunol., 2014, 7, 210–214 CrossRef CAS PubMed.
- H. Tlaskalova-Hogenova, R. Stepankova, H. Kozakova, T. Hudcovic, L. Vannucci, L. Tuckova, P. Rossmann, T. Hrncir, M. Kverka, Z. Zakostelska, K. Klimesova, J. Pribylova, J. Bartova, D. Sanchez, P. Fundova, D. Borovska, D. Srutkova, Z. Zidek, M. Schwarzer, P. Drastich and D. P. Funda, Cell. Mol. Immunol., 2011, 8, 110–120 CrossRef CAS PubMed.
- S. Hapfelmeier, M. A. Lawson, E. Slack, J. K. Kirundi, M. Stoel, M. Heikenwalder, J. Cahenzli, Y. Velykoredko, M. L. Balmer, K. Endt, M. B. Geuking, R. Curtiss III, K. D. McCoy and A. J. Macpherson, Science, 2010, 328, 1705–1709 CrossRef CAS PubMed.
- E. Holmes, J. V. Li, J. R. Marchesi and J. K. Nicholson, Cell Metab., 2012, 16, 559–564 CrossRef CAS PubMed.
- A. Khoruts and M. J. Sadowsky, Mucosal Immunol., 2011, 4, 4–7 CrossRef CAS PubMed.
- O. C. Aroniadis and L. J. Brandt, Curr. Opin. Gastroenterol., 2013, 29, 79–84 CrossRef PubMed.
- H. Diao, Z. Gao, B. Yu, P. Zheng, J. He, J. Yu, Z. Huang, D. Chen and X. Mao, J. Anim. Sci. Biotechnol., 2016, 7, 1–7 CrossRef PubMed.
- M. J. Hamilton, A. R. Weingarden, M. J. Sadowsky and A. Khoruts, Am. J. Gastroenterol., 2012, 107, 761–767 CrossRef PubMed.
- H. Diao, H. L. Yan, Y. Xiao, B. Yu, J. Yu, J. He, P. Zheng, B. H. Zeng, H. Wei and X. B. Mao, BMC Microbiol., 2016, 16, 1–16 CrossRef PubMed.
- B. Zeng, G. Li, J. Yuan, W. Li, H. Tang and H. Wei, Curr. Microbiol., 2013, 67, 313–321 CrossRef CAS PubMed.
- G. K. Hart and G. J. Dobb, JPEN, J. Parenter. Enteral Nutr., 1988, 12, 465–468 CrossRef CAS PubMed.
- X. Mao, C. Gu, H. Hu, J. Tang, D. Chen, B. Yu, J. He, J. Yu, J. Luo and G. Tian, PLoS One, 2016, 11, e0146312 Search PubMed.
- K. S. Kunert, A. S. Tisdale and I. K. Gipson, Arch. Ophthalmol., 2002, 120, 330–337 CrossRef CAS PubMed.
- H. Diao, P. Zheng, B. Yu, J. He, X. Mao, J. Yu and D. Chen, Asian-Australas. J. Anim. Sci., 2015, 28, 827–839 CrossRef CAS PubMed.
- H. Qi, Z. Xiang, G. Han, B. Yu, Z. Huang and D. Chen, Afr. J. Biotechnol., 2011, 10, 3704–3708 CAS.
- N. Fierer, J. A. Jackson, R. Vilgalys and R. B. Jackson, Appl. Environ. Microbiol., 2005, 71, 4117–4120 CrossRef CAS PubMed.
- H. Diao, P. Zheng, B. Yu, J. He, X. Mao, J. Yu and D. Chen, Livest. Sci., 2014, 167, 249–256 CrossRef.
- F. Shanahan, Best Pract. Res., Clin. Gastroenterol., 2002, 16, 915–931 CrossRef.
- O. C. Aroniadis and L. J. Brandt, Curr. Opin. Gastroenterol., 2013, 29, 79–84 CrossRef PubMed.
- H. B. Kim, K. Borewicz, B. A. White, R. S. Singer, S. Sreevatsan, Z. J. Tu and R. E. Isaacson, Vet. Microbiol., 2011, 153, 124 CrossRef PubMed.
- W. Xiao, H. Liu, H. Zhao, L. Bai, J. Peng, X. Liu, J. Wang, Y. Song and B. Cao, Chin. J. Vet. Sci., 2013, 33, 472–476 CAS.
- I. Sekirov, S. L. Russell, L. C. Antunes and B. B. Finlay, Physiol. Rev., 2010, 90, 859–904 CrossRef CAS PubMed.
- W. Yang, F. Meng, J. Peng, P. Han, F. Fang, L. Ma and B. Cao, Electron. J. Biotechnol., 2014, 17, 262–267 CrossRef CAS.
- E. Ransom-Jones, D. L. Jones, A. J. McCarthy and J. E. McDonald, Microb. Ecol., 2012, 63, 267–281 CrossRef CAS PubMed.
- W. Ikeda-Ohtsubo, N. Faivre and A. Brune, Environ. Microbiol. Rep., 2010, 2, 554–559 CrossRef PubMed.
- Q. Niu, P. Li, S. Hao, Y. Zhang, S. W. Kim, H. Li, X. Ma, S. Gao, L. He, W. Wu, X. Huang, J. Hua, B. Zhou and R. Huang, Sci. Rep., 2015, 5, 9938 CrossRef CAS PubMed.
- S. H. Duncan, R. I. Aminov, K. P. Scott, P. Louis, T. B. Stanton and H. J. Flint, Int. J. Syst. Evol. Microbiol., 2006, 56, 2437–2441 CrossRef CAS PubMed.
- C. Liu, S. M. Finegold, Y. Song and P. A. Lawson, Int. J. Syst. Evol. Microbiol., 2008, 58, 1896–1902 CrossRef CAS PubMed.
- A. Partty, L. Lehtonen, M. Kalliomaki, S. Salminen and E. Isolauri, Pediatr. Res., 2015, 78, 470–475 CrossRef CAS PubMed.
- S. K. Mazmanian, C. H. Liu, A. O. Tzianabos and D. L. Kasper, Cell, 2005, 122, 107–118 CrossRef CAS PubMed.
- I. I. Ivanov, L. Frutos Rde, N. Manel, K. Yoshinaga, D. B. Rifkin, R. B. Sartor, B. B. Finlay and D. R. Littman, Cell Host Microbe, 2008, 4, 337–349 CAS.
- T. Lähteinen, T. Rinttilä, J. M. K. Koort, R. Kant, K. Levonen, M. Jakava-Viljanen, J. Björkroth and A. Palva, Livest. Sci., 2015, 180, 164–171 CrossRef.
- M. Kverka, Z. Zakostelska, K. Klimesova, D. Sokol, T. Hudcovic, T. Hrncir, P. Rossmann, J. Mrazek, J. Kopecny, E. F. Verdu and H. Tlaskalova-Hogenova, Clin. Exp. Immunol., 2011, 163, 250–259 CrossRef CAS PubMed.
- M. H. Kim, S. G. Kang, J. H. Park, M. Yanagisawa and C. H. Kim, Gastroenterology, 2013, 145, 396–406 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra11234c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |