 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DNA interfaces with dimensional materials for biomedical applications
Narges Asefifeyzabadi,
Prabhangshu Kumer Das,
Avokerie Hillary Onorimuo,
Grace Durocher and
Mohtashim Hassan Shamsi *
*
School of Chemical and Biomolecular Sciences, Southern Illinois University, Carbondale, IL, USA. E-mail: mshamsi@siu.edu; Fax: +1-618-453-6408; Tel: +1-618-453-6461
First published on 23rd August 2021
Abstract
DNA interfaces with nano, micro, and macro materials have gained widespread attention for various applications. Such interfaces exhibit distinct functions and properties not only due to the unique properties of interfacing materials but also sequence- and conformation-dependent characteristics of the DNA. Therefore, DNA interfaces with diverse dimensional materials have advanced our understanding of the interaction mechanisms and the properties of such interfaces. The unique interfacial properties of such novel materials have applications in nanotechnology, biophysics, cell biology, biosensing, and bioelectronics. The field is growing rapidly with the frequent emergence of new interfaces carrying remarkable interfacial character. In this review article, we have classified the DNA interfaces into 0D, 1D, 2D, and 3D categories based on the types of dimensional materials. We review the key efforts made in the last five years and focus on types of interfaces, interfacing mechanisms, and their state-of-the-art applications. This review will draw a general interest because of the diversity in the DNA materials science but also the unique applications that will play a cutting-edge role in biomedical and biosensing research.
1 Introduction
DNA interactions with metals and their ions were the focus in the past for their change in structural proeprties and applications.1 However, DNA interfaces with diverse dimensional materials (i.e. 0D, 1D, 2D, and 3D), Fig. 1a, are rapidly growing having diverse interaction mechanisms, nucleic acid behavior, and interfacial properties. The DNA–dimensional materials interfaces have wide applications in nanotechnology, biophysics, cell biology, biosensing, bioelectronics biomedical research.2–5 DNA interfaces with dimensional materials, Fig. 1b, demonstrate distinct physical and chemical properties that can be promising for signal reporting, transduction mechanism, and amplification for a wide range of biological applications.6–9 Interfacing mechanisms have an important role in achieving desired properties for such materials.10 The optical properties (e.g. absorbance, reflectance, scattering, plasmon resonance, and luminescence) of the designed materials can be monitored for optical diagnostic systems.11 Electrical and electrochemical techniques monitor charge transport at the DNA–materials interfaces, which can be transformed into simple and miniaturized platforms with high sensitivity.12,13 Surface properties of the interfaces through surface probe microscopy can facilitate fabrication of smart and efficient devices.14 Development and understanding of such interfaces are critical to develop biosensors on chips,15 and integrated microfluidic platforms for rapid and low-cost detection of genetic biomarkers.16,17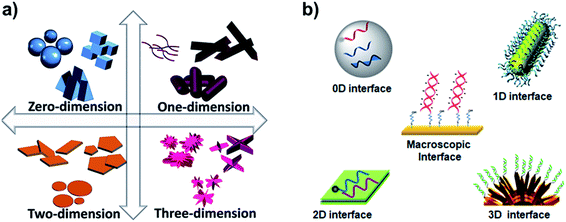 | ||
| Fig. 1 (a) Types of dimensional materials. Adapted from IntechOpen, DOI: 10.5772/intechopen.83803. (b) Types of DNA interfaces with dimensional materials. | ||
With all the growth in this research area, there is a constant need to review new developments. This review is distinct from DNA-based nanostructures (e.g. tetrahedral or origamis), which have been constantly reviewed.18–22 In the last five years, the reviews of DNA interfaces have covered only specific types of materials (e.g. gold, graphene, carbon nanotubes etc.)23–26 or a category of materials (e.g. 2D materials, nanoparticles).27–29 Here, we are reviewing all types of dimensional interfaces of the nanomaterials, except macroscopic interfaces (e.g. surface-assemblies on microbeads and macro electrode surfaces),30–32 with key developments in the biosensing applications of the DNA–materials interfaces since 2015.
1.1. 0D interfaces
Zero-dimensional materials represent a class of nanomaterials with all dimensions in nanometer scale in spherical or crystal shapes, e.g. quantum dots, nanoparticles, nanoclusters etc.33 The 0D materials have gained the major interest since the beginning of nanotechnology in the 1980s due to their powerful electronic, magnetic, and optical properties for broad applications. They differ incredibly in their properties due to their chemical makeup, size, shape, and environment. There is a plethora of applications associated with 0D materials from solar cells to drug delivery. It would be out of the scope of this review to list them all. Three major types of DNA–0D interfaces are discussed in this review because DNA interfaces with other types, e.g. polymer, metal–organic frameworks, are in their beginning stages.Liu et al. leveraged on van der Waals interaction between DNA nucleobases and AgNPs to form nucleic acid-stabilized silver nanoclusters and designed a multipurpose molecular beacon probe so called ‘activatable silver nanoclusters beacon’ (ASNCB), as shown in Fig. 2a.43 It relies on target recognition induced conformational transition of the probe and yields fluorescent signal of silver nanoclusters. With slight variation in the probe, they were able to detect influenza A virus genes, ATP, and thrombin protein. By further implementing two different color ASNCBs, the ASNCB probe was implemented for ATP imaging in living cells. Park et al. developed functional DNA-decorated Au nanomachines for triple combinatorial anti-tumor therapy, which exploited photodynamic and photothermal properties of G-quadruplex and i-motifs modified Au nanoparticles (Au-GI) shown in Fig. 2b.44 Specifically, an anti-cancerous drug doxorubicin (DOX) and a zinc phthalocyanine photosensitizer (ZnPc) were loaded onto the i-motif/cDNA duplex and G-quadruplex, respectively. When the Au-GI was internalized into cancer cells of triple negative breast cancer (TNBC) tumor model, i-motif structure constituted between the nearby Au-GIs due to the acidic environment in the cells causing aggregation of Au-GIs leading to heat generation under NIR irradiation, thus photothermal ablation of cancer cells. While simultaneously the loaded DOX is released from i-motif/cDNA duplex because of the dehybridization. The photosensitizer (ZnPc) in the G-quadruplex produces singlet oxygen under illumination by 660 nm light, contributing to the photodynamic therapy.
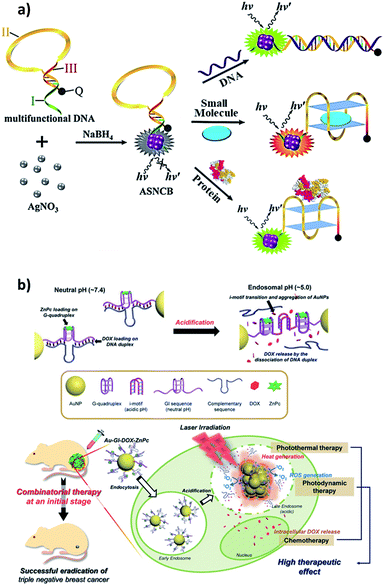 | ||
| Fig. 2 (a) Multifunctional activatable silver nanoclusters beacon (ASNCB) designed by interfacing single-stranded DNA sequences with silver for multiplex DNAs, small molecule, and protein sensing. Reprinted with permission from ref. 43 with copyright (2017) American Chemical Society. (b) Above, design of the Au-GI nanomachine and its operation mechanism depending on pH. Below, intracellular dynamic operation of the Au-GI nanomachine facilitating triple combination of photothermal, photodynamic, and chemotherapy44 with copyright (2018) Wiley and Sons. | ||
DNA–QD interfaces are unique interfaces that allow DNA-templated conjugated systems to have a wide range of applications. Recently, a novel application illustrated in Fig. 3a involved a single step incorporation of DNA-templated quantum dots (ZnS–QDH) with high quantum yield, long-term photostability and low cytotoxicity into a hydrogel network. These quantum dot DNA hydrogels were used for delivery of an anticancer drug, doxorubicin, into MDA-MA-231/Luc breast cancer cells implanted into mice. The drug delivery by Dox–ZnS–QDH increased the drug efficacy by 9-fold.53 Ma et al. developed a series of quantum dots nanobeacons (QD-NBs) for single RNA labelling and imaging in live cells.54 The QD-NBs were synthesized with controllable 1–4 valencies by conjugating a black hole quencher (BHQ1) and phosphorothioate comodified DNA onto CdTe:Zn2+ QDs via a one-pot hydrothermal method. A QD-NB with one conjugate DNA was proven to be suitable and effective for imaging single HIV-1 RNA, where target nucleic acid sequences were hybridized with the stem-loop hairpin DNA and recovered the QD fluorescence for ultrasensitive detection in live HIV-1 integrated cells (Fig. 3b).
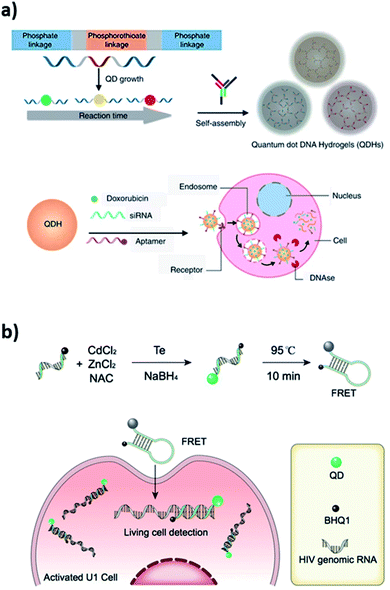 | ||
| Fig. 3 (a) Self-assembled quantum dot DNA hydrogels (QDHs) modified for cell-specific targeting with an aptamer and siRNA/drug delivery. Reprinted from ref. 53 with permission from Springer Nature. (b) Quantum dots nanobeacons (QD-NBs) comprising DNA hairpin structure for single RNA labelling and imaging in live cells. Reprinted with permission from ref. 54 with copyright (2019) American Chemical Society. | ||
1.2. 1D interface
One-dimensional or 1D materials are classified as nanomaterials having two dimensions at the nanoscale, while their third dimension is more than 100 nm. The geometric shapes proposed for these nanomaterials include nanotubes, nanofibers, nanobelts, nanorods, and nanowires as illustrated in Fig. 1a. Carbon nanotubes (CNTs) were among the most studied 1D nanomaterial when interfaced with DNA.55 However, several other types of interfaces with 1D nanostructures were developed, such as halloysite clay nanotubes (HNTs)56 and metal nanowires (e.g. silicon nanowires, SiNW).57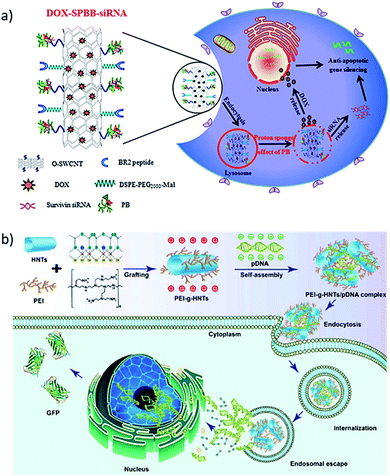 | ||
| Fig. 4 (a) Synthesis process for DOX–SPBB–siRNA, a conjugate of single-walled carbon nanotubes loaded with doxorubicin and survivin RNA, which release the drug and the RNA inside the tumor cell. Reprinted with permission from ref. 64 with copyright (2019) American Chemical Society. (b) Polyethyleneimine (PEI) grafted HNTs bind green fluorescence protein (GFP) labeled pDNA by electrostatic interaction. The PEI-g-HNTs/pDNA complex was taken by cancer cells and show higher transfection efficiency towards both 293T and HeLa cells. Reprinted with permission from ref. 72 with copyright (2017) Elsevier. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 40
1 to 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to form PEI-g-HNTs/pDNA complexes. The PEI-g-HNTs/pDNA complex showed a higher transfection efficiency of up to 46.8% towards 293T and HeLa cells compared to a 41.6% efficiency of the experimental control, which translated into 32% higher green fluorescence protein (GFP) expression in the cells because of DNA release from the PEI-g-HNTs/pDNA complex.
1 to form PEI-g-HNTs/pDNA complexes. The PEI-g-HNTs/pDNA complex showed a higher transfection efficiency of up to 46.8% towards 293T and HeLa cells compared to a 41.6% efficiency of the experimental control, which translated into 32% higher green fluorescence protein (GFP) expression in the cells because of DNA release from the PEI-g-HNTs/pDNA complex.1.3. 2D interface
Two-dimensional or 2D materials represent a class of nanomaterials having a thickness of a few nanometers or less, while their other two dimensions are more than 100 nm. They are also considered as crystalline solids comprising a single layer of atoms, but most of the studies do not follow this strict definition to explain their DNA–2D interfaces. The geometric shapes proposed for these nanomaterials include nanosheets, nanofilms, nanoflakes, and nanolayers as shown in Fig. 1a. Electrons in 2D materials are free to move in the two-dimensional plane, and the electronic structure changes with the thickness, which can be harnessed for biosensing signaling. The eminent examples of these materials are graphene and transition metal dichalcogenides (TMDs), which have been extensively interfaced with DNA among other 2D materials.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times better sensitivity through measuring the shift in Dirac potential with crumpled graphene channels with 600 zM in buffer and 20 aM in human serum, which are ∼18 and ∼600 nucleic acid molecules respectively (Fig. 5c and d). There are other derivatives of graphene, i.e. graphene oxide (GO) and reduced graphene oxide (rGO), which have also been studied in detail for interfacing DNA. We are not reviewing those derivatives as they have been reviewed a number of times elsewhere.25
000 times better sensitivity through measuring the shift in Dirac potential with crumpled graphene channels with 600 zM in buffer and 20 aM in human serum, which are ∼18 and ∼600 nucleic acid molecules respectively (Fig. 5c and d). There are other derivatives of graphene, i.e. graphene oxide (GO) and reduced graphene oxide (rGO), which have also been studied in detail for interfacing DNA. We are not reviewing those derivatives as they have been reviewed a number of times elsewhere.25
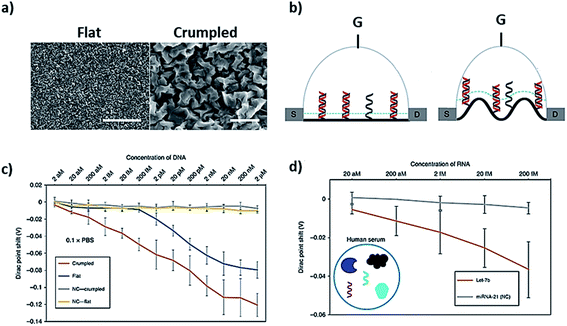 | ||
| Fig. 5 Crumpled graphene FET DNA biosensor. (a) SEM images of flat and crumpled graphene, (b) field-effect transistor made of flat and crumpled graphene to detect DNA, (c) shift in Dirac potential because of DNA hybridization, and (d) detection of DNA in human serum using G-FET. Reprinted with permission from ref. 78 with copyright (2020) Springer Nature. | ||
MoS2 is a good fluorescence quencher and has excellent electron transport properties, which allows a simple DNA biosensing application employing labelled and label-free strategies. Oudeng et al. demonstrated an interesting application of one-step in situ detection of targeted miRNAs expression in single living cancer cells via MoS2 nanosheet-based fluorescence on/off probes, Fig. 6a.85 In this application, probe ssDNA/MoS2 nanosheets were functionalized folic acid (FA)–poly(ethylene glycol). The folic acid receptors on cancer cells facilitated internalization of the probe ssDNA/MoS2 nanosheets, and the hybridization between the probes and target miRNA-21 in MCF-7 and HeLa cells caused the detection of green fluorescence following detachment of the formed duplex (Fig. 6b–d). Such non-toxic probes can potentially provide a real-time, one-step detection system for disease-relevant intracellular miRNAs. Recently, our group has demonstrated a label-free application of DNA/MoS2 system for flexible and highly sensitive electrochemical sensors dubbed as “wax-on-plastic” platforms as shown in Fig. 6e.86 These platforms can be easily fabricated through desktop wax and inkjet printers.87–89 Specifically, it was demonstrated that physisorbed DNA improves the electrochemical property of MoS2 electrodes, because it reduces the bandgap of the MoS2. The electrocatalytic current is sequence-dependent, and the hybridization event further enhances the response in the high ionic strength environment, which was translated into ultra-sensitive detection of CGG trinucleotide repeats associated with fragile X–associated tremor/ataxia syndrome. The detection limit of the platform for the prehybridized CGG duplexes was 0.4 aM and was 0.1 pM for the surface hybridization event.
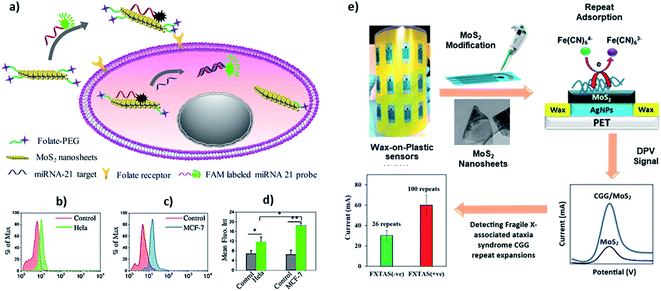 | ||
| Fig. 6 (a–d) Interface of ssDNA–MoS2–PEG–FA probe-based FRET platform for intracellular miRNA-21 detection in cancer cells, i.e. HeLa and MCF-7 cells. Reprinted with permission from ref. 85 with copyright (2018) American Chemical Society. (e) Label-free electrochemical detection of fragile X-associated repeats (CGG repeats) on MoS2 nanosheets modified wax-on-plastic platforms. Reprinted with permission from ref. 86 with copyright (2020) American Chemical Society. | ||
1.4. 3D interface
Three-dimensional (3D) materials as depicted in Fig. 1a represent a type of materials having 3D shape with overall dimensions of more than 100 nm. This class is in between macroscale and nanoscale because these materials are outgrowths of nanostructures. They can be found in a wide range of morphologies, such as dendrites, coils, hollow spheres, cubes, spindles, pillars, and nanoflowers.90–93 The performance and applications of these materials rely on their sizes, shapes, dimensions, and morphologies. For instance, due to their morphology, they can provide enough adsorption sites for involved molecules, while three dimensional access to the surface can improve mass transport of the molecules (e.g. probe and target).94–96 Therefore, 3D structured sensing electrodes allow a high surface area to immobilize a probe and a high access to target diffusion resulting in extremely low detection limits in real samples.97 Chemically, 3D structures can be comprised of metal-based,98 carbon-based, or polymer-based99 materials. The interfacing mechanisms to immobilize DNA on 3D materials can be either an adsorption or chemisorption mechanism depending on the chemical makeup of the materials. For example, graphene-based composites can interact through electrostatic attraction and strong π–π stacking with the DNA phosphate backbone and nucleobases.100 3D structures can also be functionalized with different groups to immobilize biological probes. For instance, perylene tetracarboxylic acid (PTCA) functionalized graphene surface involves π–π interaction and hydrophobic forces between graphene and perylene moiety, while providing active sites for immobilization of the 5′-NH2 modified probe DNA.101 On gold 3D structures, Au–S chemistry can be used to immobilize DNA like metallic nanoparticles.97Kelley and coworkers developed the so-called 3D nanostructured microelectrodes (NMEs) by growing 3D structures on gold microelectrode surfaces for electrochemical nucleic acid detection (Fig. 7a).97,102 These sensing electrodes offer a large surface area (Fig. 7b), which improves sample diffusion to electrode surface, facilitating binding with surface bound probe and resulting in extremely low detection limits in biological samples. For example, chip-based 3D NMEs were applied for electrochemical clamp assay for direct, rapid analysis of circulating nucleic acids in serum, Fig. 7c. Specifically, they immobilized peptide nucleic acid probes (PNA) via Au–S chemistry on the NMEs and monitored differential pulse voltammetry (DPV) response before and after exposure to the target in serum. The clamp approach before the actual measurement blocks the non-specific strands and only allows the mutated sequence to bind the surface-bound probe. The presence of mutations can be detected within 15 minutes with a limit of detection of 1 fg μL−1 and dynamic range from 1 fg μL−1 to 100 pg μL−1.102 They also found that NMEs with clutch probe assay exhibited excellent sensitivity and specificity in the detection of mutated circulatory tumor DNA (ctDNA) with detection sensitivity of 1 fg μL−1 of a target mutation in the presence of 100 pg μL−1 of wild-type DNA, approaching detecting mutations at a level of 0.01% relative to wild type.103
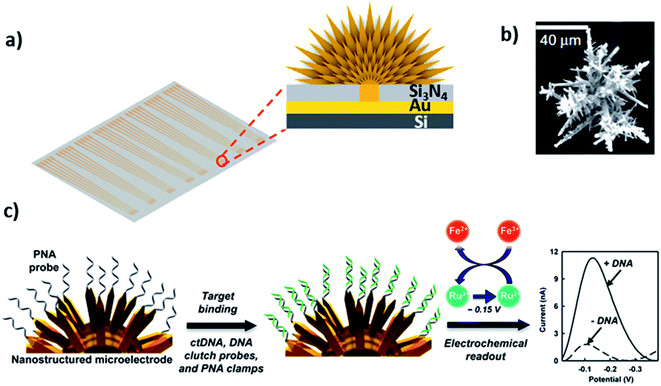 | ||
| Fig. 7 (a and b) 3D structured gold electrode so-called “nanostructured microelectrodes or NMEs” grown on silicon chip and scanning electron microscopic image of 3D structured gold electrode. Reprinted with permission from ref. 102 with copyright (2015) Springer Nature. (c) PNA probe immobilized on gold 3D structures by Au–S linkage, which allows ultrasensitive electrochemical detection of circulating tumor DNA in human serum. Reprinted with permission from ref. 103 with copyright (2016) American Chemical Society. | ||
2 Conclusion
We have reviewed here the diverse class of interfacial nanomaterials comprising DNA and dimensional nanomaterials. DNA can be directly interfaced with a range of 0D, 1D, 2D, and 3D nanomaterials through physisorption and chemisorption where the interactions induce new interfacial character to the materials. For each type of these materials, we discussed the possible and popular interfacing strategies followed by some key recent applications. Unique conformational properties and nucleobase specificity of DNA along with its ability to interact with other materials chemically and through pi-stacking, van der Waals forces, and electrostatic interactions are the primary driving forces behind the novel biomedical applications. These applications—gene therapy, drug delivery, photodynamic therapy, cell imaging, multidimensional biosensing—depend on the structural changes in the DNA triggered by its target as well as the signaling properties of the dimensional materials. Current developments in this area are pushing the boundaries and proposing applications which are specific, sensitive, and applicable for clinical use.3 Future outlook
The rapid growth in DNA–dimensional nanomaterials have opened new avenues for novel biomedical research. The biocompatibility of such materials promises their use for noninvasive surgical treatment through photothermal and photodynamic properties such as DNA–0D system, e.g. Au-GI nanomachines. While gene therapy and chemotherapy would be performed simultaneously in the future using DNA–1D interfaces, such as halloysite clay nanotubes which are inexpensive and abundant in nature. Moreover, excellent electrical and electrochemical properties of 2D graphene and MoS2 nanosheets are available to fabricate low-cost miniaturized DNA biosensing devices for point-of-care diagnostics, for e.g. wax-on-plastic flexible sensors. Nevertheless, the PNA–3D nanostructured microelectrodes are close to clinical application of the detection of genetic biomarkers in serum with high sensitivity. The field is anticipated to flourish with hybrid systems involving DNA interfacing with multiple dimensional materials directly or orthogonally for multidimensional applications.Authors contribution
NA contributed to conceptualization, investigation, validation, and writing original draft. PKD and AHO contributed to investigation. GD contributed to writing review and editing. MHS contributed to funding acquisition, supervision, writing original draft, editing, and reviewing.Permission note
Authors confirm that permissions have been received for the figures used in this article.Conflicts of interest
Authors claim no conflict of financial or intellectual conflict of interest.Acknowledgements
Authors acknowledge the funding from National Science Foundation (CBET-1940716) to support this work.References
- M. H. Shamsi and H.-B. Kraatz, J. Inorg. Organomet. Polym. Mater., 2013, 23, 4–23 CrossRef CAS.
- H. Agheli, J. Malmström, E. M. Larsson, M. Textor and D. S. Sutherland, Nano Lett., 2006, 6, 1165–1171 CrossRef CAS PubMed.
- P. M. Mendes, C. L. Yeung and J. A. Preece, Nanoscale Res. Lett., 2007, 2, 373 CrossRef CAS PubMed.
- V. N. Truskett and M. P. C. Watts, Trends Biotechnol., 2006, 24, 312–317 CrossRef CAS PubMed.
- S. Ramanathan, S. C. B. Gopinath, M. K. Md. Arshad and P. Poopalan, Biosens. Bioelectron., 2019, 141, 111434 CrossRef CAS PubMed.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. R. Lytton-Jean, M. S. Han and C. A. Mirkin, Science, 2006, 312, 1027–1030 CrossRef CAS PubMed.
- V. Pavlov, Y. Xiao, B. Shlyahovsky and I. Willner, J. Am. Chem. Soc., 2004, 126, 11768–11769 CrossRef CAS PubMed.
- I. I. Slowing, J. L. Vivero-Escoto, C.-W. Wu and V. S. Y. Lin, Adv. Drug Delivery Rev., 2008, 60, 1278–1288 CrossRef CAS PubMed.
- A.-H. Lu, E. L. Salabas and F. Schüth, Angew. Chem., Int. Ed., 2007, 46, 1222–1244 CrossRef CAS PubMed.
- K. Vikrant, N. Bhardwaj, S. K. Bhardwaj, K.-H. Kim and A. Deep, Biomaterials, 2019, 214, 119215 CrossRef CAS PubMed.
- O. S. Lee and G. C. Schatz, in Advances in Chemical Physics, ed. S. A. Rice and A. R. Dinner, 2012, vol. 149, pp. 197–249 Search PubMed.
- L. Farzin, M. Shamsipur, L. Samandari and S. Sheibani, J. Pharm. Biomed. Anal., 2018, 161, 344–376 CrossRef CAS PubMed.
- S. J. Smith, C. R. Nemr and S. O. Kelley, J. Am. Chem. Soc., 2017, 139, 1020–1028 CrossRef CAS PubMed.
- O. N. Oliveira, R. M. Iost, J. R. Siqueira, F. N. Crespilho and L. Caseli, ACS Appl. Mater. Interfaces, 2014, 6, 14745–14766 CrossRef CAS PubMed.
- S. Chen and M. H. Shamsi, J. Micromech. Microeng., 2017, 27, 083001 CrossRef.
- D. G. Rackus, M. H. Shamsi and A. R. Wheeler, Chem. Soc. Rev., 2015, 44, 5320–5340 RSC.
- A. Z. Qamar, N. Asefifeyzabadi, M. Taki, S. Naphade, L. M. Ellerby and M. H. Shamsi, J. Mater. Chem. B, 2020, 8, 743–751 RSC.
- A. Samanta and I. L. Medintz, Nanoscale, 2016, 8, 9037–9095 RSC.
- W. Wang, S. Yu, S. Huang, S. Bi, H. Han, J.-R. Zhang, Y. Lu and J.-J. Zhu, Chem. Soc. Rev., 2019, 48, 4892–4920 RSC.
- D. Han, X. Qi, C. Myhrvold, B. Wang, M. Dai, S. Jiang, M. Bates, Y. Liu, B. An, F. Zhang, H. Yan and P. Yin, Science, 2017, 358, eaao2648 CrossRef PubMed.
- P. Chidchob and H. F. Sleiman, Curr. Opin. Chem. Biol., 2018, 46, 63–70 CrossRef CAS PubMed.
- Y. Ke, C. Castro and J. H. Choi, Annu. Rev. Biomed. Eng., 2018, 20, 375–401 CrossRef CAS PubMed.
- L. Tang, Y. Wang and J. Li, Chem. Soc. Rev., 2015, 44, 6954–6980 RSC.
- H. Sun, J. Ren and X. Qu, Acc. Chem. Res., 2016, 49, 461–470 CrossRef CAS PubMed.
- B. Liu, S. Salgado, V. Maheshwari and J. Liu, Curr. Opin. Colloid Interface Sci., 2016, 26, 41–49 CrossRef CAS.
- B. Liu and J. Liu, Matter, 2019, 1, 825–847 CrossRef.
- Q. He, Q. Wu, X. Feng, Z. Liao, W. Peng, Y. Liu, D. Peng, Z. Liu and M. Mo, Int. J. Biol. Macromol., 2020, 151, 757–780 CrossRef CAS PubMed.
- S. Su, Q. Sun, X. Gu, Y. Xu, J. Shen, D. Zhu, J. Chao, C. Fan and L. Wang, TrAC, Trends Anal. Chem., 2019, 119, 115610 CrossRef CAS.
- N. Li, Y. Shang, Z. Han, T. Wang, Z.-G. Wang and B. Ding, ACS Appl. Mater. Interfaces, 2019, 11, 13835–13852 CrossRef CAS PubMed.
- N. Asefifeyzabadi, M. Taki, M. Funneman, T. Song and M. H. Shamsi, J. Mater. Chem. B, 2020, 8, 5225–5233 RSC.
- J. C. Léon, Z. She, A. Kamal, M. H. Shamsi, J. Müller and H.-B. Kraatz, Angew. Chem., Int. Ed., 2017, 56, 1–6 CrossRef.
- M. H. Shamsi, K. Choi, A. H. Ng, M. D. Chamberlain and A. R. Wheeler, Biosens. Bioelectron., 2016, 77, 845–852 CrossRef CAS PubMed.
- S. F. Shaikh, N. M. Shinde, D. Lee, A. M. Al-Enizi, K. H. Kim and R. S. Mane, in Nanostructures, IntechOpen, 2019, DOI: DOI:10.5772/intechopen.83803.
- N. Farkhari, S. Abbasian, A. Moshaii and M. Nikkhah, Colloids Surf., B, 2016, 148, 657–664 CrossRef CAS PubMed.
- H. Li and L. Rothberg, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14036–14039 CrossRef CAS PubMed.
- E. M. Nelson and L. J. Rothberg, Langmuir, 2011, 27, 1770–1777 CrossRef CAS PubMed.
- P. Gisbert-Quilis, M. Masetti, J. Morla-Folch, J. M. Fitzgerald, N. Pazos-Perez, E. Garcia-Rico, V. Giannini, R. A. Alvarez-Puebla and L. Guerrini, Adv. Mater. Interfaces, 2017, 4, 1700724 CrossRef.
- H. Kimura-Suda, D. Y. Petrovykh, M. J. Tarlov and L. J. Whitman, J. Am. Chem. Soc., 2003, 125, 9014–9015 CrossRef CAS PubMed.
- H. Pei, F. Li, Y. Wan, M. Wei, H. Liu, Y. Su, N. Chen, Q. Huang and C. Fan, J. Am. Chem. Soc., 2012, 134, 11876–11879 CrossRef CAS PubMed.
- C.-C. Wang, S.-M. Wu, H.-W. Li and H.-T. Chang, ChemBioChem, 2016, 17, 1052–1062 CrossRef CAS PubMed.
- M. S. Inkpen, Z. F. Liu, H. Li, L. M. Campos, J. B. Neaton and L. Venkataraman, Nat. Chem., 2019, 11, 351–358 CrossRef CAS PubMed.
- W. Zhou, F. Wang, J. Ding and J. Liu, ACS Appl. Mater. Interfaces, 2014, 6, 14795–14800 CrossRef CAS PubMed.
- G. Liu, J. Li, D.-Q. Feng, J.-J. Zhu and W. Wang, Anal. Chem., 2017, 89, 1002–1008 CrossRef CAS PubMed.
- H. Park, J. Kim, S. Jung and W. J. Kim, Adv. Funct. Mater., 2018, 28, 1705416 CrossRef.
- B. Liu and J. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 24833–24838 CrossRef CAS PubMed.
- L. Ma, B. Liu, P.-J. J. Huang, X. Zhang and J. Liu, Langmuir, 2016, 32, 5672–5680 CrossRef CAS PubMed.
- L. Chen, B. Liu, Z. Xu and J. Liu, Langmuir, 2018, 34, 9314–9321 CrossRef CAS PubMed.
- B. Liu and J. Liu, Langmuir, 2015, 31, 371–377 CrossRef CAS PubMed.
- B. Liu, L. Ma, Z. Huang, H. Hu, P. Wu and J. Liu, Mater. Horiz., 2018, 5, 65–69 RSC.
- A. Lopez, Y. Zhang and J. Liu, J. Colloid Interface Sci., 2017, 493, 249–256 CrossRef CAS PubMed.
- W. R. Algar and U. J. Krull, Langmuir, 2006, 22, 11346–11352 CrossRef CAS PubMed.
- A. Shahmuradyan and U. J. Krull, Anal. Chem., 2016, 88, 3186–3193 CrossRef CAS PubMed.
- L. Zhang, S. R. Jean, S. Ahmed, P. M. Aldridge, X. Li, F. Fan, E. H. Sargent and S. O. Kelley, Nat. Commun., 2017, 8, 381 CrossRef PubMed.
- Y. Ma, G. Mao, W. Huang, G. Wu, W. Yin, X. Ji, Z. Deng, Z. Cai, X.-E. Zhang, Z. He and Z. Cui, J. Am. Chem. Soc., 2019, 141, 13454–13458 CrossRef CAS PubMed.
- D. Nepal, J.-I. Sohn, W. K. Aicher, S. Lee and K. E. Geckeler, Biomacromolecules, 2005, 6, 2919–2922 CrossRef CAS PubMed.
- M. H. Shamsi and K. E. Geckeler, Nanotechnology, 2008, 19, 075604 CrossRef PubMed.
- S. K. Jat, J. Bhattacharya and M. K. Sharma, J. Mater. Chem. B, 2020, 8, 4165–4175 RSC.
- S. Iliafar, J. Mittal, D. Vezenov and A. Jagota, J. Am. Chem. Soc., 2014, 136, 12947–12957 CrossRef CAS PubMed.
- D. P. Salem, X. Gong, A. T. Liu, V. B. Koman, J. Dong and M. S. Strano, J. Am. Chem. Soc., 2017, 139, 16791–16802 CrossRef CAS PubMed.
- H. Cai, X. Cao, Y. Jiang, P. He and Y. Fang, Anal. Bioanal. Chem., 2003, 375, 287–293 CrossRef CAS PubMed.
- N. Naotoshi, T. Yasuhiko and M. Hiroto, Chem. Lett., 2002, 31, 638–639 CrossRef.
- N. W. S. Kam, Z. Liu and H. Dai, J. Am. Chem. Soc., 2005, 127, 12492–12493 CrossRef CAS PubMed.
- P. He, Y. Xu and Y. Fang, Microchim. Acta, 2006, 152, 175 CrossRef CAS.
- Y. Cao, H.-Y. Huang, L.-Q. Chen, H.-H. Du, J.-H. Cui, L. W. Zhang, B.-J. Lee and Q.-R. Cao, ACS Appl. Mater. Interfaces, 2019, 11, 9763–9776 CrossRef CAS PubMed.
- E. Joussein, S. Petit, J. Churchman, B. Theng, D. Righi and B. Delvaux, Clay Miner., 2005, 40, 383–426 CrossRef CAS.
- M. Liu, Y. Chang, J. Yang, Y. You, R. He, T. Chen and C. Zhou, J. Mater. Chem. B, 2016, 4, 2253–2263 RSC.
- T. Husain, M. H. Shoaib, F. R. Ahmed, R. I. Yousuf, S. Farooqi, F. Siddiqui, M. S. Imtiaz, M. Maboos and S. Jabeen, Int. J. Nanomed., 2021, 16, 1725–1741 CrossRef PubMed.
- Y. F. Shi, Z. Tian, Y. Zhang, H. B. Shen and N. Q. Jia, Nanoscale Res. Lett., 2011, 6, 608 CrossRef PubMed.
- M. Liu, R. He, J. Yang, W. Zhao and C. Zhou, ACS Appl. Mater. Interfaces, 2016, 8, 7709–7719 CrossRef CAS PubMed.
- M. Liu, C. Wu, Y. Jiao, S. Xiong and C. Zhou, J. Mater. Chem. B, 2013, 1, 2078–2089 RSC.
- S. Batasheva, M. Kryuchkova, R. Fakhrullin, G. Cavallaro, G. Lazzara, F. Akhatova, L. Nigamatzyanova, V. Evtugyn, E. Rozhina and R. Fakhrullin, Molecules, 2020, 25, 3557 CrossRef CAS PubMed.
- Z. Long, J. Zhang, Y. Shen, C. Zhou and M. Liu, Mater. Sci. Eng., C, 2017, 81, 224–235 CrossRef CAS PubMed.
- H. Wu, Y. Shi, C. Huang, Y. Zhang, J. Wu, H. Shen and N. Jia, J. Biomater. Appl., 2014, 28, 1180–1189 CrossRef PubMed.
- S. Zeng, L. Chen, Y. Wang and J. Chen, J. Phys. D: Appl. Phys., 2015, 48, 275402 CrossRef.
- N. Dontschuk, A. Stacey, A. Tadich, K. J. Rietwyk, A. Schenk, M. T. Edmonds, O. Shimoni, C. I. Pakes, S. Prawer and J. Cervenka, Nat. Commun., 2015, 6, 6563 CrossRef CAS PubMed.
- Y. Li, C. Wang, Y. Zhu, X. Zhou, Y. Xiang, M. He and S. Zeng, Biosens. Bioelectron., 2017, 89, 758–763 CrossRef CAS PubMed.
- S. Xu, J. Zhan, B. Man, S. Jiang, W. Yue, S. Gao, C. Guo, H. Liu, Z. Li, J. Wang and Y. Zhou, Nat. Commun., 2017, 8, 14902 CrossRef CAS PubMed.
- M. T. Hwang, M. Heiranian, Y. Kim, S. You, J. Leem, A. Taqieddin, V. Faramarzi, Y. Jing, I. Park, A. M. van der Zande, S. Nam, N. R. Aluru and R. Bashir, Nat. Commun., 2020, 11, 1543 CrossRef CAS PubMed.
- S. Manzeli, D. Ovchinnikov, D. Pasquier, O. V. Yazyev and A. Kis, Nat. Rev. Mater., 2017, 2, 17033 CrossRef CAS.
- F. Liu, Y. Zhang, H. Wang, L. Li, W. Zhao, J.-W. Shen and L. Liang, Chem. Phys., 2020, 529, 110546 CrossRef CAS.
- H. Vovusha and B. Sanyal, RSC Adv., 2015, 5, 67427–67434 RSC.
- M. Sharma, A. Kumar and P. K. Ahluwalia, RSC Adv., 2016, 6, 60223–60230 RSC.
- C. Lu, Y. Liu, Y. Ying and J. Liu, Langmuir, 2017, 33, 630–637 CrossRef CAS PubMed.
- M. Xiao, A. R. Chandrasekaran, W. Ji, F. Li, T. Man, C. Zhu, X. Shen, H. Pei, Q. Li and L. Li, ACS Appl. Mater. Interfaces, 2018, 10, 35794–35800 CrossRef CAS PubMed.
- G. Oudeng, M. Au, J. Shi, C. Wen and M. Yang, ACS Appl. Mater. Interfaces, 2018, 10, 350–360 CrossRef CAS PubMed.
- N. Asefifeyzabadi, R. Alkhaldi, A. Z. Qamar, A. A. Pater, M. Patwardhan, K. T. Gagnon, S. Talapatra and M. H. Shamsi, ACS Appl. Mater. Interfaces, 2020, 12, 52156–52165 CrossRef CAS PubMed.
- A. Z. Qamar and M. H. Shamsi, Micromachines, 2020, 11, 1–16 CrossRef PubMed.
- S. Chen, A. Z. Qamar, N. Asefifeyzabadi, M. Funneman, M. Taki, L. Elliot, M. E. Kinsel, G. R. Kinsel and M. H. Shamsi, Sci. Rep., 2019, 9, 1–9 Search PubMed.
- A. Qamar, K. Amar, F. Chowdhury, P. Kohli and M. Shamsi, RSC Adv., 2016, 6, 104919–104924 RSC.
- L. Wang and Y. Yamauchi, Chem. Mater., 2009, 21, 3562–3569 CrossRef CAS.
- J. N. Wang, L. F. Su and Z. P. Wu, Cryst. Growth Des., 2008, 8, 1741–1747 CrossRef CAS.
- J. Liu, J. Essner and J. Li, Chem. Mater., 2010, 22, 5022–5030 CrossRef CAS.
- W. Lei, D. Liu, J. Zhang, P. Zhu, Q. Cui and G. Zou, Cryst. Growth Des., 2009, 9, 1489–1493 CrossRef CAS.
- Y.-H. Jin, S.-H. Lee, H.-W. Shim, K. H. Ko and D.-W. Kim, Electrochim. Acta, 2010, 55, 7315–7321 CrossRef CAS.
- Q. Shen, L. Jiang, H. Zhang, Q. Min, W. Hou and J.-J. Zhu, J. Phys. Chem. C, 2008, 112, 16385–16392 CrossRef CAS.
- Z. Sun, J. H. Kim, Y. Zhao, D. Attard and S. X. Dou, Chem. Commun., 2013, 49, 966–968 RSC.
- L. Soleymani, Z. Fang, B. Lam, X. Bin, E. Vasilyeva, A. J. Ross, E. H. Sargent and S. O. Kelley, ACS Nano, 2011, 5, 3360–3366 CrossRef CAS PubMed.
- M. Shahi, F. Hekmat and S. Shahrokhian, Appl. Surf. Sci., 2021, 558, 149869 CrossRef CAS.
- H. Sun, L. Yang, M. P. Thompson, S. Schara, W. Cao, W. Choi, Z. Hu, N. Zang, W. Tan and N. C. Gianneschi, Bioconjugate Chem., 2019, 30, 1889–1904 CrossRef CAS PubMed.
- H.-J. Qiu, Y. Guan, P. Luo and Y. Wang, Biosens. Bioelectron., 2017, 89, 85–95 CrossRef CAS PubMed.
- H. S. Song, O. S. Kwon, J.-H. Kim, J. Conde and N. Artzi, Biosens. Bioelectron., 2017, 89, 187–200 CrossRef CAS PubMed.
- J. Das, I. Ivanov, L. Montermini, J. Rak, E. H. Sargent and S. O. Kelley, Nat. Chem., 2015, 7, 569–575 CrossRef CAS PubMed.
- J. Das, I. Ivanov, E. H. Sargent and S. O. Kelley, J. Am. Chem. Soc., 2016, 138, 11009–11016 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |
