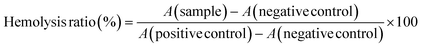Open Access Article
Open Access ArticleMetabolic labelling of cancer cells with glycodendrimers stimulate immune-mediated cytotoxicity†
David
Goyard
,
Peremobowei Iyanu
Diriwari
and
Nathalie
Berthet
 *
*
Univ. Grenoble Alpes, CNRS, DCM UMR 5250, F-38000 Grenoble, France. E-mail: nathalie.berthet@univ-grenoble-alpes.fr
First published on 9th November 2021
Abstract
The recruitment of antibody naturally present in human blood stream at the surface of cancer cells have been proved a promising immunotherapeutic strategy to fight cancer. Antibody recruiting molecules (ARMs) combining tumor and antibody binding modules have been developed for this purpose, however the formation of the interacting complex with both antibody and cell is difficult to optimize to stimulate immune-mediated cytotoxicity. To circumvent this limitation, we report herein a more direct approach combining cell metabolism of azido-sugar and bio-orthogonal click chemistry to conjugate at the cell glycocalyx structurally well-defined glycodendrimers as antibody binding module (ABM). We demonstrate that this strategy allows not only the recruitment of natural antibody at the surface of isolated cells or solid tumor models but also activate a cytotoxic response with human serum as unique source of immune effectors.
Introduction
The exploitation of endogenous antibodies naturally present in the blood stream of all individuals has recently emerged as an alternative immuno-strategy to fight cancer.1–3 To this aim, synthetic antibody recruiting molecules (ARMs) combining two binding modules, one for tumor cell (TBM) and the other for the antibody recruitment (ABM, typically dinitrophenol, αGal or Rha) have been demonstrated to successfully trigger immune cytotoxicity against cancer cells by CDC or ADCC mechanisms.4–7 From the first generation of ARMs to the more sophisticated antibody recruiting polymers (ARPs)8 or glycodendrimers (ARGs),8 significant advances have been made in the understanding of functional and structural requirements to improve immunological effects. If the multivalent presentation of ABM was clearly demonstrated as a key element to recruit endogenous antibodies,10,11 the major shortcoming of this approach concerns the TBM which has to ensure the binding of the cell surface without promoting internalisation to maintain the ABM exposure and accessibility at the cell surface. By doing so, the recruiting molecule can promote the formation of a reversible ternary interacting complex with antibodies and cancer cell. When suitable conditions are used to control this complex equilibrium process,12 the immune-mediated cytotoxic effect can be activated against the cancer cell line.13,14 To avoid the utilization of TBM and the problems associated with interacting systems involving three partners, the insertion of dinitrophenol or carbohydrate haptens in the cell membrane either with a lipid anchor or by covalent conjugation was proved to be a valuable alternative as a simplified antibody recruiting system.8,15–19In this regard, the use of well-known substrates for cell surface engineering, such as the tetraacetyl-N-azidoacetyl-mannosamine (Ac4ManNAz), represents a powerful and reliable method to modify the glycocalyx with unnatural recognition moieties. Once internalized, the intracellular metabolism of Ac4ManAz into azido sialic acid indeed leads to azido group expression onto extracellular glycans that can be engaged in bio-orthogonal reaction to further decorate the cell membrane by copper-free strain-promoted azide–alkyne cycloaddition (SPAAC).20–25 Several groups used similar approach to conjugate diverse haptens on the membrane of different cells to promote immune-mediated cytotoxicity.15,16,26 In this study, we reasoned that the conjugation of clustered ABM at the surface of cancer cells would represent a robust antibody recruiting approach. We recently identified an ARG composed of a tetravalent cluster of cRGD as TBM and an hexadecavalent dendrimer of Rha as ABM to redirect natural antibodies against cancer cells expressing αvβ3 integrins.11 This compound has been shown to stimulate immune-mediated cytotoxicity against this cell line. Among the variety of the tested compounds, we demonstrated that ARGs presenting the high rhamnose valency with a precise spatial orientation were able to recruit antibodies from human serum at the surface of cancer cells and to subsequently activate an immune response.9 Herein, we exploited a metabolic labelling strategy to attach multivalent ABMs at the cell surface. To this aim, Ac4ManNAz was first delivered to BT-549 for the surface tagging with azido group (Scheme 1, step 1). The coupling with dibenzocyclooctyne (DBCO)-bearing ABMs presenting one, four or sixteen rhamnoses to the cell membrane via copper-free chemistry was performed in the second step (Scheme 1, step 2). Finally, we evaluated the recruitment of natural antibodies present in human serum (Scheme 1, step 3) and the subsequent stimulation of the immune response (Scheme 1, step 4).
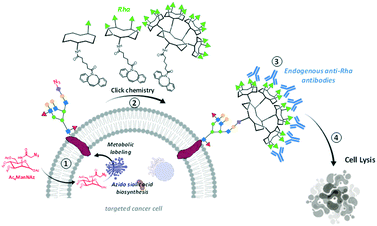 | ||
| Scheme 1 Strategy combining glycometabolism and bio-orthogonal click chemistry to label cells with clustered rhamnose antigen and activate immune response against cancer cells. | ||
Results and discussion
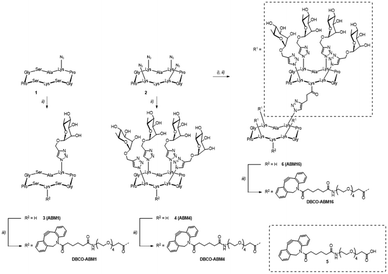
ABMs were synthesized as previously reported.11 Briefly, mono- 1 and tetraazido 2 cyclodecapeptides were functionalized via CuAAC using propargylated α-L-Rha to afford glycopeptides 3 and 4 respectively. Subsequent amide coupling between carboxylic acid-bearing DBCO derivative 5 and the free lysine side chain of compounds 3 and 4 yielded mono- and tetravalent compounds DBCO-ABM1 and DBCO-ABM4 respectively. Tetravalent glycocluster 4 was functionalized with pentynoic acid and subjected to another CuAAC reaction with scaffold 2. The resulting hexadecavalent glycodendrimer 6 was finally conjugated with DBCO derivative 5 to afford DBCO-ABM16. The synthesis route is depicted in Scheme 2. Final compounds were characterized by HRMS, 1H NMR and analytical RP-HPLC before biological studies (see ESI†).We first determined the optimal concentration of Ac4ManNAz for metabolic labelling with azido groups of the targeted cells. For that, the triple negative breast cancer cell line BT-549 was cultivated with various concentrations of Ac4ManNAz for 24 h and the conversion in azido sialic acid was followed by treatment with the commercial DBCO-PEG4-Fluor 545. The analyse by flow cytometry and confocal microscopy of the cell fluorescence enabled to determine optimal cell surface labelling for a concentration in Ac4ManNAz of 50 μM. Negligible fluorescence was observed for cells untreated with Ac4ManNAz confirming the absence of non-specific DBCO-dye binding to the cells (Fig. S1, ESI†).
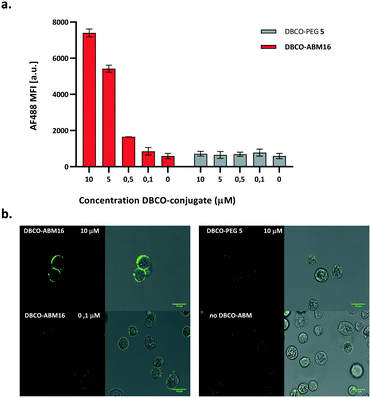
We next investigated the coupling of DBCO-conjugates (DBCO-ABM16, Fig. 1 and DBCO-ABM1–4, Fig. S2 ESI†) by SPAAC to azido-bearing cells. Extracellular Rha exposure was revealed using anti-rhamnose IgM naturally present in human serum (HS). Azido tagged BT-549 cells were treated with or without DBCO-ABM (concentrations from 0.1 to 10 μM) or DBCO-PEG 5 used as negative control. The SPAAC coupling of the DBCO-conjugates was evaluated by flow cytometry and confocal microscopy after successive incubations with HS and AlexaFluor488- anti-human IgM secondary antibody. A dose-dependent effect with a maximum fluorescence intensity at 10 μM was observed with all DBCO-ABMs (Fig. 1a, Fig. S2, ESI†) demonstrating the efficiency of the bio-orthogonal SPAAC coupling. The dose–response curve of the fluorescence of treated cells with various concentrations of DBCO-ABM16 (Fig. S3, ESI†) clearly showed a plateau at 10 μM. This result was confirmed by confocal microscopy of cells tagged with ABM16 (Fig. 1b) which revealed an intense fluorescence homogenously distributed around the cell surface at 5 and 10 μM of DBCO-conjugate, while no significant fluorescence was detected at lower concentration. Of note, no fluorescence was observed for azido-bearing cells or those treated with 10 μM of DBCO-PEG 5, thus indicating the absence of non-specific binding of anti-Rha antibodies.
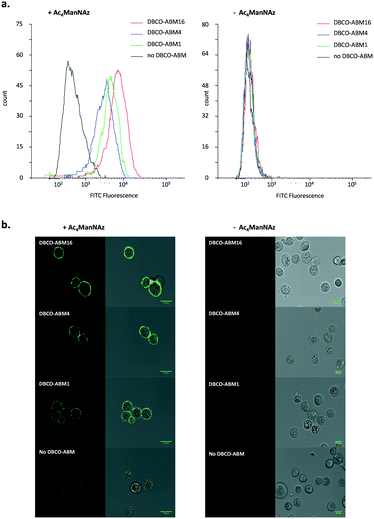
The influence of the Rha density on the recruitment of serum antibodies was next evaluated. Cells pre-incubated with or without Ac4ManNAz were treated with 10 μM of mono, tetra- and hexadecavalent DBCO-ABMs. The anti-Rha IgM recruitment was revealed by immuno-fluorescence as described earlier and analysed by flow cytometry and confocal microscopy. As shown by the histogram overlays reported in Fig. 2a, antibodies present in HS are efficiently recruited by all modified cell surface with DBCO-derivatives with a significantly higher efficiency for cells displaying the higher Rha density at their surface. Confocal microscopy confirmed this result since the highest fluorescence and homogenous dye labelling was observed with the ABM16 (Fig. 2b).
Interestingly, mono- and tetravalent ABM1 and ABM4 covalently anchored to the cell membrane were able to recruit antibodies present in HS while ARGs previously reported with low Rha density failed.11 These results demonstrate the interest of the metabolic labelling strategy to recruit antibody onto cell surface. In addition, flow cytometry analysis and confocal microscopy experiment with cells untreated with Ac4ManNAz or DBCO conjugates revealed negligible fluorescent intensity, thus suggesting the binding specificity of anti-Rha antibodies (Fig. 2b).
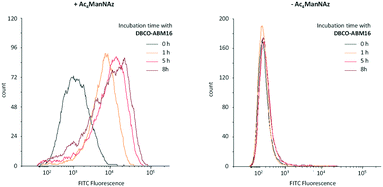
Antibody recruitment being dependent on the presence and the persistence of ABMs on the cell surface, we next studied the stability of the cell labelling with ABM. Cells pre-treated with or without Ac4ManNAz were cultured for 1–8 h with 10 μM of DBCO-ABM16 prior to being incubated with HS and the secondary antibody. The cell fluorescence was then analysed by flow cytometry. As shown in Fig. 3 persistent cell labelling with ABM16 is observed after 8 hours for cells pre-treated with Ac4ManNAz and an increase of the fluorescent intensity is also measured with increasing incubation time. These results demonstrate the stability of cell labelling with ABM over the time. However a fluorescent signal similar to the autofluorescence of the cells was observed for non azido-labelled cells incubated with the DNCO conjugate even after 8 hours showing one more time the specific ABM labelling for cells pre-treated with Ac4ManNAz.
Since DBCO-ABM conjugates were attached onto the cells for antibodies recruitment in the human serum, we also evaluated their blood compatibility in an in vitro assay. This was performed by determining the effect of DBCO-ABMs (1, 4 and 16) on human red blood cells (hRBC). Like saline treatment, incubating hRBC with all DBCO-ABMs in saline did not cause detectable changes on morphology of hRBC under light microscopy even at a concentration 10 time higher than which was showing cytotoxic effect (100 μM) (Fig. 4a). The hemolysis ratios for each DBCO-ABM treatments was calculated. Less than 2% of hemolysis was obtained for all conjugates, indicating a good blood compatibility of the DBCO conjugates (Fig. 4b).
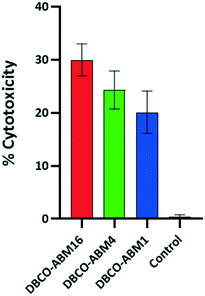
We next evaluated the ability of the recruited antibodies to promote immune response leading to cancer cell destruction. Azido tagged cells treated with or without DBCO-ABMs were incubated with human serum as unique source of antibodies and protein complement and the cell viability assay which measures the complement-dependent cytotoxicity (CDC) was examined as reported previously. The cytotoxicity effect was found to increase with the ABM valency (Fig. 5), the lowest effect being observed for cells modified with the monovalent ABM1 (20%) and the highest with the hexadecavalent ABM16 (30%). This result is in good correlation with the antibody recruitment observed by flow cytometry. By comparison with previous studies reported by other groups,16,27 the conjugation of ABM16 promote similar cytotoxic effect at much lower concentration, thus suggesting the importance of a dense presentation in hapten to the cell surface.
In order to evaluate the ability of ABMs to recruit antibody in solid tumor context, we cultured 3D-spheroid which are commonly used as in vitro model of such tumor.28,29 Spheroids grown 4 days were cultured with or without Ac4ManNAz (untreated spheroids were used as control) prior to be treated with DBCO-ABM16 and HS (as 2D culture). As described before, the recruitment of endogenous anti-Rha was revealed with AF488-conjugated anti-human secondary antibody.
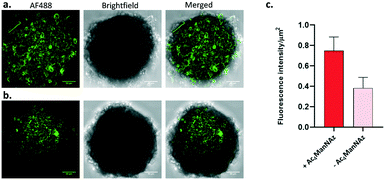
Confocal microscopy images (Fig. 6) showed high fluorescence for spheroids cultured with Ac4ManNAz and treated with DBCO-ABM16 compared to the control spheroids. The measure of the ratio of the Fluorescence intensity reported to the spheroid surface revealed a fluorescence intensity 2 times higher for cells incubated with Ac4ManNAz compared to the control spheroids. These results clearly indicate the effectiveness of the spheroid labelling and the accessibility of Rha antigens for IgM recruitment at the cell surface to induce a targeted immune response against BT-549 tumor cells.
Conclusions
In summary, we have demonstrated the robustness of cell glycometabolism and bio-orthogonal click chemistry to decorate the cell glycocalyx with antibody recruiting modules. We have clearly observed that such structural modification has the ability to redirect natural antibodies present in human serum, not only against BT-549 cell line but also against 3D-spheroid model of solid tumor. More importantly, the covalent grafting of chemically well-defined ABM with high Rha density at the cell surface was found to stimulate immune-mediated cytotoxicity of BT-549. By avoiding the use of tumor binding module which significantly complicates the utilization of bimodal antibody recruiting molecules, we believe that the present strategy combining delivery of azido sugar to cancer cell and bio-orthogonal ligation of synthetic antibody recruiting modules is highly promising in the context of cancer immunotherapy. The exploitation of this approach with azido monosaccharide embedded with targeting agent and cleavable linker is however required for a selective delivery in cancer cells. We are currently investigating strategies based on the use of receptor-mediated endocytosis or enzyme activatable azido sugar by leveraging dysregulated mechanisms leading to enzyme-overexpression in cancer cells. The results will be reported in due course.Experimental
Materials
Tetraacetylated N-azidoacetyl-D-mannosamine (Ac4ManNAz), Alexa-Fluor 488-coupled anti-human IgM were purchased from Fisher Scientific. All other chemical reagents were purchased from Aldrich (SaintQuentin Fallavier, France) or Acros (Noisy-Le-Grand, France) and were used without further purification. All protected amino acids and Fmoc-Gly-Sasrin®resin was obtained fromAdvanced ChemTech Europe (Brussels, Belgium), BachemBiochimie SARL (Voisins-Les-Bretonneux, France) and FranceBiochem S.A. (Meudon, France). Human serum (HS) were obtained from a healthy human male donor (Etablissement Français du Sang, EFS Grenoble). Cell Cytotoxicity assay kit abcam 112![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 118 was from Abcam. All experiments were performed in accordance with the Guidelines of the “Etablissement Français du Sang (EFS) Auvergne-Rhône Alpes 2017-2958”, and Experiments were approved by the ethics committee at the EFS AURA 21-033. Informed consents were obtained from human participants of this study. For peptides and glycopeptides, analytical RP-HPLC was performed on a Waters alliance 2695 separation module, equipped with a Waters 2489 UV/visible detector. Analyses were carried out at 1.23 mL min−1 (Waters XBridge Shield RP18 3.5 μM, C18, 100 × 4.6 mm) with UV monitoring at 214 nm and 250 nm using a linear A–B gradient (buffer A: 0.09% CF3CO2H in water; buffer B: 0.09%CF3CO2H in 90% acetonitrile). Preparative HPLC was performed on waters equipment consisting of a Waters 2545 controller and a Waters 2487 dual absorbance detector. Purifications were carried out at 22 mL min−1 (VP 250 × 21 mm nucleosil100-7 C18) with UV monitoring at 214 nm and 250 nm using a linear A–B gradient. 1H NMR spectra were recorded on BrukerAvanceIII 500 MHz spectrometers and chemical shifts (δ) were reported in parts per million (ppm). Spectra were referenced to the residual proton solvent peaks relative to the signal of D2O (4.79 ppm). All mass spectrometry characterizations were performed at mass spectrometry facility, PCN-ICMG, Grenoble. ESI† high resolution mass spectra of peptides and glycopeptides were measured on a LTQ Orbitrap XL spectrometer from Thermo Scientific.
118 was from Abcam. All experiments were performed in accordance with the Guidelines of the “Etablissement Français du Sang (EFS) Auvergne-Rhône Alpes 2017-2958”, and Experiments were approved by the ethics committee at the EFS AURA 21-033. Informed consents were obtained from human participants of this study. For peptides and glycopeptides, analytical RP-HPLC was performed on a Waters alliance 2695 separation module, equipped with a Waters 2489 UV/visible detector. Analyses were carried out at 1.23 mL min−1 (Waters XBridge Shield RP18 3.5 μM, C18, 100 × 4.6 mm) with UV monitoring at 214 nm and 250 nm using a linear A–B gradient (buffer A: 0.09% CF3CO2H in water; buffer B: 0.09%CF3CO2H in 90% acetonitrile). Preparative HPLC was performed on waters equipment consisting of a Waters 2545 controller and a Waters 2487 dual absorbance detector. Purifications were carried out at 22 mL min−1 (VP 250 × 21 mm nucleosil100-7 C18) with UV monitoring at 214 nm and 250 nm using a linear A–B gradient. 1H NMR spectra were recorded on BrukerAvanceIII 500 MHz spectrometers and chemical shifts (δ) were reported in parts per million (ppm). Spectra were referenced to the residual proton solvent peaks relative to the signal of D2O (4.79 ppm). All mass spectrometry characterizations were performed at mass spectrometry facility, PCN-ICMG, Grenoble. ESI† high resolution mass spectra of peptides and glycopeptides were measured on a LTQ Orbitrap XL spectrometer from Thermo Scientific.
Synthetic procedures and characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1) and purified by semi preparative RP-HPLC. Fractions containing the product were collected and lyophilized to obtain the desired compound as a white fluffy solid.
0.1) and purified by semi preparative RP-HPLC. Fractions containing the product were collected and lyophilized to obtain the desired compound as a white fluffy solid.
Instrumentation
Flow cytometry analysis was performed on a BD LSR FORTESSA Flow cytometer (laser excitation at λ = 488 nm, emission bypass filter at 525/50).Confocal microscopy images of 2D and 3D-cultures were taken with a TCS SP8 CSU Leica (HC PL APO CS2, 40×/1.30 OIL, laser excitation at λ = 488 nm and fluorescence emission collected between λ = 495 and 545 nm). Images were processed using ImageJ software.
The solution-absorbance in microplates was read on a POLARstar Omega plate reader (BMG labtech).
Cell culture method
BT-549 cells were purchased from ATCC and were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 μg mL−1 streptomycin and 100 U mL−1 penicillin (all from Sigma-Aldrich). Cells were maintained in an incubator at 37 °C in a 5% CO2 humidified atmosphere and allowed for downstream experiment until 90% confluency.Determination of the optimal concentration of Ac4ManN3 for in vitro metabolic cell labelling
BT-549 cells were seeded at a density of about 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 6-well plates and cultured for 24 h in the medium described in the cell culture method section. Various dilutions of Ac4ManNAz were prepared and add directly to the culture medium to get final concentrations from 1 μM to 50 μM and the culture was continued for 48 h. The culture medium was then removed and cells were washed 3 times with PBS prior to be incubated with 50 μM DBCO-PEG4-Fluor 545 diluted in HBSS for 1 h at 37 °C (50 mM stock solution in DMSO was prepared). Control cells without azido sugar received the same amount of DMSO. Cells were washed again 3 times with PBS then were detached by treatment with 0.5 mL per well of trypsin-EDTA for 5 min at 37 °C. Afterwards, the harvest cells were centrifuged, washed with 1 mL of PBS and resuspended in 500 μL of PBS for analysis by flow cytometry.
000 cells per well in 6-well plates and cultured for 24 h in the medium described in the cell culture method section. Various dilutions of Ac4ManNAz were prepared and add directly to the culture medium to get final concentrations from 1 μM to 50 μM and the culture was continued for 48 h. The culture medium was then removed and cells were washed 3 times with PBS prior to be incubated with 50 μM DBCO-PEG4-Fluor 545 diluted in HBSS for 1 h at 37 °C (50 mM stock solution in DMSO was prepared). Control cells without azido sugar received the same amount of DMSO. Cells were washed again 3 times with PBS then were detached by treatment with 0.5 mL per well of trypsin-EDTA for 5 min at 37 °C. Afterwards, the harvest cells were centrifuged, washed with 1 mL of PBS and resuspended in 500 μL of PBS for analysis by flow cytometry.
In vitro coupling of DBCO-ARMs to azido labeled cells and antibody recruitment analysis
BT-549 cells (150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 6-well plates) were cultured for 24 h prior to be incubated with 50 μM Ac4ManNAz as described above. After culture medium removing, cells were washed 3 times with PBS then the copper-free click chemistry of azido-labeled cells with various concentrations of each DBCO-ABMs in HBSS (0 μM, 0.1 μM, 0.5 μM, 5 μM and 10 μM) was performed for 1 h at 37 °C. Cells were then washed twice with PBS and were detached with trypsin-EDTA as described previously. Washed cells were then incubated with 50% human serum in PBS-1% BSA for 1 h at 37 °C. Following 1 wash with PBS-1% BSA, cells were incubated with 1
000 cells per well in 6-well plates) were cultured for 24 h prior to be incubated with 50 μM Ac4ManNAz as described above. After culture medium removing, cells were washed 3 times with PBS then the copper-free click chemistry of azido-labeled cells with various concentrations of each DBCO-ABMs in HBSS (0 μM, 0.1 μM, 0.5 μM, 5 μM and 10 μM) was performed for 1 h at 37 °C. Cells were then washed twice with PBS and were detached with trypsin-EDTA as described previously. Washed cells were then incubated with 50% human serum in PBS-1% BSA for 1 h at 37 °C. Following 1 wash with PBS-1% BSA, cells were incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 AF488-conjugated goat anti-human antibody for 1 h at 37 °C. Finally, the cells were washed and resuspended in 500 μL PBS and analyzed for FITC intensity by flow cytometry and confocal microscopy.
400 AF488-conjugated goat anti-human antibody for 1 h at 37 °C. Finally, the cells were washed and resuspended in 500 μL PBS and analyzed for FITC intensity by flow cytometry and confocal microscopy.
Complement-mediated cytotoxicity
BT-549 cells were plated onto the 96-well plate at the concentration of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well and left to adhere for 24 h prior to be incubated with 50 μM Ac4ManNAz for 48 h at 37 °C, 5% CO2. Plates were then washed twice with PBS and click chemistry with 10 μM DBCO-ABMs in HBSS was performed for 1 h at 37 °C. Solution was then removed and cells were incubated with 100 μL of HS 50% in HBSS for 2 h at 37 °C. Subsequently, 20% (v/v) abcam reagent was added and the plates were further incubated for 3 h at 37 °C, 5% CO2. The plates were then read at OD570 to OD605 on microplate reader. The percentage of cell cytotoxicity was calculated using the following formula:
000 cells per well and left to adhere for 24 h prior to be incubated with 50 μM Ac4ManNAz for 48 h at 37 °C, 5% CO2. Plates were then washed twice with PBS and click chemistry with 10 μM DBCO-ABMs in HBSS was performed for 1 h at 37 °C. Solution was then removed and cells were incubated with 100 μL of HS 50% in HBSS for 2 h at 37 °C. Subsequently, 20% (v/v) abcam reagent was added and the plates were further incubated for 3 h at 37 °C, 5% CO2. The plates were then read at OD570 to OD605 on microplate reader. The percentage of cell cytotoxicity was calculated using the following formula:| %cytotoxicity = [1 − (Rsample − Ro)/(Rctrl − Ro)] × 100 |
R ctrl was the absorbance ratio of OD570/OD605 for unlabelled cells incubated with HS.
R o was the averaged background (non-cell control) absorbance ratio OD570/OD605.
BT-549 tumor spheroids
Spheroids were generated by plating BT-549 cells at 5000 cells per well into ultralow adherence-96-well plates (Corning). Spheroids grew in complete medium as in 2D-cultures in the final volume of 200 μL. After 72 h, spheroids were treated with 50 μM Ac4ManNAz and 10 μM DBCO-ABMs in the same manner as in 2D-cultures. Spheroids were imaged using confocal microscope (TCS SP8 CSU, Leica, laser excitation at λ = 448 nm and fluorescence emission collected between λ = 495 and 545 nm) equipped with a 40× objective and analysis was performed with the ImageJ software.Human red blood cell hemolysis assay
All experiments were performed in accordance with the Guidelines of the “Etablissement Français du Sang (EFS) Auvergne-Rhône Alpes 2017-2958”, and Experiments were approved by the ethics committee at the EFS AURA 21-033. Informed consents were obtained from human participants of this study. Human red blood (hRBC) were obtained by centrifugation of 2 mL of whole blood collected in heparin tubes, from healthy donors (EFS Grenoble) at 800 g for 10 min at room temperature. After removal of the plasma, hRBC were further purified by washing three times with PBS and resuspended in PBS at a final concentration of 2%. The hRBC suspensions (180 μL) were incubated with 20 μL of DBCO-ABM 1, 4 or 16 (1 mM in PBS) while parallel assays were performed using PBS as negative control and a 1% (w/v) solution of Triton X-100 as positive control. After incubation at 37 °C in 5% CO2 for 2 h, each sample was centrifuged for 10 min at 800 g and 100 μL of the supernatant was transferred to a 96-well microplate. Absorbance at 540 nm was measured using a microplate reader and the results were expressed as a percentage of hemoglobin released relative to the positive control. All treatments were performed in triplicates and the hemolysis ratio (%) was calculated using equation:The integrity of the precipitated hRBC was checked by morphological observations under light microscopy.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by CNRS, Université Grenoble Alpes, ICMG FR 2607, the French ANR project Glyco@Alps (ANR-15-IDEX-02), Labex ARCANE and CBH-EUR-GS (ANR-17-EURE-0003). We acknowledges the European Research Council Consolidator Grant “LEGO” (647938). We thank Cecile Cottet (LBFA, Grenoble) for confocal microscopy experiments, the data analysis and for giving access to flow cytometer.References
- K. Sasaki, M. Harada, Y. Miyashita, H. Tagawa, A. Kishimura, T. Mori and Y. Katayama, Chem. Sci., 2020, 11, 3208 RSC.
- B. Lake, N. Serniuck, E. Kapcan, A. Wang and A. F. Rullo, ACS Chem. Biol., 2020, 15, 1089 CrossRef CAS PubMed.
- H. Hong, C. Li, L. Gong, J. Wang, D. Li, J. Shi, Z. Zhou, Z. Huang and Z. Wu, Chem. Sci., 2021, 12, 4623 RSC.
- P. J. McEnaney, C. G. Parker, A. X. Zhang and D. A. Spiegel, ACS Chem. Biol., 2012, 7, 1139 CrossRef CAS PubMed.
- P. J. McEnaney, C. G. Parker and A. X. Zhang, in Annu. Rep. Med. Chem., 2017, p. 481 Search PubMed.
- R. T. C. Sheridan, J. Hudon, J. A. Hank, P. M. Sonde and L. L. Kiessling, ChemBioChem, 2014, 15, 1393–1398 CrossRef CAS PubMed.
- S. Achilli, N. Berthet and O. Renaudet, RSC Chem. Biol., 2021, 2, 713 RSC.
- A. Uvyn, R. De Coen, M. Gruijs, C. W. Tuk, J. De Vrieze, M. van Egmond and B. G. De Geest, Angew. Chem., 2019, 131, 13122 CrossRef.
- B. Todaro, S. Achilli, B. Liet, E. Laigre, C. Tiertant, D. Goyard, N. Berthet and O. Renaudet, Biomater. Sci., 2021, 9, 4076 RSC.
- A. Uvyn and B. G. De Geest, ChemBioChem, 2020, 21, 3036 CrossRef CAS PubMed.
- B. Liet, E. Laigre, D. Goyard, B. Todaro, C. Tiertant, D. Boturyn, N. Berthet and O. Renaudet, Chem. – Eur. J., 2019, 25, 15508 CrossRef CAS PubMed.
- E. F. Douglass, C. J. Miller, G. Sparer, H. Shapiro and D. A. Spiegel, J. Am. Chem. Soc., 2013, 135, 6092 CrossRef CAS PubMed.
- C. B. Calson, P. Mowery, R. M. Owen, E. C. Dykhuizen and L. L. Kiessling, ACS Chem. Biol., 2007, 2, 119 CrossRef.
- A. F. Rullo, K. J. Fitzgerald, V. Muthusamy, M. Liu, C. Yuan, M. Huang, M. Kim, A. E. Cho and D. A. Spiegel, Angew. Chem., Int. Ed., 2016, 55, 3642 CrossRef CAS PubMed.
- A. Uvyn, R. De Coen, O. De Wever, K. Deswarte, B. N. Lambrecht and B. G. De Geest, Chem. Commun., 2019, 55, 10952 RSC.
- S. Li, B. Yu, J. Wang, Y. Zheng, H. Zhang, M. J. Walker, Z. Yuan, H. Zhu, J. Zhang, P. G. Wang and B. Wang, ACS Chem. Biol., 2018, 13, 1686 CrossRef CAS PubMed.
- C. E. Jakobsche, C. G. Parker, R. N. Tao, M. D. Kolesnikova, E. F. Douglass and D. A. Spiegel, ACS Chem. Biol., 2013, 8, 2404 CrossRef CAS PubMed.
- B. Lin, X. Wu, H. Zhao, Y. Tian, J. Han, J. Liu and S. Han, Chem. Sci., 2016, 7, 3737 RSC.
- X. Li, X. Rao, L. Cai, X. Liu, H. Wang, W. Wu, C. Zhu, M. Chen, P. G. Wang and W. Yi, ACS Chem. Biol., 2016, 11, 1205 CrossRef CAS PubMed.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046 CrossRef CAS PubMed.
- E. M. Sletten and C. R. Bertozzi, Acc. Chem. Res., 2011, 44, 666–676 CrossRef CAS PubMed.
- J. C. Jewett and C. R. Bertozzi, Chem. Soc. Rev., 2010, 39, 1272–1279 RSC.
- Y. Takayama, K. Kusamori and M. Nishikawa, Molecules, 2019, 24, 172 CrossRef.
- A. Mongis, F. Piller and V. Piller, Bioconjugate Chem., 2017, 28, 1151 CrossRef CAS.
- H. Wang and D. J. Mooney, Nat. Chem., 2020, 12, 1102 CrossRef CAS PubMed.
- S. H. Park, H. Jung, H. Lee, T. M. Kim, J. W. Cho, W. D. Jang, J. Y. Hyun and I. Shin, Chem. Commun., 2020, 56, 10650 RSC.
- X. Li, X. Xu, X. Rao, Y. Tian and W. Yi, Carbohydr. Res., 2017, 452, 25 CrossRef CAS.
- C. J. Lovitt, T. B. Shelper and V. M. Avery, Biology, 2014, 3, 345 CrossRef PubMed.
- D. Anton, H. Burckel, E. Josset and G. Noel, Int. J. Mol. Sci., 2015, 16, 5517 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1md00262g |
| This journal is © The Royal Society of Chemistry 2022 |